Abstract
The human immunodeficiency virus type 1 (HIV-1) protease (PR) makes five obligatory cleavages in the viral Gag polyprotein precursor. The cleavage events release the virion structural proteins from the precursor and allow the virion to undergo maturation to become infectious. The protease cleavage between the matrix protein (MA) domain and the adjacent capsid protein (CA) domain releases CA from the membrane-anchored MA and allows the N terminus of CA to refold into a structure that facilitates the formation of hexamer arrays that represent the structural unit of the capsid shell. In this study, we analyzed the extent to which each of the HIV-1 Gag processing sites must be cleaved by substituting the P1-position amino acid at each processing site with Ile. A mutation that blocks cleavage at the MA/CA processing site (Y132I) displayed a strong transdominant effect when tested in a phenotypic mixing strategy, inhibiting virion infectivity with a 50% inhibitory concentration of only 4% of the mutant relative to the wild type. This mutation is 10- to 20-fold more potent in phenotypic mixing than an inactivating mutation in the viral protease, the target of many successful inhibitors, and more potent than an inactivating mutation at any of the other Gag cleavage sites. The transdominant effect is manifested as the assembly of an aberrant virion core. Virus containing 20% of the Y132I mutant and 80% of the wild type (to assess the transdominant effect on infectivity) was blocked either before reverse transcription (RT) or at an early RT step. The ability of a small amount of the MA/CA fusion protein to poison the oligomeric assembly of infectious virus identifies an essential step in the complex process of virion formation and maturation. The effect of a small-molecule inhibitor that is able to block MA/CA cleavage even partially would be amplified by this transdominant negative effect on the highly orchestrated process of virion assembly.
Proteolytic cleavage of the human immunodeficiency virus type 1 (HIV-1) polyproteins Gag and Gag-Pro-Pol by the viral protease (PR) is an essential step in the maturation of the virus particle to become infectious. Proteolysis occurs concomitantly with the budding of the virus particle, and this processing releases the following mature virion structural proteins from the precursor proteins: matrix (MA), capsid (CA), spacer peptide 1 (SP1), nucleocapsid (NC), spacer peptide 2 (SP2), p6, and the viral enzymes (36). With the proteolysis of Gag, there is a dramatic structural rearrangement in which the CA proteins condense to form the cone-shaped capsid shell surrounding the NC/RNA nucleoprotein complex (43). During maturation, the released N terminus of the CA protein adopts a β-hairpin structure by forming a salt bridge between Pro1 and Asp51 of CA, which appears to be important for the assembly of conical capsid (21, 28, 38, 42). Since proteolytic processing is essential for the formation of infectious virus, PR has been the target of a very successful group of inhibitors now in clinical use.
There are five protease cleavage sites in the Gag precursor and an additional five sites in the Gag-Pro-Pol precursor. In a previous analysis using a PR inhibitor, we found that only moderate levels of inhibition of these cleavage events was necessary to ablate virion infectivity (19). This observation suggested that the processing/assembly pathway itself was a more sensitive target for inhibition than PR and raised the possibility that individual cleavage sites may not be equivalent in the extent of cleavage needed for virion infectivity, with a highly sensitive site representing a potential target for the development of an antiviral. PA-457 (Bevirimat), identified in a screen for inhibition of viral replication, inhibits the cleavage event between CA and SP1 (23, 49), although it is not clear how the drug blocks protease cleavage at this site. The drug is incorporated into immature particles, suggesting that it interacts with Gag to alter its ability to serve as a protease substrate at the site (48). Thus, it is possible to envisage inhibitors that could target specific processing sites.
Mutations that confer a dominant negative (also known as transdominant) phenotype can be a powerful way to interfere with the function of an oligomeric protein complex. Several studies have described such mutations targeting HIV-1 proteins such as Tat (17), Rev (5, 24), and Gag (15, 41), with a dominant negative Rev mutant having been tested in a gene therapy trial (1, 9, 35). In addition, an N-terminal mutation of murine leukemia virus CA functions in a transdominant manner (33). Most HIV-1 proteins function in a multimeric complex, although the virion complex of several thousand Gag proteins is by far the largest complex among the viral proteins (7, 46). This suggests that mutant Gag proteins should have the potential to display strong multiplicative effects on their inhibition of virion infectivity. In this study, we demonstrate a strongly transdominant Gag mutant, Y132I. The inclusion of only 5% of the Y132I mutant protein in a virion can inhibit more than 80% of viral infectivity. Virus containing 20% of the Y132I mutation and 80% of wild-type Y132I (W80/M20) showed aberrant and eccentric core structures, a complete loss of virion infectivity, and a large reduction in the ability to synthesize viral DNA during the subsequent round of infection. These data demonstrate a potential to identify strongly transdominant mutations in Gag that affect the assembly pathway and validate this pathway as being a highly sensitive target for the development of an antiviral.
MATERIALS AND METHODS
Constructs.
The infectious molecular clone pNL-CH, derived from the pNL4-3 clone of HIV-1, contains a silent T-to-C mutation at nucleotide 2600 to introduce an RsrII restriction enzyme site near the 5′ end of pol. Plasmid pARKz1k1-5LTRgag, containing the fragment of the 5′ long terminal repeat (LTR) and the gag region of pNL-CH, was used as a template for site-directed mutagenesis to introduce Y132I, L363I, M377I, N432I, F448I, D51A, P1F, or P1L mutations. Point mutations were generated according to the Quickchange method (Stratagene), and the mutations were confirmed by DNA sequence analysis of the entire fragment subjected to mutagenesis. The DNA fragment containing each mutation was prepared by restriction enzyme digestion with BssHII and SpeI, and the resulting fragment was cloned into pNL-CH digested with BssHII and SpeI. Each mutant clone in the pNL-CH backbone was confirmed by DNA sequence analysis.
Cell culture and DNA transfection.
293T cells and the TZM-BL cells (NIH AIDS Research and Reference Reagent Program) were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum in the presence of penicillin and streptomycin at 37°C with 5% CO2. 293T cells are derived from human embryonic kidney cells (13), and TZM-BL cells are HeLa cells which stably express CXCR4, CCR5, and CD4 (44). Human astroglioma U87 cells expressing human CD4 and CXCR4 (U87.CD4.CXCR4) (6) were obtained from the NIH AIDS Research and Reference Reagent Program and were maintained in Dulbecco's modified Eagle's medium with 15% fetal calf serum in the presence of 1 μg/ml puromycin, 300 μg/ml G418, penicillin, and streptomycin at 37°C with 5% CO2. To prepare a viral stock, 3.5 × 106 293T cells were seeded onto a 10-cm plate 24 h before transfection. The 293T cells were transfected with a total of 10 μg of each mutant or the wild-type construct using FuGENE 6 transfection reagent (Roche). For cotransfection, 0.5 × 106 293T cells were seeded onto a six-well plate 24 h before transfection. 293T cells were transfected with a total of 4 μg of wild-type and mutant constructs using FuGENE 6 transfection reagent (Roche). To prepare viruses grown in the presence of the myristoylation inhibitor 2-hydroxymyristic acid, the culture supernatant of 293T cells transfected with wild-type and mutant constructs was changed 4 h after transfection with medium containing the drug at concentrations between 0 and 30 μM.
Infectivity assay.
The culture supernatant containing virus particles was harvested 48 h after transfection and filtered through a 0.45-μm-pore-size membrane (Millipore) to remove cell debris. The culture supernatant was diluted 1:50 or 1:100 and used to infect 2 × 104 TZM-BL cells in a 96-well plate. The TZM-BL indicator cells express the luciferase gene and the lacZ gene under the control of the HIV-1 LTR. For the luciferase assay, infected TZM-BL cells were lysed 48 h postinfection. Briefly, the culture medium was removed from each well, and the cells were washed with phosphate-buffered saline. A 50-μl aliquot of 1× reporter lysis buffer (Promega) was added to the cells, and the cells were kept at −80°C. After one freeze-thaw cycle, 30 μl of cell lysate was transferred into a 96-well assay plate (Costar), and luciferase activity was measured using a luminometer (Promega).
Western analyses and EM.
Viruses harvested 48 h posttransfection were concentrated by ultracentrifugation at 24,000 rpm for 2 h at 4°C using an SW 41 Ti rotor (Beckman). Viral proteins were examined by Western analysis using polyclonal rabbit anti-HIV-1 p24/CA antibody (NIH AIDS Research and Reference Reagent Program), polyclonal goat anti-HIV-1 p7/NC antibody (kind gift from Robert Gorelick, SAIC-Frederick), or polyclonal rabbit anti-HIV-1 reverse transcriptase (RT) antibody (NIH AIDS Research and Reference Reagent Program). For electron microscopy (EM) analysis, 0.5 × 106 293T cells were seeded onto a six-well plate 24 h before transfection. 293T cells were transfected with a total of 4 μg of each construct using FuGENE 6 transfection reagent (Roche). The cells were washed with phosphate-buffered saline 48 h posttransfection, pelleted by low-speed centrifugation, and fixed in 2% glutaraldehyde in cacodylate buffer (0.1 M, pH 7.4). EM analyses were performed using thin-section electron microscopy.
Quantitative analysis of viral RNA.
Cell-free virions harvested from transfected 293T cells were treated with RNase-free DNase (Promega) at 37°C for 1 h to remove any residual plasmid DNA carried over from the transfection. Sindbis virus was also treated with RNase-free DNase and used as an internal control for real-time PCR. Viral RNA was extracted using the QIAamp viral RNA kit (Qiagen). The amount of encapsidated genomic viral RNA was detected by quantitative real-time RT-PCR using an ABI 7000 sequencer detector (Applied Biosystems) and normalized by the amount of Sindbis virus RNA. The sequences of the primers and probe to detect the HIV-1 gag region were previously described (12). Sindbis virus RNA was detected using primers SINRT-F (5′-GCCGCACACGACAATTCAC-3′) and SINRT-R (5′-GTACCCTCGTACACGGACGAA-3′) and a probe, SINRT-P (5′-6-carboxyfluorescein-CCGCATCATCTGAATTG-MGBNFQ-3′, where MGBNFQ is minor groove binder nonfluorescent quencher). Real-time PCR was performed using a TaqMan One-Step RT-PCR master mix reagent kit (Applied Biosystems).
Quantitation of viral DNA synthesis.
Filtered and DNase-treated virus supernatant was used to infect U87.CD4.CXCR4 cells seeded 24 h before infection, and total cellular DNA was isolated from the infected cells 24 h postinfection using the QIAamp DNA blood minikit (Qiagen). The amount of recovered total DNA in each sample was normalized by real-time PCR using a primer-and-probe set detecting the human RNaseP gene (Applied Biosystems). The virus input used for infection was determined by real-time RT-PCR using a primer-and-probe set described above for virion RNA. Unless specified, a normalized amount of total DNA from each sample was used as a template for quantitative PCR to measure newly synthesized viral DNA. Primers used to detect an early viral DNA product in the env gene were Henv7339F (5′-TTTTAATTGTGGAGGGGAATTTTTCTAC-3′) and Henv7666R (5′-ATATAATTCACTTCTCCAATTGTCCCTC-3′). Primers to detect HIV-1 two-LTR circles (i.e., circular DNA with two LTRs) were H9371F (5′-CCGAGAGCTGCATCCGGAGTAC-3′) and H310R (5′-GGATGCAGCTCTCGGGCCATG-3′).
RESULTS
Isoleucine mutations at the P1 positions of Gag processing sites.
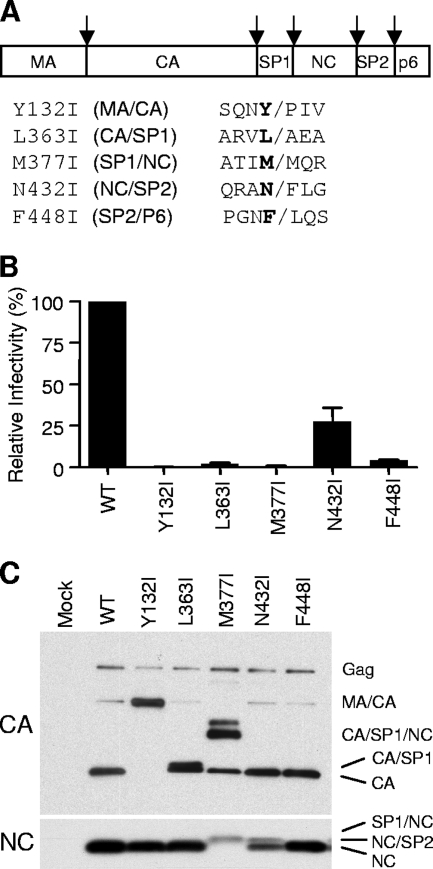
HIV-1 PR favors a large hydrophobic amino acid at the P1 position (30), however, a β-branched amino acid such as isoleucine at the P1 position strongly inhibits protease cleavage at that site (29). We created the following individual P1-position mutants to test their relative potencies for inhibiting viral infectivity in submolar amounts: Y132I, L363I, M377I, N432I, and F448I (Fig. 1A).
FIG. 1.
Ile substitutions at the P1 position of Gag cleavage sites affect Gag processing and impair viral infectivity. (A) Schematic diagram showing protein domains of the HIV-1 Gag protein. Amino acid sequences of the P1 position of the cleavage sites are shown in boldface type. The Ile substitution mutants are shown with the cleavage site. Arrows at the top indicate five different cleavage sites in Gag. (B) Relative infectivity of the P1 Ile mutant viruses. Culture supernatants of 293T cells were harvested 48 h after transfection and used to infect TZM-BL reporter cells. The TZM-BL cells were lysed and used in a luciferase assay 48 h after infection to assess the level of infection. The measurements of infectivity from the luciferase assays were normalized by the amount of viral genomic RNA in the infecting virus quantified by real-time PCR analysis. Measurements from the infectious molecular clone pNL-CH (wild type [WT]; derived from pNL4-3) were used as the wild type and were considered to be 100%. The mean data with standard errors from several experiments are shown. All the infection data shown in this paper were obtained by following the same protocol unless otherwise specified. (C) Western analysis of viral particles. Culture supernatants harvested 48 h after transfection were subjected to ultracentrifugation to concentrate the viral particles. The pelleted viral particles were analyzed by Western analysis using either anti-CA antibody (top) or anti-NC antibody (bottom) as the primary antibody.
Each of these cleavage site mutations except for the N432I mutation ablated virion infectivity in virus particles produced from 293T cells transfected with mutant viral DNA (Fig. 1B). The N432I mutant showed infectivity at about 30% of wild-type infectivity, and this is consistent with the phenotype described previously (10). Western analysis of virus particles (probed with an anti-CA antibody or an anti-NC antibody) are shown in Fig. 1C and revealed the effect of the individual mutations on Gag cleavage. Wild-type virus produced the mature CA protein (p24) with a small amount of a p40 processing intermediate and unprocessed p55 Gag protein. The Y132I mutation resulted in the production of p40 (MA/CA fusion protein) showing a complete block at the processing site between MA and CA. The L363I mutation gave a product very similar to p24, representing the CA/SP1 fusion protein (p25). The band moving slightly faster than p25 is either p24 or a p24-like product resulting from processing at minor sites in SP1 (18, 45). The M377I mutation blocked processing at the SP1/NC site and also showed a partial inhibition of cleavage at the CA/SP1 site, generating a large amount of a CA/SP1/NC product (which also bound the anti-NC antibody) (data not shown). The N432I mutation caused a partial processing defect at the NC/SP2 site, generating an NC/SP2 product and the NC protein. This analysis did not reveal processing defects with the F448I mutation (at the SP2/p6 site), although we did not probe the p6 protein. Some of the phenotypes described above are equivalent to those observed previously for other mutations at these sites (16, 45).
Transdominant negative activity of Y132I.
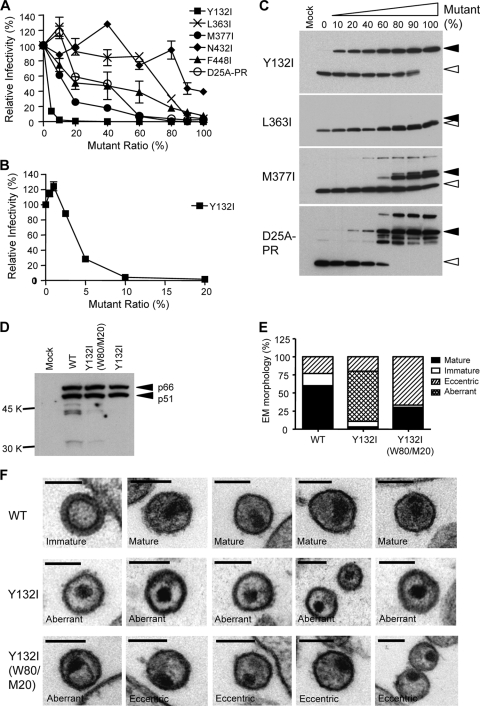
In order to modulate the extent of cleavage at a specific site, we employed a phenotypic mixing strategy by cotransfecting two DNAs, wild type and mutant, to produce virus particles that contained a mixture of proteins encoded by both genomes. Since assembly takes place with the Gag precursor, the effect of a processing defect would be apparent in the context of capsid maturation in the virion, which is a large oligomeric structure requiring the organization of several thousand CA subunits (7). To determine the potency of each processing site mutation in inhibiting infectivity, we varied the ratios of the wild-type and mutant genomes, keeping the total amount of DNA constant, and measured the infectivity of the virus produced. For a control, we included a genome that encodes a mutation at the active site of the viral protease (D25A). As can be seen in Fig. 2A, the PR mutant D25A-PR inhibited virion infectivity at a 50% level (50% inhibitory concentration [IC50]) when approximately 50% of the viral DNA encoded the D25A mutation. Decreasing amounts of protease activity resulted in the accumulation of numerous processing intermediates (Fig. 2C, bottom), analogous to the titration of a protease inhibitor.
FIG. 2.
The Y132I mutation displays a strong transdominant effect on wild-type viral infectivity in a phenotypic mixing experiment. (A) Relative potencies of Y132I, L363I, M377I, N432I, F448I, and D25A-PR mutants. Mutant and wild-type plasmid DNAs were cotransfected into 293T cells, gradually increasing the percentage of mutant from 0% to 100%. The amounts of mutant used were 0%, 10%, 20%, 40%, 60%, 80%, 90%, and 100% in each cotransfection experiment except for the Y132I mutant, where 5% mutant was added. Viral infectivity was considered to be 100% when 0% mutant was used (i.e., 100% wild type), and all other viral infectivities obtained from different mutant fractions were compared to the 100% value for the wild type. Unless specified, all the infectivity data are shown as means ± standard errors. (B) Relative viral infectivity of the Y132I mutant at a low range of mutant percentages, from 0% to 20%. (C) Western analysis of virion particles produced from transfection with wild-type and mutant DNAs. Lane 1 represents a mock transfection where no DNA was used. The percentage of mutant used for each cotransfection is shown. Open arrowheads indicate processed CA protein, and closed arrowheads in the Y132I, L363I, M377I, and D25A-PR mutants indicate the MA/CA fusion protein, CA/SP1 fusion protein, CA/SP1/NC fusion protein, and unprocessed Gag precursor, respectively. (D) Western analysis showing processed RT in viral particles of the Y132I and Y132I(W80/M20) viruses probed with an anti-HIV-1 RT antibody. Closed arrowheads indicate the completely processed p66 and p51 subunits of RT. (E) Distribution of virion morphology. Total numbers of viruses counted were 96 for the wild type (WT), 147 for the Y132I mutant, and 72 for the Y132I(W80/M20) mutant. The fractions of mature, immature, eccentric, and aberrant viruses are shown. (F) Morphology of Y132I viruses and viruses from cotransfection with a ratio of 80% wild type and 20% Y132I [Y132I(W80/M20)] assessed by thin-section EM. Examples of wild-type viruses are shown in the first row. The phenotype of each virion is labeled. The bars in each image represent 100 nm.
The five Gag cleavage site mutants (Y132I, L363I, M377I, N432I, and F448I) behaved very differently in this phenotypic mixing protocol (Fig. 2A). The Y132I mutant had a very strong negative effect on wild-type infectivity, with only 5% of the mutant DNA in the cotransfection inhibiting approximately 80% of the infectivity. In titrating down the amount of mutant DNA, we found that as little as 3 to 4% of the mutant was able to inhibit 50% of virion infectivity (IC50), and infectivity was ablated with 20% mutant DNA (Fig. 2B). The M377I mutant was also relatively potent, needing only 15% mutant to inhibit 50% of the infectivity, as was the F448I mutant, inhibiting 50% of virion infectivity with 20% mutant. The L363I and N432I mutants were the least potent, requiring approximately 70% and 90% mutant, respectively, to inhibit 50% of the infectivity (Fig. 2A). An examination of the Gag protein in the virus particles produced from the cotransfection confirmed the expected processing patterns for mixtures of wild-type and mutant Gag proteins (Fig. 2C and data not shown for N432I and F448I). Based on the quantitative analysis of the viral RNA (data not shown) and Western analysis (Fig. 2C), none of the mutants appeared to block the production of virus particles, suggesting that the loss of infectivity for each mutant was at the level of particle infectivity. We also examined the extent of processing of the Pol domain and found the appropriate processing of the RT subunits for the Y132I mutant (Fig. 2D), indicating that there were no global defects in processing. Thus, very modest inhibition of processing at the MA/CA cleavage site resulted in a dramatic loss in virion infectivity, a loss that was disproportionate compared to the other four Gag processing site mutations or compared to the inhibition of the viral protease.
We examined virion morphology using thin-section EM for the Y132I mutant virions and for noninfectious virions that contained 80% wild type and 20% of the Y132I mutant [Y132I(W80/M20)] as a measure of transdominance. Since even wild-type virions are polymorphic in structure, heterogenous structures were expected. Immature ring structures and mature cone-shaped cores are well described for wild-type HIV-1, and examples are shown in Fig. 2F. The distinctive structure that was apparent, especially in the mixed virions containing 20% of the Y132I mutant, was an eccentric core structure at the edge of the virion (Fig. 2F), suggesting residual tethering to the membrane of the viral core and a structure similar to that seen for virions produced with a limiting amount of a protease inhibitor (19, 26). In addition, some ring-shaped core structures were also observed for Y132I(W80/M20) viruses (Fig. 2F). Consistent with previously reported findings for a Y132S mutant (16), the Y132I mutant alone showed enhanced thickening near the virion membrane, presumably due to the membrane-linked MA/CA fusion protein, and a separate small dark staining core containing NC and RNA, a phenotype which we term aberrant and which is distinct from the eccentric phenotype. The relative proportions of the different forms of virions seen are shown in Fig. 2E. About 67% of the Y132I(W80/M20) viruses displayed an eccentric core, which is significantly different from the frequency of these cores in wild-type (23%) and Y132I (20%) strains (P < 0.0001).
Role of core tethering to membrane in the transdominant phenotype.
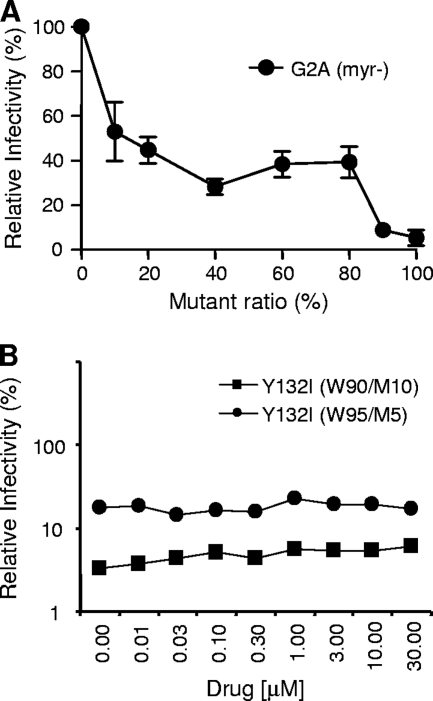
Since the MA domain of Gag is responsible for the targeting of Gag to the membrane (36), one possible explanation for the detrimental effect of the MA/CA fusion protein on viral infectivity is that the MA-linked CA protein anchored to the membrane may still participate in capsid core assembly, resulting in the entire capsid remaining tethered to the viral membrane envelope. We tested this possibility by introducing a G2A mutation into Gag to block N-terminal myristoylation in the background of the Y132I mutation so that the MA/CA fusion protein generated from the Y132I mutant genome would not be anchored in the membrane. However, when the double mutant (G2A/Y132I) was tested in the phenotypic mixing protocol, the two mutations frequently became unlinked during the cotransfection protocol by host cell-mediated recombination with wild-type DNA (data not shown; 3), precluding the use of this experimental strategy.
In an attempt to address this potential mechanism, we also used the myristoylation inhibitor 2-hydroxymyristic acid to block the myristoylation of the Gag and Gag-Pro-Pol proteins. It was previously shown that nearly complete inhibition of Gag myristoylation is required to prevent virus budding (27) and that a Gag protein that cannot be myristoylated can act as a dominant negative inhibitor (20). When we tested the G2A mutant in phenotypic mixing experiments, we also found that more than 80% of G2A mutant Gag was needed to inhibit viral infectivity to low levels (Fig. 3A). We used drug concentrations lower than the IC50 (IC50 of 30 μM) (data not shown) to permit the undermyristoylation of Gag while retaining significant infectivity. If the transdominant mechanism of the MA/CA fusion protein on viral infectivity is due to the CA protein being indirectly anchored to the membrane and affecting downstream steps, the phenotypically mixed virus particles generated in the presence of the myristoylation inhibitor should acquire some viral infectivity since the dose response to the Y132I mutant protein is much greater than the dose response to the loss of myristoylation. Y132I mutant ratios tested in this experiment were 10% [Y132I(W90/M10)] and 5% [Y132I(W95/M5)]. As shown in Fig. 3B, however, viral infectivity was essentially unchanged over a wide range of inhibitor concentrations. With this experiment strategy, we could not show a role for membrane tethering as the mechanism of the Y132I inhibition of infectivity.
FIG. 3.
Inhibition of myristoylation of the Gag protein does not relieve viral infectivity of the viruses containing either 5% or 10% of the Y132I mutant. (A) Relative potency of a mutant lacking myristoylation (G2A). (B) Relative infectivity of the virus containing either 5% or 10% of the Y132I mutant in the presence of the myristoylation inhibitor 2-hydroxymyristic acid. 293T cells were transfected with wild-type and mutant DNAs, and the drug was added into the medium 4 h after transfection. Culture supernatants were harvested 48 h after transfection, and infectivity data were obtained according to the protocol described in the legend of Fig. 2. Mean data are shown.
Effect of a D51A mutation blocking a salt bridge between Pro1 and Asp51 on virion infectivity.
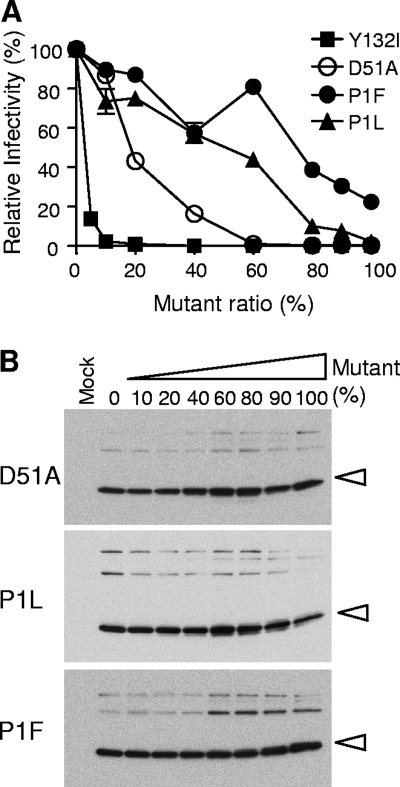
Another possibility for the transdominant mechanism of Y132I may involve the rearrangement of the N terminus of CA after the cleavage of the MA/CA processing site by PR. All orthoretroviruses encode a proline at the N terminus of CA (30). This proline plays a key structural role, as it anchors a salt bridge between its ring nitrogen and carboxylate group of a conserved aspartic acid (D51 in HIV-1 CA), creating a new loop with the N-terminal protein sequence that folds into a β-hairpin structure (21, 28, 38, 42). Preventing cleavage at the MA/CA site would sequester the proline from the formation of this loop, inhibiting proper core assembly. We attempted to mimic this effect with three different mutations by replacing the proline with either phenylalanine or leucine (P1F and P1L) or by replacing aspartic acid with alanine (D51A). P1F and P1L should permit cleavage by the protease (30) but preclude the use of a ring nitrogen in the salt bridge. As shown in Fig. 4A, the two proline substitution mutants were relatively weak in their abilities to inhibit virion infectivity in the phenotypic mixing assay; however, the D51A mutation was more potent, with an IC50 of approximately 20% mutant, although this is still less potent than the Y132I mutation. As expected, none of these mutations blocked either particle production or processing of the Gag precursor (Fig. 4B). While the D51A mutation strongly inhibits virion infectivity, it did not fully recapitulate the strong transdominant effect of the Y132I mutation. Therefore, we conclude that the Y132I mutation has a more dramatic effect on viral infectivity that may include but extends beyond disruption of this highly conserved salt bridge.
FIG. 4.
Mutations blocking salt bridge formation do not show potency equivalent to that of the Y132I mutant. (A) Relative potency of CA D51A, P1L, and P1F mutations. Phenotypic mixing experiments were performed according to the protocol described in the legend of Fig. 2. (B) Western analysis of viral particles harvested from the supernatant of 293T cells transfected with mutant and wild-type DNAs. The percentage of the mutant used for each cotransfection is shown. Open arrowheads indicate processed CA proteins.
Effect of the Y132I mutation on viral DNA synthesis.
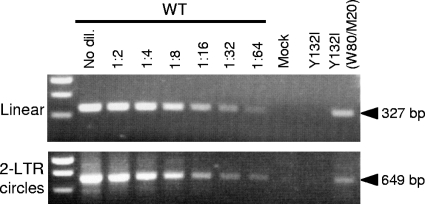
The Y132I mutation blocks infectivity without significantly inhibiting particle production (Fig. 2C), suggesting that an early step in the virus life cycle is blocked by the incorporation of the mutant protein into virions. We examined the effect of this Gag mutant on viral DNA synthesis by infecting cells with either wild-type virus, the Y132I mutant virus, or the Y132I(W80/M20) virus using equivalent amounts of virus (as measured by the amount of viral genomic RNA). To quantify the amount of an early viral DNA product and the 2-LTR circular DNA found in the nucleus, cells infected with these viruses were harvested at 24 h postinfection, and total DNA was isolated and used in semiquantitative PCR assays. The DNA isolated from the cells infected with wild-type virus was serially diluted for comparison. No detectable viral DNA was synthesized after infection with the Y132I mutant (Fig. 5) even though viral RNA was present in the virion (data not shown) and RT was present and fully processed (Fig. 2D). The Y132I(W80/M20) phenotypically mixed virions were able to synthesize an early viral DNA product at approximately 10% of the wild-type level. We also detected circular viral DNA from the Y132I(W80/M20) virus at approximately 5% of the wild-type level (Fig. 5). Thus, the results indicate that the presence of the Y132I mutant form of the Gag protein blocks detectable viral DNA synthesis either prior to or shortly after initiation, although the transdominant Y132I(W80/M20) virion allows some DNA synthesis but only at a low level, which can account for most of the transdominant phenotype. The small discordance between DNA synthesis and infectivity seen here and in the murine leukemia virus (MLV) system (33) with an analogous mutation must reflect either the stability of the viral DNA synthesized or the functioning of the preintegration complex.
FIG. 5.
Viral DNA synthesis in U87.CD4.CXCR4 cells infected with wild-type (WT) and Y132I and Y132I(W80/M20) mutant viruses. A 293T cell culture supernatant harvested at 48 h after transfection was used to infect U87.CD4.CXCR4 cells. Total DNA was isolated from the infected cells harvested at 24 h postinfection. To detect early viral DNA products, primers derived from the env gene were used. A separate set of primers was used to detect HIV-1 2-LTR circles as evidence of the synthesis of full-length viral DNA and import into the nucleus. For comparison, the PCR amplification of the wild-type sample was done in a dilution (dil.) series. Mock is a DNA sample isolated from cells infected with the culture supernatant from mock transfection (no DNA). In other experiments, we have shown that the PCR signal generated using this protocol is abolished if a viral DNA synthesis inhibitor is included at the time of infection. In this experiment, the lack of signal from the infection with the Y132I virus is consistent with a lack of transfected plasmid DNA being carried over to the infection.
DISCUSSION
In the present study, we demonstrate a potent transdominant mutation, Y132I, inhibiting wild-type infectivity with an IC50 in the range of 3 to 4% mutant to wild type. This is at least 10-fold more potent than the D25A PR mutant. However, since PR must function as a dimer, a 50:50 mix of wild-type and mutant PR results in only 25% of PR with two wild-type subunits. Thus, the Y132I mutation is actually 20-fold more potent than targeting protease activity. An analogous mutation within the Gag protein of MLV, S2D, blocking cleavage between p12 and CA showed a similar dominant-negative effect on wild-type infectivity, with 33% mutant reducing virion infectivity to 3% (33). The MA/CA processing site mutant in HIV-1 appears to be even more potent than the analogous mutation in MLV (p12/CA) since only 10% of the Y132I mutant can reduce virion infectivity to 4%.
RevM10, a transdominant mutant of the HIV-1 Rev protein, is the most extensively studied transdominant protein of HIV-1. The expression of RevM10 in human T-cell lines inhibits HIV-1 replication in vitro (2, 4, 5, 25). However, relatively high levels of RevM10, an excess of the RevM10 protein over wild-type Rev levels, appear to be required for a significant inhibition of HIV replication (31), with approximately 50% RevM10 mutant being needed to inhibit 50% of relative p24 Gag expression (24). Although HIV-1 proteins function as dimers (PR and RT), trimers (the surface and transmembrane subunits of the Env protein), tetramers (integrase), or higher oligomers (Rev), it seems unlikely that any mutation targeted outside of the multimeric interactions of Gag assembly could have an equivalently strong transdominant effect, suggesting that on a quantitative basis, the Gag assembly pathway is likely to be the most sensitive protein target in the virus life cycle. A transdominant (dominant negative) effect has also been reported for mutants of the capsid protein of poliovirus, supporting the idea that viral assembly pathways in general may be sensitive targets for inhibition (11).
An HIV-1 inhibitor currently in development provides a proof of concept for an inhibitor of a specific processing site. The maturation inhibitor PA-457 reduces cleavage at the CA/SP1 site (equivalent to the L363I mutation) (23, 49). It was previously shown that the compound is incorporated into immature HIV-1 particles, and this incorporation is largely eliminated by mutations at the CA/SP1 junction that confer PA-457 resistance (47, 48), implying a direct interaction between PA-457 and Gag. Given that the CA/SP1 cleavage site is relatively insensitive to inhibition (Fig. 2A), it seems likely that more potent inhibitors of this nature could be developed by targeting more sensitive steps in the assembly pathway, such as targeting cleavage of the MA/CA site.
Although the data presented in this paper did not reveal the exact mechanism of action of the dominant mutation, the Y132I mutation by itself retains the CA protein to the membrane-bound MA protein (Fig. 2F). In the transdominant negative setting, the viral core of Y132I(W80/M20) virus is asymmetrically tethered to the viral envelope (Fig. 2F). However, we could not rescue infectivity with a myristoylation inhibitor under conditions where the transdominant effect should have been much more sensitive to a gain of infectivity than the effect of the inhibitor on the loss of infectivity (27). Thus, we cannot account for the transdominant effect by membrane tethering. This is consistent with a major reduction in levels of viral DNA synthesis (Fig. 5), suggesting deleterious changes in the replication complex and not simply tethering of the complex to the host membrane. An MLV mutant containing an S2D Gag protein which blocks cleavage between p12 and CA also showed reductions in the amounts of viral DNA synthesized in infected cells (33), although the reductions were not as great as the one which we observed with HIV-1 Y132I Gag proteins. Mutations in HIV-1 CA affecting either core structure or core stability can lead to viruses that are defective in DNA synthesis (8, 14, 39). This suggests that the formation of a proper core is a prerequisite for downstream events such as disassembly and reverse transcription. Our EM images (Fig. 2F) clearly show that the mutant Y132I(W80/M20) virus contains an abnormal and eccentric core structure. Thus, this is likely sufficient to account for the reduction in viral DNA synthesis of the mutant virus. However, it is not clear how a small portion of the Y132I Gag proteins can poison the entire capsid assembly process. As an example that different protease-dependent steps may have different sensitivities to inhibition, a recent study demonstrated that viruses generated in the presence of a protease inhibitor had only a slight decrease in the overall thermodynamic stability of the viral RNA dimer, although the particles had incorrect core structures (26).
Our attempts to mimic the effect of a lack of processing at the MA/CA cleavage site by blocking the β-hairpin formation did not show the equivalent transdominant potency. The D51A mutation disrupts the CA Pro1-Asp51 salt bridge, rendering the virus noninfectious (Fig. 4) (42); however, this mutation was not as potent as the Y132I mutant in its transdominant effect (Fig. 4). Moreover, mutations of Pro1 were even less potent than the D51A substitution. Even though residues Pro1 and Asp 51 are highly conserved among retroviral capsid proteins, it is possible that these mutations alone do not fully block the conformational change, especially in the context of a mixture of wild-type Gag. In this regard, mutations of several amino acids within the β-hairpin have only modest effects on assembly and infectivity (43). The major difference in these mutations is that the Y132I mutation exerts its effect through a covalent bond, whereas all of the other less potent mutations exert their effects through noncovalent bonds. Due to this difference, the Y132I mutation may represent a theoretical limit on the potency of a transdominant mutation on Gag assembly/maturation.
Knowledge of a potent transdominant mutation in Gag fits in with several therapeutic paradigms. As noted above, the development of PA-457 represents a proof of concept for the inhibition of the cleavage of a specific processing site. Other inhibitors that target the complex process of Gag assembly are being developed, including a small molecule that binds to a pocket in the N-terminal domain of CA (CAP-1) (22, 37) and a helical peptide that binds to the C-terminal domain of CA (CAI) (34, 40). Finally, the utility of a protein with transdominant activity has been explored in the context of a gene therapy trial using the dominant negative mutant of Rev, RevM10 (1, 32). An important limitation in defining the MA/CA cleavage site as a drug target is the fact that in solution, this region is unstructured (38). However, the potential multiplicative effect of targeting this site argues for further efforts to exploit this highly vulnerable step in the virus life cycle.
Acknowledgments
This work was supported by an NIH award (P30-GM066524), the UNC Center For AIDS Research (NIH award P30-AI50410), and the Lineberger Cancer Center core grant (NIH award P30-CA16086).
We thank Steve Pettit for help in the early stages of this research. We also thank Eric Freed for helpful discussion.
Footnotes
Published ahead of print on 10 June 2009.
REFERENCES
- 1.Bahner, I., T. Sumiyoshi, M. Kagoda, R. Swartout, D. Peterson, K. Pepper, F. Dorey, J. Reiser, and D. B. Kohn. 2007. Lentiviral vector transduction of a dominant-negative Rev gene into human CD34+ hematopoietic progenitor cells potently inhibits human immunodeficiency virus-1 replication. Mol. Ther. 1576-85. [DOI] [PubMed] [Google Scholar]
- 2.Bahner, I., C. Zhou, X. J. Yu, Q. L. Hao, J. C. Guatelli, and D. B. Kohn. 1993. Comparison of trans-dominant inhibitory mutant human immunodeficiency virus type 1 genes expressed by retroviral vectors in human T lymphocytes. J. Virol. 673199-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bandyopadhyay, P. K., S. Watanabe, and H. M. Temin. 1984. Recombination of transfected DNAs in vertebrate cells in culture. Proc. Natl. Acad. Sci. USA 813476-3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauer, G., P. Valdez, K. Kearns, I. Bahner, S. F. Wen, J. A. Zaia, and D. B. Kohn. 1997. Inhibition of human immunodeficiency virus-1 (HIV-1) replication after transduction of granulocyte colony-stimulating factor-mobilized CD34+ cells from HIV-1-infected donors using retroviral vectors containing anti-HIV-1 genes. Blood 892259-2267. [PubMed] [Google Scholar]
- 5.Bevec, D., M. Dobrovnik, J. Hauber, and E. Bohnlein. 1992. Inhibition of human immunodeficiency virus type 1 replication in human T cells by retroviral-mediated gene transfer of a dominant-negative Rev trans-activator. Proc. Natl. Acad. Sci. USA 899870-9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bjorndal, A., H. Deng, M. Jansson, J. R. Fiore, C. Colognesi, A. Karlsson, J. Albert, G. Scarlatti, D. R. Littman, and E. M. Fenyo. 1997. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J. Virol. 717478-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Briggs, J. A., M. N. Simon, I. Gross, H. G. Krausslich, S. D. Fuller, V. M. Vogt, and M. C. Johnson. 2004. The stoichiometry of Gag protein in HIV-1. Nat. Struct. Mol. Biol. 11672-675. [DOI] [PubMed] [Google Scholar]
- 8.Cairns, T. M., and R. C. Craven. 2001. Viral DNA synthesis defects in assembly-competent Rous sarcoma virus CA mutants. J. Virol. 75242-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cordelier, P., E. Van Bockstaele, S. A. Calarota, and D. S. Strayer. 2003. Inhibiting AIDS in the central nervous system: gene delivery to protect neurons from HIV. Mol. Ther. 7801-810. [DOI] [PubMed] [Google Scholar]
- 10.Coren, L. V., J. A. Thomas, E. Chertova, R. C. Sowder II, T. D. Gagliardi, R. J. Gorelick, and D. E. Ott. 2007. Mutational analysis of the C-terminal gag cleavage sites in human immunodeficiency virus type 1. J. Virol. 8110047-10054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crowder, S., and K. Kirkegaard. 2005. Trans-dominant inhibition of RNA viral replication can slow growth of drug-resistant viruses. Nat. Genet. 37701-709. [DOI] [PubMed] [Google Scholar]
- 12.Douek, D. C., J. M. Brenchley, M. R. Betts, D. R. Ambrozak, B. J. Hill, Y. Okamoto, J. P. Casazza, J. Kuruppu, K. Kunstman, S. Wolinsky, Z. Grossman, M. Dybul, A. Oxenius, D. A. Price, M. Connors, and R. A. Koup. 2002. HIV preferentially infects HIV-specific CD4+ T cells. Nature 41795-98. [DOI] [PubMed] [Google Scholar]
- 13.DuBridge, R. B., P. Tang, H. C. Hsia, P. M. Leong, J. H. Miller, and M. P. Calos. 1987. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol. Cell. Biol. 7379-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forshey, B. M., U. von Schwedler, W. I. Sundquist, and C. Aiken. 2002. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J. Virol. 765667-5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furuta, R. A., R. Shimano, T. Ogasawara, R. Inubushi, K. Amano, H. Akari, M. Hatanaka, M. Kawamura, and A. Adachi. 1997. HIV-1 capsid mutants inhibit the replication of wild-type virus at both early and late infection phases. FEBS Lett. 415231-234. [DOI] [PubMed] [Google Scholar]
- 16.Gottlinger, H. G., J. G. Sodroski, and W. A. Haseltine. 1989. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 865781-5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Green, M., M. Ishino, and P. M. Loewenstein. 1989. Mutational analysis of HIV-1 Tat minimal domain peptides: identification of trans-dominant mutants that suppress HIV-LTR-driven gene expression. Cell 58215-223. [DOI] [PubMed] [Google Scholar]
- 18.Henderson, L. E., M. A. Bowers, R. C. Sowder II, S. A. Serabyn, D. G. Johnson, J. W. Bess, Jr., L. O. Arthur, D. K. Bryant, and C. Fenselau. 1992. Gag proteins of the highly replicative MN strain of human immunodeficiency virus type 1: posttranslational modifications, proteolytic processings, and complete amino acid sequences. J. Virol. 661856-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaplan, A. H., J. A. Zack, M. Knigge, D. A. Paul, D. J. Kempf, D. W. Norbeck, and R. Swanstrom. 1993. Partial inhibition of the human immunodeficiency virus type 1 protease results in aberrant virus assembly and the formation of noninfectious particles. J. Virol. 674050-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawada, S., T. Goto, H. Haraguchi, A. Ono, and Y. Morikawa. 2008. Dominant negative inhibition of human immunodeficiency virus particle production by the nonmyristoylated form of gag. J. Virol. 824384-4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelly, B. N., B. R. Howard, H. Wang, H. Robinson, W. I. Sundquist, and C. P. Hill. 2006. Implications for viral capsid assembly from crystal structures of HIV-1 Gag(1-278) and CA(N)(133-278). Biochemistry 4511257-11266. [DOI] [PubMed] [Google Scholar]
- 22.Kelly, B. N., S. Kyere, I. Kinde, C. Tang, B. R. Howard, H. Robinson, W. I. Sundquist, M. F. Summers, and C. P. Hill. 2007. Structure of the antiviral assembly inhibitor CAP-1 complex with the HIV-1 CA protein. J. Mol. Biol. 373355-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, F., R. Goila-Gaur, K. Salzwedel, N. R. Kilgore, M. Reddick, C. Matallana, A. Castillo, D. Zoumplis, D. E. Martin, J. M. Orenstein, G. P. Allaway, E. O. Freed, and C. T. Wild. 2003. PA-457: a potent HIV inhibitor that disrupts core condensation by targeting a late step in Gag processing. Proc. Natl. Acad. Sci. USA 10013555-13560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malim, M. H., S. Bohnlein, J. Hauber, and B. R. Cullen. 1989. Functional dissection of the HIV-1 Rev trans-activator—derivation of a trans-dominant repressor of Rev function. Cell 58205-214. [DOI] [PubMed] [Google Scholar]
- 25.Malim, M. H., W. W. Freimuth, J. Liu, T. J. Boyle, H. K. Lyerly, B. R. Cullen, and G. J. Nabel. 1992. Stable expression of transdominant Rev protein in human T cells inhibits human immunodeficiency virus replication. J. Exp. Med. 1761197-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore, M. D., W. Fu, F. Soheilian, K. Nagashima, R. G. Ptak, V. K. Pathak, and W. S. Hu. 2008. Suboptimal inhibition of protease activity in human immunodeficiency virus type 1: effects on virion morphogenesis and RNA maturation. Virology 379152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morikawa, Y., S. Hinata, H. Tomoda, T. Goto, M. Nakai, C. Aizawa, H. Tanaka, and S. Omura. 1996. Complete inhibition of human immunodeficiency virus Gag myristoylation is necessary for inhibition of particle budding. J. Biol. Chem. 2712868-2873. [DOI] [PubMed] [Google Scholar]
- 28.Mortuza, G. B., L. F. Haire, A. Stevens, S. J. Smerdon, J. P. Stoye, and I. A. Taylor. 2004. High-resolution structure of a retroviral capsid hexameric amino-terminal domain. Nature 431481-485. [DOI] [PubMed] [Google Scholar]
- 29.Pettit, S. C., G. J. Henderson, C. A. Schiffer, and R. Swanstrom. 2002. Replacement of the P1 amino acid of human immunodeficiency virus type 1 Gag processing sites can inhibit or enhance the rate of cleavage by the viral protease. J. Virol. 7610226-10233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pettit, S. C., J. Simsic, D. D. Loeb, L. Everitt, C. A. Hutchison III, and R. Swanstrom. 1991. Analysis of retroviral protease cleavage sites reveals two types of cleavage sites and the structural requirements of the P1 amino acid. J. Biol. Chem. 26614539-14547. [PubMed] [Google Scholar]
- 31.Plavec, I., M. Agarwal, K. E. Ho, M. Pineda, J. Auten, J. Baker, H. Matsuzaki, S. Escaich, M. Bonyhadi, and E. Bohnlein. 1997. High transdominant RevM10 protein levels are required to inhibit HIV-1 replication in cell lines and primary T cells: implication for gene therapy of AIDS. Gene Ther. 4128-139. [DOI] [PubMed] [Google Scholar]
- 32.Podsakoff, G. M., B. C. Engel, D. A. Carbonaro, C. Choi, E. M. Smogorzewska, G. Bauer, D. Selander, S. Csik, K. Wilson, M. R. Betts, R. A. Koup, G. J. Nabel, K. Bishop, S. King, M. Schmidt, C. von Kalle, J. A. Church, and D. B. Kohn. 2005. Selective survival of peripheral blood lymphocytes in children with HIV-1 following delivery of an anti-HIV gene to bone marrow CD34(+) cells. Mol. Ther. 1277-86. [DOI] [PubMed] [Google Scholar]
- 33.Rulli, S. J., Jr., D. Muriaux, K. Nagashima, J. Mirro, M. Oshima, J. G. Baumann, and A. Rein. 2006. Mutant murine leukemia virus Gag proteins lacking proline at the N-terminus of the capsid domain block infectivity in virions containing wild-type Gag. Virology 347364-371. [DOI] [PubMed] [Google Scholar]
- 34.Sticht, J., M. Humbert, S. Findlow, J. Bodem, B. Muller, U. Dietrich, J. Werner, and H. G. Krausslich. 2005. A peptide inhibitor of HIV-1 assembly in vitro. Nat. Struct. Mol. Biol. 12671-677. [DOI] [PubMed] [Google Scholar]
- 35.Strayer, D. S., F. Branco, J. Landre, M. BouHamdan, F. Shaheen, and R. J. Pomerantz. 2002. Combination genetic therapy to inhibit HIV-1. Mol. Ther. 533-41. [DOI] [PubMed] [Google Scholar]
- 36.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 263-334. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- 37.Tang, C., E. Loeliger, I. Kinde, S. Kyere, K. Mayo, E. Barklis, Y. Sun, M. Huang, and M. F. Summers. 2003. Antiviral inhibition of the HIV-1 capsid protein. J. Mol. Biol. 3271013-1020. [DOI] [PubMed] [Google Scholar]
- 38.Tang, C., Y. Ndassa, and M. F. Summers. 2002. Structure of the N-terminal 283-residue fragment of the immature HIV-1 Gag polyprotein. Nat. Struct. Biol. 9537-543. [DOI] [PubMed] [Google Scholar]
- 39.Tang, S., T. Murakami, N. Cheng, A. C. Steven, E. O. Freed, and J. G. Levin. 2003. Human immunodeficiency virus type 1 N-terminal capsid mutants containing cores with abnormally high levels of capsid protein and virtually no reverse transcriptase. J. Virol. 7712592-12602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ternois, F., J. Sticht, S. Duquerroy, H. G. Krausslich, and F. A. Rey. 2005. The HIV-1 capsid protein C-terminal domain in complex with a virus assembly inhibitor. Nat. Struct. Mol. Biol. 12678-682. [DOI] [PubMed] [Google Scholar]
- 41.Trono, D., M. B. Feinberg, and D. Baltimore. 1989. HIV-1 Gag mutants can dominantly interfere with the replication of the wild-type virus. Cell 59113-120. [DOI] [PubMed] [Google Scholar]
- 42.von Schwedler, U. K., T. L. Stemmler, V. Y. Klishko, S. Li, K. H. Albertine, D. R. Davis, and W. I. Sundquist. 1998. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 171555-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.von Schwedler, U. K., K. M. Stray, J. E. Garrus, and W. I. Sundquist. 2003. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J. Virol. 775439-5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei, X., J. M. Decker, H. Liu, Z. Zhang, R. B. Arani, J. M. Kilby, M. S. Saag, X. Wu, G. M. Shaw, and J. C. Kappes. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 461896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wiegers, K., G. Rutter, H. Kottler, U. Tessmer, H. Hohenberg, and H. G. Krausslich. 1998. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J. Virol. 722846-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilk, T., and S. D. Fuller. 1999. Towards the structure of the human immunodeficiency virus: divide and conquer. Curr. Opin. Struct. Biol. 9231-243. [DOI] [PubMed] [Google Scholar]
- 47.Zhou, J., C. H. Chen, and C. Aiken. 2006. Human immunodeficiency virus type 1 resistance to the small molecule maturation inhibitor 3-O-(3′,3′-dimethylsuccinyl)-betulinic acid is conferred by a variety of single amino acid substitutions at the CA-SP1 cleavage site in Gag. J. Virol. 8012095-12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou, J., L. Huang, D. L. Hachey, C. H. Chen, and C. Aiken. 2005. Inhibition of HIV-1 maturation via drug association with the viral Gag protein in immature HIV-1 particles. J. Biol. Chem. 28042149-42155. [DOI] [PubMed] [Google Scholar]
- 49.Zhou, J., X. Yuan, D. Dismuke, B. M. Forshey, C. Lundquist, K. H. Lee, C. Aiken, and C. H. Chen. 2004. Small-molecule inhibition of human immunodeficiency virus type 1 replication by specific targeting of the final step of virion maturation. J. Virol. 78922-929. [DOI] [PMC free article] [PubMed] [Google Scholar]