Abstract
Two frequently employed methods for generating well-characterized, genetically defined infectious human immunodeficiency virus type 1 in vitro include the use of infectious molecular clones (IMCs) and pseudoviruses (PVs) competent for single-round infection. We compared six matched pairs of IMCs and PVs. The relative amounts of Env incorporated and efficiency of cleavage differed substantially between the two systems. Altering the ratio of proviral genome and env expression plasmids can produce pseudovirions that are structurally more similar to the matched IMCs. Differences in Env incorporation and cleavage translated into moderate differences in assays infectivity and sensitivity to neutralizing antibodies and entry inhibitors.
In 2005, the Global Vaccine Enterprise called for the standardization of assays and put forth a strong suggestion for the use of pseudovirus (PV)-based assays (16). While there are a number of important advantages provided by the PV system (16) and significant efforts have gone into optimization and standardization (5, 8, 13, 16, 18, 23), the report highlighted a number of key areas that were in need of further study. One area was the careful comparison of replication-competent viruses and PVs with respect to envelope (Env) processing and incorporation and the effects that any differences may have on the outcome of assays.
The creation of an infectious molecular clone (IMC) that represents the full proviral genome of a human immunodeficiency virus (HIV) variant allows the precise characterization and genetic manipulation of an isolate (1). In the IMC, the HIV long terminal repeat is used as a promoter to drive expression of the genome, and mRNA quantity and splicing are therefore the same as in native virions. The creation of IMCs can be technically challenging and therefore represents a substantial bottleneck in fully characterizing the numerous variants present within an individual. The PV system provides a more manageable alternative wherein the env gene is on a separate expression vector from the rest of the genome and is expressed with an exogenous constitutive promoter, most frequently that of the cytomegalovirus immediate-early 1 gene (6).
Although previous reports have begun to compare IMCs and PVs (11, 12, 15, 20), the data remain inconclusive. To address areas where an extensive comparison has not yet been made, this study analyzed six matched pairs of HIV type 1 (HIV-1) IMCs and PVs that contain identical genomes. The study focused on four key parameters known to influence HIV-1 function and infectivity: Env cleavage into gp120 and gp41, total Env incorporated into the virion, viral infectivity, and sensitivity to inhibitors that target different steps in viral entry.
To directly compare virions produced from the IMC and PV systems, we used the env sequences of six previously described maternal primary HIV-1 isolates (22, 27). This sample set comprised three clade A isolates (208.A3, 505.H3, and 505.C2), two C/D recombinants (184.G3 and 184.E4), and one D/A recombinant (535.B1). Q23Δenv (14, 21) was used at a by-mass 20:1 backbone/env ratio (except where noted otherwise) to complement the env expression plasmids and create PV, as previously described (4). To generate proviral chimeras for making IMCs, the same Q23-17 plasmid was engineered to have an XhoI restriction site in nef at position 8360 and an XhoI site was removed from the 3′ polylinker, allowing the native env gene to be excised with the restriction enzymes SmaI and XhoI. Therefore, vpu, rev, and env were isolate specific, while the remaining genome was derived from the Q23 backbone. The Q23 derivative Q23XhoΔXho (E. M. Long and J. Overbaugh, unpublished data) was used to create IMCs by directly ligating in the env gene of interest at the SmaI/XhoI sites. Full env sequencing was performed to verify the presence of the appropriate env gene. All virus preparations were generated by transfection of the same commonly used cell line, HEK293T cells.
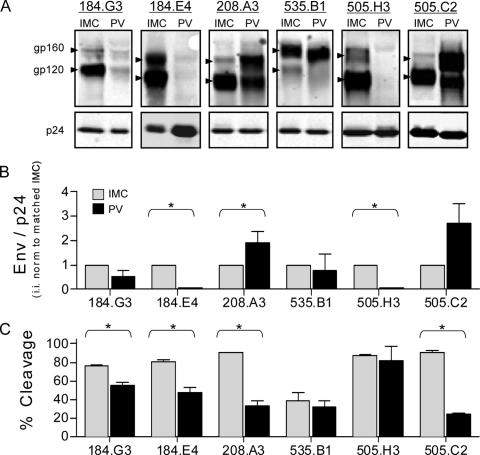
Purified virions were analyzed for Env incorporation and Env cleavage (amount of gp120 relative to that of unprocessed gp160). A compilation of quantitative Western blot assays is shown in Fig. 1A and shows representative data, obtained as previously described (4), for each matched pair. The observed variation in Env mobility between isolates is consistent with the expected sizes of the Env proteins based on the numbers of amino acids and potential N-linked glycosylation sites. A double banding pattern was present in the uncleaved gp160 form of some variants, an observation that has been consistently reported by others and is believed to represent a highly sialylated form of Env (2, 9, 11, 17, 19). The low Env expression level of PVs 184.G3, 184.E4, and 505.H3 required the exposure intensity to be optimized for each virus pair. Since the LI-COR Odyssey system has a wide linear range (25) and the calculated values rely upon the ratio of the bands within the same lane, this manipulation had no impact on the calculated values. All comparisons of matched data were analyzed on the same gel by using the same manipulations.
FIG. 1.
Env incorporation and cleavage of matched IMC-PV pairs. (A) Representative Western blot assays of six matched IMC-PV pairs. The gp160 and gp120 bands for each pair are indicated by arrowheads, as differences in variable loop length and carbohydrate numbers resulted in variations in band migration. Matched Gag p24 (p24) bands are shown along the bottom. The integrated intensity (i.i.) of each band was quantified from a minimum of three independent experiments. (B) Virion Env incorporation was calculated with the formula (gp120 i.i. + gp160 i.i.)/p24 i.i. For each variant, the IMC value was set at 1, PV was calculated relative to the matched IMC, and data were analyzed with a one-sample t test. (C) Percent cleavage was calculated with the formula gp120 i.i./(gp120 i.i. + gp160 i.i.). Matched IMC and PV values were compared by the Mann-Whitney test. An asterisk denotes P < 0.05, and error bars represent the standard error of the mean.
Each IMC-PV pair exhibited relatively equivalent levels of p24 (Fig. 1A, bottom), suggesting that the overall amounts of virions produced by the IMC and PV systems were similar (7). However, the amount of virion-associated Env differed between the two models (Fig. 1B) and was frequently lower in the PV system. The relative percentage of cleaved Env also differed significantly between the matched IMC and PV, again revealing that the PV more frequently had lower levels of cleaved gp120 Env (Fig. 1C). The most striking example of this was seen with variant 505.C2, where 90% of the total Env was in the cleaved (gp120) form in the IMC but only 24% was cleaved in the PV.
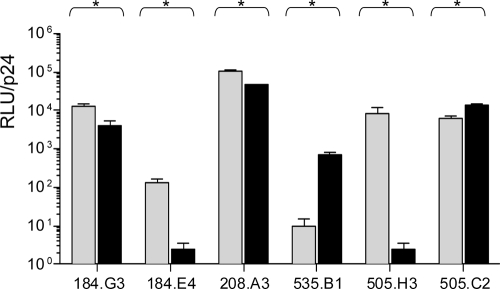
In order to determine if the observed physical differences impact measurements of biological significance, we assayed matched pairs for infectivity and neutralization sensitivity. Virus titers were determined on Tzm-bl cells by a previously described method (4) wherein relative light units from the linear part of the curve are normalized to p24 values from the same preparation. All six matched pairs exhibited statistically significant variability in infectivity, though the nature of the differences varied in direction and magnitude (Fig. 2). Neither Env incorporation nor Env cleavage alone could completely account for the direction of the difference.
FIG. 2.
Infectivity of IMC and PV matched pairs. Matched IMC (gray bars) and PV (black bars) pairs are shown. Infectivity was calculated as relative light units (RLU) normalized to p24 integrated intensity. Each sample was analyzed a minimum of four times, and mean values ± the standard errors of the means are shown. All matched pairs are significantly different by the Mann-Whitney test (P < 0.05).
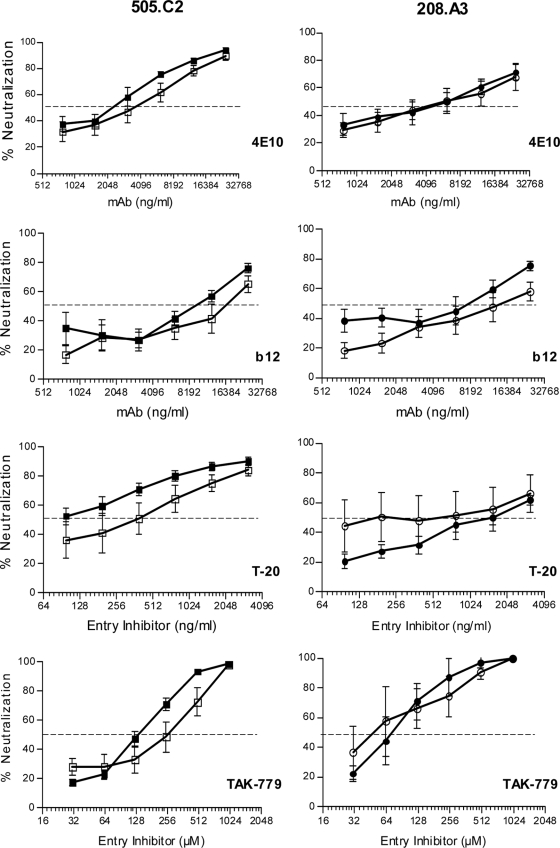
Two matched pairs, 505.C2 and 208.A3, were further tested against four entry inhibitors by using the Tzm-bl neutralization assay as previously described (4, 27). These pairs were selected because the matched IMC and PV demonstrated marked differences in Env incorporation, Env cleavage, and viral infectivity while providing robust titers to perform neutralization assays. This analysis used monoclonal antibodies (MAbs) 4E10 and IgG1-b12 (b12), the fusion inhibitor T-20, and the CCR5 inhibitor TAK-779 to target different epitopes on Env and different stages of viral fusion to the target cell as previously described (3, 10, 24, 26).
In both sets of data, the overall profile of neutralization/inhibition sensitivity was generally similar between the matched IMC and PV. The 50% inhibitory concentrations for all but one analysis were less than 2.5-fold different between IMC and PV, a benchmark frequently used to identify a significant difference in neutralization sensitivity. For isolate 505.C2, there was a modest though consistent trend for the PV to be more sensitive than the IMC to neutralization by all of the MAbs and entry inhibitors tested (Fig. 3). However, the most dramatic difference was seen with T-20, which required a concentration to neutralize the IMC that was fivefold higher than that required to neutralize the PV (50% inhibitory concentrations of 0.5 and 0.1 μg/ml, respectively), yet the difference only trended toward significance (P = 0.07, Mann-Whitney test). The 208.A3 matched pair did not exhibit any consistent patterns in sensitivity and showed an overall similar profile between the IMC and PV.
FIG. 3.
Neutralization sensitivity of matched pairs for 505.C2 and 208.A3. The matched IMC and PV for 505.C2 (left panels) and 208.A3 (right panels) were tested against MAb 4E10 (recognizes gp41), MAb b12 (recognizes CD4 binding site), the fusion inhibitor T-20, and the CCR5 inhibitor TAK-779, as indicated. The x axis is the log2 concentration of MAb (nanograms per milliliter), T-20 (nanograms per milliliter), or TAK-779 (micromolar). The y axis denotes percent neutralization calculated with the formula (background-normalized virus only − background-normalized virus with entry inhibitor)/background-normalized virus only. Each curve represents the mean ± the standard error of the mean of at least three independent experiments. None of the matched pairs reached significance by the Mann-Whitney test (P < 0.05). Open symbols denote IMC, and closed symbols denote PV. The 50% inhibitory concentration is marked with a dashed line.
Although little difference was observed between the neutralization/inhibition data obtained with either IMCs or PVs, there were unpredictable differences in infectivity combined with the marked differences in Env incorporation and cleavage. This result is likely based on the overexpression of Env in the PV system when the gene exists on a separate plasmid under the control of its own promoter, particularly a strong promoter such as that of the human cytomegalovirus immediate-early 1 gene.
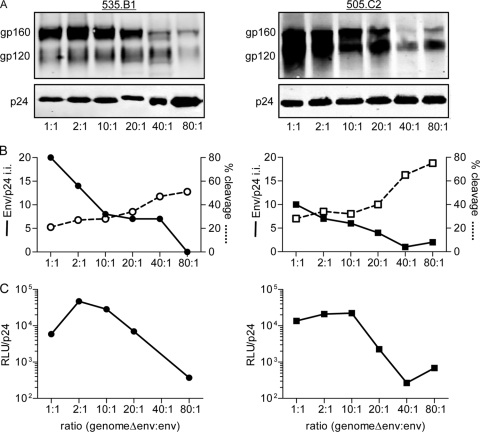
This led us to test if the amount of env plasmid present in the transfection system was responsible for the observed differences in virus outcome and to discern if the two systems could be made more congruent. A panel of PVs was created with various backbone/env plasmid ratios. The total amount of transfected DNA was held constant, while the relative amount of env plasmid was decreased. For each variant, PV reparations were made at backbone/env plasmid mass ratios of 1:1, 2:1, 10:1, 20:1, 40:1, and 80:1. The 505.C2 and 535.B1 isolates were used in this analysis.
Not surprisingly, as the backbone-to-env plasmid ratio increased (and therefore the relative amount of env plasmid decreased), the amount of virion incorporated Env also decreased dramatically (Fig. 4A). This was true for both 535.B1 and 505.C2. The total amount of p24 stayed relatively constant (Fig. 4A) despite the increase in backbone plasmid. The net result was less Env per virion as determined by the formula (gp120 i.i. + gp160 i.i.)/p24 (Fig. 4B, solid lines). Decreased amounts of env plasmid also corresponded to higher levels of Env cleavage in both of the isolates tested (Fig. 4B, dashed lines). For variant 505.C2, PV produced with the lowest amount of env plasmid (80:1 ratio) approached the level of cleavage (90%) seen in the matched 505.C2 IMC (compare Fig. 4B, dashed line, to 1C). Likewise, the 535.B1 PV produced with the lowest amount of env plasmid (80:1 ratio) exceeded the level of cleavage seen with the matched IMC (51 and 39%, respectively, compare Fig. 4B, dashed line, to 1C). For both of the 505.C2 and 535.B1 variants, the lower amount of env plasmid (80:1 ratio), which corresponded to less Env incorporation and higher Env cleavage, also resulted in decreased infectivity (Fig. 4C) spanning 2 log orders of magnitude. This lower infectivity in the PVs at the 80:1 ratio for both 535.B1 and 505.C2 is consistent with lower infectivity of the IMCs in these pairs (Fig. 2).
FIG. 4.
PV comparisons of Env incorporation, Env cleavage, and viral infectivity. A panel of PVs was created from isolates 535.B1 (left side) and 505.C2 (right side). The total amount of transfected plasmid DNA was held constant, while the relative amounts of backbone and env plasmids were varied (1:1 had equal amounts of both plasmids, while 80:1 had 80 times more backbone plasmid than env plasmid). (A) Quantitative Western blots were probed for uncleaved Env (gp160), cleaved Env (gp120), and Gag p24. (B) Env incorporation (determined with the formula [gp160 i.i. + gp120 i.i.]/p24 i.i., where i.i. is integrated intensity) is shown as a solid line, while Env cleavage (determined with the equation gp120 i.i./[gp160 i.i. + gp120 i.i.]) is shown as a dashed line. (C) Infectivity was calculated as relative light units (RLU) normalized to the number of picograms of p24. Complete analyses were repeated a minimum of two times, and representative data are shown.
In light of the need for a thorough comparison of the PV and IMC systems, this study presents a systematic analysis of four key areas in which the two systems of virus production may vary biochemically and/or phenotypically: Env virion incorporation, Env cleavage, viral infectivity, and sensitivity to entry inhibitors. This provides a direct comparison of the IMC and PV systems when virions are produced from the same frequently used producer cell line, HEK293T cells. A central finding of this study is that matched IMCs and PVs do not produce physically equivalent virions when constructed in the manner described. We found equal or significantly greater Env processing in the IMC for all six samples. The total Env incorporation was more variable; both the PV and IMC exhibited greater incorporation, depending on the isolate. Additionally, all six pairs exhibited statistically significant differences in infectivity with a range of magnitudes and directions. Despite these differences, previous work by Louder et al. (15) demonstrated no difference in neutralization sensitivity in three clade B viruses. Our study corroborates this finding with two clade A viruses, providing further validation of the PV system for determination of neutralization and inhibition sensitivity. For the characteristics in which the IMC and PV do exhibit differences, our study demonstrates a simple technique for reconciling these differences by varying the backbone/env ratio, allowing the use of the PV for biochemical analyses.
Acknowledgments
We thank Adam Parast for assistance in the construction of IMCs and Stephanie Rainwater for assistance with neutralization assays. We thank Dennis Burton for providing IgG1b12 and David Montefiori for providing HIV immunoglobulin.
This study was supported by grants P01-AI54564 (N.L.H.) and R01-AI076105 (J.O.), a Center for AIDS Research predoctoral fellowship (P30-AI27757 to W.B.P.), a grant from the Murdock Charitable Trust (N.L.H.), and donations from the James B. Pendleton Charitable Trust. The virus stocks, MAbs, and cell lines used in neutralization assays were obtained from the NIH AIDS Research and Reference Reagent Program.
Footnotes
Published ahead of print on 17 June 2009.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams, O., and A. Scheid. 2001. Stepwise deletion of the HIV type 1 glycoprotein 41 N terminus leads to an increasing export of microvesicles containing uncleaved Env glycoprotein. AIDS Res. Hum. Retrovir. 171345-1356. [DOI] [PubMed] [Google Scholar]
- 3.Baba, M., O. Nishimura, N. Kanzaki, M. Okamoto, H. Sawada, Y. Iizawa, M. Shiraishi, Y. Aramaki, K. Okonogi, Y. Ogawa, K. Meguro, and M. Fujino. 1999. A small-molecule, nonpeptide CCR5 antagonist with highly potent and selective anti-HIV-1 activity. Proc. Natl. Acad. Sci. USA 965698-5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blay, W. M., T. Kasprzyk, L. Misher, B. A. Richardson, and N. L. Haigwood. 2007. Mutations in envelope gp120 can impact proteolytic processing of the gp160 precursor and thereby affect neutralization sensitivity of human immunodeficiency virus type 1 pseudoviruses. J. Virol. 8113037-13049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, B. K., J. M. Darden, S. Tovanabutra, T. Oblander, J. Frost, E. Sanders-Buell, M. S. de Souza, D. L. Birx, F. E. McCutchan, and V. R. Polonis. 2005. Biologic and genetic characterization of a panel of 60 human immunodeficiency virus type 1 isolates, representing clades A, B, C, D, CRF01_AE, and CRF02_AG, for the development and assessment of candidate vaccines. J. Virol. 796089-6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chapman, B. S., R. M. Thayer, K. A. Vincent, and N. L. Haigwood. 1991. Effect of intron A from human cytomegalovirus (Towne) immediate-early gene on heterologous expression in mammalian cells. Nucleic Acids Res. 193979-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chertova, E., J. W. Bess, Jr., B. J. Crise, I. R. Sowder, T. M. Schaden, J. M. Hilburn, J. A. Hoxie, R. E. Benveniste, J. D. Lifson, L. E. Henderson, and L. O. Arthur. 2002. Envelope glycoprotein incorporation, not shedding of surface envelope glycoprotein (gp120/SU), is the primary determinant of SU content of purified human immunodeficiency virus type 1 and simian immunodeficiency virus. J. Virol. 765315-5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fenyö, E., A. Heath, S. Dispinseri, H. Holmes, P. Lusso, S. Zolla-Pazner, H. Donners, L. Heyndrickx, J. Alcami, V. Bongertz, C. Jassoy, M. Malnati, D. Montefiori, C. Moog, L. Morris, S. Osmanov, V. Polonis, Q. Sattentau, H. Schuitemaker, R. Sutthent, T. Wrin, G. Scarlatti, and D. Unutmaz. 2009. International network for comparison of HIV neutralization assays: the NeutNet report. PLoS ONE 4e4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freed, E. O., D. J. Myers, and R. Risser. 1989. Mutational analysis of the cleavage sequence of the human immunodeficiency virus type 1 envelope glycoprotein precursor gp160. J. Virol. 634670-4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furuta, R. A., C. T. Wild, Y. Weng, and C. D. Weiss. 1998. Capture of an early fusion-active conformation of HIV-1 gp41. Nat. Struct. Biol. 5276-279. [DOI] [PubMed] [Google Scholar]
- 11.Herrera, C., P. J. Klasse, E. Michael, S. Kake, K. Barnes, C. W. Kibler, L. Campbell-Gardener, Z. Si, J. Sodroski, J. P. Moore, and S. Beddows. 2005. The impact of envelope glycoprotein cleavage on the antigenicity, infectivity, and neutralization sensitivity of Env-pseudotyped human immunodeficiency virus type 1 particles. Virology 338154-172. [DOI] [PubMed] [Google Scholar]
- 12.Heyndrickx, L., T. Vermoesen, K. Vereecken, J. Kurth, S. Coppens, L. Aerts, A. Ohagen, Y. Van Herrewege, P. Lewi, and G. Vanham. 2008. Antiviral compounds show enhanced activity in HIV-1 single cycle pseudovirus assays as compared to classical PBMC assays. J. Virol. Methods 148166-173. [DOI] [PubMed] [Google Scholar]
- 13.Li, M., J. F. Salazar-Gonzalez, C. A. Derdeyn, L. Morris, C. Williamson, J. E. Robinson, J. M. Decker, Y. Li, M. G. Salazar, V. R. Polonis, K. Mlisana, S. A. Karim, K. Hong, K. M. Greene, M. Bilska, J. Zhou, S. Allen, E. Chomba, J. Mulenga, C. Vwalika, F. Gao, M. Zhang, B. T. M. Korber, E. Hunter, B. H. Hahn, and D. C. Montefiori. 2006. Genetic and neutralization properties of acute and early subtype C human immunodeficiency virus type 1 molecular env clones from heterosexually acquired infections in southern Africa. J. Virol. 8011776-11790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Long, E. M., S. M. Rainwater, L. Lavreys, K. Mandaliya, and J. Overbaugh. 2002. HIV type 1 variants transmitted to women in Kenya require the CCR5 coreceptor for entry, regardless of the genetic complexity of the infecting virus. AIDS Res. Hum. Retrovir. 18567-576. [DOI] [PubMed] [Google Scholar]
- 15.Louder, M. K., A. Sambor, E. Chertova, T. Hunte, S. Barrett, F. Ojong, E. Sanders-Buell, S. Zolla-Pazner, F. E. McCutchan, J. D. Roser, D. Gabuzda, J. D. Lifson, and J. R. Mascola. 2005. HIV-1 envelope pseudotyped viral vectors and infectious molecular clones expressing the same envelope glycoprotein have a similar neutralization phenotype, but culture in peripheral blood mononuclear cells is associated with decreased neutralization sensitivity. Virology 339226-238. [DOI] [PubMed] [Google Scholar]
- 16.Mascola, J. R., P. D'Souza, P. Gilbert, B. H. Hahn, N. L. Haigwood, L. Morris, C. J. Petropoulos, V. R. Polonis, M. Sarzotti, and D. C. Montefiori. 2005. Recommendations for the design and use of standard virus panels to assess neutralizing antibody responses elicited by candidate human immunodeficiency virus type 1 vaccines. J. Virol. 7910103-10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCune, J. M., L. B. Rabin, M. B. Feinberg, M. Lieberman, J. C. Kosek, G. R. Reyes, and I. L. Weissman. 1988. Endoproteolytic cleavage of gp160 is required for the activation of human immunodeficiency virus. Cell 5355-67. [DOI] [PubMed] [Google Scholar]
- 18.Moore, J. P., and D. R. Burton. 2004. Urgently needed: a filter for the HIV-1 vaccine pipeline. Nat. Med. 10769-771. [DOI] [PubMed] [Google Scholar]
- 19.Pancera, M., and R. Wyatt. 2005. Selective recognition of oligomeric HIV-1 primary isolate envelope glycoproteins by potently neutralizing ligands requires efficient precursor cleavage. Virology 332145-156. [DOI] [PubMed] [Google Scholar]
- 20.Polonis, V. R., B. K. Brown, A. Rosa Borges, S. Zolla-Pazner, D. S. Dimitrov, M. Y. Zhang, S. W. Barnett, R. M. Ruprecht, G. Scarlatti, E. M. Fenyo, D. C. Montefiori, F. E. McCutchan, and N. L. Michael. 2008. Recent advances in the characterization of HIV-1 neutralization assays for standardized evaluation of the antibody response to infection and vaccination. Virology 375315-320. [DOI] [PubMed] [Google Scholar]
- 21.Poss, M., and J. Overbaugh. 1999. Variants from the diverse virus population identified at seroconversion of a clade A human immunodeficiency virus type 1-infected woman have distinct biological properties. J. Virol. 735255-5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rainwater, S. M., X. Wu, R. Nduati, R. Nedellec, D. Mosier, G. John-Stewart, D. Mbori-Ngacha, and J. Overbaugh. 2007. Cloning and characterization of functional subtype A HIV-1 envelope variants transmitted through breastfeeding. Curr. HIV Res. 5189-197. [DOI] [PubMed] [Google Scholar]
- 23.Reynard, F., A. Fatmi, B. Verrier, and F. Bedin. 2004. HIV-1 acute infection env glycomutants designed from 3D model: effects on processing, antigenicity, and neutralization sensitivity. Virology 32490-102. [DOI] [PubMed] [Google Scholar]
- 24.Roben, P., J. P. Moore, M. Thali, J. Sodroski, C. F. Barbas III, and D. R. Burton. 1994. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J. Virol. 684821-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schutz-Geschwender, A., Y. Zhang, T. Holt, D. McDermitt, and D. M. Olive. 2004. Quantitative, two-color Western blot detection with infrared fluorescence. LI-COR Biosciences, Lincoln, NE. http://www.licor.com/bio/PDF/IRquant.pdf.
- 26.Stiegler, G., R. Kunert, M. Purtscher, S. Wolbank, R. Voglauer, F. Steindl, and H. Katinger. 2001. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 171757-1765. [DOI] [PubMed] [Google Scholar]
- 27.Wu, X., A. B. Parast, B. A. Richardson, R. Nduati, G. John-Stewart, D. Mbori-Ngacha, S. M. Rainwater, and J. Overbaugh. 2006. Neutralization escape variants of human immunodeficiency virus type 1 are transmitted from mother to infant. J. Virol. 80835-844. [DOI] [PMC free article] [PubMed] [Google Scholar]