Abstract
Hendra virus F protein-promoted membrane fusion requires the presence of the viral attachment protein, G. However, events leading to the association of these glycoproteins remain unclear. Results presented here demonstrate that Hendra virus G undergoes slower secretory pathway trafficking than is observed for Hendra virus F. This slowed trafficking is not dependent on the G protein cytoplasmic tail, the presence of the G receptor ephrin B2, or interaction with other viral proteins. Instead, Hendra virus G was found to undergo intrinsically slow oligomerization within the endoplasmic reticulum. These results suggest that the critical F-G interactions occur only after the initial steps of synthesis and cellular transport.
The Henipavirus genus of the paramyxovirus family comprises two recently emerged, zoonotic pathogens. Hendra virus, first identified in Australia in 1994, caused respiratory illness in and the subsequent death of over one dozen horses and two of the three humans infected (12, 21, 25). Nipah virus led to an outbreak of respiratory and encephalitic illnesses in Malaysia in 1999, affecting both swine and humans and leading to fatality in 105 of the 265 human cases (11). Additional periodic outbreaks of infections with these viruses have occurred (11), and evidence indicates human-to-human transmission of Nipah virus in at least one outbreak (16). Henipavirus, like many paramyxoviruses, requires the presence of two surface glycoproteins for virus-cell and cell-cell fusion (8, 9, 36): the fusion protein, F, which mediates the membrane fusion event, and the attachment protein, G, which binds cellular receptors ephrin B2 (6, 22) and ephrin B3 (23) and which is required for F-mediated membrane fusion. Interactions between the fusion and attachment proteins of a number of paramyxoviruses have been observed previously (30, 33, 35), and interactions between the henipavirus F and G proteins have been demonstrated by coimmunoprecipitation (1-5, 18). However, important questions remain concerning the timing of these interactions and the mechanism by which the attachment protein regulates F-mediated fusion. Results from studies of measles virus (30), Newcastle disease virus (33), and human parainfluenza virus (35) have suggested that the initial interaction between the two glycoproteins occurs within the endoplasmic reticulum (ER) at the time of synthesis, potentially allowing the attachment protein to hold the F protein in its prefusion conformation. In contrast, the retention of the parainfluenza virus type 5 (PIV5) F protein in the ER does not lead to the retention of the PIV5 attachment protein (29), suggesting that interaction between these glycoproteins does not occur soon after synthesis. Recently, henipavirus F proteins have been shown to undergo processing through a complex intracellular trafficking pathway, with expression on the cell surface in a nonfusogenic precursor form (F0), subsequent endocytosis, cleavage by cathepsin L into the fusogenic (F1+F2) form, and retrafficking to the plasma membrane (10, 19, 26-28). While the half-life (t1/2) of F protein uncleaved by cathepsin L is approximately 2 h (28), results from initial studies of Hendra virus G trafficking indicate much slower trafficking of this protein through the secretory pathway (37), a result inconsistent with the formation of an F-G complex in the ER.
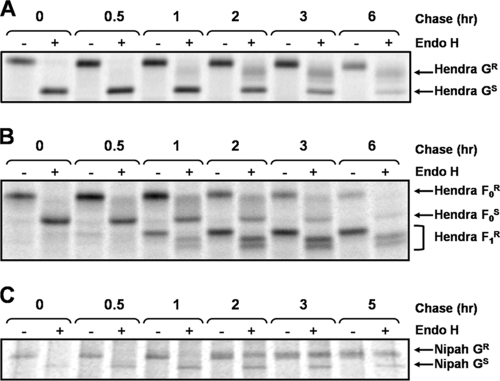
To more closely examine the trafficking of the henipavirus glycoproteins, endoglycosidase H (endo H) analysis was used as a marker for trafficking time to the medial-Golgi compartment. We previously reported that wild-type (wt) Hendra virus G protein becomes endo H resistant, with a t1/2 of between 2 and 3 h (37), suggesting slow trafficking through the secretory pathway. Porotto et al. (31) described a mutant G protein lacking the first 32 residues of the cytoplasmic tail (G-Δ32), which showed enhanced fusion promotion. This Hendra virus G-Δ32 mutant also exhibited higher overall expression than the wt Hendra virus G (S. D. Whitman and R. E. Dutch, unpublished results). To determine if this mutant exhibited altered trafficking kinetics, Vero cells transfected with pCAGGS-Hendra G-Δ32 were examined by pulse-chase analysis followed by endo H treatment as described previously (37). Trafficking kinetics similar to those of wt G were observed, as G-Δ32 became endo H resistant, with a t1/2 of approximately 2 to 3 h (Fig. 1A). In contrast, endo H analysis of Hendra virus F revealed a resistant population of F0 within 30 min (Fig. 1B), with the majority of F converted to either an F0 endo H-resistant form or to the F1 cleaved form by 2 h. The cleavage of F0 was observed concomitantly with the appearance of two partially endo H-resistant F1 bands, suggestive of differential complex sugar additions within the Golgi compartment. These data confirm that Hendra virus F and G traffic through the secretory pathway at different rates, with Hendra virus G trafficking unaffected by the 32-amino-acid deletion. Endo H analysis of the Nipah virus G dimeric form (Fig. 1C), for which the mobility shift after endo H treatment was most apparent, mirrors that of wt Hendra virus G, suggesting that slow trafficking through the secretory pathway is a property of both henipavirus G proteins.
FIG. 1.
Endo H digestion indicates differential rates of trafficking for the henipavirus F and G proteins. (A) Vero cells were transfected with pCAGGS-Hendra G-Δ32, and 24 h posttransfection, the cells were labeled for 30 min and chased for various times and Hendra virus G was immunoprecipitated using an antibody directed to a soluble form of G (7). Endo H digestion was performed as described previously (37). Proteins were separated via sodium dodecyl sulfate-polyacrylamide gel electrophoresis and visualized using the Typhoon imaging system. (B) Vero cells were transfected with pCAGGS-Hendra F. Twenty-four hours later, the cells were labeled for 30 min. Hendra virus F was immunoprecipitated with a Hendra virus F-specific antibody (28) and subjected to endo H treatment. (C) Pulse-chase analysis of Vero cells transfected with pCAGGS-Nipah G was performed. Immunoprecipitation with antibody to a soluble form of G (7) and endo H analysis were performed as described previously. R and S denote the endo H-resistant and -sensitive species, respectively. +, present; −, absent.
Slow trafficking of Hendra virus G through the secretory pathway may be caused by interactions with other viral proteins or with its receptor ephrin B2. Chinese hamster ovary (CHO) cells do not express ephrin B2 (39) and thus were utilized to examine Hendra virus G trafficking in the absence of its cognate receptor. When Hendra virus G-Δ32 was expressed in CHO cells, an endo H-resistant population appeared at 2 h (Fig. 2A) and was found to have increased at subsequent time points, consistent with results obtained using Vero cells. These data suggest that the low trafficking rate of Hendra virus G is not due to association with ephrin B2 and is not cell type specific. Coexpression of either the Hendra virus matrix (M) protein or the F protein with G-Δ32 did not alter trafficking kinetics, indicating that an interaction with either M or F is not responsible for the low rate of G-Δ32 trafficking (Whitman and Dutch, unpublished).
FIG. 2.
Endo H analysis indicates that slow trafficking through the secretory pathway is not dependent on ephrin B2 or on sequences present in the Hendra virus G cytoplasmic tail. (A) CHO cells expressing Hendra virus G-Δ32 were metabolically labeled with 35S for 30 min and chased for the times indicated. Following immunoprecipitation and endo H analysis, proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and visualized via the Typhoon imaging system. (B) Pulse-chase analysis of Vero cells expressing the Hendra virus G-Δ41 tail was performed, followed by immunoprecipitation and endo H analysis. R and S denote the endo H-resistant and -sensitive species, respectively. +, present; −, absent.
Lysine-rich motifs (KKXX or KXKXX) have been shown previously to be involved in the retention of type I glycoproteins in the ER (17), while motifs containing multiple arginines have been implicated in the retention of type II integral membrane proteins (20, 32). The type II Hendra and Nipah virus G proteins contain no and one arginine residue in their cytoplasmic domains, respectively, but do have several lysine-rich motifs. To determine if these motifs facilitated ER retention of Hendra virus G, thus resulting in a lowered trafficking rate, an additional mutant (Hendra virus G-Δ41) with the removal of the first 41 amino acids of the cytoplasmic tail, deleting both KKXX and KXKXX motifs and the majority of the cytoplasmic tail, was constructed. Consistent with the results from endo H analyses of wt Hendra virus G and Hendra virus G-Δ32, an endo H-resistant form of Hendra virus G-Δ41 appeared after only 2 h (Fig. 2B), suggesting that Hendra virus G is not retained in the ER by the putative ER retention motifs KKXX and KXKXX or by any other sequence in the cytoplasmic tail.
Endo H data strongly indicated differential rates of trafficking through the secretory pathway for Hendra virus G and Hendra virus F, suggesting that interactions between these two proteins likely occur subsequent to transport to the cell surface. To verify the different trafficking kinetics of these two proteins, the rates at which Hendra virus F and G appear on the cell surface were determined by using surface biotinylation, with the more highly expressed mutant Hendra virus G-Δ32 utilized in these experiments to facilitate visualization of the surface population. Hendra virus F was present on the cell surface as F0 at the completion of the 30-min labeling (Fig. 3A) (time zero), with F1+F2 appearing on the surface at approximately 2 h (Fig. 3A), consistent with the complex trafficking pathway observed for F. In contrast, the majority of Hendra virus G-Δ32 arrived on the cell surface after the 2-h time point (Fig. 3B). Similar cell surface profiles were observed when the proteins were coexpressed (Fig. 3C). These data confirm the differential trafficking rates and suggest that Hendra virus F and G do not interact within the secretory pathway but traffic independently to the cell surface.
FIG. 3.
Analysis of cell surface populations confirms the differential trafficking rates of the Hendra virus F and G proteins. (A) Vero cells were transfected with pCAGGS-Hendra F. Twenty-four hours later, the cells were metabolically labeled with 35S for 30 min and chased for the indicated times. Analysis of biotinylated surface proteins was performed as described previously (37). (B) Surface biotinylation of Vero cells expressing Hendra virus G-Δ32 was performed, and samples were analyzed as described previously (37). (C) Biotinylation analyses of Vero cells expressing both Hendra virus F and Hendra virus G-Δ32 were performed as described previously.
Exit out of the ER is a carefully controlled process, allowing only properly folded proteins to be transported to the Golgi compartment (15). Thus, slow folding kinetics for Hendra virus G may explain the delayed appearance of an endo H-resistant population. Unlike Hendra virus F, which folds as a trimer (13, 14), Hendra virus G is a tetramer composed of two disulfide-linked dimers (7). To examine folding kinetics, cross-linking analysis (13) was performed at various time points post-metabolic labeling. By the end of the 30-min labeling, the majority of Hendra virus F was folded into a trimeric state which could be stabilized by the addition of a cross-linker (Fig. 4A), and little of the F protein was present in the monomeric form after the addition of the cross-linker. No further increases in trimers were subsequently observed, suggesting very rapid oligomerization of Hendra virus F. Rapid oligomerization of paramyxovirus fusion proteins is consistent with the prefusion structure (38), in which monomers are tightly folded together to form the trimeric unit. The presence of a dimeric species with the addition of a cross-linker is due likely to incomplete cross-linking and does not represent an intermediate conformational population. The folding of Hendra virus G into a tetramer, however, occurs at a lower rate than that of F. In the absence of a cross-linker, monomeric G is present until 2 h postlabeling, suggesting that the formation of the disulfide-linked dimer is intrinsically slow (Fig. 4B). The formation of the disulfide-linked dimer occurs at a rate similar to that of tetramer formation, as the majority of G can be cross-linked to a tetramer by the 2-h time point. Tetramerization of other paramyxovirus attachment proteins, such as PIV5 hemagglutinin-neuraminidase, occurs more rapidly, with t1/2s for these proteins of 25 to 30 min (24), contrasting greatly with the t1/2 of 1 to 2 h observed for Hendra virus G. These results suggest that, unlike other paramyxovirus attachment proteins, Hendra virus G undergoes very slow tetramerization and that the slow trafficking of this protein through the secretory pathway is likely a direct reflection of the low intrinsic rate of folding and oligomerization.
FIG. 4.
Cross-linking analysis of Hendra virus glycoproteins indicates slow tetramerization of the Hendra virus G protein. (A) Vero cells expressing Hendra virus F were metabolically labeled for 30 min and chased for the indicated times. Cross-linking with DTSSP [3,3′-dithiobis(sulfosuccinimidyl propionate)] was performed as described previously (14). (B) Vero cells expressing Hendra virus G-Δ32 were labeled for 30 min and chased for the indicated times, and cross-linking analysis with DTSSP was performed as described previously (14). +, present; −, absent.
While the majority of paramyxovirus F proteins require their homotypic attachment protein for fusogenic activity, the role of the attachment protein in controlling F protein function remains unclear. The attachment protein has been proposed to hold the F protein in its prefusion conformation until receptor binding occurs (34), and research from several systems has suggested that this F protein-attachment protein interaction occurs in the ER during initial protein folding (30, 33, 35). Data from experiments presented here demonstrating differential rates of oligomerization and secretory pathway transport for Hendra virus F and G strongly indicate that the association of the newly synthesized proteins does not occur in the ER but that they instead traffic independently through the secretory pathway. Thus, at least for the henipaviruses, F-G interactions are unlikely to play a role in preventing premature triggering of the newly synthesized F protein. An alternative model for paramyxovirus fusion has suggested that attachment protein-fusion protein interactions occur only after the attachment protein binds the receptor. However, analysis of henipavirus G and F mutants suggests that F-G avidity inversely correlates with fusion (2, 3, 5), and Hendra virus G protein mutants deficient in receptor binding also lose the ability to coimmunoprecipitate Hendra virus F (4). These data support a model in which F-G interactions occur prior to receptor binding, with the subsequent G-ephrin B2 interaction leading to the release of the F-G interaction and the triggering of fusion. Taken together, these results support a model in which the henipavirus F and G associate only after trafficking to the cell surface. The mechanisms by which this interaction is promoted and/or regulated represent an exciting area of future research.
Acknowledgments
We thank Lin-fa Wang (Australian Animal Health Laboratory) for the Hendra and Nipah virus F and G plasmids and Christopher Broder (Uniformed Services University) for antisera to the henipavirus G protein. We are also grateful to members of the Dutch lab for critically reviewing the manuscript.
This study was supported by NIAID grant A151517 to R.E.D.
Footnotes
Published ahead of print on 24 June 2009.
REFERENCES
- 1.Aguilar, H. C., Z. A. Ataman, V. Aspericueta, A. Q. Fang, M. Stroud, O. A. Negrete, R. A. Kammerer, and B. Lee. 2009. A novel receptor-induced activation site in the Nipah virus attachment glycoprotein (G) involved in triggering the fusion glycoprotein (F). J. Biol. Chem. 2841628-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguilar, H. C., K. A. Matreyek, D. Y. Choi, C. M. Filone, S. Young, and B. Lee. 2007. Polybasic KKR motif in the cytoplasmic tail of Nipah virus fusion protein modulates membrane fusion by inside-out signaling. J. Virol. 814520-4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aguilar, H. C., K. A. Matreyek, C. M. Filone, S. T. Hashimi, E. L. Levroney, O. A. Negrete, A. Bertolotti-Ciarlet, D. Y. Choi, I. McHardy, J. A. Fulcher, S. V. Su, M. C. Wolf, L. Kohatsu, L. G. Baum, and B. Lee. 2006. N-glycans on Nipah virus fusion protein protect against neutralization but reduce membrane fusion and viral entry. J. Virol. 804878-4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bishop, K. A., A. C. Hickey, D. Khetawat, J. R. Patch, K. N. Bossart, Z. Zhu, L. F. Wang, D. S. Dimitrov, and C. C. Broder. 2008. Residues in the stalk domain of the Hendra virus G glycoprotein modulate conformational changes associated with receptor binding. J. Virol. 8211398-11409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bishop, K. A., T. S. Stantchev, A. C. Hickey, D. Khetawat, K. N. Bossart, V. Krasnoperov, P. Gill, Y. R. Feng, L. Wang, B. T. Eaton, L. F. Wang, and C. C. Broder. 2007. Identification of Hendra virus G glycoprotein residues that are critical for receptor binding. J. Virol. 815893-5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonaparte, M. I., A. S. Dimitrov, K. N. Bossart, G. Crameri, B. A. Mungall, K. A. Bishop, V. Choudhry, D. S. Dimitrov, L. F. Wang, B. T. Eaton, and C. C. Broder. 2005. Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proc. Natl. Acad. Sci. USA 10210652-10657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bossart, K. N., G. Crameri, A. S. Dimitrov, B. A. Mungall, Y. R. Feng, J. R. Patch, A. Choudhary, L. F. Wang, B. T. Eaton, and C. C. Broder. 2005. Receptor binding, fusion inhibition, and induction of cross-reactive neutralizing antibodies by a soluble G glycoprotein of Hendra virus. J. Virol. 796690-6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bossart, K. N., L. F. Wang, B. T. Eaton, and C. C. Broder. 2001. Functional expression and membrane fusion tropism of the envelope glycoproteins of Hendra virus. Virology 290121-135. [DOI] [PubMed] [Google Scholar]
- 9.Bossart, K. N., L. F. Wang, M. N. Flora, K. B. Chua, S. K. Lam, B. T. Eaton, and C. C. Broder. 2002. Membrane fusion tropism and heterotypic functional activities of the Nipah virus and Hendra virus envelope glycoproteins. J. Virol. 7611186-11198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diederich, S., M. Moll, H. D. Klenk, and A. Maisner. 2005. The Nipah virus fusion protein is cleaved within the endosomal compartment. J. Biol. Chem. 28029899-29903. [DOI] [PubMed] [Google Scholar]
- 11.Eaton, B. T., C. C. Broder, D. Middleton, and L. F. Wang. 2006. Hendra and Nipah viruses: different and dangerous. Nat. Rev. Microbiol. 423-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Field, H. E., P. C. Barratt, R. J. Hughes, J. Shield, and N. D. Sullivan. 2000. A fatal case of Hendra virus infection in a horse in north Queensland: clinical and epidemiological features. Aust. Vet. J. 78279-280. [DOI] [PubMed] [Google Scholar]
- 13.Gardner, A. E., and R. E. Dutch. 2007. A conserved region in the F2 subunit of paramyxovirus fusion proteins is involved in fusion regulation. J. Virol. 818303-8314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gardner, A. E., K. L. Martin, and R. E. Dutch. 2007. A conserved region between the heptad repeats of paramyxovirus fusion proteins is critical for proper F protein folding. Biochemistry 465094-5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hebert, D. N., and M. Molinari. 2007. In and out of the ER: protein folding, quality control, degradation, and related human diseases. Physiol. Rev. 871377-1408. [DOI] [PubMed] [Google Scholar]
- 16.Hsu, V. P., M. J. Hossain, U. D. Parashar, M. M. Ali, T. G. Ksiazek, I. Kuzmin, M. Niezgoda, C. Rupprecht, J. Bresee, and R. F. Breiman. 2004. Nipah virus encephalitis reemergence, Bangladesh. Emerg. Infect. Dis. 102082-2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson, M. R., T. Nilsson, and P. A. Peterson. 1990. Identification of a consensus motif for retention of transmembrane proteins in the endoplasmic reticulum. EMBO J. 93153-3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levroney, E. L., H. C. Aguilar, J. A. Fulcher, L. Kohatsu, K. E. Pace, M. Pang, K. B. Gurney, L. G. Baum, and B. Lee. 2005. Novel innate immune functions for galectin-1: galectin-1 inhibits cell fusion by Nipah virus envelope glycoproteins and augments dendritic cell secretion of proinflammatory cytokines. J. Immunol. 175413-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meulendyke, K. A., M. A. Wurth, R. O. McCann, and R. E. Dutch. 2005. Endocytosis plays a critical role in proteolytic processing of the Hendra virus fusion protein. J. Virol. 7912643-12649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michelsen, K., H. Yuan, and B. Schwappach. 2005. Hide and run. Arginine-based endoplasmic-reticulum-sorting motifs in the assembly of heteromultimeric membrane proteins. EMBO Rep. 6717-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murray, K., P. Selleck, P. Hooper, A. Hyatt, A. Gould, L. Gleeson, H. Westbury, L. Hiley, L. Selvey, B. Rodwell, et al. 1995. A morbillivirus that caused fatal disease in horses and humans. Science 26894-97. [DOI] [PubMed] [Google Scholar]
- 22.Negrete, O. A., E. L. Levroney, H. C. Aguilar, A. Bertolotti-Ciarlet, R. Nazarian, S. Tajyar, and B. Lee. 2005. EphrinB2 is the entry receptor for Nipah virus, an emergent deadly paramyxovirus. Nature 436401-405. [DOI] [PubMed] [Google Scholar]
- 23.Negrete, O. A., M. C. Wolf, H. C. Aguilar, S. Enterlein, W. Wang, E. Muhlberger, S. V. Su, A. Bertolotti-Ciarlet, R. Flick, and B. Lee. 2006. Two key residues in ephrinB3 are critical for its use as an alternative receptor for Nipah virus. PLoS Pathog. 2e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ng, D. T., R. E. Randall, and R. A. Lamb. 1989. Intracellular maturation and transport of the SV5 type II glycoprotein hemagglutinin-neuraminidase: specific and transient association with GRP78-BiP in the endoplasmic reticulum and extensive internalization from the cell surface. J. Cell Biol. 1093273-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Sullivan, J. D., A. M. Allworth, D. L. Paterson, T. M. Snow, R. Boots, L. J. Gleeson, A. R. Gould, A. D. Hyatt, and J. Bradfield. 1997. Fatal encephalitis due to novel paramyxovirus transmitted from horses. Lancet 34993-95. [DOI] [PubMed] [Google Scholar]
- 26.Pager, C. T., W. W. Craft, Jr., J. Patch, and R. E. Dutch. 2006. A mature and fusogenic form of the Nipah virus fusion protein requires proteolytic processing by cathepsin L. Virology 346251-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pager, C. T., and R. E. Dutch. 2005. Cathepsin L is involved in proteolytic processing of the Hendra virus fusion protein. J. Virol. 7912714-12720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pager, C. T., M. A. Wurth, and R. E. Dutch. 2004. Subcellular localization and calcium and pH requirements for proteolytic processing of the Hendra virus fusion protein. J. Virol. 789154-9163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paterson, R. G., M. L. Johnson, and R. A. Lamb. 1997. Paramyxovirus fusion (F) protein and hemagglutinin-neuraminidase (HN) protein interactions: intracellular retention of F and HN does not affect transport of the homotypic HN or F protein. Virology 2371-9. [DOI] [PubMed] [Google Scholar]
- 30.Plemper, R. K., A. L. Hammond, and R. Cattaneo. 2001. Measles virus envelope glycoproteins hetero-oligomerize in the endoplasmic reticulum. J. Biol. Chem. 27644239-44246. [DOI] [PubMed] [Google Scholar]
- 31.Porotto, M., L. Doctor, P. Carta, M. Fornabaio, O. Greengard, G. E. Kellogg, and A. Moscona. 2006. Inhibition of Hendra virus fusion. J. Virol. 809837-9849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schutze, M.-P., P. A. Peterson, and M. R. Jackson. 1994. An N-terminal double-arginine motif maintains type II membrane proteins in the endoplasmic reticulum. EMBO J. 131696-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stone-Hulslander, J., and T. G. Morrison. 1997. Detection of an interaction between the HN and F proteins in Newcastle disease virus-infected cells. J. Virol. 716287-6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takimoto, T., G. L. Taylor, H. C. Connaris, S. J. Crennell, and A. Portner. 2002. Role of the hemagglutinin-neuraminidase protein in the mechanism of paramyxovirus-cell membrane fusion. J. Virol. 7613028-13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tong, S., and R. W. Compans. 1999. Alternative mechanisms of interaction between homotypic and heterotypic parainfluenza virus HN and F proteins. J. Gen. Virol. 80107-115. [DOI] [PubMed] [Google Scholar]
- 36.Wang, L., B. H. Harcourt, M. Yu, A. Tamin, P. A. Rota, W. J. Bellini, and B. T. Eaton. 2001. Molecular biology of Hendra and Nipah viruses. Microbes Infect. 3279-287. [DOI] [PubMed] [Google Scholar]
- 37.Whitman, S. D., and R. E. Dutch. 2007. Surface density of the Hendra G protein modulates Hendra F protein-promoted membrane fusion: role for Hendra G protein trafficking and degradation. Virology 363419-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yin, H. S., X. Wen, R. G. Paterson, R. A. Lamb, and T. S. Jardetzky. 2006. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature 43938-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoneda, M., V. Guillaume, F. Ikeda, Y. Sakuma, H. Sato, T. F. Wild, and C. Kai. 2006. Establishment of a Nipah virus rescue system. Proc. Natl. Acad. Sci. USA 10316508-16513. [DOI] [PMC free article] [PubMed] [Google Scholar]