The global spread of human immunodeficiency virus (HIV) and the human suffering left in its wake have made AIDS a global heath priority. Yet the capacity of HIV to mutate and evolve rapidly has made combating this virus a tremendous challenge. While the obstacles to creating an effective HIV vaccine are formidable, advances have been made on many fronts with the development of novel vectors (62), adjuvants, and antigen design strategies. Live attenuated vaccines have been shown to confer some protection against heterologous challenge viruses in the simian immunodeficiency virus (SIV)/macaque model, providing opportunities to explore the biologic underpinnings of vaccine protection. Equally important is our growing understanding of the biology of acute infection (35, 48, 82, 84) and the role of immune responses in containing viral replication in long-term nonprogressor patients. This review focuses on summarizing strategies for contending with HIV variation in designing a T-cell-based component of a vaccine. We will summarize recent progress by many groups in a number of complementing areas, assessing the impact of diversity in a primate model system, polyepitope vaccines, conserved-region vaccines, central protein design, polyvalent natural proteins, and polyvalent central proteins. The strategies that prove most effective for priming innate cells and T and B cells ultimately could be combined with the best strategies to deal with the magnitude, character, and persistence of a vaccine-elicited immune responses to create an effective HIV vaccine candidate.
TOWARD A T-CELL-BASED HIV VACCINE
A primary motivation for working toward a T-cell-based HIV vaccine is the growing body of evidence demonstrating the importance of the CD8+ T-cell responses for controlling, if not clearing, HIV infection. This evidence includes demonstrations of the following. (i) Cytotoxic T-lymphocyte (CTL) escape is a major force driving of HIV evolution both at the population level (10, 15, 77) and within individuals (13, 14). This observation is a testament to the impact of CTL responses, as HIV mutates continually to escape CTLs. (ii) Highly functional CD8+ T-cell responses are correlated with slow AIDS disease progression (9). (iii) The most significant genetic associations with host control of viral replication in a genome-wide association study were HLA B*5701 and a site near the HLA C gene (27), both of which are associated with T-cell immunity. (iv) CD8+ T-cell responses to specific HIV epitopes may be associated with the good outcomes observed for HLA B*5701-positive (5, 46, 66, 67) and HLA B*2701-positive (4, 46) HIV-infected people. (v) There is an HLA class I heterozygote advantage as well as a rare HLA allele advantage associated with a delay of onset of AIDS (19, 86), and HLA haplotypes for which escape mutations are preserved at the population level (47) may have less impact. These observations provide indirect evidence that enabling more-extensive HLA class I-mediated responses is beneficial.
A second motivation for focusing on T-cell-based vaccines comes from nonhuman primates that have demonstrated the value of a vaccine-induced T-cell response in conferring protection against the clinical progression of disease after virus infection. Nonhuman primate studies can be used in two ways to explore the value of vaccine-elicited T-cell responses in the face of viral diversity. The first is through assessing the cross-reactive potential of HIV vaccine responses with test reagents designed to reflect the viral diversity of HIV (59, 78). The second is through heterologous challenge studies. Pathogenic SIV challenge in macaques vaccinated with live attenuated SIV generally results in an infection characterized by low levels of viremia, stable CD4+ T-cell counts, and slow progression to disease (1, 71, 97). In a recent study, a live attenuated SIV vaccine (SIVmac239deltanef) conferred significant protective immunity against the challenge with a heterologous HIV isolate; there was a 100-fold decrease in viremia over 32 weeks following infection (73). CD8+ T-cell depletion studies indicated that cellular immune responses contributed to this effect. In contrast, the role of CD8+ T cells in conferring protection against SIV replication was less clear in an examination of heterologous virus superinfection. SIVsmE660 and SIVmac251 were used to serially infect rhesus monkeys. When either strain was used as the second infecting virus, it was held in check and was only transiently detected at low levels in the plasma of the monkeys (99). The mechanism responsible for the control of the superinfecting virus was not clearly attributable to T- or B-cell immunity.
The STEP vaccine trial was a phase IIb human trial designed to explore whether an HIV T-cell vaccine could reduce infection rates or viremia upon infection (24, 64). The vaccine included HIV Gag, Pol, and Nef expressed from genes carried by the adenovirus serotype 5 (Ad5) vector, and it was hoped that vaccination with these constructs would generate immune responses that would reduce infection rates or viremia upon infection. The trial was stopped for lack of efficacy, and the outcome sent a wave of concern through the community. Not only did the vaccine not confer a benefit, the vaccine-treated group with preexisting Ad5 immunity had a higher incidence of HIV infection than the placebo-treated group (24, 64). The differences between these results and the studies done with nonhuman primates remain unresolved. However, the cumulative results of many primate studies as well as the apparent value of T-cell responses in retarding progression make it reasonable to expect that the results of the STEP trial reflect an outcome of the particular vaccine immunogens used in the trial, or of confounding variables related to certain populations of subjects in the trial, rather than a failure of the concept of a T-cell-based vaccine. Preclinical work with T-cell-based vaccine components is therefore continuing.
IMPLICATIONS OF GLOBAL HIV DIVERSITY FOR T-CELL RESPONSES
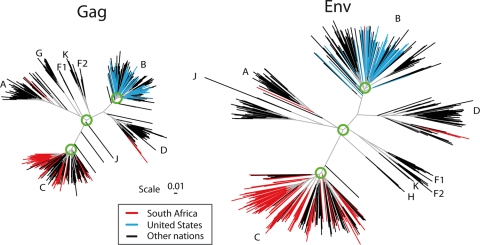
Eliciting an immune response that can protect against the extraordinary diversity of circulating HIV is a formidable challenge (34). Figure 1 illustrates two phylogenies of HIV M group variants based on HIV Gag and Env genes obtained through a global sampling of viruses. The M group, or main group, of HIV-1 viruses encompasses the viruses of the pandemic. Subtypes A to J within the M group form relatively clear genetic groups that consistently show the same clustering pattern throughout different regions in the HIV genome (74) (Fig. 1). Some of these distinct lineages may have evolved different phenotypic characteristics that impact transmission (42, 43), antigenic targets, and regions under selection (34, 37, 87) and, possibly, pathogenicity (50).
FIG. 1.
Phylogenetic trees of the major subtypes found in the HIV-1 M-group Gag and Env. These trees are based on the Los Alamos curated database of global alignment, as of December 2008, and contain one full-length gene per infected person. The trees show the relationships between the major HIV subtypes; intersubtype recombinants were excluded. The database represents global studies but not systematic sampling, as whatever sequences were published and are in GenBank are included in the HIV database. Only representative subsets of the most recently sampled B clade from the United States or C clade from South Africa were included in these trees, as these are the most heavily studied epidemics and dominate the global sampling. To illustrate how these regional single-subtype epidemics fit into the global picture, terminal lines for South African sequences are in red, those for United States sequences are in blue, and those for all other nations are in black. The green circles show the approximate region of the node selected for generation of the most recent common ancestor; in our ancestral reconstructions, we do not include the sequences that are outliers relative to the rest of the clade in these models. The trees are maximum likelihood trees using a general time-reversible model with site rate variation estimated with a gamma distribution and were generated using PhyML (40).
Phylogenetic trees such as those shown in Fig. 1 model evolution by base substitutions, and branch lengths reflect the number of mutations estimated to have occurred between the sequences used to construct the tree. HIV-1, however, has many other mechanisms to generate diversity: recombination, insertion and deletions, and gain and loss of glycosylation sites (102). Recombination occurs at high rates, and intersubtype recombination is common in populations where multiple subtypes cocirculate and the incidence of infection is high (6, 49). Recombination obscures phylogenetic relationships between viral isolates and violates the assumptions that are used to infer phylogenetic trees (53), yet recombination events play a major role in HIV evolution. Thus, trees like the ones shown in Fig. 1, that exclude intersubtype recombinants, should be interpreted with the awareness that the clades as shown can be considered basic building blocks that contribute sections to the intersubtype recombinant viruses that cocirculate with them. Within-clade phylogenetic structures are themselves muddled by recombination patterns that are not easily resolved by conventional phylogenetic methods.
Despite these limitations, phylogenetic trees that group viruses according to genetic relatedness are useful. Phylogenies provide a model for studying the timing of epidemic expansion (52, 96, 101) as well as evolutionary dynamics and immune pressure within infected individuals (48, 54). Phylogenetically defined virus lineages may have different phenotypes, as discussed above. Infections by different subtypes can result in both B- and T-cell immunological responses with subtype-related cross-reactivity patterns (8, 11, 12). There are, however, many exceptions to this. For example, the C-clade viruses from India tend to form a relatively tight genetic cluster within the C clade, but envelopes from this group of viruses have extremely diverse patterns of neutralization susceptibility when tested with a large panel of sera (57).
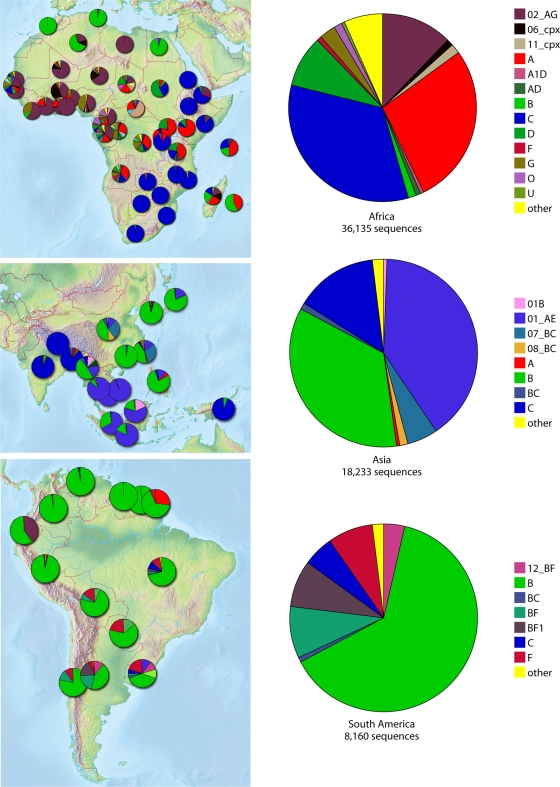
The basic structure of the phylogenetic tree provides a framework for people to think about vaccine component design while considering HIV diversity at different levels. For example, a B-clade-specific vaccine might be particularly useful in the United States, which has a primarily B-clade epidemic (Fig. 2). Similarly, a C-clade vaccine might be helpful in South Africa, and a vaccine based on the circulating recombinant form of the virus, CRF01, may be useful in parts of Asia (Fig. 2). While some vaccine designs are based on viral proteins found in particular geographic regions or nations (16, 22), others take into consideration not only the regionally circulating viral forms but also regional human population HLA allele frequencies (45, 91).
FIG. 2.
Maps showing the subtype designation of all sequences in the Los Alamos database as of 1 January 2009. These sequences are often single genes and fragments, so intersubtype recombination is underestimated. They are not sampled randomly but are the product of all HIV studies with sequences submitted to GenBank. Despite this limitation, the maps largely reflect what we know about global distributions of subtypes, but the details should be interpreted with caution. The figures were made using the HIV geography tool at the Los Alamos database (http://www.hiv.lanl.gov/components/sequence/HIV/geo/geo.comp).
In the trees shown in Fig. 1, the terminal branches that correspond to United States sequences are labeled in blue; these branches have a narrower diversity pattern than that of the full set of B viruses, which includes sequences of viruses from Asia, South America, and Europe. South African sequences are shown in red and are dispersed throughout the international C-clade sequences. A few non-C-subtype full-length gene sequences from South Africa are represented by the red lines in the A and D clades. The red and blue lines illustrate the limitations of trying to make a national vaccine, but they suggest that subtype-specific vaccine strategies may be worth exploring. If a subtype-specific vaccine was successful, it would be of immediate value at least to the target population and could lead the way for more-comprehensive strategies.
Some groups, including our own, have devised rational design strategies for a global vaccine rather than a country- or subtype-specific vaccine (34). A global vaccine is a daunting challenge given HIV diversity, but it is motivated by several factors. First, even in a regional epidemic dominated by a single subtype, such as the B-clade epidemic in the United States, pockets of HIV with greater genetic diversity can be found (61, 85). Second, most nations have complex epidemics and would not benefit from a single-subtype vaccine (Fig. 2). Finally, human testing and parallel production of multiple tracks of regional and national vaccines would be very costly and resource intensive.
The genetic distances between the viral isolates shown in Fig. 1 underlie enormous changes in HIV at the amino acid level when considered from the perspective of a T-cell response. HLA class I and class II molecules present epitopes that are contiguous peptide fragments of various lengths and are recognized by an immune response. HLA class I-presented epitopes are most commonly 9 amino acids long, but the lengths of these epitopes range from 8 to 12 amino acids. Optimal HLA class II epitopes have greater diversity in their lengths but encompass this range. Thus, a sensible way of describing HIV diversity from a vaccine perspective is not just in terms of single amino acids but also in how much variation there is in potential T-cell epitopes, protein fragments roughly nine amino acids long (59).
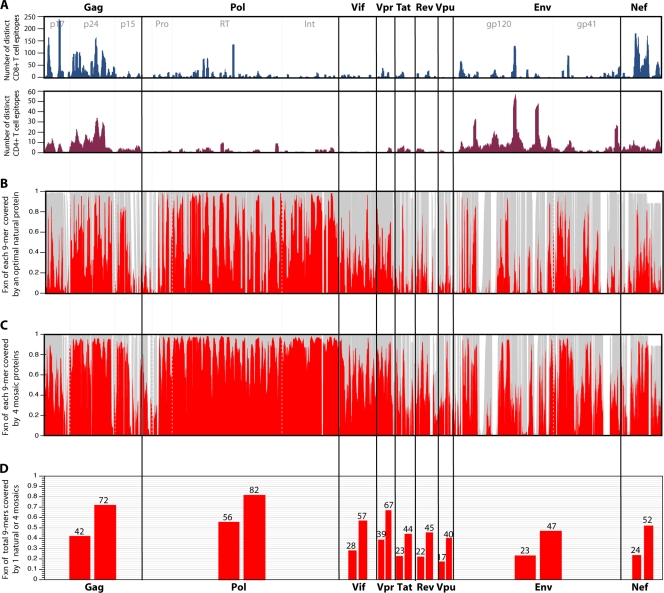
Human T-cell epitopes have been found virtually everywhere in the HIV proteome (http://www.hiv.lanl.gov/content/immunology/maps/maps.html), and a short stretch of a viral protein generally includes many distinct epitopes with different overlapping boundaries presented by different HLA molecules. Figure 3a shows the distribution of human T-cell epitopes of HIV that have been described in the literature and are assembled in the HIV immunology database. This collection is comprehensive but has biases. For example, it emphasizes the regions of the virus that have undergone the most intensive study. Also, more-variable regions are likely to be underrepresented due to mismatches with test reagents. Despite this, the ridgeline of peaks and valleys of all HIV CD8+ T-cell epitopes in the database (Fig. 3a) roughly parallels the frequency of epitope responses at the population level, which has been determined experimentally by evaluating the full proteome (2, 30). The number of perfect 9/9 potential epitope matches between a single natural putative vaccine protein and the aligned translation of the current full genome sequence M-group alignment at the Los Alamos database is shown in Fig. 3b. It shows the coverage of each 9-mer section of the alignment based on a sliding window that moves position by position through the proteome. The M-group genome alignment contains 1,206 sequences, one per infected person, and includes hundreds of recombinants as well as pure subtypes from throughout the world. The natural protein used for this comparison was selected by virtue of providing the most perfect 9/9 matches with the M-group set; the overall fraction of identical 9-mers matches between the best natural protein and the M-group set is shown in Fig. 3d.
FIG. 3.
(A) Map of known and distinctive CD4+ and CD8+ epitopes described in the literature and included in the Los Alamos database across the proteome. Experimental maps of population responses (30) are tracked, with the exception of the underrepresentation of T-cell epitope mapping of regulatory proteins in the database. This underrepresentation is a consequence of regulatory protein T-cell epitope mapping being somewhat underrepresented in the literature. (B) The fraction (Fxn) of identical matches (red) with a single natural strain that provides the optimal coverage of the M group for each 9-mer (potential epitope) in the HIV proteome is shown. The optimal natural strain is a C-subtype sequence, C.ZA.99.DU422 (GenBank accession number AY043175). This is not surprising, as C is the most common subtype in the full-length genome database. This is an alignment-based figure; the gray background illustrates how many sequences have 9-mer in a given position in the alignment, such that a section in the alignment with an insertion in only one or a few sequences will appear as a white band. (C) Illustration of the increase in the fraction of perfectly matched 9-mers at each position when a four-mosaic combination is used rather than a single natural strain. (D) Total percentage of all 9-mers covered for each protein, corresponding to the single natural strain coverage shown in panel B (lower bar for each protein) and the four-mosaic coverage (higher bar) shown in panel C. The mosaic sequences were derived, optimal natural proteins were selected, and coverage graphics for both Fig. 3 and 4 were created using the mosaic vaccine tool suite at the Los Alamos HIV database (83) (http://www.hiv.lanl.gov/content/sequence/MOSAIC/). All comparisons were made to proteins translated from the global full-length genome alignment at the Los Alamos HIV database (http://www.hiv.lanl.gov/content/sequence/NEWALIGN/align.html), so the same input set of full genome sequences were used for every protein; thus, the variability comparisons between proteins are reasonable.
What is immediately evident from the sequences shown in Fig. 3 is that not all regions within an individual viral protein are comparably distinctive between virus isolates. While most regard Gag as conserved and Env as variable, not all regions of Gag are conserved (only p24 is well conserved throughout), and not all regions of Env are variable. Overall, Pol is the most conserved protein of HIV, but even Pol is spanned by alternating short stretches of near identity and greater diversity. Nevertheless, it is the only HIV protein for which more than half (56%) of the 9-mers in the HIV database global collection (Fig. 3d) match the best natural strain. Env and the regulatory proteins typically match the best natural strain in only approximately one-quarter of the 9-mers (Fig. 3d). This means that a vaccine-elicited response to a typical epitope of a single strain of virus is more likely than not to be faced with a mismatch in the corresponding epitope of an infecting strain of virus. While older studies suggest that such single mismatches can often be tolerated by the immune system, recent detailed examinations of cross-reactivity of T-cell responses suggest that cross-reactivity detected by enzyme-linked immunospot (ELISpot) and chromium release assays may overestimate the true cross-reactivity of relevant cellular immune responses (8, 88). Epitopes that appeared to be highly promiscuous and susceptible to recognition by CD8+ T cells specific for virions of those epitopes when measured using assays that depend on an exogenously loaded peptide, such as ELISpot and chromium release assays, were not recognized by such CD8+ T cells when expressed within an infected cell in an HIV-1 inhibition assay (8, 88).
NATURAL PROTEINS AS VACCINE ANTIGENS
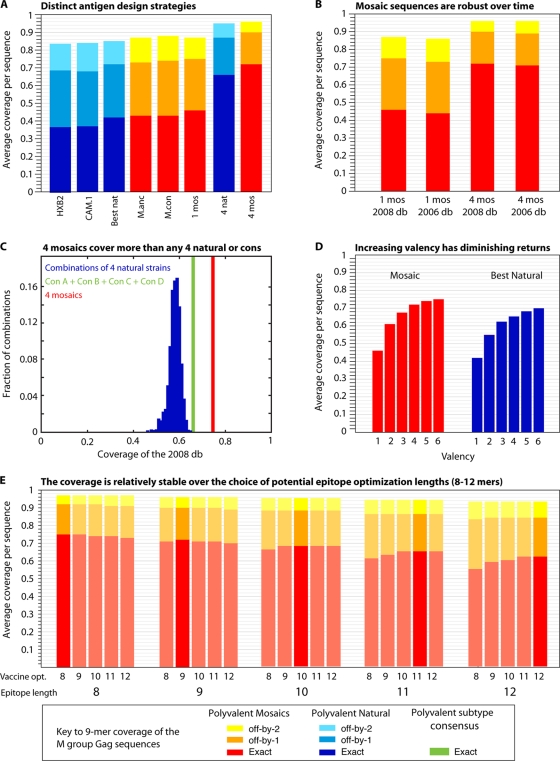
Most of the HIV vaccine prototypes developed to date have used naturally occurring HIV proteins as vaccine antigens, either unmodified or with engineered deletions introduced to improve safety or enhance immunogenicity (17, 21). A natural strain that is close to a consensus sequence (17) can be selected, or one can be selected to provide the best coverage of potential epitopes in a target population (29) (Fig. 3c and 4a). Figure 4a illustrates a comparison of the 9-mer coverage of all Gags found in the full genome alignment of 1,206 M-group sequences from the Los Alamos database based on specific vaccine antigen design concepts. These comparisons included an assessment of the coverage provided by CAM.1 and HXB2, two natural B-clade strains that have been used in vaccine trials, and the single natural HIV strain selected to give the best coverage of the entire database collection.
FIG. 4.
Comparisons that address key issues regarding mosaic and central sequence vaccine designs. All comparisons in this figure are based on Gag, but the relative rankings for Gag in this figure are paralleled, with differences in overall magnitude, by all HIV proteins, i.e., Gag is representative (B. T. Korber, data not shown). Blue represents natural proteins, and red represents mosaics. (A) Incrementally improved coverage of 9-mers in Gag in the global M-group alignment using different design strategies. From left to right, two single natural strains which have been used for vaccine studies, the single natural strain selected to provide the best 9-mer coverage, the M-group ancestor, the M-group consensus, a single mosaic, the four natural strains that in combination give the best coverage of Gag, and four mosaics. The single best natural Gag turns out to be a C-subtype sequence (see legend to Fig. 3). The four best natural Gags are a set that includes one C subtype, one B subtype, one AD recombinant type, and a CRF01 sequence. (B) Illustration of the robustness of these designs over time and with the acquisition of new data. The 2006 database mosaics were the four mosaic Gag sequences generated in a study by Fischer et al. (29) based on a set of 551 M-group sequences, one sequence per person, that were available for inclusion at the end of 2005 for the 2006 Los Alamos database. The current designs are based on the 2008 database full-length genome alignment, using sequences that were available at the end of 2007 for inclusion in the 2008 database. The 2008 database set of 1,206 Gag proteins was the test set used in both cases, and the 2006 database-derived mosaics provide almost the same coverage as the 2008 database-derived mosaics. Given the cost and time required for development and testing, and the robustness of the mosaic sequences over time and acquisition of new data, it does not make sense to continuously update mosaics during different stages of testing. (C) Comparison of the M-group 9-mer coverage based on 1,000 randomly selected sets of four natural proteins, a combination of four subtype consensus proteins (one A, one B, one C, and one D subtype consensus proteins, in green), and four mosaics. (D) Increasing the valence of a vaccine will enhance 9-mer coverage, but with diminishing returns, by inclusion of increasingly rare 9-mers. (E) Mosaic design is relatively robust against the length of potential epitopes used for the optimization (opt.) criteria.
The immunogen employed in the two first HIV vaccine efficacy trials, the VaxGen (36) and STEP (24) trials, used natural viral genes/proteins as antigens. This is the simplest model for vaccine immunogen design and the obvious thing to try first: deliver some part of HIV, either as natural protein or as a gene that can be expressed upon vaccination, and hope it elicits immune responses with enough potency, long-term memory, and cross-reactivity to confer protection. This vaccine approach has been effective for other viruses, including hepatitis B virus (103) and influenza virus (80). The intent of the VaxGen trial was to determine if vaccination with a recombinant HIV envelope protein (rgp120) vaccine could elicit beneficial neutralizing antibodies (neutralizing antibodies target the viral envelope and block viral entry into target cells). In that case, the evidence that this vaccine produced cross-reactive neutralizing antibodies was not convincingly shown prior to the initiation of the phase III trial (18, 23). Although anti-HIV antibodies were induced by the vaccine in immunized volunteers, these antibodies did not reduce the incidence of HIV-1 infection (36).
The STEP trial, on the other hand, employed immunogens that elicited T-cell responses to Gag, Pol, and Nef expressed from genes in Ad5 vectors. Of the individuals who received this vaccine, 77% mounted an HIV-specific T-cell response (64). However, many individuals responded only to one or two of the proteins. If there were only a single-epitope response to viral proteins (24), it is possible that for many vaccinees that there were mismatches between the epitope(s) recognized by the response to the vaccine and the infecting strain of virus or that naturally infected cells did not present the same epitopes that were presented in the context of the vaccine. The team of scientists analyzing specimens generated during the course of the STEP trial are sequencing the transmitted viruses and defining the reactive epitopes to assess these possibilities (24).
Another vaccination strategy being pursued employs natural antigens by use of a polyvalent vaccine that incorporates natural antigens from multiple clades of HIV. While there are well-founded concerns about the complexity and expense of generating polyvalent vaccines, such vaccines have been successfully used for other pathogens. A polyvalent vaccine is successfully used against influenza, despite the added complication of needing to frequently make new vaccines that match the changing epidemic strains (80). The current pneumococcal vaccine includes 23 capsular serotypes (44). One group of investigators is exploring the use of a polyvalent natural strain approach for HIV in a DNA prime-protein boost vaccine strategy, including one A-, two B-, two C-, and one E-clade envelope proteins and a monovalent Gag protein. This approach has been evaluated in a phase I safety and immunogenicity study (89). The vaccinees receiving the highest dose of DNA prime in this study yielded CD4+ T cells with greater polyfunctionality and some CD8+ T cells specific for Env and Gag. However, this vaccine regimen elicited an immune response that was skewed toward the induction of CD4+ T-cell responses (7), and the T-cell responses were elicited against peptides from the diverse vaccine strains of the virus (89). Another group designed a polyvalent vaccine including three Env proteins (one A, one B, and one C clade), and a single Gag, Pol, and Nef protein, all delivered in a DNA prime-recombinant Ad5 boost regimen (79). In a nonhuman primate evaluation of this vaccine strategy, a polyvalent Env vaccine elicited T-cell and neutralizing antibody responses with greater breadth than did monovalent Env immunogens; moreover, there was no evidence of antigenic interference (79). SHIV-89.6P peak viremia and set point were lower for all vaccinated animals when challenged, with a delayed decline in CD4+ T cells (79). These results in macaques led to a successful phase I trial (20), and after some debate triggered by the STEP trial, the vaccine is scheduled for advanced-phase testing in humans.
POLYEPITOPE VACCINES
The polyepitope strategy is based on investigator-designed artificial minigenes, expressed in either DNA or a viral vector, comprised of a string of epitopes lined up in a single artificial vaccine construct. The presumption is that if the protein can be expressed and is immunogenic in animal studies, these vaccine constructs will elicit the desired T-cell responses in humans. This strategy is conceptually elegant, as it allows the investigator the freedom to select epitopes that are deemed most important. For example, such a vaccine could focus the cellular immune response on the most conserved epitopes or those most frequently presented by the most common HLA class I molecules. This concept had an encouraging early precedent, given protection against a lethal challenge of the arenavirus lymphocytic choriomeningitis virus in mice (72, 94).
The utility of this approach for an HIV vaccine merited serious exploration. The first prototype for this kind of vaccine for HIV was designed by Hanke and McMichael (41). They developed a vaccine targeting the Kenyan epidemic, including the regionally prevalent clade A Gag p24 and p17, linked to a string of 22 epitopes selected by virtue of being presented by the most common HLA class I molecules in Kenya and eliciting immunodominant responses. This immunogen construct was expressed from DNA or a modified vaccinia virus Ankara (MVA), and was administered as DNA, MVA, or DNA/MVA in prime-boost combinations. The vaccines were immunogenic in mice and macaques (93), and so they progressed through human safety and immunogenicity trials (68). Most of the observed responses were to CD4+ T-cell epitopes in the Gag portion (38). Unfortunately, CD8+ T-cell responses were minimal. Five of 16 individuals had responses to the multiepitope immunogens, and 3 of these were mapped to the same epitope, suggesting that very few of the epitopes in the context of the vaccine elicited CD8+ T-cell responses. Recently completed trials of this immunization strategy in Kenya and Uganda were disappointing. The majority of vaccinated individuals did not have a detectable ELISpot response (for example, a DNA prime-recombinant MVA boost elicited ELISpot responses in only 13% of volunteers), and those that did respond showed weak and transient responses (45).
In a second HIV vaccine study of this kind, a more bioinformatic approach was applied to the polyepitope design (63). This design focused on the selection of epitopes that bind multiple HLA allelic products with the potential for presentation to the immune system by many HLA proteins. Twenty-one conserved epitopes that can be presented by HLA-A2, -A3, and -B7 supertypes were linked, and the vaccine was predicted to elicit immune responses in 85% of individuals globally. Rather than simply linking epitopes, the vaccine was specifically engineered to attempt to enhance correct processing and to minimize unnatural junctional epitopes (63). (De Groot and colleagues have proposed a similar design strategy, deriving consensus epitopes and computationally attempting to minimize junctional epitopes [25].) Studies with the polyepitope vaccine in HLA transgenic mice indicated that multiple epitopes in the construct could elicit immune responses (95). Despite encouraging preliminary results, the vaccine showed disappointing immunogenicity in human studies. Only 1 of 42 uninfected vaccinated adults made a detectable gamma interferon ELISpot response to the vaccine, and 3 had a response that could be detected by a chromium release CD8+ T-cell assay (39).
It is possible that the particular polyepitope vaccine delivery strategies and vaccination protocols could be altered to achieve better outcomes. To date, however, this interesting concept and the encouraging preliminary results of animal studies have not translated into human immunogenicity.
FOCUSING ON THE MOST CONSERVED REGIONS OF HIV
The polyepitope vaccines described in the previous section were designed to use epitopes combined into minigenes to directly focus the vaccine-induced immune response on conserved viral epitopes. Another strategy being explored to focus immune responses on conserved regions of HIV is to use immunogens created with longer sections of proteins spanning the most conserved regions of the proteome that are linked in a chimeric protein. The intent of the latter strategy is to capture the entire set of CD4+ and CD8+ T-cell epitopes harbored within these conserved regions of the virus. Vaccine-induced T-cell responses would thus have a high probability of interacting with the circulating viruses. If a vaccinee were infected, the vaccine-induced immune response might have the potential to shift the immunodominance profile and focus the initial immune response in a newly infected individual on conserved regions of the virus, where mutations that facilitate immune escape would likely have a high fitness cost (3, 76, 81, 90, 98). Since some of the benefit conferred by protective HLA alleles may be realized through the high viral fitness costs of escape mutations (67), this type of a vaccine might be able extend this benefit to infected individuals who do not carry one of the few HLA alleles associated with a good outcome.
Epitope processing and presentation of antigen to T cells remains an important issue to explore experimentally in this approach. While the long fragments in a chimeric protein may facilitate more natural processing and presentation than minigene polyepitope approaches, the basic strategy may still result in the creation of antigens that are not processed or presented optimally. Computational approaches such as those used to minimize junctional epitopes for polyepitope vaccines (63) could also be applied to the design of conserved chimeric proteins (76). The regions of HIV with the greatest sequence conservation may not be very immunogenic in naturally infected cells. For example, the most conserved regions in the proteome of HIV are found in Pol, but comparatively few CD8+ T-cell responses recognize Pol epitopes spanning these regions (Fig. 3a). Rolland et al. suggest that this may be a consequence of a lower ratio of expression of Pol relative to Gag protein (76). Furthermore, some of the most highly conserved domains of the virus may have developed strategies to avoid immune pressure long ago in evolutionary history (100) or may be seen as “self” if they are also expressed by an endogenous retrovirus. If the vaccine epitopes are not presented in a natural context, the vaccine may not confer protection.
Despite these reservations, the first mouse study of the conserved-region approach was encouraging (58). The vaccine design included the 14 most conserved regions of the HIV proteome linked in a chimeric protein. The sequence used to represent each of these regions was selected from one of four subtype consensus sequences, and the regions were globally conserved so that the M-group coverage of potential epitopes was good in the vaccine construct. BALB/c and HLA-A*0201 transgenic mice were able to generate T-cell responses to this vaccine antigen expressed as DNA, in an MVA vector, and in a human Ad5 vector (58). Thirteen of 13 HIV-infected people had memory T-cell responses to epitopes carried in the chimeric protein, and many known epitopes in the Los Alamos HIV database are harbored in these regions (58). Based on this promising start, this vaccine antigen is being moved forward into a small human safety and immunogenicity trial (T. Hanke and A. McMichael, personal communication).
CENTRAL VACCINES
Another way to address the genetic diversity of the HIV, particularly if one is aiming for the simplicity and cost-effectiveness of a monovalent vaccine component, is to design an antigen that is central to the circulating strains that the vaccine is targeting. This can be done either within an HIV subtype or at global level, including multiple subtypes of the virus (Fig. 1). Three different strategies for accomplishing this have been suggested. The first is based on a phylogenetically reconstructed ancestral sequence (26, 34, 56). An ancestral sequence is the sequence of bases that represent the most likely base in each position in an alignment at an ancestral node in a phylogenetic tree. Examples of an ancestral gene design options include reconstructions of the most likely sequence at a node near the base of a subtype-defining clade or the node that is the ancestral sequence for the entire M group of viruses (Fig. 1). The second strategy is to take the most common amino acid at every position in an alignment and concatenate those together; this is called a consensus sequence (33, 34, 56). In the third strategy, the “center-of-tree” or COT sequence approach, the point in a phylogenetic tree that minimizes the sum of the distances to all branch tips is identified (69, 75). Central sequences are all similar to each other, and the differences between them are in the range of the expected number of errors inherent in phylogenetic-based ancestral reconstruction methods (32). They will vary slightly if reestimated based on a different evolutionary model or different input data, for example, if they are recalculated as the global sampling increases from year to year.
Subtype-specific centralized protein vaccines can reduce the distance between a vaccine protein and the protein of contemporary circulating strains of HIV by roughly half relative to a typical natural vaccine strain (34). This can be visualized in phylogenetic trees (Fig. 1). If the vaccine protein is based on the model sequence near the root of a clade, one only has to traverse one long branch to be near the ancestor of the clade, and there is no need to add in the distance from the root back out to the natural strain selected for the vaccine protein. M-group central sequences bring vaccine distances to all circulating sequences approximately to the level of within-clade distances. The relatedness of a vaccine protein and the protein of a contemporary circulating strain of HIV can also be evaluated on the basis of 9-mer or potential CTL T-cell epitope coverage. Two B-clade strains that have been studied as vaccine candidates (HXB2 and CAM.1) do not match as many 9-mers in the M group as M ancestral or consensus strains do, while M-group ancestral and consensus sequences are roughly comparable (Fig. 4a).
When such vaccine design strategies were still untested, there was widespread skepticism regarding the artificial protein designs: would they fold well, be recognized by conformation sensitive antibodies, be functional, and be immunogenic? Together with our colleagues, we evaluated a reconstructed M-group consensus/ancestral Env protein. (We have tested several generations of this concept, most of our work being done with a version called CON-S) [33, 34, 92]). This was a particularly challenging central protein, and a priori was the least likely to succeed, because the sequence of the M-group consensus/ancestral Env was very distant from those of natural HIV proteins. There are two reasons for this. First, Env is the most variable protein in HIV, and second, we modeled all the way back to the center of the entire M group rather than just to the root of a single subtype (Fig. 1). The M-group consensus/ancestral protein (CON-S), however, was also particularly intriguing, as it provided a candidate protein for a global vaccine and could be evaluated as an immunogen for eliciting both B-cell and T-cell responses to a protein of a central virus strain. Despite its sequence distance from natural strains and the uncertainties inherent in modeling, CON-S proved functional in a pseudotype infection assay. When expressed as a gp140 Env oligomer, it bound antibodies that recognize conformationally determined epitopes of HIV envelopes. Most critically, it was immunogenic for both T cells and B cells when tested in small-animal studies (33, 60, 92). In fact, all consensus, ancestral, and COT sequences for M-group and subtype B and C HIV proteins that have been tested to date are well expressed and are immunogenic in small animals (33, 55, 56, 60, 75). COT sequences for B-clade Gag, Nef, and Tat all retained biological function (75). The M-group CON-S and a clade B-based Con B Env protein (55, 60) each elicited antibodies with good titers and breadths of responses against easily neutralized, so-called tier 1 HIV-1 viruses, but not against isolates that are more difficult to neutralize. It may prove useful to employ central sequences as the starting point for making strategic modifications of HIV envelopes to improve the exposure of potentially useful neutralization epitopes (55).
The CON-S envelope has also been assessed as an immunogen for eliciting T-cell responses. An M-group CON-S Env vaccine induced T cells with improved cross-reactive potential in mice (92). In a study by Santra et al., a CON-S vaccine candidate was compared with a natural B-subtype Env vaccine candidate (78) in rhesus monkeys. Both immunogens made strong autologous responses as determined by reactions with pooled peptides designed to match the vaccine protein sequences, but the responses to CON-S had much greater cross-reactivity. The number of responses to peptide series spanning 10 different natural HIV envelope proteins, including representatives of four different subtypes, was evaluated, and the CON-S Env induced three- to fourfold more responses per protein than did the natural B-clade envelope immunogen. These data indicated that the cross-reactive potential of T-cell responses to the CON-S Env was greatly enhanced relative to the responses elicited by a single natural protein vaccine (78).
Another encouraging result with central proteins was found in exploring their use as a foundation for designing peptides as reagents for ELISpot assays (31). The cross-reactivity of T-cell responses of HIV-infected humans to different variant epitopes provides indirect evidence as to whether a particular epitope variant peptide might stimulate a T-cell response that is cross-reactive. When within-subtype and M-group central sequences were used as a basis for generating peptides for use in ELISpot assays, the three strategies (consensus, ancestral, and COT) were found to be comparable for generating peptides for detecting T-cell responses. The M-group-based peptide reagents performed as well as within-subtype peptides for response detection (31).
POLYVALENT MOSAIC VACCINES
Given the emerging evidence that “central” computer model-based proteins are well expressed, immunogenic, and induce T-cell responses with improved cross-reactive potential, we decided to build on this concept and design a new generation of centralized immunogens: polyvalent protein cocktails that in combination could provide maximum coverage of potential T-cell epitopes (29). This computational polyvalent approach was motivated by the promising results observed with polyvalent natural immunogens (79). We utilized a machine-learning strategy called a genetic algorithm to computationally design sets of sequences that are similar to natural sequences but that in combination maximize the coverage of potential T-cell epitopes in the population.
The resulting mosaic sequences are derived from in silico recombinants of natural strains of HIV and are constrained in the same fashion as natural virus sequences. Boundary regions spanning recombination breakpoints are created such that all local regions of a protein are found repeated among natural sequences, and as a consequence, mosaic proteins align readily to natural viral proteins. Such a mosaic polyvalent vaccine maximizes the coverage of natural variation in all potential T-cell epitopes for a given number of proteins in a cocktail; therefore, the chance of it providing beneficial immune responses to vaccinees with diverse major histocompatibility complex class haplotypes should be improved.
Mosaic cocktails are designed in a series of steps that in many ways reflect the recombination evolutionary strategies employed by the virus itself. For example, consider the design of a mosaic cocktail created to optimize M-group coverage of Gag epitopes by three mosaic Gag proteins. The general design strategy of mosaic T-cell immunogens is as follows. The program input is the set of all M-group Gag proteins; these sequences do not have to be aligned. Three sampling pools of in silico-derived de novo recombinants (mosaics) are then created based on recombining parental strains taken from the M-group alignment. In this process, consideration is given only to those mosaics that encode intact proteins and have recombination breakpoints embedded in short regions of sequence that recur among natural strains. This avoids the creation of artificial junctional epitopes. Meanwhile, the input M-group protein alignment is fractured into all possible 9-mers (or epitope-length fragments), and the frequency of each 9-mer is tallied to provide a basis for evaluating potential epitope coverage of mosaic combinations. We then test all combinations of three mosaics, one drawn from each pool, and select the set of three that maximizes the coverage of all 9-mers in the M group of HIV sequences. Finally, we introduce new mosaics into each sample pool and iteratively repeat the optimization process until the coverage no longer improves; this typically takes 1 to 2 days of run time. In our studies, the coverage of the three mosaics approaches the coverage of the three most common 9-mers in each position of the viral protein (79), a theoretical maximum that cannot be perfectly achieved with intact proteins, as there will sometimes be contradictory amino acids in overlapping regions (the amino acid that is best for one 9-mer can be less than optimal in combination with other amino acids in an overlapping 9-mer).
Given both the extensive overlap between known epitopes presented by different HLA class I and II molecules (Fig. 3a) (http://www.hiv.lanl.gov/content/immunology/maps/maps.html) and viral sequence diversity, mosaics seem a logical strategy to create universal immunogens. By generating intact proteins that mimic natural proteins, our hope is that mosaic proteins will mimic natural processing, so epitopes that stimulate T-cell responses in the vaccine will be the same epitopes that are processed and presented in natural HIV infection, hopefully circumventing the problems in processing found with polyepitope approaches.
Mosaic polyvalent vaccines significantly improve the population coverage of potential epitopes for every protein in HIV (Fig. 3b to d). While mosaic proteins designed to optimize coverage of a single subtype provide excellent of coverage of viruses of that subtype, there is a considerable decrement of coverage of other subtypes (28, 29). In contrast, mosaics designed to optimize 9-mer coverage of the full M group of HIV isolates not only provide almost comparable coverage of a single clade of isolates compared to within-subtype mosaics but also cover all subtypes very well (29). These artificial mosaic proteins are therefore promising candidate immunogens for a global HIV vaccine. By design, mosaic proteins minimize the inclusion of rare or unique potential epitopes, preferentially cover the most common variants, and do not contain unnatural junctional epitopes. Another T-cell vaccine design strategy, COT+, also uses computational tools to maximize 9-mer coverage (70). However, this strategy does not reconstruct intact proteins. Rather, it produces a set of protein fragments that could be assembled into a polyfragment chimeric protein to complement the coverage of a single center-of-tree protein (28, 70). The mosaic strategy gave slightly enhanced coverage of 9-mers over that provided by the COT+ strategy when these two algorithms were applied to the same data set (28).
Figure 4 illustrates the application of the mosaic strategy to HIV Gag vaccine design, drawing sequences from the database of M-group Gag proteins. Figure 4a shows the incrementally improved coverage of 9-mers in Gag in the global M-group alignment using different design strategies. Figure 4b shows that the coverage of sequences is relatively constant in the face of changing input data as new HIV sequences become available each year. The M-group input data used to generate the mosaic vaccine designs and the coverage figures are based on the 2008 database. This database provided two additional years of sequence acquisition beyond that used in the study by Fischer et al. (29), and the current data set has more than doubled in size relative to the data set used in the first study. Importantly, the mosaic vaccine designed using the 2006 data set in the study by Fisher et al. gives coverage of the HIV sequences in the 2008 data set that is comparable to that provided by a mosaic vaccine designed using the 2008 data set (Fig. 4b). Figure 4c shows that the M-group 9-mer coverage provided by a combination of four different subtype consensus sequences (A, B, C, and D) approximates that conferred by the four best natural strains of HIV and that the coverage by four mosaics is substantially better than that provided by either of the other two vaccine candidates. Also, random selection of four natural strains of HIV gives a wide range of coverage, and most combinations of such strains do not approach conferment of optimal coverage. Figure 4d shows that as more sequences of HIV are included in an optimal natural immunogen set, or in a polyvalent mosaic vaccine design set, population coverage of potential epitopes increases, but by decreasing increments. Finally, we generally optimize for coverage of 9-mers, as that is the most common length of CD8+ T-cell epitopes. However, as shown in Fig. 4e, the solution the mosaic algorithm finds based on 9-mers is also a very good solution for epitopes that are 8-mers to 12-mers.
Several HIV vaccine candidates based on the mosaic approach are currently in the developmental pipeline. The pathway for these immunogens begins with the design of the mosaic protein sequences, followed by the design of a “humanized” gene to optimize their expression (65), the synthesis and expression of the genes, and then the study of the properties of the proteins, including binding to relevant antibodies. After this is accomplished, the immunogenicity of the constructs is assessed with mice. Intact Gag, Nef, Pol, and Env M-group mosaics have all been synthesized, and all are well expressed and immunogenic in mice (B. F. Haynes and N. L. Letvin, data not shown). In contrast, a Gag-partial Nef fusion protein mosaic had an immunogenic Gag portion, but the Nef portion was nonimmunogenic in mice (29). Therefore, a fusion protein strategy for these mosaics was dropped, and the two genes were expressed separately (28).
The first study of the mosaic vaccine strategy in mice was designed to test whether the breadth of the T-cell response induced by mosaic vaccines was enhanced in comparison with that induced by other vaccine approaches. That study showed a marked increase in breadth of response for both CD4+ and CD8+ T cells, with the increase being most pronounced in CD8+ T-cell responses (51). In the most striking comparison in that study, a DNA vaccine based on three natural Envs, one each from subtypes A, B, and C, was compared with a three-mosaic vaccine. The trivalent vaccine based on three natural strains elicited only two CD8+ T-cell responses to a series of peptide pools representing M-group diversity, while the trivalent mosaic vaccine elicited CD8+ T-cell responses to 10 peptide pools (59). Two macaque trials comparing mosaic designs to natural strains and consensus vaccines are currently under way.
CONCLUSIONS
There are several promising paths to follow for the development of a T-cell component of new HIV vaccine candidates. Experimental immunogens that focus on conserved regions of the HIV proteome have the potential to focus the immune response on regions of the virus where immune escape is deleterious to it and where a vaccine-elicited T-cell response is most likely to recognize disparate circulating viruses. The first study of this approach with mice generated encouraging results (58). The limited human immune responses to polyepitope vaccines (38, 39), however, suggest that vaccine candidates of this type should be carefully evaluated for immunogenicity of the multiple protein fragments and to determine if unnatural junctional epitopes are confounding their immunogenicity. By including whole conserved regions of the virus rather than just specific epitopes, CD4+ and CD8+ T-cell responses are more likely to be elicited by these vaccine candidates.
Attempting to maximize the number of cross-reactive T-cell responses through vaccination offers possibilities that are philosophically the opposite of a conserved-region approach, broadening the cross-reactive potential of all responses rather than focusing the responses. Both strategies are well reasoned, and both have valid experimental underpinnings. Whether either or both strategies will confer a benefit will require experimental resolution.
Polyclonal, central, and mosaic vaccine approaches offer strategies to increase the number of cross-reactive responses in the context of a single protein. Each of these strategies is proving to be more effective than a single natural protein approach in animal studies of immunogenicity and breadth of induced responses. This benefit is particularly marked across subtypes (51, 78, 89). In nonhuman primate vaccine-challenge studies, live attenuated vaccines can elicit protective responses against a heterologous challenge virus—better control of the virus and a better clinical outcome (73). Similarly, preexisting infection can result in control of superinfection by a heterologous strain of virus (99). One shared aspect of these experimental strategies is that they make use of immunity against the full viral proteome. These observations provide further motivation for continued exploration of the value of increasing the number of vaccine-induced T-cell responses.
A vaccine should stimulate T-cell populations that recognize virus epitopes presented by infected cells and that are not vaccine specific. A close match between the vaccine and circulating virus epitope sequences, as well as similarities between the virus and vaccine antigen processing, should enhance the likelihood of that occurring. Gag and Pol, despite being relatively conserved HIV proteins, are nonetheless still variable in the peptides that are recognized by T cells. In fact, a 10% difference between two aligned viral proteins means most epitopes from different HIV stains will be distinct. Based on this and on the fact that much of the HIV diversity seen at the population level is likely to be a consequence of immune escape (15, 77), for which observed mutations are most likely immunologically relevant, even the most conserved HIV proteins will benefit from strategies to address diversity (Fig. 3).
Polyclonal and mosaic vaccines offer potential benefits beyond improved population coverage. The most common variants of an epitope at a population level should represent relatively fit mutations that confer escape from T-cell responses. One person's virus escape form, however, can be another person's susceptible form (10), so including both in a vaccine may be helpful. If the most common forms of epitopes are presented in a vaccine immunogen, vaccine-elicited T-cell responses may effectively block the most common form and common escape routes of virus mutation in an individual, forcing the virus either to remain susceptible to the CTL response or to escape though evolution to less-fit forms. These strategies, particularly in combination with better delivery strategies and adjuvants, may improve vaccine-induced T-cell responses. A series of iterative nonhuman primate and human phase I clinical trials is needed now to define those T-cell vaccine candidates that induce the greatest breadth of T-cell responses.
Biography
 Bette Korber is a Laboratory Fellow at Los Alamos National Laboratory and an external professor at the Santa Fe Institute. She obtained her Ph.D. in Immunology from the California Institute of Technology in 1988 and was a postdoctoral fellow in retrovirology at Harvard University before joining the Theoretical Division at Los Alamos in 1990. She has led a human immunodeficiency virus (HIV) sequence and immunology database project at Los Alamos since 1996. She works with many experimentalist collaborators, developing analytical methods to help understand the impact of the human immune response on HIV evolution and HIV transmission patterns and to develop vaccine design strategies for variable pathogens. She became an Elizabeth Glaser Scientist in 1997 for her work on pediatric AIDS and mother-infant transmission and received the E. O. Lawrence Award from the Department of Energy for her achievements in the life sciences. She is an advisor for the Global Vaccine Enterprise and the Ragon Institute.
Bette Korber is a Laboratory Fellow at Los Alamos National Laboratory and an external professor at the Santa Fe Institute. She obtained her Ph.D. in Immunology from the California Institute of Technology in 1988 and was a postdoctoral fellow in retrovirology at Harvard University before joining the Theoretical Division at Los Alamos in 1990. She has led a human immunodeficiency virus (HIV) sequence and immunology database project at Los Alamos since 1996. She works with many experimentalist collaborators, developing analytical methods to help understand the impact of the human immune response on HIV evolution and HIV transmission patterns and to develop vaccine design strategies for variable pathogens. She became an Elizabeth Glaser Scientist in 1997 for her work on pediatric AIDS and mother-infant transmission and received the E. O. Lawrence Award from the Department of Energy for her achievements in the life sciences. She is an advisor for the Global Vaccine Enterprise and the Ragon Institute.
 Norman Letvin received his M.D. from Harvard Medical School in 1975, followed by a fellowship in immunology. He interned at the Hospital of the University of Pennsylvania and completed his residency at Massachusetts General Hospital. He began at the Dana Farber Cancer Institute in 1981. Dr. Letvin is a Professor of Medicine at the Harvard Medical School, Chief of the Division of Viral Pathogenesis at the Beth Israel Deaconess Medical Center, and Director of the Non-Human Primate Research Program at the NIH Vaccine Research Center. He pioneered nonhuman primates in HIV vaccine design, defined the role of cytotoxic T lymphocytes (CTLs) in controlling HIV replication, and is working toward developing vaccination strategies that protect against HIV by targeting CTLs. He has served on many scientific advisory boards, including the Research Advisory Council for the Office of AIDS Research, and is a reviewing editor for Science.
Norman Letvin received his M.D. from Harvard Medical School in 1975, followed by a fellowship in immunology. He interned at the Hospital of the University of Pennsylvania and completed his residency at Massachusetts General Hospital. He began at the Dana Farber Cancer Institute in 1981. Dr. Letvin is a Professor of Medicine at the Harvard Medical School, Chief of the Division of Viral Pathogenesis at the Beth Israel Deaconess Medical Center, and Director of the Non-Human Primate Research Program at the NIH Vaccine Research Center. He pioneered nonhuman primates in HIV vaccine design, defined the role of cytotoxic T lymphocytes (CTLs) in controlling HIV replication, and is working toward developing vaccination strategies that protect against HIV by targeting CTLs. He has served on many scientific advisory boards, including the Research Advisory Council for the Office of AIDS Research, and is a reviewing editor for Science.
 Barton Haynes received his M.D. from Baylor College of Medicine. After infectious disease as well as allergy and immunology training at the NIH, he joined the faculty at Duke University in 1980. Since 2002, he has served as the Director of the Duke Human Vaccine Institute. His work on discovery of human T-cell molecules and the biology of the human thymus led to the development of thymic transplantation as a curative treatment for DiGeorge Syndrome in children born without a thymus. He has worked toward development of an AIDS vaccine since 1986. He is the leader of the Center for HIV/AIDS Vaccine Immunology (CHAVI) at the National Institute of Allergy and Infectious Diseases and a Gates Foundation Collaboration for the AIDS Vaccine Discovery Center. Haynes is a member of the Institute of Medicine of the National Academy of Sciences and a Fellow of the American Academy of Arts and Sciences.
Barton Haynes received his M.D. from Baylor College of Medicine. After infectious disease as well as allergy and immunology training at the NIH, he joined the faculty at Duke University in 1980. Since 2002, he has served as the Director of the Duke Human Vaccine Institute. His work on discovery of human T-cell molecules and the biology of the human thymus led to the development of thymic transplantation as a curative treatment for DiGeorge Syndrome in children born without a thymus. He has worked toward development of an AIDS vaccine since 1986. He is the leader of the Center for HIV/AIDS Vaccine Immunology (CHAVI) at the National Institute of Allergy and Infectious Diseases and a Gates Foundation Collaboration for the AIDS Vaccine Discovery Center. Haynes is a member of the Institute of Medicine of the National Academy of Sciences and a Fellow of the American Academy of Arts and Sciences.
Footnotes
Published ahead of print on 13 May 2009.
REFERENCES
- 1.Abdel-Motal, U. M., J. Gillis, K. Manson, M. Wyand, D. Montefiori, K. Stefano-Cole, R. C. Montelaro, J. D. Altman, and R. P. Johnson. 2005. Kinetics of expansion of SIV Gag-specific CD8+ T lymphocytes following challenge of vaccinated macaques. Virology 333226-238. [DOI] [PubMed] [Google Scholar]
- 2.Addo, M. M., X. G. Yu, A. Rathod, D. Cohen, R. L. Eldridge, D. Strick, M. N. Johnston, C. Corcoran, A. G. Wurcel, C. A. Fitzpatrick, M. E. Feeney, W. R. Rodriguez, N. Basgoz, R. Draenert, D. R. Stone, C. Brander, P. J. Goulder, E. S. Rosenberg, M. Altfeld, and B. D. Walker. 2003. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J. Virol. 772081-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen, T. M., and M. Altfeld. 2008. Crippling HIV one mutation at a time. J. Exp. Med. 2051003-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Almeida, J. R., D. A. Price, L. Papagno, Z. A. Arkoub, D. Sauce, E. Bornstein, T. E. Asher, A. Samri, A. Schnuriger, I. Theodorou, D. Costagliola, C. Rouzioux, H. Agut, A. G. Marcelin, D. Douek, B. Autran, and V. Appay. 2007. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J. Exp. Med. 2042473-2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altfeld, M., M. M. Addo, E. S. Rosenberg, F. M. Hecht, P. K. Lee, M. Vogel, X. G. Yu, R. Draenert, M. N. Johnston, D. Strick, T. M. Allen, M. E. Feeney, J. O. Kahn, R. P. Sekaly, J. A. Levy, J. K. Rockstroh, P. J. Goulder, and B. D. Walker. 2003. Influence of HLA-B57 on clinical presentation and viral control during acute HIV-1 infection. AIDS 172581-2591. [DOI] [PubMed] [Google Scholar]
- 6.Arroyo, M. A., W. B. Sateren, D. Serwadda, R. H. Gray, M. J. Wawer, N. K. Sewankambo, N. Kiwanuka, G. Kigozi, F. Wabwire-Mangen, M. Eller, L. A. Eller, D. L. Birx, M. L. Robb, and F. E. McCutchan. 2006. Higher HIV-1 incidence and genetic complexity along main roads in Rakai District, Uganda. J. Acquir. Immune Defic. Syndr. 43440-445. [DOI] [PubMed] [Google Scholar]
- 7.Bansal, A., B. Jackson, K. West, S. Wang, S. Lu, J. S. Kennedy, and P. A. Goepfert. 2008. Multifunctional T-cell characteristics induced by a polyvalent DNA prime/protein boost human immunodeficiency virus type 1 vaccine regimen given to healthy adults are dependent on the route and dose of administration. J. Virol. 826458-6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bennett, M. S., H. L. Ng, A. Ali, and O. O. Yang. 2008. Cross-clade detection of HIV-1-specific cytotoxic T lymphocytes does not reflect cross-clade antiviral activity. J. Infect. Dis. 197390-397. [DOI] [PubMed] [Google Scholar]
- 9.Betts, M. R., M. C. Nason, S. M. West, S. C. De Rosa, S. A. Migueles, J. Abraham, M. M. Lederman, J. M. Benito, P. A. Goepfert, M. Connors, M. Roederer, and R. A. Koup. 2006. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood 1074781-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhattacharya, T., M. Daniels, D. Heckerman, B. Foley, N. Frahm, C. Kadie, J. Carlson, K. Yusim, B. McMahon, B. Gaschen, S. Mallal, J. I. Mullins, D. C. Nickle, J. Herbeck, C. Rousseau, G. H. Learn, T. Miura, C. Brander, B. Walker, and B. Korber. 2007. Founder effects in the assessment of HIV polymorphisms and HLA allele associations. Science 3151583-1586. [DOI] [PubMed] [Google Scholar]
- 11.Binley, J. M., E. A. Lybarger, E. T. Crooks, M. S. Seaman, E. Gray, K. L. Davis, J. M. Decker, D. Wycuff, L. Harris, N. Hawkins, B. Wood, C. Nathe, D. Richman, G. D. Tomaras, F. Bibollet-Ruche, J. E. Robinson, L. Morris, G. M. Shaw, D. C. Montefiori, and J. R. Mascola. 2008. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J. Virol. 8211651-11668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Binley, J. M., T. Wrin, B. Korber, M. B. Zwick, M. Wang, C. Chappey, G. Stiegler, R. Kunert, S. Zolla-Pazner, H. Katinger, C. J. Petropoulos, and D. R. Burton. 2004. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 7813232-13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borrow, P., H. Lewicki, X. Wei, M. S. Horwitz, N. Peffer, H. Meyers, J. A. Nelson, J. E. Gairin, B. H. Hahn, M. B. Oldstone, and G. M. Shaw. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3205-211. [DOI] [PubMed] [Google Scholar]
- 14.Brumme, Z. L., C. J. Brumme, J. Carlson, H. Streeck, M. John, Q. Eichbaum, B. L. Block, B. Baker, C. Kadie, M. Markowitz, H. Jessen, A. D. Kelleher, E. Rosenberg, J. Kaldor, Y. Yuki, M. Carrington, T. M. Allen, S. Mallal, M. Altfeld, D. Heckerman, and B. D. Walker. 2008. Marked epitope- and allele-specific differences in rates of mutation in human immunodeficiency type 1 (HIV-1) Gag, Pol, and Nef cytotoxic T-lymphocyte epitopes in acute/early HIV-1 infection. J. Virol. 829216-9227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brumme, Z. L., C. J. Brumme, D. Heckerman, B. T. Korber, M. Daniels, J. Carlson, C. Kadie, T. Bhattacharya, C. Chui, J. Szinger, T. Mo, R. S. Hogg, J. S. Montaner, N. Frahm, C. Brander, B. D. Walker, and P. R. Harrigan. 2007. Evidence of differential HLA class I-mediated viral evolution in functional and accessory/regulatory genes of HIV-1. PLoS Pathog. 3e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burgers, W. A., E. Shephard, J. E. Monroe, T. Greenhalgh, A. Binder, E. Hurter, J. H. Van Harmelen, C. Williamson, and A. L. Williamson. 2008. Construction, characterization, and immunogenicity of a multigene modified vaccinia Ankara (MVA) vaccine based on HIV type 1 subtype C. AIDS Res. Hum. Retrovir. 24195-206. [DOI] [PubMed] [Google Scholar]
- 17.Burgers, W. A., J. H. van Harmelen, E. Shephard, C. Adams, T. Mgwebi, W. Bourn, T. Hanke, A. L. Williamson, and C. Williamson. 2006. Design and preclinical evaluation of a multigene human immunodeficiency virus type 1 subtype C DNA vaccine for clinical trial. J. Gen. Virol. 87399-410. [DOI] [PubMed] [Google Scholar]
- 18.Burton, D. R., R. C. Desrosiers, R. W. Doms, M. B. Feinberg, R. C. Gallo, B. Hahn, J. A. Hoxie, E. Hunter, B. Korber, A. Landay, M. M. Lederman, J. Lieberman, J. M. McCune, J. P. Moore, N. Nathanson, L. Picker, D. Richman, C. Rinaldo, M. Stevenson, D. I. Watkins, S. M. Wolinsky, and J. A. Zack. 2004. Public health: a sound rationale needed for phase III HIV-1 vaccine trials. Science 303316. [DOI] [PubMed] [Google Scholar]
- 19.Carrington, M., G. W. Nelson, M. P. Martin, T. Kissner, D. Vlahov, J. J. Goedert, R. Kaslow, S. Buchbinder, K. Hoots, and S. J. O'Brien. 1999. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science 2831748-1752. [DOI] [PubMed] [Google Scholar]
- 20.Catanzaro, A. T., M. Roederer, R. A. Koup, R. T. Bailer, M. E. Enama, M. C. Nason, J. E. Martin, S. Rucker, C. A. Andrews, P. L. Gomez, J. R. Mascola, G. J. Nabel, and B. S. Graham. 2007. Phase I clinical evaluation of a six-plasmid multiclade HIV-1 DNA candidate vaccine. Vaccine 254085-4092. [DOI] [PubMed] [Google Scholar]
- 21.Chakrabarti, B. K., W.-P. Kong, B.-Y. Wu, Z.-Y. Yang, J. Friborg, X. Ling, S. R. King, D. C. Montefiori, and G. J. Nabel. 2002. Modifications of the human immunodeficiency virus envelope glycoprotein enhance immunogenicity for genetic immunization. J. Virol. 765357-5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen, Z., Y. Huang, X. Zhao, L. Ba, W. Zhang, and D. D. Ho. 2008. Design, construction, and characterization of a multigenic modified vaccinia Ankara candidate vaccine against human immunodeficiency virus type 1 subtype C/B′. J. Acquir. Immune Defic. Syndr. 47412-421. [DOI] [PubMed] [Google Scholar]
- 23.Connor, R. I., B. T. Korber, B. S. Graham, B. H. Hahn, D. D. Ho, B. D. Walker, A. U. Neumann, S. H. Vermund, J. Mestecky, S. Jackson, E. Fenamore, Y. Cao, F. Gao, S. Kalams, K. J. Kunstman, D. McDonald, N. McWilliams, A. Trkola, J. P. Moore, and S. M. Wolinsky. 1998. Immunological and virological analyses of persons infected by human immunodeficiency virus type 1 while participating in trials of recombinant gp120 subunit vaccines. J. Virol. 721552-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corey, L., M. J. McElrath, and J. G. Kublin. 2009. Post-step modifications for research on HIV vaccines. AIDS 233-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Groot, A. S., L. Marcon, E. A. Bishop, D. Rivera, M. Kutzler, D. B. Weiner, and W. Martin. 2005. HIV vaccine development by computer assisted design: the GAIA vaccine. Vaccine 232136-2148. [DOI] [PubMed] [Google Scholar]
- 26.Doria-Rose, N. A., G. H. Learn, A. G. Rodrigo, D. C. Nickle, F. Li, M. Mahalanabis, M. T. Hensel, S. McLaughlin, P. F. Edmonson, D. Montefiori, S. W. Barnett, N. L. Haigwood, and J. I. Mullins. 2005. Human immunodeficiency virus type 1 subtype B ancestral envelope protein is functional and elicits neutralizing antibodies in rabbits similar to those elicited by a circulating subtype B envelope. J. Virol. 7911214-11224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fellay, J., K. V. Shianna, D. Ge, S. Colombo, B. Ledergerber, M. Weale, K. Zhang, C. Gumbs, A. Castagna, A. Cossarizza, A. Cozzi-Lepri, A. De Luca, P. Easterbrook, P. Francioli, S. Mallal, J. Martinez-Picado, J. M. Miro, N. Obel, J. P. Smith, J. Wyniger, P. Descombes, S. E. Antonarakis, N. L. Letvin, A. J. McMichael, B. F. Haynes, A. Telenti, and D. B. Goldstein. 2007. A whole-genome association study of major determinants for host control of HIV-1. Science 317944-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fischer, W., H. X. Liao, B. F. Haynes, N. L. Letvin, and B. Korber. 2008. Coping with viral diversity in HIV vaccine design: a response to Nickle et al. PLoS Comput. Biol. 4e15. (Author reply, 4:e25.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fischer, W., S. Perkins, J. Theiler, T. Bhattacharya, K. Yusim, R. Funkhouser, C. Kuiken, B. Haynes, N. L. Letvin, B. D. Walker, B. H. Hahn, and B. T. Korber. 2007. Polyvalent vaccines for optimal coverage of potential T-cell epitopes in global HIV-1 variants. Nat. Med. 13100-106. [DOI] [PubMed] [Google Scholar]
- 30.Frahm, N., B. T. Korber, C. M. Adams, J. J. Szinger, R. Draenert, M. M. Addo, M. E. Feeney, K. Yusim, K. Sango, N. V. Brown, D. SenGupta, A. Piechocka-Trocha, T. Simonis, F. M. Marincola, A. G. Wurcel, D. R. Stone, C. J. Russell, P. Adolf, D. Cohen, T. Roach, A. StJohn, A. Khatri, K. Davis, J. Mullins, P. J. Goulder, B. D. Walker, and C. Brander. 2004. Consistent cytotoxic-T-lymphocyte targeting of immunodominant regions in human immunodeficiency virus across multiple ethnicities. J. Virol. 782187-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frahm, N., D. C. Nickle, C. H. Linde, D. E. Cohen, R. Zuniga, A. Lucchetti, T. Roach, B. D. Walker, T. M. Allen, B. T. Korber, J. I. Mullins, and C. Brander. 2008. Increased detection of HIV-specific T cell responses by combination of central sequences with comparable immunogenicity. AIDS 22447-456. [DOI] [PubMed] [Google Scholar]
- 32.Gao, F., T. Bhattacharya, B. Gaschen, J. Taylor, J. Moore, V. Novitsky, K. Yusim, D. Lang, B. Foley, S. Beddows, M. Alam, B. Haynes, B. H. Hahn, and B. Korber. 2003. Response to letter. Consensus and ancestral state HIV vaccines. Science 2991515-1518. [DOI] [PubMed] [Google Scholar]
- 33.Gao, F., E. A. Weaver, Z. Lu, Y. Li, H.-X. Liao, B. Ma, S. M. Alam, R. M. Scearce, L. L. Sutherland, J.-S. Yu, J. M. Decker, G. M. Shaw, D. C. Montefiori, B. T. Korber, B. H. Hahn, and B. F. Haynes. 2005. Antigenicity and immunogenicity of a synthetic human immunodeficiency virus type 1 group M consensus envelope glycoprotein. J. Virol. 791154-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gaschen, B., J. Taylor, K. Yusim, B. Foley, F. Gao, D. Lang, V. Novitsky, B. Haynes, B. H. Hahn, T. Bhattacharya, and B. Korber. 2002. Diversity considerations in HIV-1 vaccine selection. Science 2962354-2360. [DOI] [PubMed] [Google Scholar]
- 35.Gasper-Smith, N., D. M. Crossman, J. F. Whitesides, N. Mensali, J. S. Ottinger, S. G. Plonk, M. A. Moody, G. Ferrari, K. J. Weinhold, S. E. Miller, C. F. Reich III, L. Qin, S. Self, G. M. Shaw, T. N. Denny, L. E. Jones, D. S. Pisetsky, and B. F. Haynes. 2008. Induction of plasma (TRAIL), TNFR-2, Fas ligand and plasma microparticles after human immunodeficiency virus type 1 (HIV-1) transmission: implications for HIV-1 vaccine design. J. Virol. 827700-7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gilbert, P. B., M. L. Peterson, D. Follmann, M. G. Hudgens, D. P. Francis, M. Gurwith, W. L. Heyward, D. V. Jobes, V. Popovic, S. G. Self, F. Sinangil, D. Burke, and P. W. Berman. 2005. Correlation between immunologic responses to a recombinant glycoprotein 120 vaccine and incidence of HIV-1 infection in a phase 3 HIV-1 preventive vaccine trial. J. Infect. Dis. 191666-677. [DOI] [PubMed] [Google Scholar]
- 37.Gnanakaran, S., D. Lang, M. Daniels, T. Bhattacharya, C. A. Derdeyn, and B. Korber. 2007. Clade-specific differences between human immunodeficiency virus type 1 clades B and C: diversity and correlations in C3-V4 regions of gp120. J. Virol. 814886-4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goonetilleke, N., S. Moore, L. Dally, N. Winstone, I. Cebere, A. Mahmoud, S. Pinheiro, G. Gillespie, D. Brown, V. Loach, J. Roberts, A. Guimaraes-Walker, P. Hayes, K. Loughran, C. Smith, J. De Bont, C. Verlinde, D. Vooijs, C. Schmidt, M. Boaz, J. Gilmour, P. Fast, L. Dorrell, T. Hanke, and A. J. McMichael. 2006. Induction of multifunctional human immunodeficiency virus type 1 (HIV-1)-specific T cells capable of proliferation in healthy subjects by using a prime-boost regimen of DNA- and modified vaccinia virus Ankara-vectored vaccines expressing HIV-1 Gag coupled to CD8+ T-cell epitopes. J. Virol. 804717-4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gorse, G. J., L. R. Baden, M. Wecker, M. J. Newman, G. Ferrari, K. J. Weinhold, B. D. Livingston, T. L. Villafana, H. Li, E. Noonan, and N. D. Russell. 2008. Safety and immunogenicity of cytotoxic T-lymphocyte poly-epitope, DNA plasmid (EP HIV-1090) vaccine in healthy, human immunodeficiency virus type 1 (HIV-1)-uninfected adults. Vaccine 26215-223. [DOI] [PubMed] [Google Scholar]
- 40.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52696-704. [DOI] [PubMed] [Google Scholar]
- 41.Hanke, T., and A. J. McMichael. 2000. Design and construction of an experimental HIV-1 vaccine for a year-2000 clinical trial in Kenya. Nat. Med. 6951-955. [DOI] [PubMed] [Google Scholar]
- 42.Hu, D. J., S. Vanichseni, T. D. Mastro, S. Raktham, N. L. Young, P. A. Mock, S. Subbarao, B. S. Parekh, L. Srisuwanvilai, R. Sutthent, C. Wasi, W. Heneine, and K. Choopanya. 2001. Viral load differences in early infection with two HIV-1 subtypes. AIDS 15683-691. [DOI] [PubMed] [Google Scholar]
- 43.Hudgens, M. G., I. M. Longini, Jr., S. Vanichseni, D. J. Hu, D. Kitayaporn, P. A. Mock, M. E. Halloran, G. A. Satten, K. Choopanya, and T. D. Mastro. 2002. Subtype-specific transmission probabilities for human immunodeficiency virus type 1 among injecting drug users in Bangkok, Thailand. Am. J. Epidemiol. 155159-168. [DOI] [PubMed] [Google Scholar]
- 44.Huss, A., P. Scott, A. E. Stuck, C. Trotter, and M. Egger. 2009. Efficacy of pneumococcal vaccination in adults: a meta-analysis. CMAJ 18048-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jaoko, W., F. N. Nakwagala, O. Anzala, G. O. Manyonyi, J. Birungi, A. Nanvubya, F. Bashir, K. Bhatt, H. Ogutu, S. Wakasiaka, L. Matu, W. Waruingi, J. Odada, M. Oyaro, J. Indangasi, J. Ndinya-Achola, C. Konde, E. Mugisha, P. Fast, C. Schmidt, J. Gilmour, T. Tarragona, C. Smith, B. Barin, L. Dally, B. Johnson, A. Muluubya, L. Nielsen, P. Hayes, M. Boaz, P. Hughes, T. Hanke, A. McMichael, J. Bwayo, and P. Kaleebu. 2008. Safety and immunogenicity of recombinant low-dosage HIV-1 A vaccine candidates vectored by plasmid pTHr DNA or modified vaccinia virus Ankara (MVA) in humans in East Africa. Vaccine 262788-2795. [DOI] [PubMed] [Google Scholar]
- 46.Kaslow, R. A., M. Carrington, R. Apple, L. Park, A. Munoz, A. J. Saah, J. J. Goedert, C. Winkler, S. J. O'Brien, C. Rinaldo, R. Detels, W. Blattner, J. Phair, H. Erlich, and D. L. Mann. 1996. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat. Med. 2405-411. [DOI] [PubMed] [Google Scholar]
- 47.Kawashima, Y., K. Pfafferott, J. Frater, P. Matthews, R. Payne, M. Addo, H. Gatanaga, M. Fujiwara, A. Hachiya, H. Koizumi, N. Kuse, S. Oka, A. Duda, A. Prendergast, H. Crawford, A. Leslie, Z. Brumme, C. Brumme, T. Allen, C. Brander, R. Kaslow, J. Tang, E. Hunter, S. Allen, J. Mulenga, S. Branch, T. Roach, M. John, S. Mallal, A. Ogwu, R. Shapiro, J. G. Prado, S. Fidler, J. Weber, O. G. Pybus, P. Klenerman, T. Ndung'u, R. Phillips, D. Heckerman, P. R. Harrigan, B. D. Walker, M. Takiguchi, and P. Goulder. 2009. Adaptation of HIV-1 to human leukocyte antigen class I. Nature 458641-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Keele, B. F., E. E. Giorgi, J. F. Salazar-Gonzalez, J. M. Decker, K. T. Pham, M. G. Salazar, C. Sun, T. Grayson, S. Wang, H. Li, X. Wei, C. Jiang, J. L. Kirchherr, F. Gao, J. A. Anderson, L. H. Ping, R. Swanstrom, G. D. Tomaras, W. A. Blattner, P. A. Goepfert, J. M. Kilby, M. S. Saag, E. L. Delwart, M. P. Busch, M. S. Cohen, D. C. Montefiori, B. F. Haynes, B. Gaschen, G. S. Athreya, H. Y. Lee, N. Wood, C. Seoighe, A. S. Perelson, T. Bhattacharya, B. T. Korber, B. H. Hahn, and G. M. Shaw. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. USA 1057552-7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kijak, G. H., S. Tovanabutra, E. Sanders-Buell, V. Watanaveeradej, M. S. de Souza, K. E. Nelson, V. Ketsararat, V. Gulgolgarn, M. Wera-arpachai, S. Sriplienchan, C. Khamboonrueng, D. L. Birx, M. L. Robb, and F. E. McCutchan. 2007. Distinguishing molecular forms of HIV-1 in Asia with a high-throughput, fluorescent genotyping assay, MHAbce v. 2. Virology 358178-191. [DOI] [PubMed] [Google Scholar]
- 50.Kiwanuka, N., O. Laeyendecker, M. Robb, G. Kigozi, M. Arroyo, F. McCutchan, L. A. Eller, M. Eller, F. Makumbi, D. Birx, F. Wabwire-Mangen, D. Serwadda, N. K. Sewankambo, T. C. Quinn, M. Wawer, and R. Gray. 2008. Effect of human immunodeficiency virus Type 1 (HIV-1) subtype on disease progression in persons from Rakai, Uganda, with incident HIV-1 infection. J. Infect. Dis. 197707-713. [DOI] [PubMed] [Google Scholar]
- 51.Kong, W. P., L. Wu, T. C. Wallstrom, W. Fischer, Z. Y. Yang, S. Y. Ko, N. Letvin, B. F. Haynes, B. Hahn, B. Korber, and G. J. Nabel. 2009. Expanded breadth of the T-cell response to mosaic human immunodeficiency virus type 1 envelope DNA vaccination. J. Virol. 832201-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Korber, B., M. Muldoon, J. Theiler, F. Gao, R. Gupta, A. Lapedes, B. H. Hahn, S. Wolinsky, and T. Bhattacharya. 2000. Timing the ancestor of the HIV-1 pandemic strains. Science 2881789-1796. [DOI] [PubMed] [Google Scholar]
- 53.Korber, B., J. Theiler, and S. Wolinsky. 1998. Limitations of a molecular clock applied to considerations of the origin of HIV-1. Science 2801868-1871. [DOI] [PubMed] [Google Scholar]
- 54.Kosakovsky Pond, S. L., A. F. Poon, S. Zarate, D. M. Smith, S. J. Little, S. K. Pillai, R. J. Ellis, J. K. Wong, A. J. Leigh Brown, D. D. Richman, and S. D. Frost. 2008. Estimating selection pressures on HIV-1 using phylogenetic likelihood models. Stat. Med. 274779-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kothe, D. L., J. M. Decker, Y. Li, Z. Weng, F. Bibollet-Ruche, K. P. Zammit, M. G. Salazar, Y. Chen, J. F. Salazar-Gonzalez, Z. Moldoveanu, J. Mestecky, F. Gao, B. F. Haynes, G. M. Shaw, M. Muldoon, B. T. Korber, and B. H. Hahn. 2007. Antigenicity and immunogenicity of HIV-1 consensus subtype B envelope glycoproteins. Virology 360218-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kothe, D. L., Y. Li, J. M. Decker, F. Bibollet-Ruche, K. P. Zammit, M. G. Salazar, Y. Chen, Z. Weng, E. A. Weaver, F. Gao, B. F. Haynes, G. M. Shaw, B. T. Korber, and B. H. Hahn. 2006. Ancestral and consensus envelope immunogens for HIV-1 subtype C. Virology 352438-449. [DOI] [PubMed] [Google Scholar]
- 57.Kulkarni, S. S., A. Lapedes, H. Tang, S. Gnanakaran, M. G. Daniels, M. Zhang, T. Bhattacharya, M. Li, V. R. Polonis, F. E. McCutchan, L. Morris, D. Ellenberger, S. T. Butera, R. C. Bollinger, B. T. Korber, R. S. Paranjape, and D. C. Montefiori. 2009. Highly complex neutralization determinants on a monophyletic lineage of newly transmitted subtype C HIV-1 Env clones from India. Virology 385505-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Letourneau, S., E. J. Im, T. Mashishi, C. Brereton, A. Bridgeman, H. Yang, L. Dorrell, T. Dong, B. Korber, A. J. McMichael, and T. Hanke. 2007. Design and pre-clinical evaluation of a universal HIV-1 vaccine. PLoS One 2e984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li, F., U. Malhotra, P. B. Gilbert, N. R. Hawkins, A. C. Duerr, J. M. McElrath, L. Corey, and S. G. Self. 2006. Peptide selection for human immunodeficiency virus type 1 CTL-based vaccine evaluation. Vaccine 246893-6904. [DOI] [PubMed] [Google Scholar]
- 60.Liao, H. X., L. L. Sutherland, S. M. Xia, M. E. Brock, R. M. Scearce, S. Vanleeuwen, S. M. Alam, M. McAdams, E. A. Weaver, Z. Camacho, B. J. Ma, Y. Li, J. M. Decker, G. J. Nabel, D. C. Montefiori, B. H. Hahn, B. T. Korber, F. Gao, and B. F. Haynes. 2006. A group M consensus envelope glycoprotein induces antibodies that neutralize subsets of subtype B and C HIV-1 primary viruses. Virology 353268-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin, H. H., B. K. Gaschen, M. Collie, M. El-Fishaway, Z. Chen, B. T. Korber, S. T. Beatrice, and L. Zhang. 2006. Genetic characterization of diverse HIV-1 strains in an immigrant population living in New York City. J. Acquir. Immune Defic. Syndr. 41399-404. [DOI] [PubMed] [Google Scholar]
- 62.Liu, J., K. L. O'Brien, D. M. Lynch, N. L. Simmons, A. La Porte, A. M. Riggs, P. Abbink, R. T. Coffey, L. E. Grandpre, M. S. Seaman, G. Landucci, D. N. Forthal, D. C. Montefiori, A. Carville, K. G. Mansfield, M. J. Havenga, M. G. Pau, J. Goudsmit, and D. H. Barouch. 2009. Immune control of an SIV challenge by a T-cell-based vaccine in rhesus monkeys. Nature 45787-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Livingston, B. D., M. Newman, C. Crimi, D. McKinney, R. Chesnut, and A. Sette. 2001. Optimization of epitope processing enhances immunogenicity of multiepitope DNA vaccines. Vaccine 194652-4660. [DOI] [PubMed] [Google Scholar]
- 64.McElrath, M. J., S. C. De Rosa, Z. Moodie, S. Dubey, L. Kierstead, H. Janes, O. D. Defawe, D. K. Carter, J. Hural, R. Akondy, S. P. Buchbinder, M. N. Robertson, D. V. Mehrotra, S. G. Self, L. Corey, J. W. Shiver, and D. R. Casimiro. 2008. HIV-1 vaccine-induced immunity in the test-of-concept Step Study: a case-cohort analysis. Lancet 3721894-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Megati, S., D. Garcia-Hand, S. Cappello, V. Roopchand, A. Masood, R. Xu, A. Luckay, S. Y. Chong, M. Rosati, S. Sackitey, D. B. Weiner, B. K. Felber, G. N. Pavlakis, Z. R. Israel, L. R. Smith, J. H. Eldridge, M. K. Sidhu, and M. A. Egan. 2008. Modifying the HIV-1 env gp160 gene to improve pDNA vaccine-elicited cell-mediated immune responses. Vaccine 265083-5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Migueles, S. A., M. S. Sabbaghian, W. L. Shupert, M. P. Bettinotti, F. M. Marincola, L. Martino, C. W. Hallahan, S. M. Selig, D. Schwartz, J. Sullivan, and M. Connors. 2000. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc. Natl. Acad. Sci. USA 972709-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miura, T., M. A. Brockman, A. Schneidewind, M. Lobritz, F. Pereyra, A. Rathod, B. L. Block, Z. L. Brumme, C. J. Brumme, B. Baker, A. C. Rothchild, B. Li, A. Trocha, E. Cutrell, N. Frahm, C. Brander, I. Toth, E. J. Arts, T. M. Allen, and B. D. Walker. 2009. HLA-B57/B*5801 human immunodeficiency virus type 1 elite controllers select for rare Gag variants associated with reduced viral replication capacity and strong cytotoxic T-lymphocyte recognition. J. Virol. 832743-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mwau, M., I. Cebere, J. Sutton, P. Chikoti, N. Winstone, E. G. Wee, T. Beattie, Y. H. Chen, L. Dorrell, H. McShane, C. Schmidt, M. Brooks, S. Patel, J. Roberts, C. Conlon, S. L. Rowland-Jones, J. J. Bwayo, A. J. McMichael, and T. Hanke. 2004. A human immunodeficiency virus 1 (HIV-1) clade A vaccine in clinical trials: stimulation of HIV-specific T-cell responses by DNA and recombinant modified vaccinia virus Ankara (MVA) vaccines in humans. J. Gen. Virol. 85911-919. [DOI] [PubMed] [Google Scholar]
- 69.Nickle, D. C., M. A. Jensen, G. S. Gottlieb, D. Shriner, G. H. Learn, A. G. Rodrigo, and J. I. Mullins. 2003. Consensus and ancestral state HIV vaccines. Science 2991515-1518. (Author reply, 299:1515-1518.) [DOI] [PubMed] [Google Scholar]
- 70.Nickle, D. C., M. Rolland, M. A. Jensen, S. L. Pond, W. Deng, M. Seligman, D. Heckerman, J. I. Mullins, and N. Jojic. 2007. Coping with viral diversity in HIV vaccine design. PLoS Comput. Biol. 3e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nilsson, C., B. Makitalo, R. Thorstensson, S. Norley, D. Binninger-Schinzel, M. Cranage, E. Rud, G. Biberfeld, and P. Putkonen. 1998. Live attenuated simian immunodeficiency virus (SIV)mac in macaques can induce protection against mucosal infection with SIVsm. AIDS 122261-2270. [DOI] [PubMed] [Google Scholar]
- 72.Oldstone, M. B., A. Tishon, M. Eddleston, J. C. de la Torre, T. McKee, and J. L. Whitton. 1993. Vaccination to prevent persistent viral infection. J. Virol. 674372-4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reynolds, M. R., A. M. Weiler, K. L. Weisgrau, S. M. Piaskowski, J. R. Furlott, J. T. Weinfurter, M. Kaizu, T. Soma, E. J. Leon, C. MacNair, D. P. Leaman, M. B. Zwick, E. Gostick, S. K. Musani, D. A. Price, T. C. Friedrich, E. G. Rakasz, N. A. Wilson, A. B. McDermott, R. Boyle, D. B. Allison, D. R. Burton, W. C. Koff, and D. I. Watkins. 2008. Macaques vaccinated with live-attenuated SIV control replication of heterologous virus. J. Exp. Med. 2052537-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Robertson, D. L., J. P. Anderson, J. A. Bradac, J. K. Carr, B. Foley, R. K. Funkhouser, F. Gao, B. H. Hahn, M. L. Kalish, C. Kuiken, G. H. Learn, T. Leitner, F. McCutchan, S. Osmanov, M. Peeters, D. Pieniazek, M. Salminen, P. M. Sharp, S. Wolinsky, and B. Korber. 2000. HIV-1 nomenclature proposal. Science 28855-56. [DOI] [PubMed] [Google Scholar]
- 75.Rolland, M., M. A. Jensen, D. C. Nickle, J. Yan, G. H. Learn, L. Heath, D. Weiner, and J. I. Mullins. 2007. Reconstruction and function of ancestral center-of-tree human immunodeficiency virus type 1 proteins. J. Virol. 818507-8514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rolland, M., D. C. Nickle, and J. I. Mullins. 2007. HIV-1 group M conserved elements vaccine. PLoS Pathog. 3e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rousseau, C. M., M. G. Daniels, J. M. Carlson, C. Kadie, H. Crawford, A. Prendergast, P. Matthews, R. Payne, M. Rolland, D. N. Raugi, B. S. Maust, G. H. Learn, D. C. Nickle, H. Coovadia, T. Ndung'u, N. Frahm, C. Brander, B. D. Walker, P. J. Goulder, T. Bhattacharya, D. E. Heckerman, B. T. Korber, and J. I. Mullins. 2008. HLA class I-driven evolution of human immunodeficiency virus type 1 subtype C proteome: immune escape and viral load. J. Virol. 826434-6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Santra, S., B. T. Korber, M. Muldoon, D. H. Barouch, G. J. Nabel, F. Gao, B. H. Hahn, B. F. Haynes, and N. L. Letvin. 2008. A centralized gene-based HIV-1 vaccine elicits broad cross-clade cellular immune responses in rhesus monkeys. Proc. Natl. Acad. Sci. USA 10510489-10494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Seaman, M. S., L. Xu, K. Beaudry, K. L. Martin, M. H. Beddall, A. Miura, A. Sambor, B. K. Chakrabarti, Y. Huang, R. Bailer, R. A. Koup, J. R. Mascola, G. J. Nabel, and N. L. Letvin. 2005. Multiclade human immunodeficiency virus type 1 envelope immunogens elicit broad cellular and humoral immunity in rhesus monkeys. J. Virol. 792956-2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Skowronski, D. M., G. De Serres, J. Dickinson, M. Petric, A. Mak, K. Fonseca, T. L. Kwindt, T. Chan, N. Bastien, H. Charest, and Y. Li. 2009. Component-specific effectiveness of trivalent influenza vaccine as monitored through a sentinel surveillance network in Canada, 2006-2007. J. Infect. Dis. 199168-179. [DOI] [PubMed] [Google Scholar]
- 81.Smith, S. M. 2004. HIV CTL escape: at what cost? Retrovirology 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stacey, A. R., P. J. Norris, L. Qin, E. A. Haygreen, E. Taylor, J. Heitman, M. Lebedeva, A. DeCamp, D. Li, D. Grove, S. G. Self, and P. Borrow. 2009. Induction of a striking systemic cytokine cascade prior to peak viremia in acute human immunodeficiency virus type 1 infection, in contrast to more modest and delayed responses in acute hepatitis B and C virus infections. J. Virol. 833719-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thurmond, J., H. Yoon, C. Kuiken, K. Yusim, S. Perkins, J. Theiler, T. Bhattacharya, B. Korber, and W. Fischer. 2008. Web-based design and evaluation of T-cell vaccine candidates. Bioinformatics 241639-1640. [DOI] [PubMed] [Google Scholar]
- 84.Tomaras, G. D., N. L. Yates, P. Liu, L. Qin, G. G. Fouda, L. L. Chavez, A. C. Decamp, R. J. Parks, V. C. Ashley, J. T. Lucas, M. Cohen, J. Eron, C. B. Hicks, H. X. Liao, S. G. Self, G. Landucci, D. N. Forthal, K. J. Weinhold, B. F. Keele, B. H. Hahn, M. L. Greenberg, L. Morris, S. S. Karim, W. A. Blattner, D. C. Montefiori, G. M. Shaw, A. S. Perelson, and B. F. Haynes. 2008. Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J. Virol. 8212449-12463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tovanabutra, S., S. K. Brodine, J. R. Mascola, J. L. Sankale, E. Sanders-Buell, B. Kim, D. L. Birx, and F. E. McCutchan. 2005. Characterization of complete HIV type 1 genomes from non-B subtype infections in U.S. military personnel. AIDS Res. Hum. Retrovir. 21424-429. [DOI] [PubMed] [Google Scholar]
- 86.Trachtenberg, E., B. Korber, C. Sollars, T. B. Kepler, P. T. Hraber, E. Hayes, R. Funkhouser, M. Fugate, J. Theiler, Y. S. Hsu, K. Kunstman, S. Wu, J. Phair, H. Erlich, and S. Wolinsky. 2003. Advantage of rare HLA supertype in HIV disease progression. Nat. Med. 9928-935. [DOI] [PubMed] [Google Scholar]
- 87.Travers, S. A., M. J. O'Connell, G. P. McCormack, and J. O. McInerney. 2005. Evidence for heterogeneous selective pressures in the evolution of the env gene in different human immunodeficiency virus type 1 subtypes. J. Virol. 791836-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Valentine, L. E., S. M. Piaskowski, E. G. Rakasz, N. L. Henry, N. A. Wilson, and D. I. Watkins. 2008. Recognition of escape variants in ELISPOT does not always predict CD8+ T-cell recognition of simian immunodeficiency virus-infected cells expressing the same variant sequences. J. Virol. 82575-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang, S., J. S. Kennedy, K. West, D. C. Montefiori, S. Coley, J. Lawrence, S. Shen, S. Green, A. L. Rothman, F. A. Ennis, J. Arthos, R. Pal, P. Markham, and S. Lu. 2008. Cross-subtype antibody and cellular immune responses induced by a polyvalent DNA prime-protein boost HIV-1 vaccine in healthy human volunteers. Vaccine 263947-3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang, Y. E., B. Li, J. M. Carlson, H. Streeck, A. D. Gladden, R. Goodman, A. Schneidewind, K. A. Power, I. Toth, N. Frahm, G. Alter, C. Brander, M. Carrington, B. D. Walker, M. Altfeld, D. Heckerman, and T. M. Allen. 2009. Protective HLA class I alleles that restrict acute-phase CD8+ T-cell responses are associated with viral escape mutations located in highly conserved regions of human immunodeficiency virus type 1. J. Virol. 831845-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ward, F. E., S. Tuan, and B. F. Haynes. 1995. Analysis of HLA frequencies in population cohorts for design of HLA-based HIV vaccines, p. IV10-IV16. In B. Korber, C. Brander, B. Walker, R. Koup, J. Moore, B. Haynes, and G. Myers (ed.), HIV molecular immunology database 1995. Theoretical Biology Group, Los Alamos National Laboratory, Los Alamos, NM.
- 92.Weaver, E. A., Z. Lu, Z. T. Camacho, F. Moukdar, H. X. Liao, B. J. Ma, M. Muldoon, J. Theiler, G. J. Nabel, N. L. Letvin, B. T. Korber, B. H. Hahn, B. F. Haynes, and F. Gao. 2006. Cross-subtype T-cell immune responses induced by a human immunodeficiency virus type 1 group M consensus env immunogen. J. Virol. 806745-6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wee, E. G., S. Patel, A. J. McMichael, and T. Hanke. 2002. A DNA/MVA-based candidate human immunodeficiency virus vaccine for Kenya induces multi-specific T cell responses in rhesus macaques. J. Gen. Virol. 8375-80. [DOI] [PubMed] [Google Scholar]
- 94.Whitton, J. L., N. Sheng, M. B. Oldstone, and T. A. McKee. 1993. A “string-of-beads” vaccine, comprising linked minigenes, confers protection from lethal-dose virus challenge. J. Virol. 67348-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wilson, C. C., D. McKinney, M. Anders, S. MaWhinney, J. Forster, C. Crimi, S. Southwood, A. Sette, R. Chesnut, M. J. Newman, and B. D. Livingston. 2003. Development of a DNA vaccine designed to induce cytotoxic T lymphocyte responses to multiple conserved epitopes in HIV-1. J. Immunol. 1715611-5623. [DOI] [PubMed] [Google Scholar]
- 96.Worobey, M., M. Gemmel, D. E. Teuwen, T. Haselkorn, K. Kunstman, M. Bunce, J. J. Muyembe, J. M. Kabongo, R. M. Kalengayi, E. Van Marck, M. T. Gilbert, and S. M. Wolinsky. 2008. Direct evidence of extensive diversity of HIV-1 in Kinshasa by 1960. Nature 455661-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wyand, M. S., K. Manson, D. C. Montefiori, J. D. Lifson, R. P. Johnson, and R. C. Desrosiers. 1999. Protection by live, attenuated simian immunodeficiency virus against heterologous challenge. J. Virol. 738356-8363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yang, O. O. 2008. Retracing our STEP towards a successful CTL-based HIV-1 vaccine. Vaccine 263138-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yeh, W. W., P. Jaru-Ampornpan, D. Nevidomskyte, M. Asmal, S. S. Rao, A. P. Buzby, D. C. Montefiori, B. T. Korber, and N. L. Letvin. 2009. Partial protection of simian immunodeficiency virus (SIV)-infected rhesus monkeys against superinfection with a heterologous SIV isolate. J. Virol. 832686-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yusim, K., C. Kesmir, B. Gaschen, M. M. Addo, M. Altfeld, S. Brunak, A. Chigaev, V. Detours, and B. T. Korber. 2002. Clustering patterns of cytotoxic T-lymphocyte epitopes in human immunodeficiency virus type 1 (HIV-1) proteins reveal imprints of immune evasion on HIV-1 global variation. J. Virol. 768757-8768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yusim, K., M. Peeters, O. G. Pybus, T. Bhattacharya, E. Delaporte, C. Mulanga, M. Muldoon, J. Theiler, and B. Korber. 2001. Using human immunodeficiency virus type 1 sequences to infer historical features of the acquired immune deficiency syndrome epidemic and human immunodeficiency virus evolution. Philos. Trans. R. Soc. Lond. B 356855-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang, M., B. Gaschen, W. Blay, B. Foley, N. Haigwood, C. Kuiken, and B. Korber. 2004. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology 141229-1246. [DOI] [PubMed] [Google Scholar]
- 103.Zuckerman, J. N. 2006. Vaccination against hepatitis A and B: developments, deployment and delusions. Curr. Opin. Infect. Dis. 19456-459. [DOI] [PubMed] [Google Scholar]