Abstract
Simian virus 40 large T antigen (TAg) transforms cells in culture and induces tumors in rodents. Genetic studies suggest that TAg interaction with the chaperone hsp70 and tumor suppressors pRb and p53 may not be sufficient to elicit complete transformation of cells. In order to identify additional cellular factors important for transformation, we designed mutations on the solvent-exposed surface of TAg. We hypothesized that surface residues would interact directly with cellular targets and that the mutation of these residues might disrupt this interaction without perturbing TAg's global structure. Using structural data, we identified 61 amino acids on the surface of TAg. Each surface amino acid was changed to an alanine. Furthermore, five patches containing clusters of charged amino acids on the surface of TAg were identified. Within these patches, we selectively mutated three to four charged amino acids and thus generated five mutants (patch mutants 1 to 5). We observed that while patch mutants 3 and 4 induced foci in REF52 cells, patch mutants 1 and 2 were deficient in focus formation. We determined that the patch 1 mutant is defective in p53 binding, thus explaining its defect in transformation. The patch 2 mutant can interact with the Rb family members and p53 like wild-type TAg but is unable to transform cells, suggesting that it is defective for action on an unknown cellular target essential for transformation. Our results suggest that the histone acetyltransferase CBP/p300 is one of the potential targets affected by the mutations in patch 2.
Simian virus 40 (SV40) large T antigen is a multifunctional protein that is essential for productive viral infection and for cellular transformation (26). T antigen possesses several biochemical activities, some of which map to discrete domains that can act independently and/or coordinately. To effect replication and transformation, T antigen binds to several cellular targets via different domains/regions. For example, during replication, T antigen associates with components of the cellular replication apparatus such as DNA polymerase α, replication protein A, and topoisomerase I (11, 14, 24, 31, 39). Three regions of T antigen are essential to elicit cellular transformation (1, 2, 36). The LXCXE motif mediates binding to the members of the Rb family (pRb, p107, and p130) and in conjunction with the J domain results in the inactivation of the Rb family function. While these domains reside in the N terminus of T antigen, a third transforming function in the C terminus of T antigen is essential for inactivation of the tumor suppressor p53. Genetic studies suggest that inactivation of pRb and p53 is not always sufficient to induce T-antigen-mediated transformation (7, 30, 38), thus indicating the presence of additional targets of T antigen contributing to transformation. In the past few years, several additional targets of T antigen, including CBP/p300, Bub1, Cul7, Fbw7, and IRS-1, have been discovered (8, 9, 12, 17, 29, 40, 42); however, their roles in T-antigen-mediated transformation are not clear. T antigen also targets the DNA-damage-sensing and -processing complex Mre11-Rad50-Nbs1 and may induce genetic instability that contributes to transformation (10, 42). The issue is complicated further by the observation that T antigen has redundant functions, that is, it can act on critical targets via multiple mechanisms (7, 37).
One of the key strategies to delineate functions of T antigen required for transformation is the use of amino acid substitution and truncation mutants. However, a caveat to this approach is the production of mutants that are defective in transformation due to a loss of integrity of the secondary, or even local, structure. In this study, we combined available sequence data with structural information to design mutants. Sequence alignments allow the identification of conserved amino acid residues, while structural data provide information about amino acid residues on the surface of the molecule. This approach allows us to combine structural elements and target binding sites. In addition, identification of residues conserved across species, followed by mutation of these conserved residues, will likely yield better insights into common biological pathways. Using this method, we have generated four mutants, of which two are defective in transformation and, thus, of great interest for the identification of novel cellular pathways regulating cell growth and proliferation.
MATERIALS AND METHODS
Cells.
REF52 cells established from rat embryos were cultured in complete medium, that is, minimum essential medium (MEM) supplemented with 10% fetal bovine serum (FBS; HyClone) and 100 U/ml penicillin and 100 U/ml streptomycin (Invitrogen). Geneticin (Invitrogen) was added to the medium for cell lines expressing T antigen and its mutants.
Plasmids.
The expression of genomic clones of large T antigen and its mutants was driven by the Rous sarcoma virus promoter in constructs containing a neomycin resistance gene as described previously (33). All plasmids express small t antigen. Patch mutations and single-site alanine mutations were generated using Stratagene's multisite- and single-site-directed mutagenesis kits.
Design of mutants.
From structural information (18), we identified 61 amino acid residues on the surface of the T antigen (see Table S1 in the supplemental material) and generated 61 alanine substitution mutations using the QuikChange site-directed mutagenesis kit (Stratagene). We also noticed that of these 61 residues, 34 residues were one of the four charged amino acids (Arg, Lys, Asp, and Glu). We used the RasMol program to examine the distributions of these charged residues on the solvent-exposed surface of T antigen (Fig. 1). Within each cluster, or patch, of charged amino acids, we targeted for mutation those amino acids that were conserved in the corresponding regions of T antigens from JC, BK, and SA12 viruses. Complete protein sequences from BK, JC, and SA12 viruses were aligned with that of SV40 by using the multiple-sequence alignment program MultAlin.
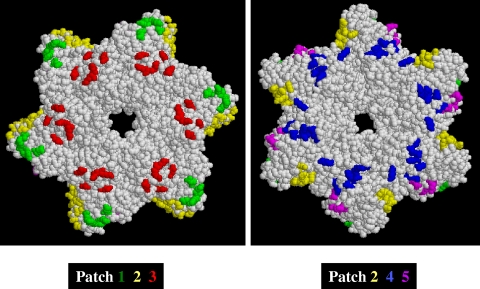
FIG. 1.
Locations of patch mutations on T antigen hexamer surface. Space-filling models of the SV40 T antigen helicase domain (amino acids 266 to 627) are shown. Clusters of charged amino acids are color coded to identify the different patches. The left panel is oriented so that the Zn domain is on top and the amino terminus is projected toward the reader. The right panel is oriented so that the ATPase domain is on top and the carboxy terminus is projected toward the reader.
Expression of T antigen mutants.
To generate cell lines stably expressing T antigen mutants, REF52 cells were transfected with DNA. After being split 1:3, cells were fed with complete medium containing Geneticin (0.4 mg/ml; Invitrogen). Drug-resistant colonies were picked and expanded into cell lines. Lysates were prepared from 2-day-postconfluence cells by incubating the cells in lysis buffer L (50 mM HEPES [pH 7.9], 400 mM KCl, 0.5 mM EDTA, 0.1% NP-40, 10% glycerol, 1 mM dithiothreitol, 0.5 mM Na3VO4, 0.5 mM NaF, 1 μg/ml pepstatin, and a protease inhibitor tablet [Roche]) for 30 min on ice and then centrifuging the lysate for 10 min at 4°C. The protein concentration was determined by the Bradford assay using a kit from Bio-Rad.
Transformation assays. (i) Focus assay.
REF52 cells were plated into 60-mm dishes such that they were 75 to 85% confluent the following day. Two micrograms of DNA was transfected into the cells by using Fugene per the instructions of the manufacturer (Roche). Twenty-four hours later, the cells were split 1:3 and fed every 3 to 4 days. After 6 weeks, the cells were stained with methylene blue and assessed for focus formation.
(ii) Anchorage independence.
REF52 cells stably expressing wild-type or mutant T antigen were tested for their ability to grow in soft agar. Two milliliters of MEM containing 0.8% agar was layered onto 60-mm dishes. Cells (5 × 104) were suspended in 2 ml of MEM containing 10% FBS and 0.4% agar, and the suspension was then layered on top of the 0.8% agar. Following cooling at room temperature for 10 min, the cells were maintained at 37°C and fed every 3 to 4 days with MEM containing 10% FBS and 0.4% agar. After 3 weeks, the cells were assessed for colony formation.
(iii) Proliferation assay.
To assay growth to high saturation density, 5 × 104 REF52 cells stably expressing wild-type or mutant T antigen were plated in triplicate into 60-mm dishes. Cells were fed every other day for 10 days. Cells in triplicate dishes were counted using a hemocytometer. To assay growth in low-serum medium, 5 × 104 cells in complete medium were plated in triplicate into 60-mm dishes. Twenty-four hours later, the medium was changed to medium containing 1% FBS. The cells were fed every other day. Cells in triplicate dishes were counted at each time point.
Plaque assays.
To perform a plaque assay, patch mutations in pRSVBneoT were subcloned into pSVB3. Single alanine substitution mutations in pSVB3 were created by using the single-site-directed mutagenesis kit (Stratagene). Plaque formation assays were performed as described previously (35).
Gel shift assay.
E2F-DNA complexes were analyzed as described previously (35). Briefly, 30 μg of lysate from REF52 cells expressing wild-type or mutant T antigen was incubated with 32P-labeled oligonucleotide containing a consensus E2F binding site. Reaction mixtures were run on 0.25× Tris-borate-EDTA-4.5% acrylamide gel to separate free E2F from E2F in complexes with Rb family proteins.
Immunoprecipitation.
For coimmunoprecipitation of pRb, p107, and p53 with T antigen, REF52 cells stably expressing T antigen or its mutants or a vector control (neo) were grown until they were 2 days past confluence. The cells were washed in phosphate-buffered saline-EDTA and then collected and lysed as described above. One hundred micrograms of lysate was immunoprecipitated with 2 μg of the appropriate antibody as described previously (33). To control for nonspecificity, an immunoprecipitation reaction mixture with no antibody (mock) was set up alongside the other reaction mixtures. Immune complexes were electrophoresed through an 8% polyacrylamide gel and then probed by standard Western blot techniques.
For coimmunoprecipitation of CBP and T antigen, we used a protocol supplied by Deppert and coworkers (personal communication). Pellets of 2-day-postconfluence cells were incubated in lysis buffer B (50 mM HEPES, pH 7.3, 150 mM NaCl, 0.1% NP-40, with 0.1% leupeptin, 1% pepstatin, 1% Pefabloc, 1% aprotinin) for 30 min on ice. The protein concentration in the clarified extract was determined as described above. Seven hundred micrograms of whole-cell lysate was added to a mixture of 5 μg of anti-CBP and 150 μl of protein G Sepharose (10% slurry) that was preincubated for 1 h at 4°C on an orbital shaker. Immune complexes were allowed to form during gentle rocking for 1.5 h at 4°C and then were washed three times in 500 μl of buffer C (buffer B supplemented with 5 mM NaF, 4 mM EGTA, pH 7.5, 1 mM EDTA, pH 7.5). Following denaturation in 2× sodium dodecyl sulfate (SDS) sample buffer (125 mM Tris-HCl, pH 6.8, 20% glycerol, 4% SDS, 200 mM dithiothreitol, 10 mg/ml bromophenol), the immunoprecipitated proteins were separated by SDS-7% polyacrylamide gel electrophoresis and then transferred onto polyvinylidene difluoride membranes (Millipore) and analyzed by standard Western blot techniques.
The antibodies used were anti-p107 (C-18; Santa Cruz), anti-p130 (C-20; Santa Cruz), anti-pRb (G-3245; BD-Pharmingen), anti-T antigen (Ab419), anti-p53 (Ab421), anti-CBP (CBP-NT) (Upstate Cell Signaling Solutions), and anti-acetylated Lys (antibody 9441; Cell Signaling).
RESULTS
Design of surface mutations based on sequence data and structural information.
We designed a series of mutations on the solvent-exposed surface of T antigen, the rationale being that surface residues, rather than residues buried within the T antigen structure, would interact directly with cellular targets. From structural information (16), we determined the distribution of charged amino acids on the hexamer surface and identified five patches on the surface of large T antigen containing a cluster of charged amino acids (Fig. 1). Within these patches, we targeted for mutation those amino acids that were conserved in the corresponding regions of the T antigens from JC, BK, and SA12 viruses (Table 1). Instead of using the traditional single-site amino acid mutagenesis approach (see Table S1 in the supplemental material), we selected three to four conserved charged amino acids in each patch and reversed the charge, i.e., Asp and Glu residues were changed to Lys and, correspondingly, Lys and Arg residues were mutated to Glu (Table 1). Thus, we designed five mutants of T antigen (termed patch mutants 1 to 5), each containing mutations in several surface positions. Our biochemical analysis is restricted to patch mutants 1 to 4, because mutagenesis in patch 5 was unsuccessful.
TABLE 1.
Distribution of charged amino acids on the surface of SV40 T antigen hexamera
| Position | Residue in:
|
Motif/domain | Residue in patch mutant:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SV40 | BK virus | JC virus | SA12 virus | 1 | 2 | 3 | 4 | 5 | ||
| 266 | K | K | K | K | Zn | |||||
| 300 | E | E | Q | E | Zn | K | ||||
| 308 | K | K | K | K | Zn | K | ||||
| 318 | E | E | E | E | Zn | K | ||||
| 319 | K | K | K | K | Zn | K | ||||
| 370 | D | D | D | D | Zn | K | ||||
| 383 | D | V | V | V | ATPase | D | ||||
| 385 | E | E | E | E | ATPase | K | ||||
| 386 | E | Q | Q | H | ATPase | E | ||||
| 400 | K | K | Q | K | ATPase | K | ||||
| 402 | D | D | D | D | ATPase | K | ||||
| 407 | D | D | D | D | ATPase | K | ||||
| 425 | K | K | K | K | ATPase | E | ||||
| 483 | R | K | R | K | ATPase | E | ||||
| 530 | E | E | E | E | ATPase | E | ||||
| 535 | K | K | R | K | ATPase | E | ||||
| 543 | K | R | R | K | ATPase | E | ||||
| 546 | D | D | D | D | ATPase | D | ||||
| 550 | K | K | K | K | ATPase | |||||
| 551 | D | I | A | I | ATPase | |||||
| 554 | K | R | R | R | ATPase | K | ||||
| 558 | E | Q | S | N | ATPase | E | ||||
| 559 | R | N | C | N | ATPase | R | ||||
| 561 | E | E | E | E | ATPase | K | ||||
| 565 | E | E | E | E | ATPase | K | ||||
| 566 | K | K | K | K | ATPase | K | ||||
| 583 | R | R | R | R | ATPase | R | ||||
| 587 | E | D | D | D | ATPase | K | ||||
| 598 | E | E | Q | Q | ATPase | K | ||||
| 601 | E | E | E | E | ATPase | K | ||||
| 604 | D | D | D | D | ATPase | K | ||||
| 605 | K | S | L | S | ATPase | K | ||||
| 606 | E | E | E | E | ATPase | K | ||||
| 614 | K | R | T | R | ATPase | K | ||||
| 627 | D | D | D | D | Variable | K | ||||
Residues in bold were selected for mutation.
Transformation and replication assays.
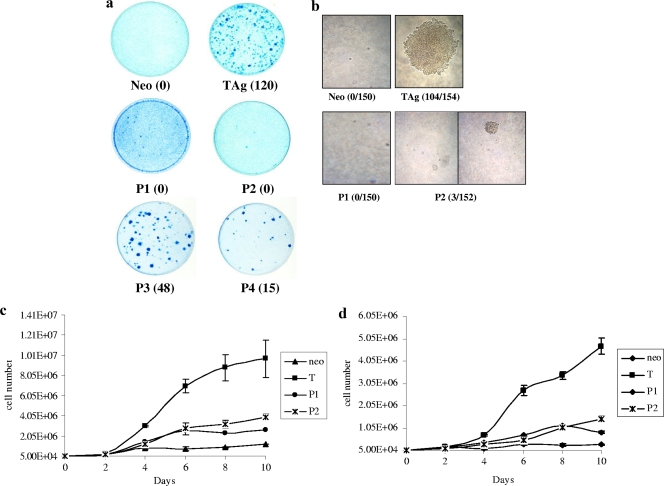
Having generated the patch mutants, we ascertained their abilities to transform cells. REF52 cells were transfected with each mutant, and the mutant was then examined for its ability to induce foci over a monolayer of untransformed cells (Fig. 2a). We saw that patch mutants 3 and 4 were able to form foci, albeit at reduced efficiency relative to wild-type T antigen; however, patch mutants 1 and 2 were defective in focus formation. We observed that patch mutants 1 and 2 in primary mouse embryo fibroblasts were also defective in focus formation (data not shown). We selected two clones each of patch mutants 1 and 2 for further biochemical analysis. REF52 cells stably expressing patch mutant 1 or 2 were examined for their ability to grow under growth-restrictive conditions, which is the hallmark of transformed cells. First, cells were seeded at low density into soft agar and their growth into colonies was monitored for 3 weeks. While T-antigen-expressing cells were able to form colonies, neither of the patch mutants was able to support growth in soft agar (Fig. 2b). Next, cells were seeded at low density and their growth was monitored for 10 days. Cells expressing patch mutant 1 or 2 were unable to grow to high saturation densities, in contrast to cells expressing wild-type T antigen (Fig. 2c). Numbers of mutant-expressing cells plateaued at 1 × 106 to 2 × 106, while T-antigen-expressing cells continued to proliferate. Finally, cells were seeded at low density and then 1 day later the serum content was lowered to 1%. Cell growth was monitored for 10 days by counting cells in triplicate dishes every other day. While cells expressing wild-type T antigen were able to proliferate in low-serum medium, neither of the patch mutants was able to support growth under these conditions (Fig. 2d). Altogether, these results support the observation that the patch mutants 1 and 2 are transformation defective.
FIG. 2.
Patch mutants 1 and 2 are transformation defective. (a) REF52 cells were transfected with wild-type T antigen (TAg), patch mutants 1 to 4 (P1 to P4), or a vector control (neo). Six weeks later, dense foci were visualized by methylene blue staining. Numbers in parentheses are the average numbers of foci counted. Similar results were obtained in three different experiments. (b) REF52 cells stably expressing wild-type or patch mutant T antigen or the vector control were suspended in 0.4% agar and then layered over 0.8% agar in p60 dishes. Each clone was studied in triplicate. At the end of 3 weeks, colonies larger than 4 mm were counted. Numbers in parentheses are the ratios of the total number of colonies observed to the total number of cells counted. (c) REF52 cells expressing a patch mutant (P1 or P2) or wild-type T antigen (T) were seeded in triplicate, and their growth over 10 days was monitored. (d) REF52 cells expressing a patch mutant or wild-type T antigen were seeded in triplicate into normal serum, and then 1 day later the serum content was reduced to 1%. Growth over 10 days was monitored.
We tested the patch mutants for their abilities to replicate in BSC40 cells, which are permissive for viral replication. Viability was assessed by a plaque formation assay. We observed that all four patch mutants were defective for viral replication (data not shown). One possible explanation for this replication defect is that within the cluster of mutations in each patch mutant, one or several specific mutations may negatively affect T antigen's interaction with the cellular replication apparatus. Indeed, when we checked single-alanine-substitution mutants with amino acid residues in these patches mutated, we found only six of these mutants to be replication defective (see Table S1 in the supplemental material). For example, patch 1 contains five amino acid residues, D598, E601, D604, K605, and E606. The mutation of D604 alone among these residues resulted in the inhibition of productive viral replication. Each patch mutant appears to contain at least one residue that is key for viral viability. The mutation of this residue may explain why each of the patch mutants was defective for replication.
Interaction with p53.
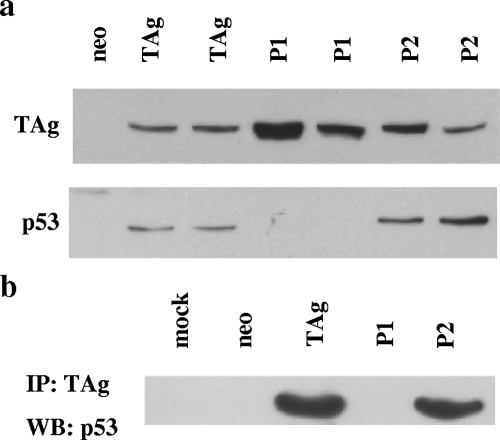
T antigen inactivates both the Rb family and p53 in order to elicit transformation. Since patch mutants 1 and 2 were transformation defective, we tested their abilities to inactivate these proteins. First, we determined whether or not the patch mutants could stabilize p53. Whole-cell lysates from REF52 cells stably expressing wild-type or mutant T antigen were subjected to Western blot analysis. As can be seen in Fig. 3a, both clones of patch mutants 1 and 2 are expressed at the same levels as, or slightly higher levels than, wild-type T antigen. Interestingly, like cells expressing T antigen, cells expressing the patch 2 mutant have increased steady-state levels of p53 compared to cells expressing the vector control; however, patch 1 mutant-expressing cells do not accumulate p53. This observation suggests that the patch 2 mutant, but not the patch 1 mutant, is able to stabilize p53 like wild-type T antigen. In order to determine whether or not the patch mutants can bind to p53, we immunoprecipitated wild-type or mutant T antigen by using an antibody to T antigen and then probed the immune complex for p53. As can be seen in Fig. 3b, p53 coimmunoprecipitates with both wild-type T antigen and the patch 2 mutant. Thus, the patch 2 mutant can bind and stabilize p53 as well as wild-type T antigen. However, the patch 1 mutant is unable to coimmunoprecipitate p53. This result suggests that the patch 1 mutant cannot bind to p53 and is likely the reason that it is unable to stabilize p53. It is important that p53 in the patch 1 mutant-expressing REF52 cells is latent but, like that in normal REF52 cells, can be activated upon DNA damage (data not shown).
FIG. 3.
The patch 2 mutant, but not the patch 1 mutant, can stabilize and bind p53. (a) Western blot analysis of whole-cell lysates from REF52 cells stably expressing wild-type T antigen (TAg), patch mutant 1 or 2 (P1 or P2), or the vector control (neo) using T-antigen- and p53-specific antibodies. (b) Results of a coimmunoprecipitation experiment. The above-mentioned lysates were incubated with anti-T antigen antibody, and the immune complexes were probed for the presence of p53. “Mock” indicates an immunoprecipitation reaction mixture containing no anti-T antigen antibody. IP, immunoprecipitation; WB, Western blotting.
Interaction with the Rb family.
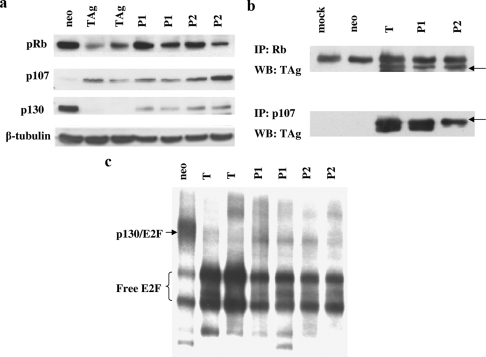
The patch 2 mutant is defective for transformation even though it is able to bind and stabilize p53. We determined whether or not it was capable of interacting with the Rb family members like wild-type T antigen. We first examined the steady-state levels of the Rb family members in lysates from REF52 cells stably expressing patch mutant 1 or 2 or wild-type T antigen (Fig. 4a). Vector control (neo)-expressing cells are growth arrested and thus have no p107 expression but have high levels of p130. In contrast, both patch mutants, like wild-type T antigen, induce p107 expression, indicating that the cells are still cycling. T antigen induces the degradation of p130, and so do patch mutants 1 and 2, albeit with reduced efficiency. Steady-state levels of pRb are higher in the mutant-expressing cells than in the cells expressing wild-type T antigen.
FIG. 4.
Patch mutants 1 and 2 (P1 and P2) interact with the Rb family like wild-type T antigen (TAg). (a) Western blot analysis of lysates from REF52 stable transfectants using antibodies specific to pRb, p107, or p130. (b) Results from coimmunoprecipitation experiments with pRb or p107 and wild-type or mutant T antigen. Arrows indicate large T antigen. IP, immunoprecipitation; WB, Western blotting. (c) Results from a gel shift assay using lysates prepared from 2-day-postconfluence REF52 stable transfectants. Lysates were incubated with a 32P-labeled oligonucleotide containing an E2F consensus site.
Next, we performed coimmunoprecipitation experiments to assess whether or not the mutations in patches 1 and 2 affected pRb binding. Lysates from REF52 cells stably expressing wild-type T antigen or patch mutants were immunoprecipitated with anti-T antigen antibody, and the immune complexes were examined for the presence of the Rb family proteins. As can be seen from Fig. 4b, patch mutants 1 and 2 were able to immunoprecipitate pRb and p107. The immune complexes were not assessed for p130, as it is degraded in wild-type and mutant T antigen cell lysates. Thus, the charge reversal mutations in the carboxy terminus of T antigen do not affect association with the Rb family.
Wild-type T antigen drives cells into S phase by binding to members of the Rb family, thus releasing E2F transcription factors from Rb-mediated repression. We next addressed the question of whether or not the patch mutants also were able to disrupt Rb-E2F complexes. Gel shift assays were performed with lysates of REF52 cells that were 2 days postconfluence. Normal REF52 cells are growth arrested past confluence and contain a high level of the p130-E2F complex and low levels of free E2Fs (Fig. 4c). As expected, T-antigen-expressing REF52 cells do not have this slower migrating species but have high levels of free E2Fs. Interestingly, lysates from cells expressing patch mutants 1 and 2 behave similarly to T antigen lysates, indicating that the patch mutants are capable of disrupting Rb-E2F complexes.
Interaction with CBP/p300.
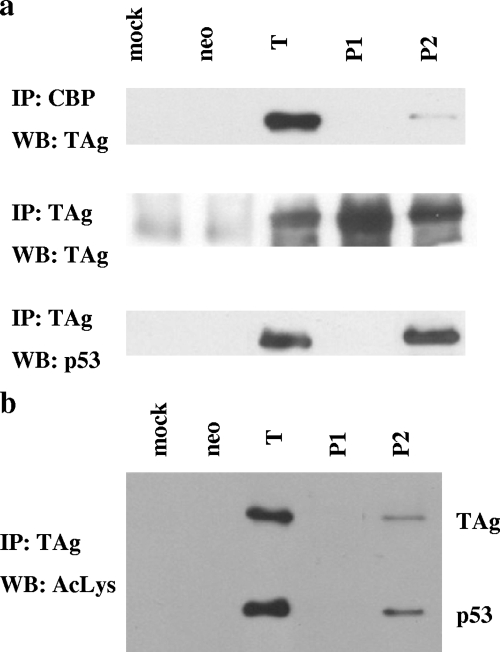
The patch 2 mutant is defective for transformation even though it targets p53 and pRb as well as T antigen does. This observation implies that the mutations in patch 2 affect the interaction with some other cellular factor that is also important for transformation. One possible target is the histone acetyltransferase (HAT) CBP/p300. T antigen associates with CBP/p300 through p53 (29), and the carboxy terminus of T antigen has been implicated previously in binding to this HAT. In addition, interaction with CBP/p300 is essential for adenovirus E1A-mediated cellular transformation (13, 41). We performed coimmunoprecipitation experiments to determine whether or not the patch 2 mutant could interact with CBP/p300. Lysates from REF52 cells stably expressing wild-type or mutant T antigen were incubated with anti-CBP antibody, and the immune complexes were then examined for the presence of T antigen (Fig. 5a). Wild-type T antigen coimmunoprecipitates with CBP, but the amount of immunoprecipitated T antigen is very small, indicating that the majority of T antigen is free of CBP. The patch 1 mutant does not coimmunoprecipitate with CBP. This observation is consistent with the facts that CBP/p300 can interact with T antigen only via p53 and that the patch 1 mutant does not bind and stabilize p53 (Fig. 3b). The patch 2 mutant also does not coimmunoprecipitate with CBP even though it is able to bind p53 (Fig. 5a, bottom). Thus, the association is probably inhibited by the charge reversal mutations in patch 2.
FIG. 5.
Mutations in patch 2 inhibit the association with CBP. (a) Lysates from REF52 stable transfectants were immunoprecipitated with anti-CBP antibody (top) or anti-T antigen (middle and bottom), and the immune complexes were probed for the presence of T antigen (top and middle) or p53 (bottom). T, wild-type T antigen; P1 and P2, patch mutants 1 and 2; IP, immunoprecipitation; WB, Western blotting. (b) The acetylation statuses of wild-type or mutant T antigen and the associated p53 were examined by first immunoprecipitating lysates from REF52 stable transfectants with anti-T antigen antibody conjugated to protein G Sepharose beads and then probing the immune complexes with anti-acetylated Lys (AcLys) antibody.
One of the known outcomes of T antigen interaction with CBP/p300 is the acetylation of lysine 697 of T antigen and the associated p53 (29). Since the patch 2 mutant is defective for CBP binding, we predicted that the acetylation of this mutant would be greatly reduced. Indeed, as illustrated in Fig. 5b, the patch 2 mutant is acetylated to a much lesser extent than wild-type T antigen. Interestingly, p53 associated with the patch 2 mutant is also acetylated to a lesser extent than p53 in T-antigen-expressing cells.
DISCUSSION
Mutational analysis of SV40 large T antigen has provided many insights into pathways regulating cell proliferation and cell growth. The investigation of new mutants should allow the identification of novel T-antigen-interacting partners involved in cellular replication and transformation. In the present study, we have designed T antigen mutants by combining sequence and structural information. The potential advantage of this approach is that amino acid residues on the solvent-exposed surface of the T antigen molecule, rather than those residues that are buried within its three-dimensional structure, can be targeted. Mutating surface residues will most likely disrupt interaction with cellular targets without perturbing the global structure of T antigen. We chose to make charge reversal mutations to maximize the chances of disrupting interactions with cellular targets. We generated four mutants (patch mutants 1 to 4) that contain a cluster of amino acid substitutions on the solvent-exposed surface of T antigen (Table 2 gives a summary of results). Two of these mutants, patch mutants 3 and 4, were able to transform cells, although with reduced efficiency in comparison to wild-type T antigen. On the other hand, patch mutants 1 and 2 were defective for transformation.
TABLE 2.
Biochemical analysis of patch mutants 1 to 4a
| Patch mutant | Amino acids and mutationsb in patch | Replication result | Focus formation result | Anchorage independence result | Biological targets |
|---|---|---|---|---|---|
| 1 | E598K, E601K, D604K, K605, E606K | −ve | −ve | −ve | p53, CBP/p300 |
| 2 | D383, E385K, E386, K554, E558, R559, E561K, E565K, K566, K614, D627K | −ve | −ve | −ve | CBP/p300 |
| 3 | E300K, K308, E318K, K319, D370K | −ve | +ve | ND | Unknown |
| 4 | K425E, R483E, E530, K535E, K543E, D546 | −ve | +ve | ND | Unknown |
−ve, negative; +ve, positive; ND, not determined.
Mutations are highlighted in bold.
The patch 1 mutant did not transform cells probably because it is unable to bind and inactivate p53. Indeed, examination of the p53-T antigen complex shows that the mutations in patch 1 map to the p53-T antigen interface (20). Additionally, one residue altered in patch 5, D402, has previously been shown to be critical for binding of T antigen to p53 (20, 21). Thus, the p53 binding site on the patch 1 mutant is perturbed by the charge reversal mutations.
Patch 2 is located near patch 1 (Fig. 1a); however, it does not affect the interaction with p53, suggesting that the integrity of the T antigen structure is maintained using the charge reversal mutant design. The patch 2 mutant interacts with the Rb family and with p53 like wild-type T antigen, yet it is transformation defective. This suggests that the patch 2 mutations interfere with a transforming activity of T antigen other than interactions with Rb and p53. In support of this finding, genetic evidence also suggests that binding with Rb and p53 is not sufficient to completely transform cells (30).
Several reports implicate the transcriptional adapter protein CBP/p300 in T-antigen-mediated transformation. For example, truncation mutants of T antigen lacking the first 82 amino acids are transformation defective, but their transformation activity is restored upon the coexpression of wild-type adenovirus E1A but not a p300 binding mutant of E1A (43). The requirement for CBP/p300 in adenovirus E1A-mediated transformation is well established (12, 13, 41) and may involve E1A inhibition of CBP/p300 function (32). An important distinction between the interactions of E1A and T antigen with CBP/p300 is that while E1A binds to CBP/p300 directly (3, 23, 34), T antigen does so indirectly via p53 (5, 19). Indeed, our results with the patch 1 mutant are consistent with this finding. The patch 1 mutant is deficient for p53 binding and, thus, is unable to coimmunoprecipitate CBP (Fig. 5a).
We took two approaches to determine if the patch 2 mutations interfere with T antigen-CBP/p300 interaction. First, we examined whether or not the patch 2 mutant could interact with CBP/p300 (Fig. 5a). Second, we examined the acetylation status of the T antigen K697 residue on the patch 2 mutant (Fig. 5b). Our results provide evidence that CBP binds to the area of T antigen identified by the residues of patch 2 (Table 2), as the mutation of these residues results in decreased binding to CBP. It is possible that not all of these residues are critical for CBP binding; alternatively, they may all be part of the binding interface. In the future, mutations will be made singly and in combination to ascertain the exact CBP/p300 binding requirements. Thus, our data suggest that CBP/p300 makes contact with both p53 and large T antigen and that both of these interactions are required to secure CBP/p300 to the complex.
CBP/p300 is endowed with HAT activity. In the trimeric complex, CBP/p300 acetylates both T antigen and p53 (29). The functional consequences of T antigen acetylation are as yet unknown. We found that the patch 2 mutant is acetylated to a lesser extent than wild-type T antigen, ostensibly a consequence of the weakened interaction between the patch 2 mutant and CBP/p300. Acetylation of T antigen Lys697 per se is probably not required for transformation, as C-terminal truncation mutants lacking the host range domain are still capable of transforming cultured cells (25, 27, 28). It is possible that acetylation is only a marker for the presence of CBP/p300 and does not have any biological significance of its own. On the other hand, it is intriguing that the acetylation site is conserved on T antigens of the human and the baboon polyomaviruses (6, 29). Thus, acetylation may serve as a signal for the recruitment of one or more cellular factors important for all the viruses. Additionally, reduced acetylation on p53 or other substrates might have more important biological consequences. p53 acetylation stabilizes the protein and increases its DNA binding and transcriptional activity (4, 15, 16, 22).
At this stage, we cannot be sure that the transformation defect observed with the patch 2 mutant is due to its decreased binding of CBP. The reason for this uncertainty is that the patch 2 mutant is replication defective. The D402N mutant, which cannot bind to p53 and therefore is unable to interact with CBP/p300, is, however, able to support viral replication, suggesting that interaction with CBP is not required for viral replication (21). Based on this observation, we assume that some other activity of T antigen which is important for replication is perturbed by the patch 2 mutations, in addition to interaction with CBP.
Supplementary Material
Acknowledgments
This work was supported by NIH grants CA40586 and CA120997 to J.M.P. and by NIH grant AI055926 to X.S.C.
We also thank the Deppert laboratory (Heinrich-Pette Institute) for their assistance with the CBP-T antigen coimmunoprecipitation protocol. We thank Alison Roos for substantial confirmation of some of the data presented in the manuscript.
Footnotes
Published ahead of print on 24 June 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Ahuja, D., M. T. Saenz-Robles, and J. M. Pipas. 2005. SV40 large T antigen targets multiple cellular pathways to elicit cellular transformation. Oncogene 247729-7745. [DOI] [PubMed] [Google Scholar]
- 2.Ali, S. H., and J. A. DeCaprio. 2001. Cellular transformation by SV40 large T antigen: interaction with host proteins. Semin. Cancer Biol. 1115-23. [DOI] [PubMed] [Google Scholar]
- 3.Arany, Z., D. Newsome, E. Oldread, D. M. Livingston, and R. Eckner. 1995. A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature 37481-84. [DOI] [PubMed] [Google Scholar]
- 4.Barlev, N. A., L. Liu, N. H. Chehab, K. Mansfield, K. G. Harris, T. D. Halazonetis, and S. L. Berger. 2001. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol. Cell 81243-1254. [DOI] [PubMed] [Google Scholar]
- 5.Borger, D. R., and J. A. DeCaprio. 2006. Targeting of p300/CREB binding protein coactivators by simian virus 40 is mediated through p53. J. Virol. 804292-4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cantalupo, P., A. Doering, C. S. Sullivan, A. Pal, K. W. Peden, A. M. Lewis, and J. M. Pipas. 2005. Complete nucleotide sequence of polyomavirus SA12. J. Virol. 7913094-13104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cavender, J. F., A. Conn, M. Epler, H. Lacko, and M. J. Tevethia. 1995. Simian virus 40 large T antigen contains two independent activities that cooperate with a ras oncogene to transform rat embryo fibroblasts. J. Virol. 69923-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cotsiki, M., R. L. Lock, Y. Cheng, G. L. Williams, J. Zhao, D. Perera, R. Freire, A. Entwistle, E. A. Golemis, T. M. Roberts, P. S. Jat, and O. V. Gjoerup. 2004. Simian virus 40 large T antigen targets the spindle assembly checkpoint protein Bub1. Proc. Natl. Acad. Sci. USA 101947-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeAngelis, T., J. Chen, A. Wu, M. Prisco, and R. Baserga. 2006. Transformation by the simian virus 40 T antigen is regulated by IGF-I receptor and IRS-1 signaling. Oncogene 2532-42. [DOI] [PubMed] [Google Scholar]
- 10.Digweed, M., I. Demuth, S. Rothe, R. Scholz, A. Jordan, C. Grotzinger, D. Schindler, M. Grompe, and K. Sperling. 2002. SV40 large T-antigen disturbs the formation of nuclear DNA-repair foci containing MRE11. Oncogene 214873-4878. [DOI] [PubMed] [Google Scholar]
- 11.Dornreiter, I., A. Hoss, A. K. Arthur, and E. Fanning. 1990. SV40 T antigen binds directly to the large subunit of purified DNA polymerase alpha. EMBO J. 93329-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Egan, C., S. T. Bayley, and P. E. Branton. 1989. Binding of the Rb1 protein to E1A products is required for adenovirus transformation. Oncogene 4383-388. [PubMed] [Google Scholar]
- 13.Egan, C., T. N. Jelsma, J. A. Howe, S. T. Bayley, B. Ferguson, and P. E. Branton. 1988. Mapping of cellular protein-binding sites on the products of early-region 1A of human adenovirus type 5. Mol. Cell. Biol. 83955-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gannon, J. V., and D. P. Lane. 1987. p53 and DNA polymerase alpha compete for binding to SV40 T antigen. Nature 329456-458. [DOI] [PubMed] [Google Scholar]
- 15.Gu, W., and R. G. Roeder. 1997. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90595-606. [DOI] [PubMed] [Google Scholar]
- 16.Ito, A., C. H. Lai, X. Zhao, S. Saito, M. H. Hamilton, E. Appella, and T. P. Yao. 2001. p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. EMBO J. 201331-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kasper, J. S., H. Kuwabara, T. Arai, S. H. Ali, and J. A. DeCaprio. 2005. Simian virus 40 large T antigen's association with the CUL7 SCF complex contributes to cellular transformation. J. Virol. 7911685-11692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, D., R. Zhao, W. Lilyestrom, D. Gai, R. Zhang, J. A. DeCaprio, E. Fanning, A. Jochimiak, G. Szakonyi, and X. S. Chen. 2003. Structure of the replicative helicase of the oncoprotein SV40 large tumour antigen. Nature 423512-518. [DOI] [PubMed] [Google Scholar]
- 19.Lill, N. L., S. R. Grossman, D. Ginsberg, J. DeCaprio, and D. M. Livingston. 1997. Binding and modulation of p53 by p300/CBP coactivators. Nature 387823-827. [DOI] [PubMed] [Google Scholar]
- 20.Lilyestrom, W., M. G. Klein, R. Zhang, A. Joachimiak, and X. S. Chen. 2006. Crystal structure of SV40 large T-antigen bound to p53: interplay between a viral oncoprotein and a cellular tumor suppressor. Genes Dev. 202373-2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin, J. Y., and D. T. Simmons. 1991. The ability of large T antigen to complex with p53 is necessary for the increased life span and partial transformation of human cells by simian virus 40. J. Virol. 656447-6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, L., D. M. Scolnick, R. C. Trievel, H. B. Zhang, R. Marmorstein, T. D. Halazonetis, and S. L. Berger. 1999. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol. Cell. Biol. 191202-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lundblad, J. R., R. P. Kwok, M. E. Laurance, M. L. Harter, and R. H. Goodman. 1995. Adenoviral E1A-associated protein p300 as a functional homologue of the transcriptional co-activator CBP. Nature 37485-88. [DOI] [PubMed] [Google Scholar]
- 24.Melendy, T., and B. Stillman. 1993. An interaction between replication protein A and SV40 T antigen appears essential for primosome assembly during SV40 DNA replication. J. Biol. Chem. 2683389-3395. [PubMed] [Google Scholar]
- 25.Pipas, J. M. 1985. Mutations near the carboxyl terminus of the simian virus 40 large tumor antigen alter viral host range. J. Virol. 54569-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pipas, J. M. 2009. SV40: cell transformation and tumorigenesis. Virology 384294-303. [DOI] [PubMed] [Google Scholar]
- 27.Pipas, J. M., K. W. Peden, and D. Nathans. 1983. Mutational analysis of simian virus 40 T antigen: isolation and characterization of mutants with deletions in the T-antigen gene. Mol. Cell. Biol. 3203-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polvino-Bodnar, M., and C. N. Cole. 1982. Construction and characterization of viable deletion mutants of simian virus 40 lacking sequences near the 3′ end of the early region. J. Virol. 43489-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poulin, D. L., A. L. Kung, and J. A. DeCaprio. 2004. p53 targets simian virus 40 large T antigen for acetylation by CBP. J. Virol. 788245-8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sachsenmeier, K. F., and J. M. Pipas. 2001. Inhibition of Rb and p53 is insufficient for SV40 T-antigen transformation. Virology 28340-48. [DOI] [PubMed] [Google Scholar]
- 31.Simmons, D. T., T. Melendy, D. Usher, and B. Stillman. 1996. Simian virus 40 large T antigen binds to topoisomerase I. Virology 222365-374. [DOI] [PubMed] [Google Scholar]
- 32.Smits, P. H., L. de Wit, A. J. van der Eb, and A. Zantema. 1996. The adenovirus E1A-associated 300 kDa adaptor protein counteracts the inhibition of the collagenase promoter by E1A and represses transformation. Oncogene 121529-1535. [PubMed] [Google Scholar]
- 33.Srinivasan, A., A. J. McClellan, J. Vartikar, I. Marks, P. Cantalupo, Y. Li, P. Whyte, K. Rundell, J. L. Brodsky, and J. M. Pipas. 1997. The amino-terminal transforming region of simian virus 40 large T and small t antigens functions as a J domain. Mol. Cell. Biol. 174761-4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stein, R. W., M. Corrigan, P. Yaciuk, J. Whelan, and E. Moran. 1990. Analysis of E1A-mediated growth regulation functions: binding of the 300-kilodalton cellular product correlates with E1A enhancer repression function and DNA synthesis-inducing activity. J. Virol. 644421-4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sullivan, C. S., P. Cantalupo, and J. M. Pipas. 2000. The molecular chaperone activity of simian virus 40 large T antigen is required to disrupt Rb-E2F family complexes by an ATP-dependent mechanism. Mol. Cell. Biol. 206233-6243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sullivan, C. S., and J. M. Pipas. 2002. T antigens of simian virus 40: molecular chaperones for viral replication and tumorigenesis. Microbiol. Mol. Biol. Rev. 66179-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tevethia, M. J., H. A. Lacko, and A. Conn. 1998. Two regions of simian virus 40 large T-antigen independently extend the life span of primary C57BL/6 mouse embryo fibroblasts and cooperate in immortalization. Virology 243303-312. [DOI] [PubMed] [Google Scholar]
- 38.Wei, W., W. A. Jobling, W. Chen, W. C. Hahn, and J. M. Sedivy. 2003. Abolition of cyclin-dependent kinase inhibitor p16Ink4a and p21Cip1/Waf1 functions permits Ras-induced anchorage-independent growth in telomerase-immortalized human fibroblasts. Mol. Cell. Biol. 232859-2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weisshart, K., P. Taneja, and E. Fanning. 1998. The replication protein A binding site in simian virus 40 (SV40) T antigen and its role in the initial steps of SV40 DNA replication. J. Virol. 729771-9781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Welcker, M., and B. E. Clurman. 2005. The SV40 large T antigen contains a decoy phosphodegron that mediates its interactions with Fbw7/hCdc4. J. Biol. Chem. 2807654-7658. [DOI] [PubMed] [Google Scholar]
- 41.Whyte, P., H. E. Ruley, and E. Harlow. 1988. Two regions of the adenovirus early region 1A proteins are required for transformation. J. Virol. 62257-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu, X., D. Avni, T. Chiba, F. Yan, Q. Zhao, Y. Lin, H. Heng, and D. Livingston. 2004. SV40 T antigen interacts with Nbs1 to disrupt DNA replication control. Genes Dev. 181305-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yaciuk, P., M. C. Carter, J. M. Pipas, and E. Moran. 1991. Simian virus 40 large-T antigen expresses a biological activity complementary to the p300-associated transforming function of the adenovirus E1A gene products. Mol. Cell. Biol. 112116-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.