Abstract
The hepatitis C virus NS2 protein has been recently implicated in virus particle assembly. To further understand the role of NS2 in this process, we conducted a reverse genetic analysis of NS2 in the context of a chimeric genotype 2a infectious cell culture system. Of 32 mutants tested, all were capable of RNA replication and 25 had moderate-to-severe defects in virus assembly. Through forward genetic selection for variants capable of virus spread, we identified second-site mutations in E1, E2, NS2, NS3, and NS4A that suppressed NS2 defects in assembly. Two suppressor mutations, E1 A78T and NS3 Q221L, were further characterized by additional genetic and biochemical experiments. Both mutations were shown to suppress other NS2 defects, often with mutual exclusivity. Thus, several NS2 mutants were enhanced by NS3 Q221L and inhibited by E1 A78T, while others were enhanced by E1 A78T and inhibited by NS3 Q221L. Furthermore, we show that the NS3 Q221L mutation lowers the affinity of native, full-length NS3-NS4A for functional RNA binding. These data reveal a complex network of interactions involving NS2 and other viral structural and nonstructural proteins during virus assembly.
Hepatitis C virus (HCV) is a major cause of acute and chronic liver disease and contributes to the development of hepatocellular carcinoma. HCV is an enveloped, positive-strand RNA virus, the type member of the Hepacivirus genus in the family Flaviviridae (43). HCV exhibits high levels of sequence diversity that cluster into seven major genotypes and numerous subtypes (21).
HCV genomes are 9.6 kb and encode a single long open reading frame of ∼3,011 codons (43). Translation of this genome produces a large polyprotein that is co- and posttranslationally processed by viral and host proteases into 10 distinct products. The N-terminal one-third of the polyprotein encodes the structural proteins, which are thought to compose the virus particle. These include an RNA-binding nucleocapsid protein, core (C), and two viral envelope glycoproteins, E1 and E2. E1 and E2 are type I membrane proteins that coordinately fold into a heterodimer complex (36). The remainder of the genome encodes the nonstructural (NS) proteins NS2, NS3, NS4A, NS4B, NS5A, and NS5B, which mediate the intracellular aspects of the viral life cycle. In addition, a small viroporin-like protein, p7, resides between the structural and NS genes.
HCV encodes two proteases, the NS2-NS3 cysteine autoprotease and the NS3-NS4A serine protease. The only known substrate of the NS2-NS3 autoprotease is the NS2/3 junction. This enzyme is encoded by the C-terminal 121 amino acids (aa) of NS2, which forms a homodimer with twin composite active sites composed of two residues from one chain and one residue from the other (45). In addition, the serine protease domain of NS3 plays a noncatalytic role in stimulating NS2/3 cleavage (69). Upstream of the cysteine protease domain, the N-terminal hydrophobic region of NS2 mediates interaction with cellular membranes. While the membrane topology of NS2 is not yet fully known (67, 80), N-terminal cleavage by endoplasmic reticulum-resident signal peptidase and C-terminal cleavage by the cytosolic NS2-NS3 cysteine protease indicate that NS2 likely contains one or three transmembrane (TM) domains.
The NS3-NS4A serine protease is encoded by the N-terminal domain of NS3 and is responsible for downstream cleavages at the NS3/4A, NS4A/B, NS4B/5A, and NS5A/B junctions. NS4A, a small (54-aa), membrane-anchored protein, acts as a cofactor for the serine protease activity by helping to complete the chymotrypsin-like fold of NS3 (14, 46). In addition to polyprotein processing, NS3-NS4A serine protease helps to dampen the innate antiviral response by cleaving cellular proteins involved in signal transduction (65).
The C-terminal region of NS3 encodes an RNA helicase/NTPase activity that is essential for viral replication, although it is not yet clear which specific step(s) of the replication cycle requires this activity (29, 33). Interestingly, the NS3 serine protease and RNA helicase domains enhance each other's activities, suggesting that proteolysis and RNA replication may be functionally coordinated (5, 6). In addition, NS4A helps to promote RNA-stimulated ATP hydrolysis by the NS3 helicase domain (4).
In addition to their role in polyprotein processing, emerging evidence indicates that NS2 and NS3-NS4A participate in virus particle assembly (52). Prior work showed that NS2 is not essential for RNA replication of subgenomic replicons engineered to express NS3 through NS5B (44). The first evidence for an additional function of NS2 came from the construction of improved chimeric genotype 2a cDNA clones that replicated to high titers in cell culture (HCVcc). Pietschmann and colleagues showed that the Jc1 chimera containing a J6-JFH1 junction between the first and second putative TM domains of NS2 yielded higher-titer viruses than the original infectious J6/JFH chimera (41, 58). Furthermore, a number of adaptive mutations that improve virus production have been mapped to NS2 and NS3 (22, 23, 27, 53, 64, 68, 82). By using bicistronic constructs to express NS2 and NS3 independently of NS2/3 cleavage, two groups showed that full-length NS2, but not uncleaved NS2-NS3 or the NS2 cysteine protease active sites, was required for virus production (24, 25). Moreover, a limited number of mutations in NS2 were shown to inhibit virus assembly (24, 79, 83).
Despite these observations, the role of NS2 in virus assembly remains unclear. We have therefore undertaken a genetic analysis to target conserved residues in NS2 for site-directed mutagenesis and identified a number of key residues that are important for virus assembly. Further analysis revealed that a network of genetic interactions among NS2, E1-E2, and NS3-NS4A helps to direct virus assembly. Finally, a suppressor mutation in NS3 was shown to influence functional RNA binding by the RNA helicase/ATPase.
MATERIALS AND METHODS
Computation and nomenclature.
An NS2 protein sequence alignment was constructed by using the ClustalW2 web server (35). To minimize phylogenetic bias, 14 NS2 sequences representing all seven major genotypes were used in the alignment. The HCV strains used (and their accession numbers) were H77 (NC_004102), HCV-1 (M62321), Con1 (AJ238799), HC-J6 (D00944), JFH1 (AB047639), MD2b-1 (AF238486), CB (AF046866), Tr Kj (D49374), ED43 (NC_009825), SA13 (AF064490), EUH1480 (NC_009826), EUHK (Y12083), Th580 (NC_009827), and QC69 (EF108306).
The membrane topology of NS2 was predicted by using the PolyPhobius and Philius web servers (26, 62), which produced results in close agreement. The SCAMPI, OCTOPUS, and PRO/PRODIV servers yielded similar results for TM1 (7, 75, 76) but differed in their predictions for downstream regions of NS2. Given that NS2 likely contains one or three TM domains, some of these predictions could be discounted. We therefore consider the PolyPhobius prediction to be the best current working model for NS2 topology.
Molecular models were rendered by using PyMol (http://www.pymol.org). The level of NS2 sequence conservation was mapped onto the crystal structure of the NS2 cysteine protease domain (Protein Data Bank entry 2HD0 [45]) by using the ConSurf server (34). A model of NS3 with bound nucleic acid was created by performing a structural alignment of a single-chain NS3-NS4A (Protein Data Bank entry 1CU1 chain A [81]) and an NS3 helicase domain DNA (Protein Data Bank entry 1A1V [28]). For clarity in rendering this model, loop regions were smoothed in PyMol.
Graphs and curve fittings were performed by using Prism 4 (GraphPad Software, La Jolla, CA). Figures were assembled by using Adobe Creative Suite 4 (Adobe Systems, San Jose, CA).
In keeping with recently adapted conventions of HCV nomenclature (31), relative numbering of protein sequences is used throughout this work. For clarity, the absolute positions of all NS2 mutations and their suppressors are provided relative to the polyprotein of reference strain H77.
Molecular biology.
Standard molecular biology methods were used throughout (66). Plasmid sequences were verified through DNA sequencing at the W. M. Keck Foundation Biotechnology Resource Center at Yale University. The pFL-J6/JFH1 chimeric genotype 2a HCV cDNA clone was reconstructed by gene synthesis of the JFH1 replicon (Codon Devices, Cambridge, MA) and insertion of the HC-J6 core-NS2 region (a kind gift of Jens Bukh, NIH) as previously described (41). The Jc1 chimera (58) was then reconstructed by gene synthesis of JFH1 NS2 codons 30 to 217 and inserting this region into pJ6/JFH through mutually primed DNA synthesis, yielding pJc1.
The Jc1/GLuc2A reporter virus was constructed in a manner similar to that used for J6/JFH(p7-Rluc2A) (25). First, a silent MluI site was inserted at the p7-NS2 junction of pJc1 via QuikChange site-directed mutagenesis (Stratagene, La Jolla, CA) with primers YO-0178 (5′-TGC TTA TGA CGC GTC TGT GCA TGG-3′) and YO-0179 (5′-CCA TGC ACA GAC GCG TCA TAA GCA-3′), yielding pJc1•MluI. The secreted fragment (codons 18 to 185) of Gaussia princeps luciferase (GLuc) was then amplified from pCMV-GLuc (New England BioLabs, Ipswich, MA) by using primers YO-0180 (5′-GAC GCG TCT AAG CCC ACC GAG AAC AAC GAA GAC-3′) and YO-0181 (5′-CAG CAA ATC GAA ATT GAG AAG CTG TTT TCC GTC ACC ACC GGC CCC CTT GAT-3′) and then fused to 21 codons of the foot-and-mouth disease virus (FMDV) 2A gene by amplification with YO-0180 and YO-0181 (5′-TAC GCG TCA TAT GGT CCT GGG TTA CTC TCT ACG TCG CCT GCC AGC TTC AGC AAA TCG AAA TTG A-3′). The resulting product was cloned into pCR2.1-TOPO (Invitrogen, Carlsbad, CA), sequenced, and subcloned as an MluI fragment into pJc1•MluI, yielding pJc1/GLuc2A.
To facilitate site-directed mutagenesis, a truncated form of pJc1•MluI was created by ligating 3,113-bp EcoRI/EcoRV and 4,123-bp EcoRI/ScaI fragments of this plasmid to create pJc1•MluΔNSP. Mutations were then introduced into the NS2 gene by QuikChange site-directed mutagenesis with primers listed in Table 1 and a complementary primer. Following sequence verification, the mutations were subcloned into pJc1/GLuc2A as either a 1,078-bp MluI/AvrII fragment ligated to 8,479-bp EcoRI/AvrII and 3,377-bp EcoRI/partial MluI fragments of pJc1/GLuc2A or as a 906-bp NotI/AvrII fragment to a 12,040-bp NotI/AvrII fragment of pJc1/GLuc2A.
TABLE 1.
Site-directed mutagenesis of NS2
| NS2 mutation | Jc1 codon | H77 codon | Primer sequence (5′-3′)a |
|---|---|---|---|
| G10A | 823 | 819 | TCTGTGCATGGCCAGATAGCCGCGGCTCTGCTGGTAATG |
| P24A | 837 | 833 | CACTCTCTTTACTCTCACCGCCGGGTATAAGACCCT |
| K27A | 840 | 836 | CTCTCACCCCCGGGTATGCCACCCTCCTCGGCCAGTGT |
| W35A | 848 | 844 | TCCTCGGCCAGTGTCTGGCCTGGTTGTGCTATCTCCTG |
| W36A | 849 | 845 | TCCTCGGCCAGTGTCTGTGGGCCTTGTGCTATCTCCTG |
| Y39A | 852 | 848 | TGTCTGTGGTGGTTGTGCGCCCTCCTGACCCTGGGGGAA |
| E45A | 858 | 854 | TCTCCTGACCCTGGGGGCCGCCATGATTCAGGAGT |
| E45K | 858 | 854 | TCTCCTGACCCTGGGGAAGGCCATGATTCAGGAGT |
| P53A | 866 | 862 | TGATTCAGGAGTGGGTAGCCCCCATGCAGGTGCG |
| R58A | 871 | 867 | TACCACCCATGCAGGTGGCCGGCGGCCGCGATGGCAT |
| R61A | 874 | 870 | ATGCAGGTGCGCGGCGGCGCCGATGGCATCGCGTGG |
| D62A | 875 | 871 | AGGTGCGCGGCGGCCGCGCCGGCATCGCGTGGGCCGT |
| P73A | 886 | 882 | CCGTCACTATATTCTGCGCCGGTGTGGTGTTTGACATT |
| F77A | 890 | 886 | TCTGCCCGGGTGTGGTGGCCGACATTACCAAATGGCTT |
| D78A | 891 | 887 | TGCCCGGGTGTGGTGTTTGCCATTACCAAATGGCTTT |
| D78K | 891 | 887 | TGCCCGGGTGTGGTGTTTAAGATTACCAAATGGCTTT |
| K81A | 894 | 890 | TGGTGTTTGACATTACCGCCTGGCTTTTGGCGTTGCT |
| K81E | 894 | 890 | TGGTGTTTGACATTACCGAATGGCTTTTGGCGTTGCT |
| P89A | 902 | 898 | CTTTTGGCGTTGCTTGGGGCCGCTTACCTCTTAAGGGCCGCTT |
| R111A | 924 | 920 | GAGCTCACGCTCTGATAGCCGTATGCGCTTTGGTGAA |
| Y124A | 937 | 933 | AAGCAGCTCGCGGGGGGTAGGGCCGTTCAGGTGGCGCTATT |
| R156A | 969 | 965 | ACTGGGCCGCTAGCGGCCTGGCCGACTTAGCGGTCGCCGT |
| D157A | 970 | 966 | ACTGGGCCGCTAGCGGCCTGCGCGCCTTAGCGGTCGCCGT |
| V160A | 973 | 969 | TGACACATGTGCCGTACGCCGTCAGAGCTCACGCTCTG |
| S168A | 981 | 977 | TGGAACCCATCATCTTCGCCCCGATGGAGAAGAAGGT |
| S168D | 981 | 977 | TGGAACCCATCATCTTCGACCCGATGGAGAAGAAGGT |
| S168E | 981 | 977 | TGGAACCCATCATCTTCGAGCCGATGGAGAAGAAGGT |
| E171A | 984 | 980 | TCATCTTCAGTCCGATGGCCAAGAAGGTCATCGTCTG |
| K173A | 986 | 982 | TCAGTCCGATGGAGAAGGCCGTCATCGTCTGGGGAGC |
| D180A | 993 | 989 | TCATCGTCTGGGGAGCGGCCACGGCTGCATGTGGGGA |
| D181A | 994 | 990 | ATCGTCTGGGGAGCGGAGGCCGCTGCATGTGGGGACA |
| R196A | 1009 | 1005 | ACTTCCCGTGTCCGCCGCCCTCGGCCAGGAGATCCTC |
Forward primers are shown; complementary reverse primers were also used in site-directed mutagenesis. Mutation sites are underlined.
pJc1/GLuc2A(Δcore) was made by first creating an in-frame deletion within the core gene of pJc1. This was accomplished by ligating 543-bp EcoRI/SfoI, 2,168-bp Ecl136II/NotI, and 9,389-bp NotI/EcoRI fragments of pJc1. The resulting deletion was then subcloned as a 2,319-bp EcoRI/BsaBI fragment into pJc1/GLuc2A. The polymerase-defective control, pJc1/GLuc2A(GNN), was made by site-directed mutagenesis of pJ6/JFH with primers RU-O-7907 (5′-ACA ATG CTG GTA TGC GGC AAC AAC CTA GTA GTC ATC TCA GAA AG-3′) and RU-O-7908 (5′-CTT TCT GAG ATG ACT ACT AGG TTG TTG CCG CAT ACC AGC ATT GT-3′). The resulting mutation in NS5B was then subcloned as a 2,215-bp RsrII/XbaI fragment into pJc1/GLuc2A.
RNA transcription and cleanup were performed as previously described (41).
Cell culture and RNA transfection.
All cell culture work was performed in an enhanced biosafety level 2+ suite licensed by the State of Connecticut Department of Public Health. Huh-7.5 cells (8) were maintained in Dulbecco's modified Eagle medium (Invitrogen) containing 10% fetal calf serum (HyClone, Logan, UT) and 1 mM nonessential amino acids (Invitrogen). Cells were transfected by electroporation as previously described (41) or by using the TransIT-mRNA Transfection Kit (Mirus Bio, Madison, WI).
GLuc activity.
Conditioned cell culture medium was collected at various times postelectroporation or at 72 h postinfection, clarified by centrifugation (16,000 × g for 5 min), and mixed with 0.25 volume of Renilla 5× lysis buffer (Promega, Madison, WI) to kill HCV infectivity. GLuc activity was measured on a Berthold Centro LB 960 luminescent plate reader with 20 μl sample injected with 50 μl Gaussia luciferase assay reagent (New England BioLabs), integrated over 10 s.
Infectivity measurements.
Absolute values of viral infectivity were determined by using an endpoint dilution assay as previously described (40). Briefly, virus was serially diluted into complete growth medium and each dilution was used to infect multiple wells of a 96-well plate. At 4 days postinfection, the cells were washed with phosphate-buffered saline (PBS), fixed with ice-cold methanol, and immunostained for NS5A expression by using the 9E10 monoclonal antibody (41). Fifty percent endpoints were calculated by using the method of Reed and Muench (61).
To measure the relative infectivity of GLuc2A reporter viruses, conditioned cell culture medium was collected at various times postelectroporation, clarified by centrifugation (16,000 × g for 5 min), and used to infect naïve cells seeded at 6.4 × 103/well in 96-well plates. Following a 4-h adsorption period, cells were washed three times with Dulbecco's PBS and incubated with complete medium for an additional 72 h.
To measure the relative intracellular infectivity of GLuc2A reporter viruses, cells were harvested by trypsinization at 48 h posttransfection, centrifuged (1,200 × g, 5 min), resuspended in a small volume of complete medium, snap-frozen in liquid nitrogen, and stored at −80°C. Cells were subjected to three rounds of thawing (37°C) and refreezing (−196°C) in liquid nitrogen. Cellular debris was removed by centrifugation (16,000 × g, 5 min), and the supernatants were tested for infectivity as described above.
RNA quantitation.
RNA was extracted from conditioned cell culture medium by using the QIAamp Viral RNA Mini Kit (Qiagen, Valencia, CA). HCV RNA levels were measured by real-time reverse transcription (RT)-PCR as previously described (41). Briefly, reactions were run with the LightCycler RNA amplification HybProbe kit (Roche Applied Sciences, Mannheim, Germany) containing 2 μl RNA sample or RNA quantitation standards, 8 mM MgCl2, 375 nM each primer, 250 nM probe, and 1 U RNase inhibitor (United States Biochemical, Cleveland, OH). Reactions were run on a Roche LightCycler 480.
Identification of suppressor mutations.
To select for variants that had regained infectivity, RNA-transfected cells were seeded onto 10-cm dishes and serially passed 1:3 every 3 days. During bulk passage, the conditioned cell culture medium was clarified and stored frozen and a subset of cells was seeded onto 12- or 24-well dishes and then fixed and stained for NS5A as described above. For Jc1/GLuc2A, nearly all of the cells were NS5A positive within 48 h postelectroporation; for Jc1/GLuc2A(Δcore), about 30% of the cells were NS5A positive at short intervals after electroporation and this declined to a few percent with serial passage. No increases in Jc1/GLuc2A(Δcore)-positive cells were ever noted. In contrast, NS2 mutants that had defects in virus assembly frequently gave rise to variants that could spread within four to six passages. Alternatively, to limit competition within the viral population, RNA-transfected cells were seeded onto 96-well plates and serially passed 1:3 every 3 days. Again, cell culture medium was collected, clarified by centrifugation, and tested for infectivity in naïve cells by either GLuc2A assay or NS5A immunostaining as described above. Positive wells were then expanded for further analysis.
To biologically clone revertant viruses, the conditioned cell culture medium was serially diluted to the limiting endpoint and used to infect naïve cells in 96-well plates. Conditioned cell culture medium was collected, and the cells were stained for NS5A as described above. Positive wells were then expanded for recloning and further analysis.
Viral RNAs were extracted from conditioned cell culture medium by using the QIAamp Viral RNA Mini Kit (Qiagen) and used as templates to synthesize random hexamer-primed cDNAs with SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer's instructions. The Jc1/GLuc2A coding regions were amplified by using specific primers listed in Table 2 and illustra PuReTaq Ready-To-Go PCR beads (GE Healthcare, Buckinghamshire, United Kingdom). PCR products were purified by using QIAquick PCR spin columns and sequenced at the Keck Center. Mutations of interest were cloned by using pCR2.1-TOPO (Invitrogen), their sequences were verified, and they were subcloned back into pJc1/GLuc2A by using common restriction sites.
TABLE 2.
Primers used to amplify the HCV coding region for sequencing
| Amplicon | Orientation | Sequence (5′-3′) |
|---|---|---|
| 1 | Forward | TCCCGGGAGAGCCATAGTGGT |
| Reverse | AGCCATCCTGCCCAGCCGAGTC | |
| 2 | Forward | CCAGCCCATCCCTAAAGATCGG |
| Reverse | CACGGCCACATTCGGTGAGA | |
| 3 | Forward | CGGCTACATGGTGACTAACGAC |
| Reverse | GCCAACAGAAGGATGACAACGA | |
| 4 | Forward | GCTCTGCTCTGCCCTCTACGTG |
| Reverse | CGGACATGCGTTCGGGACA | |
| 5 | Forward | TCCTTGCACACCGGCTTTA |
| Reverse | GCCGCATTTGAGGTAAGTGGTA | |
| 6 | Forward | TTCAACGCCAGCACGGACCTGT |
| Reverse | TCCATAAGCAGGCGCAAAC | |
| 7 | Forward | CAATTCATGTATGGCCTATCA |
| Reverse | CTCTTTGTCGCCTTCGT | |
| 8 | Forward | CACCAGGGGCTGTCTGATCT |
| Reverse | CGCCACCTGAACATACCTACC | |
| 9 | Forward | AGGGCCGCTTTGACACA |
| Reverse | TCGACGGCTCCACACTTGCAC | |
| 10 | Forward | TGCTGAGGGGGACTTGGTAG |
| Reverse | GCCGTAGCCAGCACAGTTAGTCTGA | |
| 11 | Forward | CGATGAATGCCACGCTGTGGATGCT |
| Reverse | GGGAGAGGAAGTGGGCGTCTA | |
| 12 | Forward | CCGTCAGGCTTAGAGCGTATTTCA |
| Reverse | AGTGTTGACAATCCTGCGAGGTATT | |
| 13 | Forward | AGCAGGCCCAGGACATACAAC |
| Reverse | CAGCTTGGGGAACAATTTAGAGGTC | |
| 14 | Forward | GTGTGGGACTGGGTTTGCACCATCT |
| Reverse | GGCTCGAGAAAGTCCAGAACGG | |
| 15 | Forward | TGACGTGGACATGGTCGATGCCAAC |
| Reverse | CCGCTAGCTTGATGTCCTTTAAGAC | |
| 16 | Forward | TCACAGAGGGCTAAAAAGGTAACT |
| Reverse | TCCCCTGGCTTTCTGAGATGACTAC | |
| 17 | Forward | CCATGTTCAACAGCAAGGGTCAAAC |
| Reverse | CGACGGTGAACCAACTGGATAAGTC | |
| 18 | Forward | GAGAGGTTACACGGGCTTGAC |
| Reverse | GGACCTTTCACAGCTAGCCGTGACT |
Protein expression.
Huh-7.5 cells were seeded at 5 × 105/well into six-well plates the day before use. Cells were infected for 1 h at a multiplicity of infection (MOI) of 10 with vTF7-3 (19) and transfected with pJc1/GLuc2A derivatives by using Mirus LT1 reagent according to the manufacturer's recommendations. At 24 h postinfection, cells were washed twice with Dulbecco's PBS, lysed in 0.3 ml sample buffer (50 mM Tris [pH 6.8], 100 mM dithiothreitol [DTT], 2% [wt/vol] sodium dodecyl sulfate [SDS], 10% [vol/vol] glycerol, 0.1% [wt/vol] bromophenol blue), and homogenized by multiple passes through a 27-gauge needle. Samples (5 μl) were denatured by boiling, resolved by SDS-polyacrylamide gel electrophoresis (PAGE), and transferred to Immobilon P membranes (Millipore, Bedford, MA). Membranes were blocked in PBS-T (PBS containing 0.1% [vol/vol] Tween 20 [polyoxyethylene sorbitan monolaurate]) containing 5% (wt/vol) dry milk and probed with this blocking buffer containing primary monoclonal antibodies against core (C7-50, Affinity BioReagents, Golden, CO), E2 (3/11; a kind gift from Jane McKeating) (15), NS2 (6H6; a kind gift from Charles M. Rice) (T. Dentzer, I. Lorenz, and C. M. Rice, unpublished data), NS3 (9G2; Virogen, Watertown, MA), NS5A (9E10; a kind gift from Charles M. Rice and Timothy Tellinghuisen) (41), or ß-actin (AC-15; Sigma-Aldrich, St. Louis, MO). Following several washes with PBS-T, membranes were probed with horseradish peroxidase-conjugated secondary antibodies and washed repeatedly and antigens were detected with SuperSignal West Pico chemiluminescent substrate (Pierce, Rockford, IL).
Characterization of NS3-NS4A enzymology.
Wild-type (WT) and mutant forms of HCV genotype 2a (strain JFH1) NS3-NS4A were amplified by PCR and cloned into the pET-SUMO vector (Invitrogen) according to the manufacturer's suggestions. Proteins were purified from Escherichia coli as previously described (6). Briefly, His6-SUMO fusion proteins were purified through a nickel column (Qiagen) and then through a HiLoad Superdex 200 16/60 gel filtration column (GE Healthcare), treated with 10 U of SUMO protease (Invitrogen) to remove the His6-SUMO fusion, and subsequently purified in the flowthrough from a second nickel column. Protein preparations were divided into 10-μl aliquots and stored at −80°C.
Protease assays were performed as previously described (5). Briefly, 60-μl reaction mixtures contained 40 mM NS3-NS4A, 5 μM RET-S1 substrate (70) (obtained from AnaSpec [San Jose, CA] and provided as a kind gift from Anna Pyle), and helicase assay buffer (25 mM morpholinepropanesulfonic acid [MOPS]-NH4+ [pH 6.5], 3 mM MgCl2, 1% [vol/vol] glycerol, 2 mM DTT, 30 mM NaCl, 0.2% [vol/vol] Triton X-100). Fluorescence readings (excitation, 350 nm; emission, 440 nm) were collected at 37°C in a temperature-controlled cuvette on a Cary Eclipse spectrophotometer (Varian, Inc., Palo Alto, CA).
RNA filter-binding assays were performed with 5′-end 32P-labeled RNA oligonucleotide RNA1-TS34 as previously described (6). Briefly, 20 fmol labeled, gel-purified RNA1-TS34 (200 pM, final concentration) was incubated with various concentrations of NS3-NS4A protein for 1 h at 37°C in 100-μl reaction mixtures containing helicase assay buffer and then bound to a nitrocellulose (Pierce)/nylon (Nytran; Whatman, Kent, United Kingdom) membrane sandwich with a dot blot apparatus. The membrane was then washed with ice-cold helicase assay buffer and exposed to a PhosphorImager (Molecular Dynamics). The amount of RNA bound by NS3-NS4A was determined, and the Kd was calculated by using the Hill equation (Ys = [S]n/Kd + [S]n).
RNA-unwinding (helicase) assays were performed as previously described (3-5). Briefly, 10 pmol NS3-NS4A (100 nM, final concentration) was preincubated with 0.2 nM duplex substrate for 1 h at 37°C in 100-μl reaction mixtures containing helicase assay buffer. Helicase reactions were then initiated by adding 6 μl of the preincubation mixtures to 2 μl of ATP-trap mix (25 mM MOPS-NH4+ [pH 6.5], 3 mM MgCl2, 30 mM NaCl, 1 μM top-strand oligonucleotide, 16 mM ATP). The unwinding substrate used in these assays was RNA2, a 34-bp duplex with a 20-nucleotide (nt) 3′ overhang (4).
ATPase activity was monitored as previously described (6). Briefly, 1 pmol NS3-NS4A (50 nM, final concentration) was preincubated with various concentrations of RNA1-TS34 for 1 h at 37°C in 18-μl reaction mixtures containing 25 mM MOPS-NH4+ (pH 6.5), 0.75 mM MgCl2, 1% (vol/vol) glycerol, 2 mM DTT, 30 mM NaCl, and 0.2% (vol/vol) Triton X-100. Reactions were then initiated by the addition of ATP to 1 mM (2 μl of 10 mM unlabeled ATP, 3.75 μCi of [γ-32P]ATP [Perkin-Elmer, Waltham, MA]). At various time points, the free 32Pi was separated from [γ-32P]ATP by thin-layer chromatography and quantified with a PhosphorImager. The amount of ATP hydrolysis was calculated as a function of time and RNA concentration. Initial velocities were calculated from a linear regression of the data, and the initial reaction velocities versus the RNA concentration was plotted and fitted to the Michaelis-Menten equation (v0 = Vmax[S]/Km + [S]). The RNA stimulation factors were calculated by dividing the ATPase velocities determined in the presence of saturating RNA levels (i.e., the Vmax value) by the basal ATPase velocity in the absence of RNA.
RESULTS
We conducted reverse genetic analysis of NS2 by using a chimeric genotype 2a cDNA clone that efficiently replicates and produces high titers of infectious virus in cell culture (HCVcc). Since Pietschmann and colleagues showed that the Jc1 “C3” chimeric junction within NS2 produced higher levels of infectious virus than our original J6/JFH1 chimera (41, 58), we considered this to be the most suitable genetic background for our studies on NS2. We therefore reconstructed the Jc1 J6/JFH chimera and showed that it produced high infectivity titers (∼106 50% tissue culture infective doses [TCID50]/ml) within the first 48 h posttransfection (data not shown).
GLuc2A as a sensitive reporter of HCVcc replication and infectivity.
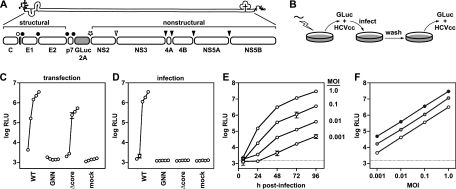
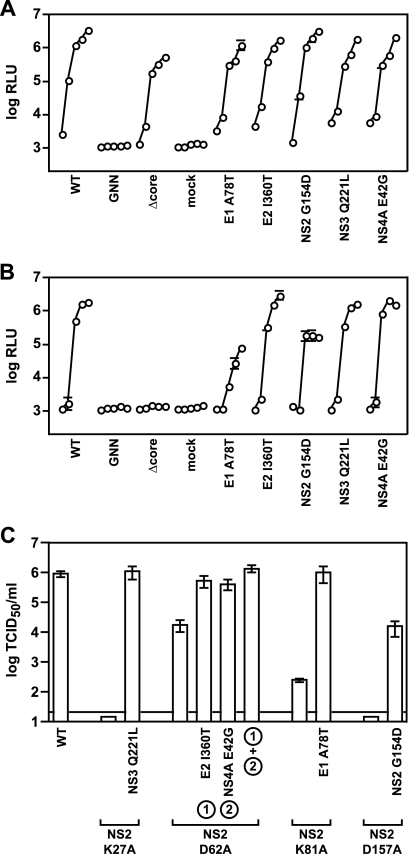
To facilitate the rapid measurement of HCVcc replication and infectivity, we constructed a Jc1-based reporter virus similar to the monocistronic J6/JFH reporter virus encoding Renilla reniformis luciferase (Rluc) described by Jones et al. (25). The GLuc gene and a 21-aa “autocleaving” 2A peptide of FMDV were encoded between p7 and NS2 of the Jc1 chimeric infectious cDNA clone (Fig. 1A). Advantages of the GLuc reporter include its smaller gene size (0.5 kb) and an enzymatic activity >2,000-fold higher than that of Rluc (71). This design led to the secretion of GLuc2A enzyme and infectious reporter virus into the culture medium of Jc1/GLuc2A-transfected Huh-7.5 cells. Thus, HCV replication could be noninvasively monitored over time by measuring secreted GLuc2A activity (Fig. 1B and C). A polymerase-defective control, Jc1/GLuc2A(GNN), showed that GLuc2A expression was dependent on HCV replication (Fig. 1C and D). Furthermore, the level of GLuc2A expression correlated over time with the level of intracellular HCV RNA as determined by quantitative RT-PCR (data not shown).
FIG. 1.
The GLuc reporter virus is a sensitive reporter of relative HCV replication and infectivity. (A) The Jc1/GLuc2A reporter construct used in these studies. The HCV genome and polyprotein processing scheme, including a GLuc-FMDV 2A insertion, are shown. Signal peptidase cleavages are indicated by filled circles, signal peptide peptidase cleavage by an open circle, FMDV 2A “autocleavage” by a star, NS2-NS3 cysteine autoprotease cleavage by an open arrowhead, and NS3-NS4A serine protease cleavages by filled arrowheads. (B) The work flow of our reporter virus assays is diagrammed. RNA was electroporated into Huh-7.5 cells, and culture medium was harvested at various times posttransfection. Secreted HCVcc was used to infect naïve cells; secreted GLuc activity was assayed as detailed in Materials and Methods. (C) Time course of HCV replication-dependent GLuc2A secretion. Read from left to right, each curve represents relative light units (log RLU) at 8, 24, 48, 72, and 96 h posttransfection with the indicated construct. Values shown are averages from three independent transfections; error bars represent the standard deviation of the mean. (D) Time course of infectious HCVcc production. Medium collected at 8, 24, 48, 72, and 96 h posttransfection was used to infect naïve cells. Following a 4-h period of virus adsorption, the cells were washed and incubated in fresh medium for an additional 72 h and the medium was collected and assayed for GLuc activity. Values represent the averages from medium collected from the transfections shown in panel C; error bars represent the standard deviation of the mean. (E) GLuc activity correlates with the amount of input reporter virus. Parallel cell cultures were infected with Jc1/GLuc2A in quadruplicate at the indicated MOIs. The medium was harvested at the indicated times and assayed for GLuc activity. Values are averages from four independent infections from a representative experiment; error bars represent the standard deviation of the mean. Dotted line, limit of detection. (F) Secreted GLuc2A provides linear responses to virus input over time. The data in panel E were transformed to plot GLuc activity (log RLU) at 48 h (open circles), 72 h (gray circles), and 96 h (filled circles) versus the input MOI (shown on a log scale).
The production of infectious reporter virus was monitored over time by infecting naïve cells with the conditioned medium from RNA-transfected cells and measuring newly secreted GLuc2A at a fixed endpoint (Fig. 1B and D). GLuc2A expression was inhibited by neutralizing virus particles with an E2-specific monoclonal antibody or by blocking the viral coreceptor with anti-CD81 (data not shown), indicating that reporter gene expression was dependent on HCV entry. Furthermore, a mutant containing an in-frame deletion within the core gene, Jc1/GLuc2A(Δcore), replicated but was unable to produce infectious reporter virus (Fig. 1C and D). At later time points, Jc1/GLuc2A(Δcore)-transfected cells produced lower levels of GLuc2A than did WT Jc1/GLuc2A-transfected cells, most likely because this virus was unable to spread within the transfected cell population. This was confirmed by fixing the transfected cells and immunostaining for HCV NS5A over time (data not shown). Jc1/GLuc2A produced high levels of infectious HCVcc, typically, 2 × 105 to 5 × 105 TCID50/ml, which were only two- to fivefold lower than those of unmodified Jc1. Furthermore, the relative levels of GLuc activity correlated with the amount of input virus (Fig. 1E) and gave linear responses over several orders of magnitude at multiple time points (Fig. 1F).
As described below, the GLuc2A insertion was stable in unselected virus populations for up to 4 weeks. Thus, the Jc1/GLuc2A reporter virus provides a sensitive, genetically stable, and reliable method to measure relative levels of HCVcc replication and infectivity.
Mutagenesis of conserved NS2 residues.
We hypothesized that NS2 contributes to virus particle production by interacting with other molecules, most likely via conserved residues on the surface of NS2. To help identify these determinants, we conducted site-directed mutagenesis of the NS2 gene within the Jc1/GLuc2A infectious clone, targeting conserved structural features of this protein. To guide our mutagenesis strategy, we constructed an alignment of 14 NS2 genes representing seven HCV genotypes (see Materials and Methods).
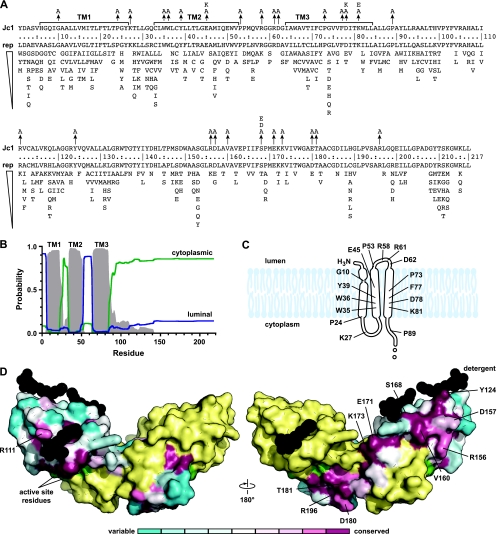
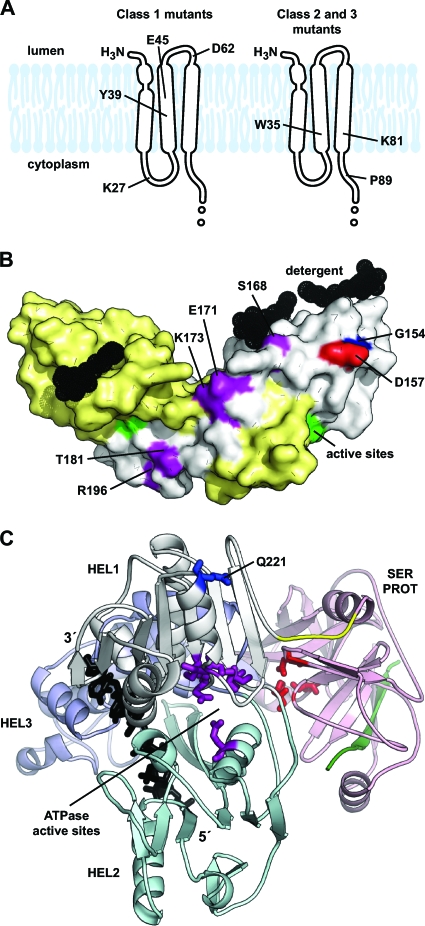
The precise membrane topology of NS2 has not yet been defined. Based on N-terminal cleavage of NS2 by the ER-resident signal peptidase and C-terminal cleavage by the cytoplasmic NS2-NS3 autoprotease (2), the hydrophobic N-terminal region of NS2 likely contains one or three TM domains. In support of this model, Jirasko et al. recently showed that NS2 residues 3 to 23 form an interrupted alpha helix that likely serves as the first TM domain (24). While putative TM domains 2 and 3 remain to be defined, we used a homology-based prediction as a working model of NS2 topology (Fig. 2A and B). A number of interesting features are apparent in this region of the protein, including several highly conserved helix-breaking (Gly and Pro) residues, charged residues (Arg, Asp, Glu, and Lys), and aromatic residues (Phe, Trp, Tyr) within the putative TM domains and loop regions. We therefore targeted these residues for mutagenesis to Ala, since this residue is least likely to disrupt protein structure. In addition, several charge reversal mutations were constructed by substituting Lys for acidic residues and Glu for basic residues.
FIG. 2.
Summary of NS2 structure and mutations. (A) The sequence of Jc1 NS2 is shown on the top line. TM domain 1 (TM1) and putative TM domains 2 and 3 (TM2 and TM3) are shown. Arrows indicate substitution mutations described in this work. Below the residue numbers is the repertoire of residues found at each position, ranked in order of prevalence. (B) NS2 membrane topology as predicted from our alignment by PolyPhobius (26). (C) A model of the NS2 TM domains with mutagenesis target sites indicated. (D) Structure of the dimeric NS2 cysteine protease domain from HCV strain H77. The solvent-excluded surface of Protein Data Bank entry 2HD0 chain A is shown colored by conservation at each residue; chain B is yellow, cysteine protease active site residues are green, and detergent molecules are black. Mutagenesis target sites are indicated on chain A.
The structure of the NS2 cysteine protease domain (residues 94 to 217) was recently solved by X-ray crystallography, revealing an unusual dimeric protease containing two composite active sites (45). Interestingly, a number of residues on the surface of NS2 are highly conserved (Fig. 2D), suggesting that they may be involved in intermolecular interactions. We therefore targeted these residues for site-directed mutagenesis to Ala. However, we did not specifically target conserved internal residues or residues involved in NS2 dimer formation or proteolysis. Since it has been proposed that phosphorylation of residue S168 regulates NS2 stability (17), we also constructed phosphomimetic Asp and Glu substitutions at this position.
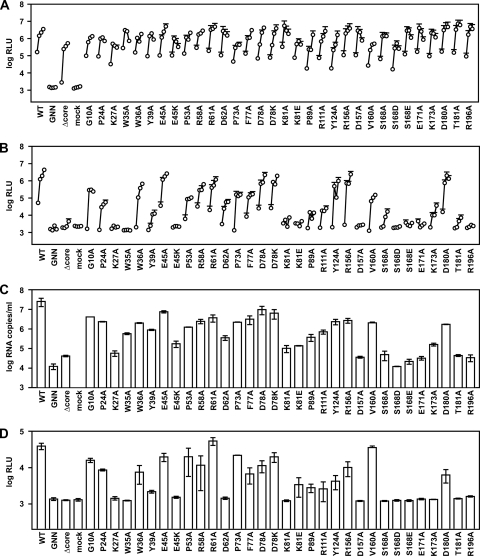
A summary of NS2 mutants is provided in Table 1. The panel of Jc1/ΔGLuc2A NS2 mutants was transfected into Huh-7.5 cells to determine their ability to replicate and produce infectious virus. As shown in Fig. 3A, all of the NS2 mutants efficiently replicated and secreted GLuc2A similar to WT Jc1/GLuc2A and Jc1/GLuc2A(Δcore), while the polymerase-defective control Jc1/GLuc2A(GNN) did not. When assayed for relative infectivity, a number of NS2 mutants showed severe defects in infectious particle production (Fig. 3B). These included the NS2 K27A, W35A, Y39A, E45K, K81A, K81E, P89A, R111A, D157A, S168A, S168D, S168E, E171A, K173A, T181A, and R196A mutants. Several mutants (G10A, P24A, W36A, P53A, R58A, D62A, P73A, F77A, and V160A) showed intermediate phenotypes, while a few NS2 mutants (E45A, R61A, D78A, D78K, Y124A, R156A, and D180A) produced high levels of infectious virus. These data are consistent with an essential role for NS2 in producing infectious virus particles and define a number of residues involved in this process.
FIG. 3.
NS2 mutant phenotypes. (A) Replication of NS2 mutants. Cells were transfected with each indicated mutant, and medium was collected at 24, 48, 72, and 96 h posttransfection. Values represent the time course of secreted GLuc activity for each transfection, expressed as an average of at least three independent transfections; error bars represent the standard deviation of the mean. For legibility, only the top error bars are shown. (B) Infectivity of NS2 mutants. Relative infectivity levels in the medium from panel A were quantified by infecting naïve cells and monitoring secreted GLuc activity as in Fig. 1. (C) RNA release of NS2 mutants. The amount of HCV RNA present in the medium at 72 h posttransfection was determined by quantitative RT-PCR as described in Materials and Methods. Values represent average RNA quantities from at least three independent transfections; error bars represent the standard deviation of the mean. (D) Intracellular infectivity of NS2 mutants. Cells were lysed at 48 h posttransfection and used to infect naive cells as described in Materials and Methods. Values represent average relative infectivity measurements from at least three independent transfections; error bars represent the standard deviation of the mean.
We considered that NS2 mutants could be blocked in releasing virus particles or may release virus particles that are not infectious. To discern between these possibilities, we measured the amount of viral RNA released into the cell culture medium, which serves as a marker for virus particles (Fig. 3C). WT Jc1/GLuc2A efficiently released HCV RNA into the cell culture medium, while Jc1/GLuc2A(GNN) and Jc1/GLuc2A(Δcore) did not. The low level of HCV RNA present in the culture medium of these mutants was due to residual RNA left over from transfection. In addition, replication-competent HCV subgenomes have been shown to release small amounts of viral RNA into the cell culture medium, independent of virus particle production (41, 59, 77). For the panel of NS2 mutants, the amount of RNA released into the cell culture medium strongly correlated with the amount of relative infectivity released into the cell culture medium (Fig. 3C). The only notable exceptions were the NS2 W35A, W39A, and R111A mutants, which appeared to secrete particles with low specific infectivity, albeit at reduced levels. These data indicate that NS2 is required prior to or at virus release.
Gastaminza et al. recently demonstrated that intracellular infectious HCVcc particles, which likely represent nascent virus particles undergoing maturation, can be released from virus-producing cells by freeze-thawing (20). To determine whether NS2 is required for the intracellular formation of virus particles or for virus particle release, we examined the relative infectivity present in the lysates of cells transfected with the panel of NS2 mutants. As shown in Fig. 3D, the intracellular relative infectivity strongly correlated with the amount of relative infectivity released into the cell culture medium. These data are consistent with a role for NS2 at an early stage of virus assembly, prior to or at the formation of intracellular infectious particles.
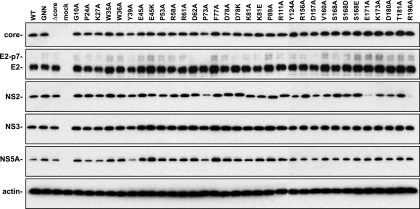
Polyprotein processing and stability of NS2 mutants.
NS2 is required for processing of the full-length polyprotein. We therefore examined whether mutations in NS2 disrupted the accumulation of NS2 or other viral proteins by Western blot analysis. Initial results with RNA-transfected cells indicated that HCV proteins were detectable by 48 h posttransfection and that by this time point, viral genomes that produced infectious particles gave rise to higher levels of HCV proteins (data not shown). Given the kinetics of Jc1/GLuc2A virus production, this increased expression could be attributed to the spread of these viruses in the transfected cell populations. Since the purpose of these studies was to analyze HCV polyprotein expression and processing irrespective of virus production, we analyzed HCV protein expression in Huh-7.5 cells infected with a vaccinia virus expressing T7 RNA polymerase and transfected with the WT or mutant HCV cDNA clones under the control of a T7 promoter. As shown in Fig. 4, there were no major differences in the accumulation of core, E2, NS3, or NS5A across our panel of NS2 mutants. One noticeable exception was the Y39A mutation, which led to a small but reproducible decrease in NS5A accumulation. A majority of the mutants produced WT levels of NS2 protein with some differences in NS2 migration in SDS-PAGE (Fig. 4). However, mutants P73A, E171A, and R196A produced reduced amounts of detectable NS2. Since it has been reported that phosphorylation of NS2 residue S168 leads to degradation by the proteasome (17), we examined whether treatment with MG132, a potent inhibitor of the proteasome, would stabilize WT or mutant forms of NS2. Treating cells with 20 μM MG132 for 4 h caused an approximately twofold increase in WT NS2 accumulation but did not stabilize the E171A or R196A mutant form of NS2 (data not shown), suggesting that these mutants were not degraded by the proteasome. Furthermore, it is notable that mutations S168A, S168D, and S168E had little effect on NS2 accumulation (Fig. 4) and were also unaffected by MG132 (data not shown).
FIG. 4.
Polyprotein processing of NS2 mutants. Parallel cultures of Huh-7.5 cells were infected with vaccinia virus vTF7-3 for 1 h, transfected with the indicated Jc1/GLuc2A cDNA clones, and lysed at 24 h postinfection. Proteins were separated by SDS-PAGE and detected by Western blotting as described in Materials and Methods. Each image is a composite of two blots run in parallel.
Selection of NS2 mutants capable of virus spread.
We reasoned that NS2 mutants unable to produce virus particles might accumulate additional mutations that restored virus particle production. Furthermore, such revertants would naturally become more abundant due to their ability to spread within transfected cell populations. We therefore transfected Huh-7.5 cells with NS2 mutants that were defective in virus production, serially passed the transfected cells, and monitored for spread of the virus within the transfected cell population by immunostaining for NS5A. Initially, cell populations were passaged in bulk. In several cases, we were able to select for viruses capable of spread within four to six cell passages. To determine the genetic basis of these events, we biologically cloned individual viruses by endpoint dilution, expanded them, and sequenced them by RT-PCR. In a few cases, the original NS2 mutation was retained and second-site changes were identified. However, in many cases, the original NS2 mutation had reverted back to the WT sequence. We reasoned that such revertants would likely dominate a virus population and hinder our ability to identify second-site suppressor mutations. We therefore modified our selection protocol, subdividing transfected cell populations into 96-well plates and serially passing them in the 96-well format in order to limit competition between different viral lineages. Viruses were then biologically cloned from positive wells and sequenced (see Materials and Methods). This strategy enhanced our ability to identify NS2 mutants that retained the original mutation and contained novel second-site changes.
During the course of our forward genetic analysis, the GLuc2A insertion was sequenced in 17 virus clones that had been selected for virus growth—but not for GLuc activity—by up to 4 weeks of continuous cell passage. The GLuc2A insertion was retained in all of the virus clones, and only a single point mutation, a silent change, was detected within the GLuc gene of one isolate. Thus, the GLuc2A insertion was stably maintained for at least several weeks of passage.
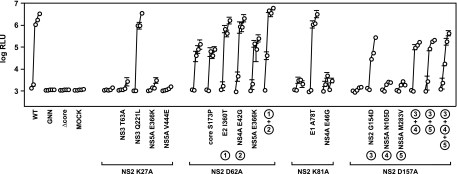
A summary of our forward genetic analysis of NS2 mutants is presented in Table 3. As noted, a large number of clones simply reverted back to the WT sequence and were not sequenced further. However, several clones retained the original mutation in NS2 and contained novel second-site changes in the core, E1, E2, NS2, NS3, NS4A, or NS5A gene. To determine whether these second-site changes were able to suppress defects in virus assembly, we reconstructed them in the context of the original NS2 mutants. As shown in Fig. 5, the second-site NS3 mutation Q221L (H77 codon 1247) fully suppressed the severe virus assembly defect caused by NS2 K27A, while the NS3 T63A, NS5A E366K, or NS5A V444E mutation did not. Similarly, the moderate defect in virus assembly caused by NS2 mutation D62A was fully suppressed by E2 mutation I360T (H77 codon 739) or NS4A mutation E42G (H77 codon 1699), either alone or in combination, but not by the core S173P or NS5A E366K mutation. The severe virus assembly defect caused by NS2 mutation K81A was fully suppressed by E1 mutation A78T (H77 codon 269) but not by NS4A mutation E46G. Finally, the severe defect in virus assembly caused by NS2 mutation D157A was partially suppressed by the intragenic mutation G154D (H77 codon 963) but not by NS5A mutation N105D or M283V, either alone or in combination with NS2 mutation G154D. All of these constructs were fully competent for RNA replication and GLuc2A expression (data not shown). These data indicate that NS2 defects could be suppressed by mutations in E1, E2, NS2, NS3, and NS4A.
TABLE 3.
Sequence analysis of NS2 mutants selected after serial passage in Huh-7.5 cells
| NS2 mutant | No. of clones sequenced | No. of revertants | Additional mutations detecteda |
|---|---|---|---|
| K27A | 2 | 0 | Clone 1, NS3 Q221L, NS5A E366K and V444E; clone 2, NS3 T63A and Q221L |
| E45A | 5 | 5 | ND |
| D62A | 2 | 0 | Clone 1, core S173P, E2 I360T, NS4A E42G; clone 2, E2 I360T, NS4A E42G, NS5A E366K |
| K81A | 2 | 1 | E1 A78T, NS4A E46G |
| K81E | 2 | 2 | ND |
| D157A | 4 | 3 | NS2 G154D, NS5A N105D and M283V |
| S168D | 13 | 13 | ND |
| S168E | 2 | 2 | ND |
| R196A | 2 | 2 | ND |
ND, not determined.
FIG. 5.
Suppressors of NS2 defects in infectious virus production. Viral infectivities were measured in medium collected at 8, 24, 48, 72, and 96 h posttransfection. Values represent average relative infectivity measurements from at least three independent transfections; error bars represent the standard deviation of the mean.
To determine whether the suppressors were general enhancers of virus production, we examined their effects on the replication and assembly of WT Jc1/GLuc2A. All of the suppressors were fully competent for RNA replication and GLuc2A expression (Fig. 6A). Furthermore, the E2 I360T, NS3 Q221L, and NS4A E42G mutations had little effect on WT virus production, while the E1 A78T and NS2 G154D mutations decreased WT virus production (Fig. 6B). These data indicate that the suppressors were not general enhancers of HCV particle production.
FIG. 6.
NS2 suppressors in the WT background. (A) Replication of Jc1/GLuc2A containing the indicated second-site changes at 8, 24, 48, 72, and 96 h posttransfection. Values represent RLU averages from at least three independent transfections; error bars represent the standard deviation of the mean. (B) The medium collected in panel A was used to infect naive cells and calculate relative infectivities as in Fig. 1. Values represent average relative infectivity measurements from at least three independent transfections; error bars represent the standard deviation of the mean. (C) NS2 mutations and suppressors function in the absence of the GLuc2A insert. Infectious virus production was measured for Jc1 NS2 mutants, which lack the GLuc2A insert, by limiting dilution of the cell culture medium harvested at 48 h posttransfection. Values represent TCID50 calculated from three independent transfections; error bars represent the standard error of the mean.
We further examined the phenotypes of NS2 mutants and their suppressors in the context of a Jc1 viral genome lacking GLuc. As shown in Fig. 6C, NS2 mutations had similar effects on infectious virus production and were suppressed in a manner similar to that of the GLuc-expressing virus. These data indicate that defects caused by the NS2 mutations, and their suppression by second-site changes in other parts of the genome, were fully reproduced outside the context of the reporter construct.
It was interesting that the NS3 Q221L mutation was identified as a suppressor of the NS2 K27A mutant, since NS3 Q221L was previously found as an adaptive mutation that enhanced the production of a chimeric H77/JFH virus in cell culture (49). We therefore wondered if this mutation would suppress other NS2 defects. Similarly, we examined whether the A78T mutation in E1, which was a strong suppressor of the NS2 K81A mutation, was a general suppressor of other NS2 defects. To test these hypotheses, the NS3 Q221L and E1 A78T mutations were reconstructed in the context of other NS2 mutants with defects in virus assembly. All double mutants replicated and efficiently expressed GLuc2A in transfected cells (data not shown).
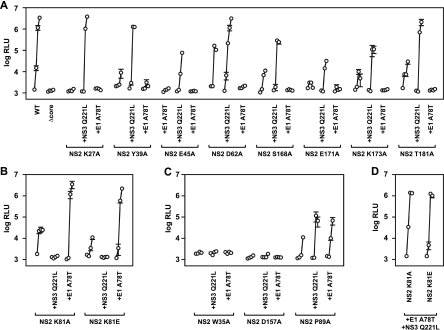
Surprisingly, the relative infectivities of the double mutants allowed us to segregate them into three phenotypic classes. (i) For class 1 NS2 mutants, defects in virus assembly were partially or fully suppressed by NS3 mutation Q221L. These included the NS2 K27A, Y39A, E45K, D62A, S168A, E171A, K173A, and T181A mutants (Fig. 7A), as well as the NS2 S168D, S168E, and R196A mutants (data not shown). Interestingly, all class 1 NS2 mutants containing the E1 A78T mutation had severe infectivity defects, indicating that E1 A78T either inhibited or did not enhance virus assembly. (ii) In class 2 NS2 mutants, both the NS2 K81A and K81E mutants were fully rescued by the E1 A78T mutation but strongly inhibited by the NS3 Q221L mutation (Fig. 7B). (iii) In class 3 NS2 mutants, the NS3 Q221L and E1 A78T mutations behaved similarly. NS2 W35A and D157A mutants were not enhanced by either NS3 Q221L or E1 A78T (Fig. 7C). Similarly, the NS2 R111A mutant was not enhanced by either second-site mutation (data not shown). Only the NS2 P89A mutant was moderately enhanced by both NS3 Q221L and E1 A78T (Fig. 7C).
FIG. 7.
Distinct epistatic interactions among NS2, E1, and NS3. Relative viral infectivities were measured in medium collected at 8, 24, 48, and 96 h posttransfection. Values represent average relative infectivity measurements from at least three independent transfections; error bars represent the standard deviation of the mean. (A) Class 1 mutants were suppressed by NS3 Q221L and inhibited by E1 A78T. (B) Class 2 mutants were suppressed by E1 A78T and inhibited by NS3 Q221L. (C) Class 3 mutants were affected by NS3 Q221L and E1 A78T equally. (D) Class 3 triple mutants containing both E1 A78T and NS3 Q221L.
Since the E1 A78T and NS3 Q221L mutations had opposite effects on many NS2 mutants, we wanted to determine if one mutation was dominant over the other. We therefore constructed NS2 K81A and K81E mutants containing both E1 A78T and NS3 Q221L. These triple mutants replicated (data not shown) and efficiently produced infectious virus (Fig. 7D), indicating that the effect of the E1 A78T mutation (suppression) was dominant over the effect of the NS3 Q221L mutation (inhibition) in the NS2 K81A and K81E mutant backgrounds. Similarly, the E1 A78T mutation phenotype (inhibited virus assembly) was dominant over the NS3 Q221L phenotype in the WT background or in several other representative class 1 NS2 mutants (data not shown).
Enzymatic activities of NS3-NS4A Q221L.
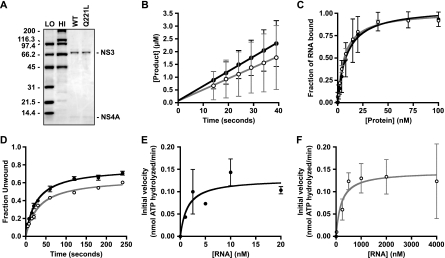
The biochemical properties of NS3 Q221L have been partially characterized in vitro by using a glutathione S-transferase-NS3 fusion protein and a peptide fragment of NS4A (49). We and others previously showed that the NS3 serine protease and RNA helicase domains coregulate each other's activity (4, 6, 18, 30), that full-length NS4A contributes to the RNA helicase activity of the NS3-NS4A holoenzyme (4), and that various affinity purification tags can influence the RNA-binding properties and subsequent activities of NS3 or NS3-NS4A (5, 6). To gain further insight into how the NS3 Q221L mutation influences biochemical properties of NS3-NS4A, we purified native, untagged WT or Q221L mutant NS3-NS4A protein complexes to characterize their enzymatic activities (Fig. 8A).
FIG. 8.
Enzymatic activities of WT and Q221L mutant NS3-NS4A. (A) The WT and Q221L mutant NS3-NS4A proteins were purified as described in Materials and Methods, and 3.75 pmol of each protein preparation was separated by 4 to 12% SDS-PAGE and stained with Coomassie brilliant blue. Bio-Rad low (LO)- and high (HI)-molecular-weight standards are shown. The values on the left are molecular sizes in kilodaltons. (B) Serine protease activities of WT (filled circles) and Q221L mutant (open circles) NS3-NS4A. (C) RNA-binding activities of WT (filled circles) and Q221L mutant (open circles) NS3-NS4A. (D) RNA helicase activities of WT (filled circles) and Q221L mutant (open circles) NS3-NS4A. (E) RNA-stimulated ATPase activity of WT NS3-NS4A. (F) RNA-stimulated ATPase activity of Q221L mutant NS3-NS4A. All values represent the average of three independent experiments; error bars represent the standard deviation of the mean. Note that for panels E and F, data were normalized by subtracting basal ATPase activities in order to examine only the RNA-stimulated ATPase activities.
As shown in Fig. 8B, both the WT and Q221L mutant NS3-NS4A proteins exhibited efficient serine protease activity against a model NS4A/4B peptide substrate in fluorescence cleavage assays. The rate constants of cleavage of WT and Q221L mutant NS3-NS4A were 58.0 ± 0.5 and 43.6 ± 0.4 nM/s, respectively. In addition, the burst kinetics indicated that the fractions of the WT and Q221L mutant NS3-NS4A proteins exhibiting serine protease activity were 95% ± 5% and 98% ± 2%, respectively. These data indicate that both protein complexes had high specific protease activity and that the Q221L mutation in NS3 had minimal effects on this activity.
We examined the RNA-binding activities of WT and Q221L mutant NS3-NS4A in filter-binding assays with a 34-nt single-stranded RNA (Fig. 8C). The two protein complexes bound this RNA with similar affinities (WT Kd = 4.0 ± 0.5 nM; Q221L mutant Kd = 9.0 ± 0.8 nM). We next examined the RNA-unwinding activities of WT and Q221L mutant NS3-NS4A by using a 34-bp model RNA duplex with a 20-nt 3′ overhang (4). Reactions were performed under single-cycle conditions by using an excess of enzyme and an unlabeled template strand “trap,” which prevented reannealing of the labeled template strand after unwinding and minimized the rebinding of NS3-NS4A that dissociated from its template strand (3). Both WT and Q221L mutant NS3-NS4A unwound the substrate to similar amplitudes (WT, 0.67 ± 0.09; Q221L mutant, 0.55 ± 0.01) and with similar rate constants (WT kobs, 0.030 ± 0.001 s−1; Q221L mutant kobs, 0.030 ± 0.002 s−1) (Fig. 8D). At lower protein concentrations, this difference in amplitude was more noticeable. For instance, at 5 nM enzyme, NS3-NS4A Q221L unwound 0.06 ± 0.01 of the RNA while WT NS3-NS4A unwound 0.24 ± 0.01 of the RNA. These data indicate that the Q221L mutation in NS3 causes a small but discernible defect in NS3-NS4A RNA helicase activity.
To further characterize the effects of the NS3 Q221L mutation, we examined the RNA-stimulated ATPase activities of WT and Q221L mutant NS3-NS4A complexes. This assay measures functional RNA binding, i.e., the binding of single-stranded RNA that stimulates ATPase activity. Based on the initial velocity of ATP hydrolysis versus the RNA concentration, WT NS3-NS4A reached its maximal ATP hydrolysis at much lower concentrations of RNA than did NS3-NS4A containing the Q221L mutation (Fig. 8E and F). For WT NS3-NS4A, the Km for RNA-stimulated ATPase activity was 2.0 nM, while for Q221L mutant NS3-NS4A, the Km was 200 nM (Table 4). These results indicate that the Q221L mutation lowered the efficiency with which RNA binding led to stimulation of ATPase activity. The rate constants of basal ATPase activity for WT and Q221L NS3-NS4A were 0.13 ± 0.03 and 0.11 ± 0.02 nmol ATP/s, respectively, indicating that the Q221L mutation had no effect on the basal ATPase activity. Furthermore, once sufficient RNA was added, it stimulated the WT and Q221L mutant NS3-NS4A ATPase activities to similar extents, 2.2- and 2.4-fold, respectively (Table 4). These data indicate that while the NS3 Q221L mutation lowered the affinity of functional RNA binding, it did not alter the inherent ATPase activity of NS3-NS4A.
TABLE 4.
RNA-stimulated NTPase activities of WT and Q221L mutant NS3-NS4Aa
| NS3-NS4A protein | Km (nM) | Vmaxb | kcatc | Catalytic efficiencyd | RNA stimulation factor |
|---|---|---|---|---|---|
| WT | 2.0 ± 1.0 | 0.29 ± 0.03 | 0.16 ± 0.03 | 80 | 2.2 |
| Q221L | 200 ± 60 | 0.26 ± 0.01 | 0.15 ± 0.01 | 0.8 | 2.4 |
The data shown are averages ± standard deviations or averages from three experiments.
Values were calculated before normalization and are expressed as nanomoles of ATP hydrolyzed per minute.
Values are expressed as nanomoles of ATP per minute per picomole of NS3-NS4A.
Values were calculated as kcat/Km and are expressed as nanomoles of ATP hydrolyzed per minute per picomole of protein per μM of RNA.
DISCUSSION
The first evidence that NS2 contributes to HCV particle assembly came from Pietschmann et al., who showed that the Jc1 chimeric junction within NS2 coordinately increased virus titers and core release from HCVcc-producing cells (58). One implication of their data is that the first TM domain of NS2 functions optimally with the viral structural proteins, while downstream regions of NS2 sequences function optimally with other regions of the viral genome. Starting from the Jc1 background, our mutagenesis revealed several key residues in NS2 whose mutation inhibited virus production (Fig. 3B to D). Furthermore, we identified several second-site changes in the E1, E2, NS2, NS3, and NS4A genes that could suppress NS2 defects in virus assembly (Fig. 5 to 7). Two of these suppressors, NS3 Q221L and E1 A78T, were characterized further and revealed additional layers of genetic interaction. Taken together, our data indicate that NS2 contributes to virus assembly in concert with the E1-E2 glycoprotein and NS3-NS4A enzyme complexes.
The NS3 Q221L suppressor mutation.
The NS3 Q221L mutation was initially selected in the NS2 K27A mutant background (Table 3 and Fig. 5) but also suppressed assembly defects in other class 1 NS2 mutants (Fig. 7). The class 1 NS2 mutations were not clustered but included residues within the putative TM domain and a discontinuous strip of residues on the surface of the cysteine protease domain (Fig. 9A and B). Thus, it is doubtful that these residues directly interact with NS3 residue 221. The NS3 Q221L mutation was previously shown to act as a cell culture-adaptive mutation that enhanced virus particle production by a genotype 1a/2a chimeric genome (49). Given that this chimera contained the core-NS2 region of H77S, it is tempting to speculate that NS3 Q221L might simultaneously compensate for multiple genetic differences in NS2.
FIG. 9.
Summary of NS2 suppressors. (A) NS2 TM domain target sites that were suppressed by NS3 Q221L (left) and E1 A78T (right). (B) NS2 cysteine protease domain target sites that were suppressed by NS3 Q221L (purple) and NS2 G154D (red). The location of NS2 G154 is shown in blue. NS2 chain A is white, chain B is yellow, and detergent molecules are black. (C) Ribbon model of NS3 (see Materials and Methods) showing the serine protease domain (SER PROT) in pink, the protease-helicase linker in yellow, and helicase domains (HEL1, HEL2, and HEL3) in white, cyan, and blue, respectively. The NS4A cofactor peptide is green, serine protease active site residues are red, ATPase active site residues are purple, nucleic acid is black, and NS3 Q221 is blue.
To understand how the Q221L mutation affected the biochemical properties of NS3-NS4A, we purified untagged native WT and mutant NS3-NS4A and characterized their enzymatic activities. To our knowledge, this is the first characterization of native untagged NS3-NS4A of strain JFH1. Overall, WT NS3-NS4A exhibited biochemical properties similar to those of genotype 1b HCV-N and genotype 1a HCV-H77 NS3-NS4A (4-6). The NS3 Q221L mutation had only minimal effects on the serine protease, RNA binding, and basal ATPase activities of NS3-NS4A (Fig. 8). These results were in agreement with those of Ma et al., who previously characterized the NS3 Q221L mutation in the context of a glutathione S-transferase-tagged NS3 protein containing the central cofactor peptide of NS4A (49). However, we also found that NS3 Q221L caused a small but detectable decrease in RNA unwinding and a strong decrease in functional RNA binding, i.e., RNA binding that stimulated NS3-NS4A ATPase activity (Fig. 8E and F). This may seem counterintuitive, since the ATPase activity provides the power to drive RNA helicase activity, which was close to normal. However, the NS3 Q221L mutation did not affect the levels of basal or stimulated ATPase activity, only the amount of RNA needed to achieve this stimulation. Moreover, we previously noted that other replication-competent forms of NS3-NS4A exhibit a range of RNA-stimulated ATPase Km values (4; R. K. F. Beran, unpublished data). Finally, we cannot rule out the possibility of a larger effect of the NS3 Q221L mutation on helicase activity at low RNA concentrations, since an excess of unlabeled top-strand RNA was required to perform unwinding assays under single-cycle conditions. Nevertheless, our data demonstrate that NS3 Q221L alters the RNA binding of NS3-NS4A in a functionally significant manner.
NS3 Q221 (H77 codon 1247) is highly conserved, with 866 of 868 NS3 sequences in the euHCVdb database encoding Gln at this position. On a structural model of NS3-NS4A with a bound tracking strand, Q221 is predicted to lie on the surface of helicase subdomain 1, distal to the major RNA-binding groove and ATPase-active sites (Fig. 9C). Thus, it is not obvious how the Q221L mutation affects functional RNA binding. It is notable, however, that NS3 residue Q221 resides at one end of an alpha helix, the opposite end of which is the ATP-binding Walker A motif-containing P loop. In crystal structures of the related dengue virus NS3 helicase domain obtained in the presence and absence of RNA ligand, RNA binding helps to order the ATP-binding site and actually tugs on this alpha helix (47). Thus, NS3 Q221 may help the RNA-stimulated ordering of the ATP-binding pocket through an allosteric effect transmitted along this helix. It is also notable that two additional cell culture-adaptive mutations in the NS3 helicase domain (I286V and I399V) could functionally substitute for Q221L and restore H77S/JFH1 virus assembly (49). These residues face into the RecA-like folds of helicase subdomains 1 and 2, respectively, further suggesting that conformational differences in helicase structure can impact virus assembly.
It is not yet clear whether the decrease in functional RNA binding observed in the NS3 Q221L mutant directly relates to virus assembly or whether it is a side effect of altered NS3-NS4A structure. It is relevant to note that other RNA viruses utilize virus-encoded helicases/NTPases during genome packaging. Helicases/NTPases play a role in packaging eukaryotic and bacterial double-stranded RNA virus genomes into preformed capsids (60, 72). Furthermore, the poliovirus 2C protein has NTPase activity and has been implicated in virus assembly (39, 74).
Other members of the Flaviviridae family utilize an NS3 serine protease-NTPase/RNA helicase in concert with an NS2 ortholog during virus assembly. Similar to HCV, the pestiviruses encode an NS2 cysteine protease and an NS3 serine protease-RNA helicase that utilizes an NS4A cofactor (43). Pestivirus particle assembly requires an uncleaved form of NS2-NS3, although the enzymatic activities encoded by NS2-NS3 are dispensable for this function (1, 51). Among the flaviviruses, NS3 is also a serine protease-RNA helicase, but it utilizes an upstream protein, NS2B, as a serine protease cofactor (43). Although they are not similar to HCV NS2, the yellow fever virus (YFV) and Kunjin virus NS2A proteins are multi-TM-domain proteins required for virus assembly (32, 38). In the case of YFV, defects in NS2A can be suppressed by second-site changes in the helicase domain of NS3 (32). Further genetic analysis confirmed that a packaging function of YFV NS3 can be supplied in trans, independent of its serine protease and RNA helicase activities (56). However, in this case, the YFV replicon still encoded an active RNA helicase. Thus, it appears that functional links between NS3 and NS2 or an NS2-like protein are conserved among the members of the family Flaviviridae, although it remains unclear whether NS3 NTPase/RNA helicase activity is involved in virus assembly.
The E1 A78T suppressor mutation.
The E1 A78T suppressor mutation strongly enhanced NS2 K81A and K81E mutant virus assembly and partially suppressed that of the NS2 P89A mutant (Fig. 7 and 9A). The suppressive effects of this mutation were highly specific, inhibiting the infectivity of WT virus and several class 1 NS2 mutants (Fig. 6B and 7). E1 A78 (H77 codon 269) is highly conserved, containing Ala in 1,728 of the 1,742 HCV sequences in the euHCVdb sequence database. Furthermore, this position is located within a conserved hydrophobic region (H77 codons 262 to 290) that is postulated to act as a fusion peptide. Flint and colleagues first noted that this region shows limited sequence similarity to fusion peptides of the flavivirus E and paramyxovirus F proteins (16). Furthermore, this region can induce membrane permeability in E. coli and model membrane systems (9, 57). Mutations within this region were shown to inhibit the entry or fusion of HCVcc and HCV-pseudotyped retrovirus particles (12, 37, 63). However, other mutations in this region inhibited the incorporation of properly folded and glycosylated E1-E2 heterodimers into HCV-pseudotyped retrovirus particles, suggesting that this region may also be involved in stabilizing protein folding or heterodimer formation (37, 63). Our data indicate a functional link between this region of E1 and NS2 in mediating virus assembly, which could involve either direct or indirect interactions. While NS2 K81 lies within the third putative TM domain and P89A lies within the cytoplasm, it has been proposed that E1 may adopt alternate topologies such that the internal hydrophobic region makes intramembrane contact with core protein (48, 50, 54). Such models await rigorous examination with a functional virus replication system.
The E2 I360T, NS4A E42G, and NS2 G154D suppressor mutations.
The NS2 D62A mutant exhibited a moderate defect in virus assembly and expressed a class 1 suppression phenotype (Fig. 7A), being enhanced and inhibited by the NS3 Q221L and E1 A78T mutations, respectively. In addition, this mutant was also suppressed by second-site mutation E2 I360T or NS4A E42G. Individually, E2 I360T or NS4A E42G each restored virus assembly to near-WT levels (Fig. 5 and 6C), but neither enhanced WT virus assembly (Fig. 6B), suggesting that they were not simply cell culture-adaptive mutations. When combined, these mutations restored virus assembly to levels even slightly above that of the WT (Fig. 5 and 6C), further suggesting functional links between NS2 and the HCV glycoprotein and NS3-NS4A enzyme complexes.
E2 residue I360 (H77 codon 739) lies within the C-terminal TM domain, downstream of the central region that mediates E1-E2 interaction (10, 11, 55). Thus, this residue could potentially interact with the NS2 TM domains. NS4A residue E42 (H77 codon 1699) lies in the C-terminal acidic domain of NS4A. Our prior mutational analysis of NS4A indicated that E1699 was tolerant of Ala substitution in the context of the Con1 genotype 1b subgenomic replicon (42). However, mutation of nearby residues caused major defects in viral genome replication and NS5A hyperphosphorylation, and several NS4A defects were suppressed by second-site mutations in NS3. Additional experiments indicated that the C-terminal region of NS4A contributes to functional RNA binding and NS3-NS4A RNA helicase activity (4). Taken together, these data further support the model in which interactions among NS2, the viral glycoproteins, and the NS3-NS4A enzyme complex contribute to virus assembly.
The NS2 D157A mutant exhibited a severe assembly defect that was not suppressed by either the E1 A78T or the NS3 Q221L mutation (Fig. 7C). However, this defect was partially suppressed by a nearby mutation in NS2, G154D. Given the similar location of these residues in the membrane-proximal region of the NS2 cysteine protease domain (Fig. 9B), there appears to be a preference for a negatively charged residue on this surface. One possibility is that NS2 D157 needs to interact with a positively charged surface on another protein or polar lipid head groups at the membrane surface. This could account for the inhibitory effect of the NS2 G154D mutation on the WT virus (Fig. 6B). Similarly, another class 3 mutation, NS2 R111A, is well positioned to interact with the polar head group of lipids (Fig. 2D).
Other notable NS2 mutants.
It was surprising that the NS2 G10A mutation had only a moderate effect on virus assembly, since Jirasko et al. recently showed that this mutation caused severe assembly defects in JFH1 or a Con1/JFH1 chimera (24). Thus, the difference in genetic background could explain our different findings. In addition, these authors also noted inefficient signal peptidase cleavage at the p7/NS2 junction in the G10A mutant. We did not observe a difference in N-terminal processing for the NS2 G10A mutant (Fig. 4 and data not shown). However, in the context of our Jc1/GLuc2A reporter, this cleavage is mediated by the FMDV 2A “autocleaving” peptide. Thus, inefficient N-terminal processing of NS2 may have also contributed to the severe phenotype of the G10A mutant observed by Jirasko et al.
Franck et al. showed that NS2 from genotype 1 viruses is rapidly degraded by the proteasome and that this degradation is dependent on phosphorylation of NS2 residue S168 by casein kinase II (17). In our chimeric genotype 2a culture system, the NS2 S168A, S168D, and S168E mutants showed strong-to-severe defects in virus assembly (Fig. 3 and 7). Our data are in agreement with those of two other groups, who recently showed that the NS2 S168A mutation inhibits virus assembly (24, 83). However, we did not detect differences in the stability of these mutant NS2 proteins, with or without a proteasome inhibitor (Fig. 4 and data not shown). Furthermore, J6 NS2 stability was unaffected by the casein kinase II inhibitor 2-dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole (73). Thus, NS2 S168 does not appear to be a major determinant of NS2 stability in the J6 and JFH1 genetic backgrounds.
We did, however, note lower NS2 accumulation with the NS2 P73A, E171A, and R196A mutants (Fig. 4 and data not shown). Since the NS2 E171A and R196A defects in virus assembly were suppressed by NS3 Q221L, we examined whether these mutant forms of NS2 were stabilized by the NS3 Q221L mutation. However, NS3 Q221L did not stabilize NS2 E171A or NS2 R196A (data not shown). This could indicate that the NS3 Q221L mutation allows virus assembly to occur even when NS2 concentrations are limiting. Alternatively, it is notable that the R196A mutation lies immediately adjacent to the epitope determined for the anti-NS2 monoclonal antibody used in our studies (Dentzer et al., unpublished). Thus, we cannot, at present, discern whether NS2 D196A was unstable or simply undetectable with this reagent.
Although NS2 residue S168 does not appear to regulate NS2 stability in genotype 2a virus culture systems, phosphorylation at this site could serve to regulate NS2 function. As noted previously, residue S168 appears to face the membrane (Fig. 9B) and phosphorylation of this site would likely destabilize interaction of the cysteine protease domain with cellular membranes (24). The NS2 S168D and S168E mutations, which were designed to mimic S168 phosphorylation, caused severe defects in virus assembly. Thus, phosphorylation of NS2 at residue S168, if it occurs, likely serves to inhibit virus assembly. In this regard, it is notable that Ser is highly conserved at NS2 position 168 (H77 codon 977) for all HCV strains in the euHCVdb database, except for genotype 4a viruses, which encode Thr at this position. Interestingly, NS2 mutation T168S was identified as a cell culture-adaptive mutation for a genotype 4a/2a chimeric virus, ED43/JFH1 (68), further suggesting that NS2 residue 168 may be in epistasis with downstream NS sequences.
Positive and negative epistatic interactions among NS2, the viral glycoproteins, and NS3-NS4A.
One striking observation was that the NS3 Q221L and E1 A78T mutations enhanced or inhibited virus production in different genetic backgrounds. Specifically, the NS3 Q221L mutation, which enhanced virus production of the class 1 NS2 mutants, inhibited residual virus assembly in the class 2 NS2 mutants (Fig. 7). Conversely, the E1 A78T mutation, which enhanced virus production of the class 2 NS2 mutants, inhibited WT virus assembly, as well as residual virus assembly in the class 1 NS2 mutants (Fig. 6 and 7). These data indicate that both positive and negative epistatic interactions occur among NS2, E1, and NS3. One possibility is that NS2 may function during virus assembly by bringing together viral envelope glycoprotein complexes and replicase complexes through separate sets of interactions. Thus, the antagonism we observed between some sequence combinations (e.g., NS2 K81A and NS3 Q221L) may reflect the strengthening of one set of interactions at the expense of the other.
Our genetic data demonstrate that functional interactions occur among NS2, E1-E2, and NS3-NS4A during virus assembly. These data imply that there may be direct or indirect physical interactions among these protein complexes that await further inquiry. In addition, NS2 has been reported to interact with cellular proteins Hsp90 and CIDE-B, which could contribute to virus assembly (13, 78). Our panel of NS2 mutants and their suppressors provide powerful new tools to help dissect NS2 interactions and their role in virus assembly.
Acknowledgments
We thank J. Bloom, N. Counihan, Y. Modis, A. M. Pyle, and P. Turner for helpful discussions and comments on the manuscript. We thank C. M. Rice for providing reagents and many enjoyable years of mentorship, C. Jones for advice on GLuc construct design, J. Bukh for the pJ6CF plasmid, and J. McKeating for providing the anti-E2 3/11 antibody.
This work was funded through U.S. Public Health Service grant KO1 CA107092 (to B.D.L.); an Edward Mallinckrodt, Jr., Foundation award (to B.D.L.); and a Kingsley Fellowship in Medical Research (to B.D.L.). T.P. was funded by the Anna Fuller Foundation Postdoctoral Research Program in Infectious Causes of Cancer. R.K.F.B. was a Gilead Sciences postdoctoral fellow.
Footnotes
Published ahead of print on 10 June 2009.
REFERENCES
- 1.Agapov, E. V., C. L. Murray, I. Frolov, L. Qu, T. M. Myers, and C. M. Rice. 2004. Uncleaved NS2-3 is required for production of infectious bovine viral diarrhea virus. J. Virol. 782414-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartenschlager, R., M. Frese, and T. Pietschmann. 2004. Novel insights into hepatitis C virus replication and persistence. Adv. Virus Res. 6371-180. [DOI] [PubMed] [Google Scholar]
- 3.Beran, R. K., M. M. Bruno, H. A. Bowers, E. Jankowsky, and A. M. Pyle. 2006. Robust translocation along a molecular monorail: the NS3 helicase from hepatitis C virus traverses unusually large disruptions in its track. J. Mol. Biol. 358974-982. [DOI] [PubMed] [Google Scholar]
- 4.Beran, R. K., B. D. Lindenbach, and A. M. Pyle. 2009. The NS4A protein of hepatitis C virus promotes RNA-coupled ATP hydrolysis by the NS3 helicase. J. Virol. 833268-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beran, R. K., and A. M. Pyle. 2008. Hepatitis C viral NS3-4A protease activity is enhanced by the NS3 helicase. J. Biol. Chem. 28329929-29937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beran, R. K., V. Serebrov, and A. M. Pyle. 2007. The serine protease domain of hepatitis C viral NS3 activates RNA helicase activity by promoting the binding of RNA substrate. J. Biol. Chem. 28234913-34920. [DOI] [PubMed] [Google Scholar]
- 7.Bernsel, A., H. Viklund, J. Falk, E. Lindahl, G. von Heijne, and A. Elofsson. 2008. Prediction of membrane-protein topology from first principles. Proc. Natl. Acad. Sci. USA 105:7177-7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blight, K. J., J. A. McKeating, and C. M. Rice. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 7613001-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciccaglione, A. R., A. Costantino, C. Marcantonio, M. Equestre, A. Geraci, and M. Rapicetta. 2001. Mutagenesis of hepatitis C virus E1 protein affects its membrane-permeabilizing activity. J. Gen. Virol. 822243-2250. [DOI] [PubMed] [Google Scholar]
- 10.Ciczora, Y., N. Callens, C. Montpellier, B. Bartosch, F. L. Cosset, A. Op de Beeck, and J. Dubuisson. 2005. Contribution of the charged residues of hepatitis C virus glycoprotein E2 transmembrane domain to the functions of the E1E2 heterodimer. J. Gen. Virol. 86:2793-2798. [DOI] [PubMed] [Google Scholar]
- 11.Ciczora, Y., N. Callens, F. Penin, E. I. Pecheur, and J. Dubuisson. 2007. Transmembrane domains of hepatitis C virus envelope glycoproteins: residues involved in E1E2 heterodimerization and involvement of these domains in virus entry. J. Virol. 812372-2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drummer, H. E., I. Boo, and P. Poumbourios. 2007. Mutagenesis of a conserved fusion peptide-like motif and membrane-proximal heptad-repeat region of hepatitis C virus glycoprotein E1. J. Gen. Virol. 881144-1148. [DOI] [PubMed] [Google Scholar]
- 13.Erdtmann, L., N. Franck, H. Lerat, J. Le Seyec, D. Gilot, I. Cannie, P. Gripon, U. Hibner, and C. Guguen-Guillouzo. 2003. The hepatitis C virus NS2 protein is an inhibitor of CIDE-B-induced apoptosis. J. Biol. Chem. 27818256-18264. [DOI] [PubMed] [Google Scholar]
- 14.Failla, C., L. Tomei, and R. De Francesco. 1994. Both NS3 and NS4A are required for proteolytic processing of hepatitis C virus nonstructural proteins. J. Virol. 683753-3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flint, M., C. Maidens, L. D. Loomis-Price, C. Shotton, J. Dubuisson, P. Monk, A. Higginbottom, S. Levy, and J. A. McKeating. 1999. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J. Virol. 736235-6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flint, M., J. M. Thomas, C. M. Maidens, C. Shotton, S. Levy, W. S. Barclay, and J. A. McKeating. 1999. Functional analysis of cell surface-expressed hepatitis C virus E2 glycoprotein. J. Virol. 736782-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franck, N., J. Le Seyec, C. Guguen-Guillouzo, and L. Erdtmann. 2005. Hepatitis C virus NS2 protein is phosphorylated by the protein kinase CK2 and targeted for degradation to the proteasome. J. Virol. 792700-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frick, D. N., R. S. Rypma, A. M. Lam, and B. Gu. 2004. The nonstructural protein 3 protease/helicase requires an intact protease domain to unwind duplex RNA efficiently. J. Biol. Chem. 2791269-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 838122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gastaminza, P., S. B. Kapadia, and F. V. Chisari. 2006. Differential biophysical properties of infectious intracellular and secreted hepatitis C virus particles. J. Virol. 8011074-11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gottwein, J. M., and J. Bukh. 2008. Cutting the Gordian knot—development and biological relevance of hepatitis C virus cell culture systems. Adv. Virus Res. 7151-133. [DOI] [PubMed] [Google Scholar]
- 22.Gottwein, J. M., T. K. Scheel, T. B. Jensen, J. B. Lademann, J. C. Prentoe, M. L. Knudsen, A. M. Hoegh, and J. Bukh. 2009. Development and characterization of hepatitis C virus genotype 1-7 cell culture systems: role of CD81 and scavenger receptor class B type I and effect of antiviral drugs. Hepatology 49364-377. [DOI] [PubMed] [Google Scholar]
- 23.Jensen, T. B., J. M. Gottwein, T. K. Scheel, A. M. Hoegh, J. Eugen-Olsen, and J. Bukh. 2008. Highly efficient JFH1-based cell-culture system for hepatitis C virus genotype 5a: failure of homologous neutralizing-antibody treatment to control infection. J. Infect. Dis. 1981756-1765. [DOI] [PubMed] [Google Scholar]
- 24.Jirasko, V., R. Montserret, N. Appel, A. Janvier, L. Eustachi, C. Brohm, E. Steinmann, T. Pietschmann, F. Penin, and R. Bartenschlager. 2008. Structural and functional characterization of non-structural protein 2 for its role in hepatitis C virus assembly. J. Biol. Chem. 28328546-28562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones, C. T., C. L. Murray, D. K. Eastman, J. Tassello, and C. M. Rice. 2007. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J. Virol. 818374-8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Käll, L., A. Krogh, and E. L. Sonnhammer. 2005. An HMM posterior decoder for sequence feature prediction that includes homology information. Bioinformatics 21(Suppl. 1)i251-i257. [DOI] [PubMed] [Google Scholar]
- 27.Kato, T., Y. Choi, G. Elmowalid, R. K. Sapp, H. Barth, A. Furusaka, S. Mishiro, T. Wakita, K. Krawczynski, and T. J. Liang. 2008. Hepatitis C virus JFH-1 strain infection in chimpanzees is associated with low pathogenicity and emergence of an adaptive mutation. Hepatology 48732-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, J. L., K. A. Morgenstern, J. P. Griffith, M. D. Dwyer, J. A. Thomson, M. A. Murcko, C. Lin, and P. R. Caron. 1998. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure 689-100. [DOI] [PubMed] [Google Scholar]
- 29.Kolykhalov, A. A., K. Mihalik, S. M. Feinstone, and C. M. Rice. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 742046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuang, W. F., Y. C. Lin, F. Jean, Y. W. Huang, C. L. Tai, D. S. Chen, P. J. Chen, and L. H. Hwang. 2004. Hepatitis C virus NS3 RNA helicase activity is modulated by the two domains of NS3 and NS4A. Biochem. Biophys. Res. Commun. 317211-217. [DOI] [PubMed] [Google Scholar]
- 31.Kuiken, C., C. Combet, J. Bukh, I. T. Shin, G. Deleage, M. Mizokami, R. Richardson, E. Sablon, K. Yusim, J. M. Pawlotsky, and P. Simmonds. 2006. A comprehensive system for consistent numbering of HCV sequences, proteins and epitopes. Hepatology 441355-1361. [DOI] [PubMed] [Google Scholar]
- 32.Kümmerer, B. M., and C. M. Rice. 2002. Mutations in the yellow fever virus nonstructural protein NS2A selectively block production of infectious particles. J. Virol. 764773-4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lam, A. M., and D. N. Frick. 2006. Hepatitis C virus subgenomic replicon requires an active NS3 RNA helicase. J. Virol. 80404-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landau, M., I. Mayrose, Y. Rosenberg, F. Glaser, E. Martz, T. Pupko, and N. Ben-Tal. 2005. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 33W299-W302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 232947-2948. [DOI] [PubMed] [Google Scholar]
- 36.Lavie, M., A. Goffard, and J. Dubuisson. 2007. Assembly of a functional HCV glycoprotein heterodimer. Curr. Issues Mol. Biol. 971-86. [PubMed] [Google Scholar]
- 37.Lavillette, D., E. I. Pecheur, P. Donot, J. Fresquet, J. Molle, R. Corbau, M. Dreux, F. Penin, and F. L. Cosset. 2007. Characterization of fusion determinants points to the involvement of three discrete regions of both E1 and E2 glycoproteins in the membrane fusion process of hepatitis C virus. J. Virol. 818752-8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leung, J. Y., G. P. Pijlman, N. Kondratieva, J. Hyde, J. M. Mackenzie, and A. A. Khromykh. 2008. Role of nonstructural protein NS2A in flavivirus assembly. J. Virol. 824731-4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li, J. P., and D. Baltimore. 1990. An intragenic revertant of a poliovirus 2C mutant has an uncoating defect. J. Virol. 641102-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lindenbach, B. D. 2009. Measuring HCV infectivity produced in cell culture and in vivo. Methods Mol. Biol. 510329-336. [DOI] [PubMed] [Google Scholar]
- 41.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309623-626. [DOI] [PubMed] [Google Scholar]
- 42.Lindenbach, B. D., B. M. Pragai, R. Montserret, R. K. Beran, A. M. Pyle, F. Penin, and C. M. Rice. 2007. The C terminus of hepatitis C virus NS4A encodes an electrostatic switch that regulates NS5A hyperphosphorylation and viral replication. J. Virol. 818905-8918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lindenbach, B. D., H. J. Thiel, and C. M. Rice. 2007. Flaviviridae: the viruses and their replication, p. 1101-1152. In D. M. Knipe and P. M. Howley (ed.), Fields virology, fifth ed., vol. 1. Lippincott-Raven Publishers, Philadelphia, PA. [Google Scholar]
- 44.Lohmann, V., F. Korner, J. O. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285110-113. [DOI] [PubMed] [Google Scholar]
- 45.Lorenz, I. C., J. Marcotrigiano, T. G. Dentzer, and C. M. Rice. 2006. Structure of the catalytic domain of the hepatitis C virus NS2-3 protease. Nature 442831-835. [DOI] [PubMed] [Google Scholar]
- 46.Love, R. A., H. Parge, J. A. Wickersham, Z. Hostomsky, N. Habuka, E. W. Moomaw, T. Adachi, and Z. Hostomska. 1996. The crystal structure of hepatitis C virus NS3 proteinase reveals a trypsin-like fold and a structural zinc binding site. Cell 87331-342. [DOI] [PubMed] [Google Scholar]
- 47.Luo, D., T. Xu, C. Hunke, G. Gruber, S. G. Vasudevan, and J. Lescar. 2008. Crystal structure of the NS3 protease-helicase from dengue virus. J. Virol. 82173-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma, H. C., C. H. Ke, T. Y. Hsieh, and S. Y. Lo. 2002. The first hydrophobic domain of the hepatitis C virus E1 protein is important for interaction with the capsid protein. J. Gen. Virol. 833085-3092. [DOI] [PubMed] [Google Scholar]
- 49.Ma, Y., J. Yates, Y. Liang, S. M. Lemon, and M. Yi. 2008. NS3 helicase domains involved in infectious intracellular hepatitis C virus particle assembly. J. Virol. 827624-7639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Migliaccio, C. T., K. E. Follis, Y. Matsuura, and J. H. Nunberg. 2004. Evidence for a polytopic form of the E1 envelope glycoprotein of hepatitis C virus. Virus Res. 10547-57. [DOI] [PubMed] [Google Scholar]
- 51.Moulin, H. R., T. Seuberlich, O. Bauhofer, L. C. Bennett, J. D. Tratschin, M. A. Hofmann, and N. Ruggli. 2007. Nonstructural proteins NS2-3 and NS4A of classical swine fever virus: essential features for infectious particle formation. Virology 365376-389. [DOI] [PubMed] [Google Scholar]
- 52.Murray, C. L., C. T. Jones, and C. M. Rice. 2008. Architects of assembly: roles of Flaviviridae non-structural proteins in virion morphogenesis. Nat. Rev. Microbiol. 6699-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murray, C. L., C. T. Jones, J. Tassello, and C. M. Rice. 2007. Alanine scanning of the hepatitis C virus core protein reveals numerous residues essential for production of infectious virus. J. Virol. 8110220-10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakai, K., T. Okamoto, T. Kimura-Someya, K. Ishii, C. K. Lim, H. Tani, E. Matsuo, T. Abe, Y. Mori, T. Suzuki, T. Miyamura, J. H. Nunberg, K. Moriishi, and Y. Matsuura. 2006. Oligomerization of hepatitis C virus core protein is crucial for interaction with the cytoplasmic domain of E1 envelope protein. J. Virol. 8011265-11273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Op De Beeck, A., R. Montserret, S. Duvet, L. Cocquerel, R. Cacan, B. Barberot, M. Le Maire, F. Penin, and J. Dubuisson. 2000. The transmembrane domains of hepatitis C virus envelope glycoproteins E1 and E2 play a major role in heterodimerization. J. Biol. Chem. 27531428-31437. [DOI] [PubMed] [Google Scholar]
- 56.Patkar, C. G., and R. J. Kuhn. 2008. Yellow fever virus NS3 plays an essential role in virus assembly independent of its known enzymatic functions. J. Virol. 823342-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pérez-Berná, A. J., M. R. Moreno, J. Guillén, A. Bernabeu, and J. Villalaín. 2006. The membrane-active regions of the hepatitis C virus E1 and E2 envelope glycoproteins. Biochemistry 453755-3768. [DOI] [PubMed] [Google Scholar]
- 58.Pietschmann, T., A. Kaul, G. Koutsoudakis, A. Shavinskaya, S. Kallis, E. Steinmann, K. Abid, F. Negro, M. Dreux, F. L. Cosset, and R. Bartenschlager. 2006. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc. Natl. Acad. Sci. USA 1037408-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pietschmann, T., V. Lohmann, A. Kaul, N. Krieger, G. Rinck, G. Rutter, D. Strand, and R. Bartenschlager. 2002. Persistent and transient replication of full-length hepatitis C virus genomes in cell culture. J. Virol. 764008-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Poranen, M. M., R. Tuma, and D. H. Bamford. 2005. Assembly of double-stranded RNA bacteriophages. Adv. Virus Res. 6415-43. [DOI] [PubMed] [Google Scholar]
- 61.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent end points. Am. J. Hyg. 27493-497. [Google Scholar]
- 62.Reynolds, S. M., L. Kall, M. E. Riffle, J. A. Bilmes, and W. S. Noble. 2008. Transmembrane topology and signal peptide prediction using dynamic Bayesian networks. PLoS Comput. Biol. 4:e1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Russell, R. S., K. Kawaguchi, J. C. Meunier, S. Takikawa, K. Faulk, J. Bukh, R. H. Purcell, and S. U. Emerson. 13 March 2009. Mutational analysis of the hepatitis C virus E1 glycoprotein in retroviral pseudoparticles and cell-culture-derived H77/JFH1 chimeric infectious virus particles. J. Viral Hepatitis [Epub ahead of print.] doi: 10.1111/j.1365-2893.2009.01111.x. [DOI] [PMC free article] [PubMed]
- 64.Russell, R. S., J. C. Meunier, S. Takikawa, K. Faulk, R. E. Engle, J. Bukh, R. H. Purcell, and S. U. Emerson. 2008. Advantages of a single-cycle production assay to study cell culture-adaptive mutations of hepatitis C virus. Proc. Natl. Acad. Sci. USA 1054370-4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saito, T., and M. Gale, Jr. 2008. Regulation of innate immunity against hepatitis C virus infection. Hepatol. Res. 38:115-122. [DOI] [PubMed] [Google Scholar]
- 66.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, third ed., vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 67.Santolini, E., L. Pacini, C. Fipaldini, G. Migliaccio, and N. Monica. 1995. The NS2 protein of hepatitis C virus is a transmembrane polypeptide. J. Virol. 697461-7471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Scheel, T. K., J. M. Gottwein, T. B. Jensen, J. C. Prentoe, A. M. Hoegh, H. J. Alter, J. Eugen-Olsen, and J. Bukh. 2008. Development of JFH1-based cell culture systems for hepatitis C virus genotype 4a and evidence for cross-genotype neutralization. Proc. Natl. Acad. Sci. USA 105997-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schregel, V., S. Jacobi, F. Penin, and N. Tautz. 2009. Hepatitis C virus NS2 is a protease stimulated by cofactor domains in NS3. Proc. Natl. Acad. Sci. USA 1065342-5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Taliani, M., E. Bianchi, F. Narjes, M. Fossatelli, A. Urbani, C. Steinkuhler, R. De Francesco, and A. Pessi. 1996. A continuous assay of hepatitis C virus protease based on resonance energy transfer depsipeptide substrates. Anal. Biochem. 24060-67. [DOI] [PubMed] [Google Scholar]
- 71.Tannous, B. A., D. E. Kim, J. L. Fernandez, R. Weissleder, and X. O. Breakefield. 2005. Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol. Ther. 11435-443. [DOI] [PubMed] [Google Scholar]
- 72.Taraporewala, Z. F., and J. T. Patton. 2004. Nonstructural proteins involved in genome packaging and replication of rotaviruses and other members of the Reoviridae. Virus Res. 10157-66. [DOI] [PubMed] [Google Scholar]
- 73.Tellinghuisen, T. L., K. L. Foss, and J. Treadaway. 2008. Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog. 4:e1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vance, L. M., N. Moscufo, M. Chow, and B. A. Heinz. 1997. Poliovirus 2C region functions during encapsidation of viral RNA. J. Virol. 718759-8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Viklund, H., and A. Elofsson. 2004. Best alpha-helical transmembrane protein topology predictions are achieved using hidden Markov models and evolutionary information. Protein Sci. 131908-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Viklund, H., and A. Elofsson. 2008. OCTOPUS: improving topology prediction by two-track ANN-based preference scores and an extended topological grammar. Bioinformatics 241662-1668. [DOI] [PubMed] [Google Scholar]
- 77.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Waxman, L., M. Whitney, B. A. Pollok, L. C. Kuo, and P. L. Darke. 2001. Host cell factor requirement for hepatitis C virus enzyme maturation. Proc. Natl. Acad. Sci. USA 9813931-13935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Welbourn, S., V. Jirasko, V. Breton, S. Reiss, F. Penin, R. Bartenschlager, and A. Pause. 2009. Investigation of a role for lysine residues in non-structural proteins 2 and 2/3 of the hepatitis C virus for their degradation and virus assembly. J. Gen. Virol. 901071-1080. [DOI] [PubMed] [Google Scholar]
- 80.Yamaga, A. K., and J. H. Ou. 2002. Membrane topology of the hepatitis C virus NS2 protein. J. Biol. Chem. 27733228-33234. [DOI] [PubMed] [Google Scholar]
- 81.Yao, N., P. Reichert, S. S. Taremi, W. W. Prosise, and P. C. Weber. 1999. Molecular views of viral polyprotein processing revealed by the crystal structure of the hepatitis C virus bifunctional protease-helicase. Structure Fold Des. 71353-1363. [DOI] [PubMed] [Google Scholar]
- 82.Yi, M., Y. Ma, J. Yates, and S. M. Lemon. 2007. Compensatory mutations in E1, p7, NS2, and NS3 enhance yields of cell culture-infectious intergenotypic chimeric hepatitis C virus. J. Virol. 81629-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yi, M., Y. Ma, J. Yates, and S. M. Lemon. 2009. trans-Complementation of an NS2 defect in a late step in hepatitis C virus (HCV) particle assembly and maturation. PLoS Pathog. 5e1000403. [DOI] [PMC free article] [PubMed] [Google Scholar]