Abstract
The papillomavirus (PV) E2 protein is an important regulator of the viral life cycle. It has diverse roles in viral transcription, DNA replication, and genome maintenance. Our laboratory has previously identified the cellular bromodomain protein Brd4 as a key interacting partner of E2. Brd4 mediates the transcriptional activation function of E2 and plays an important role in viral genome maintenance in dividing cells. E2 interacts with the C-terminal domain (CTD) of Brd4, and the CTD functions in a dominant-negative manner through binding E2 and interfering with E2's interaction with the full-length Brd4 protein. Previous studies have shown that PV E2 proteins are short lived; however, the mechanisms regulating their stability and degradation have not yet been well established. In this study, we explored the role of Brd4 in the regulation of bovine PV 1 (BPV1) and human PV 16 (HPV16) E2 stability. Expression of the Brd4 CTD dramatically increases E2 levels. Both BPV1 E2 and HPV16 E2 are regulated by ubiquitylation, and Brd4 CTD expression blocks this ubiquitylation, thus stabilizing the E2 protein. Furthermore, we have identified the cullin-based E3 ligases and specifically cullin-3 as potential components of the ubiquitylation machinery that targets both BPV1 and HPV16 E2 for ubiquitylation. Expression of the Brd4 CTD blocks the interaction between E2 and the cullin-3 complex. In addition to Brd4's role in mediating E2 transcription and genome tethering activities, these data suggest a potential role for Brd4 in regulating E2 stability and protein levels within PV-infected cells.
Papillomaviruses (PVs) are small, double-stranded DNA tumor viruses that infect squamous epithelial cells. Their life cycle is closely linked to the differentiation program of these cells (15). The full-length PV E2 protein has several roles in the viral life cycle (15). Together with the viral E1 helicase, E2 initiates origin-specific viral DNA replication. Through its ability to link viral genomes to host chromosomes during mitosis, E2 is also required for viral genome maintenance. In addition, E2 is an important regulator of viral transcription. The structure of the E2 protein resembles that of a prototypical transcription factor, with an amino-terminal transcriptional activation domain and a carboxy-terminal DNA binding and dimerization domain separated by a less well-conserved hinge region. Depending upon the location of its cognate DNA binding sites within the promoter region, E2 can act either as a transcriptional activator or as a repressor. For the high-risk human PVs (HPVs), including HPV16 and HPV18, E2 represses expression from the LCR promoter that controls expression of the E6 and E7 viral oncogenes (33). During progression to cervical cancer, the HPV DNA is frequently integrated into the host cellular chromosomes in a manner that abolishes E2 expression, thus leading to increased E6 and E7 expression. The deregulated expression of the E6 and E7 oncogenes is thought to be a critical step in HPV-mediated transformation and carcinogenesis.
E2 proteins are unstable and degraded in a proteasome-dependent manner (4, 23, 24). However, the mechanisms regulating E2 degradation are not well understood. It has been shown that BPV1 E2 is phosphorylated by casein kinase II at serine 301 within the hinge region, promoting its degradation (23). It has been suggested that this phosphorylation might induce a conformational change, allowing recognition by the ubiquitin/proteasome degradation machinery. Interestingly, mutation of serine 301 not only stabilizes BPV1 E2 but also leads to an increase in viral DNA copy number (24), raising the possibility that the regulation of E2 stability might play a role in the switch from episomal maintenance to productive replication. The hinge region, however, is not conserved across different PV types, suggesting that the mechanisms regulating the stability of different E2 proteins may vary.
Our laboratory has previously identified the cellular bromodomain protein Brd4 as a key interacting partner of E2 (40). Brd4 is a BET family protein (7, 8) that regulates both cell growth and cellular transcription. In a gene knockout mouse model, Brd4 heterozygotes display growth defects associated with a reduced proliferation rate and embryos nullizygous for Brd4 are not viable (14). Several studies have also linked Brd4 to certain cancers, further supporting its role in cellular growth control (5, 9, 10). Brd4's roles in transcriptional regulation and cell growth seem to be coupled; Brd4 is recruited to a number of G1 genes during G0-G1 progression and stimulates binding of positive transcription elongation factor b and RNA polymerase II to the promoters, resulting in transcriptional elongation (21, 39). As such, Brd4 stimulates G1 gene transcription and promotes progression to S phase (21, 39).
The C-terminal domain (CTD) of Brd4 mediates binding to E2 (40), and Brd4 is involved in several E2 functions. For BPV1, Brd4 has been shown to be an important component of the viral genome maintenance machinery that tethers the viral genome to a host chromosome during mitosis (3, 40). The Brd4 CTD functions in a dominant-negative manner and releases viral episomes from mitotic chromosomes by competing with the full-length Brd4 protein for binding to E2 (40, 41). Brd4 also mediates E2-dependent transcription activation as the Brd4 CTD or small interfering RNA against Brd4 abolishes the trans-activation function of E2 (20, 30, 31). One study also implicates Brd4 in the transcriptional repression function of E2 (37), although evidence exists for Brd4-independent repression mechanisms (29). In the present study, we have explored whether the Brd4 CTD affects E2 protein stability. We have found that overexpression of the Brd4 CTD can stabilize both BPV1 E2 and HPV16 E2 by blocking their ubiquitylation. We have also identified cullin-3 as a potential component of the E3 ubiquitin ligase machinery involved in E2 protein ubiquitylation. Our results demonstrate that the Brd4 CTD and cullin-3 compete with one another for the ability to complex with E2. Together, our findings illustrate that Brd4 not only contributes to several E2 functions throughout the viral life cycle but also regulates E2 stability.
MATERIALS AND METHODS
Cell culture and plasmids.
The human cervical cancer cell line C33A was cultured in Dulbecco's minimal essential medium supplemented with 10% fetal bovine serum, 100 units/ml of penicillin, and 100 μg/ml of streptomycin. Single C33A-pOZN-E2TA clones were derived from pooled cell lines previously described (40). The codon-optimized pMEP4-HPV16 E2 (for mammalian expression) and pMEP4-BPV1 E2 plasmids were generously provided by Alison McBride (NIH, Bethesda, MD) (24). pOZN-HA-FLAG-BPV1 E2 (p5066), pCDNA4C-SV40NLS-Xpress-Brd4-CTD (p4948), and pcDNA4C-Xpress-Brd4 (p4947) were described previously (40). pOZN-HA-BPV1 E2 (p6075) was derived from pOZN-HA-FLAG-BPV1 E2 by mutating the FLAG tag coding sequence. pCDNA4C-SV40NLS-Xpress-Brd4-CTD F1349A (p5943) and pCDNA4C-SV40NLS-Xpress-Brd4-CTD D1352A (p5944) were derived from pCDNA4C-SV40NLS-Xpress-Brd4-CTD by single-nucleotide mutagenesis. pOZ-N-HPV16 E2 (p6072) was generated by subcloning the HPV16 E2 coding sequence into the pOZ-N vector by using XhoI and NotI sites. Expression plasmids for dominant-negative cullins [pCDNA-HCUL1-N452-FLAG (p6066), pCDNA-HCUL2-N427-FLAG (p6067), pCDNA-HCUL3-N418-FLAG (p6068), pCDNA-HCUL4A-N440-FLAG (p6069), pCDNA-HCUL4B-N594-FLAG (p6070), and pCDNA-HCUL5-N441-FLAG (p6071)] have previously been described (6, 17). pCDNA-HA-ubiquitin (p6075) was previously described (13). pCDNA4C-Xpress-DN-CUL3 (p6082) was generated by subcloning the DN-CUL3 coding sequence into pCDNA4C by using EcoRV and XhoI sites. pDest-FLAG-BPV1 E2 (p6083) was generated by subcloning the BPV1 E2 coding sequence into the pDest-FLAG vector by using the gateway system (Invitrogen).
Transient transfections, analysis of protein expression, and immunoprecipitation.
For the analysis of total cell protein expression, C33A cells were plated in 10-cm plates at densities resulting in 60 to 70% confluent monolayers at the end of the experiment. After adhering overnight at 37°C, cells were transfected with plasmids by using FuGENE 6 (Roche) according to the manufacturer's instructions. At 48 h posttransfection, cells were washed with phosphate-buffered saline (PBS) and then harvested with 2% sodium dodecyl sulfate (SDS) lysis buffer (50 mM Tris, pH 6.8, 2% SDS). After 8 s of sonication at 35% intensity (Branson digital sonifier), lysates were cleared by centrifugation at 18,000 × g for 10 min at 4°C. Lysates were normalized for protein concentration (BCA protein assay kit; Thermo Scientific) and E2 protein levels analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblotting. The primary antibodies used in these experiments were antihemagglutinin (anti-HA) (no. 12013819001; Roche), anti-FLAG (F7425; Sigma), anti-Xpress (46-0528; Invitrogen), and antiactin (MAB1501; Chemicon). Secondary anti-mouse and anti-rabbit antibodies were purchased from Millipore. For ubiquitylation assays, C33A cells were plated in 10-cm plates at densities resulting in 60% confluence at the end of the experiment. After overnight incubation at 37°C, cells were transfected with the plasmids encoding HA-tagged ubiquitin and FLAG-tagged E2 in different combinations as described in the figure legends. At 48 h posttransfection, cells were washed with PBS and harvested in Tris-buffered-saline-based lysis buffer (100 mM Tris, pH 7.5, 150 mM NaCl, 1% NP-40, 1% TritonX-100, 1 mM dithiothreitol [DTT], and 1× protease inhibitor cocktail [BD Biosciences]). After 8 s of sonication at 35% intensity, cell lysates were clarified by centrifugation at 18,000 × g for 10 min at 4°C. Anti-HA agarose beads (10-μl bead volume, no. A2095; Sigma) were added to the supernatants and incubated for 3 h at 4°C. The anti-HA agarose beads were then washed with DTT-free Tris-buffered-saline-based lysis buffer four times. SDS-PAGE and immunoblotting with anti-FLAG antibodies were then performed to detect the ubiquitin-modified E2 species. MG132 (C4859; Sigma) was used in this study at a concentration of 30 μM.
Protein half-life assay.
C33A cells were seeded in 35-mm plates at densities resulting in 60 to 70% confluent monolayers at the end of the experiment. After overnight incubation at 37°C, cells were transfected with the indicated plasmid DNA via FuGENE6. At 48 h posttransfection, cells were treated with 40 μg/ml cycloheximide and harvested with 2% SDS lysis buffer at the time points indicated in the figure legends. Cell lysates were normalized for protein concentration and E2 protein levels determined by SDS-PAGE and immunoblotting. For the purpose of quantitating E2 protein levels, different dilutions of a known cell lysate sample with E2 were included in each gel and corresponding immunoblot. Image quantitation was performed using ImageQuant TL version 2005.04 software (Molecular Probes), data analysis was done with GraphPad Prism 4, and the half-lives of E2 proteins were calculated using a one-phase exponential-decay model.
Cullin binding assays.
Cells were plated as described above for transient transfections, analysis of total protein levels, and immunoprecipitations and then transfected with plasmids encoding FLAG-tagged DN-CUL3 or DN-CUL1 with HA-tagged E2 in different combinations as described in the figure legends. At 48 h posttransfection, cells were washed with PBS and harvested with PBS-based lysis buffer (PBS plus 1% NP-40, 1% TritonX-100, 1 mM DTT, and 1× protease inhibitor cocktail). After sonication at 35% intensity for 8 s, cell lysates were clarified at 18,000 × g for 10 min at 4°C. Anti-HA agarose beads (10-μl bead volume) were added to the cleared lysates and incubated overnight at 4°C. DTT-free PBS-based lysis buffer was used to wash the anti-HA agarose beads four times. SDS-PAGE and immunoblot analysis with anti-FLAG and anti-HA antibodies were performed to detect any complex formation between E2 and DN-cullin proteins.
Quantification of E2 mRNA expression by real-time PCR.
Total cell RNA was extracted 48 h after transfection by using an RNeasy RNA isolation kit (Qiagen) according to the manufacturer's protocol. Total RNA was reverse transcribed by using a reverse transcription system (Promega) with oligo(dT) primers. To quantify the relative abundance of HPV16 E2 RNA, a real-time system (LightCycler 2.0; Roche) was used. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) served as an endogenous control used to normalize expression data. Each sample was analyzed in duplicate. The relative expression levels and standard errors were calculated by LightCycler software.
RESULTS
The Brd4 CTD increases E2 protein levels.
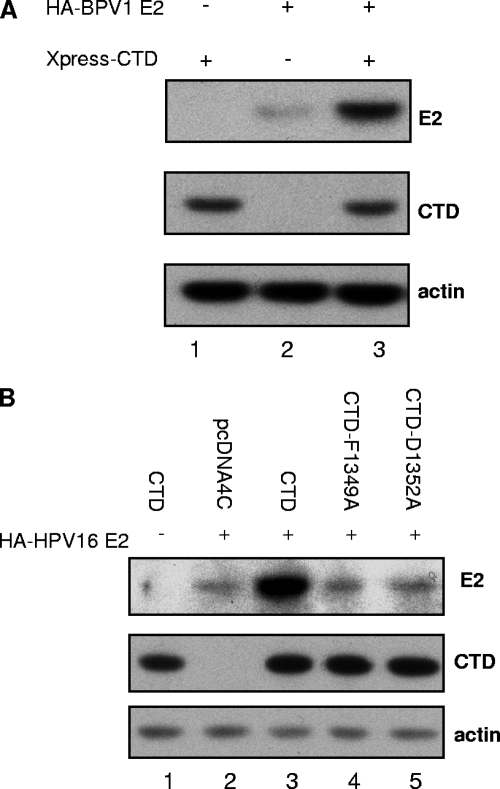
It has previously been shown by Ilves and colleagues that coexpression of BPV1 E2 and the Brd4 CTD results in an increased level of BPV1 E2 protein (16). We have confirmed this finding (Fig. 1A) and extended the observation to HPV16 E2 (Fig. 1B, compare lanes 2 and 3). The studies described herein were designed to further explore the consequences of Brd4 CTD expression on E2 protein levels and determine the mechanism by which E2 proteins are increased when the Brd4 CTD is also expressed. We next asked whether the Brd4 CTD-dependent increase in E2 protein required a direct interaction between the Brd4 CTD and HPV16 E2. We utilized two Brd4 CTD point mutants, Brd4 CTD(D1352A) and Brd4 CTD(F1349A), which are unable to bind E2(1). In contrast to what was observed with the wild-type (wt) Brd4 CTD, coexpression of these Brd4 CTD mutant proteins did not lead to an increase in HPV16 E2 protein levels (Fig. 1B, compare lanes 4 and 5 with lane 3). Based upon these results, we conclude that the Brd4 CTD must be able to bind E2 in order for increased protein levels to be observed.
FIG. 1.
The Brd4 CTD increases E2 protein levels. (A) C33A cells were transfected with 0.5 μg of plasmids expressing HA-tagged BPV1 E2 and 0.5 μg XPress-tagged Brd4 CTD in the indicated combinations. Cells were harvested after 48 h and total protein levels analyzed by SDS-PAGE and immunoblotting with antibodies against the HA tag (detection of E2), Xpress tag (Brd4 CTD), and actin. (B) C33A cells were transfected with 0.5 μg of plasmids expressing HPV16 E2 or the empty control pcDNA4C vectors (pcDNA4C) and plasmids expressing the wt Brd4 CTD (CTD-wt) or mutant forms of the Brd4 CTD (CTD-F1349A and CTD-D1352A) as indicated. Cells were harvested after 48 h, and protein levels were analyzed as described for panel A with antibodies against the HA tag (HPV16 E2), the Xpress tag (Brd4 CTD), and actin.
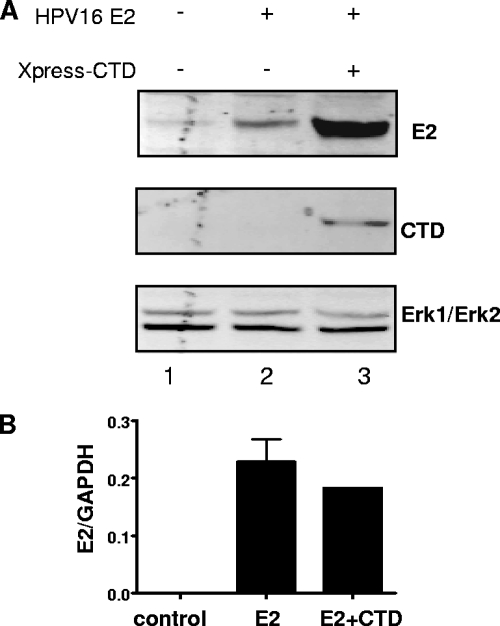
Since Brd4 has known transcriptional activation functions that can affect both cellular and viral promoters (21, 30, 39), it was possible that the increased E2 protein was a consequence of Brd4 CTD-mediated increase in transcription from the murine leukemia virus (MLV) long terminal repeat (LTR) or cytomegalovirus (CMV) promoters driving BPV1 and HPV16 E2 expression, respectively. To rule out this possibility, we tested the effect of Brd4 CTD expression on CMV-luciferase and MLV LTR-luciferase reporter plasmids. Luciferase levels were unaffected by coexpression of the Brd4 CTD (data not shown), indicating that the increase in E2 was not due to increased CMV- or MLV LTR-driven transcription. In addition, real time-PCR measuring HPV16 E2 mRNA levels directly confirmed that the Brd4 CTD increases HPV16 E2 protein level (Fig. 2A, lane 2 versus lane 3) without affecting E2 mRNA levels (Fig. 2B).
FIG. 2.
The Brd4 CTD does not increase the transcription of HPV16 E2. (A) C33A cells were transfected with 2.6 μg of plasmids expressing HPV16 E2 or the empty control vectors and plasmids expressing the wt Brd4 CTD as indicated. Cells were harvested after 48 h for whole-cell protein extraction and RNA preparation. Protein levels were analyzed by Western blotting with antibodies against the FLAG tag (HPV16 E2), the Xpress tag (Brd4 CTD), and Erk1/Erk2 (as a loading control). The weak band in lane 1 of the top panel is a nonspecific background band running at the same position as E2. (B) RNA was prepared from the cells as described above and used for reverse transcription. Real-time PCR was performed with primers specific for HPV16 E2. Primers for the GAPDH gene were used as an internal control, and values for HPV16 E2 were normalized accordingly.
The Brd4 CTD stabilizes E2.
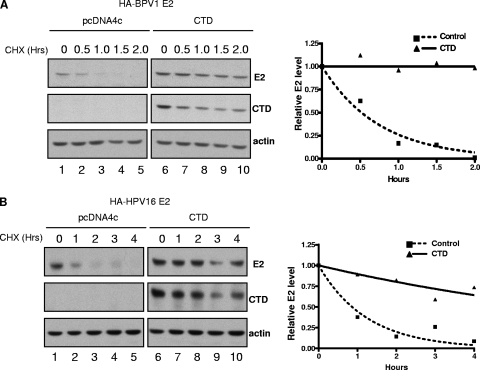
Since it has been reported that E2 proteins are short lived, with a half-life of approximately 30 to 60 min (4, 24), we hypothesized that the Brd4 CTD might somehow mediate a posttranslational stabilization of BPV1 E2 and HPV16 E2. To test this, we examined how Brd4 CTD expression affected the half-lives of the E2 proteins. C33A cells were transfected with HA-tagged BPV1 or HPV16 E2 and pCDNA4c plasmid with or without the Brd4 CTD. At 48 h posttransfection, cells were treated with cycloheximide to block de novo protein synthesis. Cells were harvested at the indicated times after cycloheximide treatment, and E2 protein levels were determined by Western blot analysis. As shown in Fig. 3A and B, the half-lives of BPV1 E2 and HPV16 E2 measured 0.51 ± 0.10 h and 0.78 ± 0.10 h, respectively. In the presence of the Brd4 CTD, however, the half-lives of these E2 proteins were significantly prolonged. The Brd4 CTD extended the half-life of BPV1 E2 to more than 9 h in each of three independent experiments; for HPV16 E2, the half-life was extended by the Brd4 CTD to more than 6 h in each of two independent experiments. To rule out the possibility that the protein stabilization effects of the Brd4 CTD were nonspecific, we examined the impact of the Brd4 CTD on another short-lived viral protein, HPV16 E6, and found no stabilization (data not shown). We conclude that the Brd4 CTD-mediated increase in E2 levels is specific and is consequential to protein stabilization.
FIG. 3.
The Brd4 CTD stabilizes E2 proteins. C33A cells were cotransfected with 0.5 μg of plasmids expressing HA-tagged BPV1 E2 (A) or HPV16 E2 (B) and pcDNA4C with or without 0.5 μg of plasmids expressing the Brd4 CTD as indicated. Cells were treated with 40 μg/ml cycloheximide (CHX) 48 h after transfection for the lengths of time indicated. Cell lysates were harvested and E2 protein levels analyzed by SDS-PAGE and immunoblotting. Protein bands from the immunoblots were quantitated with ImageQuant, using the actin signal for normalization.
The Brd4 CTD blocks E2 ubiquitylation.
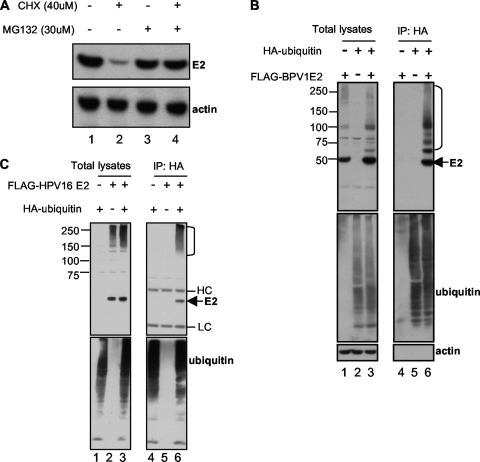
Previous studies have reported that the BPV1 and HPV18 E2 proteins are regulated by proteasome-mediated degradation, possibly through ubiquitylation (4, 24), and this has recently been extended to the HPV11 and HPV31 E2 proteins (11). We therefore tested whether Brd4 CTD expression affected BPV1 and HPV16 E2 ubiquitylation. We first confirmed that BPV1 E2 degradation was mediated by the proteasome. As shown in Fig. 4A, the degradation of BPV1 E2 in a cycloheximide chase experiment could be blocked by the addition of the proteasome inhibitor MG132. To determine if BPV1 E2 and HPV16 E2 were being ubiquitylated, we coexpressed FLAG-tagged BPV1 or HPV16 E2 protein with a HA-tagged ubiquitin or an empty pcDNA plasmid control. In this experiment, we expressed E2 from a cadmium-inducible metallothionein promoter (24). We chose these E2 expression constructs because more-physiologically relevant (not overexpressed) levels of E2 could be observed in cells after 5 h of low-concentration CdCl2 treatment (24). At 48 h posttransfection, cells were treated with 1 μM CdCl2 for 5 h and then harvested. Ubiquitylated proteins were immunoprecipitated using anti-HA antibodies, and the immunoprecipitates were analyzed by immunoblotting for E2 or ubiquitin. As shown in Fig. 4B, ubiquitylated forms of BPV1 E2 were clearly detected in the presence of HA-tagged ubiquitin (lane 6). In this experiment, the nonubiquitylated form of BPV1 E2 was also present in the HA-ubiquitin immunoprecipitates, likely due to the fact that E2 dimerizes and nonubiquitylated E2 was brought down in the immunoprecipitation in complex with ubiquitylated E2. Similarly, we also detected ubiquitylated forms of HPV16 E2 (Fig. 4C). These results confirm that E2 proteins are ubiquitylated and degraded in a proteasome-dependent manner in vivo.
FIG. 4.
E2 proteins are ubiquitylated and degraded in a proteasome-dependent manner. (A) C33A cells stably expressing BPV1 E2 were treated for 4 h with 40 μg/ml cycloheximide (CHX) and 30 μM MG132 in the indicated combinations. Cells were then harvested and E2 protein levels analyzed by SDS-PAGE and immunoblot analysis with antibodies against the HA tag of E2 and actin. (B and C) C33A cells were transfected with 1 μg of a plasmid expressing HA-ubiquitin and 3 μg of pMEP4-FLAG-BPV1 E2 (B) or pMEP4-FLAG-16 E2 (C) in the indicated combinations. E2 expression was induced at 48 h posttransfection with 1 μM CdCl2 for 5 h, and then cells were harvested. To detect E2 ubiquitylation, cell lysates were immunoprecipitated (IP) with anti-HA antibodies and immunoprecipitated proteins as well as total cell lysates were analyzed by SDS-PAGE and immunoblot analyses using antibodies against the FLAG tag (E2), the HA tag (ubiquitin), and actin. E2 ubiquitylation signals are indicated by the right bracket.
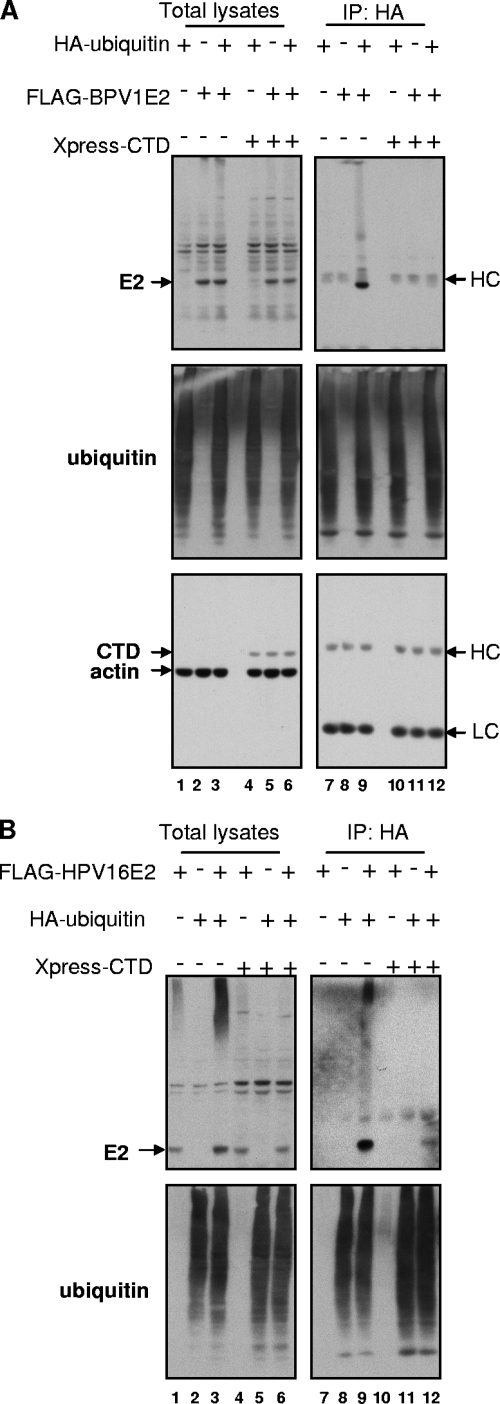
We next asked how Brd4 CTD expression affected E2 ubiquitylation. The experiments were performed as described above, but samples were also transfected with the Xpress-tagged Brd4 CTD or the empty pcDNA4C control plasmid. Although the ubiquitylated forms of BPV1 E2 were readily detected in the absence of the Brd4 CTD (Fig. 5A, lane 9), no ubiquitylated forms of BPV1 E2 were detected in cells expressing the Brd4 CTD (Fig. 5A, lane 12). Similarly, the Brd4 CTD greatly reduced the ubiquitylation of HPV16 E2 (Fig. 5B, lane 9 versus lane 12). Of note, in these experiments, the expression of the Brd4 CTD did not affect the overall levels of total protein ubiquitylation in the cells. In the experiments whose results are shown in Fig. 5A and B, E2 was expressed from an inducible metallothionein promoter in the pMEP4 vector for 5 h with cadmium. Under these conditions of relatively short induction, no E2 stabilization effect by the Brd4 CTD was observed compared to the marked stabilization achieved when both E2 and the Brd4 CTD are coexpressed for 48 h as seen in Fig. 1 and 2. Together, these results suggest that the Brd4 CTD prevents E2 degradation by the proteasome through blocking of its ubiquitylation.
FIG. 5.
The Brd4 CTD blocks E2 ubiquitylation. C33A cells were transfected with 1 μg of a plasmid expressing HA-ubiquitin, 3 μg of pMEP4-FLAG-BPV1 E2 (A) or pMEP4-FLAG-HPV16 E2 (B), and 2 μg of a plasmid expressing Xpress-CTD in the indicated combinations. E2 expression was induced at 48 h posttransfection with 1 μM CdCl2 for 5 h, and then cells were harvested. To detect ubiquitylated E2, cell lysates were immunoprecipitated (IP) with anti-HA antibodies and immunoprecipitated proteins analyzed by SDS-PAGE and immunoblots with antibodies against the FLAG tag (E2), the HA tag (ubiquitin), the Xpress tag (Brd4 CTD), and actin. HC, heavy chain; LC, light chain.
Cullin-based E3 ubiquitin ligases regulate PV E2 stability.
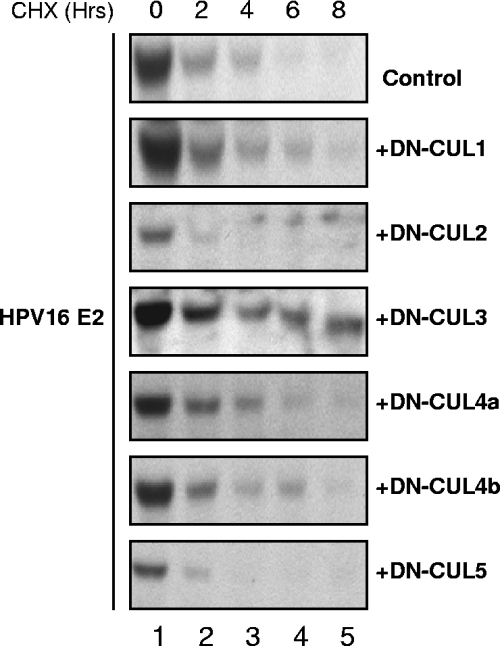
Ubiquitylation requires the activities of an E1 activation enzyme, E2 conjugating enzymes, and E3 ubiquitin ligases. Of these, the E3 ligases are responsible for substrate recognition. The cullin-based E3 ubiquitin ligases have been implicated in the ubiquitylation of cell cycle-regulated proteins (2). In some cases, phosphorylation of the substrate has been shown to serve as a trigger for cullin-dependent ubiquitylation (32). Given that the ubiquitylation and degradation of the PV E2 proteins have been reported to be regulated by phosphorylation (24) and occur in a cell cycle-dependent manner (4), we speculated that cullin-based E3 ubiquitin ligases would be good candidates for the PV E2 proteins. To test this hypothesis and determine whether any of the cullin-based ubiquitin E3 ligases were involved in regulating E2 stability, we utilized dominant-negative, truncated forms of the cullin proteins. Cullin-based E3 proteins are macromolecular complexes containing at least three subunits: a cullin protein; a ring finger protein, Rbx1; and a substrate recognition linker protein (25). The N-terminal fragments of cullin proteins act in a dominant-negative manner by forming enzymatically inactive complexes with the linker proteins and substrates but not with the essential Rbx1 catalytic core (2, 6). C33A cells were transfected with HA-tagged HPV16 E2 expression plasmid in combination with a pcDNA4C empty control plasmid or FLAG-tagged expression plasmids for DN-cullins in an initial screen to determine whether any of the cullins might be involved in regulating E2 stability. The expression of each of the DN-cullin proteins was confirmed by Western blot analysis (data not shown). At 48 h posttransfection, cells were treated with cycloheximide and harvested at the indicated times posttreatment. Under the conditions of this initial screen, the half-life of HPV16 E2 was approximately 1.7 h. The expression of most of the DN-cullin proteins had either no or only moderate impact on HPV16 E2 stability; however, a significant HPV16 E2 stabilization was observed in cells that expressed DN-CUL3, which extended the half-life to 6.5 h (Fig. 6). This result suggested that cullin-3 might be part of the enzymatic machinery responsible for the ubiquitylation of HPV16 E2.
FIG. 6.
Stabilization of HPV16 E2 by dominant-negative cullins. C33A cells were transfected with 0.5 μg of a plasmid expressing HA-tagged HPV16 E2 alone or combined with 0.5 μg of plasmids expressing DN-CUL1, DN-CUL2, DN-CUL3, DN-CUL4A, DN-CUL4B, or DN-CUL5. At 48 h posttransfection, cells were treated with 40 μg/ml cycloheximide (CHX). Cell lysates were harvested at the indicated times posttreatment, and E2 protein levels were analyzed by SDS-PAGE and immunoblotting using antibodies against the HA tag of E2. Under the conditions of this experiment, the half-life of HPV16 E2 alone was 1.7 h. The calculated half-lives of HPV16 E2 in the presence of the dominant-negative cullins were as follows: for DN-CUL1, 2.5 h; for DN-CUL2, 2.7 h; for DN-CUL3, 6.5 h; for DN-CUL4A, 3.8 h; for DN-CUL4B, 4.0 h; and for DN-CUL5, 1.8 h.
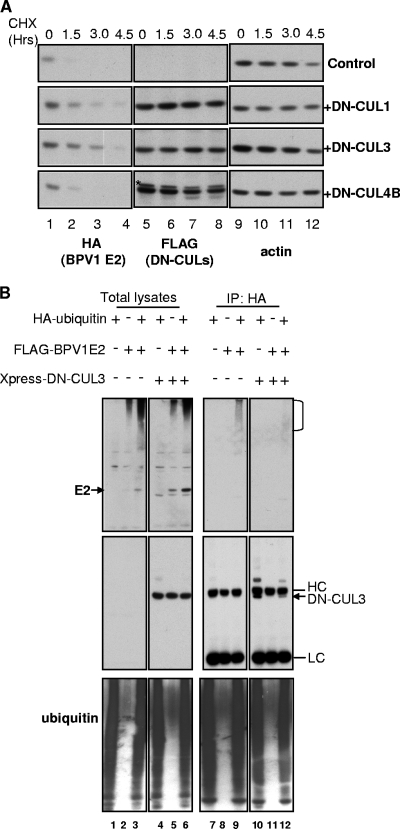
We then tested whether the cullin-based E3 ligases were also involved in the stabilization of the BPV1 E2 protein. In this experiment, we examined DN-CUL1 and DN-CUL3 as well as DN-CUL4B. Similar to what was observed with HPV16 E2, the coexpression of DN-CUL3 led to the greatest stabilization of the BPV1 E2 protein (Fig. 7A). The half-life of HPV16 E2 in the control experiment was 0.5 h. DN-CUL3 extended the half-life to 2.0 h. We also observed moderate stabilization by DN-CUL1 (half-life, 1.4 h) but not with DN-CUL4B (half-life, 0.5 h). We next examined whether DN-CUL3 affects BPV1 E2 ubiquitylation. As shown in Fig. 7B, the ubiquitylated forms of BPV1 E2 were readily detected in the absence of DN-CUL3 (top panel, lane 9), and the ubiquitylation of BPV1 E2 was markedly reduced in cells expressing DN-CUL3 (top panel, lane 12). From these experiments, we concluded that cullin-3 was involved in the ubiquitylation of both the BPV1 and the HPV16 E2 proteins. The modest stabilization of E2 observed with some of the other DN-cullin proteins also suggests that other cullin-based E3 ligase complexes might also have roles in regulating E2 protein stability.
FIG. 7.
Stabilization of BPV1 E2 by dominant-negative cullins. (A) C33A cells were transfected with 0.5 μg HA-BPV1 E2 alone or combined with 0.5 μg of a plasmid that expressed either DN-CUL1, DN-CUL3, or DN-CUL4B. Cells were treated and analyzed as described for Fig. 5A. An anti-HA antibody was used to detect the HA tag on BPV1 E2. Expression of the DN-CUL constructs was assessed using an antibody against its FLAG tag, and an antiactin antibody was utilized for visualization of actin. The star in the Flag blot for DN-CUL4B indicates a nonspecific band recognized by the anti-FLAG antibody. CHX, cycloheximide. (B) C33A cells were transfected with 1 μg of a plasmid expressing HA-ubiquitin, 3 μg of pDest-FLAG-BPV1 E2, and 2 μg of a plasmid expressing Xpress-DN-CUL3 in the indicated combinations. Cells were harvested at 48 h posttransfection. To detect ubiquitylated E2, cell lysates were immunoprecipitated (IP) with anti-HA antibodies and immunoprecipitated proteins analyzed by SDS-PAGE and immunoblots with antibodies against the FLAG tag (E2), HA tag (ubiquitin), and Xpress tag (DN-CUL3). E2 ubiquitylation signals are indicated by the right bracket. HC, heavy chain; LC, light chain.
E2 and DN-CUL3 can form a complex.
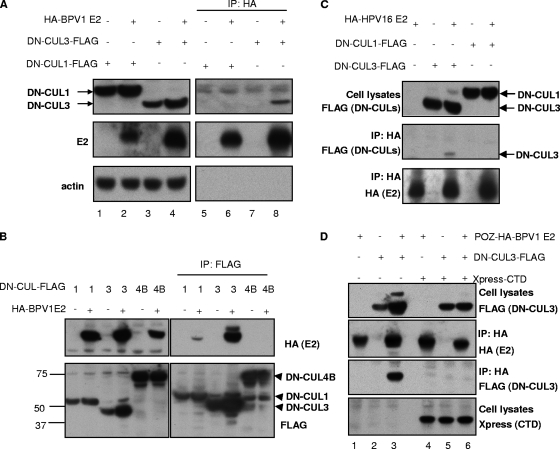
To further evaluate the role of a cullin-3-based E3 ligase in the ubiquitylation and degradation of E2, we tested whether BPV1 E2 and DN-CUL3 could be found in a complex together. Catalytically inactive forms of E3 ligases are often used to examine substrate binding since the ligase/substrate interactions are usually transient and ligase binding promotes the degradation of the substrate. C33A cells were therefore transfected with HA-tagged BPV1 E2 alone or combined with FLAG-tagged DN-CUL3. In this experiment, we also examined the binding of DN-CUL1 since it resulted in a modest stabilization of BPV1 E2. Cell lysates were harvested at 48 h posttransfection and subjected to immunoprecipitation with an anti-HA antibody. Total cell as well as the precipitated proteins were separated by SDS-PAGE and analyzed by immunoblotting. As shown in Fig. 8A, the anti-HA immunoprecipitation of BPV1 E2 pulled down DN-CUL3 (upper panel, lane 8). In contrast, we observed no major band for DN-CUL1; however, a nonspecific protein band in all lanes ran at about the position DN-CUL1 (lanes 5 to 8) and would have obscured a minor band for the DN-CUL1 protein. The interaction of BPV1 E2 with DN-CUL3 was confirmed by a reciprocal immunoprecipitation by anti-FLAG immunoprecipitation (Fig. 8B) for the DN-CULs and Western blot analysis for HA-tagged E2. In this experiment, we observed minor binding of E2 with DN-CUL1 but not with DN-CUL4B. We also confirmed that HPV16 E2 bound DN-CUL3 by immunoprecipitation and immunoblot analysis (Fig. 8C). These data indicate that the PV E2 proteins and cullin-3 (and possibly also cullin-1 for BPV1 E2) can form a complex in vivo. We also note that coexpression of E2 appears to stabilize DN-CUL3, a reproducible phenomenon that we do not yet fully understand.
FIG. 8.
The Brd4 CTD blocks E2/cullin-3 complex formation. (A) C33A cells were transfected with 3 μg of pOZN-HA-BPV1 E2 in combination with 3 μg of a control pcDNA plasmid or plasmids expressing either FLAG-DN-CUL1 or FLAG-DN-CUL3 as indicated. Cell lysates were harvested at 48 h posttransfection and immunoprecipitated (IP) with anti-HA antibodies. Total and immunoprecipitated proteins were analyzed by SDS-PAGE and immunoblot analysis using antibodies against the FLAG tag (DN-CUL1 and DN-CUL3), the HA tag (E2), and actin. (B) C33A cells were transfected with 3 μg of pOZN-HA-BPV1 E2 alone or combined with 3 μg of plasmids expressing FLAG-tagged DN-CUL1, DN-CUL3, or DN-CUL4B as indicated. Cell lysates were harvested at 48 h posttransfection and immunoprecipitated with anti-FLAG antibodies. Total and immunoprecipitated proteins were analyzed by SDS-PAGE and immunoblot analysis using antibodies against FLAG (DN-CULs), HA (BPV1 E2), and actin. (C) C33A cells were transfected with 3 μg of pOZN-HA-HPV16 E2 alone or combined with 3 μg of plasmids expressing FLAG-tagged DN-CUL1 or DN-CUL3 as indicated. Cell lysates were harvested at 48 h posttransfection and immunoprecipitated with anti-HA antibodies. Total and immunoprecipitated proteins were analyzed by SDS-PAGE and immunoblot analysis using antibodies against FLAG (DN-CULs) and HA (BPV1 E2). (D) C33A cells were transfected with 3 μg of plasmid pOZN-HA-BPV1 E2, 3 μg of a plasmid expressing FLAG-DN-CUL3, and pcDNA4C with or without the Xpress-tagged Brd4 CTD as indicated. Cell lysates were harvested at 48 h posttransfection and immunoprecipitated with anti-HA antibodies. Immunoprecipitated proteins were separated by SDS-PAGE and analyzed by immunoblotting against FLAG (DN-CUL3), HA (BPV1 E2), and Xpress (Brd4 CTD).
The Brd4 CTD blocks binding of cullin-3 to BPV1 E2.
Given that the Brd4 CTD abolished the ubiquitylation and degradation of E2, we speculated that the Brd4 CTD might block the interaction between E2 and the cullin-based E3 ligase involved in its degradation. To test this hypothesis, C33A cells were transfected with HA-tagged BPV1 E2, FLAG-tagged DN-CUL3, and the Xpress-tagged Brd4 CTD in the indicated combinations (Fig. 8D). We used DN-CUL3 in this experiment since it was the most effective in stabilizing both HPV16 and BPV1 E2 proteins and because its binding with E2 could be readily detected. Cells were lysed at 48 h posttransfection, and the binding of E2 to DN-CUL3 was assessed by immunoprecipitation and immunoblot analysis. Examination of proteins that immunoprecipitated with the anti-HA antibody for E2 confirmed that BPV1 E2 interacts with DN-CUL3 (third panel, lane 3) in the absence of the Brd4 CTD. However, the interaction between BPV1 E2 and DN-CUL3 was abolished in cells also expressing the Brd4 CTD (lane 6). These results indicate that the Brd4 CTD may stabilize E2 by blocking its interaction with a cullin-3 ubiquitin protein ligase complex that ubiquitylates PV E2 and targets it for degradation by the proteasome.
DISCUSSION
Brd4 was identified as a major interacting partner of the PV E2 protein in a proteomic analysis of BPV1 E2 (40). A number of laboratories have now confirmed that the binding of Brd4 to E2 is a general property of most if not all E2 proteins and that this binding is mediated though a highly conserved region in the N-terminal transactivation domain of E2 (1, 3, 30). Our initial studies indicated that Brd4 played a role in the tethering of BPV1 E2 and viral episomes to cellular chromosomes during mitosis (40) and that disruption of this interaction resulted in the loss of viral episomes (41). Factors in addition to Brd4 have now been implicated in E2's episome maintenance function (22), and it is clear that different PVs rely on distinct cellular proteins to ensure plasmid maintenance in dividing cells (28). Brd4 also mediates the transcriptional activation function of E2, a characteristic that has now been validated for all PVs examined to date (20, 30, 31). In addition, Brd4 has been implicated in the ability of E2 to silence transcription (37).
The Brd4 CTD has been an important tool utilized in studies to understand how Brd4 affects the various functions of E2. The Brd4 CTD can be stably expressed in cells and, by binding to E2, competes away interaction of E2 with endogenous Brd4 (40). Although expression of the Brd4 CTD has no obvious cell cycle effects and can be well tolerated (30, 40, 41), it was previously noted that its expression resulted in increased levels of BPV1 E2 protein (16). This observation prompted our current study for examining the mechanism by which the Brd4 CTD increased E2 protein levels. We have confirmed that the Brd4 CTD indeed increases E2 levels and does so by stabilizing E2 through blocking of E2 ubiquitylation. This mechanism is distinct from that of TaxBP1, which has been reported to stabilize PV E2 proteins without affecting their ubiquitylation status (36). Furthermore, our studies have identified cullin-3 as a component of the E3 ligase machinery that binds E2. The Brd4 CTD blocks the interaction between this ligase and PV E2, thereby providing a mechanism for the observed stabilization.
There are at least six cullin genes in eukaryotes (18). Cullin-based E3 ubiquitin ligases are macromolecular complexes sharing similar architectures (25). The CTD of a cullin associates with a Ring finger protein, called Rbx1, which interacts with the E2 ubiquitin conjugating enzyme and is essential for catalytic activity. The cullin N terminus is responsible for associating with substrates, and substrate specificity is achieved through adaptor proteins capable of directly interacting with both cullins and the substrates. The SCF (Skp1-Cul1-F-box) complex is the best-characterized cullin-based ubiquitin ligase (2, 25), in which the F-box protein acts as the adaptor protein to recognize substrates. It is generally agreed that BTB domain proteins function as the substrate-specific adaptors for cullin-3-containing E3 ligases (12, 26, 27, 34, 38). The interaction between PV E2 and cullin-3 that we describe in this study is most likely indirect and mediated by a BTB domain adaptor protein that is involved in recognizing E2. It is still not clear which BTB domain proteins are responsible for E2 ubiquitylation, and we are actively pursuing this question.
The effect of the Brd4 CTD on E2 stability suggested that full-length Brd4 might influence the stability of E2 in PV-infected cells. Indeed, two recent studies have implicated Brd4 in E2 stability (11, 19). Thus, in addition to mediating some of E2's functions as an effector, Brd4 may actually be involved upstream of E2 as a regulator. In preliminary studies, we have found that BPV1 E2 is stabilized in small interfering RNA knockdown studies of Brd4, supporting a role for endogenous Brd4 in regulating E2 stability. However, we have not discerned whether this effect is direct or indirect, since knockdown of Brd4 in our hands has cell cycle effects.
Many transcription factors, including Myc, are unstable and regulated through the ubiquitin-proteasome pathway (35). For some of these factors, the region for ubiquitylation overlaps with their transcription activation domains, indicating that the proteasome pathway plays an important role in their transcription activities (20). Given that Brd4 is required for PV E2-mediated transcriptional activation (20, 30, 31), our finding that Brd4 also regulates E2 stability raises the possibility that, at least for BPV1 E2, ubiquitylation and transactivation are coupled and ubiquitylation is essential for E2's transcriptional activation. A study with HPV18 E2, however, did show that ubiquitylation and transactivation are separable; a mutant E2 protein whose transactivation function is impaired is as unstable as the wt E2 protein (4). However, it has not been tested whether ubiquitylation-deficient E2 has the same transactivation function as wt E2, which will be important to investigate once the ubiquitylation sites are mapped and the ubiquitylation machinery for E2 is identified.
The life cycle of PV is closely linked with the differentiation of epithelial cells. The triggers which induce the change from episomal maintenance to productive replication in the viral life cycle are unknown. E2 regulates viral replication, and it was suggested that the level of E2 increases during differentiation, in turn affecting viral DNA replication (24). It is likely that changes in cellular proteins regulate E2 stability and protein level, resulting in the switch of the viral life cycle. Based upon the results presented in this work, it will be interesting to study whether modulation of E2's interaction with Brd4 and the cullin-based E3 ubiquitin-ligase complexes contributes to the changes in E2 levels observed during epithelial differentiation.
Acknowledgments
We are grateful to members of our laboratories for helpful discussions.
This work has been supported by grants R01CA116720 to P.M.H., RO1AG011085 and RO1GM070565 to J.W.H., and T32CA009361-27 to J.A.S. from the National Institutes of Health.
Footnotes
Published ahead of print on 24 June 2009.
REFERENCES
- 1.Abbate, E. A., C. Voitenleitner, and M. Botchan. 2006. Structure of the papillomavirus DNA-tethering complex E2:Brd4 and a peptide that ablates HPV chromosomal association. Mol. Cell 24877-889. [DOI] [PubMed] [Google Scholar]
- 2.Ang, X. L., and J. W. Harper. 2005. SCF-mediated protein degradation and cell cycle control. Oncogene 242860-2870. [DOI] [PubMed] [Google Scholar]
- 3.Baxter, M. K., M. G. McPhillips, K. Ozato, and A. A. McBride. 2005. The mitotic chromosome binding activity of the papillomavirus E2 protein correlates with interaction with the cellular chromosomal protein, Brd4. J. Virol. 794806-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellanger, S., C. Demeret, S. Goyat, and F. Thierry. 2001. Stability of the human papillomavirus type 18 E2 protein is regulated by a proteasome degradation pathway through its amino-terminal transactivation domain. J. Virol. 757244-7251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crawford, N. P., J. Alsarraj, L. Lukes, R. C. Walker, J. S. Officewala, H. H. Yang, M. P. Lee, K. Ozato, and K. W. Hunter. 2008. Bromodomain 4 activation predicts breast cancer survival. Proc. Natl. Acad. Sci. USA 1056380-6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cullinan, S. B., J. D. Gordan, J. Jin, J. W. Harper, and J. A. Diehl. 2004. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol. Cell. Biol. 248477-8486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dey, A., F. Chitsaz, A. Abbasi, T. Misteli, and K. Ozato. 2003. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc. Natl. Acad. Sci. USA 1008758-8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dey, A., J. Ellenberg, A. Farina, A. E. Coleman, T. Maruyama, S. Sciortino, J. Lippincott-Schwartz, and K. Ozato. 2000. A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G2-to-M transition. Mol. Cell. Biol. 206537-6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.French, C. A., I. Miyoshi, J. C. Aster, I. Kubonishi, T. G. Kroll, P. Dal Cin, S. O. Vargas, A. R. Perez-Atayde, and J. A. Fletcher. 2001. BRD4 bromodomain gene rearrangement in aggressive carcinoma with translocation t(15;19). Am. J. Pathol. 1591987-1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.French, C. A., I. Miyoshi, I. Kubonishi, H. E. Grier, A. R. Perez-Atayde, and J. A. Fletcher. 2003. BRD4-NUT fusion oncogene: a novel mechanism in aggressive carcinoma. Cancer Res. 63304-307. [PubMed] [Google Scholar]
- 11.Gagnon, D., S. Joubert, H. Senechal, A. Fradet-Turcotte, S. Torre, and J. Archambault. 2009. Proteasomal degradation of papillomavirus E2 protein is inhibited by overexpression of the bromodomain-containing protein 4. J. Virol. 834127-4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geyer, R., S. Wee, S. Anderson, J. Yates, and D. A. Wolf. 2003. BTB/POZ domain proteins are putative substrate adaptors for cullin 3 ubiquitin ligases. Mol. Cell 12783-790. [DOI] [PubMed] [Google Scholar]
- 13.Hattori, T., K. Kitagawa, C. Uchida, T. Oda, and M. Kitagawa. 2003. Cks1 is degraded via the ubiquitin-proteasome pathway in a cell cycle-dependent manner. Genes Cells 8889-896. [DOI] [PubMed] [Google Scholar]
- 14.Houzelstein, D., S. L. Bullock, D. E. Lynch, E. F. Grigorieva, V. A. Wilson, and R. S. Beddington. 2002. Growth and early postimplantation defects in mice deficient for the bromodomain-containing protein Brd4. Mol. Cell. Biol. 223794-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Howley, P. M., and D. R. Lowy. 2007. Papillomaviruses, p. 2299-2354. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed., vol. 2. Lippincott, Williams and Wilkins, Philadelphia, PA. [Google Scholar]
- 16.Ilves, I., K. Maemets, T. Silla, K. Janikson, and M. Ustav. 2006. Brd4 is involved in multiple processes of the bovine papillomavirus type 1 life cycle. J. Virol. 803660-3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin, J., X. L. Ang, T. Shirogane, and J. W. Harper. 2005. Identification of substrates for F-box proteins. Methods Enzymol. 399287-309. [DOI] [PubMed] [Google Scholar]
- 18.Kipreos, E. T., L. E. Lander, J. P. Wing, W. W. He, and E. M. Hedgecock. 1996. cul-1 is required for cell cycle exit in C. elegans and identifies a novel gene family. Cell 85829-839. [DOI] [PubMed] [Google Scholar]
- 19.Lee, A. Y., and C. M. Chiang. 2009. Chromatin adaptor Brd4 modulates E2 transcription activity and protein stability. J. Biol. Chem. 2842778-2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McPhillips, M. G., J. G. Oliveira, J. E. Spindler, R. Mitra, and A. A. McBride. 2006. Brd4 is required for E2-mediated transcriptional activation but not genome partitioning of all papillomaviruses. J. Virol. 809530-9543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mochizuki, K., A. Nishiyama, M. K. Jang, A. Dey, A. Ghosh, T. Tamura, H. Natsume, H. Yao, and K. Ozato. 2008. The bromodomain protein Brd4 stimulates G1 gene transcription and promotes progression to S phase. J. Biol. Chem. 2839040-9048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parish, J. L., A. M. Bean, R. B. Park, and E. J. Androphy. 2006. ChlR1 is required for loading papillomavirus E2 onto mitotic chromosomes and viral genome maintenance. Mol. Cell 24867-876. [DOI] [PubMed] [Google Scholar]
- 23.Penrose, K. J., M. Garcia-Alai, G. de Prat-Gay, and A. A. McBride. 2004. Casein kinase II phosphorylation-induced conformational switch triggers degradation of the papillomavirus E2 protein. J. Biol. Chem. 27922430-22439. [DOI] [PubMed] [Google Scholar]
- 24.Penrose, K. J., and A. A. McBride. 2000. Proteasome-mediated degradation of the papillomavirus E2-TA protein is regulated by phosphorylation and can modulate viral genome copy number. J. Virol. 746031-6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petroski, M. D., and R. J. Deshaies. 2005. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 69-20. [DOI] [PubMed] [Google Scholar]
- 26.Pintard, L., A. Willems, and M. Peter. 2004. Cullin-based ubiquitin ligases: Cul3-BTB complexes join the family. EMBO J. 231681-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pintard, L., J. H. Willis, A. Willems, J. L. Johnson, M. Srayko, T. Kurz, S. Glaser, P. E. Mains, M. Tyers, B. Bowerman, and M. Peter. 2003. The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature 425311-316. [DOI] [PubMed] [Google Scholar]
- 28.Poddar, A., S. C. Reed, M. G. McPhillips, J. E. Spindler, and A. A. McBride. 2009. The human papillomavirus type 8 E2 tethering protein targets the ribosomal DNA loci of host mitotic chromosomes. J. Virol. 83640-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schweiger, M. R., M. Ottinger, J. You, and P. M. Howley. 2007. Brd4-independent transcriptional repression function of the papillomavirus E2 proteins. J. Virol. 819612-9622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schweiger, M. R., J. You, and P. M. Howley. 2006. Bromodomain protein 4 mediates the papillomavirus E2 transcriptional activation function. J. Virol. 804276-4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Senechal, H., G. G. Poirier, B. Coulombe, L. A. Laimins, and J. Archambault. 2007. Amino acid substitutions that specifically impair the transcriptional activity of papillomavirus E2 affect binding to the long isoform of Brd4. Virology 35810-17. [DOI] [PubMed] [Google Scholar]
- 32.Spruck, C. H., and H. M. Strohmaier. 2002. Seek and destroy: SCF ubiquitin ligases in mammalian cell cycle control. Cell Cycle 1250-254. [PubMed] [Google Scholar]
- 33.Thierry, F., J. Heard, K. Dartmann, and M. Yaniv. 1987. Characterization of a transcriptional promoter of human papillomavirus 18 and modulation of its expression by simian virus 40 and adenovirus early antigens. J. Virol. 61134-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van den Heuvel, S. 2004. Protein degradation: CUL-3 and BTB—partners in proteolysis. Curr. Biol. 14R59-R61. [PubMed] [Google Scholar]
- 35.von der Lehr, N., S. Johansson, and L. G. Larsson. 2003. Implication of the ubiquitin/proteasome system in Myc-regulated transcription. Cell Cycle 2403-407. [PubMed] [Google Scholar]
- 36.Wang, X., S. R. Naidu, F. Sverdrup, and E. J. Androphy. 2009. Tax1BP1 interacts with papillomavirus E2 and regulates E2-dependent transcription and stability. J. Virol. 832274-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu, S. Y., A. Y. Lee, S. Y. Hou, J. K. Kemper, H. Erdjument-Bromage, P. Tempst, and C. M. Chiang. 2006. Brd4 links chromatin targeting to HPV transcriptional silencing. Genes Dev. 202383-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu, L., Y. Wei, J. Reboul, P. Vaglio, T. H. Shin, M. Vidal, S. J. Elledge, and J. W. Harper. 2003. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature 425316-321. [DOI] [PubMed] [Google Scholar]
- 39.Yang, Z., N. He, and Q. Zhou. 2008. Brd4 recruits P-TEFb to chromosomes at late mitosis to promote G1 gene expression and cell cycle progression. Mol. Cell. Biol. 28967-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.You, J., J. L. Croyle, A. Nishimura, K. Ozato, and P. M. Howley. 2004. Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell 117349-360. [DOI] [PubMed] [Google Scholar]
- 41.You, J., M. R. Schweiger, and P. M. Howley. 2005. Inhibition of E2 binding to Brd4 enhances viral genome loss and phenotypic reversion of bovine papillomavirus-transformed cells. J. Virol. 7914956-14961. [DOI] [PMC free article] [PubMed] [Google Scholar]