Abstract
Rotaviruses have a genome composed of 11 segments of double-stranded RNA (dsRNA) surrounded by three protein layers. The virus contains an RNA-dependent RNA polymerase that synthesizes RNA transcripts corresponding to all segments of the viral genome. These transcripts direct the synthesis of the viral proteins and also serve as templates for the synthesis of the complementary strand to form the dsRNA genome. In this work, we analyzed the kinetics of transcription and replication of the viral genome throughout the replication cycle of the virus using quantitative reverse transcription-PCR. The role of the proteins that form double-layered particles ([DLPs] VP1, VP2, VP3, and VP6) in replication and transcription of the viral genome was analyzed by silencing their expression in rotavirus-infected cells. All of them were shown to be essential for the replication of the dsRNA genome since in their absence there was little synthesis of viral mRNA and dsRNA. The characterization of the kinetics of RNA transcription and replication of the viral genome under conditions where these proteins were silenced provided direct evidence for a second round of transcription during the replication of the virus. Interestingly, despite the decrease in mRNA accumulation when any of the four proteins was silenced, the synthesis of viral proteins decreased when VP2 and VP6 were knocked down, whereas the absence of VP1 and VP3 did not have a severe impact on viral protein synthesis. Characterization of viral particle assembly in the absence of VP1 and VP3 showed that while the formation of triple-layered particles and DLPs was decreased, the amount of assembled lower-density particles, often referred to as empty particles, was not different from the amount in control-infected cells, suggesting that viral particles can assemble in the absence of either VP1 or VP3.
The family Reoviridae includes viruses that have a genome composed of 9 to 12 segments of double-stranded RNA (dsRNA). Rotaviruses belong to the most medically significant genus of the family since they are the main cause of infantile gastroenteritis, causing approximately 500,000 deaths per year in children less than 5 years of age (23). These viruses have a genome composed of 11 segments of dsRNA, which is enclosed in a capsid formed by three concentric layers of protein (33). The innermost layer, formed by VP2, contains the viral genome and 12 copies each of the virus RNA-dependent RNA polymerase (RdRP; VP1) and the guanylyltransferase and methylase enzyme (VP3); these viral elements constitute the core of the virus. The addition of VP6 on top of the VP2 layer produces double-layered particles (DLPs). The outermost layer, characteristic of infectious, triple-layered particles (TLPs), is composed of two proteins, VP4 and VP7.
During or shortly after cell entry, the infecting TLP uncoats, loosing the two proteins of the outer layer and yielding a DLP, which is transcriptionally active (14). The nascent transcripts are extruded into the cell's cytoplasm through channels located near the icosahedral fivefold vertices of the particle (32). The viral mRNAs contain 5′-methylated cap structures but lack the poly(A) tails characteristic of most cellular mRNAs (11). The viral RNA transcripts direct the synthesis of six structural (VP1 to VP4, VP6, and VP7) and six nonstructural (NSP1 to NSP6) proteins (4). In addition to their function as mRNAs, the viral transcripts also serve as RNA templates (positive-strand RNA) for the synthesis of negative-strand RNA to form the dsRNA genome segments. The segmented dsRNA genome of rotaviruses is never detected free in the cytoplasm but is transcribed and replicated within viral capsids by the RdRP. The synthesis of negative-strand RNA has been proposed to occur in perinuclear nonmembranous, electrodense cytoplasmic structures, known as viroplasms, concurrently with the packaging of positive-strand RNA into core replication intermediate (RI) particles (30). It has been proposed that this process leads to the production of new transcriptionally active, dsRNA-containing double-layered RI particles, which are thought to be responsible for an enhanced second round of transcription, resulting in a second wave of assembly of double-layered RI particles (4). The occurrence of this second round, however, has not been directly demonstrated.
The participation of several viral proteins in the replication process of rotavirus has been studied using different approaches; the characterization of temperature-sensitive mutants underlined the importance of VP1, VP2, and VP3 in the replication process (17, 39). In a different approach, an in vitro replication assay was developed (2); in this system it has been observed that VP1 and VP2 are necessary and sufficient to replicate the genome and that no other structural or nonstructural protein was necessary although the lack of genome encapsidation in this system indicated the possible role of other viral proteins in coordinating the process of replication and sorting of the RNA segments in equimolar amounts during an infection (24). Finally, the relevance of NSP5 and NSP2 in the replication of the virus has been shown in vivo using RNA interference. It was found that genome replication, viral mRNA translation, and viroplasm formation were severely affected in rotavirus-infected cells expressing low levels of NSP5 or NSP2 (1, 16, 35).
In this report we determined the kinetics of replication and transcription of the rotaviral genome during the replication cycle using real-time quantitative reverse transcription-PCR (qRT-PCR). The role of the proteins that constitute the DLPs (VP1, VP2, VP3, and VP6) in an infected cell was analyzed by silencing the expression of each of these proteins in rotavirus-infected cells. We found that all of them are essential for the replication of the viral genome since in their absence there is little synthesis of viral mRNA, and the second round of transcription does not take place. Interestingly, despite the decrease in transcription when any of the four proteins was silenced, only VP2 and VP6 seem to be important for the translation of the viral proteins, whereas the absence of VP1 and VP3 does not have a severe impact on the translation of viral proteins. The assembly of viral particles in the absence of VP1 and VP3 was assessed by CsCl gradients. As expected, we found that there was little assembly of TLPs and DLPs in the absence of these proteins, but the amount of lower-density particles formed, often referred to as empty particles (to indicate that they are devoid of viral dsRNA), was not different from the quantity in the nonsilenced control, suggesting that the assembly of the viral particles can proceed in the absence of either VP1 or VP3. The stability of a transfected viral exogenous reporter mRNA in uninfected or infected cells was measured by qRT-PCR; when VP2 was silenced, we found a decrease in the half-life of the reporter rotavirus mRNA, suggesting that, besides its role as a capsid protein during the replication process, VP2 might also participate in stabilizing the mRNAs committed to translation.
MATERIALS AND METHODS
Cells and viruses.
The rhesus monkey epithelial cell line MA104 was grown in Eagle's minimal essential medium (MEM) supplemented with 10% fetal bovine serum and was used for all experiments carried out in this work. Rhesus rotavirus RRV was obtained from H. B. Greenberg (Stanford University, Stanford, CA); simian rotavirus SA11 was obtained from H. H. Malherbe, and porcine rotavirus YM was isolated in our laboratory. All rotavirus strains were propagated in MA104 cells as described previously (22).
Antibodies.
Monoclonal antibodies (MAbs) to VP2 (3A8), VP6 (255/60), and NSP4 (B4) were kindly provided by H. B. Greenberg. MAb HS2 directed to VP4 (21), the rabbit antirotavirus polyclonal serum raised against purified RRV TLPs, the rabbit anti-VP1, and antivimentin serum were produced in our laboratory. Rabbit polyclonal antisera to NSP2 and NSP3 have been described previously (9). Fab fragments to digoxigenin (DIG)-peroxidase were purchased from Roche, Inc. Alexa Fluor 488- and 647-conjugated secondary antibodies were purchased from Molecular Probes (Eugene, OR). Horseradish peroxidase-conjugated goat anti-rabbit polyclonal antibody was from Perkin Elmer Life Sciences (Boston, MA), and horseradish peroxidase-conjugated rabbit anti-mouse immunoglobulin G was from Zymed (San Francisco, CA).
Real-time PCR.
Reverse transcriptase and PCR primers were designed using the Primer3 software (http://fokker.wi.mit.edu/cgi-bin/primer3/primer3_www_slow.cgi). The sequences of the primers used are shown in Table 1. The amplification efficiency of each pair of primers was determined by using the slope value of the standard curve in the following formula: efficiency = (10(−1/slope) − 1) ×100. All the primers used had an amplification efficiency greater than 90% (data not shown). To specifically determine the levels of each RNA strand, RT-PCR was performed in two separate steps. The first step was performed using only one of the primers (depending on the strand to be quantitated) in the following mixture: 12.5 μl of 2× SYBR green Master Mix (Applied Biosystems), 0.125 μl of reverse transcriptase (50 U/μl), 0.25 μl of RNase inhibitor (20 U/μl), and 1 μl of either forward or reverse primer (2.5 pmol/μl), plus RNA (1 ng; boiled for 5 min and chilled on ice) in a total volume of 24 μl. The reaction mixture was incubated for 30 min at 48°C, and the reverse transcriptase was inactivated by incubating the samples at 90°C for 10 min and immediately chilled on ice. Then, for the PCR step, 1 μl of the complementary primer (2.5 pmol/μl) was added to the reaction mixture (for a final volume of 25 μl), and the samples were amplified by PCR in an ABI Prism 7500 Sequence Detector System (Applied Biosystems) with the following thermal protocol: 95°C for 10 min and then 40 cycles of 95°C for 15 s and 60°C for 1 min, followed by dissociation phase of 60°C to 95°C for 30 min.
TABLE 1.
Sequence for primers used in this work
| Gene | Forward primer (5′ → 3′) | Reverse primer (5′ → 3′) | Amplicon (nt)a |
|---|---|---|---|
| 1 | GGACGCCGCACTAGATCAATT | GGTCATCATGCTTTACTGGTTCC | 1038-1168 |
| 6 | CCACTTGGTATCCGACTTTGA | GAATACGTGGACGCATCCTT | 1228-1309 |
| 10 | TCCTGGAATGGCGTATTTTC | GAGCAATCTTCATGGTTGGAA | 122-214 |
| 10Mut | TCCTGGAATGGCGTATTTTC | GAGCAATCTTCATGGTTCTGC | 122-214* |
| GAPDH | ACCTGACCTGCCGTCTAGAAA | CCTGCTTCACCACCTTCTTGAT | 783-836 |
| Luciferase | GCCTGAAGTCTCTGATTAAGT | ACACCTGCGTCGAAGATGT | 1359-1456 |
nt, nucleotide; *, same nucleotide positions of gene 10 primer, with changes indicated in bold.
A standard curve was generated by amplifying known amounts of dsRNA using either forward or reverse primers (for negative- or positive-strand amplification, respectively), during the RT step. After the PCR amplification, the ABI Prism software was used to set a cutoff line with the obtained fluorescence values (y axis) for all the samples between the logarithmic phases of the amplification curves. Then, the logarithm of concentration of each sample was plotted against the cycle number where the amplification curve of the sample reached the cutoff line (cycle threshold [CT]). The amount of either positive- or negative-strand RNA from unknown samples was determined by extrapolating the CT value onto the corresponding standard curve. The amount of negative-strand RNA is referred to as dsRNA (since it is only present in this form), whereas the amount of mRNA was calculated by subtracting the amount of negative-strand RNA from the amount of positive-strand RNA obtained in an assay.
Relative quantification.
Negative- or positive-strand RNA was amplified, as described above, and the CT for each amplified sample was calculated. The relative increase was determined by the 2−ΔΔCT method (15). Data are expressed as the relative difference in either negative- or positive-strand RNA with respect to the total amount of negative-strand RNA present at time zero postinfection (assigning this sample the arbitrary value of 1). GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA was used as an internal control for all the samples.
siRNA transfection.
Duplex small interfering RNAs (siRNAs) were obtained from Dharmacon Research (Lafayette, CO), and the target sequences used to knock down VP1, VP2, VP3, VP4, and VP6 genes were previously described (20). As an irrelevant control, a previously reported siRNA to firefly luciferase was used (19). Transfection of siRNAs into MA104 cells was performed as previously described (3). Briefly, the siRNAs were transfected using Lipofectamine (Invitrogen). The transfection mixture was added to confluent cell monolayers and incubated for 8 h at 37°C. After this time, the mixture was removed, and the cells were kept in MEM for 48 h at 37°C prior to virus infection.
Infection of cells and titration of viral progeny.
Transfected cell monolayers, in 24- or 48-well plates, were infected with 3 focus-forming units of virus per cell and then incubated for 24 h at 37°C. At this time the cells were then lysed by two freeze-thaw cycles, and the lysates were treated with 10 μg/ml of trypsin for 30 min at 37°C. The infectious titers of the viral preparations were obtained by an immunoperoxidase focus assay as described previously (22). Briefly, confluent MA104 cells in 96-well plates were washed twice with phosphate-buffered saline (PBS), and twofold serial dilutions of the above-mentioned viral lysate were adsorbed to the cells for 60 min at 37°C. After the adsorption period, the virus inoculum was removed, the cells were washed once with PBS, MEM was added, and the infection was left to proceed for 14 h for RRV at 37°C. RRV-infected cells were detected by an immunoperoxidase focus detection assay using a rabbit hyperimmune serum to rotavirus, as described previously (22). The numbers of focus-forming units were counted with the help of a Visiolab 1000 station (Biocom, France) as previously reported (10).
Immunoblots.
Cells were transfected with siRNAs and infected with rotavirus RRV as described above. The cells were lysed with Laemmli sample buffer, and the proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to nitrocellulose membranes (Millipore, Bedford, MA). Membranes were blocked with 5% nonfat dried milk in PBS and incubated for 1 h at room temperature with primary antibodies in PBS containing 1% milk, followed by an incubation with secondary, species-specific, horseradish peroxidase-conjugated antibodies or with Alexa Fluor-488- and -647-conjugated secondary antibodies. The peroxidase activity was developed by a Western Lightning Chemiluminescence Reagent Plus (Perkin Elmer Life Sciences), following the manufacturer's instructions, and the fluorescently labeled antibodies were visualized on a Typhoon Trio (Amersham) and quantified using ImageQuant TL software.
Immunofluorescence.
MA104 cells grown on glass coverslips were transfected as previously mentioned, and at 48 h posttransfection the cells were infected with RRV at a multiplicity of infection (MOI) of 3. At 8 h postinfection (hpi), the cells were fixed with 2% paraformaldehyde in PBS for 20 min at room temperature. After this time the cells were washed twice with PBS containing 50 mM NH4Cl, permeabilized by incubation with PBS-0.5% Triton X-100-50 mM NH4Cl for 15 min at room temperature, and washed twice with PBS with gentle swirling. The coverslips were then incubated for 1 h at room temperature with primary antibodies diluted in blocking buffer (50 mM NH4Cl, 1% bovine serum albumin [BSA] in PBS) and then rinsed four times with PBS. The coverslips were then incubated with the appropriate Alexa Fluor-labeled secondary antibodies in blocking buffer for 1 h at room temperature. The cells were washed four times with PBS and mounted on glass slides with Fluoprep (BioMérieux). The slides were analyzed with a Nikon E600 epifluorescence microscope coupled to a DXM1200 digital still camera (Nikon). The images were then digitally captured and prepared in Adobe Photoshop, version 7.0.
Radiolabeling, isolation, and analysis of viral particles.
Cells grown in 48-well plates were transfected with siRNAs and infected with rotavirus RRV as described above. At 7 hpi, the medium was replaced by MEM without methionine, supplemented with 25 μCi/ml of Easy Tag EXPRESS-35S labeling mix (Dupont-NEN) and incubated for 1 h; after this period the cells were washed and lysed with Laemmli sample buffer. For isolation and purification of viral particles, cells grown in 48-well plates were transfected with siRNAs and infected with rotavirus RRV as described above. At 6 hpi, the medium was replaced by MEM without methionine, supplemented with 25 μCi/ml of Easy Tag EXPRESS-35S labeling mix (Dupont-NEN). At 12 hpi the cells were frozen and thawed twice, the viral lysate (from 12 wells/condition) was extracted with Freon 113 (Dupont, Wilmington, DE), CsCl was added to the aqueous phase to obtain a density of 1.36 g/cm3, the mixture was centrifuged for 18 h at 35,000 rpm in an SW40Ti rotor, and fractions (400 μl) were collected from the bottom. The protein composition of each fraction was analyzed by SDS-PAGE and autoradiography. A densitometric analysis of the protein bands in the autoradiogram was done using the ImageQuant TL software.
Preparation of the RNA probe.
To prepare an RNA probe complementary to viral mRNA gene 10, a cDNA copy of RRV gene 10 was amplified using primers T7-10 forward (5′-CGCGGCGCCTAATACGACTCACTATAGGCTTTTAAAAGTTCTG-3′) and YM10-3′ reverse (5′-CAGACCCGGGCCGCGGTCACATTAAGACCGTTC-3′), cleaved with KasI and AvaI, and ligated into the KasI and AvaI sites of plasmid pGEM-3Z (pGEM-NSP4). In this construct, the gene for NSP4 was cloned downstream of the T7 promoter and upstream of the SP6 promoter. This plasmid was linearized with EcoRI, which cleaves gene 10 at nucleotide position 427, and purified by phenol-chloroform extraction and ethanol precipitation. RNA probes were generated by in vitro transcription of the linearized plasmid using a Megascript SP6 kit (Ambion) in the presence of DIG-UTP (Roche) according to the manufacturer's instructions. The SP6 polymerization mix contained 1 μg of linearized DNA; 5 mM each of ATP, GTP, and CTP; 1.66 mM UTP; and 0.66 mM DIG-UTP. The RNA probe produced was 323 nucleotides in length, and its concentration was determined by spectrophotometry.
FISH.
To detect by hybridization the RNA segment 10 positive strand, the protocol of fluorescence in situ hybridization (FISH) as previously described by Mingle et al. (18) was followed. Briefly, MA104 cells were grown in coverslips and infected with RRV at an MOI of 3. At 6 hpi cells were fixed with 2% paraformaldehyde in PBS for 15 min at room temperature, washed twice with 50 mM NH4Cl in PBS, permeabilized with 0.5% Triton X-100, and incubated for 30 min at room temperature with 1.5% H2O2. Fixed cells were incubated overnight at 58°C with 120 μl (0.2 ng/μl) of an RNA probe complementary to the positive strand of rotavirus RNA gene 10 (see above) in hybridization buffer (50% formamide, 5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 50 mg/ml heparin, 0.1% Tween 20, and 1 μg/ml of total yeast RNA). Then, the cells were washed three times with hybridization buffer at 70°C and three times with 0.1 × SSC and blocked with blocking buffer (1% BSA, 0.2% nonfat milk, 0.3% Triton X-100) for 1 h at room temperature. The RNA probe was detected by incubating the cells with a monoclonal anti-DIG antibody conjugated with peroxidase for 1 h at room temperature. After this step, the fluorescence signal for RNA detection was obtained by using the tyramide signal amplification system (TSA-Plus Fluorescence Palette System; Perkin Elmer) as recommended by the manufacturer. For simultaneous protein immunodetection, specific antibodies were used during the anti-DIG antibody incubation (all antibodies were diluted 1:1,000); cells were washed five times with TNT buffer (0.1 M Tris pH 7.5, 0.15 M NaCl, and 0.05% Tween 20), incubated with the corresponding secondary antibody for 1 h at 37°C, and washed five times with TNT buffer. Finally, the cells were mounted on glass slides with Fluoprep (Bio Mérieux) and observed as previously described.
RNA stability assays.
Gene 10 cloned in plasmid pGEM3Z (pGEM-NSP4) was mutagenized at nucleotide position 190 and positions 193 to 196 using a QuikChange kit (Stratagene), and the resultant plasmid (pGEM-NSP4mut) was sequenced to verify the introduced mutations. As a control mRNA, we used a luciferase reporter gene to which an AclI restriction site after the poly(A) tail sequence was introduced by PCR in the luciferase T7 control plasmid (Promega) using the primers T7-forward (5′-TAATACGACTCACTATAG-3′) and reverse (5′-AAAAAAAAAAAAAACGTTATTGGCATCACCGGC-3′). The resulting PCR product was cleaved with AclI, and blunt ends were generated by micrococcal nuclease digestion. The SacII-linearized pGEM-NSP4mut plasmid and the poly(A)-luciferase PCR product were purified by phenol-chloroform extraction and ethanol purification. The gene 10 segment was positioned such that the T7 transcript derived from plasmid pGEM-NSP4mut (reporter gene 10Mut) digested with SacII was predicted to have the same 5′ and 3′ terminal ends of the corresponding rotavirus gene 10 sequence (GenBank accession number L41247). Capped RNAs were synthesized using a MegaScript T7 transcription system (Ambion) in the presence of a cap analog (New England Biolabs). The T7 polymerization mixture contained 1 μg of linearized DNA; 7.5 mM each of ATP, CTP, and UTP; 1.5 mM GTP; and 6 mM cap analog (New England Biolabs). RNA was purified by phenol-chloroform extraction, its concentration was determined by spectrophotometry, and aliquots were stored at −80°C.
To measure the half-life of the reporter mRNAs, 1 μg of RNA was transfected for 2 h into 5 × 105 MA104 cells, which were left uninfected or were previously infected with rotavirus for 1 h at 37°C, using Lipofectamine 2000 (Invitrogen). The transfection mixture was removed, and the cells were washed four times with MEM and incubated for 30 min at 37°C with 10 mg/ml RNase A in MEM to remove the extracellular nonspecifically bound RNA. Then, the cells were washed four times with 1% BSA in MEM and four times with MEM and incubated at 37°C for different times. Aliquots of the cells were harvested using Trizol at different times postinfection, and the RNA was purified as previously described. The reporter RNA gene 10Mut and the luciferase RNA were quantitated by real-time RT-PCR as described above, using GAPDH as an internal control. Primer 10Mut (Table 1) was used as a reverse primer to amplify the reporter for gene 10; in control assays this primer did not amplify the wild-type rotavirus gene 10 (data not shown). Primers used to amplify GAPDH and luciferase RNAs are also indicated in Table 1.
RESULTS
Standardization of real-time RT-PCR assay for specific quantitation of negative and positive strands of viral RNA.
To determine if it was possible to follow the kinetics of transcription and replication of the rotavirus genome using qRT-PCR, we standardized a protocol to independently quantify both strands (negative and positive) of the dsRNA produced during a viral infection. A set of primers (Table 1) was designed to amplify rotavirus genes 1, 6, and 10 as representatives of the different sizes of the viral genes (large, medium, and small, respectively). The efficiency of the primers was measured by generating a standard curve from twofold serial dilutions (from 0.05 to 150 ng) of a known amount of rotavirus dsRNA (see Materials and Methods). Given the double-stranded nature of the rotaviral genome, adding both primers to the RT-PCR would result in the amplification of both strands simultaneously. To avoid this problem, only one of the primers (depending on the strand to be quantitated) was used for RT; the complementary primer was added before the PCR was started and after a 10-min denaturation step to eliminate the reverse transcriptase activity. In this way, the cDNA synthesized from only one of the viral RNA strands was quantified.
To test if this assay was specific to quantify each viral RNA strand in a mix of negative and positive strands, a constant amount of viral mRNA (positive-strand RNA) was mixed with serial dilutions of viral dsRNA, and each strand was quantified in separate reactions by qRT-PCR. We found that the amount of negative-strand RNA was equivalent to the amount of dsRNA added and that the difference between positive- and negative-strand RNAs was equivalent to the amount of mRNA added to the reaction mixture (data not shown).
Kinetics of dsRNA and mRNA accumulation during the virus replication cycle.
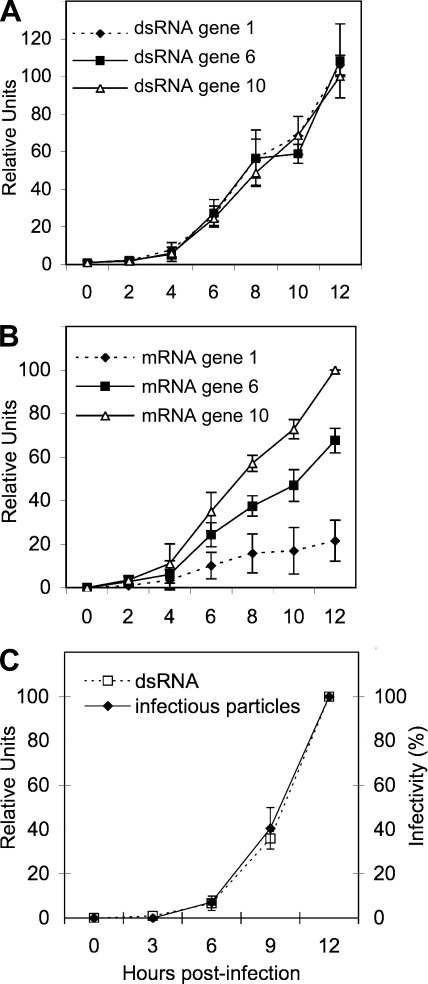
Once we established that the real-time RT-PCR assay was sufficiently sensitive and specific to separately quantify the positive and negative strands of the viral RNA, we decided to characterize the kinetics of synthesis of both types of RNAs produced during a 12-h infection course. MA104 cells were infected at an MOI of 0.1 to minimize the possibility of having more than one infecting virus per cell. After the adsorption period, noninternalized virus was washed off, total RNA from infected cultures was harvested every 2 h up to 12 hpi, and the amounts of both viral mRNA and dsRNA from viral segments 1, 6, and 10 were determined by qRT-PCR. The results shown in Fig. 1A and B indicate that while the dsRNA accumulation kinetics for the three RNA segments evaluated seemed equimolar, despite their differences in size, the amount of mRNA accumulated during the infection period was not equimolar. The amount of transcript detected was apparently size dependent since the amount of mRNA from RNA segment 10 produced at 12 hpi was 4.5 and 2 times more abundant than that for RNA segments 1, and 6, respectively, which is in agreement with the results reported by Stacy-Phipps and Patton, who quantitated the total amount of viral mRNA and dsRNA using an electrophoretic system (38). Interestingly, and also as previously reported (38), the amount of mRNA accumulated at 12 hpi was, on average, about six times more abundant than the dsRNA accumulated during that period of time.
FIG. 1.
Kinetics of transcription and replication of the viral RNA. Confluent MA104 cells were infected with RRV at an MOI of 0.1 by adsorbtion for 30 min at 4°C and then for 30 min at 37°C; the noninternalized virions were washed off with 3 mM EGTA in PBS, and MEM at 37°C was added. At the indicated times postinfection, RNAs were Trizol extracted and quantified by RT-PCR as described in Materials and Methods, or with 0.1% Triton X-100 for infectious virus titration. (A) RT-PCR of the negative strand of genes 1, 6, and 10. (B) Quantitation of the mRNAs corresponding to viral segments 1, 6, and 10. (C) RT-PCR of the negative strand of RNA segment 10 and titration of infectious particles by an immunoperoxidase focus assay as described in Materials and Methods. In panels A and B the qRT-PCR results are expressed as the increase relative to the amount of mRNA or negative strand from RNA segment 10 accumulated at 12 hpi, which was taken as 100%. Data shown represent the arithmetic means ± standard deviation of three independent experiments. In panel C data are expressed as percentage of total RNA synthesis or infectious particles produced at 12 hpi, which was taken as 100%. Data shown represent the arithmetic means ± standard deviation of three independent experiments.
To determine if there was a correlation between the production of viral dsRNA and the production of infectious viral particles, MA104 cells were infected as previously described, and the cells were harvested at different times. The viral dsRNA for gene 10 was quantitated by qRT-PCR, and the amount of infectious viral progeny produced was quantitated by an immunoperoxidase focus forming assay (22). The kinetics of both assays displayed the same tendency: a slight linear increase during the first 4 h and then a logarithmic increase at later times (Fig. 1C). These results suggest that the assembly of infectious viral particles parallels the replication of the viral genome.
Role of VP1, VP2, VP3, and VP6 in the viral replication cycle. (i) siRNAs directed to the DLP proteins specifically knock down their expression.
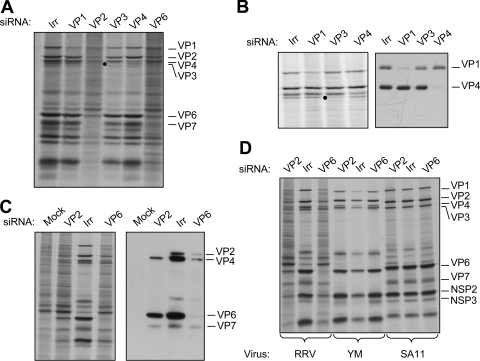
To study the role of DLP proteins in the transcription and replication of the viral genome during infection and to determine if it was possible to experimentally dissociate these two events, we silenced the expression of VP1, VP2, VP3, VP4, and VP6 by RNA interference. The cells were transfected with siRNAs that target each one of the above-mentioned RRV genes or with an siRNA complementary to the firefly luciferase gene as an irrelevant control. In these assays, MA104 cells were transfected with each siRNA, infected with rotavirus RRV at 48 h posttransfection, and metabolically labeled for 60 min at 7 hpi. The synthesis of viral and cellular proteins was assessed by SDS-PAGE and autoradiography (Fig. 2). The knockdown of every viral protein was apparent in the autoradiography shown in Fig. 2A. The effectiveness of each siRNA was also confirmed by Western blot analysis using specific antibodies against VP1, VP2, VP6, and VP4, which was used as a silencing control (Fig. 2B and C). The silencing of VP3 was assessed only by SDS-7% PAGE of 35S-labeled infected cells since specific antibodies against this protein were not available (Fig. 2B). When VP1 was silenced, a cellular protein that comigrates with VP1 was apparent in the autoradiogram shown in Fig. 2A and B; however, the Western blot with an antibody against VP1 confirmed that this protein was efficiently silenced.
FIG. 2.
Silencing the expression of the proteins that form the DLP has a differential effect on viral translation. MA104 cells in 48-well plates were transfected with the indicated siRNA as described in Materials and Methods. At 48 h posttransfection, the cells were infected with RRV at an MOI of 3 (A, B, and C) or with rotavirus strains with RRV, YM, and SA11 (D), and at 7 hpi the cells were radiolabeled for 60 min with 25 μCi/ml of Easy-tag Express-35S and then lysed. The labeled proteins were resolved by SDS-10% PAGE (A, B, and D) or by SDS-7% PAGE (B) and detected by autoradiography (left panels) or by immunoblot analysis (right panels) using either a mixture of antibodies to VP1 and VP4 (B) or a polyclonal antirotavirus antibody (C). Irr, irrelevant control siRNA.
Silencing the expression of the VP1, VP3, and VP4 genes did not affect significantly the amount of total viral protein synthesized, as we have previously observed (20). In contrast, silencing the expression of the VP2 and VP6 genes caused a general reduction in the synthesis of the viral proteins, which also correlated with an increased synthesis of cellular proteins (Fig. 2A).
It has been reported that some siRNAs can activate the interferon system, causing a nonspecific inhibition of cell protein synthesis (31, 37). Since the effect of the siRNAs directed to VP2 and to VP6 was more severe than expected, we analyzed the effect of these two siRNAs on cells infected with two different strains of rotavirus, a porcine rotavirus (YM) and a simian rotavirus (SA11), so that we could discard the possibility that these two siRNAs induced a nonspecific, generalized shutoff of protein synthesis. The sequences of the VP6 and VP2 genes from these two strains differ in 4 nucleotides and more than 10 nucleotides, respectively, from the sequences of the siRNAs used to knock down the expression of the corresponding RRV genes. Figure 2D shows that while the siRNAs to VP2 and VP6 almost completely inhibited the synthesis of RRV viral proteins, they did not affect the synthesis of any of the YM and SA11 proteins, indicating that the effect observed with the siRNAs directed to RRV VP6 and VP2 was specific and did not directly affect any other cellular or viral targets.
(ii) Silencing the expression of DLP proteins decreases the yield of infectious virus.
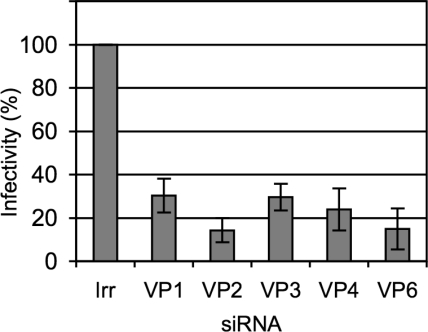
We then tested the effect of knocking down the expression of the DLP proteins on the production of infectious rotavirus progeny. siRNAs directed to VP1 to VP6 were transfected into MA104 cells, and 48 h after transfection the cells were infected with rotavirus RRV; 12 h later the progeny virus produced under these conditions was recovered, and its titer was determined by a focus-forming assay. Transfection with any of the siRNAs tested decreased the yield of viral progeny by about 80% while the irrelevant siRNA used as control did not affect the production of virus (Fig. 3). The inhibition of RRV viral yield by the siRNAs to VP2 and VP6 was shown to be specific since these siRNAs did not affect the production of infectious viral particles when the same assay was performed with the porcine rotavirus YM or the simian rotavirus SA11 (data not shown).
FIG. 3.
The yield of infectious progeny virus is decreased in cells expressing low levels of VP1 to VP6. MA104 cells in 48-well plates were transfected with the indicated siRNAs, and at 48 h posttransfection the cells were infected with RRV at an MOI of 3. The cells were harvested at 12 hpi, and the infectious virus produced was determined by an immunoperoxidase assay, as described in Materials and Methods. Data are expressed as the percentage of the infectivity obtained when the cells were transfected with an irrelevant (Irr) siRNA. Data shown represent the arithmetic mean ± standard deviation of three independent experiments.
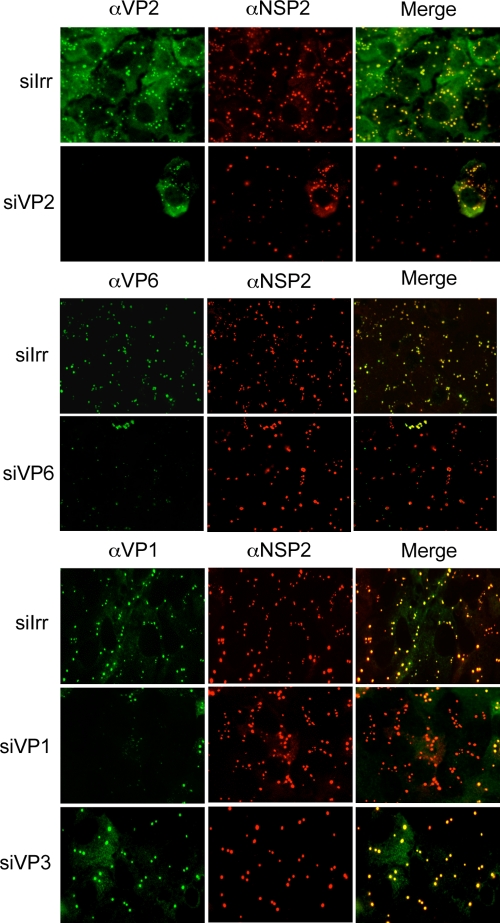
(iii) Viroplasms are formed in the absence of the DLP proteins.
Viroplasms are electrodense, nonmembranous structures that form upon rotavirus infection. It has been shown that viroplasms contain the proteins that constitute the DLPs, the viral RNA, and the nonstructural proteins NSP2 and NSP5 that are sufficient to form these structures when transiently expressed in transfected cells (5). To determine if, besides these two nonstructural proteins, any of the DLP proteins was necessary to form viroplasms in an infected cell, we used immunofluorescence to detect viroplasms in MA104 cells in which VP1, VP2, VP3, or VP6 was silenced. We found that viroplasms were formed in all cases irrespective of the siRNA used; however, the number and size of the viroplasms varied depending on the protein silenced (Fig. 4). When the expression levels of VP2 and VP6 were knocked down, the viroplasms appeared smaller and there were fewer of them than in control transfected cells. The siRNA directed to VP3 resulted in the formation of fewer but larger viroplasms. In general, even though the morphology and size of these structures varied depending on the protein silenced, none of these proteins seems to be essential for the formation of the viroplasms.
FIG. 4.
The formation of viroplasms is not impaired when the DLP proteins are knocked down. MA104 cells grown in coverslips were transfected with the indicated siRNAs; at 48 h posttransfection the cells were infected with RRV, and at 8 hpi the cells were fixed and immunostained as indicated in Materials and Methods. Antibodies against VP1, VP2, and VP6 were used, depending on the siRNA used, to assess the silencing of the corresponding protein of interest; antibodies to NSP2, VP6, and VP2 were used as markers of viroplasm formation, as indicated. α, anti; siIrr, irrelevant siRNA.
(iv) The synthesis of viral mRNA and dsRNA is inhibited when the expression of any of the DLP proteins is knocked down.
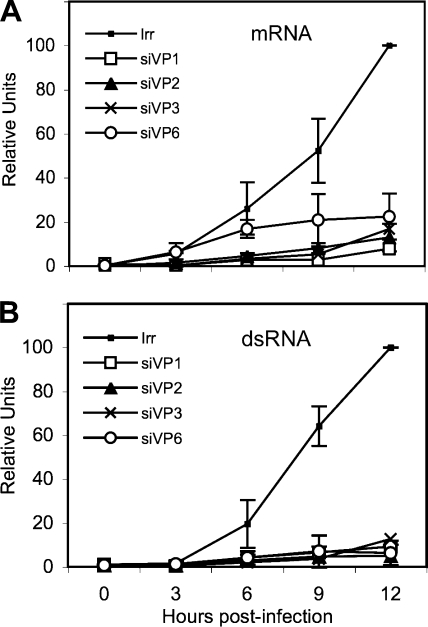
Once the qRT-PCR method was standardized and once the kinetics of accumulation of viral dsRNA and mRNA during a rotavirus infection was determined (Fig. 1), we decided to analyze the effect of silencing the expression of the DLP proteins in the synthesis of viral mRNA and dsRNA. MA104 cells were transfected with the siRNAs directed to VP1, VP2, VP3, or VP6; 48 h later the cells were infected with RRV, and total RNA was harvested with Trizol at different times postinfection. The amount of viral mRNA and dsRNA from viral segment 10 (which was used as a reporter gene) was determined by qRT-PCR as described above. Knocking down the expression of either VP1, VP2, VP3, or VP6 resulted in a marked decrease in the production of both dsRNA and mRNA (Fig. 5), which was more evident after 3 hpi, when there was a change in the kinetics of accumulation of RNA from being linear to an almost logarithmic increase. At 12 hpi the amount of dsRNA detected was about 1/10 of the amount of dsRNA produced in infected cells transfected with an irrelevant siRNA; a similar decrease in the amount of mRNA was detected when the expression of any of the DLP proteins was silenced. To discard the possibility that the mRNA and dsRNA from segment 10 behaved differently from the rest of the viral genes (given the fact that this mRNA, which encodes NSP4, is translated in the rough endoplasmic reticulum), we also determined the accumulation kinetics of the mRNA and dsRNA from gene segment 6 in cells in which either VP1 or VP2 was silenced. The results obtained using gene segment 6 were very similar to those found for gene segment 10 (data not shown), suggesting that the accumulation kinetics of at least these two genes are similar, irrespective of their site of translation.
FIG. 5.
Kinetics of transcription and replication of the viral RNA in the absence of the DLP proteins. Confluent MA104 cells were transfected with the siRNAs directed to VP1, VP2, VP3, or VP6 (designated as siVP1, for example), as indicated, and 48 h later the cells were infected with RRV at an MOI of 0.1 for 1 h at 37°C; the noninternalized virions were washed off with 3 mM EGTA in PBS, and at the indicated times postinfection, RNAs were Trizol extracted and quantified by RT-PCR as described in Materials and Methods using primer pairs specific for gene segment 10. (A) RT-PCR of the mRNA corresponding to viral segment 10. (B) Quantitation of the negative strand of gene 10. The qRT-PCR results are expressed as the increase relative to the amount of mRNA or negative strand produced in cells transfected with the control siRNA, which was taken as 100%. Data shown represent the arithmetic means ± standard deviation of three independent experiments.
Reduced levels of VP1 or VP3 decrease the formation of DLPs and TLPs, whereas the amount of low-density particles remains similar.
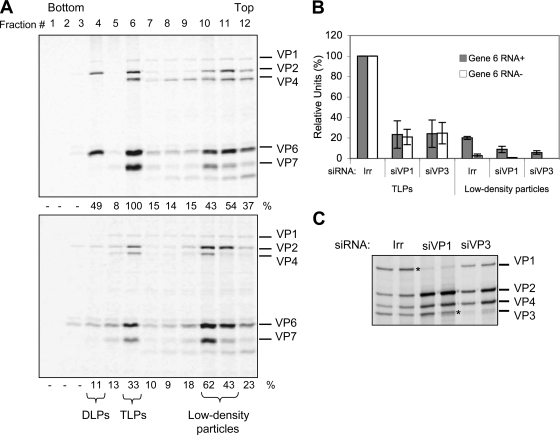
Since the synthesis of viral proteins was not affected when either VP1 or VP3 was silenced, we evaluated the effect of the absence of these proteins on virus particle assembly. For this, cells transfected with either a control siRNA or with siRNAs to VP1 and to VP3 were infected with RRV and labeled with an 35S labeling mix, and at 12 hpi the virus particles produced were purified by CsCl density gradients. The gradients were fractionated, and the protein composition of each fraction was analyzed by SDS-PAGE and autoradiography (Fig. 6A). The protein distribution obtained from the gradient of the control transfected cells shows that the DLPs and TLPs are distributed in fractions 4 to 6 while lower-density particles, also referred to as empty particles, are distributed near the top of the gradient in fractions 10 and 11. When the fractions from the gradients where VP3 or VP1 (not shown) were silenced, a decrease in the amounts of viral proteins present in the fractions corresponding to DLPs or TLPs was apparent while the viral proteins present in the upper fractions did not seem to be affected. To quantify this observation, the amount of VP6 in each fraction was determined by densitometry, and the amount of viral RNA present in fractions 6 and 10 was also determined by qRT-PCR (Fig. 6B).
FIG. 6.
Reduced levels of VP1 or VP3 decrease the formation of DLPs and TLPs, whereas the amounts of low-density particles remain similar. MA104 cells were transfected with the indicated siRNAs, and at 72 hpi the cells were infected with RRV at an MOI of 3. At 6 hpi cells were radiolabeled for 6 h with 25 μCi/ml of Easy-tag Express-35S and then harvested. Viral particles from each condition were separated by CsCl gradients (see Materials and Methods). Twelve 400-μl fractions were collected from each gradient and analyzed by PAGE and autoradiography (A). The amount of VP6 present in each fraction was calculated by densitometric analysis. The numbers below each lane represent the relative amounts of VP6 with respect to the amounts of VP6 present in the TLP fraction of the control gradient, which was taken as 100%. (B) Total RNA from the TLPs and low-density particle fractions was Trizol extracted and quantified by qRT-PCR as described in Materials and Methods using primer pairs specific for the positive (Gene 6 RNA+) and negative (Gene 6 RNA−) strands of rotavirus gene segment 6. The qRT-PCR results are expressed as the increase relative to the amount of positive or negative strand found in the TLP fraction produced in cells transfected with the control siRNA, which was taken as 100%. Data shown represent the arithmetic means ± standard deviation of three independent experiments. (C). Fractions 10 and 11 (low-density fractions) from gradients of the indicated conditions were resolved by SDS-7% PAGE and detected by autoradiography. An asterisk indicates the position of the silenced proteins. For siRNAs, siVP1 represents the siRNA directed against VP1; other designations follow the same form. Irr, irrelevant control siRNA.
Typically, density gradients of rotavirus particles yield two bands near the middle of the tube that correspond to DLPs and TLPs (3) and two additional lighter bands near the top of the gradient that contain particles with a protein composition very similar to DLPs and TLPs but devoid of dsRNA. In the experiments shown here, the viral proteins were labeled with 35S, and the gradients were fractioned and analyzed by gel electrophoresis. Figure 6A shows a representative PAGE gel of the gradient fractions obtained from a control infection and of an infection where VP3 was silenced. The amounts of TLPs produced when VP3 was knocked down were decreased by about 70% compared to the control transfected cells while the amount of protein present in the lighter fractions stayed in similar proportions. When the RNA present in the TLPs obtained from cells with knocked down expression of VP1 or VP3 was quantified, a similar result was obtained (Fig. 6B). Interestingly, when the amount of positive and negative strands of RNA segment 6 were quantified in the upper bands of the control infection, it was found that there was about seven times more positive- than negative-strand RNA, suggesting that at least part of these lighter particles are associated to transcripts of positive polarity. The amount of plus-sense gene 6 RNA present in the low-density particles from the conditions where VP1 or VP3 was silenced was reduced to about 8% and 6%, respectively, compared to the corresponding fractions of the control infection. Figure 6C shows an autoradiography of a 7% PAGE gel in which fractions 10 and 11 of each gradient were analyzed. The decreased amounts of VP1 and VP3 demonstrate the effectiveness of the siRNA treatment and also show that low-density particles can be assembled in the absence of either protein.
Cosilencing VP1 and VP6 partially restores the translation of viral proteins.
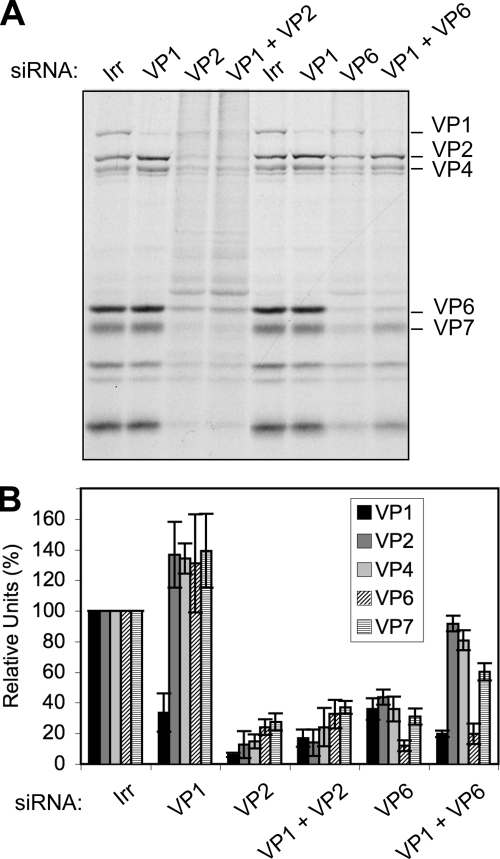
The finding that viral protein synthesis was differently affected when VP1, VP2, VP3, or VP6 was knocked down, despite the fact that the amount of viral mRNA synthesized in the infected cells was small and that the quantity was very similar in all cases, was unexpected. It has been proposed that during infection VP1 and VP3 bind to the 3′- and 5′-terminal ends of viral mRNAs, respectively (26, 27) and that, in turn, these interactions favor the formation of a core RI containing VP1, VP2, and VP3 that precludes the replication of the RNA. One possible explanation of the results previously described could be that when the expression of VP2 or VP6 is knocked down, the viral mRNAs may still be bound by VP1 and VP3; thus, the translation of the viral mRNAs would be prevented, and they could not be replicated in the absence of VP2 or VP6. On the other hand, in the absence of either VP1 or VP3, the viral mRNAs could not be bound to form the precore RI, and thus the mRNAs would remain available for translation. If this were the case, it would be expected that silencing the expression of either VP1 or VP3 in combination with either VP2 or VP6 would result in the partial rescue of viral protein synthesis. To test this hypothesis, cells were transfected with combinations of siRNAs directed to VP1 and VP2 or to VP1 and VP6; 48 h later the cells were infected, and the amount of viral protein synthesis was assessed by autoradiography of 35S-labeled lysates (Fig. 7A) and by densitometric analysis of Western blots (Fig. 7B). We found that viral protein synthesis was severely inhibited in cells where VP2 was silenced either alone or in combination with VP1. In contrast, the translation of viral protein synthesis was partially restored when VP6 was silenced in combination with VP1.
FIG. 7.
Cosilencing VP1 and VP6 partially recovers the translation of viral proteins. Cells were transfected with a mixture of siRNAs against VP1 and either VP6 or VP2, as indicated, and 48 h later they were infected with RRV at an MOI of 3. At 7 hpi cells were radiolabeled for 60 min with 25 μCi/ml of Easy-tag Express-35S and then lysed. Protein synthesis was visualized by autoradiography (A) or proteins were transferred to a nitrocellulose membrane for Western blot assays using MAb 3A8 to stain VP2 and polyclonal rabbit antibodies to VP1 and TLPs. As secondary antibodies, Alexa Fluor-conjugated antibodies were used. (B) The relative amount of each viral protein was calculated by densitometry of the fluorescent bands using ImageQuant TL software (Amersham Biosciences), and it is expressed as a percentage of the corresponding protein obtained when the irrelevant control siRNA(Irr) was used, which was taken as 100%. Data represent the arithmetic means ± standard deviation of at least three independent experiments.
Viral mRNA is more stable in infected cells.
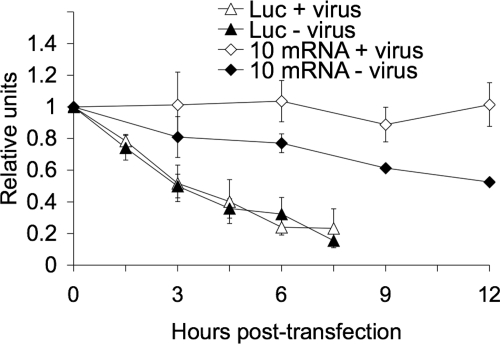
Since the efficient translation of the viral proteins was not restored when either VP1 (Fig. 7) or VP3 (data not shown) was silenced in VP2-knockdown cells, it could be possible that the inhibition of viral protein synthesis in VP2-silenced cells is due to a decreased stability of the viral mRNAs. To test this possibility, we measured the half-life of viral mRNAs in rotavirus-infected cells. Since it is not possible to completely stop viral transcription in an infected cell and since, therefore, it is not possible to reliably measure the decay of viral mRNAs using a radioactive method, we measured the stability of a reporter viral mRNA using qRT-PCR. In this case we measured the half-life of an in vitro T7 transcript of RRV gene 10 containing bona fide viral 5′- and 3′-terminal ends (see Materials and Methods). We also introduced into this construct five changes at nucleotide position 190 and positions 193 to 196 that allowed the differential amplification of this in vitro transcript using a reverse primer (Table 1, 10Mut) that hybridized specifically with the mutant transcript and not with the endogenous wild-type gene 10 mRNA (data not shown). The in vitro transcribed RNA was transfected into mock-infected or RRV-infected cells, the excess of noninternalized RNA was removed by RNase A treatment, total RNA was harvested at different times posttransfection, and the mutant transcript was quantified by qRT-PCR. As a control we used an in vitro transcribed firefly luciferase gene that contained cap at the 5′ end and a poly(A) tail at its 3′ end. The primers used to quantitate this gene are shown in Table 1. We found that the reporter viral mRNA had a half-life of about 12 h in uninfected cells while it remained essentially undegraded during the 12-h assay period when transfected in rotavirus-infected cells (Fig. 8). According to this method, the half-life of the control luciferase mRNA was about 240 min (Fig. 8), in agreement with previous reports (6), and it did not change in RRV-infected cells, suggesting that the change in stability observed for the gene 10 reporter viral mRNA was virus specific.
FIG. 8.
Rotavirus mRNA half-life increases in infected cells. MA104 cells (5 × 105 cells/assay) were infected (+ virus) or not (− virus) with RRV at an MOI of 3. At 1 hpi, 1 μg of either luciferase (Luc) or gene 10 reporter mRNA was transfected using Lipofectamine 2000 for 2 h. At the indicated times posttransfection, cells were lysed, and the transfected mRNAs were quantified by RT-PCR using primers for luciferase or the reporter gene 10Mut (Table 1), as described in Materials and Methods. The half-lives of the exogenous capped RNAs were calculated by a linear regression analysis of the results from the qRT-PCR at different times posttransfection. The half-life of rotavirus gene 10 mRNA in mock-infected cells was about 12 h, while in infected cells no significant decay was observed during the 12 h of the assay. The luciferase mRNA half-life (240 min) remained the same in uninfected cells and cells infected with rotavirus. Data represent the arithmetic means ± standard deviation of three independent experiments.
The core protein VP2 is involved in the stability of viral mRNAs.
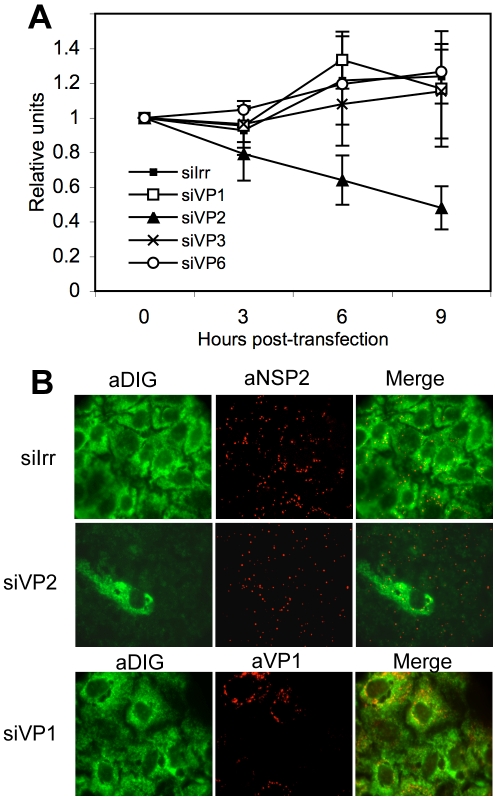
To determine if the stability of the reporter viral mRNA depended on the presence of some of the DLP proteins, we silenced the expression of VP1, VP2, VP3, or VP6 and determined the half-life of the reporter viral mRNA by RT-PCR. In these experiments MA104 cells were transfected with the siRNAs directed to the DLP proteins, and 48 h later the cells were infected with RRV. At 2 hpi the reporter gene 10Mut mRNA was transfected, and its stability was determined by qRT-PCR. We found that the half-life of the reporter was the same in the cells that were transfected with the siRNAs directed to VP1, VP3, and VP6 as the half-life obtained in the control transfected cells (Fig. 9A). Interestingly, the stability of the reporter gene 10Mut decreased in the cells where VP2 was silenced, in which at 9 hpi there was about 50% less reporter gene 10Mut than in the control transfected cells. These results suggest that in the absence of VP2 or with only small amounts of VP2, the viral mRNAs are less stable in MA104 cells.
FIG. 9.
VP2 has a role in the stability of viral mRNA. (A) MA104 cells in 24-well plates were transfected with the indicated siRNAs, and at 48 hpi the cells were infected with RRV at an MOI of 3. At 1 hpi, 1 μg of gene 10 reporter mRNA was transfected using Lipofectamine 2000 for 2 h. At the indicated times after RNA transfection, cells were lysed, and the transfected mRNA was quantified by RT-PCR using primers for reporter gene 10Mut (Table 1), as described in Materials and Methods. The half-life of the transfected RNA was calculated using a linear regression of the results of qRT-PCR at the different times posttransfection. The half-life of rotavirus gene 10 mRNA in cells transfected with an irrelevant siRNA (siIrr) and siRNAs directed against VP1 (siVP2), VP3 (siVP3), and VP6 (siVP6) was longer than 12 h. The half-life of rotavirus mRNA when in cells transfected with the siVP2 was about 9 h. (B) MA104 cells grown in coverslips were transfected with the indicated siRNAs; at 48 h posttransfection the cells were infected with RRV, and at 6 hpi the cells were fixed, permeabilized, and hybridized with a DIG-labeled RNA probe as indicated in Materials and Methods. Viral proteins or bound probe were detected using the indicated antibodies (indicated by the a- prefix; diluted 1:1,000) and secondary Alexa Fluor-conjugated antibodies. The fluorescence signal corresponding to the bound probe was developed using a tyramide signal amplification system (Perkin Elmer).
To further confirm this result, we performed a FISH assay to visualize the cellular distribution of the viral mRNA. In these experiments, MA104 cells were transfected with the siRNAs to VP1 or VP2, infected with RRV, and fixed and hybridized with a DIG-labeled RNA probe that was complementary to the 3′-terminal 300 nucleotides of gene 10. At the same time the cells were immunostained with antibodies directed to viral proteins to observe the formation of viroplasms. Figure 9B shows that while the viral RNA was homogeneously distributed in the cytoplasm of infected cells transfected with a control siRNA or with the siRNA directed to VP1, there was almost no detectable RNA in the cells where VP2 was silenced even though the presence of viroplasms was clear (thus confirming that the cells were infected). The panel where the siRNA directed to VP2 was used shows one cell in which probably VP2 was not silenced, and this single cell indeed shows the presence of viral RNA throughout the cytoplasm in contrast to the surrounding cells in which no hybridization signal was detected. The same results were obtained when a DIG-labeled RNA probe complementary to the 3′-terminal nucleotides of gene 6 were used for the FISH assays (data not shown). These assays confirm the previous results indicating that the presence of VP2 is important for the stability of the viral mRNA.
DISCUSSION
The kinetics of replication and transcription of the rotaviral RNA during the replicative cycle of the virus have been investigated in the past using radioactively labeled viral RNA and either a hybridization assay in solution or PAGE under conditions that allowed the separation of viral dsRNA from mRNA (12, 38). When characterizing the production of different mRNA segments from rotavirus UK, Johnson and McCrae (12) found that at 8.5 hpi there were more RNA-positive molecules for RNA segment 1 than for RNA segment 10 while the levels of accumulation of mRNA of RNA segment 6 and gene 10 were the same. They also found that the production of the 11 dsRNA segments was not equimolar. On the other hand, Stacy-Phipps and Patton found that there was an equimolar production of all the dsRNA segments during the infection of rotavirus SA11 while the transcription seemed to be size dependent (38).
In this work we used a sensitive qRT-PCR protocol to quantify rotavirus mRNA and dsRNA. This method has been previously used to detect and quantify rotavirus load, detecting as few as 10 RNA molecules per reaction (7, 34). In order to establish in more detail the kinetics of rotavirus mRNA and dsRNA production during the infection process, we adjusted the qRT-PCR protocol to differentially quantify the positive and negative viral RNA strands. When analyzing the kinetics of viral mRNA and dsRNA of rotavirus RRV, we found that there was apparently a size-dependent transcription that corresponded to the difference in sizes of the genes, at least for the three RNA segments analyzed. We also found that the synthesis of dsRNA was equimolar at all times for the genes quantified. In fact, a controlled replication of the viral RNA segments has been supported by in vitro and in vivo experiments (28).
Despite the differences in the amounts of mRNAs transcribed from the different viral genes, it is clear that the kinetics of viral mRNA and dsRNA production followed a characteristic and similar pattern: there was a small linear increment of plus- and minus-strand RNA during the first 4 hpi and then a logarithmic increase at later times of infection. This behavior suggests that the entering infectious particles begin to produce a small amount of mRNA, which is translated and replicated (since a small increase in the dsRNA accumulation and infectious progeny was detected as early as 4 hpi); as soon as new DLPs are assembled, these particles begin to transcribe their genomes, initiating a secondary wave of transcription that increases considerably the amount of viral mRNA and dsRNA. This idea is supported by the observation that when any of the viral proteins that constitute the DLP (VP1, VP2, VP3, and VP6) is knocked down, the synthesis of plus- and minus-strand RNA remained linear throughout the infection course, and the logarithmic increase in RNA synthesis observed in the control cells was absent. Taken together, these results directly demonstrate the existence of a second wave of RNA transcription and replication carried out by newly assembled DLPs. Even though this second wave has been presumed to exist for a long time, direct evidence for its occurrence was lacking.
Silencing the expression of the DLP proteins also allowed us to study their role during the replication cycle of the virus. The importance of these structural proteins has been previously assessed using in vitro replication assays and through the characterization of the phenotype of temperature-sensitive mutants. By means of these strategies, it was shown that VP1 and VP2 are necessary and sufficient to replicate the viral RNA in a cell-free system using baculovirus-expressed proteins (29). However, the newly synthesized dsRNA was not packaged, even when VP3 was added to the assay, so it was concluded that, in vivo, some other viral proteins, such as NSP2 and NSP5, should be needed for the encapsidation process and/or for the selection of the RNAs to be replicated. Using a temperature-sensitive virus with a lesion in VP3, Vásquez et al. (39) demonstrated that this protein plays an important role during the RNA replication process since only empty, single-shelled particles were assembled at the nonpermissive temperature, suggesting that VP3 might have another function besides adding the cap structure to mRNAs. Additionally, Mansell and Patton (17) used a temperature-sensitive mutant with a lesion in VP6 to gain insight into the role of this protein in the replication of the virus. They found that at the nonpermissive temperature the mutant virus decreased its transcription rate by 20-fold, but the replication of the viral genome only decreased 2-fold; the investigators thus concluded that VP6 was required for the transcriptional activity but that it was not needed for the replicase activity of the DLPs.
In this work we found that even though the synthesis of viral mRNA was severely inhibited when VP1, VP2, VP3, or VP6 was silenced, the synthesis of viral proteins was differentially affected. When VP1 or VP3 was knocked down, the level of viral proteins synthesized was not affected. In fact, when VP1 was silenced, there was an increase of about 30% in the synthesis of viral protein. These results suggest that only a small proportion of the mRNA produced in the infected cell is sufficient to maintain the translation of viral proteins at maximum levels since we found that the same amount of protein was produced when there was 10 times less mRNA (when VP1 or VP3 was knocked down) compared to control, infected cells. One possible explanation of these observations is that the cell translation machinery is already saturated with the viral mRNAs produced at early hours postinfection.
In a recent work, the expression of VP1 from the simian rotavirus strain SA11-5N was silenced by RNA interference (36). In that work, the effect observed when VP1 was silenced was different from our findings. The main difference is that while in our hands the synthesis of viral protein was not altered when VP1 was knocked down, in the work by Silvestri et al. there was a reduction in the overall protein synthesis. These differences might be the result of the different rotavirus strains used. Alternatively, the differences observed might be due to a differential silencing efficiency of the siRNAs used. We have found that there are variable effects between different siRNAs directed to the same viral protein (16) such that some siRNAs are less potent in silencing the expression of a given protein.
In contrast to the results obtained when VP1 or VP3 were silenced, in the absence of VP2 or VP6 or in the presence of small amounts of these proteins, the translation of viral mRNAs was severely decreased. If the mRNA produced by the entering particles is enough to produce the same level of viral proteins observed in a normal infection, why is viral translation decreased when VP2 or VP6 is knocked down? It has been reported that the simplest RI consists of the viral mRNA associated to VP1 and VP3 (8, 25), which was proposed as the first step in the assembly of the replicative particle. VP2 then becomes associated with this RI, forming the core particle competent in replication, and finally VP6 is added to the core to form DLPs, which are competent to initiate the second round of transcription. Our results could possibly indicate that early in the infection the newly synthesized VP1 and VP3 bind to most of the available mRNA to form RIs so that the positive-sense RNA can be used as a template for the synthesis of the genomic dsRNA. When either of these two proteins is silenced, the mRNA might not be sequestered to form RIs, and then it could be available for translation, whereas when VP2 or VP6 is silenced, most of the mRNA produced by the entering viral particle is sequestered by VP1 and VP3 and cannot be translated. To test this hypothesis, we silenced either VP1 and VP2 or VP1 and VP6 at the same time and quantified the production of viral proteins. Cosilencing of VP1 and VP6 partially restored the synthesis of viral proteins, suggesting that at least part of the inhibition of protein synthesis when VP6 is silenced could be due to the interaction of VP1 with the viral mRNAs. The fact that viral mRNA translation was only partially restored when VP1 and VP6 were cosilenced suggests that VP6 might have another function that could be relevant during the translation of the viral proteins.
While it was expected that in the absence of VP1, the viral RdRp, the transcription and replication of the viral RNAs were going to be strongly inhibited, the observation of the same phenotype when VP3 was silenced was completely unexpected, given its reported activity as the guanylyl- and methyltransferase. Why does the capping enzyme of the virus prevent the transcription and replication of the genome? One possibility is that when VP3 is knocked down, the assembly of viral particles cannot take place, and thus replication is blocked. To test this possibility, the assembly of viral particles under conditions where VP1 or VP3 was silenced was determined. The finding that in the absence of either of these proteins the formation of DLPs and TLPs was decreased was not surprising, given the lack of replication of the viral RNA observed; however, it was interesting to find that the amounts of lower-density particles formed were very similar in the control infected cells and in cells where VP1 or VP3 was silenced. The presence of these less-dense particles, often referred as to empty particles, is frequently observed during virus purification; their protein content is very similar to that of TLPs and DLPs although on some occasions NSP2 and NSP5 have been found to be associated to them; thus, they might represent a mix of RIs and spontaneously assembled empty viral particles. The finding that these particles are associated with positive-strand viral RNA suggests that at least some of them represent true RIs. From the results obtained here, it was not possible to ascertain whether the detected plus-sense RNA was packaged inside the viral capsid or whether it was only associated to the capsid in a different form. The observation that these types of particles can be formed when either VP1 or VP3 is knocked down suggests that their assembly can proceed in the absence of either protein. These results are in agreement with those reported by Vasquez et al. (39). Taken together, these results suggest that the decrease in viral replication observed when VP3 was silenced is most probably due to an RNA replication defect rather than to an assembly problem, and they suggest that VP3 may have a novel and thus far undetermined role during in vivo replication of the virus.
In contrast, when VP2 and VP1 were cosilenced, the translation of the viral proteins was not restored, suggesting that VP2 could have a role in the metabolism of viral RNA in addition to being required for the replication of the genome. To explore the direct or indirect participation of VP2 in the stability of the viral RNAs, the half-life of a reporter viral mRNA was determined in infected cells in which VP2 was silenced (Fig. 9A). The decrease in the half-life observed (9 h) under these conditions does not seem to explain the severe decrease in protein translation attained. Thus, to determine if in the absence of VP2 there was a change in the cellular distribution of the viral mRNA that could contribute to the decreased viral mRNA translation, we utilized FISH to localize the distribution of viral mRNA in infected cells. As expected, the distributions of gene 10 (Fig. 9B) and gene 6 (data not shown) mRNAs were cytoplasmic with a strong perinuclear signal, close to viroplasms, which corresponds to the sites where the mRNA is produced (35). The same distribution of the mRNA was found when VP1 was silenced although a slight decrease in the fluorescent signal was observed. In contrast, when VP2 was silenced, there was a weak hybridization signal throughout the cell's cytoplasm, showing that little viral mRNA was present in the cells. These results suggest that in the absence of VP2 either there is a severe degradation of the viral mRNA or the transcriptional activity of the entering particles is blocked; this latter possibility seems less likely.
Silvestri et al. (35) characterized the cellular distribution of a bromodeoxyuridine-labeled rotavirus mRNA that was transfected into rotavirus-infected MA104 cells and found that at 9 hpi the labeled RNA collected into punctuate centers that did not colocalize with viroplasms. In this work we used a different strategy to visualize the viral RNA. In our case we infected MA104 cells, and the endogenously transcribed viral RNA (genes 10 or 6) was visualized by hybridization using a DIG-labeled probe complementary to a region of the mRNAs. The differences in the patterns observed by us and by Silvestri et al. may be due to the amount of RNA that can be transfected, in contrast to the amount of viral RNA that accumulates at 6 hpi. We think that either the viral mRNA, after been synthesized in the viroplasms, diffuses to the cytoplasm or that a protein (viral or cellular) actively pulls out the mRNAs committed for translation.
Silencing the expression of the viral proteins in the context of an infected cell allows us to learn about the roles of each individual protein during the replicative cycle of rotavirus. We have found that some viral proteins are somehow involved in regulating the translation of the viral mRNAs or their half-lives. For instance, VP2 seems to be important for the stability of the viral mRNAs while VP6 might have a role in the translation of viral proteins. It is also clear that there are many protein-protein and protein-RNA interactions that take place during this process that make the interpretation of the knocked-down phenotypes not straightforward. Nevertheless, the characterization of these phenotypes in conjunction with the previous findings with rotavirus variants, and hopefully with the reverse genetic system recently developed (13), will help us to unravel the details of the replicative cycle of this virus and its interactions with the host cell.
Acknowledgments
We are grateful to Mery Piña for the sequence of RRV gene 3 and to Paul Gaytan and Eugenio Lopez for their support with the synthesis of oligonucleotides.
This work was supported by grants 55005515 from the Howard Hughes Medical Institute, IN212288 from DGAPA-UNAM, and 60025 from CONACyT. C.A.-B. is a recipient of a scholarship from CONACYT.
Footnotes
Published ahead of print on 24 June 2009.
REFERENCES
- 1.Campagna, M., C. Eichwald, F. Vascotto, and O. R. Burrone. 2005. RNA interference of rotavirus segment 11 mRNA reveals the essential role of NSP5 in the virus replicative cycle. J. Gen. Virol. 861481-1487. [DOI] [PubMed] [Google Scholar]
- 2.Chen, D., C. Q. Zeng, M. J. Wentz, M. Gorziglia, M. K. Estes, and R. F. Ramig. 1994. Template-dependent, in vitro replication of rotavirus RNA. J. Virol. 687030-7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dector, M. A., P. Romero, S. Lopez, and C. F. Arias. 2002. Rotavirus gene silencing by small interfering RNAs. EMBO Rep. 31175-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Estes, M. a. K. A. 2007. Rotaviruses, p. 1917-1974. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 5.Fabbretti, E., I. Afrikanova, F. Vascotto, and O. R. Burrone. 1999. Two non-structural rotavirus proteins, NSP2 and NSP5, form viroplasm-like structures in vivo. J. Gen. Virol. 80333-339. [DOI] [PubMed] [Google Scholar]
- 6.Fan, X., E. Roy, L. Zhu, T. C. Murphy, M. Kozlowski, M. S. Nanes, and J. Rubin. 2003. Nitric oxide donors inhibit luciferase expression in a promoter-independent fashion. J. Biol. Chem. 27810232-10238. [DOI] [PubMed] [Google Scholar]
- 7.Fenaux, M., M. A. Cuadras, N. Feng, M. Jaimes, and H. B. Greenberg. 2006. Extraintestinal spread and replication of a homologous EC rotavirus strain and a heterologous rhesus rotavirus in BALB/c mice. J. Virol. 805219-5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gallegos, C. O., and J. T. Patton. 1989. Characterization of rotavirus replication intermediates: a model for the assembly of single-shelled particles. Virology 172616-627. [DOI] [PubMed] [Google Scholar]
- 9.Gonzalez, R. A., M. A. Torres-Vega, S. Lopez, and C. F. Arias. 1998. In vivo interactions among rotavirus nonstructural proteins. Arch. Virol. 143981-996. [DOI] [PubMed] [Google Scholar]
- 10.Guerrero, C. A., S. Zarate, G. Corkidi, S. Lopez, and C. F. Arias. 2000. Biochemical characterization of rotavirus receptors in MA104 cells. J. Virol. 749362-9371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imai, M., K. Akatani, N. Ikegami, and Y. Furuichi. 1983. Capped and conserved terminal structures in human rotavirus genome double-stranded RNA segments. J. Virol. 47125-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson, M. A., and M. A. McCrae. 1989. Molecular biology of rotaviruses. VIII. Quantitative analysis of regulation of gene expression during virus replication. J. Virol. 632048-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Komoto, S., J. Sasaki, and K. Taniguchi. 2006. Reverse genetics system for introduction of site-specific mutations into the double-stranded RNA genome of infectious rotavirus. Proc. Natl. Acad. Sci. USA 1034646-4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lawton, J. A., M. K. Estes, and B. V. Prasad. 1997. Three-dimensional visualization of mRNA release from actively transcribing rotavirus particles. Nat. Struct. Biol. 4118-121. [DOI] [PubMed] [Google Scholar]
- 15.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25402-408. [DOI] [PubMed] [Google Scholar]
- 16.Lopez, T., M. Rojas, C. Ayala-Breton, S. Lopez, and C. F. Arias. 2005. Reduced expression of the rotavirus NSP5 gene has a pleiotropic effect on virus replication. J. Gen. Virol. 861609-1617. [DOI] [PubMed] [Google Scholar]
- 17.Mansell, E. A., and J. T. Patton. 1990. Rotavirus RNA replication: VP2, but not VP6, is necessary for viral replicase activity. J. Virol. 644988-4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mingle, L. A., N. N. Okuhama, J. Shi, R. H. Singer, J. Condeelis, and G. Liu. 2005. Localization of all seven messenger RNAs for the actin-polymerization nucleator Arp2/3 complex in the protrusions of fibroblasts. J. Cell Sci. 1182425-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montero, H., C. F. Arias, and S. Lopez. 2006. Rotavirus nonstructural protein NSP3 is not required for viral protein synthesis. J. Virol. 809031-9038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montero, H., M. Rojas, C. F. Arias, and S. Lopez. 2008. Rotavirus infection induces the phosphorylation of eIF2α but prevents the formation of stress granules. J. Virol. 821496-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Padilla-Noriega, L., R. Werner-Eckert, E. R. Mackow, M. Gorziglia, G. Larralde, K. Taniguchi, and H. B. Greenberg. 1993. Serologic analysis of human rotavirus serotypes P1A and P2 by using monoclonal antibodies. J. Clin. Microbiol. 31622-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pando, V., P. Isa, C. F. Arias, and S. Lopez. 2002. Influence of calcium on the early steps of rotavirus infection. Virology 295190-200. [DOI] [PubMed] [Google Scholar]
- 23.Parashar, U. D., E. G. Hummelman, J. S. Bresee, M. A. Miller, and R. I. Glass. 2003. Global illness and deaths caused by rotavirus disease in children. Emerg. Infect. Dis. 9565-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patton, J. T. 1990. Evidence for equimolar synthesis of double-strand RNA and minus-strand RNA in rotavirus-infected cells. Virus Res. 17199-208. [DOI] [PubMed] [Google Scholar]
- 25.Patton, J. T. 2001. Rotavirus RNA replication and gene expression. Novartis Found. Symp. 23864-81. [DOI] [PubMed] [Google Scholar]
- 26.Patton, J. T. 1996. Rotavirus VP1 alone specifically binds to the 3′ end of viral mRNA, but the interaction is not sufficient to initiate minus-strand synthesis. J. Virol. 707940-7947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patton, J. T., and D. Chen. 1999. RNA-binding and capping activities of proteins in rotavirus open cores. J. Virol. 731382-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patton, J. T., and C. O. Gallegos. 1990. Rotavirus RNA replication: single-stranded RNA extends from the replicase particle. J. Gen. Virol. 711087-1094. [DOI] [PubMed] [Google Scholar]
- 29.Patton, J. T., M. T. Jones, A. N. Kalbach, Y. W. He, and J. Xiaobo. 1997. Rotavirus RNA polymerase requires the core shell protein to synthesize the double-stranded RNA genome. J. Virol. 719618-9626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patton, J. T., and E. Spencer. 2000. Genome replication and packaging of segmented double-stranded RNA viruses. Virology 277217-225. [DOI] [PubMed] [Google Scholar]
- 31.Persengiev, S. P., X. Zhu, and M. R. Green. 2004. Nonspecific, concentration-dependent stimulation and repression of mammalian gene expression by small interfering RNAs (siRNAs). RNA 1012-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pesavento, J. B., S. E. Crawford, M. K. Estes, and B. V. Prasad. 2006. Rotavirus proteins: structure and assembly. Curr. Top. Microbiol. Immunol. 309189-219. [DOI] [PubMed] [Google Scholar]
- 33.Prasad, B. V., G. J. Wang, J. P. Clerx, and W. Chiu. 1988. Three-dimensional structure of rotavirus. J. Mol. Biol. 199269-275. [DOI] [PubMed] [Google Scholar]
- 34.Schwarz, B. A., R. Bange, T. W. Vahlenkamp, R. Johne, and H. Muller. 2002. Detection and quantitation of group A rotaviruses by competitive and real-time reverse transcription-polymerase chain reaction. J. Virol. Methods 105277-285. [DOI] [PubMed] [Google Scholar]
- 35.Silvestri, L. S., Z. F. Taraporewala, and J. T. Patton. 2004. Rotavirus replication: plus-sense templates for double-stranded RNA synthesis are made in viroplasms. J. Virol. 787763-7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silvestri, L. S., M. A. Tortorici, R. Vasquez-Del Carpio, and J. T. Patton. 2005. Rotavirus glycoprotein NSP4 is a modulator of viral transcription in the infected cell. J. Virol. 7915165-15174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sledz, C. A., M. Holko, M. J. de Veer, R. H. Silverman, and B. R. Williams. 2003. Activation of the interferon system by short-interfering RNAs. Nat. Cell Biol. 5834-839. [DOI] [PubMed] [Google Scholar]
- 38.Stacy-Phipps, S., and J. T. Patton. 1987. Synthesis of plus- and minus-strand RNA in rotavirus-infected cells. J. Virol. 613479-3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vásquez, M., A. M. Sandino, J. M. Pizarro, J. Fernandez, S. Valenzuela, and E. Spencer. 1993. Function of rotavirus VP3 polypeptide in viral morphogenesis. J. Gen. Virol. 74937-941. [DOI] [PubMed] [Google Scholar]