Abstract
Sensory neurons within trigeminal ganglia (TG) are the primary site for bovine herpesvirus 1 (BHV-1) latency. During latency, viral gene expression is restricted to the latency-related (LR) gene and the open reading frame ORF-E. We previously constructed an LR mutant virus that expresses LR RNA but not any of the known LR proteins. In contrast to calves latently infected with wild-type (wt) BHV-1 or the LR rescued virus, the LR mutant virus does not reactivate from latency following dexamethasone (DEX) treatment. In this study, we demonstrated that bICP0, but not bICP4, transcripts were consistently detected in TG of calves infected with the LR mutant or LR rescued virus following DEX treatment. Calves latently infected with the LR rescued virus but not the LR mutant virus expressed late transcripts, which correlated with shedding of infectious virus following DEX treatment. The bICP4 and bICP0 genes share a common immediate-early promoter, suggesting that this promoter was not consistently activated during DEX-induced reactivation from latency. The bICP0 gene also contains a novel early promoter that was activated by DEX in mouse neuroblastoma cells. Expression of a cellular transcription factor, C/EBP-alpha, was stimulated by DEX, and C/EBP-alpha expression was necessary for DEX induction of bICP0 early promoter activity. C/EBP-alpha directly interacted with bICP0 early promoter sequences that were necessary for trans activation by C/EBP-alpha. In summary, DEX treatment of latently infected calves induced cellular factors that stimulated bICP0 early promoter activity. Activation of bICP0 early promoter activity does not necessarily lead to late gene expression and virus shedding.
Bovine herpesvirus 1 (BHV-1) is a significant viral pathogen of cattle that is responsible for a variety of disease conditions, which include conjunctivitis, pneumonia, genital disorders, or abortions. BHV-1 also causes rhinotracheitis, a serious upper respiratory tract infection, and infection can initiate shipping fever, a potentially fatal polymicrobial disease (42). Like other members of the Alphaherpesvirinae subfamily, BHV-1 establishes lifelong latency in trigeminal ganglionic neurons following acute replication in the mucosal epithelium. Reactivation from latency occurs periodically, resulting in virus shedding and spread to susceptible cattle. Reactivation can occur after stress or corticosteroid treatment, which mimics stress (35, 40).
Viral gene expression is temporally regulated in three distinct phases: immediate early (IE), early (E), or late (L) (reviewed in references 18 and 19). IE gene expression is stimulated by a virion component, bTIF, which interacts with a cellular transcription factor (Oct-1) to transactivate IE gene expression (29, 30). Two IE transcription units exist: IE transcription unit 1(IEtu1) and IEtu2 (48-50). IEtu1 encodes functional homologues of two herpes simplex virus type 1 (HSV-1) proteins, ICP0 and ICP4. IEtu2 encodes a protein that is similar to the HSV-1 IE gene, ICP22 (39). BHV-1-encoded ICP0 (bICP0) is translated from an IE (IE/2.9) or E mRNA (E/2.6) because an IE promoter (IEtu1 promoter) and E promoter regulate bICP0 RNA expression (6, 48-50). The IE promoter regulates IE expression of bICP4 and bICP0. Expression of bICP4 leads to repression of IEtu1 promoter activity, whereas bICP0 activates its own E promoter and all other viral promoters. Thus, bICP0 is considered to be the major regulatory protein that stimulates productive infection (6, 48-50).
The latency-related (LR) gene is abundantly transcribed in trigeminal ganglia (TG) of latently infected calves (23, 35, 36). LR RNA is antisense with respect to the bICP0 gene (18, 19, 22) and encodes at least three proteins. LR gene products inhibit mammalian cell growth (8, 38), productive infection (1, 9, 38), and apoptosis (3, 12). While expression of LR proteins is necessary for inhibiting apoptosis (14), protein expression is not necessary for inhibiting cell growth or productive infection. The LR gene has two open reading frames (ORF1 and ORF2), and two reading frames that lack an initiating ATG (RF-B and RF-C). A mutant BHV-1 virus with three stop codons at the beginning of ORF2 was constructed to test whether LR protein expression regulates the latency reactivation cycle in cattle (15). The LR mutant virus grows to similar titers as wild-type (wt) BHV-1 or the LR rescued virus in cultured bovine cells, indicating that LR gene products are not required for growth. ORF2 and RF-B are expressed when bovine cells are infected with the wt or the LR rescued virus but not with the LR mutant virus (13, 16, 17). ORF1 expression is reduced but not totally eliminated following infection of cultured cells with the LR mutant virus (26). Calves infected with the LR mutant virus exhibit diminished clinical symptoms and reduced shedding of infectious virus in the eye, tonsil, or TG (14, 15, 33). The LR mutant virus prematurely expresses LR RNA relative to rescued or wt BHV-1, which correlates with an enhanced interferon response in cells infected with the LR mutant virus (31).
Stress, in part due to increased corticosteroid levels, and/or immune suppression can initiate reactivation from latency. During reactivation from latency, productive viral gene expression is readily detected in sensory neurons, LR gene expression decreases, and infectious virus can be detected in nasal or ocular swabs (18, 19, 22). Administration of dexamethasone (DEX), a synthetic corticosteroid, to calves or rabbits latently infected with BHV-1 reproducibly leads to activation of viral gene expression and reactivation from latency (14, 18, 19, 21, 22, 35). After DEX treatment, many neurons express lytic viral genes, but only a small subset of the neurons that express viral genes appear to produce infectious virus (35). Thus, the ability of DEX to consistently initiate reactivation from latency makes BHV-1 a useful model to identify cellular factors that stimulate reactivation from latency.
In this study, we tested whether calves latently infected with the LR mutant virus express viral genes in TG following DEX treatment. Expression of bICP0 but not bICP4 RNA was consistently detected in TG following DEX treatment of calves infected with the LR mutant virus or wt BHV-1. In transient transfection assays, DEX stimulated bICP0 E promoter activity. The ability of DEX to induce expression of a cellular transcription factor (C/EBP-alpha) was important for stimulating bICP0 E promoter activity.
MATERIALS AND METHODS
Cells.
Murine neuroblastoma 2A (Neuro-2A) and rabbit skin (RS) cells were grown in Earle's modified Eagle's medium (EMEM) supplemented with 5% fetal calf serum (FCS). Bovine kidney (CRIB) cells were grown in EMEM supplemented with 10% FCS. All media contained penicillin (10 U/ml) and streptomycin (100 μg/ml).
Viruses.
The Cooper strain of BHV-1 (wt virus) was obtained from the National Veterinary Services Laboratory, Animal and Plant Health Inspection Services, Ames, IA. For the construction of the LR mutant virus, 25 bp of the LR gene sequence from the Cooper strain were replaced with an oligonucleotide that contains a unique EcoRI restriction site and three stop codons to prevent protein expression (15). The LR mutant virus was previously characterized both in vitro and in vivo (14, 15, 25, 31-33). Stock cultures of the respective viruses were prepared in bovine cells (MDBK).
Animals.
BHV-1-free crossbred calves (∼200 kg) were used for this study. Calves were inoculated with 107 PFU of the indicated virus into each nostril and eye, for a total of 4 × 107 PFU/animal, as described previously (14, 15, 25, 31-33). Calves were housed under strict isolation and given antibiotics before and after BHV-1 infection to prevent secondary bacterial infection. Experiments were performed in accordance with the American Association of Laboratory Animal Care guidelines and the University of Nebraska IACUC. At 60 days postinfection, calves were injected intravenously with 100 mg of DEX. Three calves/time point/virus were used for these studies.
Tissue samples.
TG from the respective infected calves were collected at necropsy at 60 days postinfection and at 24 and 48 h after DEX treatment. TG were stored at −80°C for nucleic acid extraction or processed for histological procedures. Part of the TG used for this study was also used for previous studies (14, 31-33).
RNA extraction and preparation for reverse transcription-PCR (RT-PCR).
RNA extraction was performed as described by Chomeczynski and Sacchi (2). TG were minced, placed into 10 ml of solution D (4 M guanidine thiocyanate, 25 mM sodium citrate [pH 7.0], 0.5% sarcosyl, and 14 mM β-mercaptoethanol) and homogenized. Two phenol extractions were performed. RNA concentrations were determined spectrophotometrically (260 nm), and RNA was reprecipitated in 3 volumes of ethanol.
Three micrograms of RNA was treated with 1 U of DNase I (RNase free; Invitrogen, Life Technologies) for 15 min at room temperature in the presence of RNase inhibitor (RNAsin; Promega, Madison, WI). After DNase I treatment, samples were incubated at 65°C for 7.5 min in the presence of 2 mM EDTA to eliminate DNase I activity. RT reactions for analysis of IEtu1, bICP4, bICP22, thymidine kinase (TK), bTIF, and gC genes were performed with a poly(dT) primer at 65°C for 7.5 min and chilled on ice. RT reactions for bICP0 were performed with a strand-specific primer (CGTCAGGTCTATCACTGTGGAGAT). Sixteen microliters of ice-cold RT-PCR mix (20 mM Tris-HCl [pH 8.3], 50 mM KCl, 2.5 mM MgCl2, 100 μg of bovine serum albumin per ml, 1 mM dithiothreitol, a 0.5 mM concentration of each deoxynucleoside triphosphate, and 10 U of RNAsin) was added. The reaction mixture was incubated for 10 min at 25°C and then for 50 min at 42°C. When strand-specific primers were used, the 25°C incubation was omitted. As a control of DNA contamination in the RNA samples, 2 μg of RNA (DNase I treated) was mixed with ice-cold RT mix lacking reverse transcriptase.
PCR.
An aliquot (2 μl) of the RT reaction mixture or DNA was used for each PCR, using primers specific for BHV-1 genes. IEtu1, bICP0, bICP4, bICP22, and gC primers were previously described (37). IEtu1 primer pairs amplify bICP0, bICP4 and bcirc. bICP22 is a dual-kinetic transcript (IE/L), TK is an E gene, and gC and bTIF are L genes. β-Actin primers were used as internal controls. PCRs were carried out in 50 μl of 10× commercial PCR buffer, 5 mM MgCl2, 200 μM concentration of each deoxynucleoside triphosphate, 1 μM concentration of each primer, and Taq polymerase. Amplification was carried out for 37 cycles by denaturing at 95°C for 1 min, annealing at 65°C for 1 min, and extension at 72°C for 2 min. Amplification of bICP0 required an annealing temperature of 66°C. Upon completion of the last cycle, reaction mixtures were further incubated at 72°C for 7 min to ensure complete extension of the amplified product. PCR products were electrophoresed on 2% agarose gels and stained with ethidium bromide.
Plasmids.
The C/EBP-alpha wt plasmid contains the wt sequence of mouse C/EBP-alpha in an adenoviral vector (pAdTrack) that contains mammalian promoter/enhancer sequences (44). The C/EBP-alpha mutant plasmid contains a mutation in the C/EBP-alpha ORF that produces a single amino acid change (R290A) in C/EBP-alpha, resulting in the loss of DNA binding activity (43). The empty vector pcDNA3.1 was purchased from Invitrogen.
Six DNA reporter constructs were generated by PCR using the wt BHV-1 genome as a template and a common 3′ primer (5′-ctcgagCCTGCTGGGCGACACAAACAACAGA-3′) with the following 5′ primers: EP-943, 5′-ggtaccGCGACGGCGGCAATAAAGACGAGTC-3′; EP-638, 5′-ggtaccGCCCTCGGTCTCGGTCGGAG-3′; EP-172, 5′-gggtaccGCCTTGCGTGGGGGGTTTCG-3′; EP-143, 5′-gggtaccAGCCGGGGGGTGCGGGCC-3′; EP-133, 5′-gggtaccTGCGGGCCTTTCGCCG-3′; or EP-71, 5′-gggtaccGCTCCCGGCGCGTCA-3′). The promoter fragments were cloned into the promoterless vector pCAT-Basic (E1871; Promega) at the unique XhoI and KpnI sites to generate plasmids EP-943, EP-638, EP-172, EP-143, EP-133, and EP-71 (see Fig. 2). The numbers in the plasmid name refer to the length of the bICP0 E promoter fragment inserted into the chloramphenicol acetyltransferase (CAT) vector. Two additional constructs, EP-50 and EP-42, were created using synthesized duplex sequences (IDT). Duplex oligonucleotides were digested with XhoI and KpnI and cloned into the promoterless vector pCAT-Basic. E promoter inserts were confirmed by DNA sequencing (Genomics Core Research Facility-UNL). Plasmids were prepared from bacterial cultures by alkaline lysis and two rounds of cesium chloride centrifugation.
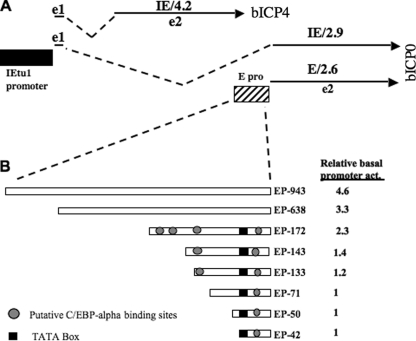
FIG. 2.
Schematic of IEtu1 and bICP0 E promoter constructs used in this study. (A) Positions of bICP4 and bICP0 transcripts are shown. IEtu1 encodes bICP4 (IE/4.2 transcript) and bICP0 (IE/2.9) (49, 50). The IEtu1 promoter activates IE expression of IE/4.2 and IE/2.9 (denoted by the black rectangle). E/2.6 is the early transcript that encodes bICP0, and an early promoter (E pro) activates expression of this transcript (48). Exon 2 (e2) of bICP0 contains all of the protein coding sequences of bICP0. The dashed lines are intron sequences. (B) bICP0 E promoter constructs were prepared as described in Materials and Methods. Position of putative C/EBP-alpha binding sites and TATA box are shown. Basal promoter activity (act) was measured in Neuro-2A cells.
Measurement of CAT activity.
Neuro-2A cells grown in 60-mm dishes were cotransfected with the designated plasmids as indicated in the respective figure legends. Neuro-2A cells were transfected with NeuroTransIt (MIR2145; Mirus), according to the manufacturer's instructions. Where indicated, cells were cultured in the presence of 1 μM water-soluble DEX (D2915; Sigma) at the time of transfection. After 48 h, cell extract was prepared by three freeze-thaw cycles in 0.25 M Tris-HCl, pH 7.4. Cell debris was pelleted by centrifugation, and protein concentrations were determined. CAT activity was measured in the presence of 0.1 μCi of [14C]chloramphenicol (CFA754; Amersham Biosciences) and 0.5 mM acetyl-coenzyme A (A2181; Sigma). The reaction mixture was incubated at 37°C for 15 min to 2 h, depending on the activator used. All forms of chloramphenicol were separated by thin-layer chromatography. CAT activity was quantified using a Bio-Rad Molecular Imager FX (Molecular Dynamics, CA). Levels of CAT activity are expressed as the induction relative to the vector control.
SDS-PAGE and Western blotting.
At 48 h after transfection, whole-cell lysate was prepared. Cells were washed with phosphate-buffered saline (PBS) and suspended in NP-40 lysis buffer (100 mM Tris [pH 8.0], 1 mM EDTA, 100 mM NaCl, 1% NP-40, 1 mM phenylmethylsulfonyl fluoride, and one tablet of complete protease inhibitor [Roche Molecular Biochemicals] per 10 ml). Cell lysate was incubated on ice for 30 min, sonicated, and then clarified by centrifugation at 10,000 × g at 4°C for 15 min. Protein concentrations were quantified by the Bradford assay. For sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), proteins were mixed with an equal amount of 1× sample loading buffer (62.5 mM Tris-HCl [pH 6.8], 2% SDS, 50 mM dithiothreitol, 0.1% bromophenol blue, 10% glycerol) and boiled for 5 min. Proteins were separated in a 12% SDS-PAGE gel. After electrophoresis, proteins were transferred onto a polyvinylidene difluoride membrane (Immobilon-P; Millipore) and blocked for 4 h in 5% nonfat dry milk with Tris-buffered saline-0.1% Tween 20 (TBS-T). Membranes were then incubated with primary antibody overnight at 4°C. The C/EBP-alpha antibody (14AA; Santa Cruz Biotechnology) was diluted 1:500 in the blocking solution. An antibody directed against β-actin (Santa Cruz Biotechnology, Santa Cruz, CA) was used as a loading control. After 45 min of washing with TBS-T, the blots were incubated with donkey anti-rabbit horseradish peroxidase-conjugated immunoglobulin G (Amersham Biosciences), which was diluted 1:1,000 in 5% nonfat milk in TBS-T. Blots were washed for 45 min with TBS-T and exposed to Amersham ECL reagents, and then autoradiography was performed.
siRNA transfection for Western blotting and CAT assay.
Mouse C/EBP-alpha small interfering RNA (siRNA) containing a pool of three target-specific 20- to 25-oligonucleotide siRNAs designed to reduce gene expression (sc-37048; Santa Cruz Biotechnology) were used in this study. The Block-iT-Fluorescent Oligo was used as a control siRNA (44-2926; Invitrogen). It is a fluorescence-conjugated control containing a scrambled sequence that does not degrade any known cellular RNA.
Neuro-2A cells grown in 60-mm dishes were cotransfected with 1 μg of EP-172 with either 100 nM mouse C/EBP-alpha siRNA or the fluorescent control siRNA. After 4 h, the transfection mixture was removed and replaced by fresh medium. At 48 h posttransfection, cell extract was collected for Western blot analysis or CAT assays.
Measurement of virus titers in bovine cells.
RS cells grown in 60-mm dishes were transfected with 100 nM mouse C/EBP-alpha siRNA, the fluorescent control siRNA, or transfection reagent alone. After 4 h, the complex was removed and replaced by fresh medium. After 24 h, cells were infected with wt BHV-1 at a multiplicity of infection of 0.1 and incubated for an additional 24 h. Medium and cells were collected and subjected to two freeze-thaw cycles. Cell debris was pelleted, and the supernatant was titrated on CRIB cells (bovine kidney) that were plated onto six-well plates 24 h prior to virus infection (90% confluence at the time of infection). After 1 h of adsorption at 37°C, cells were rinsed with PBS and overlaid with a 50:50 mixture of 1.4% SeaPlaque Agar in PBS and EMEM supplemented with 10% FCS.
EMSA.
Neuro-2A whole-cell lysate was prepared by lysing cells with NP-40 lysis buffer. Thirty micrograms of protein extract was incubated in a volume of 16 μl of binding buffer (10 mM Tris-HCl, pH 8, 150 mM KCl, 0.5 mM EDTA, 0.1% Triton X-100, 12.5% glycerol) in the presence of 1 μg of poly(dI-dC) (P4929; Sigma), and 0.5 pmols of double-stranded DNA probe labeled with 10 μCi of [γ-32P]ATP. Incubation was for 1 h at room temperature. For supershift experiments, 5 μg of antibody (for C/EBP-alpha, sc-61X; for C/EBP-beta, sc-150X; Santa Cruz Biotechnology) was added to the reaction mixture after 30 min and allowed to incubate an additional 30 min. The DNA-protein complexes were electrophoresed on a 5% polyacrylamide gel in 0.5× Tris-borate-EDTA buffer for 3 h at 160 V. To improve band resolution, 1.5 M sodium acetate, pH 5.3, was added to the lower buffer chamber during electrophoresis. The gel was exposed to a phosphorimager plate and analyzed using a Bio-Rad Molecular Imager FX. The bICP0 E probes used for electrophoretic mobility shift assay (EMSA) were the following: C1, GCCTTGCGTGGGGGGTTTCGCCTTGGGGCAGC; C2, TGCGGGCCCTTTCGCCGCCCG; and C3, TCTGTTGTTTGTGTCGCCCA. An oligonucleotide containing three consensus C/EBP-alpha binding sites (underlined), CGCAATATTGCGCAATATTGCAAT, was used as a positive control for binding to C/EBP-alpha.
RESULTS
Analysis of viral transcripts during reactivation from latency.
Calves latently infected with the LR mutant virus do not shed detectable levels of infectious virus after DEX treatment (14). At a molecular level, two possibilities seemed feasible to explain this observation. First, TG neurons latently infected with the LR mutant virus may express a subset of lytic viral genes after DEX treatment, but infectious virus is not produced. Second, DEX treatment of calves latently infected with the LR mutant virus does not stimulate lytic viral gene expression.
For these studies, total RNA was prepared from TG of calves latently infected with the LR mutant virus or the LR rescued virus after DEX treatment to initiate reactivation from latency. Previous studies demonstrated that the LR rescued virus but not the LR mutant virus reactivated from latency after DEX treatment (14, 31-33). To test whether IE, E, or L genes were expressed in TG following DEX treatment, RT-PCR was performed using primers that specifically amplify viral genes. Specific oligonucleotide primers (Fig. 1A) were used to test whether bICP0, bICP4, TK, or gC was expressed during DEX-induced reactivation from latency. These primers were previously used to analyze viral gene expression in TG during acute infection of calves (37). Similar levels of bICP0, bICP4, TK, or gC cDNA-amplified products were detected following infection of MDBK cells with the LR mutant virus or the LR mutant rescued virus (Fig. 1B). Only when reverse transcriptase was included in the cDNA reaction mixtures were amplified products detected, confirming that amplification was due to cDNA, not DNA contamination.
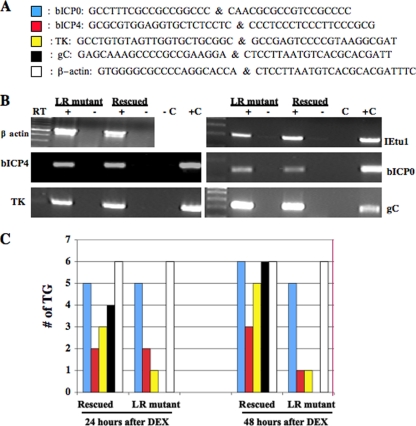
FIG. 1.
Examination of BHV-1 transcripts during reactivation from latency. (A) The primers shown were previously used to detect viral transcripts in TG of infected calves (37). (B) RNA was prepared from MDBK cells infected with the LR mutant virus or the LR rescued virus (6 h after infection). cDNA synthesis was performed as previously described (37). RT reactions with (+) or without (−) reverse transcriptase are shown. DNA from BHV-1- and mock-infected MDBK cells were used as positive (C+) and negative (C−) controls, respectively, for the PCRs. The first lane contains a 100-bp ladder (NE Biolabs). Primers used for these studies are shown in panel A. (C) To initiate reactivation from latency, calves latently infected (60 days after infection) with the LR rescued virus or the LR mutant virus (LR−) were given a single intravenous injection (jugular vein) of DEX (100 mg). At 24 or 48 h after DEX treatment, total RNA was prepared from each TG as previously described (14, 37, 45-47). cDNA synthesis was primed using poly(dT), and then PCR was performed with the designated primers listed in panel A. In a separate cDNA synthesis reaction, the bICP0 primer was used to examine bICP0 expression. The results are summarized in the graphs from TG of three calves/time point. Each ganglion was treated separately.
bICP0 transcripts were detected in five of six TG following 24 h of DEX treatment, regardless of whether calves were infected with the LR mutant virus or the LR rescued virus (Fig. 1C). Forty-eight hours after DEX treatment six of six TG prepared from calves latently infected with the LR rescued virus or five of six TG latently infected with the LR mutant virus expressed bICP0 RNA. In contrast, only two or three of the six TG in each group (LR mutant virus or LR rescued virus, respectively) expressed bICP4 at 24 or 48 h after DEX treatment. These studies also indicated that three or five TG prepared from calves latently infected with the LR rescued virus expressed detectable levels of TK mRNA (an E transcript) at 24 or 48 h after DEX treatment, respectively (Fig. 1C). Only one TG was positive for the TK transcript when calves were infected with the LR mutant virus after DEX treatment. At 48 h after DEX treatment, six of six TG prepared from calves latently infected with the LR rescued virus were positive for gC transcripts (Fig. 1C). gC transcription was not detected in TG of calves latently infected with the LR mutant virus after DEX treatment. In summary, DEX treatment consistently activated bICP0 expression, regardless of whether calves were latently infected with the LR mutant virus or LR rescued virus. DEX treatment of calves latently infected with the LR mutant virus did not lead to detectable levels of gC transcription, a prototype L gene.
DEX activates the bICP0 E promoter.
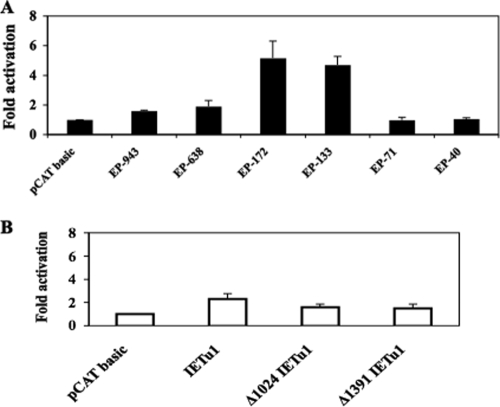
The results shown in Fig. 1 demonstrated that the bICP0 transcript, but not the bICP4 transcript, was consistently activated in TG following treatment of latently infected calves with DEX. bICP0 expression is regulated by IE and E promoters (6, 48-50) (Fig. 2A), suggesting that DEX treatment stimulated expression of cellular factors that subsequently activated the bICP0 E promoter regardless of the outcome of reactivation from latency. To test this hypothesis, the bICP0 promoter construct was transfected into mouse neuroblastoma cells (Neuro-2A) and treated with DEX after transfection. For these studies, eight constructs containing upstream sequences of the bICP0 E promoter were used (Fig. 2B). The basal level of promoter activity for EP-943 was slightly higher than that of EP-638 and at least two times higher than that of EP-172 or EP-133. Basal promoter activity of EP-71, EP-50, and EP-42 was at least fourfold less than that of EP-943.
Two E promoter constructs, EP-172 and EP-133, were activated by DEX more than fourfold in Neuro-2A cells (Fig. 3A). More extensive deletions (constructs EP-71 or EP-40) were not activated by DEX. The larger E promoter constructs, EP-943 and EP-638, were activated less than twofold by DEX. As a comparison, three IEtu1 promoter constructs were tested for DEX induction (Fig. 3B). None of the constructs was activated by DEX more than twofold. As expected, the empty vector (pCAT basic) was not activated by DEX (Fig. 3A and B).
FIG. 3.
DEX induces bICP0 E promoter activity. (A) Neuro-2A cells were transfected with 1 μg of the designated bICP0 E promoter construct. Cells were cultured in EMEM containing 1% FBS and 1 μM DEX at the time of transfection. At 48 h posttransfection, cells were collected and processed for CAT activity as described in Materials and Methods. CAT activity of cells treated with EMEM containing no DEX was set to 1. All other values are expressed as the relative increase in activation with respect to their controls. The results are the average of three independent experiments. (B) IEtu1 promoter constructs were assessed for DEX induction as described in panel A. Description of the IEtu1 promoter constructs was described previously (7, 28, 29). IEtu1-CAT contains IEtu1 promoter sequences that were cloned upstream of pSV0CAT (a promoter minus CAT expression vector; this plasmid was provided by V. Misra, Saskatoon, Saskatchewan, Canada). Two deletion constructs, Δ1024 IEtu1 and Δ1391 IEtu1, have 1024 or 1391 bp removed from the 5′ terminus, respectively.
DEX activates a cellular transcription factor, C/EBP-alpha, in Neuro-2A cells.
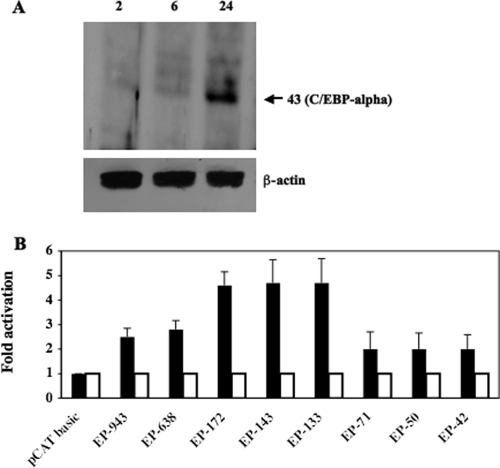
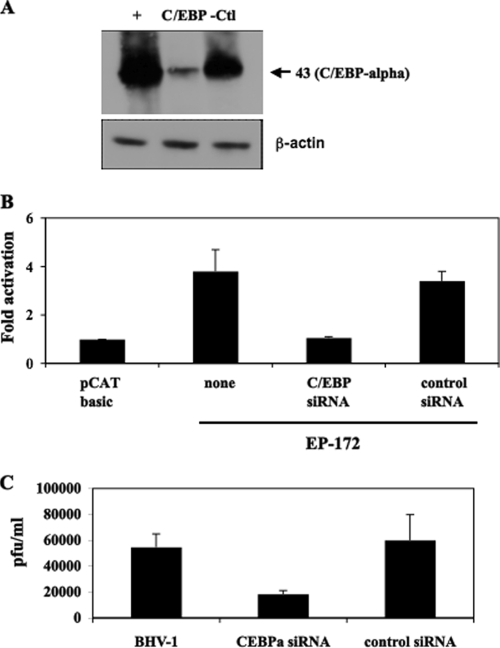
A previous study demonstrated that the cellular transcription factor C/EBP-alpha is stimulated when latently infected calves are treated with DEX (27). Furthermore, C/EBP-alpha stimulates productive infection when overexpressed (27), and C/EBP-alpha plus bICP0 or bTIF stimulates IEtu1 promoter activity more efficiently than just the viral trans activator alone (28). To test whether DEX induced C/EBP-alpha protein expression in Neuro-2A cells, cells were treated with 1 μM DEX, and Western blotting was performed. Twenty-four hours after DEX treatment, C/EBP-alpha protein expression was consistently induced (Fig. 4A). Conversely, β-actin protein levels were similar regardless of DEX treatment.
FIG. 4.
DEX induces C/EBP-alpha expression in cultured cells, and C/EBP-alpha activates the bICP0 early promoter. (A) Neuro-2A cells were treated with 1 μM DEX, and cell lysate was collected at various times after treatment (hours). A total of 100 μg of protein was electrophoresed, and Western blot analysis was performed using a C/EBP-alpha or β-actin antiserum that was diluted 1:500. The molecular mass is in kilodaltons. (B) Neuro-2A cells were cotransfected with 1 μg of the designated bICP0 E promoter construct and 0.1 μg of wt C/EBP-alpha (filled columns) or the DNA binding mutant of C/EBP-alpha (open columns). Constant amounts of DNA were used for all transfections by adding pcDNA3.1, a blank expression vector. At 48 h posttransfection, cells were collected and processed for CAT activity as described in Materials and Methods. CAT activity of the cells transfected with the control pCAT basic vector was given a value of 1. All other values are expressed as the relative increase in activation with respect to the control. The results are the average of four independent experiments.
A plasmid that expresses C/EBP-alpha, but not a DNA binding mutant of C/EBP-alpha, transactivated three bICP0 E promoter constructs (EP-172, EP-133, and EP-143) four- to fivefold (Fig. 4B). Constructs with additional deletions (EP-71, EP-50, and EP-42) were transactivated only twofold. The larger bICP0 E promoter constructs, EP-943 and EP-638, were transactivated two- to threefold by C/EBP-alpha. As expected, pCAT-basic was not transactivated by C/EBP-alpha.
C/EBP-alpha expression is important for DEX-induced activation of the bICP0 E promoter.
To test whether induction of C/EBP-alpha by DEX was important for activating bICP0 E promoter activity, Neuro-2A cells were transfected with an siRNA directed against C/EBP-alpha, and then the ability of DEX to induce bICP0 E promoter activity was measured. A C/EBP-alpha siRNA (Fig. 5A) but not a control siRNA reduced C/EBP-alpha protein levels after DEX treatment. As expected, protein levels of β-actin were similar regardless of the siRNA transfected into Neuro-2A cells. bICP0 E promoter activity (EP-172) was not stimulated by DEX when cells were transfected with the C/EBP-alpha siRNA (Fig. 5B). Conversely, the control siRNA had little or no effect on DEX induction of the bICP0 E promoter.
FIG. 5.
The C/EBP-alpha siRNA inhibits the ability of DEX to stimulate bICP0 E promoter activity. (A) Neuro-2A cells were cotransfected with 1 μg of C/EBP-alpha and either 100 nM C/EBP-alpha siRNA or the control siRNA. At 48 h posttransfection, cells were collected and lysed with NP-40 lysis buffer, and 30 μg protein was electrophoresed by 12% SDS-PAGE. Proteins in the gel were transferred onto a polyvinylidene difluoride membrane and probed with the C/EBP-alpha antiserum that was diluted 1:500. The molecular mass marker is in kilodaltons. (B) Neuro-2A cells were cotransfected with 1 μg of EP-172 and with 100 nM C/EBPα siRNA or a control siRNA, as described in Materials and Methods. Four hours later the transfection complex was removed and replaced by fresh medium containing 1 μM DEX. At 48 h posttransfection, cells were collected and processed for CAT activity. CAT activity of cells transfected with the control pCAT basic vector was given a value of 1. All other values are expressed as the relative increase in activation with respect to the control. The results are the average of three independent experiments. (C) RS cells were transfected with the C/EBP-alpha siRNA (100 nm), the control siRNA (100 nm), or no siRNA. Twenty-four hours after transfection, cultures were infected with wt BHV-1 at a multiplicity of infection of 0.1. At 24 h after infection, plaque assays were performed as described in Materials and Methods.
Overexpression of C/EBP-alpha stimulates plaque formation three- to fivefold (27), suggesting that silencing C/EBP-alpha expression would reduce the levels of plaquing efficiency. In several independent studies, the C/EBP-alpha siRNA (Fig. 5C) but not the control siRNA inhibited productive infection approximately threefold. In summary, these studies suggested that C/EBP-alpha expression was necessary for DEX-induced bICP0 E promoter activity and efficient productive infection.
C/EBP-alpha interacts with the bICP0 E promoter.
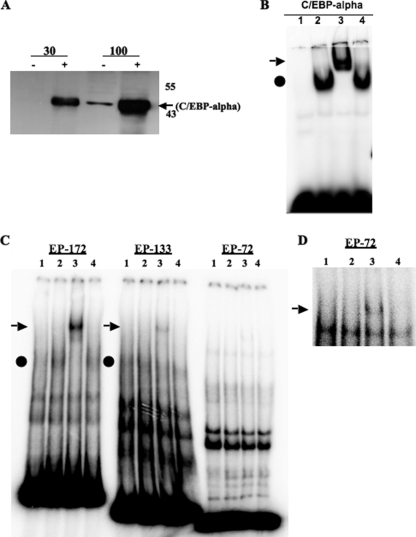
EMSAs were performed to determine whether C/EBP-alpha directly interacted with sequences in the bICP0 E promoter. Since C/EBP-alpha was expressed at low levels in Neuro-2A cells, a C/EBP-alpha expression plasmid was transfected into Neuro-2A cells to induce protein levels (Fig. 6A), and then extracts were prepared to test whether C/EBP-alpha interacted with the bICP0 E promoter. As a control, an oligonucleotide that contained three consensus C/EBP-alpha binding sites was used. As expected, binding to the C/EBP-alpha consensus oligonucleotide was enhanced when C/EBP-alpha was overexpressed in Neuro-2A cells (Fig. 6B, lane 2). An antibody that recognizes C/EBP-alpha (lane 3) but not C/EBP-beta (lane 4) “supershifted” the DNA-protein complex, confirming that a stable interaction occurred.
FIG. 6.
C/EBP-alpha directly interacts with the bICP0 E promoter. (A) Neuro-2A cells were transfected with 1 μg of C/EBPα. At 48 h posttransfection, cells were collected and lysed with NP-40 lysis buffer. Thirty or 100 μg of protein were electrophoresed by 12% SDS-PAGE. Proteins in the gel were transferred onto a polyvinylidene difluoride membrane and probed with the C/EBP-alpha antiserum that was diluted 1:500. The molecular mass marker is in kilodaltons. (B) EMSA was performed using a double-stranded DNA probe containing three consensus C/EBP-alpha binding sites. The probe was incubated with 5 μg of Neuro-2A cell lysate (lane 1) or Neuro-2A cell lysate in which C/EBP-alpha was overexpressed (lanes 2 to 4). The supershift was visualized by adding C/EBP-alpha (lane 3) or C/EBP-beta antibodies (lane 4) to the binding reaction as described in Materials and Methods. The arrow denotes the position of the supershift while the circle represents the C/EBP-alpha inducible band. (C) EMSA was performed using the bICP0 E promoter fragments derived from EP-172, EP-133, or EP-72. The lanes are the same as described in panel B. (D) To better visualize the super-shift for EP-72, autoradiography was performed for 24 h versus 4 as shown in panel C.
Enhanced binding to the EP-172 promoter was also detected after overexpression of C/EBP-alpha (Fig. 6C, lane 2 versus 1). Addition of a specific antibody directed against C/EBP-alpha (lane 3) but not C/EBP-beta (lane 4) supershifted the induced band. Although the C/EBP-alpha antibody supershifted a band when the EP-133 fragment was used as a probe (Fig. 6C, lane 3), binding was less efficient than with the EP-172 probe. When the EP-72 fragment was used as a probe, a faint supershifted band was detected (Fig. 6C, lane 3) that was readily detected when the gel was exposed for longer times (Fig. 6D, lane 3).
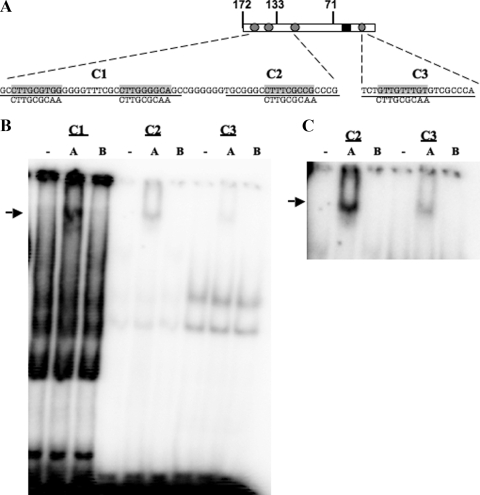
The bICP0 E promoter contains three C/EBP-alpha-like binding sites clustered near the 5′ terminus of EP-172 and a putative site downstream of the TATA box (Fig. 7A). To test whether C/EBP-alpha interacted with these individual domains, oligonucleotides spanning these domains (Fig. 7A, C1 to C3) were synthesized, and EMSAs were performed (Fig. 7B). Supershift assays using a C/EBP-alpha-specific antibody (Fig. 7B, lanes A) and the C/EBP-beta antibody (lanes B) suggested that C/EBP-alpha specifically interacted with the C1 and C2 oligonucleotides. Prolonged exposure of the autoradiograph revealed that C/EBP-alpha interacted less efficiently with the C3 olignonucleotide (Fig. 7C). In several independent studies, the band supershifted by the C/EBP-alpha antibody was more distinct than that of samples not treated with antibody, suggesting that the antibody stabilized the interaction between C/EBP-alpha and bICP0 E promoter sequences. As expected, the consensus C/EBP-alpha probe interfered with C/EBP-alpha-specific binding (data not shown). In summary, this study indicated that at least three different elements within the bICP0 E promoter interacted with C/EBP-alpha.
FIG. 7.
Interactions between segments of the bICP0 E promoter and C/EBP-alpha. (A) Schematic of EMSA probes C1, C2, and C3. C1 contains two putative C/EBP-alpha binding sites found within EP-172. C2 contains the other putative C/EBP-alpha binding site found upstream of the TATA box. C3 contains a putative C/EBP-alpha binding site downstream of the TATA box. The consensus C/EBP binding site in the rat CYPD5 promoter (24) is shown as a comparison to the putative sites in the bICP0 E promoter. (B) EMSA using C1, C2, or C3 probes. Probes were incubated with 30 μg of Neuro-2A cell lysate. Antibodies directed against C/EBP-alpha (A) or C/EBP-beta (B) were used in supershift assays as described in Materials and Methods. (C) To better visualize the supershift for C2 and C3, autoradiography was performed for 24 h versus 4 h as shown in panel B.
DISCUSSION
In this study, we demonstrated that viral gene expression was stimulated in TG of calves latently infected with the LR mutant virus after DEX treatment. The inability to detect infectious virus in ocular or nasal swabs after calves latently infected with the LR mutant virus were treated with DEX (14) correlated with the lack of gC transcription after DEX treatment. These findings implied that the LR mutant virus was unable to infect neurons that support reactivation from latency or that the LR mutant virus killed neurons that support reactivation from latency. Interestingly, higher rates of neuronal apoptosis occur in TG neurons near the end of acute infection (establishment of latency) following infection with the LR mutant virus (25). Furthermore, in situ hybridization revealed that a small percentage of viral genome-positive neurons are present in TG of calves infected with wt or the LR rescued virus but not the LR mutant virus (14). These observations suggested that the antiapoptosis activity of a protein encoded by the LR gene promotes survival of a specific subset of neurons that can yield infectious virus after DEX treatment (3, 5, 10, 25, 41, 46). Neurons that contain few copies of viral genomes do not support extensive viral gene expression during acute infection, and consequently they survive acute infection and permit establishment of viral latency, regardless of whether LR gene products are expressed. The finding that different populations of neurons in TG of mice are more permissive for HSV-1 (51) is consistent with the prediction that a subset of neurons in TG contain specific cellular factors that support reactivation from latency.
Relative to other alphaherpesvirus members, the organization of the BHV-1 ICP4 and ICP0 genes is unique because a common IE promoter drives expression of bICP0 and bICP4 (48) (Fig. 2A). bICP0 also contains an E promoter located near the 5′ end of the coding exon of bICP0. Although it was not well understood why the bICP0 gene contains two promoters, we assumed this was necessary to ensure that constitutive expression of bICP0 occurred during productive infection.
DEX treatment appeared to stimulate cellular factors, which then activated the bICP0 E promoter in TG of latently infected calves (Fig. 1C) and mouse neuroblastoma (Neuro-2A) cells (Fig. 3). Previous studies demonstrated that DEX treatment of calves latently infected with BHV-1 leads to C/EBP-alpha expression (27), which correlated with reactivation from latency and activation of the bICP0 E promoter. The finding that C/EBP-alpha siRNAs inhibited the ability of DEX to activate the bICP0 E promoter supported the conclusion that C/EBP-alpha was necessary for stimulating bICP0 E promoter activity after DEX treatment. DEX can activate C/EBP-alpha in several distinct cell types (4, 11, 34), including Neuro-2A (Fig. 4) and TG neurons (27). Three distinct regions of the bICP0 E promoter interacted with C/EBP-alpha, as judged by EMSA and supershift studies. Based on transient transfection assays, we suggest that at least two of the C/EBP-alpha binding sites were necessary for efficiently trans activation by C/EBP-alpha. The weak binding site in the C3 oligonucleotide (Fig. 7A) was not sufficient for activation by C/EBP-alpha because EP-71, EP-50, and EP-42 were not transactivated by C/EBP-alpha (Fig. 4B). The C/EBP-alpha binding sites in the bCP0 E promoter appeared to be low-affinity binding sites compared to the consensus site because 30 μg of nuclear extract was necessary to observe C/EBP-alpha-induced binding with EP-172, whereas only 5 μg of protein in nuclear extracts was necessary to detect inducible binding to an oligonucleotide containing three consensus C/EBP-alpha sites (Fig. 6). The weak C/EBP-alpha binding sites in the bICP0 E promoter may inhibit activation of the E promoter during latency unless the neuronal environment is conducive for reactivation from latency.
The larger bICP0 E promoter constructs (EP-943 and EP-638) were not efficiently activated by C/EBP-alpha or by DEX treatment. However, EP-943 and EP-638 promoter constructs had two- to threefold-higher levels of basal promoter activity. We suggest there may be cis-acting negative regulatory elements located in the upstream region of the bICP0 early promoter that interferes with the ability of C/EBP-alpha to activate promoter activity. Furthermore, cellular transcription factors that stimulate the larger promoter constructs may interfere with the ability of C/EBP-alpha to activate E promoter activity. Since the bICP0 E promoter overlaps bICP4 protein coding sequences, these additional bICP4 protein-coding sequences may not be a part of the true bICP0 E promoter and, consequently, may interfere with transient transfection reporter assays.
A protein encoded by an alternatively spliced LR RNA stably interacts with C/EBP-alpha (26). The finding that C/EBP-alpha stimulates productive infection (26), the IEtu1 promoter (28), and the bICP0 E promoter suggests that interactions between the LR protein and C/EBP-alpha help to extinguish viral transcription, thus promoting the establishment and maintenance of latency. DEX represses LR RNA (35) and LR promoter activity (20), suggesting that LR gene products do not directly stimulate reactivation from latency. However, reduced levels of LR proteins following DEX induction of reactivation may allow C/EBP-alpha to stimulate viral gene expression. Although C/EBP-alpha appears to be an important component in the DEX-induced signaling cascade that stimulates viral gene expression, it seems clear that additional DEX-inducible cellular proteins are required for infectious virus to be produced.
Acknowledgments
This research was supported by two USDA grants (08-00891 and 06-01627) and a NIAID grant (R21AI069176). A grant to the Nebraska Center for Virology (1P20RR15635) also supported certain aspects of these studies. Aspen Workman was partially supported by a fellowship from a Ruth L. Kirschstein National Research Service Award 1 T32 AIO60547 (National Institute of Allergy and Infectious Diseases) and by the National Center for Research Services (NIH; P20 RRO 16469).
Footnotes
Published ahead of print on 24 June 2009.
REFERENCES
- 1.Bratanich, A. C., N. D. Hanson, and C. Jones. 1992. The latency-related gene of bovine herpesvirus 1 inhibits the activity of immediate-early transcription unit 1. Virology 191988-991. [DOI] [PubMed] [Google Scholar]
- 2.Chomczynski, P., and N. Sacchi. 1987. Single step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162156-159. [DOI] [PubMed] [Google Scholar]
- 3.Ciacci-Zanella, J., M. Stone, G. Henderson, and C. Jones. 1999. The latency-related gene of bovine herpesvirus 1 inhibits programmed cell death. J. Virol. 739734-9740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cram, E. J., R. A. Ramos, E. C. Wang, H. H. Cha, Y. Nishio, and G. L. Firestone. 1998. Role of the CCAAT/Enhancer binding protein-alpha transcription factor in the glucocorticoid stimulation of p21waf1/cip1 gene promoter activity in growth arrested rat hepatoma cells. J. Biol. Chem. 2732008-2014. [DOI] [PubMed] [Google Scholar]
- 5.Devireddy, L. R., and C. Jones. 1999. Activation of caspases and p53 by bovine herpesvirus 1 infection results in programmed cell death and efficient virus release. J. Virol. 733778-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fraefel, C., J. Zeng, Y. Choffat, M. Engels, M. Schwyzer, and M. Ackermann. 1994. Identification and zinc dependence of the bovine herpesvirus 1 transactivator protein BICP0. J. Virol. 683154-3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geiser, V., and C. Jones. 2003. Stimulation of bovine herpesvirus 1 productive infection by the adenovirus E1A gene and a cell cycle regulatory gene, E2F-4. J. Gen. Virol. 84929-938. [DOI] [PubMed] [Google Scholar]
- 8.Geiser, V., and C. Jones. 2005. The latency related gene encoded by bovine herpesvirus 1 encodes a small regulatory RNA that inhibits cell growth. J. Neurovirol. 11563-570. [DOI] [PubMed] [Google Scholar]
- 9.Geiser, V., M. Inman, Y. Zhang, and C. Jones. 2002. The latency related (LR) gene of bovine herpes virus 1 (BHV-1) can inhibit the ability of bICP0 to activate productive infection. J. Gen. Virol. 832965-2971. [DOI] [PubMed] [Google Scholar]
- 10.Geiser, V., S. Rose, and C. Jones. 2008. Bovine herpes virus type 1 induces cell death by a cell-type-dependent fashion. Microb. Pathog. 44459-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hausman, G. J. 2000. The influence of dexamethasone and insulin on expression of CCAAT/enhancer binding protein isoforms during preadipocyte differentiation in porcine stromal-vascular cell cultures: evidence for very early expression of C/EBPalpha. J. Anim. Sci. 781227-1235. [DOI] [PubMed] [Google Scholar]
- 12.Henderson, G., G.-C. Perng, A. Nesburn, S. Wechsler, and C. Jones. 2004. The latency related gene of bovine herpesvirus 1 can suppress caspase 3 and caspase 9 during productive infection. J. Neurovirol. 1064-70. [DOI] [PubMed] [Google Scholar]
- 13.Hossain, A., L. M. Schang, and C. Jones. 1995. Identification of gene products encoded by the latency-related gene of bovine herpesvirus 1. J. Virol. 695345-5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inman, M., L. Lovato, A. Doster, and C. Jones. 2002. A mutation in the latency related gene of bovine herpesvirus 1 interferes with the latency-reactivation cycle of latency in calves. J. Virol. 766771-6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inman, M., L. Lovato, A. Doster, and C. Jones. 2001. A mutation in the latency-related gene of bovine herpesvirus 1 leads to impaired ocular shedding in acutely infected calves. J. Virol. 758507-8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang, Y., A. Hossain, M. T. Winkler, T. Holt, A. Doster, and C. Jones. 1998. A protein encoded by the latency-related gene of bovine herpesvirus 1 is expressed in trigeminal ganglionic neurons of latently infected cattle and interacts with cyclin-dependent kinase 2 during productive infection. J. Virol. 728133-8142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang, Y., M. Inman, Y. Zhang, N. A. Posadas, and C. Jones. 2004. A mutation in the latency related gene of bovine herpesvirus 1 (BHV-1) inhibits protein expression of a protein from open reading frame 2 (ORF-2) and an adjacent reading frame during productive infection. J. Virol. 783184-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones, C. 1998. Alphaherpesvirus latency: its role in disease and survival of the virus in nature. Adv. Virus Res. 5181-133. [DOI] [PubMed] [Google Scholar]
- 19.Jones, C. 2003. Herpes simplex virus type 1 and bovine herpesvirus 1 latency. Clin. Microbiol. Rev. 1679-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones, C., G. Delhon, A. Bratanich, G. Kutish, and D. Rock. 1990. Analysis of the transcriptional promoter which regulates the latency-related transcript of bovine herpesvirus 1. J. Virol. 641164-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones, C., T. J. Newby, T. Holt, A. Doster, M. Stone, J. Ciacci-Zanella, C. J. Webster, and M. W. Jackwood. 2000. Analysis of latency in cattle after inoculation with a temperature sensitive mutant of bovine herpesvirus 1 (RLB106). Vaccine 183185-3195. [DOI] [PubMed] [Google Scholar]
- 22.Jones, C., V. Geiser, G. Henderson, Y. Jiang, F. Meyer, S. Perez, and Y. Zhang. 2006. Functional analysis of bovine herpesvirus 1 (BHV-1) genes expressed during latency. Vet. Microbiol. 113199-210. [DOI] [PubMed] [Google Scholar]
- 23.Kutish, G., T. Mainprize, and D. Rock. 1990. Characterization of the latency-related transcriptionally active region of the bovine herpesvirus 1 genome. J. Virol. 645730-5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, Y.-H., S. C. Willims, M. Baer, E. Sterneck, F. J. Gonzalez, and P. F. Johnson. 1997. The ability of C/EBPβ but not C/EBPα to synergize with an Sp1 protein is specified by the leucine zipper and activation domain. Mol. Cell. Biol. 172038-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lovato, L., M Inman, G Henderson, A Doster, and C. Jones. 2003. Infection of cattle with a bovine herpesvirus 1 strain that contains a mutation in the latency related gene leads to increased apoptosis in trigeminal ganglia during the transition from acute infection to latency. J. Virol. 774848-4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer, F., S Perez, Y Jiang, Y Zhou, G Henderson, and C. Jones. 2007. Identification of a novel protein encoded by the latency-related gene of bovine herpesvirus 1. J. Neurovirol. 13569-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyer, F., S. Perez, V. Geiser, M. Sintek, M. Inman, and C. Jones. 2007. A protein encoded by the bovine herpes virus 1 latency related gene interacts with specific cellular regulatory proteins, including the CCAAT enhancer binding protein alpha. J. Virol. 8159-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer, F., and C. Jones. 2009. C/EBP-alpha cooperates with bTIF to activate the bovine herpesvirus 1 immediate early transcription unit 1 promoter. J. Neurovirol. 15123-130. [DOI] [PubMed] [Google Scholar]
- 29.Misra, V., A. C. Bratanich, D. Carpenter, and P. O'Hare. 1994. Protein and DNA elements involved in transactivation of the promoter of the bovine herpesvirus (BHV) 1 IE-1 transcription unit by the BHV αgene trans-inducing factor. J. Virol. 684898-4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Misra, V., S. Walker, S. Hayes, and P. O'Hare. 1995. The bovine herpesvirus alpha gene trans-inducing factor activates transcription by mechanisms different from those of its herpes simplex virus type 1 counterpart VP16. J. Virol. 695209-5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perez, S., F. Meyer, K. Saira, A. Doster, and C. Jones. 2008. Premature expression of the latency-related RNA encoded by bovine herpesvirus 1 correlates with higher levels of beta interferon RNA expression in productively infected cells. J. Gen. Virol. 891338-1345. [DOI] [PubMed] [Google Scholar]
- 32.Perez, S., L. Lovato, J. Zhou, A. Doster, and C. Jones. 2006. Comparison of inflammatory infiltrates in trigeminal ganglia of cattle infected with wild type BHV-1 versus a virus strain containing a mutation in the LR (latency-related) gene. J. Neurovirol. 12392-397. [DOI] [PubMed] [Google Scholar]
- 33.Perez, S., M. Inman, A. Doster, and C. Jones. 2005. Latency-related gene encoded by bovine herpesvirus 1 promotes virus growth and reactivation from latency in tonsils of infected calves. J. Clin. Microbiol. 43393-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramos, R., Y. Nishio, A. C. Maiyar, K. E. Simon, C. C. Ridder, Y. Ge, and G. L. Firestone. 1996. Glucocorticoid-stimulated CCAAT/enhancer-binding protein alpha expression is required for steroid-induced G1 cell cycle arrest of minimal-deviation rat hepatoma cells. Mol. Cell. Biol. 165288-5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rock, D., J. Lokensgard, T. Lewis, and G. Kutish. 1992. Characterization of dexamethasone-induced reactivation of latent bovine herpesvirus 1. J. Virol. 662484-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rock, D. L., S. L. Beam, and, and J. E. Mayfield. 1987. Mapping bovine herpesvirus type 1 latency-related RNA in trigeminal ganglia of latently infected rabbits. J. Virol. 613827-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schang, L., and C. Jones. 1997. Analysis of bovine herpesvirus 1 transcripts during a primary infection of trigeminal ganglia of cattle. J. Virol. 716786-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schang, L. M., A. Hossain, and C. Jones. 1996. The latency-related gene of bovine herpesvirus 1 encodes a product which inhibits cell cycle progression. J. Virol. 703807-3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwyzer, M., U. V. Wirth, B. Vogt, and C. Fraefel. 1994. BICP22 of bovine herpesvirus 1 is encoded by a spliced 1.7 kb RNA which exhibits immediate early and late transcription kinetics. J. Gen. Virol. 751703-1711. [DOI] [PubMed] [Google Scholar]
- 40.Sheffy, B. E., and D. H. Davies. 1972. Reactivation of a bovine herpesvirus after corticosteroid treatment. Proc. Soc. Exp. Biol. Med. 140974-976. [DOI] [PubMed] [Google Scholar]
- 41.Shen, W., and C. Jones. 2008. Open reading frame 2 encoded by the latency related gene of bovine herpesvirus 1 has antiapoptotic activity in transiently transfected neuroblastoma cells. J. Virol. 8210940-10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tikoo, S. K., M. Campos, and L. A. Babiuk. 1995. Bovine herpesvirus 1 (BHV-1): biology, pathogenesis, and control. Adv. Virus Res. 45191-223. [DOI] [PubMed] [Google Scholar]
- 43.Wang, G., and N. A. Timchenko. 2005. Dephosphorylated C/EBPα accelerated cell proliferation through sequestering retinoblastoma protein. Mol. Cell. Biol. 251325-13538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, S. E., F. Y. Wu, Y. Yu, and G. S. Hayward. 2003. CCAAT/enhancer-binding protein-α is induced during the early stages of Kaposi's sarcoma-associated herpesvirus (KSHV) lytic cycle reactivation and together with the KSHV replication and transcription activator (RTA) cooperatively stimulates the viral RTA, MTA, and PAN promoters. J. Virol. 779590-9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winkler, M. T., A. Doster, J. H. Sur, and C. Jones. 2002. Analysis of bovine trigeminal ganglia following infection with bovine herpesvirus 1. Vet. Microbiol. 86139-155. [DOI] [PubMed] [Google Scholar]
- 46.Winkler, M. T., A. Doster, and C. Jones. 1999. Bovine herpesvirus 1 can infect CD4+ T lymphocytes and induce programmed cell death during acute infection of cattle. J. Virol. 738657-8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winkler, M. T. C., A. Doster, and C. Jones. 2000. Persistence and reactivation of bovine herpesvirus 1 in the tonsil of latently infected calves. J. Virol. 745337-5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wirth, U. V., C. Fraefel, B. Vogt, C. Vlcek, V. Paces, and M. Schwyzer. 1992. Immediate-early RNA 2.9 and early RNA 2.6 of bovine herpesvirus 1 are 3′ coterminal and encode a putative zinc finger transactivator protein. J. Virol. 662763-2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wirth, U. V., K. Gunkel, M. Engels, and M. Schwyzer. 1989. Spatial and temporal distribution of bovine herpesvirus 1 transcripts. J. Virol. 634882-4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wirth, U. V., B. Vogt, and M. Schwyzer. 1991. The three major immediate-early transcripts of bovine herpesvirus 1 arise from two divergent and spliced transcription units. J. Virol. 65195-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang, L., C. C. Voytek, and T. P. Margolis. 2000. Immunohistochemical analysis of primary sensory neurons latently infected with herpes simplex virus type 1. J. Virol. 74209-217. [DOI] [PMC free article] [PubMed] [Google Scholar]