Abstract
Helper-dependent adenovirus (hdAd) vectors have shown great promise as therapeutic gene delivery vehicles in gene therapy applications. However, the level and duration of gene expression from hdAd can differ considerably depending on the nature of the noncoding stuffer DNA contained within the vector. For example, an hdAd containing 22 kb of prokaryotic DNA (hdAd-prok) expresses its transgene 60-fold less efficiently than a similar vector containing eukaryotic DNA (hdAd-euk). Here we have determined the mechanistic basis of this phenomenon. Although neither vector was subjected to CpG methylation and both genomes associated with cellular histones to similar degrees, hdAd-prok chromatin was actively deacetylated. Insertion of an insulator element between the transgene and the bacterial DNA derepressed hdAd-prok, suggesting that foreign DNA nucleates repressive chromatin structures that spread to the transgene. We found that Sp100B/Sp100HMG and Daxx play a role in repressing transgene expression from hdAd and act independently of PML bodies. Thus, we have identified nuclear factors involved in recognizing foreign DNA and have determined the mechanism by which associated genes are repressed.
Efficient delivery and expression of foreign genes are of great importance in medicine and basic science. In many gene therapy applications, expression of the therapeutic gene would be required for the lifetime of the patient, yet many vector systems display only transient expression, lasting as little as a few days or weeks. Helper-dependent adenovirus (hdAd) vectors can enhance the duration of expression of a therapeutic gene; studies of mice and nonhuman primates have yielded several years of gene expression after a single administration (28). Indeed, several studies have described lifelong expression of a gene and persistent phenotypic correction in mouse models of human disease (18, 26, 42).
Most hdAds contain noncoding “stuffer” DNA to maintain the size of the vector within appropriate limits for efficient DNA packaging; vectors constructed below ∼27 kb undergo DNA rearrangement in order to increase the size of the genome to 27 to 38 kb (31, 38). Interestingly, the nature of the stuffer DNA included in the hdAd has a significant effect on the function of the vector. An hdAd vector containing 22 kb of eukaryotic DNA (hdAd-euk) expressed a transgene to a higher level and for a longer duration than a vector containing 22 kb of prokaryotic DNA (hdAd-prok), both in vitro and in vivo (29). The genomes of the two vectors persisted at similar levels within the livers of transduced mice, suggesting that incorporation of prokaryote-derived stuffer DNA into an hdAd leads to the shutoff of associated transgenes. As a result of these observations, most current hdAd vectors are constructed using stuffer DNA derived from eukaryotic sources (27).
Silencing of transgenes associated with prokaryotic DNA is not unique to hdAd. Removal of the bacterial origin of replication and antibiotic resistance gene from herpes simplex virus (HSV) amplicons resulted in a 20-fold improvement in gene expression in normal human fibroblasts in vitro, and more-persistent reporter gene expression in nude mice, compared to amplicons retaining the bacterial elements (39). Similarly, removal of bacterial sequences from plasmids results in significantly improved transgene expression in vitro and in vivo (2, 3, 34). For both plasmid and HSV amplicons, the mechanisms by which the bacterial sequences impair transgene expression are not fully understood. However, the bacterial sequences appear to nucleate the formation of a repressive chromatin structure(s) that spreads to the transgene (4, 39).
In this study, we experimentally address the mechanism behind the repressive effects of prokaryotic DNA on gene expression in hdAd vectors. We found that prokaryotic DNA inhibits eukaryotic gene expression in cis, via induction of histone deacetylation, which is independent of DNA methylation. Furthermore, our data indicate that Sp100 and Daxx are involved in repressing the expression of genes associated with prokaryotic DNA.
MATERIALS AND METHODS
Cells and virus culture.
Propagation of 293 cells (12) (a kind gift from Frank Graham, McMaster University, Hamilton, Ontario, Canada), 293-N3S cells (Microbix), A549 cells (ATCC), and 116 cells (27) (a kind gift from Philip Ng, Baylor College of Medicine, Houston, TX) was performed as described previously (35). HeLa cells (ATCC) were maintained in Dulbecco's modified Eagle's medium (Sigma-Aldrich) supplemented with 10% fetal calf serum, 2 mM GlutaMAX, and 1× antibiotic/antimycotic (Invitrogen).
hdAd-prok (AdRP1030), hdAd-euk (AdRP1050) (29), hdAd-PGK-mSEAP (21, 24), and AdRP2050 (37) have been described previously. hdAd-prok-ins and hdAd-euk-ins, which contain the HS4 double-core insulator element (32) (a kind gift of Gary Felsenfeld) between the eukaryotic expression cassettes and stuffer DNA, were generated using standard molecular cloning protocols (Fig. 1A). The helper virus AdRP2050 was propagated, and titers were determined, as described previously (35). hdAd-prok and hdAd-euk were propagated in 116 cells using AdRP2050 as the helper virus, as described previously (27, 30). A second pair of hdAd vectors containing stuffer DNAs derived from prokaryotic and eukaryotic stuffer DNAs were also utilized and are designated hdAd-prok2 and hdAd-euk2, respectively. hdAd-euk2 is HDΔ28E4LacZ (a kind gift from Philip Ng, Baylor College of Medicine) and contains the native Escherichia coli β-galactosidase (β-Gal) gene under the regulation of the murine cytomegalovirus (mCMV) immediate-early promoter/enhancer and the simian virus 40 polyadenylation sequence, stuffer DNA derived from the human hypoxanthine-guanine phosphoribosyltransferase (HPRT1) gene, and genomic DNA derived from the C346 cosmid (locus DXS455A), as previously described (27, 43). hdAd-prok2 contains an identical β-Gal expression cassette and two randomly cloned fragments of prokaryotic DNA derived from the E. coli DH10B genome (GenBank accession number NC_010473; fragments comprising nucleotides 3845034 to 3861440 and 4233399 to 4240086). hdAd-prok2 and hdAd-euk2 were amplified using helper virus AdNG163 (27). Infectious hdAd particles were scored as infectious units (IU), as determined on 293 cells using previously established protocols (9, 30). Due to the reduced intensity of staining of cells infected with hdAd-prok, the titers of this vector were determined in cells treated with 250 ng/ml trichostatin A (TSA; Upstate), which resulted in a 1.5- to 2-fold increase in the vector titer over that obtained on cells not treated with TSA. Virus particle counts were determined as previously described (35). The ratios of total particles to infectious particles for hdAd-prok, -prok2, -euk, and -euk2 were 11:1, 7:1, 6:1, and 3:1, respectively. Contamination of hdAd vector stocks with helper virus was determined as PFU on 293 cells (35), and all hdAd vectors had similar ratios of IU to PFU (approximately 0.05% helper virus contamination). Infections were performed using standard methods (35). Reporter gene expression was determined using a chemiluminescent β-Gal assay kit (Roche). Data were analyzed by a t test or analysis of variance with a Tukey test, with significance defined as a P value less than 0.05.
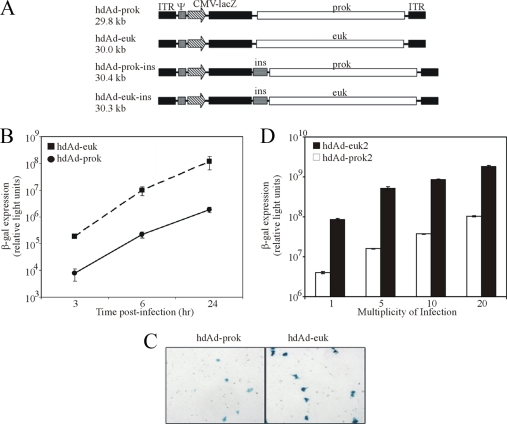
FIG. 1.
hdAd-prok expresses its transgene less efficiently than hdAd-euk. (A) Schematic representation of the hdAd vectors used in this study. All vectors encode the Ad serotype 5 inverted terminal repeats (ITR) and packaging signal (ψ), the murine cytomegalovirus immediate-early promoter and enhancer (CMV), the lacZ open reading frame, and the simian virus 40 polyadenylation signal (not shown). hdAd-prok and hdAd-prok-ins encode 22 kb of prokaryotic DNA (prok), whereas hdAd-euk and hdAd-euk-ins encode 22 kb of eukaryotic DNA (euk). hdAd-prok-ins and hdAd-euk-ins each have the HS4 double-core insulator element (ins) inserted between the expression cassette and the stuffer DNA. hdAd-prok2 (not shown) is similar in structure to hdAd-prok but contains a stuffer element derived from E. coli genomic DNA. hdAd-euk2 is identical to HDΔ28E4LacZ (27). (B) HeLa cells were infected with hdAd-prok or -euk (MOI, 1 IU/cell). At 3, 6, or 24 h postinfection, cell lysates were prepared and assayed for β-Gal activity (means ± standard errors; n = 4). (C) HeLa cells were infected with hdAd-prok or -euk (MOI, 0.1 IU/cell) and were stained with X-Gal 24 h later. (D) HeLa cells were infected with hdAd-prok2 or -euk2 at varying MOIs, and 24 h later, cell lysates were assayed for β-Gal activity. Error bars represent the ranges of values obtained from two duplicate samples. Representative data from three independent experiments are presented.
Plasmids and siRNA.
Plasmids encoding FLAG-tagged Sp100A, Sp100B, and Sp100HMG were kindly provided by Hans Will (Heinrich-Pette Institute for Experimental Virology, Hamburg, Germany). Plasmids encoding hemagglutinin-tagged pp71 and pp71delDID2/3 were kindly provided by Robert Kalejta (University of Wisconsin, Madison). Plasmids expressing wild-type and mutant Gam1 were obtained from Susanna Chiocca (European Institute of Oncology, Milan, Italy). Pooled small interfering RNAs (siRNA) targeting Sp100 and Daxx were obtained from Dharmacon. All transfections were performed using Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions.
ChIP.
Chromatin immunoprecipitation (ChIP) assays were performed using a modified version of the Upstate Biotechnology protocol. Lysis buffers and ChIP dilution buffer were supplemented with 1 mM phenylmethylsulfonyl fluoride (OmniPur), 10 μg/ml leupeptin (Roche), and 10 μg/ml aprotinin(Roche). Confluent HeLa or A549 cells in 15-cm-diameter dishes (∼2.5 × 107 cells) were infected with the indicated virus (multiplicity of infection [MOI], 10 IU/cell); 24 h later, formaldehyde was added to the culture medium to a concentration of 1%, and the plates were incubated for 10 min at room temperature. After a 5-min incubation in 0.125 M glycine, monolayers were rinsed twice with ice-cold phosphate-buffered saline, scraped into phosphate-buffered saline supplemented with 1 mM phenylmethylsulfonyl fluoride, and transferred to 1.5-ml microcentrifuge tubes. Cells were pelleted by centrifugation (500 × g, 5 min, 4°C), suspended in 0.5 ml cell lysis buffer [5 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES; pH 8), 85 mM KCl, 0.5% NP-40], and incubated for 45 min at 4°C with constant rotation. The nuclei were pelleted by centrifugation (1,250 × g, 5 min, 4°C) and lysed in 0.25 ml nuclear lysis buffer (50 mM Tris-HCl [pH 8], 1% sodium dodecyl sulfate [SDS], 10 mM EDTA) for 10 min. Chromatin was sheared to an average size of 200 to 1,000 bp by sonication (5 to 10 pulses of 10 s with a Vibra-Cell VCX 600 ultrasonic processor [Sonics & Materials] equipped with a stepped microtip at a setting of 30% of maximal amplitude). Sheared chromatin was diluted 10-fold with ChIP dilution buffer supplemented with 22.2 μg/ml sheared herring sperm DNA and 1.11 mg/ml bovine serum albumin. Dilute chromatin was precleared with 50 μl of a 50% slurry of protein G beads (Upstate) (which had been preincubated for 1 h in the presence of 200 μg/ml sheared herring sperm DNA and 1 mg/ml bovine serum albumin) for 2 h at 4°C with constant rotation. Precleared chromatin was then incubated overnight at 4°C with constant rotation with the indicated antibody and 50 μl of a 50% slurry of protein G beads. The antibodies used in these experiments were anti-adenovirus E1A (M58; Neomarkers) or horseradish peroxidase-conjugated goat-anti mouse immunoglobulin G (170-6516; Bio-Rad) (negative controls), anti-unmodified H3 (ab1791; Abcam), anti-pan-acetyl H3 (06-599; Upstate), anti-pan-acetyl H4 (06-866; Upstate), anti-RNA polymerase II (N-20; Santa Cruz), anti-2meK9H3 (07-441; Upstate), and anti-3meK27H3 (Ab6002; Abcam).
After immunoprecipitation, the beads were rinsed sequentially with low-salt immune complex wash buffer (ICWB), high-salt ICWB, and lithium chloride ICWB; then they were rinsed twice with Tris-EDTA. The beads were then incubated twice for 15 min with 0.25 ml immune complex elution buffer (100 mM sodium bicarbonate, 1% SDS). Eluates were combined; NaCl was added to 200 mM; and the samples were incubated for 4 to 6 h at 65°C to reverse the cross-links. Next, 10 μl 0.5 mM EDTA, 20 μl 1 M Tris-HCl (pH 6.5), and 1 μl 20-mg/ml proteinase K (Sigma) were added to the samples, which were then incubated for 2 h at 42°C. DNA was purified by phenol-chloroform extraction and precipitated with isopropanol and 20 μg glycogen (Roche). ChIP DNA was suspended in 20 μl H2O and analyzed by semiquantitative PCR using Taq polymerase (Invitrogen). Preliminary semiquantitative PCR analyses were performed using varying volumes of input DNA (typically representing 0.05%, 0.1%, and 0.2% of input) and varying cycle numbers to establish the PCR conditions that would yield endpoints in the linear range of amplification for all primer sets used. Semiquantitative analyses typically involved 25 to 30 cycles of amplification, and annealing temperatures for all primer sets are displayed in Table 1. PCR products were subjected to electrophoresis on 1.5 or 2% agarose gels and were stained with ethidium bromide.
TABLE 1.
Oligonucleotides used in this study
| Application and primer name | Primer sequence | Annealing temp (°C)a | Referenceb |
|---|---|---|---|
| ChIP | |||
| β-Actin FWD | CAAAACTCTCCCTCCTCCTCTTC | 60 | 40 |
| β-Actin REV | GAGCCATAAAAGGCAACTTTCGGAA | 60 | 40 |
| IFN-β FWD | CCTCGAGTCCCAAGTCTTGTTTTACAATTTGC | 60 | |
| IFN-β REV | CAAGCTTTTGACAACACGAACAGTGTCGC | 60 | |
| Myt1 FWD | CAGGAAGACACCTCTCACAC | 65 | 19 |
| Myt1 REV | ACAGTGTCCAGGGGCTTTGC | 65 | 19 |
| mCMV FWD | GTTCTTCGAGCCAATACACGTCAATG | 56 | 41 |
| mCMV REV | GTACCGACGCTGGTCGCGCC | 56 | |
| lacZ FWD | TGAAGCAGAAGCCTGCGATGTCGG | 60 | |
| lacZ REV | CACAGCGGATGGTTCGGATAAAGCG | 60 | |
| hdAd-delta stuffer FWD | GTAAAGCCGAACCCGGGAAACTG | 56.5 | |
| hdAd-delta stuffer REV | CGTGTGGGAGAAGGGCAGGATC | 56.5 | |
| Bisulfite conversion | |||
| mCMV bis con FWD | AATCCATAACAAAATCCTCTAACAACTTAAATT | 65 | |
| mCMV bis con REV | GTATATAAGGTTAATAGGGGTGAGTTATTGGG | 65 | |
| prok stuf mid FWD | GCGAAGCTTATCCCTTCTAATACTATCATCAACATTAC | 50, 65 | |
| prok stuf mid REV | GCGCTCGAGTTTATGATGTTTTGTTGGATATGTATTT | 50, 65 |
For PCR with the oligonucleotide pair. The two temperatures displayed for prok stuf mid FWD and prok stuf mid REV were used for the first 10 and last 30 rounds of amplification, respectively.
Where references are not provided, the oligonucleotides were unique to this study.
Analysis of DNA methylation.
To examine the status of CpG methylation prior to host cell transduction, DNA was isolated from 1 × 1011 hdAd-prok particles by SDS-proteinase K extraction (35), digested with MspI or HpaII, separated on a 1.0% agarose gel, and visualized by staining with ethidium bromide. To examine the methylation status of vector DNA after cell transduction, A549 cells were infected at an MOI of 250 IU/cell, and at 1 or 7 days postinfection, total cellular DNA was isolated and digested with MspI, HpaII, SphI, or McrB/C (New England Biolabs). The resulting DNA was separated on a 1% agarose gel, transferred to a nylon membrane, and probed with digoxigenin (DIG)-labeled hdAd-prok plasmid DNA. DNA labeling and probe detection were performed using the DIG High Prime DNA labeling and detection starter kit II (Roche). The DNA methylation status of hdAd-prok and -euk was also analyzed using bisulfite conversion under previously established conditions (10, 13). Converted DNA was suspended in 20 μl H2O, and 5 μl was used for PCR with 40 amplification cycles and the primer sets shown in Table 1. The resulting PCR fragments were cloned into pBluescript KS(+) (Stratagene) and sequenced by StemCore Laboratories (Ottawa, Ontario, Canada).
Immunoblotting.
At the indicated time points after infection, whole-cell extracts were obtained in 2× protein sample buffer (62.5 mM Tris-HCl [pH 6.8], 25% glycerol, 2% SDS, 0.01% bromophenol blue, 0.715 M β-mercaptoethanol). Samples were boiled for 5 min; insoluble debris was pelleted by centrifugation (20,000 × g, 3 min, 22°C); and 10 to 30% of each sample was resolved on SDS-polyacrylamide gels. Protein was transferred to a polyvinylidene fluoride membrane (Bio-Rad) using standard techniques. The membranes were probed with anti-FLAG M2 (1:10,000) (Sigma), anti-α-tubulin (1:5,000) (Oncogene), or anti-Sp100 (1:2,000) (Chemicon).
RESULTS
Prokaryotic DNA represses transgene expression.
We previously showed that transgene expression from an hdAd is significantly influenced by the nature of the stuffer DNA (29). An hdAd vector containing approximately 22 kb of prokaryotic DNA (hdAd-prok) expressed its transgene ∼60-fold less efficiently than a similar vector containing eukaryotic DNA (hdAd-euk) at all time points examined (Fig. 1B). The reduced expression of hdAd-prok was on a per-cell basis; individual transduced cells stained less intensely with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (Fig. 1C). This effect was not caused by a global reduction in the level of gene expression within the cell, since coinfection of hdAd-prok with a second hdAd had no effect on the ability of the second hdAd vector to express its transgene (data not shown). Therefore, the prokaryotic DNA present in hdAd-prok represses transgene expression in cis, not via global downregulation of gene expression within the host cell.
To determine whether the reduced transgene expression for the hdAd containing prokaryotic DNA was a general phenomenon or whether it was specific to the bacteriophage lambda DNA contained in hdAd-prok, we constructed a second vector containing two nonconsecutive fragments of DH10B genomic DNA, designated hdAd-prok2. We compared the expression from this vector to that from a second hdAd (here designated hdAd-euk2) containing an identical lacZ expression cassette and eukaryotic stuffer DNA derived from the human HPRT and DXS455A loci, as previously described (27). As with our first set of vectors, we observed an ∼20-fold-higher level of transgene expression from hdAd-euk2 than from hdAd-prok2 (Fig. 1D). The difference in transgene expression between these two vectors occurred regardless of the MOI. Taken together, these data suggest that reduced transgene expression is a common feature of hdAd containing prokaryote-derived stuffer DNA.
The hdAd-prok transgene is associated with hypoacetylated histones.
We recently determined that histones are deposited on hdAd vectors shortly after nuclear translocation and that the DNA is assembled into physiologically spaced nucleosomes (P. J. Ross and R. J. Parks, unpublished data). To examine the chromatin state of hdAd-prok and -euk, we employed ChIP with antibodies raised against markers of transcriptionally repressed (dimethyl K9 H3 and trimethyl K27 H3) or transcriptionally active (acetylated histone H3, acetylated histone H4, and RNA polymerase II) chromatin. Although the two vectors exhibited similar levels of association with unmodified histone H3, neither vector exhibited any significant association with histone H3 methylated at K9 or K27 within the mCMV promoter or lacZ open reading frame (Fig. 2A). However, compared to that for hdAd-euk, we observed decreased association of the hdAd-prok mCMV promoter with acetylated histones H3 and H4 (Fig. 2B). Consistent with our expression data (Fig. 1B), we also observed reduced association of RNA polymerase II with hdAd-prok (Fig. 2B). ChIP specificity and efficiency were confirmed by analyzing cellular loci. As expected, the Myt1 promoter, which is a known target of polycomb silencing in HeLa cells (19), was associated with dimethyl K9 H3 and trimethyl K27 H3 (Fig. 2A). The constitutively active β-actin promoter was associated with acetylated histones H3 and H4 but not with H3 methylated at K9 or K27 (Fig. 2A and B), and the uninduced beta interferon promoter was not associated with acetylated histones (Fig. 2B). Taken together, these data suggest that prokaryotic DNA represses gene expression independently of histone methylation, likely via the recruitment of factors that deacetylate the transgene promoter.
FIG. 2.
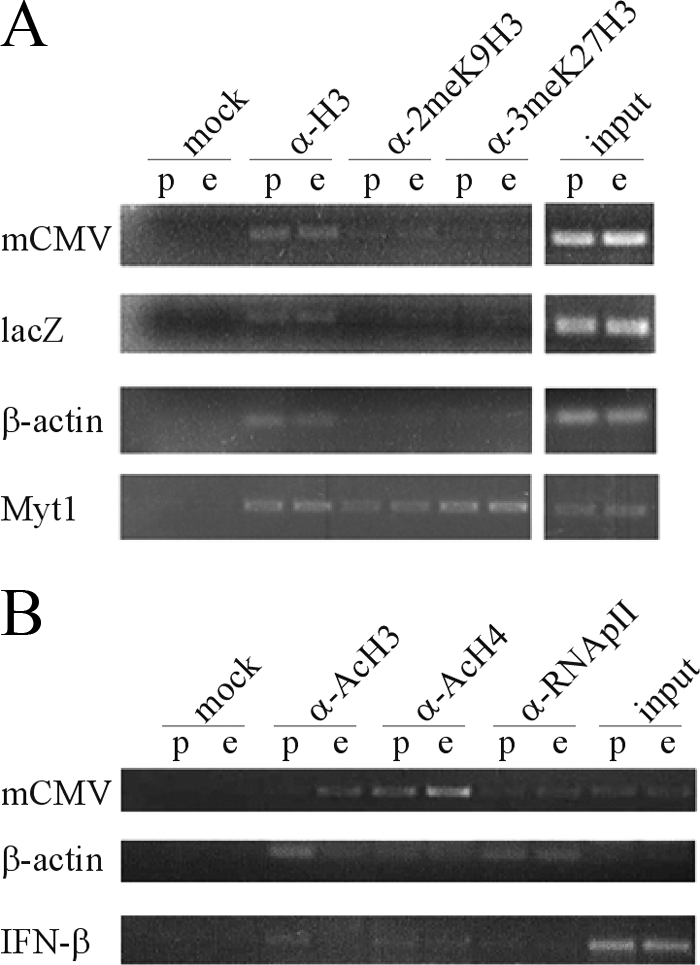
hdAd-prok exhibits reduced association with markers of transcriptionally active chromatin compared to hdAd-euk. HeLa cells were infected with hdAd-prok (p) or hdAd-euk (e) (MOI, 10 IU/cell), and 24 h postinfection, the cells were processed for ChIP with the indicated antibodies. ChIP DNA was analyzed by semiquantitative PCR using vector-specific (mCMV promoter and lacZ open reading frame) or host-specific (β-actin, beta interferon [IFN-β], and Myt1) primers. PCR products were resolved on agarose gels and visualized by ethidium bromide staining. (A) ChIP was performed with antibodies raised against heterochromatin-associated histone modifications. In HeLa cells, Myt1 is constitutively inactive (positive control), whereas β-actin is constitutively active (negative control). α, anti-; 2meK9H3, histone H3 dimethylated at K9; 3meK27H3, histone H3 trimethylated at K27. (B) ChIP was performed with antibodies raised against markers of transcriptionally active chromatin. In HeLa cells, β-actin is constitutively active (positive control), whereas IFN-β is inactive in unstimulated cells (negative control). Ac, acetylated; RNApII, RNA polymerase II.
Inhibition of histone deacetylation relieves the repressive effects of prokaryotic DNA.
Transcriptionally silent promoters are typically associated with deacetylated histones (17). Deacetylation is performed by the histone deacetylase (HDAC) family of enzymes. Having determined that hdAd-prok transgene DNA is associated with hypoacetylated histones, we next examined the role of HDACs in the repressive effects of prokaryotic DNA. Administration of TSA, a microbial small molecule that inhibits class I and II HDACs, resulted in a dose-dependent increase in transgene expression from both vectors, although hdAd-prok exhibited a larger increase in expression than hdAd-euk (40- and 10-fold, respectively) (Fig. 3A). Therefore, we conclude that hdAd-prok is targeted for repression by cellular HDACs.
FIG. 3.
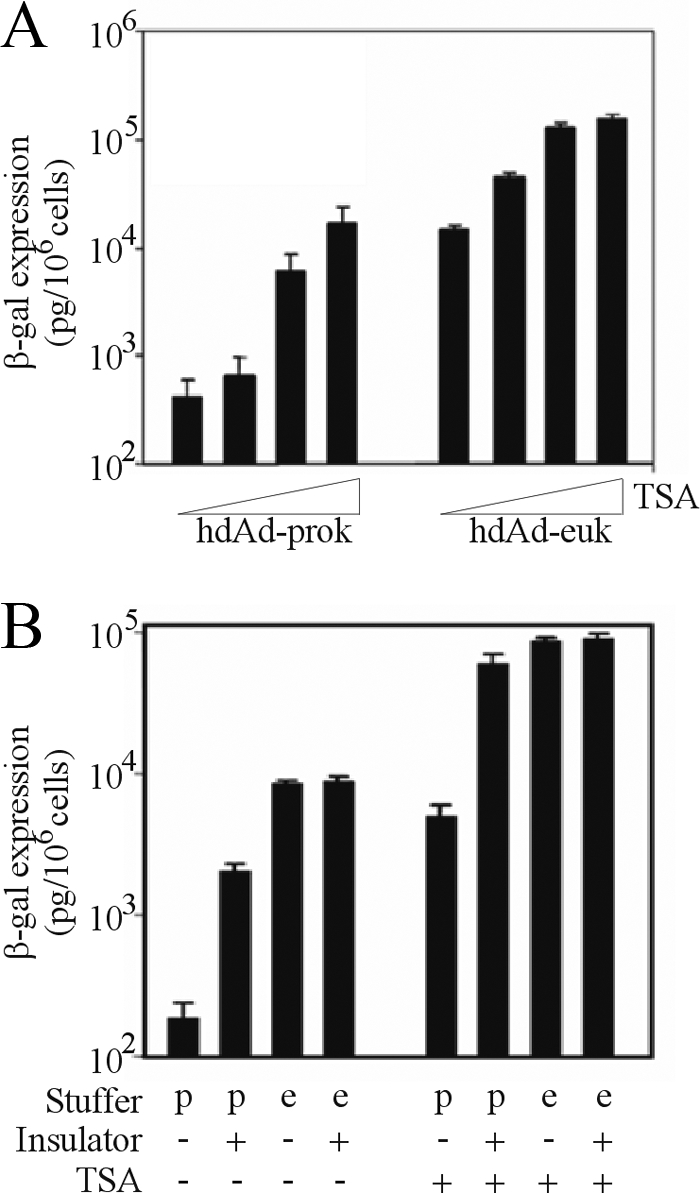
Inhibition of histone deacetylation or spread of chromatin modifications from stuffer DNA to the transgene abrogates the repressive effects of prokaryotic DNA. (A) HeLa cells were pretreated for 5 h with TSA and then infected with hdAd-prok or -euk (MOI, 1 IU/cell). At 18 h postinfection, cell lysates were harvested and assayed for β-Gal activity (means ± standard errors; n = 3). (B) HeLa cells were either left untreated or pretreated for 5 h with 250 ng/ml TSA; subsequently, they were infected with hdAd-prok (p) or -euk (e) or with similar vectors containing the HS4 insulator element. Eighteen hours postinfection, cell lysates were harvested and assayed for β-Gal activity (means ± standard errors; n = 3).
Our studies suggest that prokaryotic DNA nucleates the formation of hypoacetylated chromatin, which results in the repression of associated transgenes. Thus, the insertion of an insulator element, which blocks the spread of transcriptionally repressive chromatin (32), should alleviate the repressive effects of prokaryotic stuffer DNA. To test this hypothesis, we inserted the chicken beta-globin locus-derived HS4 double-core insulator element between the transgene and the stuffer DNA of hdAd-prok and hdAd-euk (generating hdAd-prok-ins and hdAd-euk-ins, respectively) (Fig. 1A). Insertion of the insulator element had no effect on hdAd-euk transgene expression (Fig. 3B); however, inclusion of the insulator element in hdAd-prok-ins resulted in a 33-fold increase in transgene expression relative to that of the parental vector, hdAd-prok (Fig. 3B). To confirm that this effect was due to the barrier element function of the HS4 insulator, we generated an additional hdAd-prok vector that contained a 1-kb segment of eukaryotic DNA (derived from the stuffer DNA immediately adjacent to the lacZ expression cassette in hdAd-euk) between the lacZ expression cassette and the lambda stuffer DNA in hdAd-prok; however this had no effect on the level of transgene expression from hdAd-prok (data not shown). Recent work has shown that additional elements not included in the minimal HS4 double-core insulator are required for its optimal function as a barrier element (1), likely explaining why we did not see complete recovery of hdAd-prok expression to hdAd-euk levels. Consistent with this, treatment of cells with TSA in combination with the HS4 insulator element completely recovered expression from hdAd-prok (Fig. 3B). These data clearly show that blocking the action of HDACs restores hdAd-prok expression to the level of hdAd-euk. Moreover, these data reveal that prokaryotic DNA is targeted as a nucleation site of transcriptionally repressive chromatin, which then spreads to the transgene.
hdAd-prok vector DNA is not methylated.
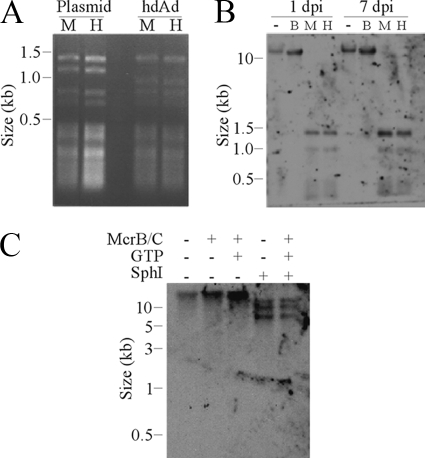
In eukaryotic cells, silenced genes are frequently associated with enhanced DNA methylation at CpG dinucleotides. We next evaluated the methylation status of the hdAd DNA before and after host cell transduction. To determine whether hdAd-prok DNA is methylated prior to entering the host cell, purified virus DNA was digested with the isoschizomer MspI or HpaII. MspI digestion is methylation independent, whereas HpaII digestion is blocked by CpG methylation. Both enzymes were able to fully digest parental plasmid DNA (which is not CpG methylated, due to the absence of bacterial CpG methyltransferases) and capsid-derived DNA (Fig. 4A), indicating that the viral DNA is unmethylated at CpG residues prior to host cell transduction. To determine whether hdAd-prok DNA is methylated after host cell transduction, we isolated DNA from infected cells and digested it with MspI, HpaII, or the endonuclease McrB/C (with or without codigestion with SphI). McrB/C digests DNA between (A/G) methyl-C half-sites (therefore excluding the MspI/HpaII recognition sequences, CCGG). Southern blot analysis showed no significant McrB/C cleavage of DNA obtained 1 or 7 days after infection and no difference in the digestion pattern with HpaII or MspI at either time point (Fig. 4B). Since the hdAd-prok genome is ∼30 kb, modest digestion of a small number of methylated CpG motifs by McrB/C may be difficult to discern. Therefore, we digested genomic DNA from hdAd-prok-infected cells with SphI (which digests the hdAd-prok genome into three fragments that are easily resolved by conventional agarose gel electrophoresis) with or without McrB/C. The results of this experiment clearly showed no difference in the hdAd-prok banding pattern upon the addition of McrB/C (Fig. 4C). Therefore, hdAd-prok DNA remains unmethylated, even as long as 1 week after infection.
FIG. 4.
hdAd-prok DNA is unmethylated. (A) hdAd-prok plasmid DNA or DNA from purified hdAd-prok virions was digested with MspI (M) or HpaII (H). The digested DNA was resolved on an agarose gel and visualized by ethidium bromide staining. (B) A549 cells were infected with hdAd-prok (MOI, 250 IU/cell), and genomic DNA was harvested at the indicated days postinfection (dpi). Genomic DNA was either left undigested (−) or digested with McrB/C (B), MspI (M), or HpaII (H). (C) A549 cells were infected with hdAd-prok. Genomic DNA was harvested 24 h postinfection and digested with the indicated enzymes with or without GTP. The digested DNA was analyzed by Southern blotting with DIG-labeled hdAd-prok plasmid DNA.
The analyses described above excluded global hypermethylation of hdAd-prok DNA. However, to analyze specific regions of the viral genome in greater detail, we performed high-resolution bisulfite sequencing of DNA from infected cells. We sequenced a 380-bp fragment of the mCMV promoter containing 22 CpG motifs and a 287-bp fragment from the central region of the prokaryotic stuffer DNA containing 20 CpG motifs. Sequence analysis of eight clones from the mCMV promoter and stuffer DNA detected no methylation of the 42 CpG motifs examined (data not shown). Together, our global and site-specific analyses failed to detect any evidence of hdAd-prok DNA methylation. Therefore, we conclude that DNA methylation is not involved in mediating the repressive effects of prokaryotic DNA on transgene expression.
PML and PML bodies are unnecessary for the repression of hdAd-prok.
Having determined that methylation of CpG motifs in prokaryotic stuffer DNA was not required for the repression of hdAd-prok (Fig. 4), we sought to identify cellular factors that recognize prokaryotic DNA and mediate the deacetylation of associated histones, leading to the repression of associated transgenes. PML bodies assemble de novo near the genomes of large DNA viruses soon after host cell transduction (6). These viruses, which include CMV, HSV, and Ad, harbor genes that reorganize PML bodies, which is necessary for productive infection. PML bodies also contain the cellular proteins Sp100 and Daxx, which have been implicated in repressing viral gene expression (25, 36). Therefore, we hypothesized that PML bodies may be involved in repressing genes associated with prokaryotic DNA.
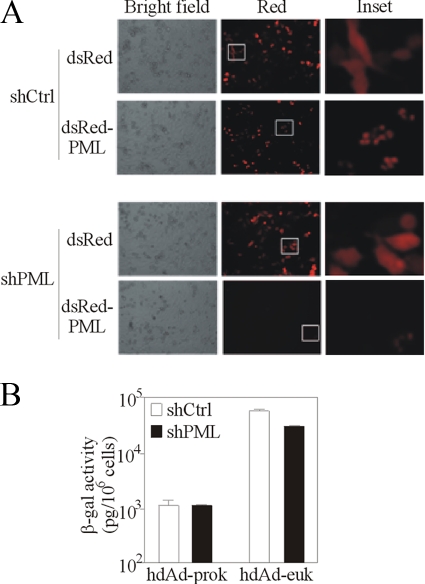
To examine the role of PML in the repression of hdAd-prok, we generated a plasmid carrying a short hairpin RNA (shRNA) that was shown previously to knock down the expression of PML (8). Transfection of cells with this plasmid blocked ectopic expression of dsRed-PML (Fig. 5A), demonstrating the efficacy of this plasmid in our hands. However, depletion of PML had no effect on the expression of hdAd-prok and only modestly affected the expression of hdAd-euk (Fig. 5B). In agreement with this observation, we found that overexpression of the chicken Ad protein Gam1, which blocks the SUMOylation of PML and prevents the formation of PML bodies (5), did not affect hdAd transgene expression (data not shown). Therefore, PML is unnecessary for the repression of hdAd-prok.
FIG. 5.
PML and PML bodies are unnecessary for the repression of hdAd-prok. HeLa cells were transfected with a plasmid carrying a shRNA that targeted a sequence in the PML mRNA (shPML) or with a plasmid carrying a control shRNA (shCtrl) and were analyzed for the efficiency of knockdown and the effect on hdAd gene expression. (A) Forty-eight hours after shRNA delivery, the cells were transfected with plasmids expressing either dsRed or dsRed-PML. One day later, the cells were visualized using bright-field or fluorescence microscopy. (B) Forty-eight hours after shRNA delivery, the cells were infected with hdAd-prok or -euk (MOI, 1 IU/cell); 24 h later, cell lysates were harvested and assayed for β-Gal activity (means ± ranges of values from duplicate samples). These data are representative of two experiments.
Sp100B and Sp100HMG have a modest effect on hdAd transgene expression.
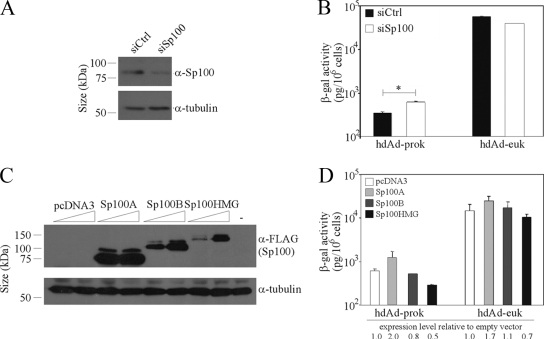
Although PML is essential for the formation of PML bodies, several constituents of PML bodies—Sp100, Daxx, and ATRX—accumulate on incoming HSV genomes in the absence of PML (7). The major Sp100 isoform, Sp100A, is associated exclusively with PML bodies in vivo; in contrast, Sp100B and Sp100HMG exhibit a diffuse staining pattern and repress HSV type 1 gene expression and replication (25). To determine if Sp100 plays a role in the regulation of hdAd-prok expression, we transfected cells with a pool of siRNA that target mRNA sequences common to all four Sp100 isoforms. Transfection with Sp100 siRNA resulted in an ∼65% knockdown of Sp100A, the most prevalent isoform in HeLa cells (Fig. 6A). Sp100 knockdown resulted in a modest decrease in hdAd-euk expression while increasing hdAd-prok expression 2.2-fold (Fig. 6B), suggesting that Sp100 repressor isoforms may contribute to the repression of hdAd-prok transgene expression. To determine whether distinct Sp100 isoforms exhibited differential effects on hdAd-prok gene expression, we transiently overexpressed Sp100A, Sp100B, or Sp100HMG in hdAd-prok- or -euk-infected cells (Fig. 6C and D). Overexpression of Sp100A resulted in ∼2-fold increases in the expression of both vectors (Fig. 6D), consistent with the role of this isoform in gene activation. In contrast, overexpression of Sp100B had no effect on hdAd-euk but decreased hdAd-prok expression by 20%. Although Sp100HMG decreased the expression of both vectors, hdAd-prok was repressed to a greater degree than hdAd-euk (50% and 30%, respectively) (Fig. 6D). Therefore, Sp100 isoforms that have previously been implicated in innate defense against viral intruders influence the expression of the transgenes carried by hdAd.
FIG. 6.
Sp100B and Sp100HMG influence expression from hdAd vectors. (A and B) HeLa cells were transfected with control siRNA (siCtrl) or pooled siRNA targeting sequences in the mRNAs of all Sp100 isoforms (siSp100). (A) Forty-eight hours later, whole-cell lysates were harvested for immunoblot analysis with anti-Sp100 or anti-α-tubulin. (B) Forty-eight hours after transfection, the cells were infected with hdAd-prok or -euk (MOI, 1 IU/cell). The next day, cell lysates were assayed for β-Gal activity (means ± standard errors; n = 3). The asterisk indicates a significant difference. (C and D) HeLa cells were transfected with pcDNA3 or with plasmids carrying FLAG-tagged Sp100A, Sp100B, or Sp100HMG. (C) Twenty-four hours later, whole-cell lysates were harvested for immunoblot analysis with anti-FLAG and anti-α-tubulin. (D) One day after transfection, the cells were infected with hdAd-prok or -euk (MOI, 1 IU/cell). The next day, cell lysates were assayed for β-Gal activity (means ± standard errors; n = 2).
Daxx mediates the repression of hdAd-prok.
Repressive isoforms of Sp100 reduce expression from hdAd-prok to a greater degree than that from hdAd-euk, suggesting some selectivity, although the effect is very modest. Therefore, we focused on a third PML body constituent, Daxx, a transcriptional corepressor that interacts with HDACs (15) and represses the expression of human (36) and murine (41) CMV immediate-early genes. To determine whether Daxx plays a role in the repression of hdAd-prok, we examined transgene expression from hdAd-prok in cells transiently transfected with pp71, a CMV tegument protein that induces proteosomal degradation of Daxx (36), or a mutated form of pp71 that is unable to bind Daxx (pp71delDID2/3) (14). Transgene expression by hdAd-prok and -euk was unaffected by pp71delDID2/3; however, expression of pp71 resulted in an 8.5-fold increase in transgene expression by hdAd-prok and only a 2.5-fold increase in expression from hdAd-euk (Fig. 7A). We also examined transgene expression in cells in which Daxx expression had been knocked down; although siRNA-mediated depletion of Daxx was not as effective as pp71 in relieving the repressive effects of prokaryotic DNA, similar trends were observed (3.5- and 1.5-fold induction of hdAd-prok and -euk, respectively) (Fig. 7B). Importantly, Daxx-mediated repression of gene expression in our system was due to action against the prokaryotic DNA contained in our construct and not to direct action against the CMV promoter, since inclusion of an insulator element between the stuffer and the expression cassette abrogated repression (Fig. 3B). Therefore, in addition to Sp100 repressor isoforms, Daxx plays a role in repressing the expression of genes associated with prokaryotic DNA.
FIG. 7.
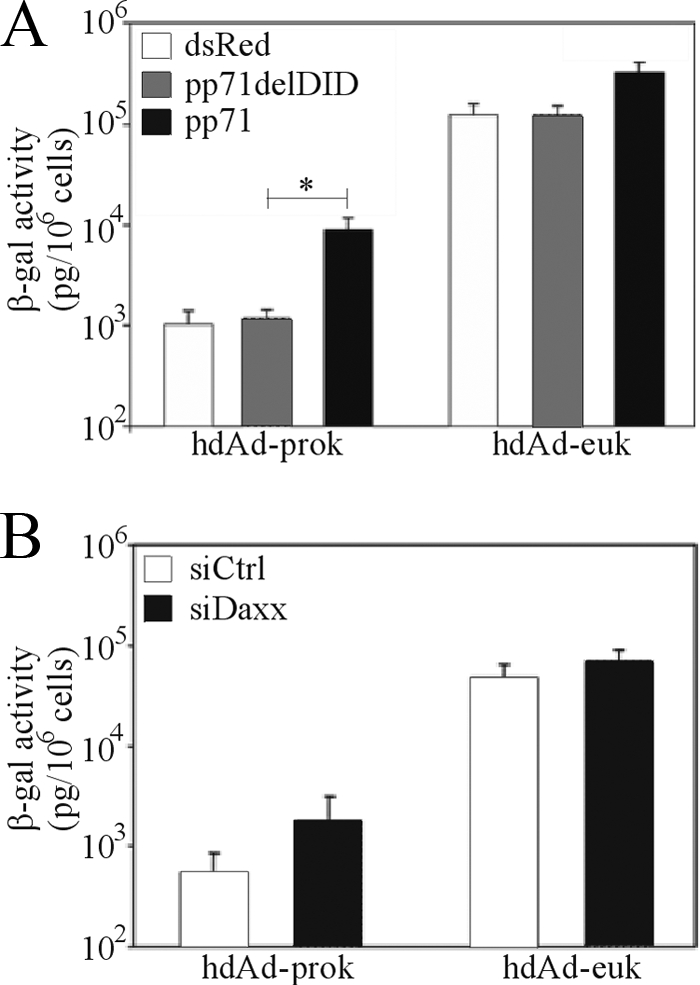
Daxx contributes to the repression of hdAd-prok transgene expression. (A) HeLa cells were transfected with an empty vector or with plasmids encoding wild-type or mutant (delDID) pp71. Next day, the cells were infected with hdAd-prok or -euk (MOI, 1 IU/cell); 24 h later, they were assayed for β-Gal activity (means ± standard errors; n = 3). The asterisk indicates a significant difference. (B) HeLa cells were transfected with control siRNA (siCtrl) or with pooled siRNA targeting sequences in the Daxx mRNA (siDaxx). Forty-eight hours posttransfection, the cells were infected with hdAd-prok or -euk (MOI, 1 IU/cell); 24 h later, they were assayed for β-Gal activity (means ± standard errors; n = 2).
DISCUSSION
We have used hdAd vectors containing prokaryotic DNA to address the mechanistic basis of how the covalent linkage of prokaryotic DNA represses eukaryotic gene expression. The repressive effects of prokaryotic DNA were abrogated by inhibition of histone deacetylation or by insertion of an insulator element between the transgene and the stuffer DNA (Fig. 3). Although we saw no evidence of heterochromatin (i.e., methylation of H3 K9 or K27), we observed reduced association of the hdAd-prok expression cassette with acetylated histones H3 and H4 (Fig. 2). We did not detect methylation of prokaryotic stuffer DNA or the hdAd-prok chromatin (Fig. 2 and 4). These data suggest that prokaryotic DNA recruits HDACs, which induce local histone hypoacetylation that spreads to the expression cassette, resulting in reduced transgene expression. Importantly, we have identified Sp100 repressive isoforms and Daxx as playing a role in inhibiting expression from hdAd (Fig. 6 and 7).
The repressive effects of prokaryotic DNA sequences on the level and duration of transgene expression have been noted for several vector systems, including Ad (29), HSV amplicons (39), and plasmids (2, 3, 34). Kay and colleagues observed that prokaryotic DNA sequences in plasmid DNA exclusively repressed gene expression in cis (3). Furthermore, their recent results support our conclusion that DNA methylation is unnecessary for the repression of genes linked to prokaryotic DNA (4). However, results from this group (33) and another analyzing HSV amplicon vectors (39) suggested that prokaryotic DNA results in the assembly of associated transgenes into heterochromatin. In contrast to both of these studies, we detected no evidence of association with methylated H3 K9 (Fig. 2A). Although Suzuki et al. (39) observed enhanced methylation of H3 K9 on the prokaryotic sequences contained in their HSV amplicon as early as 1 day postinfection, methylation of the transgene-associated histones was not observed until 6 days after gene delivery, despite obvious differences in expression at much earlier time points. Our results suggest that soon after gene delivery, the repressive effects of prokaryotic DNA are mediated by histone deacetylation. Assembly into heterochromatin at later time points may be a consequence of transcriptional inactivation and not the causative factor.
Having excluded an involvement of DNA methylation in the repressive effects of prokaryotic DNA (Fig. 4), we searched for cellular factors that may recognize and repress the expression of genes associated with foreign DNA. The Sp100 gene encodes four isoforms, of which three (B, C, and HMG) have been implicated in transcriptional repression and antiviral defense (16, 25). Depletion of all Sp100 isoforms increased the expression of hdAd-prok (Fig. 6B), and overexpression of repressive Sp100 isoforms repressed both hdAd vectors, although hdAd-prok appeared to be affected to a greater degree (Fig. 6D). The Sp100 repressor isoforms each contain a DNA binding domain that binds unmethylated CpG motifs (which are prevalent in the hdAd-prok backbone and also in the lacZ open reading frame present in both vectors) in a cooperative manner (16). All Sp100 isoforms interact with heterochromatin protein 1 (HP1), a potent repressor of gene expression (45). However, transcriptional repression by HP1 is thought to be mediated by recruitment of the H3 K9-specific histone methyltransferase Suv39H1 and DNA methyltransferases (11). Our data do not support this mechanism, since neither hdAd vector was associated with methylated histones (Fig. 2A) or DNA (Fig. 4). In our system, Sp100 may be repressing gene expression via a chromatin-independent process, perhaps by HP1-mediated DNA compaction (46).
Identification of Daxx as a repressor of hdAd gene expression reveals a mechanism for the recruitment of HDACs to the hdAd genome. Daxx is a transcriptional corepressor that binds to HDAC2 (15) and represses the expression of CMV genes (36, 41). Recently, Daxx has been shown to inhibit the replication of Ad deficient in E4orf3, suggesting that wild-type Ad has evolved a method to antagonize the effects of Daxx (44). Daxx does not contain a DNA binding domain (20), indicating that another factor must recruit Daxx to the hdAd genome. One of the few known cellular targets of Daxx-mediated repression is c-Met, and the region of the mouse c-Met promoter that recruits Daxx is a 65-bp fragment that is 78% GC and contains 10 CpG motifs (22, 23). Thus, Daxx may be recruited to CpG-containing DNA by an as yet unknown protein that binds GC-rich DNA or CpGs. However, it should be noted that knockdown of neither Sp100 nor Daxx restored transgene expression from hdAd-prok to the level of hdAd-euk, suggesting that there are likely other cellular factors that are involved in the selective repression of vectors containing prokaryotic DNA.
In summary, our results provide insight into the mechanism of identification and repression of foreign DNA, which may represent an innate mechanism of nuclear defense that suppresses the expression of potentially harmful genes of foreign intruders.
Acknowledgments
We thank Kathy Poulin, Adam Smith, Catherine Barrett, Robert Meulenbroek, and Karen Powell for excellent technical assistance; Jeffrey Dilworth, David Picketts, and Michael Rudnicki for helpful suggestions and critical evaluation of the manuscript; and Frank Graham, Gary Felsenfeld, Hans Will, Robert Kalejta, Susanna Chiocca, and Philip Ng for supplying reagents.
This work was supported by grants from the Canadian Institutes of Health Research, the Muscular Dystrophy Association of Canada, the Amyotrophic Lateral Sclerosis Society of Canada, the Jesse Davidson Foundation for Gene and Cell Therapy, and the Cancer Research Society (Canada). P.J.R. was supported by Post-Graduate and Canada Graduate Scholarships from the Natural Science and Engineering Research Council of Canada.
Footnotes
Published ahead of print on 10 June 2009.
REFERENCES
- 1.Aker, M., J. Tubb, A. C. Groth, A. A. Bukovsky, A. C. Bell, G. Felsenfeld, H.-P. Kiem, G. Stamatoyannopoulos, and D. W. Emery. 2007. Extended core sequences from the cHS4 insulator are necessary for protecting retroviral vectors from silencing position effects. Hum. Gene Ther. 18333-343. [DOI] [PubMed] [Google Scholar]
- 2.Chen, Z. Y., C. Y. He, A. Ehrhardt, and M. A. Kay. 2003. Minicircle DNA vectors devoid of bacterial DNA result in persistent and high-level transgene expression in vivo. Mol. Ther. 8495-500. [DOI] [PubMed] [Google Scholar]
- 3.Chen, Z. Y., C. Y. He, L. Meuse, and M. A. Kay. 2004. Silencing of episomal transgene expression by plasmid bacterial DNA elements in vivo. Gene Ther. 11856-864. [DOI] [PubMed] [Google Scholar]
- 4.Chen, Z. Y., E. Riu, C. Y. He, H. Xu, and M. A. Kay. 2008. Silencing of episomal transgene expression in liver by plasmid bacterial backbone DNA is independent of CpG methylation. Mol. Ther. 16548-556. [DOI] [PubMed] [Google Scholar]
- 5.Colombo, R., R. Boggio, C. Seiser, G. F. Draetta, and S. Chiocca. 2002. The adenovirus protein Gam1 interferes with sumoylation of histone deacetylase 1. EMBO Rep. 31062-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Everett, R. D., and M. K. Chelbi-Alix. 2007. PML and PML nuclear bodies: implications in antiviral defence. Biochimie 89819-830. [DOI] [PubMed] [Google Scholar]
- 7.Everett, R. D., J. Murray, A. Orr, and C. M. Preston. 2007. Herpes simplex virus type 1 genomes are associated with ND10 nuclear substructures in quiescently infected human fibroblasts. J. Virol. 8110991-11004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Everett, R. D., S. Rechter, P. Papior, N. Tavalai, T. Stamminger, and A. Orr. 2006. PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J. Virol. 807995-8005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fields-Berry, S. C., A. L. Halliday, and C. L. Cepko. 1992. A recombinant retrovirus encoding alkaline phosphatase confirms clonal boundary assignment in lineage analysis of murine retina. Proc. Natl. Acad. Sci. USA 89693-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frommer, M., L. E. McDonald, D. S. Millar, C. M. Collis, F. Watt, G. W. Grigg, P. L. Molloy, and C. L. Paul. 1992. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. USA 891827-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuks, F., P. J. Hurd, R. Deplus, and T. Kouzarides. 2003. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 312305-2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graham, F. L., J. Smiley, W. C. Russell, and R. Nairn. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 3659-74. [DOI] [PubMed] [Google Scholar]
- 13.Herman, J. G., J. R. Graff, S. Myohanen, B. D. Nelkin, and S. B. Baylin. 1996. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl. Acad. Sci. USA 939821-9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hofmann, H., H. Sindre, and T. Stamminger. 2002. Functional interaction between the pp71 protein of human cytomegalovirus and the PML-interacting protein human Daxx. J. Virol. 765769-5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hollenbach, A. D., C. J. McPherson, E. J. Mientjes, R. Iyengar, and G. Grosveld. 2002. Daxx and histone deacetylase II associate with chromatin through an interaction with core histones and the chromatin-associated protein Dek. J. Cell Sci. 1153319-3330. [DOI] [PubMed] [Google Scholar]
- 16.Isaac, A., K. W. Wilcox, and J. L. Taylor. 2006. SP100B, a repressor of gene expression, preferentially binds to DNA with unmethylated CpGs. J. Cell. Biochem. 981106-1122. [DOI] [PubMed] [Google Scholar]
- 17.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 2931074-1080. [DOI] [PubMed] [Google Scholar]
- 18.Kim, I. H., A. Jozkowicz, P. A. Piedra, K. Oka, and L. Chan. 2001. Lifetime correction of genetic deficiency in mice with a single injection of helper-dependent adenoviral vector. Proc. Natl. Acad. Sci. USA 9813282-13287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirmizis, A., S. M. Bartley, A. Kuzmichev, R. Margueron, D. Reinberg, R. Green, and P. J. Farnham. 2004. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev. 181592-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindsay, C. R., V. M. Morozov, and A. M. Ishov. 2008. PML NBs (ND10) and Daxx: from nuclear structure to protein function. Front. Biosci. 137132-7142. [DOI] [PubMed] [Google Scholar]
- 21.Maelandsmo, G. M., P. J. Ross, M. Pavliv, R. A. Meulenbroek, C. Evelegh, D. A. Muruve, F. L. Graham, and R. J. Parks. 2005. Use of a murine secreted alkaline phosphatase as a non-immunogenic reporter gene in mice. J. Gene Med. 7307-315. [DOI] [PubMed] [Google Scholar]
- 22.Michaelson, J. S., and P. Leder. 2003. RNAi reveals anti-apoptotic and transcriptionally repressive activities of DAXX. J. Cell Sci. 116345-352. [DOI] [PubMed] [Google Scholar]
- 23.Morozov, V. M., N. A. Massoll, O. V. Vladimirova, G. G. Maul, and A. M. Ishov. 2008. Regulation of c-met expression by transcription repressor Daxx. Oncogene 272177-2186. [DOI] [PubMed] [Google Scholar]
- 24.Muruve, D. A., M. J. Cotter, A. K. Zaiss, L. R. White, Q. Liu, T. Chan, S. A. Clark, P. J. Ross, R. A. Meulenbroek, G. M. Maelandsmo, and R. J. Parks. 2004. Helper-dependent adenovirus vectors elicit intact innate but attenuated adaptive host immune responses. J. Virol. 785966-5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Negorev, D. G., O. V. Vladimirova, A. Ivanov, F. Rauscher III, and G. G. Maul. 2006. Differential role of Sp100 isoforms in interferon-mediated repression of herpes simplex virus type 1 immediate-early protein expression. J. Virol. 808019-8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oka, K., L. Pastore, I. H. Kim, A. Merched, S. Nomura, H. J. Lee, M. Merched-Sauvage, C. Arden-Riley, B. Lee, M. Finegold, A. Beaudet, and L. Chan. 2001. Long-term stable correction of low-density lipoprotein receptor-deficient mice with a helper-dependent adenoviral vector expressing the very low-density lipoprotein receptor. Circulation 1031274-1281. [DOI] [PubMed] [Google Scholar]
- 27.Palmer, D., and P. Ng. 2003. Improved system for helper-dependent adenoviral vector production. Mol. Ther. 8846-852. [DOI] [PubMed] [Google Scholar]
- 28.Palmer, D. J., and P. Ng. 2005. Helper-dependent adenoviral vectors for gene therapy. Hum. Gene Ther. 161-16. [DOI] [PubMed] [Google Scholar]
- 29.Parks, R. J., J. L. Bramson, Y. Wan, C. L. Addison, and F. L. Graham. 1999. Effects of stuffer DNA on transgene expression from helper-dependent adenovirus vectors. J. Virol. 738027-8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parks, R. J., L. Chen, M. Anton, U. Sankar, M. A. Rudnicki, and F. L. Graham. 1996. A helper-dependent adenovirus vector system: removal of helper virus by Cre-mediated excision of the viral packaging signal. Proc. Natl. Acad. Sci. USA 9313565-13570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parks, R. J., and F. L. Graham. 1997. A helper-dependent system for adenovirus vector production helps define a lower limit for efficient DNA packaging. J. Virol. 713293-3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Recillas-Targa, F., M. J. Pikaart, B. Burgess-Beusse, A. C. Bell, M. D. Litt, A. G. West, M. Gaszner, and G. Felsenfeld. 2002. Position-effect protection and enhancer blocking by the chicken beta-globin insulator are separable activities. Proc. Natl. Acad. Sci. USA 996883-6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riu, E., Z. Y. Chen, H. Xu, C. Y. He, and M. A. Kay. 2007. Histone modifications are associated with the persistence or silencing of vector-mediated transgene expression in vivo. Mol. Ther. 151348-1355. [DOI] [PubMed] [Google Scholar]
- 34.Riu, E., D. Grimm, Z. Huang, and M. A. Kay. 2005. Increased maintenance and persistence of transgenes by excision of expression cassettes from plasmid sequences in vivo. Hum. Gene Ther. 16558-570. [DOI] [PubMed] [Google Scholar]
- 35.Ross, P. J., and R. J. Parks. 2007. Construction of first-generation adenoviral vectors, p. 149-166. In T. Friedmann and J. Rossi (ed.), Gene transfer: delivery and expression of DNA and RNA. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 36.Saffert, R. T., and R. F. Kalejta. 2006. Inactivating a cellular intrinsic immune defense mediated by Daxx is the mechanism through which the human cytomegalovirus pp71 protein stimulates viral immediate-early gene expression. J. Virol. 803863-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sargent, K., P. Ng, C. Evelegh, F. L. Graham, and R. J. Parks. 2004. Development of a size restricted pIX deleted helper virus for amplification of helper dependent adenovirus vectors. Gene Ther. 11504-511. [DOI] [PubMed] [Google Scholar]
- 38.Smith, A. C., K. L. Poulin, and R. J. Parks. 2009. DNA genome size affects the stability of the adenovirus virion. J. Virol. 832025-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzuki, M., K. Kasai, and Y. Saeki. 2006. Plasmid DNA sequences present in conventional herpes simplex virus amplicon vectors cause rapid transgene silencing by forming inactive chromatin. J. Virol. 803293-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takahashi, Y., J. B. Rayman, and B. D. Dynlacht. 2000. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 14804-816. [PMC free article] [PubMed] [Google Scholar]
- 41.Tang, Q., and G. G. Maul. 2003. Mouse cytomegalovirus immediate-early protein 1 binds with host cell repressors to relieve suppressive effects on viral transcription and replication during lytic infection. J. Virol. 771357-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Toietta, G., V. P. Mane, W. S. Norona, M. J. Finegold, P. Ng, A. F. McDonagh, A. L. Beaudet, and B. Lee. 2005. Lifelong elimination of hyperbilirubinemia in the Gunn rat with a single injection of helper-dependent adenoviral vector. Proc. Natl. Acad. Sci. USA 1023930-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Toietta, G., L. Pastore, V. Cerullo, M. Finegold, A. L. Beaudet, and B. Lee. 2002. Generation of helper-dependent adenoviral vectors by homologous recombination. Mol. Ther. 5204-210. [DOI] [PubMed] [Google Scholar]
- 44.Ullman, A. J., and P. Hearing. 2008. The cellular proteins PML and Daxx mediate an innate antiviral defense antagonized by the adenovirus E4 ORF3 protein. J. Virol. 827325-7335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van der Vlag, J., J. L. den Blaauwen, R. G. Sewalt, R. van Driel, and A. P. Otte. 2000. Transcriptional repression mediated by polycomb group proteins and other chromatin-associated repressors is selectively blocked by insulators. J. Biol. Chem. 275697-704. [DOI] [PubMed] [Google Scholar]
- 46.Zhao, T., T. Heyduk, C. D. Allis, and J. C. Eissenberg. 2000. Heterochromatin protein 1 binds to nucleosomes and DNA in vitro. J. Biol. Chem. 27528332-28338. [DOI] [PubMed] [Google Scholar]