Abstract
One of the two SP1 sites in the proximal enhancer of the human cytomegalovirus (HCMV) major immediate-early (MIE) promoter is essential for transcription in human fibroblast cells (H. Isomura, M. F. Stinski, A. Kudoh, T. Daikoku, N. Shirata, and T. Tsurumi, J. Virol. 79:9597-9607, 2005). Upstream of the two SP1 sites to −223 relative to the +1 transcription start site, there are an additional five DNA binding sites for eukaryotic transcription factors. We determined the effects of the various transcription factor DNA binding sites on viral MIE RNA transcription, viral gene expression, viral DNA synthesis, or infectious virus production. We prepared recombinant HCMV bacterial artificial chromosome (BAC) DNAs with either one site missing or one site present upstream of the two SP1 sites. Infectious recombinant HCMV BAC DNAs were transfected into various cell types to avoid the effect of the virion-associated transactivators. Regardless of the cell type, which included human fibroblast, endothelial, and epithelial cells, the CREB site had the most significant and independent effect on the MIE promoter. The other sites had a minor independent effect. However, the combination of the different transcription factor DNA binding sites was significantly stronger than multiple duplications of the CREB site. These findings indicate that the CREB site in the presence of the other sites has a major role for the replication of HCMV.
Human cytomegalovirus (HCMV) replicates in cells of the endoderm, mesoderm, and ectoderm, which include macrophages and dendritic, epithelial, endothelial, fibroblast, smooth muscle, neuronal, and glial cells (35-37). Disease in humans occurs in multiple organs, such as the lung, liver, retina, gastrointestinal tract, etc. However, the virus fails to replicate in the progenitor cells of the bone marrow and monocytes of the blood (37). In the permissive cells, the viral major immediate-early (MIE) promoter is associated with chromatin characteristic of activation of transcription (28). In the nonpermissive cells, the MIE promoter is associated with chromatin characteristic of repression of transcription (32). While expression of the viral MIE genes is a prerequisite to reactivation from latency, without cellular differentiation there is little to no viral replication (42).
The viral MIE enhancer-containing promoter can respond to cellular signal transduction events and is a focal point for starting viral replication because it regulates the levels of transcription of two MIE downstream genes, designated IE1 (UL123) and IE2 (UL122) (5, 41). The IE1 gene product, which enhances viral replication, is not essential. The essential IE2 gene product is a multifunctional protein that regulates cellular transcription, viral transcription, and movement of the cell cycle (for a review, see reference 40). Therefore, the MIE enhancer-containing promoter has one of the key roles in the genesis of disease by regulating MIE gene expression.
Upstream of the MIE promoter is an array of cis-acting binding sites that are repetitive and bind cellular eukaryotic transcription factors (for a review, see reference 41). These cis-acting sites are between approximately −550 and −39 relative to the transcription start site, +1. It has been proposed that the array of different transcription factor binding sites might facilitate the rate of viral transcription and the range of cell type specificity for viral replication. The role of the duplication of the transcription factor binding sites is not understood. Transient transfection experiments indicated that the 18-bp repeat containing the NF-κB site and the 19-bp repeat containing the CREB site contributed to the strength of the MIE promoter (9). Surprisingly, deletion of all the NF-κB sites or all the CREB sites in the context of the entire enhancer of the viral genome did not affect viral replication in human foreskin fibroblast (HFF) or murine cells after infection at a high or low multiplicity of infection (MOI) (1, 16). While it is possible that the other sites compensated for the loss of either the NF-κB sites or the CREB sites, it is more likely that the virion-associated transactivators, such as glycoprotein gB and tegument protein pp71, had a very strong positive effect on the mutated MIE enhancer-containing promoter and consequently viral replication in permissive HFF cells appeared normal.
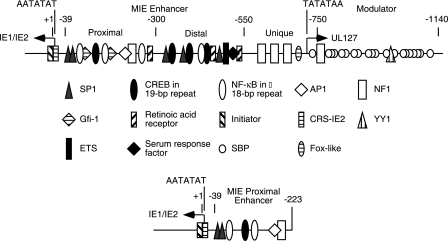
The HCMV MIE regulatory region is very complicated and consists of the MIE promoter, the enhancer, the unique region, and the modulator (Fig. 1). The enhancer has been analyzed as two components, designated the distal and proximal enhancers. The distal enhancer, between −550 and −300, contains multiple transcription factor binding sites, including one NF-κB site within an 18-bp repeat, three CREB sites within 19-bp repeats, three SP1 sites within 21-bp repeats, plus retinoic acid receptor, ETS, and serum response factor sites. The modulator, unique region, and distal enhancer could be deleted without an effect on viral replication after infection of HFF cells at a high MOI (26). At a low MOI, there was at least a 100-fold reduction in viral replication efficiency and a small-plaque phenotype when the distal enhancer was deleted with or without the unique region and the modulator (13, 26). However, the phenotype of the recombinant virus could be reversed by the addition of UV-inactivated virus, suggesting an important effect of the virion-associated transactivators on the proximal enhancer between approximately −300 and −39. The region between −300 and −223, which contains an NF-κB site and a retinoic acid receptor site, had little to no effect on expression from the MIE proximal enhancer-containing promoter (13). Successively larger 5′-end truncations from −223 to −55 of the proximal enhancer resulted in recombinant viruses that replicate slower and to lower titers (13). For recombinant HCMV bacterial artificial chromosome (BAC) DNA replication in HFF cells, one of the two SP1 binding sites at either −75 or −55 upstream of the MIE promoter was required (12). Recombinant viral BAC DNA with deletion to −39, with just the MIE TATA box containing the promoter element, did not replicate in HFF cells (13). These results indicated that an SP1 transcription factor binding site is essential and one or more of the upstream transcription factor binding sites are important for the efficiency of viral replication. Last, the region between the TATA box and the transcription start site also has an effect on viral replication (11). Immediately downstream of the transcription start site is an initiator-like sequence that binds a cellular protein that contacts the viral DNA upstream and downstream of the transcription start site (25). Site-specific mutation in this region affects the efficiency of transcription from the MIE promoter in vitro and in vivo at the earliest stages of viral infection. As the viral IE86 protein accumulates, it binds to this region, referred to as the cis repression sequence, and represses transcription from the MIE promoter (3, 22, 31).
FIG. 1.
Diagram of the HCMV MIE promoter and regulatory control region. The enhancer between −550 and −39 has been investigated as a proximal enhancer between −300 and −39 and a distal enhancer between −550 and −300. The unique region has a repressive effect on the divergent UL127 promoter, unless sequences upstream of the TATA box are deleted. The modulator has a positive effect on the MIE promoter in transient transfection assays, but this effect was not detected with recombinant viruses. The MIE promoter and proximal enhancer to −223 used in this study are diagramed below.
Upstream of the MIE promoter in the proximal enhancer to −223, there are two SP1 sites (−75 and −55), one CREB site within a 19-bp repeat (−137), two NF-κB sites within an 18-bp repeat (−98 and −161), one AP1 site (−171), and one NF1 site within a 16-bp repeat (−200) (Fig. 1). A significant role for the two NF-κB sites was questioned when human fibroblast cells expressing dominant-negative NF-κB had only minor effects on the level of viral replication for recombinant virus with just a proximal enhancer (13). Whether an individual transcription factor binding site, a duplication of individual sites, or a combination of different transcription factor binding sites was the most critical factor for efficient viral replication is not known. To answer these questions, we constructed recombinant viruses with either one type of transcription factor binding site missing or one type present upstream of the essential SP1 sites and the MIE promoter. We assayed for the effects of these mutations on viral gene expression and viral replication in different cell types permissive for HCMV. To avoid the effect of the virion-associated transactivators and cellular signal transduction pathways activated by viral attachment, cells were transfected with the various recombinant HCMV BAC DNAs and viral gene expression and viral replication were assayed. Because the laboratory-adapted Towne virus does not spread to cells of nonfibroblast origin, the mutations in the various transcription factor binding sites were also assayed in a viral BAC from a clinical isolate.
Here we report that the CREB site upstream of the two SP1 sites in the proximal enhancer has the greatest independent effect on viral replication in different cell types. The other transcription factor binding sites upstream of the two SP1 sites have a lower level of independent effect on transcription from the MIE promoter. The CREB binding site among the other different transcription factor binding sites has the greatest effect on the production of infectious virus. The CREB site is stronger among the other sites than duplications of the CREB sites without any other sites present. How the CREB site in the proximal enhancer serves as a focal point for cooperative interaction with the other transcription factor binding sites to enhance transcription from the MIE promoter is discussed below. The CREB binding site among the other different transcription factor binding sites has the greatest effect on the production of infectious virus.
MATERIALS AND METHODS
Cells.
Primary HFF cells were maintained in Eagle's minimal essential medium supplemented with 10% newborn calf serum (Sigma, St. Louis, MO). Retinal pigmented epithelial (RPE) cells and h-TERT RPE-1 cells from American Type Culture Collection (ATCC) (Manassas, VA) were grown in a Dulbecco's minimal essential medium-F12 (50/50) mixture containing 10% fetal bovine serum (FBS) (HyClone Logan, UT). Human aortic endothelial (HAOE) cells were grown in endothelial cell basal medium 2 (Lonza, Walkersville, MD) supplemented with growth factors, cytokines, and 10% FBS. Primary hepatocyte cells were grown in hepatocyte basal medium with ultra-glutamine 1 supplemented with growth factors (Lonza) and 10% FBS. All media contained penicillin (100 U/ml) and streptomycin (100 μg/ml). All cells were cultured at 37°C in 5% CO2.
Enzymes.
Restriction endonucleases were purchased from New England Biolabs Inc. (Beverly, MA). RNasin and RNase-free DNase were purchased from Promega (Madison, WI). Expanded high-fidelity Taq DNA polymerase for PCR was purchased from Invitrogen (Carlsbad, CA).
Plasmids, BACs, and recombinant viruses.
The plasmid pdlMCATdl-694/-223 (dlE-694/-223), containing the UL121-to-UL128 open reading frames, including the early UL127 viral promoter upstream of the chloramphenicol acetyltransferase (CAT) gene, was derived from pdlMCAT-694/-640R (20). The region between −694 and −223 was deleted by endonuclease digestion with BsrGI and NcoI, blunted with Klenow, and religated.
The Stratagene QuikChange XL mutagenesis kit (Stratagene, La Jolla, CA) was used to mutate the known transcription factor binding sites upstream of the SP1 sites at −55 and −75. The CREB (−137), AP1 (−171), 2NF-κB (−98 and −161), and NF1 (−200) sites were mutated by substitution with the restriction endonuclease sites BamHI, PstI, EcoRI, EcoRV, and EcoNI, respectively, using the oligonucleotides 5′-GCCAAAACAAACTCCCATTGgatcCAATGGGGTGGAGACTTGG-3′, 5′-GGAGACTTGGAAATCCCCGctgcagAAACCGCTATCCACGC-3′, 5′-GGGTTATTACGACATTTTGGAAgaattCGTTGATTTTGGTGCCAAAAC-3′, 5′-CAATGGGGTGGAGACTTGcAAgatatCGTGAGTCAAACCGCTAT-3′, and 5′-CCGCTATCCACGCCCATaccTGTAgTtgggAAACCGCATCACCATG-3′, respectively, and their complementary oligonucleotides. Lowercase letters represent mutated bases. To generate the mutation in the AP1 site when the NF-κB site at −161 was already mutated, the primer 5′-GGAGACTTGcAAgatatCGctgcagAAACCGCTATCCACGC-3′ and its complement were used. The 19-bp repeat sequence between −74 and −56 does not have a palindromic CREB site and was not mutated. The UL123 exon 1-UL128 DNA fragment was amplified from the shuttle vectors by PCR with the primers 5′-TCACCAATAGGGGAGTGGTC-3′ and 5′-CCCCTCCGTGTTGTAGGTTA-3′ and was used for homologous recombination with either Towne BAC from F. Liu (University of California, Berkeley) or FIX BAC from T. Shenk (Princeton University, Princeton, NJ) in DY 380 cells as described previously (13). HCMV BAC DNAs were selected for kanamycin resistance (Kanr). BAC DNA was isolated using the Nucleobond BAC Maxiprep kit (BD Biosciences, Palo Alto, CA) according to the manufacturer's instructions. All recombinant HCMV BAC DNAs were digested with the endonuclease HindIII, fractionated in a 0.6% agarose gel, and analyzed for any spurious viral genome deletions or rearrangements. All site-specific mutations in recombinant HCMV BAC DNAs were confirmed by DNA sequencing (University of Iowa DNA Core Facility) and by restriction endonuclease digestion (see Fig. 2 and 7). HFF cells were transfected with 1 or 3 μg of each recombinant BAC DNA and 1 μg of the plasmid pSVpp71 using the calcium phosphate precipitation method (6). Recombinant viruses were isolated, propagated, and maintained as described previously (13).
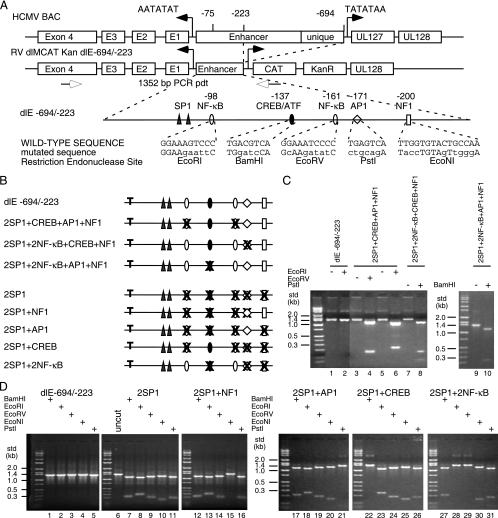
FIG. 2.
Construction of HCMV BAC mutations in the MIE proximal enhancer. (A) Diagram of the MIE enhancer/unique region flanked by the IE1 (UL123) and UL127 promoters. The distal enhancer and unique region were deleted between −694 and −223 relative to the IE1 transcription start site (+1) to isolate the proximal enhancer to −223. The UL127 open reading frame/modulator region was substituted by the CAT and kanamycin resistance (KanR) genes to generate RV dlMCAT Kan dl-694/-223 (dlE-694/-223). The known eukaryotic transcription-factor binding sites between −39 and −223 are designated, along with the mutated sequences and the newly generated restriction endonuclease sites. Each type of transcription factor binding site is designated by a symbol. (B) Summary diagram of mutations in the proximal enhancer with either one transcription factor binding site missing upstream of the two SP1 sites or one transcription factor binding site present. “T” designates the TATA box. The symbols for each type of transcription factor binding site are defined in panel A. (C and D) After PCR to generate the 1,352-bp DNA product (pdt) of the wild type or the recombinant HCMV DNAs with various mutations in the proximal enhancer region, the DNAs were digested with restriction endonucleases to prove an altered transcription factor binding site between −223 and −39. (D) HCMV BAC mutations with one transcription factor binding site present upstream of the two SP1 sites were constructed in both Towne BAC and FIX BAC. std, size standard.
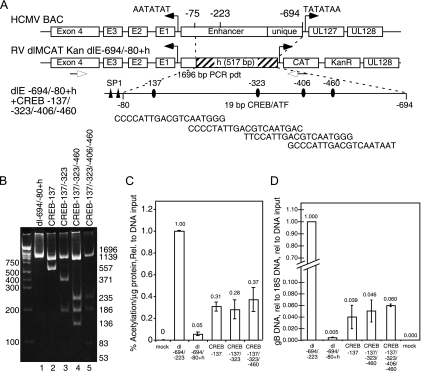
FIG. 7.
A variety of different transcription factor binding sites upstream of two SP1 sites has a greater effect on early viral gene expression than multiple CREB sites. Transfection of recombinant HCMV Towne BAC DNAs and the measurement of CAT activity or viral DNA synthesis were as described in Materials and Methods. (A) Diagram of the HCMV enhancer/unique region substituted by heterologous DNA with one or more CREB sites within a 19-bp repeat sequence upstream of two SP1. (B) Restriction endonuclease AatII digestion of the 1,696-bp PCR DNA product detecting the insertion of one or more 19-bp repeat sequences containing a consensus CREB site inserted into heterologous DNA at positions −137, −323, −406, and −460 relative to the transcription start site (+1). (C) Viral gene expression as measured by CAT activity at 14 days after transfection. Rel., relative. (D) Quantitative PCR of gB DNA at 14 days after transfection. rel, relative.
The enhancer region between −694 and −80 was replaced by heterologous DNA to generate the plasmid pdlMCATdl-694/-80+h. The heterologous DNA sequence was amplified from the US3 open reading frame with the primers 5′-GAGtGtACAGACCTACGAGCAGGGTTCTG-3′ and 5′-TCGGtcaacggggcggGGTGTTGGTGCTCGCGATCC-3′. Following restriction endonuclease digestion with BsrGI and HincII, the 501-bp fragment was ligated into the same sites in pdlMCATdl-694/-640R. The 19-bp repeats containing CREB sites were introduced into the heterologous region at the same spacing as in the wild-type enhancer (centered at −137, −323, −406, and −460) using the Stratagene QuikChange XL mutagenesis kit and the oligonucleotides 5′-CTCCGATACCACCAccccattgacgtcaatgggCGAGTCCGCGAGTC-3′, 5′-GCTCCGCGCTCATCAcccctattgacgtcaatgacCACCTGCAGTCTGTC-3′, 5′-CTCCCCACCTCGTAGAttccattgacgtcaatgggTCCACGGTGTGCGGC-3′, and 5′-GAGCTATAGTCCATCgcccattgacgtcaataatCGGCATATTTCTTGG-3′ and their complements, respectively. Shuttle vectors with multiple 19-bp insertions were generated using successive rounds of QuikChange mutagenesis.
Transfection assays.
Recombinant HCMV BACs were assayed by transfection and not by infection because the virion-associated transactivators, such as viral glycoprotein gB and tegument protein pp71, activate the proximal enhancer-containing viruses as described previously (23, 43). The various cell types were transfected by the calcium phosphate precipitation method (6). The cells were transfected in quadruplicate using 3 μg of HCMV BAC DNA and 2 μg beta-galactosidase (β-Gal) expression vector (simian virus 40 promoter-β-Gal). Transfection efficiency was normalized to either the cell-associated viral BAC DNA at 1 day after transfection or the β-Gal activity at 14 days after transfection.
To deplete the level of a transcription factor in HFF cells, 3 μg of plasmid DNA for the expression of either a nonspecific short hairpin RNA (shRNA) or a specific shRNA (SA Biosciences Corp., Frederick, MD) for a cellular transcription factor was transfected with 3 μg of recombinant HCMV Towne dlE-694/-223.
CAT assay.
We used a modified UL127 promoter to measure activation of the early UL127-CAT reporter by CAT assay, as described previously (20). The percentage of acetylated [C14]chloramphenicol (Perkin-Elmer Life Sciences, Waltham, MA) was determined by image analysis. CAT activity was normalized to micrograms of protein and either β-Gal activity or viral DNA input as described previously (29, 30).
Western blotting.
Cell lysates were fractionated by sodium dodecyl sulfate-containing polyacrylamide gel electrophoresis and blotted onto nitrocellulose membranes as described previously (29, 30). The CREB or ATF transcription factor was detected with affinity-purified goat anti-human CREB antibody (R&D Systems, Minneapolis, MN) or affinity-purified goat anti-human/mouse ATF1 antibody (R&D Systems, Inc.). Mouse anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Cemicon International, Temecula, CA) and mouse anti-β-tubulin (Cell Signaling Technology, Beverly, MA) were used to detect cellular GAPDH and β-tubulin, respectively, which served as loading controls. The secondary antibodies conjugated with horseradish peroxidase (HRP) were goat anti-mouse immunoglobulin G HRP (R&D Systems) and sheep anti-mouse immunoglobulin G HRP (GE Healthcare UK Limited, Little Chalfont, Buckinghamshire, United Kingdom), respectively. All antibodies were used according to the manufacturer's instructions.
Real-time PCR analysis.
The HCMV gB DNA was detected and quantified using multiplex real-time PCR for either viral DNA input or viral DNA synthesis. Whole-cell DNA was harvested at 1 day posttransfection for viral DNA input assay or at various days posttransfection for viral DNA synthesis assay as described previously (29, 30). Real-time PCR was performed using 0.4 μg DNA in a final volume of 20 μl of the Platinum PCR Supermix-UDG cocktail (Invitrogen, Carlsbad, CA). HCMV gB primers and 6-carboxyfluorescein-6-carboxytetramethylrhodamine probe, and cellular 18S DNA primers and VIC-6-carboxytetramethylrhodamine probe (Applied Biosystems, Foster City, CA) were used simultaneously. Thermal cycling conditions and quantification of gB DNA were as described previously (11).
Viral RNA was also quantified using a multiplex real-time PCR. Whole-cell RNA was harvested using Tri Reagent (Molecular Research Center, Cincinnati, OH) and treated with RNase-free DNase, and 2 μg of RNA was converted to cDNA using a random hexamer (Invitrogen) primer and reverse transcriptase (RT) as described previously (11). RNA lacking RT served as a control to detect any undegraded input viral DNA. Primers and probes for the detection of HCMV MIE, UL37X1 IE, and UL44 early gene expression and 18S rRNA expression were as described previously (29, 30, 42). Thermal cycling conditions were performed as described previously (29, 30, 42). Quantitation of relative viral RNA was done according to a standard curve analysis. Threshold cycle values of samples not treated with reverse transcriptase but analyzed in parallel to the RT-treated samples did not differ appreciably from baseline values.
RESULTS
Recombinant viruses.
We have reported that the strength of the HCMV proximal enhancer upstream of the viral MIE promoter was directly related to the viral DNA sequence between −223 and −39 relative to the transcription start site at +1 (13). We also reported that the minimal eukaryotic transcription factor binding site upstream of the MIE promoter for viral replication in HFF cells is one of the SP1 sites at either −75 or −55 (12). Between −223 and the two SP1 sites are the following known transcription factor binding sites: CREB at −137; AP1 at −171; two NF-κB sites at −98 and −161; and NF1 at −200 (Fig. 2A). The 19-bp repeat sequence at −62 in HCMV Towne has a nonconsensus CREB site. To determine which eukaryotic transcription factor binding sites upstream of the SP1 sites in the proximal enhancer contribute most significantly to viral replication in cell culture, we constructed recombinant viruses with either one site missing or one site present upstream of the two SP1 sites. Recombinant viruses for HCMV Towne or FIX were constructed as described in Materials and Methods. All mutations were confirmed by DNA sequencing. In addition, each transcription factor binding site was substituted with a restriction endonuclease site (Fig. 2A). The presence of the mutated site was determined by specific restriction endonuclease digestion of PCR DNA fragments spanning the region. Figure 2C and D compare wild-type viral DNA with mutant viral DNA after digestion with the various restriction endonucleases. These results substantiated the DNA sequence analysis and confirmed the mutated site. Last, all recombinant viral BAC DNAs were digested with the restriction endonuclease HindIII to check for any spurious deletions or rearrangements within the viral genome (data not shown). There were two separate isolations of each recombinant HCMV BAC with identical properties. The designations of the various recombinant HCMV BACs and the locations of the mutations are summarized in Fig. 2B.
Absence of CREB site affects level of viral gene expression.
To determine which sites in the proximal enhancer have the greatest effect upstream of the two SP1 sites in the absence of the virion-associated transactivators, we transfected HFF cells with HCMV BAC DNA as described in Materials and Methods. We assayed for either CAT expression from the viral UL127 promoter or viral DNA synthesis as described in Materials and Methods. The UL127 promoter without the upstream repressor is a highly inducible viral promoter after expression of the MIE genes (13, 24, 42). To quantify the activation of multiple early viral genes, we assayed for viral DNA synthesis. Replicated viral DNA (gB target) was quantified relative to input viral BAC DNA and to cellular 18S DNA.
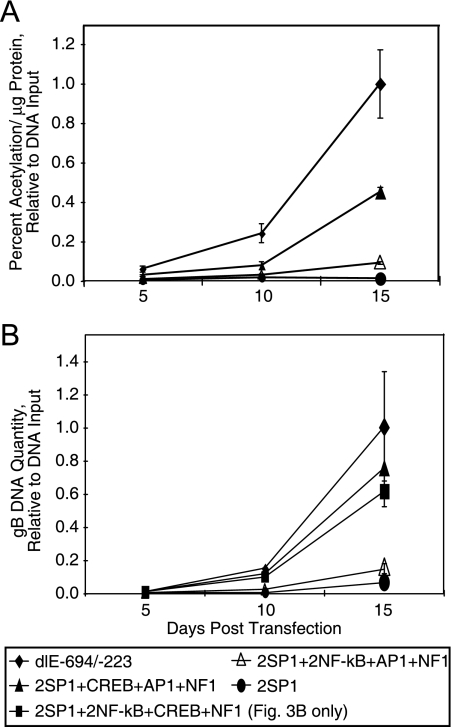
At 5 days posttransfection, there were small differences between the various recombinant viruses. At 10 days posttransfection, recombinant viral BACs having a CREB site had slightly higher levels of both CAT activity and viral DNA synthesis. Figure 3A shows that the proximal enhancer with a CREB site among the other binding sites had approximately a fivefold-stronger effect on viral gene expression at 15 days posttransfection as measured by CAT assay. The presence of the other sites upstream of the two SP1 sites, such as AP1, NF-κB, and NF1, had a slightly positive effect relative to that of the two SP1 sites alone (Fig. 3A). Figure 3B shows that the same recombinant viral BACs with a CREB site had an eight- to ninefold-stronger effect on viral gene expression as measured by viral DNA synthesis assay. In contrast, recombinant viral BACs with a single AP1 site, two NF-κB sites, and a single NF1 site upstream of the two SP1 sites were only slightly stronger in viral expression than the two SP1 sites alone. However, recombinant viruses containing the proximal enhancer to −223 (dlE-694/-223) had the highest level of viral gene expression as measured by CAT activity and viral DNA synthesis. These data indicated that recombinant HCMV BAC DNAs with a CREB site in the proximal enhancer upstream of the two SP1 sites and the MIE promoter expressed viral genes more significantly than the other transcription factor binding sites without a CREB site.
FIG. 3.
The effect of various transcription factor binding sites upstream of two SP1 binding sites on early viral gene expression when one transcription factor binding site is mutated. Transfection of recombinant HCMV Towne BAC DNAs and the measurement of CAT activity or viral DNA synthesis were as described in Materials and Methods. (A) Viral gene expression as measured by CAT activity at various days after transfection. (B) Quantitative PCR of gB DNA at various days after transfection.
CREB site upstream of two SP1 sites has greatest effect on viral gene expression.
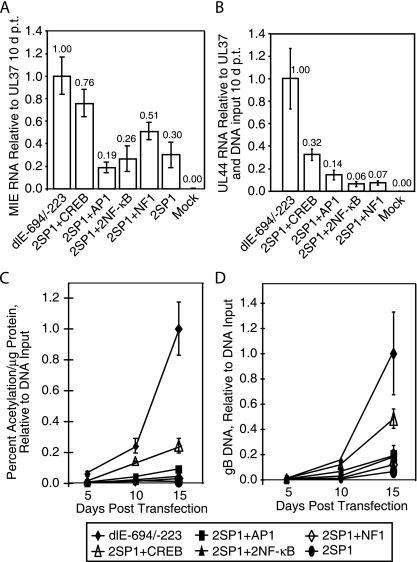
To determine which individual transcription factor binding site was most effective in the absence of the other sites upstream of the two SP1 sites and the MIE promoter, recombinant HCMV BAC DNAs were constructed and isolated as described in Materials and Methods and as shown in Fig. 2B and D. To demonstrate that the transcription factor binding sites were affecting transcription from the MIE promoter, we analyzed the level of IE1/IE2 transcript relative to that of UL37X1 transcripts by real-time RT-PCR as described in Materials and Methods. The levels of UL37X1 RNA were approximately equivalent among the various HCMV BAC DNA recombinants at 10 days posttransfection. The level of MIE RNA at 10 days posttransfection was approximately threefold higher with recombinant viral BAC 2SP1 + CREB than with recombinant viral BAC 2SP1. However, the level with 2SP1 + CREB was approximately 1.3-fold lower than that with the recombinant viral BAC dlE-694/-223. A single AP1 or two NF-κB sites upstream of two SP1 sites did not have a significant effect on the relative amount of IE1/IE2 RNA. The NF1 site had a slightly larger amount of IE1/IE2 RNA than that with the recombinant viral BAC with just two SP1 sites (Fig. 4A). However, the amount with 2SP1 + CREB was significantly higher from that with the other HCMV BAC DNA recombinants (P < 0.03).
FIG. 4.
The CREB transcription factor binding site upstream of two SP1 sites has the greatest effect on early viral gene expression. Transfection of HCMV Towne BAC DNAs and the measurement of either IE1/IE2 RNA, UL44 early RNA, CAT activity, or viral DNA were as described in Materials and Methods. (A) Quantitative RT-PCR of IE1/IE2 RNA at 10 days after transfection (d p.t.). (B) Quantitative RT-PCR of UL44 RNA at 10 days posttransfection. (C) Viral gene expression as measured by CAT activity at various days after transfection. (D) Quantitative PCR of gB DNA at various days after transfection.
To demonstrate that an increase in MIE transcription was directly related to early viral gene expression, we determined the amount of UL44 RNA relative to 18S RNA, UL37X1 IE RNA, and the viral DNA input at 10 days posttransfection. While UL44 RNA levels for 2SP1 + CREB were approximately 1/3 those for the proximal enhancer to −223, UL44 RNA levels for 2SP1 + CREB were 2.3 (P < 0.02)- to 5.3-fold higher than those for 2SP1 + AP1, 2SP1 + 2NF-κB, and 2SP1 + NF1 (Fig. 4B). We also used CAT expression from the UL127 promoter as a measure of viral gene expression. Figure 4C shows that the recombinant viral BAC with a proximal enhancer containing just a CREB site upstream of the two SP1 sites had approximately a fourfold-stronger effect on early viral gene expression at 15 days posttransfection as measured by CAT assay. However, without the other transcription factor binding sites, one CREB site was approximately fivefold weaker than the recombinant BAC containing a proximal enhancer at −223 (dlE-694/-223). Figure 4D confirms the above results and shows that viral DNA synthesis was approximately fivefold higher at 15 days posttransfection with a recombinant viral BAC containing one CREB site upstream of the two SP1 sites but approximately twofold weaker than that with the proximal enhancer at −223 (dlE-694/-223). The recombinant viral BAC with an AP1, NF-κB, or NF1 site upstream of two SP1 sites had slightly more early viral gene expression than that with just two SP1 sites alone.
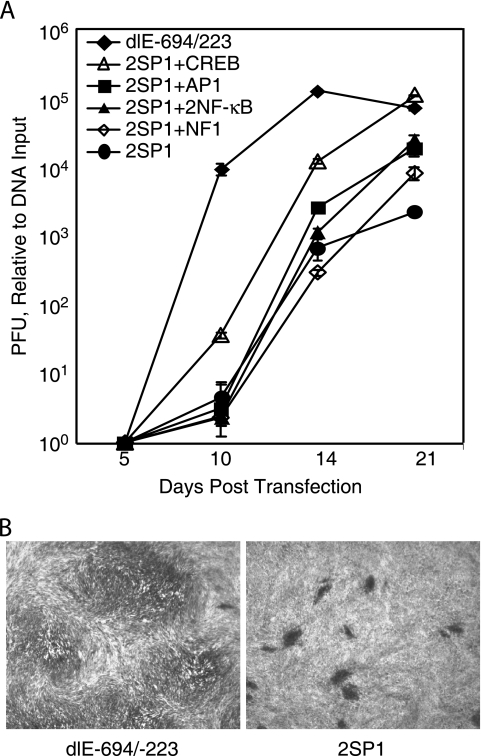
To confirm that higher levels of early viral gene expression resulted in infectious virus production, the effect of one transcription factor binding site upstream of the two SP1 sites in recombinant viral BAC DNAs was assayed for infectious virus production. Cells plus cell culture media were harvested various days after transfection of the recombinant BAC DNAs, and the total yield of infectious virus was determined by plaque assay. The recombinant viral BAC with one CREB site upstream of the two SP1 sites replicated at an approximately 10-fold higher level than the recombinant viral BACs with a single AP1 site, two NF-κB sites, or a single NF1 site at 10 and 14 days posttransfection (Fig. 5A). However, the recombinant viral BAC with the proximal enhancer to −223 (dlE-694/-223) replicated at a more than 100-fold greater level at 10 days posttransfection and at a 10-fold-greater level at 14 days postinfection than the recombinant viral BAC with one CREB site upstream of two SP1 sites (Fig. 5A). By 21 days posttransfection, the recombinant viral BAC with one CREB site upstream of the two SP1 sites was approximately 10-fold higher in titer than the recombinant viral BAC containing one AP1 site, two NF-kB sites, or one NF1 site upstream of two SP1 sites and almost 100-fold higher than the recombinant viral BAC with two SP1 sites (Fig. 5A). The recombinant viral BAC with an AP1 site, two NF-κB sites, or an NF1 site upstream of two SP1 sites replicated to slightly higher titers than that with two SP1 sites alone and rendered a small-plaque phenotype. While there was little difference in the small-plaque phenotype between viruses containing the proximal enhancer to −223 and either a CREB, AP1, two NF-κB, or NF1 transcription factor binding site upstream of the two SP1 sites, the recombinant viral BAC with just two SP1 sites upstream of the MIE promoter had a minute-plaque-size phenotype (Fig. 5B). The recombinant virus with just two SP1 sites replicated slower and spread less to adjacent cells. These data indicate that in the absence of the virion-associated transactivators and the additional transcription factor binding sites in the distal and proximal enhancers, the CREB transcription factor binding site contributes most significantly to viral replication in HFF cells. However, without the other transcription factor binding sites, the enhancer with the single CREB site upstream of the two SP1 sites has less of an effect on the MIE promoter in HFF cells. These data suggest that the other sites have a synergistic effect on transcription.
FIG. 5.
The CREB transcription factor binding site upstream of two SP1 has the greatest effect on infectious viral replication. Transfection of HCMV Towne BAC DNA and plaque assay in HFF cells were as described in Materials and Methods and elsewhere in the text. (A) Yield of infectious virus at various days after transfection. (B) Comparison of plaque sizes for recombinant viruses dlE-694/-223 and 2SP1 at 21 days posttransfection.
CREB site upstream of the two SP1 sites has greatest effect on viral gene expression in different cell types.
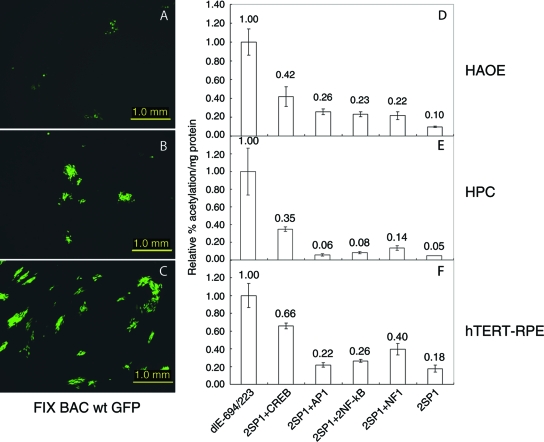
While it is possible to transfect Towne BAC DNA into various cell types like endothelial or epithelial cells, the virus does not spread to adjacent cells to form a focus of infection. HCMV Towne is defective in the UL128 to UL131 genes, which affects entry into endothelial or epithelial cells as described previously (7). To assay the effects of the various transcription factor binding sites in the proximal enhancer upstream of the two SP1 sites in endothelial or epithelial cell types, we transferred the mutated MIE enhancer from Towne BAC to the clinical isolate FIX BAC. We chose FIX BAC because it has more sequence similarities to Towne than the other clinical viral isolates in BAC DNA (27). To monitor the spread of FIX BAC in the various cell types after transfection of the viral DNA, we used wild-type FIX BAC expressing enhanced green fluorescent protein (GFP). The development of green fluorescent foci was used as an indicator for the efficiency of transfection and the spread of virus to adjacent cells. The HAOE cells were transfected less efficiently with FIX BAC DNA than the hepatocyes and hTERT RPE cells. FIX BAC replicated less efficiently in endothelial cells with smaller and less-intense foci relative to replication in the epithelial cells (compare Fig. 6A, B, and C). Regardless of the cell type, viral gene expression, as determined by CAT assay, was highest with recombinant viral BAC dlE-694/-223 and lowest with 2SP1 (Fig. 6D, E, and F). While the difference between 2SP1 + CREB and the other FIX BAC DNA recombinants were small, they were significant (P < 0.03). A single CREB site upstream of 2SP1 increased viral gene expression two- to sixfold more than a single AP1 site, two NF-κB sites, or a single NF1 site. Similar results were obtained with a viral DNA synthesis assay (data not shown). However, the effect of a single CREB site upstream of two SP1 sites was two- to threefold less than that of dlE-694/-223 in all cell types. The CREB site in combination with the two SP1 sites contributes to the highest level of transcription from the MIE promoter in different cell types.
FIG. 6.
The CREB transcription factor binding site upstream of two SP1 sites has the greatest effect on early viral gene expression in different HCMV permissive cell types. Transfection of wild-type FIX BAC GFP or recombinant FIX BAC DNAs dlE-694/-223, 2SP1 + CREB, 2SP1 + AP1, 2SP1 + 2NF-κB, 2SP + NF1, and 2SP1 into endothelial and epithelial cells, respectively, and viral gene expression as measured by CAT activity were as described in Materials and Methods. (A, B, and C) FIX BAC GFP fluorescence. (D, E, and F) CAT assay of FIX BAC recombinant DNAs in various cell types. (A and D) HAOE cells; (B and E) hepatocytes; (C and F) hTERT-RPE cells.
Multiple CREB sites upstream of the two SP1 sites have less effect on early viral expression than a combination of different sites.
Since the CREB site had a key role in activation of transcription from the MIE promoter, we determined whether the other transcription factor binding sites could be substituted by duplication of the CREB site. Additional recombinant BACs were constructed with one to four CREB sites within heterologous DNA by inserting the 19-bp repeat sequence at locations identical to the positions in the wild-type virus relative to the transcription start site (+1). Figure 7A is a diagram showing the position of the CREB sites upstream of the two SP1 sites and the MIE promoter. Figure 7B is a restriction endonuclease digestion of a PCR DNA product spanning the region of mutagenesis and establishing the construction of the various recombinant HCMV BACs with various insertions of the 19-bp repeat sequence containing the CREB sites. The presence of the CREB sites in heterologous DNA upstream of the two SP1 sites was also confirmed by DNA sequencing (data not shown). Two separate recombinant HCMV BACs with different numbers of CREB sites upstream of the two SP1 sites gave similar results. After transfection of HFF cells with the various recombinant Towne BAC DNAs, cells were harvested at 14 days posttransfection and assayed for either CAT expression from the viral UL127 promoter or viral DNA synthesis as described in Materials and Methods. Although the CREB site insertion upstream of the two SP1 sites increased viral gene expression approximately five- to sevenfold with one, two, or three CREB sites upstream, the maximal level of expression as measured by CAT activity was approximately fourfold less than that from the proximal enhancer at −223 (dlE-694/-223) (Fig. 7C). Viral DNA synthesis with the proximal enhancer at −223 (dlE-694/-223) was approximately 16-fold higher than that with one, three, or four CREB sites upstream of the two SP1 sites (Fig. 7D).
Although the CREB binding site has a key role in viral MIE and early gene expression in HFF, epithelial, and endothelial cells, the combination of a single CREB site with the other transcription factor binding sites, such as an AP1 site, two NF-κB sites, and an NF1 site upstream of 2SP1, had a greater effect than one to four CREB sites alone. The presence of the other transcription factor binding sites in the presence of the CREB site contributed more significantly in productive viral infection than multiple CREB sites. While the CREB site is a focal point for the initiation of viral MIE transcription in a latency model cell culture system (15), the combination of different transcription factor binding sites is stronger than the duplication of an individual CREB site.
shRNA inhibitory to CREB expression has significant effect on viral gene expression.
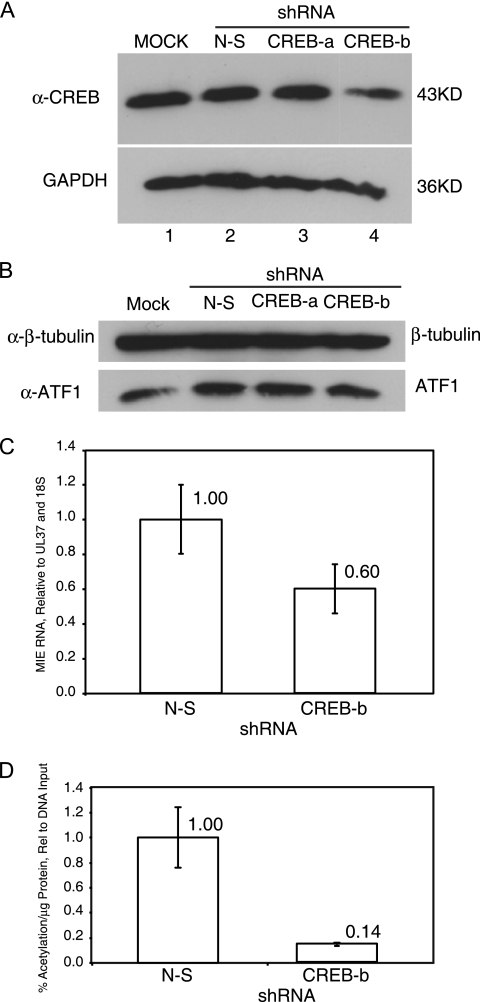
To determine the role of the CREB site in the presence of the other transcription factor DNA binding sites in the proximal enhancer, we used shRNAs to deplete the level of CREB transcription factors in HFF cells. After transfection of plasmids expressing shRNAs, the level of the cellular transcription factor was determined by Western blotting at 3 days posttransfection as described in Materials and Methods. Two different shRNAs specific to CREB were used. One had a minor effect (shRNA-a) on the level of CREB relative to nonspecific shRNA and the other a major effect (shRNA-b), which reduced the level of CREB more than twofold (63%) relative to the control (Fig. 8A). These shRNAs had no effect on the level of ATF (Fig. 8B).
FIG. 8.
shRNA inhibitory to CREB expression has a significant effect on viral early gene expression. HFF cells were transfected with plasmids expressing different shRNAs specific to CREB and analyzed for the steady-state level of CREB or ATF-1 by Western blotting 3 days posttransfection. GAPDH or β-tubulin served as a loading control. Viral gene expression was measured at 10 days posttransfection for MIE IE1/IE2 RNA and at 14 days posttransfection for chloramphenicol acetylation per microgram of protein relative to dlE-694/-223 DNA input as described in Materials and Methods. (A) Western blot with anti-CREB antibodies. Lanes: 1, mock-transfected cells; 2, nonspecific shRNA (N-S); 3, shRNA specific to CREB-a; 4, shRNA specific to CREB-b. (B) Western blot with anti-ATF-1 antibodies. Lanes: 1, mock-transfected cells; 2, nonspecific shRNA (N-S); 3, shRNA specific to CREB-a; 4, shRNA specific to CREB-b. (C) RT-PCR of MIE IE1/1E2 RNA at 10 days posttransfection with nonspecific shRNA (N-S) or shRNA-b specific to CREB. (D) CAT activity with nonspecific shRNA (N-S) or shRNA-b specific to CREB.
Plasmid DNAs (3 μg) expressing either nonspecific shRNA or specific shRNAs were cotransfected with recombinant HCMV FIX BAC dlE-694/-223 DNA (3 μg) as described in Materials and Methods. We analyzed the level of IE1/IE2 transcript relative to UL37X1 RNA and cellular 18S RNA by real-time RT-PCR as described in Materials and Methods. The MIE RNA at 10 days posttransfection was reduced approximately 40% (P < 0.02) with shRNA-b for CREB relative to results with the nonspecific shRNA (Fig. 8C). The level of viral gene expression was also determined by CAT assay at 14 days posttransfection as described above. The shRNA (shRNA-b) specific to CREB that had a significant effect on the steady-state level of CREB in the HFF cells at 14 days posttransfection reduced CAT expression from the viral UL127 promoter by approximately 85% relative to results with the nonspecific shRNA (Fig. 8D). In addition, shRNA-b specific to CREB significantly reduced viral DNA synthesis and foci of infection relative to those with nonspecific shRNA in the HFF cells (data not shown). While these data suggest that an active CREB transcription factor has a major and critical role for transcription from the proximal enhancer-containing MIE promoter, there are early viral promoters regulated by CREB, such as the UL112 promoter (19, 34). Therefore, shRNA-b specific to CREB likely has multiple effects on the viral replication cycle.
DISCUSSION
We know very little about the events necessary to activate the latent HCMV genome toward productive viral replication. A better understanding of the non-virion-induced signal transduction events that activate transcription from the HCMV MIE promoter during latency would shed additional light on the life cycle of the virus. Cellular signal transduction events that activate cellular transcription factors play an important role in reactivation from latency. In a latency cell culture model system, the transcription factor CREB was critical for reactivation of the quiescent viral genomes (15). In this study, the most responsive transcription factor in the various cell types transfected with HCMV BAC DNA was also CREB. CREB was most responsive in the presence of multiple different transcription factor binding sites, suggesting synergy between the sites.
Mitogen-activated protein kinases (MAPKs) activate CREB by phosphorylation, which facilitates an interaction with the CREB binding protein (CBP). CBP is a histone acetyltransferase (HAT) that interacts with TFIIB, which is a part of the transcription initiation complex. CBP also interacts with other transcription factors, such as AP1, NF-κB, SP1, etc. (18, 21, 28). The cooperative interaction of CREB/CBP with the other transcription factors in the proximal promoter may have affected the efficiency of HCMV BAC DNA replication more than multiple duplications of just CREB transcription factor binding sites. This may explain why the HCMV BAC dlE-694/-223 was stronger for viral gene expression than the recombinant HCMV BACs containing multiple CREB sites inserted into heterologous DNA. When the steady-state level of CREB in HFF cells was depleted with shRNA specific to CREB, there was approximately a 40% reduction in IE1/IE2 RNA and an 85% reduction in viral gene expression in HCMV BAC dlE-694/-223-infected cells. These data suggest that the CREB site in the proximal enhancer is critical for initiation of significant transcription from the MIE promoter and potentially other viral promoters.
The HAT function of CBP establishes a strong link between signaling and chromatin modifications. Murphy et al. (28) detected chromatin modifications at the HCMV MIE promoter after reactivation from latency, consistent with chromatin remodeled by HAT. Early during productive infection, the viral genome is associated with HATs that acetylate the histone and open up the chromatin structure for transcription (10, 28, 42). Sp1 may also have an effect on the acetylation of histones at the MIE promoter. HCMV infection induces an increase in Sp1 DNA binding activity, and the viral glycoproteins gB and gH in the viral envelope are responsible for this induction (43). Although AP1 is also activated by MAPK, the AP1 site had less of a significant and independent effect than the CREB site. Similar results were detected for NF-κB and NF1.
Enhancerless HCMV with just the TATA-box-containing promoter element does not replicate in human fibroblast cells (13). The minimal enhancer element for HCMV replication in human fibroblast cell culture is an Sp1 binding site at either −75 or −55 (12, 13). Sp1 or Sp3 transcription factors bind the GC boxes at −75 and −55 (12). Sp1 is reported to interact with TAF4 (33) and possibly factors binding to the initiator sequence at the transcription start site (4). Viral replication is greatly diminished with one or two Sp1 sites, and there is a characteristic minute-plaque phenotype.
When using infectious viral DNA in the absence of the distal enhancer and the virion envelope and tegument proteins, the CREB, AP1, NF-κB, NF1, and Sp1 cis-acting sites in the proximal enhancer cumulatively affect transcription from the MIE promoter and recombinant virus replication. The NF-κB sites, as well as the AP1 and NF1 sites, have a moderate effect in the presence of the two Sp1 sites, but a single CREB site has a more significant effect. The CREB site may act as a focal point for the activation of the other transcription factors that bind to the proximal enhancer. Although the CREB site contributes the most to the MIE promoter with two SP1 sites, the other transcription factor binding sites also contribute independently, resulting in the small-plaque phenotype. In contrast, with just two SP1 sites upstream of the MIE promoter, there was a minute-plaque phenotype. Although phosphatidylinositol 3-kinase [PI(3)K] activity, which activates NF-κB, is stimulated during HCMV infection (2), the NF-κB sites in the proximal enhancer had a less significant role than the CREB site for productive viral replication. The PI(3)K activity may have more of an effect on the infected cell than on the viral genome. In monocytes, the induction of NF-κB activity has a significant role in monocyte programming for viral dissemination (2, 38). PI(3)K activity induces a proinflammatory response exhibited by increased cytokine/chemokine secretion, motility, endothelial cell adhesion, transendothelial cell migration, and monocyte-to-macrophage differentiation. In addition, differentiation of monocytes to macrophages promotes productive virus replication (39).
CREB is activated by multiple factors, including growth factors, hormones, cyclic nucleotides, etc. (14). It is possible that the viral MIE proteins activate a stress pathway involving p38/MAPK, which in turn activates transcription from the MIE promoter. In addition, the IE86 protein of HCMV is reported to bind CREB and stimulate CREB-dependent gene transcription (19). The viral IE72 protein is reported to activate AP1 (17), but an AP1 site upstream of two SP1 sites had only a small effect on the MIE promoter and viral gene expression. The UL97 viral kinase of HCMV phosphorylates Rb, which contributes to movement of the cell cycle toward the G1/S transition point and activation of multiple cellular enzymes for viral DNA synthesis (8). It is not known whether the UL97 viral kinase contributes to CREB phosphorylation and activation. Why CREB plays a critical role in the activation of the MIE promoter is currently being investigated.
ADDENDUM IN PROOF
While the manuscript was under review, J. Yuan et al. (J. Virol. 83:6391-6403, 2009) reported that HCMV could be reactivated from quiescently infected pluripotent embryonal NTera2 cells by stimulation of the PKA-CREB-TORC2 signaling cascade.
Acknowledgments
We thank the members of the Stinski laboratory and Jeffery Meier for critical readings of the manuscript.
This work was supported by grant AI-13562 from the National Institutes of Health (to M.F.S.), Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports, Culture and Technology of Japan (grants no. 20012056, 19041078, and 20390137 to Tatsuya Tsurumi and no. 19590487 to H.I.), Research on Health Sciences focusing on Drug Innovation (grant no. SH54412 to H.I.), and a Grant-in-Aid for Cancer Research (no. 19-01 to H.I.) from the Ministry of Health, Labor and Welfare.
Footnotes
Published ahead of print on 24 June 2009.
REFERENCES
- 1.Benedict, C. A., A. Angulo, G. Patterson, S. Ha, H. Huang, M. Messerle, C. F. Ware, and P. Ghazal. 2004. Neutrality of the canonical NF-kB-dependent pathway for human and murine cytomegalovirus transcription and replication in vitro. J. Virol. 78741-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan, G., E. R. Bivins-Smith, M. S. Smith, and A. D. Yurochko. 2008. Transcriptome analysis of NF-kappa B and phosphatidylinositol 3-kinase-regulated genes in human cytomegalovirus-infected monocytes. J. Virol. 821040-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cherrington, J. M., E. L. Khoury, and E. S. Mocarski. 1991. Human cytomegalovirus ie2 negatively regulates α gene expression via a short target sequence near the transcription start site. J. Virol. 65887-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Emami, F. L., and W. W. Navarre. 1995. Core promoter specificities of the Sp1 and VP16 transcriptional activation domains. Mol. Cell Biol. 155906-5916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gealy, C., C. Humphreys, M. F. Stinski, and R. Caswell. 2007. An activation-defective mutant of the HCMV IEp86 protein inhibits NF-kappa B-mediated stimulation of the human IL-6 promoter. J. Gen. Virol. 882435-2440. [DOI] [PubMed] [Google Scholar]
- 6.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of adenovirus 5 DNA. Virology 52456-467. [DOI] [PubMed] [Google Scholar]
- 7.Hahn, G., M. F. Ewcwllo, M. Patrone, E. Percivalle, G. Campanini, A. Sarasini, M. Wagner, A. Gallina, G. Milanesi, U. Koszinowski, F. Baldanti, and G. Gerna. 2004. Human cytomegalovirus UL131-128 genes are indispensable for virus growth in endothelial cells and virus transfer to leukocytes. J. Virol. 7810023-10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hume, A. J., J. S. Finkel, J. P. Kamil, D. M. Coen, M. R. Culbertson, and R. F. Kalejta. 2008. Phosphorylation of retinoblastoma protein by viral protein with cyclin-dependent kinase function. Science 320797-799. [DOI] [PubMed] [Google Scholar]
- 9.Hunninghake, G. W., M. M. Monick, B. Liu, and M. F. Stinski. 1989. The promoter-regulatory region of the major immediate-early gene of human cytomegalovirus responds to T-lymphocyte stimulation and contains functional cyclic AMP-response elements. J. Virol. 633026-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ioudinkova, E., M. C. Arcangeletti, A. Rynditch, F. De Conto, F. Motta, S. Covan, F. Pinardi, S. V. Razin, and C. Chezzi. 2006. Control of human cytomegalovirus gene expression by differential histone modifications during lytic and latent infection of a monocytic cell line. Gene 384120-128. [DOI] [PubMed] [Google Scholar]
- 11.Isomura, H., M. F. Stinski, A. Kudoh, S. Nakayama, T. Murata, Y. Sato, S. Iwahori, and T. Tsurumi. 2008. A cis-acting element between the TATA box and the transcription start site of the major immediate-early (MIE) promoter of human cytomegalovirus determines efficiency of viral replication. J. Virol. 82849-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Isomura, H., M. F. Stinski, A. Kudoh, T. Daikoku, N. Shirata, and T. Tsurumi. 2005. Two Sp1/Sp3 binding sites in the major immediate-early proximal enhancer of human cytomegalovirus have a significant role in viral replication. J. Virol. 799597-9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Isomura, H., T. Tatsuya, and M. F. Stinski. 2004. The role of the proximal enhancer of the major immediate-early promoter in human cytomegalovirus replication. J. Virol. 7812788-12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johannessen, M., M. P. Delghandi, and U. Moens. 2004. What turns CREB on? Cell Signal. 161211-1227. [DOI] [PubMed] [Google Scholar]
- 15.Keller, M. J., A. W. Wu, J. I. Andrews, P. W. McGonagill, E. E. Tibesar, and J. L. Meier. 2007. Reversal of human cytomegalovirus major immediate-early enhancer/promoter silencing in quiescently infected cells via the cyclic AMP signaling pathway. J. Virol. 816669-6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keller, M. J., D. G. Wheeler, E. Cooper, and J. L. Meier. 2003. Role of the human cytomegalovirus major immediate-early promoter's 19-base-pair-repeat cyclic AMP-response element in acutely infected cells. J. Virol. 776666-6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, S. J., S. S. Yu, I. S. Lee, S. Ohno, J. Yim, S. Kim, and H. S. Kang. 1999. Human cytomegalovirus IE1 protein activates AP1 through a cellular protein kinase. J. Gen. Virol. 80961-969. [DOI] [PubMed] [Google Scholar]
- 18.Lalli, E., and P. Sassone-Corsi. 1994. Signal transduction and gene regulation: the nuclear response to cAMP. J. Biol. Chem. 26917359-17362. [PubMed] [Google Scholar]
- 19.Lang, D., S. Gebert, H. Arlt, and T. Stamminger. 1995. Functional interaction between the human cytomegalovirus 86-kilodalton IE2 protein and the cellular transcription factor CREB. J. Virol. 696030-6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lashmit, P. E., C. A. Lundquist, J. L. Meier, and M. F. Stinski. 2004. Cellular repressor inhibits human cytomegalovirus transcription from the UL127 promoter. J. Virol. 785113-5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, D. C., D. F. Carmichael, E. G. Krebs, and G. S. McKnight. 1983. Isolation of a cDNA clone for the type I regulatory subunit of bovine cAMP-dependent protein kinase. Proc. Natl. Acad. Sci. USA 803608-3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, B., T. W. Hermiston, and M. F. Stinski. 1991. A cis-acting element in the major immediate-early (IE) promoter of human cytomegalovirus is required for negative regulation by IE2. J. Virol. 65897-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, B., and M. F. Stinski. 1992. Human cytomegalovirus contains a tegument protein that enhances transcription from promoters with upstream ATF and AP-1 cis-acting elements. J. Virol. 664434-4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lundquist, C. A., J. L. Meier, and M. F. Stinski. 1999. A strong negative transcriptional regulatory region between the human cytomegalovirus UL127 gene and the major immediate-early enhancer. J. Virol. 739039-9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Macias, M. P., L. Huang, P. E. Lashmit, and M. F. Stinski. 1996. Cellular and viral protein binding to a cytomegalovirus promoter transcription initiation site: effects on transcription. J. Virol. 703628-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meier, J. L., and J. A. Pruessner. 2000. The human cytomegalovirus major immediate-early distal enhancer region is required for efficient viral replication and immediate-early expression. J. Virol. 741602-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murphy, E., Y. Dong, J. Grimwood, J. Schmutz, M. Dickson, M. A. Jarvis, G. Hahn, J. A. Nelson, R. M. Myers, and T. E. Shenk. 2003. Coding potential of laboratory and clinical stains of human cytomegalovirus. Proc. Natl. Acad. Sci. USA 10014976-14981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murphy, J., W. Fischle, E. Verdin, and J. Sinclair. 2002. Control of cytomegalovirus lytic gene expression by histone acetylation. EMBO J. 211112-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petrik, D. T., K. P. Schmitt, and M. F. Stinski. 2006. Inhibition of cellular DNA synthesis by the human cytomegalovirus IE86 protein is necessary for efficient virus replication. J. Virol. 803872-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petrik, D. T., K. P. Schmitt, and M. F. Stinski. 2007. The autoregulatory and transactivating functions of the human cytomegalovirus IE86 protein use independent mechanisms for promoter binding. J. Virol. 815807-5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pizzorno, M. C., and G. S. Hayward. 1990. The IE2 gene products of human cytomegalovirus specifically down-regulate expression from the major immediate-early promoter through a target located near the cap site. J. Virol. 646154-6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reeves, M. B., P. A. MacAry, P. J. Lehner, J. G. Sissons, and J. H. Sinclair. 2005. Latency, chromatin remodeling, and reactivation of human cytomegalovirus in the dendritic cells of healthy carriers. Proc. Natl. Acad. Sci. USA 1024140-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saluja, D., M. F. Vassallo, and N. Tanese. 1998. Distinct subdomains of human TAFII130 are required for interactions with glutamine-rich transcriptional activators. Mol. Cell. Biol. 185734-5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwartz, R., B. Helmich, and D. H. Spector. 1996. CREB and CREB-binding proteins play an important role in the IE2 86-kilodalton protein-mediated transactivation of the human cytomegalovirus 2.2-kilobase RNA promoter. J. Virol. 706955-6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sinzger, C., A. Grefte, B. Plachter, A. S. H. Gouw, T. Hauw The, and G. Jahn. 1995. Fibroblasts, epithelial cells, endothelial cells, and smooth muscle cells are the major targets of human cytomegalovirus infection in lung and gastrointestinal tissues. J. Gen. Virol. 76741-750. [DOI] [PubMed] [Google Scholar]
- 36.Sinzger, C., and G. Jahn. 1996. Human cytomegalovirus cell tropism and pathogenesis. Intervirology 39302-319. [DOI] [PubMed] [Google Scholar]
- 37.Sinzger, C., B. Plachter, A. Grefte, A. S. H. Gouw, T. H. The, and G. Jahn. 1996. Tissue macrophages are infected by human cytomegalovirus. J. Infect. Dis. 173240-245. [DOI] [PubMed] [Google Scholar]
- 38.Smith, M. S., G. L. Bentz, P. M. Smith, E. R. Bivins, and A. D. Yurochko. 2004. HCMV activates PI(3)K in monocytes and promotes monocyte motility and transendothelial migration in a PI(3)K-dependent manner. J. Leukoc. Biol. 7665-76. [DOI] [PubMed] [Google Scholar]
- 39.Soderberg-Naucler, C., D. N. Streblow, K. N. Fish, J. Allan-Yorke, P. P. Smith, and J. A. Nelson. 2001. Reactivation of latent human cytomegalovirus in CD14+ monocytes is differentiation dependent. J. Virol. 757543-7554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stinski, M. F., and D. T. Petrik. 2008. Functional roles of the human cytomegalovirus essential IE86 protein. Curr. Top. Microbiol. Immunol. 325133-152. [DOI] [PubMed] [Google Scholar]
- 41.Stinski, M. F., and H. Isomura. 2008. Role of the cytomegalovirus major immediate early enhancer in acute infection and reactivation from latency. Med. Microbiol. Immunol. 197223-232. [DOI] [PubMed] [Google Scholar]
- 42.Yee, L.-F., P. L. Lin, and M. F. Stinski. 2007. Ectopic expression of HCMV IE72 and IE86 proteins is sufficient to induce early gene expression but not production of infectious virus in undifferentiated promonocytic THP-1 cells. Virology 363174-188. [DOI] [PubMed] [Google Scholar]
- 43.Yurochko, A. D., E.-S. Hwang, L. Rasmussen, S. Keay, L. Pereira, and E.-S. Huang. 1997. The human cytomegalovirus UL55 (gB) and UL75 (gH) glycoprotein ligands initiate the rapid activation of Sp1 and NF-κB during infection. J. Virol. 715051-5059. [DOI] [PMC free article] [PubMed] [Google Scholar]