Abstract
Papillomaviruses (PVs) are a large family of small DNA viruses infecting mammals, reptiles, and birds. PV infection induces cell proliferation that may lead to the formation of orogenital or skin tumors. PV-induced cell proliferation has been related mainly to the expression of two small oncoproteins, E6 and E7. In mammalian PVs, E6 contains two 70-residue zinc-binding repeats, whereas E7 consists of a natively unfolded N-terminal region followed by a zinc-binding domain which folds as an obligate homodimer. Here, we show that both the novel francolin bird PV Francolinus leucoscepus PV type 1 (FlPV-1) and the chaffinch bird PV Fringilla coelebs PV contain unusual E6 and E7 proteins. The avian E7 proteins contain an extended unfolded N terminus and a zinc-binding domain of reduced size, whereas the avian E6 proteins consist of a single zinc-binding domain. A comparable single-domain E6 protein may have existed in a common ancestor of mammalian and avian PVs. Mammalian E6 C-terminal domains are phylogenetically related to those of single-domain avian E6, whereas mammalian E6 N-terminal domains seem to have emerged by duplication and subsequently diverged from the original ancestral domain. In avian and mammalian cells, both FlPV-1 E6 and FlPV-1 E7 were evenly expressed in the cytoplasm and the nucleus. Finally, samples of full-length FlPV-1 E6 and the FlPV-1 E7 C-terminal zinc-binding domain were prepared for biophysical analysis. Both constructs were highly soluble and well folded, according to nuclear magnetic resonance spectroscopy measurements.
Papillomaviruses (PVs) are nonenveloped, epitheliotropic, double-stranded DNA viruses that cause a variety of diseases in a multitude of hosts. Based on available whole-genome sequences and subgenomic amplicons, more than 200 human and over 55 nonhuman mammalian PV types have been described (7, 34, 35, 37, 38). To date, two avian PV types have been characterized (37, 38).
The genomic organizations of the PVs are remarkably similar. The genome is ca. 8 kb in length and comprises an upstream regulatory region (URR), the early genes (E1, E2, E4, E6, and E7), and the late genes that encode the capsid proteins (L1 and L2). Although most PVs code for these seven open reading frames (ORFs), only the URR, the replicative proteins E1 and E2 (and possibly the E4 gene), and the capsid proteins L1 and L2 are strictly conserved in all PVs (11).
Upon infection of the stratified squamous epithelia, PV gene expression is linked to the differentiation state of the infected epithelium cells. The expression of early PV proteins, in particular E6 and E7, primes the proliferation of the infected epithelium. This proliferation, which is absolutely required for viral replication, may become malignant depending on the PV strain considered. Several “high-risk” mucosal human PV (HPV) strains (predominantly HPV type 16 [HPV-16], HPV-18, and HPV-45) have been shown to be responsible for cervical cancer (19).
The ability of PVs to induce proliferation of the infected cells has been attributed mainly to two small “oncoproteins,” E6 and E7. In genital high-risk HPVs, these proteins play a prominent role in cell immortalization and transformation (31). In most mammalian PVs, E6 is a small protein of about 150 amino acids, with two conserved N- and C-terminal zinc-binding domains, E6N and E6C, respectively (12). The solution structure of the HPV-16 E6C domain was recently determined (23). The sequence alignments pointed to a structural similarity between the E6C and E6N domains, suggesting that a single-domain protein possessing the same fold might have once existed. Earlier phylogenetic studies had suggested that gene duplication may have given rise to the current double-domain E6 proteins (5). Interestingly, although the E6 ORF has been found in most mammalian PVs (with the exception of bovine papillomavirus type 3 [BPV-3], BPV-4, BPV-6, HPV-101, and HPV-103 [3, 7]), it was not detected in the two avian PVs previously sequenced (37, 38).
In this study, we present the full sequence of the genome of a novel PV from a francolin bird (Francolinus leucoscepus PV type 1 [FlPV-1]) and compare it to the two other avian PV genomes known to date (Psittacus erithacus PV [PePV] and Fringilla coelebs PV [FPV]). In light of recent structural data, we compare the unusual avian E6 and E7 ORFs to their mammalian orthologs. We describe the expression and purification of recombinant avian PV E6 and E7 proteins, their biophysical characterization, and cellular localization. Finally, we use phylogenetic techniques to investigate the evolutionary history of the E6 protein family.
MATERIALS AND METHODS
Origin and processing of the samples.
Samples were collected with prewetted (0.9% NaCl solution) cotton-tipped swabs that were drawn back and forth over the healthy skin of a francolin bird (Francolinus leucoscepus) and then suspended in 1 ml of 0.9% NaCl solution. After removal of the swabs, the samples were stored at −80°C until further analysis. DNA was extracted from the samples using the QIAamp DNA blood minikit (Qiagen) by following the manufacturer's protocol. This procedure yielded 36.5 μg/ml total DNA.
Cloning and sequencing of the FlPV-1 genome.
Cloning and sequencing were performed as described elsewhere (39). Briefly, the papillomaviral DNA was amplified by using rolling circle amplification (RCA), using the TempliPhi 100 amplification kit (Amersham Biosciences), by following a protocol that was recently optimized for amplification of complete PV genomic DNA (29).
To investigate whether PV DNA was amplified, 2 μl of the RCA product was digested with a panel of restriction enzymes. After digestion, the products were run on a 0.8% agarose gel to check for the presence of a DNA band consistent with full-length PV DNA (circa 8 kb) or multiple bands, with sizes adding up to this length. Digestion of the RCA product with XbaI resulted in two DNA fragments of approximately 4 and 3.2 kb. Both fragments were cloned into the pUC18 vector. The sequence of the 3.2-kb fragment was obtained by primer walking, starting with the M13 primer set. The EZ::TN <KAN-2> insertion kit (Epicentre) was used to retrieve the sequence of the 4-kb fragment, according to the manufacturer's protocol. The reaction product was used to transform One Shot MAX Efficiency DH5α-T1R competent cells (Invitrogen). Twenty-four colonies were selected, and the provided primers were used to sequence the insertion clones bidirectionally from primer binding sites at the 5′ and 3′ ends of the inserted transposon. The remaining gaps in the sequence were determined by primer walking. Sequencing was performed on an ABI Prism 3100 genetic analyzer (Perkin-Elmer Applied Biosystems, Foster City, CA). Chromatogram sequencing files were inspected with Chromas 2.2 (Technelysium, Helensvale, Australia), and contigs were compiled using SeqMan II (DNASTAR, Madison, WI).
Cell culture, transfections, and immunofluorescence.
To express FlPV-1 E6 and E7 proteins in eukaryotic cells fused to either the C terminus of enhanced green fluorescent protein (EGFP) (EGFP-E6, EGFP-E7), the C terminus of the myc epitope (myc-E6, myc-E7), or the N terminus of the myc epitope (E6-myc, E7-myc), the coding sequences of FlPV-1 E6 and FlPV-1 E7 were individually PCR amplified using the primers listed in Table S1 in the supplemental material and inserted between the HindIII and KpnI restriction sites of pEGFP-C3 (Clontech, Ozyme, St. Quentin-les-Yvelines, France) or between the NcoI and NotI restriction sites of the pEF/myc/cyto plasmid (Invitrogen, Cergy-Pontoise, France). All constructs were checked by sequencing.
HaCaT and HeLa cells were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum and 50 μg/ml gentamicin. QT6 quail fibroblasts were cultivated in McCoy medium supplemented with 9% fetal calf serum, 1% chicken serum, and 50 μg/ml gentamicin. Culture media and additives were obtained from Invitrogen (Cergy-Pontoise, France). The two cell lines were maintained at 37°C under 5% CO2. Transfections were carried out with jetPEI reagent (Polyplus Transfection, Illkirch, France), according to the manufacturer's instructions. Briefly, 3 × 105 HeLa or HaCaT cells were transfected with 3 μg of plasmid and 6 μl of jetPEI in a well of a six-well plate. QT6 was transfected similarly, except for the amount of the plasmid (5 μg) and the volume of jetPEI (10 μl). Forty-eight hours after transfection, cells were fixed and analyzed by EGFP fluorescence and immunofluorescence using an anti-myc monoclonal antibody and an Alexa Fluor 488 goat anti-mouse secondary antibody (Invitrogen, Cergy-Pontoise, France), as previously described (14).
DNA and protein sequence analysis.
The putative ORFs were predicted with either the ORF Finder tool on the NCBI server of the National Institutes of Health (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) or the equivalent tool on the ExPASy server (http://www.expasy.ch/tools/dna.html). To evaluate the tendency for globularity rather than native disorder within the PV ORFs, we used GLOBPLOT (http://globplot.embl.de) (17) and IUPRED (http://iupred.enzim.hu) (8). Using conserved cysteine and hydrophobic residues as anchoring points, we manually aligned the E6 and E7 protein sequences of FlPV-1 to HPV E6 and E7 domains, for which the three-dimensional structure has been solved.
Phylogenetic analyses.
We downloaded 131 whole-genome nucleotide sequences from GenBank. The entire sequences were aligned using the default parameters in Muscle 3.6 (9). Next, we performed phylogenetic analysis using the ratchet method described by Nixon (20), as implemented in PAUPRat (33).
To explore the possibility of a duplication event, we constructed a maximum parsimony tree in which we used each separate Zn finger domain as a separate taxon. To do this, we clipped the amino acid sequences in between the Zn finger domains (relative to position 86 in HPV-16 E6). The rabbit PVs were separated into three Zn finger domains. These separate domains were all aligned in Muscle using the default parameters (9). PAUPRat (33) was used to implement parsimony ratchets (20) in PAUP* version 4.0, beta 10 (36). PAUP* was used to calculate the strict consensus tree shown below (see Fig. 8). Mesquite version 2.1 (http://mesquiteproject.org) was used to visualize the trees. In this phylogram, the length of the branches represents the number of evolutionary changes in that lineage and may be interpreted as an indication of time and rate of evolution. Partition homogeneity test was performed, as implemented in PAUP* (36).
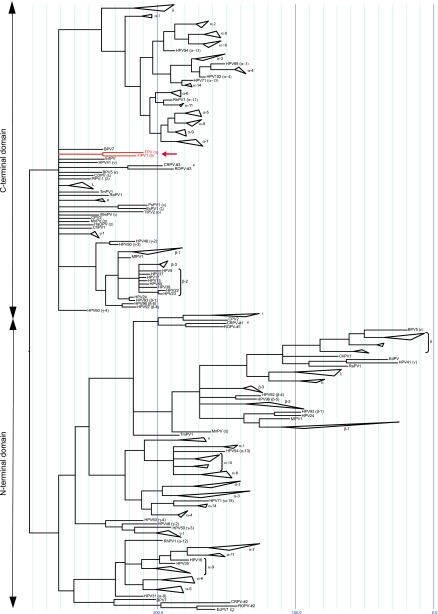
FIG. 8.
Maximum parsimony phylogram of separate E6 domains based on 128 clipped E6 protein sequences, as described in Materials and Methods. For clarity, only the PV species are shown (the complete tree showing all PV strains used for the calculation is provided in Fig. S1 in the supplemental material). The different rabbit E6 domains are numbered one to three. The branch lengths are proportional to the amount of changes acquired over time. The vertical lines serve as a ruler to measure the acquired changes.
Production of 15N-labeled FlPV-1 E6 and FlPV-1 E7 C terminus.
The full-length ORF of FlPV-1 E6 (83 residues) and the last 59 C-terminal residues of FlPV-1 E7 (containing the putative zinc-binding region) were cloned into the vector PET-M 41 (kindly provided by Gunter Stier, EMBL), allowing overexpression of proteins fused to the C terminus of maltose binding protein (MBP) via a TEV protease-sensitive linker (22). Overnight cultures of Escherichia coli BL21 cells containing the expression constructs in LB medium plus 35 μg/ml kanamycin were diluted 1:10 in 500 ml of M9 15N-labeled minimal medium (21) and grown at 37°C until A600 equaled 0.6. Cultures were supplemented with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside), further grown overnight at 18°C, and pelleted by centrifugation. To minimize oxidation problems, all purification buffers were degassed using a vacuum pump and then bubbled extensively with argon. The pellets were resuspended in buffer A (Tris-HCl [pH 6.8], NaCl 400 mM, 2 mM dithiothreitol [DTT]) containing 5% glycerol, 1 mg/ml DNase I, 1 mg/ml RNase I, and complete EDTA-free protease inhibitor cocktail. Cells were broken by sonication on ice and then centrifuged at 18,000 × g at 6°C for 30 min. The supernatant was filtered (0.22 mm; Millipore) and loaded onto a 50-ml column of amylose resin (New England Biolabs) preequilibrated with buffer A. After being extensively washed with buffer A, the MBP-fused constructs were eluted with buffer A supplemented with 10 mM maltose. The eluates were incubated for 12 to 24 h at 6°C with recombinant TEV protease (21) until full release of the MBP tag was achieved. The TEV cleavage site results in two additional residues (Gly-Ala) on the N terminus of the construct, prior to the methionine residue. The digestion products were concentrated and applied on a HiLoad 16/60 Superdex 75 gel filtration column (Amersham Biosciences) preequilibrated with buffer A. Pure monomeric FlPV-1 E6 eluted as a single peak at the volume expected for a 9-kDa monomer, according to the calibration of the column. The pure FlPV-1 E7 C-terminal domain eluted as a single peak at the volume expected for a 13-kDa protein. This suggests that the E7 C terminus behaves as a dimer (the theoretical molecular mass of the construct was 6.5 kDa). The samples were adjusted to 20 mM phosphate buffer, 50 mM NaCl, and 2 mM DTT, pH 6.8, by performing dilution/concentration steps using a 15-ml Ultrafree Biomax 5K NMWL membrane (Millipore). The final concentration was raised to 0.3 to 0.4 mM.
Analytical ultracentrifugation.
Sedimentation velocity experiments were done at 4°C using two-channel charcoal centerpieces and a velocity of 46,000 rpm in a Beckman Optima XL-A centrifuge fitted with a four-hole AN-60 rotor. Sedimentation velocity profiles were collected by monitoring the absorbance signal at 280 nm in buffer A. Sedimentation coefficient and molecular weight distributions were analyzed by the c(s) method implemented in the Sedfit software package (30). Buffer density and viscosity corrections were made, according to the data published by Laue et al. (15).
NMR spectroscopy.
Nuclear magnetic resonance (NMR) samples were dissolved in a buffer containing 20 mM phosphate, 50 mM NaCl, 2 mM DTT, and 10% 2D2O at pH 6.8. All spectra were acquired on a Bruker DRX600 spectrometer equipped with a z-gradient triple-resonance cryoprobe. In all spectra, the water signal was suppressed using the WATERGATE sequence (26). Data were processed using either NMRPipe (6) or UXNMR (Bruker) and analyzed with XEASY (1).
Nucleotide sequence accession number.
The nucleotide sequence of the FlPV-1 genome reported in this article was deposited in GenBank under accession number EU188799.
RESULTS
Genomic structure of a novel avian PV, FlPV-1.
We isolated a novel avian PV from the healthy skin of a francolin bird (Francolinus leucoscepus). The FlPV-1 was cloned and sequenced. The complete FlPV-1 genome is 7,498 bp and has a GC content of 52.93%.
All known PVs have their ORFs on the sense strand of their circular double-stranded DNA genome. Most PVs have seven ORFs coding for five early (E1, E2, E4, E6, and E7) and two late capsid proteins (L1 and L2). Like the other two avian PVs characterized so far (37, 38), FlPV-1 has an unusual organization of the early region (Fig. 1). The E6 and E7 ORFs have original structures, which are discussed in a separate paragraph in Results. The early region of FlPV-1 also harbors a putative E9 (X-) ORF. This ORF is well conserved in all three avian PVs and was consistently predicted as a folded globular protein by the online programs GLOBPLOT (17) and IUPRED (8). However, neither SMART (16) nor Pfam (10) were able to identify any known modular domain in the putative E9 protein. Furthermore, the putative E9 protein is not homologous to any other protein, either in mammalian PVs or in any other living organism. Since the E9-X ORF is fully imbedded within the E1 ORF, it is possible that it does not code for a truly expressed and functional protein.
FIG. 1.
Location of the predicted ORFs of FlPV-1, FPV, and PePV. Each ORF is represented as a rectangle. Numbers show the nucleotide positions of the start and stop codons. The start codon for the E6 ORF (FlPV-1 and FPV) is used as position 1. HPV-6 is shown for comparison of the early regions. In this graphical representation, the ATG of the E7 ORF for PePV was arbitrarily chosen to be preceded by 250 nt of the URR. This allows for a better comparison of the downstream genomic organizations, highlighting the lack of the E6 ORF in PePV.
The other early ORFs of avian PVs are more reminiscent of the usual ORFs in mammalian PVs. The E1 ORF codes for the largest FlPV-1 protein (659 amino acids) and contains the conserved ATP-binding site for the ATP-dependent helicase (GVPDSGKS) in its C-terminal part. The FlPV-1 E2 ORF shares high similarity with its mammalian counterparts (Table 1) and presents the canonical organization with an N-terminal activation domain and a C-terminal DNA-binding domain. Completely contained within the E2 gene, but read in another frame, lies the putative E4 ORF. Like most E4 ORFs, the FlPV-1 E4 contains a high concentration of cytosine di-, tri-, or tetranucleotides, which are responsible for its typical high proline content (13 proline residues out of 132 amino acids or 9.9%).
TABLE 1.
Percentages of similarity for FIPV-1, FPV, and PePV ORFs mutually and to other PVsa
| FIPV-1 ORF | % nt (aa) similarity of FlPV-1 ORFs to those ofb:
|
||||
|---|---|---|---|---|---|
| FPV | PePV | HPV-1a | HPV-16 | BPV-1 | |
| E6c | 48 (39) | 43 (29) | 44 (24) | 37 (24) | |
| E6 (N-term)d | NAf | 37 (20) | 32 (18) | 34 (22) | |
| E6 (C-term)e | NAf | 43 (32) | 38 (25) | 35 (22) | |
| E7 | 40 (31) | 42 (29)g | 40 (39) | 41 (27) | 35 (21) |
| E9/Xh | 39 (27) | 43 (30) | |||
| E1 | 53 (48) | 54 (49) | 46 (37) | 43 (32) | 47 (38) |
| E2 | 49 (45) | 52 (47) | 45 (35) | 41 (29) | 40 (34) |
| E4 | — | 39 (30) | 32 (19) | 34 (22) | 34 (19) |
| L2 | 46 (41) | 48 (42) | 41 (27) | 43 (29) | 39 (26) |
| L1 | 56 (55) | 61 (64) | 50 (47) | 53 (46) | 49 (42) |
Percentage of nucleotide (amino acid) sequence similarity based on the pairwise comparison of the indicated ORFs of FlPV-1 and the indicated ORFs of FPV, PePV, HPV-1a (prototype skin type), HPV-16 (prototype HR genital type), and BPV-1 (bovine fibropapilloma). Percentages of nucleotide (amino acid) identity were calculated by pairwise alignments. N-term, N terminus; C-term, C terminus.
NA, not alignable because of insufficient similarity; —, an E4 ORF was not reported in the FPV, and we were unable to identify a putative E4 ORF.
The E6 ORF of FlPV-1 aligned with full-length E6 of FPV, HPV1-a, HPV-16, and BPV-1.
The E6 ORF of FlPV-1 aligned with E6N domains of HPV-1a, HPV-16, and BPV-1.
The E6 ORF of FlPV-1 aligned with E6C domain of HPV-1a, HPV-16, and BPV-1.
The FPV genome has a single-domain E6 protein (see the text).
The E7 ORF of FIPV-1 aligned with E8 ORF of PePV.
The ORF is labeled E9 in PePV and X in FPV.
The late region codes for the major (L1) and minor (L2) capsid protein genes. Both L1 and L2 contain a series of arginine and lysine residues at their C-terminal ends, which is likely to function as a nuclear localization signal.
The classic noncoding region (NCR) between the stop codon of L1 and the start codon of E6 counts 456 bp (nucleotides [nt] 7053 to 7498). The NCR contains several regulators of papillomaviral replication; this NCR usually contains an E1 recognition site flanked by two E2-binding sites. This conformation allows for the binding of an E1/E2 complex in order to activate the origin of replication. An E1 recognition site (E1BS, ATATCGGCGTAGAGTAT) is present at nt 7349 to 7365. Two putative E2-binding sites (E2BS) are found at nt 7391 to 7402 (GCC-N6-GGC) and at nt 7441 to 7451 (ACG-N6-GGT). At its 5′ end, the NCR also contains a polyadenylation site (AATAAA, nt 7465 to 7470), 18 bp upstream of a CA dinucleotide and the G/T cluster, necessary for the processing of the L1 and L2 capsid mRNA transcript (2).
In Table 1, we compare the different ORFs of FlPV-1 with FPV, PePV, HPV-1a (a benign cutaneous PV; GenBank accession number NC_001356), and HPV-16 (a prototype high-risk mucosal PV; GenBank accession number NC_001526). The L1 sequence of FlPV-1 shares 61.1% and 55.5% nucleotide similarity with PePV and FPV, respectively. In agreement with the PV classification system, this places FlPV-1 in the theta-PV genus together with PePV (7).
Presence of an E6 ORF in avian PVs.
ORF analysis of the FlPV-1 genome revealed the presence of a putative E6 ORF in the early region. E6 was previously reported to be absent in the two other avian PVs, FPV and PePV (37, 38). This prompted us to reanalyze these genomes and their putative ORFs. The PePV genome (7,304 bp) is notably shorter than the genomes of FPV (7,729 bp) and FlPV-1 (7,498 bp) (Fig. 1). However, we observed that the NCR separating the stop codon of the last late ORF (L1) and the start codon of the first early protein (E6 in FlPV-1, E7 in FPV, and E8 in PePV) was considerably longer in FPV (730 bp) than in PePV (460 bp) and FlPV-1 (446 bp). Closer inspection of the extended NCR of FPV revealed a putative E6 ORF which had been previously unnoticed. This E6 ORF runs from base 7483 to base 8 of the FPV genome sequence currently deposited in GenBank (accession number AY057109). When we take this putative FPV E6 into account, the NCRs of the three avian PVs have comparable lengths (446, 460, or 482 bp). However, PePV clearly differs from FPV and FlPV-1 by the absence of an E6 ORF. The shorter size of the PePV genome, combined with the position of PePV in the PV phylogenetic tree (see Fig. 7) supports the possibility that the PePV E6 has been lost via a deletion event.
FIG. 7.
Maximum parsimony tree based on the whole-genome alignment of 133 available PVs. This tree shows the different PV genera clustering together, as reported previously (7). In this tree, the novel FlPV-1 (boxed) clusters with PePV in the theta-PV genus. The presence/absence of the E6 ORF was traced on this tree (red, absent; green, one zinc-binding domain [avian PVs]; blue, two domains [normal mammalian situation]; yellow, four domains [two rabbit PVs, which each have an extra Zn-binding domain]).
Structure-based sequence analysis of avian E6 and E7 ORFs.
Avian E6 ORFs encode small proteins (83 and 85 residues for FlPV-1 E6 and FPV E6, respectively, compared to 158 residues for HPV-16 E6). In striking contrast to mammalian E6 ORFs, they contain only one repeat of the conserved zinc-binding domain. The amino acid sequences of FlPV-1 and FPV E6 are 39% identical. We investigated their structural relationship to HPV E6 proteins by aligning their sequences with those of the E6C and E6N domains of HPV-16 E6 in the context of the structural data available for the HPV-16 E6C domain (Fig. 2A and C). The figure highlights conserved positions of the E6C and E6N domains which play important structural roles, such as the four cysteine residues involved in zinc binding, core-buried residues (characterized by a low percentage of exposure), and helix caps (i.e., H-bond acceptor or donor side chains situated at the N termini or C termini of helices, respectively) (27). Most of these key structural positions are conserved within the two avian E6 ORFs, suggesting they may adopt a fold reminiscent of the E6C and E6N domains. Interestingly, single-domain avian E6 proteins appear as a mixed blend of E6N and E6C domains: some regions are more similar to HPV-16 E6N, while other sites are more similar to HPV-16 E6C. In addition, avian E6 proteins have two insertions in the H2-S2 and S2-S3 loops compared to those of the HPV-16 E6C sequence (Fig. 2C). These insertions may either behave as extended loops or contribute additional secondary-structure elements, such as an extended helix H2 or a fourth beta strand. The proposed positions for these insertions within the HPV-16 E6C fold are indicated in Fig. 2A.
FIG. 2.
Structure-based analysis of the putative zinc-binding domains of avian PV E6 and E7 ORFs. (A) General folding of an E6 zinc-binding domain (here, HPV-16 E6C) (23). Secondary-structure elements are indicated. Arrows indicate proposed locations for inserted sequences within avian PV E6. C-ter, C terminus; N-ter, N terminus. (B) General folding of an E7 zinc-binding dimeric domain (here, HPV-45 E7C) (25), with the secondary-structure elements indicated. (C) Alignment of the full FlPV-1 and FPV E6 ORFs with HPV E6 zinc-binding domains HPV-16 E6C and HPV-16 E6N, in the context of structural information derived from the published structure of HPV-16 E6C (23). Plotted above the alignment is the percentage of accessibility to the solvent for each structured residue within the published structure of HPV-16 E6C (23), calculated as described by Chothia (4) and using the WHATIF program (40). Arrows and cylinders denote beta-strands and α-helices, respectively. Positions playing a key structural role within the HPV-16 E6C structure are colored as follows: magenta, hydrophobic (W, F, Y, L, I, V, M); green, basic (K, R, H); red, acidic (E, D); orange, polar (Q, N, T, S); brown, cysteine (C); and cyan, small (G, A, P). Boxed, boldfaced characters pointed out by arrowheads indicate presumably exposed hydrophobic positions of FPV E6 substituted by hydrophilic positions in FlPV-1 E6. (D) Alignment of the zinc-binding C-terminal domains of FlPV-1 FPV and PePV E7 ORFs with the equivalent domains in HPV, for which structural data are available, as follows: HPV-1a E7 (18) and HPV-45 E7 (25). Structural information is derived from the HPV-45 E7 structure.
Most mammalian PV E7 proteins consist of a natively unfolded 60-residue N-terminal region which includes a conserved pRb-binding motif (consensus LXCXE), followed by a 50-residue zinc-binding domain which folds as an obligate homodimer (18, 25) The avian PV E7 proteins also comprise a long N-terminal region (from 60 to 100 residues, depending on which ATG codon is used for the protein's translation), consistently predicted to be natively unfolded by programs such as GLOBPLOT (17) and IUPRED (8) and containing the conserved LXCXE motif. The C-terminal region of avian PV E7 is a putative zinc-binding domain with two pairs of CXXC motifs, displaying similarity with the equivalent region in mammalian PV E7. We analyzed the sequences of this region in the context of the crystal structure of the zinc-binding domain of HPV-1a E7 (18) and the NMR structure of HPV-45 E7 (25) (Fig. 2). The spacer between the zinc-liganding cysteine pairs is shorter in avian PV E7 (23 residues in FlPV-1 and PePV and 21 residues in FPV) than in mammalian PV E7 (29 to 30 residues). This likely results in the main secondary-structure elements (one helix and two beta-strands) being shorter in avian PV E7 proteins.
Cellular localization of avian E6 and E7.
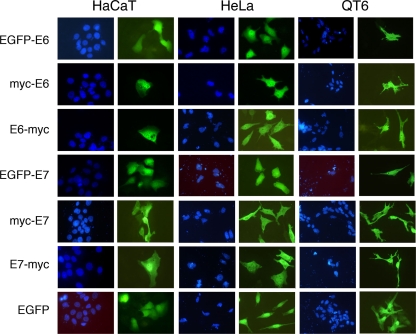
To analyze the expression and localization of avian E6 and E7 proteins in living cells, the ORF of FlPV-1 E6 (83 residues) and the long ORF of FlPV-1 E7 (148 residues) were both cloned in eukaryotic expression plasmids, allowing for their fusion individually to the C terminus of EGFP, the C terminus of the myc tag, or the N terminus of the myc tag. Constructs were transfected, allowing for heterologous expression of the fusion proteins in HaCaT and HeLa cells as well as in the avian cell line QT6. The localization of the constructs was analyzed by either green fluorescent protein (GFP) fluorescence microscopy or myc tag-based immunofluorescence (Fig. 3). All fusion proteins were equally distributed in the cytoplasm and nucleus. The localization of either E6 or E7 was essentially not influenced either by the position (N or C terminal) of the detection tag or by its nature (short myc peptide or large GFP protein). Finally, the detected signal for all fusion constructs tested was very homogeneous, suggesting that both proteins were folded and did not undergo aggregation. Upon cellular overexpression, misfolded aggregated proteins generate higher-intensity spots, localized mostly in cytoplasm (13, 32).
FIG. 3.
Expression of FlPV-1 E6 and FlPV-1 E7 proteins in HaCaT, HeLa, and QT6 cells. Cells were transfected with pEGFP-E6, pEF-myc-E6, pEF-E6-myc, pEGFP-E7, pEF-myc-E7, pEF-E7-myc, and the empty pEGFP-C3 vector. The different E6 and E7 fusion proteins and EGFP were detected by fluorescence either directly (EGFP) or after immunofluorescence using an anti-myc monoclonal antibody. DNA was stained with DAPI (4′,6-diamidino-2-phenylindole) to visualize nuclei.
Recombinant FlPV-1 E6 is a highly soluble monomeric protein.
We cloned and expressed FlPV-1 E6 as a fusion to MBP. The fusion protein was affinity purified and processed by the TEV protease to separate MBP and E6 moieties that were subsequently resolved by gel filtration chromatography (Fig. 4A). In contrast to samples of full-length HPV E6, which are generally prone to aggregation (24, 41), this sample of wild-type FlPV-1 E6 could be concentrated up to 300 μM without undergoing any detectable aggregation.
FIG. 4.
Expression, purification, and size characterization of FlPV-1 E6. (A) Expression and purification. The MBP-FlPV-1 E6 fusion was overexpressed in bacteria (lane 1). The expression pellet was resuspended and sonicated, and the cleared supernatant was loaded onto MBP-specific amylose resin (lane 2, cleared supernatant input; lane 3, MBP-E6 eluate) and subjected to TEV digestion, yielding MBP and FlPV-1 E6 proteins (lane 4). FlPV-1 E6 was then separated from MBP and TEV by gel filtration (lane 5). (B) Analytical gel filtration. Purified FlPV-1 E6 was loaded onto an analytical Superdex 75 10/30 gel filtration column. The elution peak of FlPV-1 E6 (9.2 kDa) is indicated, together with those of myoglobin (17.6 kDa) and HPV-16 E6C (8.9 kDa), utilized as size markers. mAU, milli-absorbance units. (C) Analytical ultracentrifugation. The molecular weight distribution of purified FlPV-1 E6 was derived from sedimentation velocity experiments. The distribution corresponds to a protein of approximately 10 kDa, and no larger oligomeric form is detected.
Next we analyzed the oligomeric state of FlPV-1 E6 (theoretical mass, 9.5 kDa). Upon analytical gel filtration (Fig. 4B), FlPV-1 E6 eluted as a single peak, corresponding to a 9.2-kDa monomer. Analytical ultracentrifugation measurements performed on concentrated FlPV-1 E6 also revealed a perfectly monomeric species of approximately 10 kDa, without any detectable trace of further oligomers (Fig. 4C). Finally, NMR relaxation experiments (data not shown) indicated that the protein tumbles with a correlation time of 7 ns at 298 K, which is consistent with a 9-kDa monomer. Under our experimental conditions, gel filtration, NMR, and ultracentrifugation data are all in agreement with FlPV-1 E6 being a monomeric protein.
We also attempted to produce samples of the FPV E6 ORF by applying the same strategy. However, FPV E6 turned out to have poor solubility, and so far, we have not been able to obtain sufficiently concentrated samples for structural analysis. This might be due to a defect in FPV E6 folding or to poor solubility of the folded FPV E6 peptide. By comparing FPV E6 ORFs to those of FlPV-1, we counted that six presumably exposed positions occupied by polar residues in FlPV-1 E6 are replaced by hydrophobic residues in FPV E6. The occurrence of six additional hydrophobic residues on the surface of the folded FPV E6 compared to the folded FlPV-1 E6 provides the most likely explanation for the lower solubility of FPV E6.
The C terminus of FlPV-1 E7 is a soluble dimeric domain.
The last 59 residues of FlPV-1 E7 were cloned and expressed as a fusion to MBP and purified by following the same procedure as described for FlPV-1 E6 protein (Fig. 5A). When subjected to preparative gel filtration chromatography, the E7 C-terminal construct (theoretical mass, 6.5 kDa) eluted earlier than the 9.5-kDa E6 monomer at the volume expected for a 13-kDa protein (Fig. 5B). This suggested that the FlPV-1 E7 C-terminal construct behaved as a dimeric domain, as previously observed for the C-terminal domain of HPV E7 (18, 25). The sample was concentrated up to 300 μM without aggregation.
FIG. 5.
Expression, purification, and size characterization of the FlPV-1 E7 C terminus. (A) Expression and purification. The last 59 C-terminal residues of FlPV-1 E7 were fused to the C terminus of MBP and overexpressed in bacteria (lane 1). Pellets were sonicated, and the centrifugation-cleared supernatant was purified by affinity on MBP-specific amylose resin (lanes 2, flowthrough; lane 3, maltose eluate) and subjected to TEV digestion, yielding MBP and the FlPV-1 E7 C terminus (lane 4). The FlPV-1 E7 C terminus was then separated from MBP and TEV by gel filtration (lane 5). Due to its small size (6.5 kDa) and its high concentration, the sodium dodecyl sulfate-denatured construct was visualized in the migration front of the gel. (B) Preparative gel filtration. Upon preparative gel filtration, the FlPV-1 E7 C terminus (theoretical size of monomer, 6.5 kDa) shows an elution peak corresponding to a 13-kDa dimer, preceding the peak of the FlPV-1 E6 monomer (9.2 kDa). mAU, milli-absorbance units.
FPV E6 and FPV E7 C termini are folded according to HSQC NMR measurements.
MBP-FlPV-1 E6 and MBP-FlPV-1 E7 fusion constructs were expressed in minimal medium, allowing for 15N labeling. Samples were subsequently purified and concentrated up to 300 μM. Standard 1H-15N heteronuclear single-quantum correlation (HSQC) NMR experiments were recorded. The spectrum of FlPV-1 E6 (Fig. 6A) is typical for a folded protein; i.e., resonances are spread over a large frequency scale along the 1H-labeled axis. In addition, most peaks display a homogeneous line width consistent with the size of the protein, suggesting that the protein does not undergo aggregation. Approximately 80 amide resonances can be counted in the HSQC spectrum. This number is close to that predicted from the sequence of the construct. For the FlPV-1 E7 C-terminal construct (Fig. 6B), we observe approximately 35 to 40 peaks, with homogeneous line widths and wide spectral dispersion, which are characteristic of a well-folded region. We also observe a number of additional peaks displaying inhomogeneous shapes and line widths and occupying a very narrow frequency shift range at the center of the spectrum. The latter peaks may represent unfolded regions flanking the folded zinc-binding region, as previously observed in an NMR study of the HPV-45 E7 protein (25).
FIG. 6.
(A) 1H-15N HSQC NMR spectra of 15N-labeled FlPV-1 E6 (300 μM), recorded at pH 6.5, 20°C, and 600 MHz (1H frequency). The widespread repartition of the N-H correlation peaks as well as their homogeneity in size and intensity are indicative of a well-folded monodisperse protein. (B) 1H-15N HSQC NMR spectra of the 15N-labeled FlPV-1 E7 C terminus (C-term) (300 μM), recorded at pH 6.5, 20°C, and 600 MHz (1H frequency). A total of 35 to 40 widespread and homogeneous N-H correlation peaks indicate a well-folded region, whereas the inhomogeneous peaks at the center of the spectrum suggest the presence of nonfolded flanking sequences.
Phylogenetic analysis of the avian PVs.
Figure 7 shows a maximum parsimony tree based on the complete genome alignments of 135 animal PVs. To make visual inspection of this tree easier, we opted to collapse the human PV genera with more than two taxa. We chose this option over the use of representative PVs because it allows for the inclusion of all the available information during the tree construction process.
The resulting tree clusters all known PVs into the previously reported PV genera (7). FlPV-1 clusters with the other avian PVs at the root of the mammalian PV tree in the theta-PV genus. We also traced the state of the E6 ORF. The avian PVs have one Zn-binding domain, the taxa with the normal mammalian situation have 2 domains, and the two rabbit PVs each have an extra Zn-binding domain.
Phylogenetic analysis of separate E6 zinc-binding domains.
Earlier phylogenetic studies suggested that the mammalian E6 proteins may have resulted from the duplication of a single-domain ancestor (5). To further investigate the possibility of a duplication event in the E6 ORF, we constructed a tree in which all of the separate E6 domains were used as separate taxa (Fig. 8). This phylogenetic tree clusters all C-terminal and N-terminal domains together. To facilitate visual inspection, only the PV species (according to the classification by de Villiers et al. [7]) have been represented in Fig. 8. A more complete tree showing the domains of most PV strains is provided in Fig. S1 in the supplemental material. Although both halves of the tree cluster the PVs mostly like the tree shown in Fig. 7, there is a significant polytomy at the root of the C-terminal tree. Interestingly, the two avian PVs (FPV and FlPV-1) cluster at the root of the C-terminal tree, whereas the extra duplication in the rabbit PVs clusters with the N-terminal domains. The most likely explanation for these placements is the hypothesis that the C-terminal domains are most closely related to the original single-domain E6 ORF and that the extra domains in the rabbit PVs are the results of a more recent duplication of the E6N domain. Also, the differences in branch lengths in this phylogram suggest that the N-terminal domains are evolving faster than the C-terminal domain. To quantify the differences between both trees, we calculated Rohlf's consistency index. The obtained Rohlf consistency index (1) of 0.122 supports the hypothesis of different evolutionary histories for both halves of this tree. Next, we also performed a partition homogeneity test (as implemented in PAUP*) which returned a P value of 1 to 100/99 (0.01). Due to the unresolved node at the root of the N-terminal half of the tree, only the HPV types were included in this analysis. Nevertheless, the obtained P value strongly supports the different evolutionary histories of both halves of this protein. These observations support the hypothesis that a duplication event in the ancestral PV gave rise to the two-mammalian-domain E6 proteins. The possibility for this duplication event is further illustrated by the extra duplication in the rabbit PVs.
DISCUSSION
In this paper, we report on the characterization of the third avian PV known to date. Although the avian PVs share the “core” ORFs (i.e., E1, E2, L1, and L2) with the mammalian PVs, we observed striking differences in the organizations of the early regions. First, the three avian PVs all contain a putative E9 (X-ORF), not found in the mammalian PVs. In addition, avian E7 ORFs present a dimeric C-terminal zinc-binding domain more compact than that of mammalian E7, while the presumably natively unfolded N-terminal region of avian E7 appears to be longer than those in the mammalian orthologs. Finally, avian E6 ORFs (when present) are composed of only one zinc-binding domain, in striking contrast with all mammalian E6 proteins, which always contain at least a pair of such domains. This single-domain avian E6 appears to contain extended secondary structures/loops compared to those of the mammalian E6N and E6C domains.
Both biophysical analysis of purified samples and immunofluorescence analysis in living cells demonstrated that avian E6 and the C terminus of E7 are highly soluble and folded. ORFs coding for soluble globular folded proteins are submitted to a very high selection pressure for conserving a defined set of buried residues (mostly hydrophobics and zinc-binding cysteins in the cases of PV E6 and E7), which dictate the protein's fold, and surface residues (preferentially polar or charged), which rule the degree of solubility of the protein. The conservation of folding and solubility propensities in the avian E6 and E7 ORFs leaves little doubt that both ORFs code for functional proteins.
Remarkably, the identification of a single-domain E6 protein in avian PVs provides the first experimental data in support of the hypothesis proposed in 1987 by Cole and Danos (5). These authors suggested that the two domains of mammalian PV E6 proteins may be the result of a duplication of a common single-domain ancestor. This duplication event may have taken place during the 310 million years separating birds and mammals from their common ancestor. In our phylogenetic analysis, mammalian C-terminal domains cluster with avian E6, suggesting that they are more closely related to the postulated ancestral single-domain E6, whereas the N-terminal domains have evolved faster, thereby acquiring more differentiated features. Recently, both Garcia-Vallve et al. and Rector et al. independently showed that the E6 and E7 proteins appear to mutate twice as fast as the rest of the viral genome (11, 28). It is feasible that a (recent) duplication event can explain this increased mutation rate. Whether the observed differences in evolutionary history of these two halves of mammalian E6 proteins have phenotypic importance remains to be addressed.
We previously proposed that the putative ancestral single-domain E6 might have existed as a homodimer (23). Here, we have carefully analyzed the oligomeric state of the single-domain FlPV-1 E6. This protein is a perfect monomer, even at high concentrations. This argues against the notion that the ancestral single-domain E6 existed as a dimer. However, it remains possible that avian E6 might dimerize upon binding to particular targets. This scenario would explain the evolutionary advantage of having both domains connected by a linker, as is the case in all mammalian PVs with an E6 ORF. Further functional and structural studies of avian E6 might provide interesting insights on these questions.
Avian PVs, especially in their early region, therefore provide a unique perspective on the evolution of a very ancient family of viruses. The single-domain E6 protein, the “downsized” zinc-binding domain of E7, and the putative E9 ORF may represent ancient features inherited from a putative common ancestral PV. This study has also provided insights into the occurrence and the history of a duplication event in the E6 protein of current mammalian PVs. Due to the importance of E6 in HPV-induced pathogenesis, it remains to be answered how this duplication event altered the biology of mammalian PVs. But it is likely that the proposed duplication had important consequences for the fitness and evolutionary success of these ancient viruses.
Supplementary Material
Acknowledgments
This work was supported by CNRS, University of Strasbourg, Association de Recherche contre le Cancer (ARC), Ligue Nationale contre le Cancer, Agence Nationale de la Recherche (ANR), and the Flemish Fund for Scientific Research (FWO) grant G.0513.06. A.O.M.O.S. was supported by a grant from the ARC and A.R. by a postdoctoral fellowship from the K. U. Leuven Research Fund.
Footnotes
Published ahead of print on 24 June 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Bartels, C., T. H. Xia, M. Billeter, P. Guntert, and K. Wuhtrich. 1995. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR 61-10. [DOI] [PubMed] [Google Scholar]
- 2.Birnstiel, M. L., M. Busslinger, and K. Strub. 1985. Transcription termination and 3′ processing: the end is in site! Cell 41349-359. [DOI] [PubMed] [Google Scholar]
- 3.Chen, Z., M. Schiffman, R. Herrero, R. DeSalle, and R. D. Burk. 2007. Human papillomavirus (HPV) types 101 and 103 isolated from cervicovaginal cells lack an E6 open reading frame (ORF) and are related to gamma-papillomaviruses. Virology 360447-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chothia, C. 1984. Principles that determine the structure of proteins. Annu. Rev. Biochem. 53537-572. [DOI] [PubMed] [Google Scholar]
- 5.Cole, S. T., and O. Danos. 1987. Nucleotide sequence and comparative analysis of the human papillomavirus type 18 genome. Phylogeny of papillomaviruses and repeated structure of the E6 and E7 gene products. J. Mol. Biol. 193599-608. [DOI] [PubMed] [Google Scholar]
- 6.Delaglio, F., S. Grzesiek, G. W. Vuister, G. Zhu, J. Pfeifer, and A. Bax. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6277-293. [DOI] [PubMed] [Google Scholar]
- 7.de Villiers, E. M., C. Fauquet, T. R. Broker, H. U. Bernard, and H. zur Hausen. 2004. Classification of papillomaviruses. Virology 32417-27. [DOI] [PubMed] [Google Scholar]
- 8.Dosztanyi, Z., V. Csizmok, P. Tompa, and I. Simon. 2005. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 213433-3434. [DOI] [PubMed] [Google Scholar]
- 9.Edgar, R. C. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finn, R. D., J. Mistry, B. Schuster-Bockler, S. Griffiths-Jones, V. Hollich, T. Lassmann, S. Moxon, M. Marshall, A. Khanna, R. Durbin, S. R. Eddy, E. L. Sonnhammer, and A. Bateman. 2006. Pfam: clans, web tools and services. Nucleic Acids Res. 34D247-D251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia-Vallve, S., A. Alonso, and I. G. Bravo. 2005. Papillomaviruses: different genes have different histories. Trends Microbiol. 13514-521. [DOI] [PubMed] [Google Scholar]
- 12.Grossman, S. R., and L. A. Laimins. 1989. E6 protein of human papillomavirus type 18 binds zinc. Oncogene 41089-1093. [PubMed] [Google Scholar]
- 13.Kopito, R. R. 2000. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 10524-530. [DOI] [PubMed] [Google Scholar]
- 14.Lagrange, M., S. Charbonnier, G. Orfanoudakis, P. Robinson, K. Zanier, M. Masson, Y. Lutz, G. Trave, E. Weiss, and F. Deryckere. 2005. Binding of human papillomavirus 16 E6 to p53 and E6AP is impaired by monoclonal antibodies directed against the second zinc-binding domain of E6. J. Gen. Virol. 861001-1007. [DOI] [PubMed] [Google Scholar]
- 15.Laue, T. M., B. D. Shah, T. M. Ridgeway, and S. L. Pelletier. 1992. Analytical ultracentrifugation, p. 90-125. In S. E. Harding, A. J. Rowe, and J. C. Horton (ed.), Biochemistry and polymer science. The Royal Society of Chemistry, Cambridge, United Kingdom.
- 16.Letunic, I., R. R. Copley, B. Pils, S. Pinkert, J. Schultz, and P. Bork. 2006. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 34D257-D260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Linding, R., R. B. Russell, V. Neduva, and T. J. Gibson. 2003. GlobPlot: exploring protein sequences for globularity and disorder. Nucleic Acids Res. 313701-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, X., A. Clements, K. Zhao, and R. Marmorstein. 2006. Structure of the human papillomavirus E7 oncoprotein and its mechanism for inactivation of the retinoblastoma tumor suppressor. J. Biol. Chem. 281578-586. [DOI] [PubMed] [Google Scholar]
- 19.Munoz, N., X. Castellsague, A. B. de Gonzalez, and L. Gissmann. 2006. HPV in the etiology of human cancer. Vaccine 24(Suppl. 3)S1-S10. [DOI] [PubMed] [Google Scholar]
- 20.Nixon, K. C. 1999. The parsimony ratchet, a new method for rapid parsimony analysis. Cladistics 15407-414. [DOI] [PubMed] [Google Scholar]
- 21.Nomine, Y., S. Charbonnier, L. Miguet, N. Potier, D. A. Van, R. A. Atkinson, G. Trave, and B. Kieffer. 2005. 1H and 15N resonance assignment, secondary structure and dynamic behaviour of the C-terminal domain of human papillomavirus oncoprotein E6. J. Biomol. NMR 31129-141. [DOI] [PubMed] [Google Scholar]
- 22.Nomine, Y., S. Charbonnier, T. Ristriani, G. Stier, M. Masson, N. Cavusoglu, A. Van Dorsselaer, E. Weiss, B. Kieffer, and G. Trave. 2003. Domain substructure of HPV E6 oncoprotein: biophysical characterization of the E6 C-terminal DNA-binding domain. Biochemistry 424909-4917. [DOI] [PubMed] [Google Scholar]
- 23.Nomine, Y., M. Masson, S. Charbonnier, K. Zanier, T. Ristriani, F. Deryckere, A. P. Sibler, D. Desplancq, R. A. Atkinson, E. Weiss, G. Orfanoudakis, B. Kieffer, and G. Trave. 2006. Structural and functional analysis of E6 oncoprotein: insights in the molecular pathways of human papillomavirus-mediated pathogenesis. Mol. Cell 21665-678. [DOI] [PubMed] [Google Scholar]
- 24.Nomine, Y., T. Ristriani, C. Laurent, J. F. Lefevre, E. Weiss, and G. Trave. 2001. Formation of soluble inclusion bodies by hpv e6 oncoprotein fused to maltose-binding protein. Protein Expr. Purif. 2322-32. [DOI] [PubMed] [Google Scholar]
- 25.Ohlenschlager, O., T. Seiboth, H. Zengerling, L. Briese, A. Marchanka, R. Ramachandran, M. Baum, M. Korbas, W. Meyer-Klaucke, M. Durst, and M. Gorlach. 2006. Solution structure of the partially folded high-risk human papilloma virus 45 oncoprotein E7. Oncogene 255953-5959. [DOI] [PubMed] [Google Scholar]
- 26.Piotto, M., V. Saudek, and V. Sklenar. 1992. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR 2661-665. [DOI] [PubMed] [Google Scholar]
- 27.Presta, L. G., and G. D. Rose. 1988. Helix signals in proteins. Science 2401632-1641. [DOI] [PubMed] [Google Scholar]
- 28.Rector, A., P. Lemey, R. Tachezy, S. Mostmans, S. J. Ghim, D. K. Van, M. Roelke, M. Bush, R. J. Montali, J. Joslin, R. D. Burk, A. B. Jenson, J. P. Sundberg, B. Shapiro, and R. M. Van. 2007. Ancient papillomavirus-host co-speciation in Felidae. Genome Biol. 8R57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rector, A., R. Tachezy, and M. Van Ranst. 2004. A sequence-independent strategy for detection and cloning of circular DNA virus genomes by using multiply primed rolling-circle amplification. J. Virol. 784993-4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schuck, P. 2000. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 781606-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwarz, E., U. K. Freese, L. Gissmann, W. Mayer, B. Roggenbuck, A. Stremlau, and H. zur Hausen. 1985. Structure and transcription of human papillomavirus sequences in cervical carcinoma cells. Nature 314111-114. [DOI] [PubMed] [Google Scholar]
- 32.Sibler, A. P., A. Nordhammer, M. Masson, P. Martineau, G. Trave, and E. Weiss. 2003. Nucleocytoplasmic shuttling of antigen in mammalian cells conferred by a soluble versus insoluble single-chain antibody fragment equipped with import/export signals. Exp. Cell Res. 286276-287. [DOI] [PubMed] [Google Scholar]
- 33.Sikes, D. S., and P. O. Lewis. 2001. PAUPRat: PAUP* implementation of the parsimony ratchet. Beta software, version 1. Department of Ecology and Evolutionary Biology, University of Connecticut, Storrs.
- 34.Sundberg, J. P., M. Van Ranst, R. D. Burk, and A. B. Jenson. 1997. The nonhuman (animal) papillomaviruses: host range, epitope conservation, and molecular diversity, p. 47-68. In G. von Krogh and G. Gross (ed.), Human papillomavirus infections in dermatovenereology. CRC Press, Boca Raton, FL.
- 35.Sundberg, J. P., M. Van Ranst, and A. B. Jenson. 2001. Papillomavirus infections, p. 223-231. In E. S. Williams and I. K. Barker (ed.), Infectious diseases of wild mammals. Iowa State University Press, Ames.
- 36.Swofford, D. L. 1998. PAUP* 4.0: phylogenetic analysis using parsimony. Sinauer Associates, Sunderland, MA.
- 37.Tachezy, R., A. Rector, M. Havelkova, E. Wollants, P. Fiten, G. Opdenakker, B. Jenson, J. Sundberg, and M. Van Ranst. 2002. Avian papillomaviruses: the parrot Psittacus erithacus papillomavirus (PePV) genome has a unique organization of the early protein region and is phylogenetically related to the chaffinch papillomavirus. BMC Microbiol. 219-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Terai, M., R. DeSalle, and R. D. Burk. 2002. Lack of canonical E6 and E7 open reading frames in bird papillomaviruses: Fringilla coelebs papillomavirus and Psittacus erithacus timneh papillomavirus. J. Virol. 7610020-10023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Doorslaer, K., A. Rector, P. Vos, and M. Van Ranst. 2006. Genetic characterization of the Capra hircus papillomavirus: a novel close-to-root artiodactyl papillomavirus. Virus Res. 118164-169. [DOI] [PubMed] [Google Scholar]
- 40.Vriend, G. 1990. Parameter relation rows: a query system for protein structure function relationships. Protein Eng. 4221-223. [DOI] [PubMed] [Google Scholar]
- 41.Zanier, K., Y. Nomine, S. Charbonnier, C. Ruhlmann, P. Schultz, J. Schweizer, and G. Trave. 2007. Formation of well-defined soluble aggregates upon fusion to MBP is a generic property of E6 proteins from various human papillomavirus species. Protein Expr. Purif. 5159-70. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.