Abstract
Specific therapy is not available for hantavirus cardiopulmonary syndrome caused by Andes virus (ANDV). Peptides capable of blocking ANDV infection in vitro were identified using antibodies against ANDV surface glycoproteins Gn and Gc to competitively elute a cyclic nonapeptide-bearing phage display library from purified ANDV particles. Phage was examined for ANDV infection inhibition in vitro, and nonapeptides were synthesized based on the most-potent phage sequences. Three peptides showed levels of viral inhibition which were significantly increased by combination treatment with anti-Gn- and anti-Gc-targeting peptides. These peptides will be valuable tools for further development of both peptide and nonpeptide therapeutic agents.
Andes virus (ANDV), an NIAID category A agent linked to hantavirus cardiopulmonary syndrome (HCPS), belongs to the family Bunyaviridae and the genus Hantavirus and is carried by Oligoryzomys longicaudatus rodents (11). HCPS is characterized by pulmonary edema caused by capillary leak, with death often resulting from cardiogenic shock (9, 16). ANDV HCPS has a case fatality rate approaching 40%, and ANDV is the only hantavirus demonstrated to be capable of direct person-to-person transmission (15, 21). There is currently no specific therapy available for treatment of ANDV infection and HCPS.
Peptide ligands that target a specific protein surface can have broad applications as therapeutics by blocking specific protein-protein interactions, such as preventing viral engagement of host cell receptors and thus preventing infection. Phage display libraries provide a powerful and inexpensive tool to identify such peptides. Here, we used selection of a cyclic nonapeptide-bearing phage library to identify peptides capable of binding the transmembrane surface glycoproteins of ANDV, Gn and Gc, and blocking infection in vitro.
To identify peptide sequences capable of recognizing ANDV, we panned a cysteine-constrained cyclic nonapeptide-bearing phage display library (New England Biolabs) against density gradient-purified, UV-treated ANDV strain CHI-7913 (a gift from Hector Galeno, Santiago, Chile) (17, 18). To increase the specificity of the peptides identified, we eluted phage by using monoclonal antibodies (Austral Biologicals) prepared against recombinant fragments of ANDV Gn (residues 1 to 353) or Gc (residues 182 to 491) glycoproteins (antibodies 6B9/F5 and 6C5/D12, respectively). Peptide sequences were determined for phage from iterative rounds of panning, and the ability of phage to inhibit ANDV infection of Vero E6 cells was determined by immunofluorescent assay (IFA) (7). Primary IFA detection antibodies were rabbit polyclonal anti-Sin Nombre hantavirus (SNV) nucleoprotein (N) antibodies which exhibit potent cross-reactivity against other hantavirus N antigens (3). ReoPro, a commercially available Fab fragment which partially blocks infection of hantaviruses in vitro by binding the entry receptor integrin β3 (5), was used as a positive control (80 μg/ml) along with the original antibody used for phage elution (5 μg/ml). As the maximum effectiveness of ReoPro in inhibiting hantavirus entry approaches 80%, we set this as a threshold for maximal expected efficacy for normalization. The most-potent phage identified by elution with the anti-Gn antibody 6B9/F5 bore the peptide CPSNVNNIC and inhibited hantavirus entry by greater than 60% (61%) (Table 1). From phage eluted with the anti-Gc antibody 6C5/D12, those bearing peptides CPMSQNPTC and CPKLHPGGC also inhibited entry by greater than 60% (66% and 72%, respectively).
TABLE 1.
Peptide-bearing phage eluted from ANDV
| Phage | % Inhibition (SD)a | P valueb |
|---|---|---|
| Phage bearing the following peptides eluted with anti-Gn antibody 6B9/F5 | ||
| Group 1 (<30% inhibition) | ||
| CDQRTTRLC | 8.45 (15.34) | 0.0002 |
| CPHDPNHPC | 9.94 (7.72) | 0.333 |
| CQSQTRNHC | 11.76 (13.25) | 0.0001 |
| CLQDMRQFC | 13.26 (9.92) | 0.0014 |
| CLPTDPIQC | 15.70 (14.05) | 0.0005 |
| CPDHPFLRC | 16.65 (15.22) | 0.8523 |
| CSTRAENQC | 17.56 (16.50) | 0.0004 |
| CPSHLDAFC | 18.98 (20.06) | 0.0017 |
| CKTGHMRIC | 20.84 (7.47) | 0.0563 |
| CVRTPTHHC | 20.89 (27.07) | 0.1483 |
| CSGVINTTC | 21.57 (19.61) | 0.0643 |
| CPLASTRTC | 21.65 (5.98) | 0.004 |
| CSQFPPRLC | 22.19 (8.26) | 0.0004 |
| CLLNKQNAC | 22.34 (7.78) | 0.001 |
| CKFPLNAAC | 22.89 (6.15) | 0.0001 |
| CSLTPHRSC | 23.63 (16.74) | 0.0563 |
| CKPWPMYSC | 23.71 (6.68) | 0.0643 |
| CLQHDALNC | 24.01 (7.60) | 1 |
| CNANKPKMC | 24.67 (11.67) | 0.0004 |
| CPKHVLKVC | 25.30 (28.36) | 0.0003 |
| CTPDKKSFC | 26.91 (11.15) | 0.399 |
| CHGKAALAC | 27.22 (32.53) | 0.005 |
| CNLMGNPHC | 28.08 (21.35) | 0.0011 |
| CLKNWFQPC | 28.64 (18.49) | 0.0016 |
| CKEYGRQMC | 28.76 (29.33) | 0.0362 |
| CQPSDPHLC | 29.44 (31.22) | 0.0183 |
| CSHLPPNRC | 29.70 (17.37) | 0.0061 |
| Group 2 (30-59% inhibition) | ||
| CSPLLRTVC | 33.05 (20.26) | 0.0023 |
| CHKGHTWNC | 34.17 (12.50) | 0.0795 |
| CINASHAHC | 35.62 (13.03) | 0.3193 |
| CWPPSSRTC | 36.75 (26.95) | 0.0006 |
| CPSSPFNHC | 37.78 (7.11) | 0.0001 |
| CEHLSHAAC | 38.47 (7.60) | 0.0115 |
| CQDRKTSQC | 38.74 (9.12) | 0.1802 |
| CTDVYRPTC | 38.90 (25.03) | 0.006 |
| CGEKSAQLC | 39.11 (27.52) | 0.0013 |
| CSAAERLNC | 40.13 (6.33) | 0.0033 |
| CFRTLEHLC | 42.07 (5.01) | 0.0608 |
| CEKLHTASC | 43.60 (27.92) | 0.1684 |
| CSLHSHKGC | 45.11 (49.81) | 0.0864 |
| CNSHSPVHC | 45.40 (28.80) | 0.0115 |
| CMQSAAAHC | 48.88 (44.40) | 0.5794 |
| CPAASHPRC | 51.84 (17.09) | 0.1935 |
| CKSLGSSQC | 53.90 (13.34) | 0.0145 |
| Group 3 (60-79% inhibition) | ||
| CPSNVNNIC | 61.11 (25.41) | 0.1245 |
| Negative control | 0 (6.15) | |
| 6B9/F5 (5 μg/ml) | 26.77 (5.33) | |
| ReoPro (80 μg/ml) | 79.86 (4.88) | |
| Phage bearing the following peptides eluted with anti-Gc antibody 6C5/D12 | ||
| Group 1 (<30% inhibition) | ||
| CHPGSSSRC | 1.01 (7.03) | 0.0557 |
| CSLSPLGRC | 10.56 (13.62) | 0.7895 |
| CTARYTQHC | 12.86 (3.83) | 0.3193 |
| CHGVYALHC | 12.91 (7.32) | 0.0003 |
| CLQHNEREC | 16.79 (13.72) | 0.0958 |
| CHPSTHRYC | 17.23 (14.53) | 0.0011 |
| CPGNWWSTC | 19.34(9.91) | 0.1483 |
| CGMLNWNRC | 19.48 (19.42) | 0.0777 |
| CPHTQFWQC | 20.44 (13.65) | 0.0008 |
| CTPTMHNHC | 20.92 (11.68) | 0.0001 |
| CDQVAGYSC | 21.79 (23.60) | 0.0063 |
| CIPMMTEFC | 24.33 (9.28) | 0.2999 |
| CERPYSRLC | 24.38 (9.09) | 0.0041 |
| CPSLHTREC | 25.06 (22.78) | 0.1202 |
| CSPLQIPYC | 26.30 (34.29) | 0.4673 |
| CTTMTRMTC (×2) | 29.27 (8.65) | 0.0001 |
| Group 2 (30-59% inhibition) | ||
| CNKPFSLPC | 30.09 (5.59) | 0.4384 |
| CHNLESGTC | 31.63 (26.67) | 0.751 |
| CNSVPPYQC | 31.96 (6.51) | 0.0903 |
| CSDSWLPRC | 32.95 (28.54) | 0.259 |
| CSAPFTKSC | 33.40 (10.64) | 0.0052 |
| CEGLPNIDC | 35.63 (19.90) | 0.0853 |
| CTSTHTKTC | 36.28 (13.42) | 0.132 |
| CLSIHSSVC | 36.40 (16.44) | 0.8981 |
| CPWSTQYAC | 36.81 (32.81) | 0.5725 |
| CTGSNLPIC | 36.83 (31.64) | 0.0307 |
| CSLAPANTC | 39.73 (4.03) | 0.1664 |
| CGLKTNPAC | 39.75 (16.98) | 0.2084 |
| CRDTTPWWC | 40.08 (18.52) | 0.0004 |
| CHTNASPHC | 40.26 (4.77) | 0.5904 |
| CTSMAYHHC | 41.89 (8.61) | 0.259 |
| CSLSSPRIC | 42.13 (29.75) | 0.2463 |
| CVSLEHQNC | 45.54 (6.55) | 0.5065 |
| CRVTQTHTC | 46.55 (8.45) | 0.3676 |
| CPTTKSNVC | 49.28 (14.00) | 0.3898 |
| CSPGPHRVC | 49.50 (42.60) | 0.0115 |
| CKSTSNVYC | 51.20 (4.60) | 0.0611 |
| CTVGPTRSC | 57.30 (11.31) | 0.0176 |
| Group 3 (60-79% inhibition) | ||
| CPMSQNPTC | 65.60 (13.49) | 0.014 |
| CPKLHPGGC | 71.88 (27.11) | 0.0059 |
| Negative control | 0.26 (4.53) | |
| 6C5/D12 (5 μg/ml) | 22.62 (8.40) | |
| ReoPro (80 μg/ml) | 80.02 (76.64) |
Standard deviations of four experiments are shown in parentheses. Peptide-bearing phage were added at 109 phage/μl.
P values for the pairwise amino acid alignment score of each peptide versus that of integrin β3 were determined using an unpaired Student's t test. P values considered statistically significant are shown in bold.
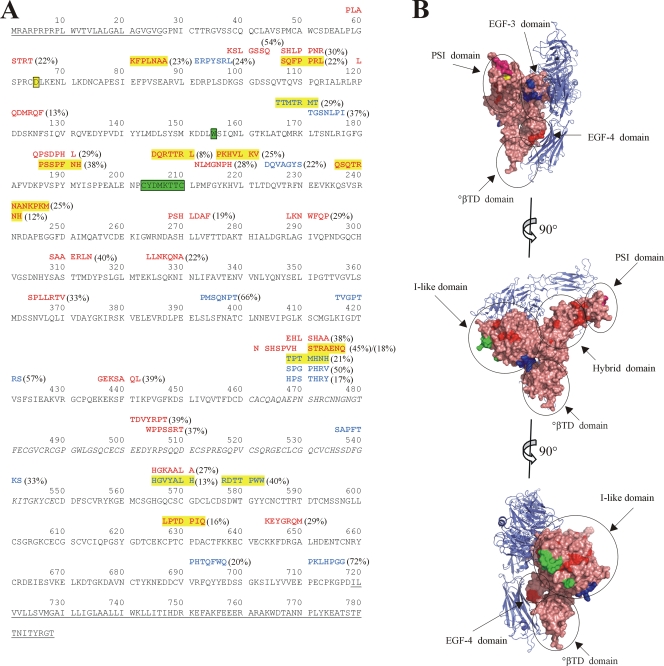
To determine whether the peptide sequences of any of the identified inhibitory phage showed homology to integrin β3, a known entry receptor for pathogenic hantaviruses (6, 7), we used the Gap program to perform a pairwise amino acid alignment of each peptide versus the extracellular portion of integrin β3 and determined P values for the alignments. Of 45 phage eluted with the anti-Gn antibody, 6B9/F5, 27 of the peptide sequences showed homology to integrin β3 (P < 0.05), and 9 were highly significant (P ≤ 0.0005) (Fig. 1A). Of the latter, CKFPLNAAC and CSQFPPRLC map to the hybrid domain (Fig. 1B), which is proximal to the plexin-semaphorin-integrin domain (PSI) containing residue D39, shown to be critical for viral entry in vitro (19). Five sequences (CPSSPFNH, CPKHVLKVC, CNANKPKMC, CQSQTRNHC, and CDQRTTRLC) map to the I-like (or βA) domain near the binding site of ReoPro (2). Finally, CLPTDPIQC maps to the epidermal growth factor 4 (EGF-4) domain, and CSTRAENQC aligns to a portion of β3 untraceable in the crystal structure, specifically the linker region between the hybrid domain and EGF-1. Although this represents a disordered portion of the protein (22), the location of this loop proximal to the PSI domain is worth noting, due to the role of the PSI domain in facilitating viral entry (19). Therefore, 60% of phage eluted with the anti-Gn antibody showed some homology to integrin β3, and those with highly significant P values predominantly mapped to or proximal to regions of known interest in viral entry.
FIG. 1.
Inhibitory peptides identified through phage panning against ANDV show homology to integrin β3. (A) Alignment of phage peptide sequences with P values for integrin β3 pairwise alignment of less than 0.05. Residues comprising the signal peptide, transmembrane, and cytoplasmic domains, which were not included during pairwise alignment, are underlined. Residues 461 to 548, which are missing in the crystal structure, are italicized. Residues involved in the ReoPro binding site are highlighted in green (2). Residue D39 of the PSI domain is highlighted in yellow (19). Peptides are shown above the sequence of integrin β3, with antibody 6C5/D12-eluted sequences shown in blue text and sequences eluted with antibody 6B9/F5 shown in red. Peptide sequences with alignment P values of ≤0.0005 are highlighted in yellow. Percent inhibition of the peptide-bearing phage is shown in parentheses. (B) View of integrin αvβ3 (PDB ID 1U8C [23]). αv is shown in blue ribbon diagram, and β3 is shown in salmon-colored surface representation, with specific domains circled. Residues corresponding to the ReoPro binding site are shown in green, as in panel A, and D39 is shown in yellow. Regions corresponding to 6C5/D12-eluted peptides with P values of ≤0.0005 for alignment with integrin β3 (highlighted in panel A) are shown in blue, and those corresponding to 6B9/F5-eluted peptides with P values of ≤0.0005 for alignment with integrin β3 are shown in red. Alignment of peptide PLASTRT (P value of 0.0040) adjacent to D39 of the PSI domain is shown in magenta. Graphics were prepared using Pymol (DeLano Scientific LLC, San Carlos, CA).
Of the 41 peptide-bearing phage eluted with the anti-Gc antibody 6C5/D12, 14 showed sequence homology to integrin β3 (P < 0.05), 4 of which had P values of ≤0.0005 (Fig. 1A). Of the latter, sequence CTTMTRMTC mapped to the base of the I-like domain (Fig. 1B), while CHGVYALHC and CRDTTPWWC mapped to the EGF-3 domain. Finally, sequence CTPTMHNHC mapped to the linker region untraceable in the crystal structure. Therefore, in contrast to peptide sequences identified by competition with the anti-Gn antibody, sequences identified by competition with the anti-Gc antibody 6C5/D12 appear to be mostly unrelated to integrin β3.
As a low level of pathogenic hantavirus infection can be seen in cells lacking integrin β3, such as CHO cells (19), we asked if any of the identified peptide sequences could represent a previously unidentified receptor. We used the Basic Local Alignment Search Tool to search a current database of human protein sequences for potential alternate receptors represented by these peptides. However, none of the alignments identified proteins that are expressed at the cell surface, eliminating them as potential candidates for alternate viral entry receptors. This suggests that the majority of the peptides identified here likely represent novel sequences for binding ANDV surface glycoproteins.
To determine whether synthetic peptides would also block infection, we synthesized cyclic peptides based on the 10 most-potent peptide-bearing phage. These peptides, in the context of phage presentation, showed levels of inhibition ranging from 44 to 72% (Table 2). When tested by IFA at 1 mM, four of the synthetic peptides showed inhibition levels significantly lower than those of the same peptide presented in the context of phage. This is not surprising, as steric factors due to the size of the phage and the multivalent presentation of peptide in the context of phage may both contribute to infection inhibition (8). However, there was no significant difference in inhibition by synthetic peptide versus peptide-bearing phage for six of the sequences, implying that inhibition in the context of phage was due solely to the nature of the peptide itself and not to steric factors or valency considerations contributed by the phage, which contrasts with our previous results, determined by using phage directed against αvβ3 integrin (10).
TABLE 2.
Synthetic cyclic peptides inhibit ANDV infection
| Target | Sample | % Inhibition bya:
|
|
|---|---|---|---|
| Peptide-bearing phage | Synthetic peptide | ||
| Gn | CMQSAAAHC | 48.88 (44.40) | 59.66 (11.17) |
| Gc | CTVGPTRSC | 57.30 (11.31) | 46.47 (7.61) |
| Gn | CPSNVNNIC | 61.11 (25.41) | 44.14 (10.74) |
| Gn | CEKLHTASC | 43.60 (27.92) | 34.87 (9.26) |
| Gc | CPKLHPGGC | 71.88 (27.11) | 30.95 (7.73)b |
| Gn | CSLHSHKGC | 45.11 (49.81) | 29.79 (9.34) |
| Gc | CPMSQNPTC | 65.60 (13.49) | 18.19 (8.55)b |
| Gn | CKSLGSSQC | 53.90 (13.34) | 18.10 (7.55)b |
| Gn | CNSHSPVHC | 45.40 (28.80) | 15.52 (10.48) |
| Gn | CPAASHPRC | 51.84 (17.09) | 0 (10.72)b |
| Integrin β3 | ReoPro | 80.10 (7.72) | |
| Gn | 6B9/F5 antibody | 42.72 (6.75) | |
| Gc | 6C5/D12 antibody | 31.04 (7.81) | |
Standard deviations of the results of at least four experiments are shown in parentheses.
Mean percent inhibition between phage and synthetic peptide differs significantly (P < 0.05).
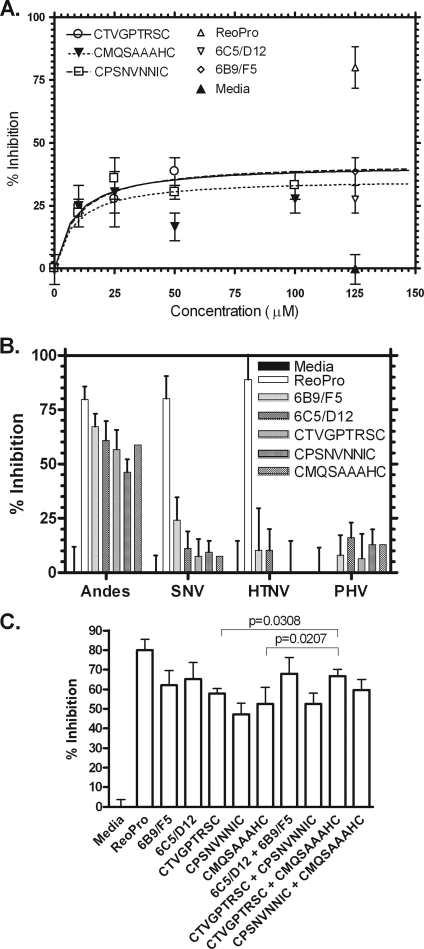
The three most-potent synthetic peptides were examined for their ability to inhibit ANDV entry in a dose-dependent manner. The concentration of each peptide that produces 50% of its maximum potential inhibitory effect was determined. As shown in Fig. 2A, the 50% inhibitory concentration for each of the peptides was in the range of 10 μM, which from our experience is a reasonable potency for a lead compound to take forward for optimization.
FIG. 2.
Activities of synthetic peptides in inhibition of ANDV infection in vitro. (A) Peptides were examined for their ability to block ANDV infection of Vero E6 cells in a dose-dependent manner by IFA. (B) Peptides were tested in parallel for the ability to block infection of Vero E6 cells by ANDV, SNV, HTNV, and PHV. (C) Peptides were tested, singly or in combination, for the ability to block ANDV infection of Vero E6 cells. For all experiments, controls included media, ReoPro at 80 μg/ml, and monoclonal antibodies 6C5/D12 and 6B9/F5 at 5 μg/ml. All peptides were used at 1 mM. Data points represent n = 2 to 6, with error bars showing the standard errors of the means. Statistical analyses were performed on replicate samples using an unpaired Student's t test.
In order to determine the specificity of the three most-potent synthetic cyclic peptides in blocking ANDV, we examined them for inhibition of ANDV infection versus two other pathogenic hantaviruses, SNV and Hantaan virus (HTNV), or the nonpathogenic hantavirus Prospect Hill virus (PHV). As shown in Fig. 2B, ReoPro, which binds integrin β3, showed inhibition of infection by each of the pathogenic hantavirus strains, known to enter cells via β3, but not the nonpathogenic PHV, which enters via integrin β1 (6, 7). In contrast, peptides selected for the ability to bind ANDV were highly specific inhibitors of ANDV versus SNV, HTNV, or PHV. The specificities of peptides eluted by the anti-Gn monoclonal antibody are not surprising, as they are likely due to global differences in the Gn amino acid sequence. Specifically, sequence homologies between ANDV and SNV, HTNV, and PHV are 61%, 36%, and 51%, respectively, for the region corresponding to the immunogen for antibody 6B9/F5. Although homology between the immunogen for antibody 6C5/D12 and the corresponding Gc region of these viruses is somewhat higher (82% with SNV, 63% with HTNV, and 71% with PHV), the possibility that the monoclonal antibody used here recognizes a three-dimensional epitope lends itself to the high specificity of the peptides.
The current model for cellular infection by hantaviruses (14) is as follows. Viral binding of the host cell surface target integrin is followed by receptor-mediated endocytosis and endosome acidification. Lowered pH induces conformational changes in Gn and/or Gc, which facilitate membrane fusion and viral release into the cytosol. As there is currently little information available about whether one glycoprotein is dominant in mediating infection, and as neutralizing epitopes have been found on both Gn and Gc glycoproteins (1, 4, 12, 13, 20), we examined whether combining anti-Gn- and anti-Gc-targeted synthetic peptides would lead to an increased infection blockade compared to those for single treatments. As shown in Fig. 2C, the combination of anti-Gn and anti-Gc peptides CMQSAAAHC and CTVGPTRSC resulted in a significant increase in infection inhibition (P = 0.0207 for CMQSAAAHC, and P = 0.0308 for CTVGPTRSC) compared to that resulting from single treatments. Although the high specificity of the peptides for ANDV makes it unlikely that this combination treatment will lead to more cross-reactivity with other pathogenic hantaviruses, this can be determined only by additional testing. Regardless, these data suggest a unique role for each of these viral proteins in the infection process as well as the benefits of targeting multiple viral epitopes for preventing infection.
To our knowledge, the peptides reported here are the first identified that directly target ANDV, and this work further illustrates the power of coupling phage display and selective elution techniques in the identification of novel peptide sequences capable of specific protein-protein interactions from a large, random pool of peptide sequences. These novel peptide inhibitors (R. S. Larson, P. R. Hall, H. Njus, and B. Hjelle, U.S. patent application 61/205,211) provide leads for the development of more-potent peptide or nonpeptide organics for therapeutic use against HCPS.
Acknowledgments
This work was supported by the NCMR grant “Integrated Network of Ligand-based Autonomous Bioagent Detectors” and by Public Health Service grants U01AI56618 (B.H.), U01AI054779 (B.H.), R56AI034448 (R.S.L.), and 1CO6RR012511. P.R.H. was supported by National Institute of Allergy and Infectious Diseases grant T32AI07538-06 and by NIH Ruth L. Kirschstein National Research Service Awards grant F32AI074246-01A1.
Footnotes
Published ahead of print on 10 June 2009.
REFERENCES
- 1.Arikawa, J., A. L. Schmaljohn, J. M. Dalrymple, and C. S. Schmaljohn. 1989. Characterization of Hantaan virus envelope glycoprotein antigenic determinants defined by monoclonal antibodies. J. Gen. Virol. 70615-624. [DOI] [PubMed] [Google Scholar]
- 2.Artoni, A., J. Li, B. Mitchell, J. Ruan, J. Takagi, T. A. Springer, D. L. French, and B. S. Coller. 2004. Integrin beta3 regions controlling binding of murine mAb 7E3: implications for the mechanism of integrin alphaIIbbeta3 activation. Proc. Natl. Acad. Sci. USA 10113114-13120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bharadwaj, M., C. R. Lyons, I. A. Wortman, and B. Hjelle. 1999. Intramuscular inoculation of Sin Nombre hantavirus cDNAs induces cellular and humoral immune responses in BALB/c mice. Vaccine 172836-2843. [DOI] [PubMed] [Google Scholar]
- 4.Dantas, J. R., Jr., Y. Okuno, H. Asada, M. Tamura, M. Takahashi, O. Tanishita, Y. Takahashi, T. Kurata, and K. Yamanishi. 1986. Characterization of glycoproteins of viruses causing hemorrhagic fever with renal syndrome (HFRS) using monoclonal antibodies. Virology 151379-384. [DOI] [PubMed] [Google Scholar]
- 5.Gauvreau, G. M., A. B. Becker, L. P. Boulet, J. Chakir, R. B. Fick, W. L. Greene, K. J. Killian, P. M. O'byrne, J. K. Reid, and D. W. Cockcroft. 2003. The effects of an anti-CD11a mAb, efalizumab, on allergen-induced airway responses and airway inflammation in subjects with atopic asthma. J. Allergy Clin. Immunol. 112331-338. [DOI] [PubMed] [Google Scholar]
- 6.Gavrilovskaya, I. N., E. J. Brown, M. H. Ginsberg, and E. R. Mackow. 1999. Cellular entry of hantaviruses which cause hemorrhagic fever with renal syndrome is mediated by β3 integrins. J. Virol. 733951-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gavrilovskaya, I. N., M. Shepley, R. Shaw, M. H. Ginsberg, and E. R. Mackow. 1998. Beta3 integrins mediate the cellular entry of hantaviruses that cause respiratory failure. Proc. Natl. Acad. Sci. USA 957074-7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hall, P. R., B. Hjelle, D. C. Brown, C. Ye, V. Bondu-Hawkins, K. A. Kilpatrick, and R. S. Larson. 2008. Multivalent presentation of antihantavirus peptides on nanoparticles enhances infection blockade. Antimicrob. Agents Chemother. 522079-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hallin, G. W., S. Q. Simpson, R. E. Crowell, D. S. James, F. T. Koster, G. J. Mertz, and H. Levy. 1996. Cardiopulmonary manifestations of hantavirus pulmonary syndrome. Crit. Care Med. 24252-258. [DOI] [PubMed] [Google Scholar]
- 10.Larson, R. S., D. C. Brown, C. Ye, and B. Hjelle. 2005. Peptide antagonists that inhibit Sin Nombre virus and Hantaan virus entry through the β3-integrin receptor. J. Virol. 797319-7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.López, N., P. Padula, C. Rossi, M. E. Lazaro, and M. T. Franze-Fernandez. 1996. Genetic identification of a new hantavirus causing severe pulmonary syndrome in Argentina. Virology 220223-226. [DOI] [PubMed] [Google Scholar]
- 12.Lundkvist, A., J. Horling, L. Athlin, A. Rosen, and B. Niklasson. 1993. Neutralizing human monoclonal antibodies against Puumala virus, causative agent of nephropathia epidemica: a novel method using antigen-coated magnetic beads for specific B cell isolation. J. Gen. Virol. 741303-1310. [DOI] [PubMed] [Google Scholar]
- 13.Lundkvist, A., and B. Niklasson. 1992. Bank vole monoclonal antibodies against Puumala virus envelope glycoproteins: identification of epitopes involved in neutralization. Arch. Virol. 12693-105. [DOI] [PubMed] [Google Scholar]
- 14.Mackow, E. R., and I. N. Gavrilovskaya. 2001. Cellular receptors and hantavirus pathogenesis. Curr. Top. Microbiol. Immunol. 25691-115. [DOI] [PubMed] [Google Scholar]
- 15.Martinez, V. P., C. Bellomo, J. San Juan, D. Pinna, R. Forlenza, M. Elder, and P. J. Padula. 2005. Person-to-person transmission of Andes virus. Emerg. Infect. Dis. 111848-1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mertz, G. J., L. Miedzinski, D. Goade, A. T. Pavia, B. Hjelle, C. O. Hansbarger, H. Levy, F. T. Koster, K. Baum, A. Lindemulder, W. Wang, L. Riser, H. Fernandez, and R. J. Whitley. 2004. Placebo-controlled, double-blind trial of intravenous ribavirin for the treatment of hantavirus cardiopulmonary syndrome in North America. Clin. Infect. Dis. 391307-1313. [DOI] [PubMed] [Google Scholar]
- 17.Prescott, J., P. R. Hall, V. S. Bondu-Hawkins, C. Ye, and B. Hjelle. 2007. Early innate immune responses to Sin Nombre hantavirus occur independently of IFN regulatory factor 3, characterized pattern recognition receptors, and viral entry. J. Immunol. 1791796-1801. [DOI] [PubMed] [Google Scholar]
- 18.Prescott, J., C. Ye, G. Sen, and B. Hjelle. 2005. Induction of innate immune response genes by Sin Nombre hantavirus does not require viral replication. J. Virol. 7915007-15015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raymond, T., E. Gorbunova, I. N. Gavrilovskaya, and E. R. Mackow. 2005. Pathogenic hantaviruses bind plexin-semaphorin-integrin domains present at the apex of inactive, bent alphavbeta3 integrin conformers. Proc. Natl. Acad. Sci. USA 1021163-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vapalahti, O., A. Lundkvist, and A. Vaheri. 2001. Human immune response, host genetics, and severity of disease. Curr. Top. Microbiol. Immunol. 256153-169. [DOI] [PubMed] [Google Scholar]
- 21.Vial, P. A., F. Valdivieso, G. Mertz, C. Castillo, E. Belmar, I. Delgado, M. Tapia, and M. Ferres. 2006. Incubation period of hantavirus cardiopulmonary syndrome. Emerg. Infect. Dis. 121271-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiong, J. P., T. Stehle, B. Diefenbach, R. Zhang, R. Dunker, D. L. Scott, A. Joachimiak, S. L. Goodman, and M. A. Arnaout. 2001. Crystal structure of the extracellular segment of integrin alphaV beta3. Science 294339-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiong, J. P., T. Stehle, S. L. Goodman, and M. A. Arnaout. 2004. A novel adaptation of the integrin PSI domain revealed from its crystal structure. J. Biol. Chem. 27940252-40254. [DOI] [PubMed] [Google Scholar]