Abstract
The envelope protein E of flaviviruses mediates both receptor-binding and membrane fusion. At the virion surface, 180 copies of E are tightly packed and organized in a herringbone-like icosahedral structure, whereas in noninfectious subviral particles, 60 copies are arranged in a T=1 icosahedral symmetry. In both cases, the basic building block is an E dimer which exposes the binding sites for neutralizing antibodies at its surface. It was the objective of our study to assess the dependence of the antigenic structure of E on its quaternary arrangement, i.e., as part of virions, recombinant subviral particles, or soluble dimers. For this purpose, we used a panel of 11 E protein-specific neutralizing monoclonal antibodies, mapped to distinct epitopes in each of the three E protein domains, and studied their reactivity with the different soluble and particulate forms of tick-borne encephalitis virus E protein under nondenaturing immunoassay conditions. Significant differences in the reactivities with these forms were observed that could be related to (i) limited access of certain epitopes at the virion surface; (ii) limited occupancy of epitopes in virions due to steric hindrance between antibodies; (iii) differences in the avidity to soluble forms compared to the virion, presumably related to the flexibility of E at its domain junctions; and (iv) modulations of the external E protein surface through interactions with its stem-anchor structure. We have thus identified several important factors that influence the antigenicity of the flavivirus E protein and have an impact on the interaction with neutralizing antibodies.
Flaviviruses form a genus in the family Flaviviridae (52) and comprise a number of important human pathogens such as yellow fever, dengue, Japanese encephalitis, West Nile, and tick-borne encephalitis (TBE) viruses (30). They are small, enveloped viruses with only three structural proteins, designated C (capsid), M (membrane), and E (envelope). The E protein is oriented parallel to the viral membrane and forms a head-to-tail homodimeric complex (Fig. 1A and B). The structure of the E ectodomain (soluble E [sE])—consisting of about 400 amino acids and lacking the 100 C-terminal amino acids (including the so-called stem and two transmembrane helices)—has been determined by X-ray crystallography for several flaviviruses (Fig. 1A) (25, 34, 36, 38, 44, 55). Both of the essential entry functions—receptor-binding and membrane fusion after uptake by receptor-mediated endocytosis—are mediated by E, which is therefore the primary target for virus-neutralizing antibodies (11, 42, 43, 45).
FIG. 1.
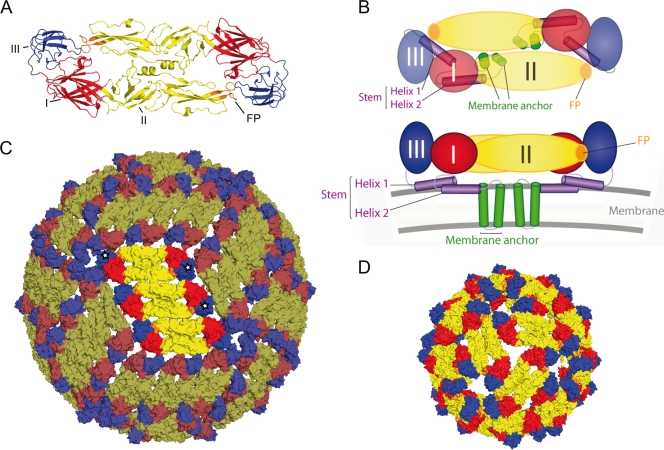
Structures and schematic representations of the TBE virus E protein, virions, and RSPs. In all panels, DI, DII, and DIII of the E protein are shown in red, yellow, and blue, respectively, and the fusion peptide (FP) is in orange. (A) Ribbon diagram of the sE dimer (top view). (B) Schematic of the full-length E dimer in a top view (upper panel) and side view (lower panel). The position of the two transmembrane helices of the membrane anchor and the two helices of the stem are based on Zhang et al. (54) and are shown in green and purple, respectively. (C) Pseudo-atomic structure of the virion based on cryo-EM reconstructions of dengue and West Nile viruses (27, 37, 54). One of the 30 rafts, each consisting of three parallel dimers, is highlighted. DIIIs of three monomers belonging to one icosahedral asymmetric unit are labeled by white stars. (D) Pseudo-atomic structure of RSP based on cryo-EM reconstructions (12).
As revealed by cryo-electron microscopy (cryo-EM), mature infectious virions have smooth surfaces, comparable to a golf ball (27, 37). Their envelopes are icosahedrally symmetric and consist of a closed shell of 180 E monomers that are arranged in a herringbone-like pattern of 30 rafts of three dimers each (Fig. 1C) (27). On the other hand, capsid-lacking subviral particles, which can be produced in recombinant form by the coexpression of prM and E, have a different symmetry, with 30 E dimers in a T=1 icosahedral structure (Fig. 1D) (12, 49).
The peculiar organization of E in virions is reminiscent of the tight packing of capsid proteins in nonenveloped viruses, for which it was shown that the native antigenic structure is strongly dependent on the intact capsid structure and not completely represented by isolated forms of capsid proteins (1, 41, 53). Such modulations of antigenic structure may be due to conformational changes in the course of packaging the capsid proteins into virions and/or to the fact that antibody binding sites at the virion surface are composed of residues that come together only through the juxtaposition of capsid proteins or neighboring protein subunits. Even in the case of spiky viral envelope proteins, the dependence of certain epitopes on the quaternary organization of the envelope glycoproteins has been described (8, 47).
For flaviviruses, structural studies provide evidence for the considerable flexibility of E, especially at the junctions between the individual domains I, II, and III (DI, DII, and DIII) (7, 35, 55), suggesting that soluble forms may display differences in antigenic structure compared to those fixed in the closed envelope shell of whole virions. Furthermore, because of the tight packing of E at the virion surface, certain epitopes may be cryptic in the context of whole virus particles but accessible in soluble forms of E (40, 51).
Studies on the antigenic structure of flaviviruses have used different antigen preparations including virions, recombinant subviral particles (RSPs), and soluble forms and subunits of E (10, 15-17, 32, 39, 40, 46, 49, 51), but so far no systematic comparative analysis of E in different physical forms and quaternary arrangements has been conducted. It was therefore the objective of our study, using TBE virus as a model, to investigate possible structural and/or antigenic differences between (i) soluble dimeric forms of E, including C-terminally truncated sE and detergent-solubilized full-length E (Fig. 1A and B); (ii) E in the context of whole virions (Fig. 1C); and (iii) E in the context of RSPs (Fig. 1D). For this purpose we used, and further characterized, a set of monoclonal antibodies (MAbs) directed to each of the three domains of E. All of these MAbs have neutralizing activity (17, 24) and therefore, by definition, react with infectious virions.
Through these analyses, we demonstrate that the reactivity of several MAbs is significantly dependent on the quaternary arrangement of E and differs between virions, RSPs, and/or sE dimers. We thus provide evidence for previously unrecognized structural factors that have an impact on the antigenicity of the flavivirus E protein.
MATERIALS AND METHODS
Production of TBE virus and sE dimers.
TBE virus strain Neudoerfl (33) was grown in primary chicken embryo cells, harvested 48 h after infection, concentrated by ultracentrifugation, and purified by rate-zonal sucrose density gradient centrifugation and equilibrium sucrose density gradient centrifugation, as described previously (19). This procedure led to the complete separation of virions from subviral particles. sE dimers were produced by limited digestion of purified TBE virions with trypsin at 0°C and subsequent purification by anion exchange chromatography (21).
Mutagenesis and production of subviral particles (RSPs).
For the generation of wild-type and mutant RSPs, we used the cDNA clone SV-PEwt (2) of TBE virus strain Neudoerfl, containing the prM and E genes flanked by parts of the C and NS1 genes, under the control of the simian virus 40 early promoter. Single-site mutations in E were introduced using a GeneTailor Site-Directed Mutagenesis System (Invitrogen). RSPs were produced by transfecting COS-1 cells (ATCC CRL 1650) with recombinant plasmids by electroporation (49). After 48 h, the RSPs in clarified cell culture supernatants were pelleted by ultracentrifugation. The pellet was resuspended in TAN buffer (50 mM triethanolamine, 100 mM NaCl, pH 8.0), and the concentration of E protein was determined by enzyme-linked immunosorbent assay (ELISA) after incubation for 30 min at 65°C in the presence of 0.4% sodium dodecyl sulfate (SDS) (22). The maturation state of RSPs (extent of prM cleavage) was analyzed by quantitative Western blotting as described previously (14).
Standardization of E protein contents in different antigen preparations.
The protein concentration of highly purified virus preparations (purity verified by SDS-polyacrylamide gel electrophoresis ) was determined according to Schaffner and Weissmann (48). E accounts for approximately 70% of the total proteins in virions. The E protein concentration in RSPs was determined by SDS-ELISA as described in Heinz et al. (22) using purified virions as a standard. Under the conditions of this assay, all E molecules are solubilized into monomers, and the result is therefore not influenced by possible differences in antigen recognition of the native E protein in virus and RSPs. Purified preparations of sE (purity verified by SDS-PAGE) were the same as used for crystallographic analysis (21, 44), and the concentration of sE was identical to the total protein concentration of the preparation, as determined according to Schaffner and Weissmann (48).
MAbs.
A panel of previously described TBE virus E protein-specific MAbs (17, 24) was used. Antibodies were purified from murine ascitic fluids using protein A-Sepharose High Performance columns (GE Healthcare Life Sciences) according to the manufacturer's recommendations. Fab fragments were generated from purified MAbs by papain cleavage as described previously (20) and purified by affinity chromatography (isotype immunoglobulin G2a [IgG2a]) or ion exchange chromatography (isotype IgG1).
Epitope mapping by four-layer ELISA.
A four-layer ELISA was carried out as described previously (49). Briefly, wild-type and mutant RSPs at a concentration of 0.5 μg/ml E protein were captured by guinea pig anti-TBE virus Ig-coated plates and incubated with predetermined dilutions of each of 22 neutralizing and nonneutralizing E protein-specific MAbs. For the detection of bound antibodies, we used peroxidase-labeled rabbit anti-mouse IgG. Connecting the absorbance values obtained with each of the MAbs yields characteristic reactivity profiles that distinguish wild-type and mutant RSPs.
Neutralization assay.
TBE virus neutralization assays were performed in baby hamster kidney cells (BHK-21) as described previously (51). Serial dilutions of MAbs were mixed with 10 times the 50% tissue culture infective dose of virus and incubated for 1 h at 37°C. BHK-21 cells were added, and incubation was continued for three days. The presence of virus in the supernatant was determined using a four-layer ELISA. The concentration of each MAb at which 50% neutralization was achieved (NT50) was determined by nonlinear regression analysis using a variable slope (GraphPad Prism, version 5; GraphPad Software Inc., San Diego, CA).
Blocking ELISA.
In the blocking ELISA, in principle performed as described by Stiasny et al. (51), equimolar concentrations of the antigen (E) in different physical forms (virion, RSP, and sE) were preincubated in serial dilutions with fixed concentrations of each MAb or Fab fragment. This step was carried out in the absence of detergent to maintain the lipid membranes of virions and RSPs. After incubation for 90 min at 37°C, the proportion of antibodies that were not bound to the antigens during this preincubation step was determined by transferring the mixtures to microtiter plates coated with 1 μg/ml purified TBE virus and detecting these residual antibodies by using peroxidase-labeled rabbit anti-mouse IgG as described previously (18). For the comparison of full-length E (produced by Triton X-100 solubilization of purified virus) and sE, the antigen-antibody preincubation mixtures contained 2% Tween 20 in order to prevent aggregation of the full-length E protein. In all instances, at the highest concentration of blocking antigen, the molar excess of E protein over MAb was at least 300-fold. The results were expressed as percent reactivity of MAb, defined as the quotient of absorbance in the presence of the blocking antigen and the absorbance in the absence of the blocking antigen: (A490 nm in the presence of blocking antigen/A490 nm in the absence of blocking antigen) × 100.
Determination of antibody avidities in blocking ELISA.
For estimating the avidity of MAbs and Fab fragments with E protein-containing TBE virus antigens, the dissociation constant in solution (KD) was calculated using the data of the blocking ELISA according to a method described by Friguet et al. (13) and modified by F. J. Stevens (50) and S. A. Bobrovnik (5). Briefly, the association constant in solution (KA) was calculated by the use of the following equation: li × KA = (A0 − Ai)/Ai + 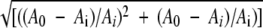 (5), where li is the antigen concentration at each measurement point of the blocking ELISA, A0 is the absorbance at 490 nm in the absence of blocking antigen, and Ai is the absorbance at 490 nm in the presence of blocking antigen at each measurement point of the blocking ELISA. The slope of this linear relationship corresponds to KA, while KD was calculated using the equation KD = 1/KA. At least three independent experiments were used for the calculation of the mean KD as well as the corresponding 95% confidence intervals for each MAb-antigen combination. Two selected examples of such an analysis are shown in the Fig. S1 in the supplemental material.
(5), where li is the antigen concentration at each measurement point of the blocking ELISA, A0 is the absorbance at 490 nm in the absence of blocking antigen, and Ai is the absorbance at 490 nm in the presence of blocking antigen at each measurement point of the blocking ELISA. The slope of this linear relationship corresponds to KA, while KD was calculated using the equation KD = 1/KA. At least three independent experiments were used for the calculation of the mean KD as well as the corresponding 95% confidence intervals for each MAb-antigen combination. Two selected examples of such an analysis are shown in the Fig. S1 in the supplemental material.
Determination of antibody occupancy.
For determining the number of MAbs bound per virion at saturation conditions, 5 μg of purified TBE virus was incubated with serial dilutions of MAbs or Fabs (5-, 10-, 15-, and 20-fold molar excess of antibody molecules) overnight at 4°C in a total volume of 100 μl. The virion-antibody complexes were subjected to ultracentrifugation (2 h at 35,000 rpm in a Beckmann SW 40 rotor) through a 2-ml sucrose cushion (10% sucrose in TAN buffer, pH 8.0). The pellet was resuspended in SDS-gel sample buffer and analyzed by SDS-PAGE according to Laemmli (28). The proteins were stained with Coomassie brilliant blue, and the relative intensity of the corresponding bands was measured by densitometry using Scion image software. Quantitative evaluations were based on standard curves of the E protein as well as of the MAb A4. Evidence for saturation was obtained by the demonstration that an increase in MAb concentration did not result in a further increase of bound antibodies. Two selected examples are shown in the Fig S2 in the supplemental material.
Statistical analysis.
Data were analyzed with GraphPad Prism, version 5 (GraphPad Software Inc., San Diego, CA). Two-tailed t tests were used to compare ELISA avidities, which were considered significantly different when the P value was less than 0.05.
RESULTS
Epitope mapping of neutralizing MAbs by mutagenesis of RSPs.
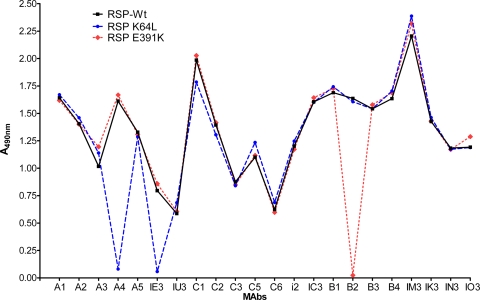
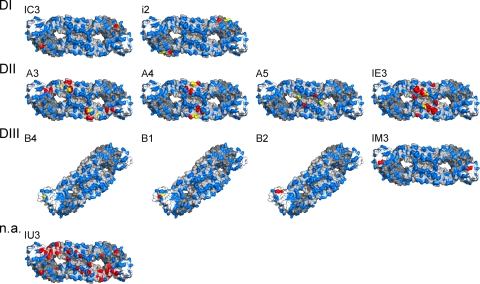
In this study, we used a set of 11 TBE virus E protein-specific and neutralizing antibodies, as shown in Table 1. In the case of 8 of the 11 antibodies, information on the location of the corresponding binding sites in E had been obtained previously employing sequence analysis of neutralization escape mutants (23, 32). In order to obtain a more complete picture of these eight epitopes and also of the three additional epitopes defined by the antibodies B2, IU3, and IM3, we used a mutagenesis approach employing RSPs and exchanged surface-exposed residues all over the external surface of E. A total of 51 individual mutations were introduced (Table 2) that affected neither secretion nor maturation of RSPs (see Materials and Methods). The reactivity of the RSP mutants with each of the 11 antibodies was tested in a four-layer ELISA (see Materials and Methods). Two examples of the reactivity profiles obtained are shown in Fig. 2, and all of the results are summarized graphically in Fig. 3. Overall, most of the mutations affecting MAb binding clustered at sites compatible with the size of individual Fab footprints (6, 29). The mutagenesis approach thus provided additional valuable information on the location and, in some cases, the extension of the epitopes. However, in three instances (MAbs A3, IE3, and IU3), mutations affecting the binding of the MAb were too distant to be part of the same Fab footprint. MAb IU3 presents an extreme example of this category as mutations affecting its binding form a continuous patch along DII and also extend to adjacent regions in DI and DIII. Therefore, indirect conformational effects caused by these mutations have to be postulated, and the nature and precise location of this specific epitope remain undefined.
TABLE 1.
TBE virus neutralizing MAbs with domain specificity and corresponding escape mutations
TABLE 2.
Amino acid residues of E mutated in TBE virus RSPs
| Domain | Amino acid mutations in RSPs |
|---|---|
| DI | R2D, A47K, D149E, T150K, E155A, S158N, T162G, S164D, T166E, I173S, T175K, K296R, K298R, K300R |
| DII | N52E, K64L, S66K, D67G, K69R, A71V, R73D, M77K, A83K, Q87D, H104G, L107F, K118D, A120K, E122G, A123K, K124D, K126R, K136A, K204D, T205D, E207K, E230K, G231T, Q233A, V250I, E277S |
| DIII | T303S, T305S, R316L, T331K, H347E, N366A, N367A, Y384A, H390K, E391K |
FIG. 2.
Binding profiles of monoclonal antibodies in four-layer ELISA. The reactivities of 22 neutralizing and nonneutralizing MAbs specific for the TBE virus E protein were analyzed with wild-type (Wt) RSPs, mutant K64L, and mutant E391K.
FIG. 3.
Surface representations of the TBE virus sE dimer and binding sites of 11 neutralizing MAbs, as indicated at the top of each structure. The position of all 51 amino acids replaced for epitope mapping by mutagenesis (Table 2) are highlighted in blue, those affecting the binding of individual MAbs are shown in red, and mutations found in neutralization escape mutants (Table 1) are shown in yellow. The domain specificities of the antibodies (DI, DII, DIII, and not assignable [n.a.]) are indicated on the left.
Antigenic reactivity of MAbs with virions, RSPs, and sE in blocking ELISA.
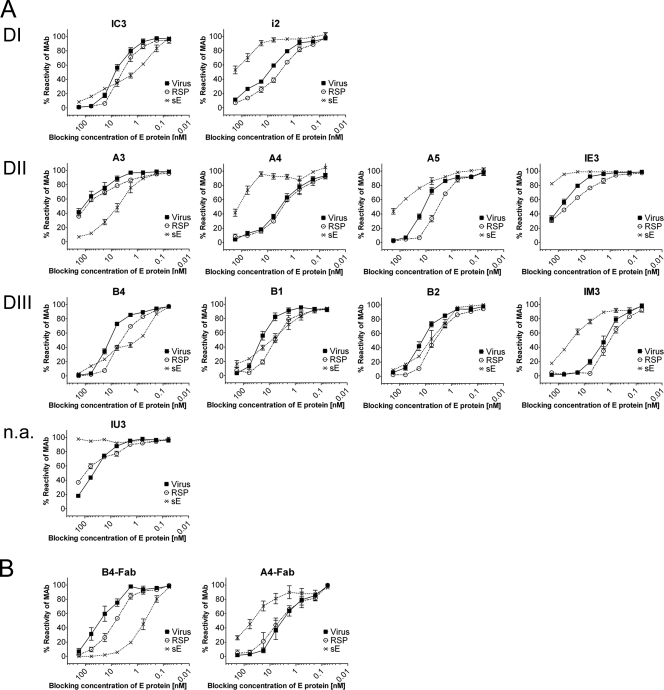
To assess possible antigenic differences between E in soluble and in particulate forms, the reactivities of each of the MAbs with serial dilutions of purified preparations of virions, RSPs, and sE were standardized to equimolar concentrations of E, as described in Materials and Methods. The antigenic reactivities of these standardized preparations were compared in a blocking ELISA format in the absence of detergent to maintain the structural integrity of virions and RSPs (see Materials and Methods) (Fig. 4). The information contained in the blocking curves displayed in Fig. 4 was also used to calculate the relative ELISA avidity of each MAb with respect to its binding to the virus, RSP, and sE (see Materials and Methods) (Table 3).
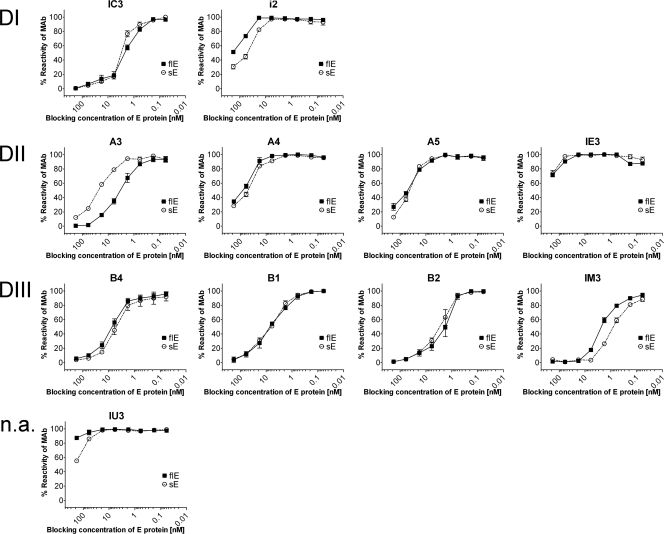
FIG. 4.
Blocking ELISA in the absence of detergent with TBE virus, RSPs, and sE dimers (standardized to equimolar concentrations of E) and 11 TBE virus-neutralizing MAbs (A) as well as two Fab fragments (B). The domain specificities of the antibodies (DI, DII, and DIII and not assignable [n.a.]) are indicated on the left side, and the designations of the antibodies are indicated at the top of each panel. Serial dilutions of the antigens were preincubated with a predetermined concentration of the respective MAb, and the fraction of MAb not blocked by its reaction with antigen was detected in ELISA with TBE virus-coated plates (see Materials and Methods). The data shown represent the mean of three independent experiments, and the standard error of the mean is indicated by error bars.
TABLE 3.
Neutralizing activities of MAbs and their ELISA avidities to different forms of the TBE virus E protein
| MAb or MAb-Fabg | E domain | NT50 (μg/ml)a | ELISA reactivityb
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Virus
|
RSP
|
sE dimer
|
||||||||
| KD (nM) | 95% CI (nM) | KD (nM) | 95% CI (nM) | P (virion vs RSP)c | KD (nM) | 95% CI (nM) | P (virion vs sE dimer)c | |||
| MAb | ||||||||||
| IC3 | DI | 3.5e | 1.0 | 0.6-1.5 | 1.6 | 0.9-2.2 | NS | NC | NC | |
| i2 | DI | 10.1e | 11.6 | 10.0-13.2 | 6.6 | 4.7-8.5 | 0.007 | 74.6 | 36.1-113.0 | 0.004 |
| A3 | DII | 23.3e | 41.0 | 39.3-42.6 | 50.1 | 36.0-64.2 | NS | 4.8 | 4.6-5.0 | <0.0001 |
| A4 | DII | 1.6e | 4.4 | 3.2-5.6 | 4.6 | 4.2-5.1 | NS | 54.9 | 39.5-68.3 | <0.0001 |
| A5 | DII | 41.1e | 2.5 | 2.0-3.1 | 3.0 | 1.5-4.6 | NS | 57.2 | 43.7-70.7 | <0.0001 |
| IE3 | DII | 273.1f | 40.0 | 23.4-55.9 | 44.0 | 25.8-62.9 | NS | >100 | ||
| B4 | DIII | 70.9f | 1.4 | 0.9-1.9 | 1.5 | 1.2-1.8 | NS | NC | NC | |
| B1 | DIII | 21.9f | 3.0 | 0.3.6-5.6 | 2.8 | 2.0-3.6 | NS | 10.8 | 8.9-12.7 | 0.0005 |
| B2 | DIII | 9.6f | 4.9 | 3.0-6.8 | 2.0 | 1.8-2.3 | 0.0029 | 5.9 | 3.0-8.9 | NS |
| IM3 | DIII | 77.9f | 3.0 | 2.3-3.6 | 2.9 | 2.2-3.7 | NS | 25.4 | 17.6-33.3 | 0.0003 |
| IU3 | NAd | 15.3f | 15.8 | 13.4-18.2 | NC | NC | >100 | |||
| MAb-Fab | ||||||||||
| A4-Fab | 2.1 | 1.3-2.8 | 1.4 | 1.3-1.5 | NS | 29.1 | 17.5-42.5 | 0.0065 | ||
| B4-Fab | 5.2 | 3.9-6.5 | 4.5 | 1.2-7.7 | NS | 0.29 | 0.28-0.30 | <0.0001 | ||
Concentration of antibodies at which 50% neutralization was achieved.
KD and 95% confidence interval (CI) values are the means of at least three independent experiments. NC, not calculable due to an irregular binding curve.
KD values were compared by t tests. P values of <0.05 were rated as significant and are highlighted in bold. NS, not significant.
NA, not assignable to a single domain.
Data are the means of three independent experiments.
Data are the means of two independent experiments.
MAb-Fab, Fab fragment of a specific MAb.
(i) Comparison of the MAb reactivities with virions and RSPs.
With 8 of the 11 MAbs (A5, B1, B2, and B4 and, to a lesser extent, IE3, IC3, i2, and IM3) the blocking capacity of RSPs was higher than that of the virus despite the presence of equimolar concentrations of E. Virus and RSPs yielded identical or nearly identical results (for MAbs A4, A3, and IU3) only in a few instances. For MAbs B2 and i2, the higher blocking activity of RSPs might be explained by the higher ELISA avidity of these MAbs with RSPs than with virions (Table 3). This explanation, however, cannot apply to the other MAbs because they have identical ELISA avidities with both particulate forms (Table 3). It is therefore suggested that the reactivity differences observed in the blocking ELISA are due to the fact that the occupancy of E molecules in virions was lower than in RSPs in these cases.
(ii) Comparison of the MAb reactivities with virions and sE.
Two distinct patterns were obtained: MAbs A4 (including its Fab fragment), A5, IE3, IU3, i2, and IM3 exhibited both lower reactivities and lower avidities to sE than to virions (Fig. 4 and Table 3), thus providing evidence for differences in the structure of these epitopes. However, for the remaining MAbs (MAbs A3, IC3, B4, B1, and B2) sE had stronger blocking activity than virions, especially at low concentrations of E (Fig. 4A). In the specific case of A3, this was consistent with an approximately eightfold higher avidity to sE than to virions (Table 3). Avidity calculations were not possible for MAbs IC3 and B4 due to bisigmoidally shaped dose-response curves of sE, indicating differences in avidity at different E/IgG ratios (Fig. 4A). Such a pattern might be caused by antibody-mediated cross-linking of different sE dimers at high concentrations. To assess this hypothesis, a comparison was undertaken of the binding characteristics of the whole MAb B4 with that of its Fab fragment. As can be seen from Fig. 4B, the Fab displayed no irregularities in the sigmoidal binding curve and thus allowed the estimation of Fab affinity, which was significantly higher with sE than with whole virus (Table 3). In contrast to MAb A3 and Fab B4, MAbs B1 and B2 had lower and similar avidities for sE compared to virions, respectively, but nevertheless were blocked more efficiently by sE than by virions at low antigen concentrations. This comparison suggests a submaximal occupancy of potential binding sites for these two antibodies on virions.
Occupancy of virions at antibody saturation.
Since some of the results obtained in the binding experiments might best be interpreted by a submaximal occupancy of epitopes on virus particles, the maximum achievable occupancy at antibody saturation was determined for 9 of the 11 MAbs (no conclusive results were obtained with MAbs i2 and IE3). For that purpose, purified virus preparations were mixed with serial dilutions of antibodies, and virus-MAb complexes were separated from unbound antibodies by ultracentrifugation through a sucrose cushion. The pelleted material was subjected to SDS-PAGE and Coomassie staining, and the ratio of E to MAb in the immunocomplexes was determined densitometrically (see Materials and Methods). The results (Table 4) revealed that, depending on the MAb, only ∼30 to 90 antibody molecules were bound per virion at saturation. This limitation in the occupancy of epitopes could most likely be explained by the tight packing of the E proteins at the virion surface and steric hindrance between the relatively bulky whole antibody molecules. If this interpretation is valid, a higher degree of occupancy should be achievable with the corresponding Fab fragment in such cases. This proved to be the case with Fab A4 (used as an example), which was able to occupy not only 90 but 150 of the total 180 theoretical binding sites present per virion (Table 4).
TABLE 4.
Average number of IgG molecules bound per TBE virion under saturation conditions (occupancy)
| MAb or Mab-Faba | E domain | No. of bound IgG molecules per virionc |
|---|---|---|
| MAb | ||
| IC3 | DI | 34.3 ± 4.0 |
| i2 | DI | — |
| A3 | DII | 57.5 ± 0.7 |
| A4 | DII | 88.2 ± 4.5 |
| A5 | DII | 26.3 ± 4.6 |
| IE3 | DII | — |
| B4 | DIII | 44.7 ± 5.3 |
| B1 | DIII | 34.8 ± 5.2 |
| B2 | DIII | 37.8 ± 2.9 |
| IM3 | DIII | 35.6 ± 4.4 |
| IU3 | NAb | 37.0 ± 11.3 |
| MAb-Fab | ||
| A4-Fab | 150.0 ± 38.6 |
MAb-Fab, Fab fragment of a specific MAb.
NA, not assignable to a single domain.
Data shown in this table are the results from at least four independent experiments (mean ± standard deviation). —, no conclusive results obtained.
Comparison of full-length and truncated E dimers.
The differences observed between virus and sE might be related not only to the quaternary arrangement of E at the virion surface and/or differences in the accessibility of epitopes but also to the lack of the stem-anchor region in sE. Therefore, a comparative analysis of full-length and sE dimers in a blocking assay format was undertaken, similar to that used for the experiments depicted in Fig. 4A except for the addition of the detergent Tween 20, necessary for keeping full-length E in solution (Fig. 5). Although 6 of the 11 MAbs reacted identically with both forms of E, significant differences in reactivity and avidity were observed with MAbs A3, IM3, and i2. MAbs IU3 and IE3 did not yield conclusive results because of their low reactivity. Two patterns were observed with the MAbs that differentiated between the two forms of E. A3 had a much higher blocking activity and ELISA avidity for the truncated protein (KD of 3.4 ± 0.3 nM) than for the full-length E (KD of 24.4 ± 4.3 nM), and the opposite was true for the MAbs IM3 (KD of 46.3 ± 12.3 nM for sE and 2.7 ± 0.1 nM for full-length E) and i2 (KD of >100 nM for sE and 52.3 ± 3.2 nM for full-length E). These data indicate that the presence of the stem-anchor region has a significant impact on the antigenic structure of the E protein ectodomain.
FIG. 5.
Blocking ELISA in the presence of detergent with TBE virus sE dimers and full-length E dimers (solubilized from virions with Triton X-100) and 11 TBE virus-neutralizing MAbs. The domain specificities of the antibodies (DI, DII, and DIII and not assignable [n.a.]) are indicated on the left side, and the designations of the antibodies are indicated at the top of each panel. Serial dilutions of the antigens were preincubated with a predetermined concentration of the respective MAb, and the fraction of MAb not blocked by its reaction with antigen was detected in an ELISA with TBE virus-coated plates (see Materials and Methods). The data shown represent the mean of three independent experiments, and the standard error of the mean is indicated by error bars.
DISCUSSION
In this work, we have investigated the dependence of the antigenic structure of the TBE virus E protein on its quaternary arrangement. Using 11 E-specific MAbs as probes, we found significant differences in the antigenicity of soluble and particulate forms of E. The phenomena observed could be related to several factors including epitope accessibility, structural flexibility of E, and different degrees of occupancy by antibodies.
It is an important finding of our study that some of the antibodies displayed a higher avidity for sE than for E in the context of virions (and to a certain extent also RSPs). This was especially striking in the case of MAb A3, which reacts with an epitope at the edge of DII, and for Fab B4, which reacts with the lateral side of DIII (Fig. 3). In both cases, the results obtained can best be explained by a limited accessibility of these epitopes in the context of particles due to lateral contacts between E dimers that characterize the specific icosahedral arrangements of particulate forms (Fig. 1C and D). Such a partially cryptic nature has been described for other epitopes of the flavivirus E protein, including those involving the highly conserved fusion peptide loop at the tip of DII (40, 51). Avidity, however, would not be influenced in a situation in which a subset of a given MAb-defined epitope is totally cryptic while the rest is fully accessible for antibody binding, as described for DIII-specific epitopes at the fivefold symmetry axis of virions (26).
The differences in avidity observed between soluble and particulate forms could also reflect true structural differences of E that relate to its flexibility at the junctions between the three domains and the constraints imposed on individual E dimers through their tight packing in virions. The best evidence for such structural differences was obtained with MAb A4 and its derived Fab fragment, both of which have an at least 10-fold higher affinity for E as part of virions or RSPs than as sE (Table 3). This epitope has been mapped to the edge of DII by both mutagenesis and neutralization escape approaches (Fig. 3 and Table 1), and one could argue that the full epitope in virions is composed of amino acid residues from different E subunits, brought together through their specific quaternary arrangement in virions. Such “overlapping” contributions would of course be absent in sE and could provide an alternative explanation for the avidity/affinity differences observed with the two forms. If this were true, the reactivity of virions and RSPs should also be different because the arrangement of E dimers, and therefore the E dimer contacts in these two particulate structures, are unrelated (herringbone-like in virions and T=1 icosahedral in RSPs) (Fig. 1C and D). However, the binding curves of MAb A4 and Fab A4 and their deduced avidities/affinities with virions and RSPs are virtually identical. These data therefore suggest true structural differences between sE and E in particles, probably due to the flexibility of E at its domain interfaces (7, 35, 55).
The higher avidity of some MAbs/Fabs for RSPs than virions may indeed be related to the different icosahedral structures and chemical environments of E dimers in these particles. Examples are MAb B2 and Fab B4 (both directed to the lateral side of DIII) and also MAb i2 that has been mapped to the side of DI (Fig. 3). Cryo-EM has demonstrated an extremely tight packing of E dimers at the surface of virions (27, 37), whereas a somewhat looser arrangement has been revealed for RSPs (12). We therefore propose that, even though the E dimer building blocks are the same in virions and RSPs, the differences in quaternary arrangement account for the variations in antigenic reactivity observed between the two forms. Alternatively, the dynamic nature of the flavivirus surface, as first demonstrated for mature dengue virus particles (31), could also account for some of the differences observed.
All of the atomic structures of flavivirus E proteins determined so far consist of the ectodomain only and lack the so-called stem and the double membrane anchor (Fig. 1A). Although cryo-EM revealed that the stem-anchor of the protein is not accessible at the virion surface (54), our data provide evidence for a contribution of these sequences to the reactivity of some neutralizing MAbs. Specifically, the DIII-specific MAb IM3 and the DI-specific MAb i2 had significantly stronger reactions with the full-length protein, whereas, in contrast, the DII-specific MAb A3 had much higher avidity for the truncated form. The structural basis of these observations is not clear at present and requires the structural determination of the full-length flavivirus E protein. Given the flexibility of E at its domain junctions, it is quite possible, however, that interactions of the stem-anchor at the bottom of the molecule can have subtle effects on the hinges between the domains and thus influence epitopes recognized by antibodies at the surface.
Blocking ELISAs as performed in this study offer the advantage that the antigen-antibody reaction takes place in solution and is therefore not influenced by denaturation artifacts through the coating of antigens to solid phases. However, it is important to note that the extent of blocking not only is a function of antibody avidity but can also reflect differences in the antibody occupancy of the antigens compared. This was clearly shown through the results obtained with some MAbs that, although they had a higher avidity for the virus, were blocked significantly better by equimolar concentrations of sE. Indeed, a careful analysis of the number of antibody molecules bound to virions under different antibody/virus ratios revealed that only between ∼30 and 90 molecules, depending on the specific antibody, could be bound to the available 180 epitopes per virion at saturation. One explanation for this limitation is certainly steric hindrance between the bulky antibody molecules. Consistent with this interpretation is the observation that this number could be significantly increased through the use of the corresponding Fab fragment (MAb A4). In addition to such steric hindrance factors, however, the inaccessibility of a subfraction of the potential 180 epitopes at the virion surface (e.g., at the fivefold symmetry axis) (26, 31) could also contribute to the differences observed. In any case, comparative antigenic analyses of the same protein at equimolar concentrations, with the protein presented in different quaternary arrangements and/or physical forms, have to be treated with great caution in order to avoid misleading interpretations.
RSPs have been valuable tools for studying the properties of flavivirus glycoproteins, especially with respect to particle assembly (4), membrane fusion (3, 9, 14, 49), and antigenic structure (3, 49). As part of the present work, we have also carried out an extensive mutagenesis study with TBE virus RSPs to delineate the epitopes of the 11 neutralizing MAbs used. While in most cases the results were consistent with the possible sizes of Fab-covered areas on protein antigens, some antibodies were affected by mutations at quite distant locations on the E protein surface. In such instances, indirect effects of the mutations on the corresponding epitope have to be postulated. The most extreme example in this context was that of MAb IU3 (Fig. 3). Its reactivity was abolished or impaired by mutations in all three E protein domains, making a location of the actual binding site by the use of this technology virtually impossible. In the absence of other information (e.g., from neutralization escape mutations and/or structures of antibody-antigen complexes), the data obtained in such mutagenesis approaches can therefore lead to erroneous interpretations.
In conclusion, our data provide experimental evidence for several factors that modulate the antigenic structure of the flavivirus E protein. These are most likely related to the flexibility of E at its domain junctions, influences of the stem-anchor, and constraints imposed on E through its assembly into particles. The differences in antigenic reactivity observed between particulate and soluble forms of E are especially relevant with respect to the standardization of antigenic contents and immunogenicity of vaccines that contain different physical forms of flavivirus antigens.
Supplementary Material
Acknowledgments
We thank Walter Holzer for help with virus production and the generation of sE dimers and Gabriel O'Riordain for critical reading of the manuscript.
This work was supported by the Austrian Science Fund (Fonds zur Foerderung der wissenschaftlichen Forschung), FWF project number P17035-B09.
Footnotes
Published ahead of print on 24 June 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Acharya, R., E. Fry, D. Stuart, G. Fox, D. Rowlands, and F. Brown. 1989. The three-dimensional structure of foot-and-mouth disease virus at 2.9 Å resolution. Nature 337709-716. [DOI] [PubMed] [Google Scholar]
- 2.Allison, S. L., C. W. Mandl, C. Kunz, and F. X. Heinz. 1994. Expression of cloned envelope protein genes from the flavivirus tick-borne encephalitis virus in mammalian cells and random mutagenesis by PCR. Virus Genes 8187-198. [DOI] [PubMed] [Google Scholar]
- 3.Allison, S. L., J. Schalich, K. Stiasny, C. W. Mandl, and F. X. Heinz. 2001. Mutational evidence for an internal fusion peptide in flavivirus envelope protein E. J. Virol. 754268-4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allison, S. L., Y. J. Tao, G. O'Riordain, C. W. Mandl, S. C. Harrison, and F. X. Heinz. 2003. Two distinct size classes of immature and mature subviral particles from tick-borne encephalitis virus. J. Virol. 7711357-11366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bobrovnik, S. A. 2003. Determination of antibody affinity by ELISA. Theory. J. Biochem. Biophys. Methods 57213-236. [DOI] [PubMed] [Google Scholar]
- 6.Braden, B. C., and R. J. Poljak. 1995. Structural features of the reactions between antibodies and protein antigens. FASEB J. 99-16. [DOI] [PubMed] [Google Scholar]
- 7.Bressanelli, S., K. Stiasny, S. L. Allison, E. A. Stura, S. Duquerroy, J. Lescar, F. X. Heinz, and F. A. Rey. 2004. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J. 23728-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, L. E., J. M. Murray, D. O. White, and D. C. Jackson. 1990. An analysis of the properties of monoclonal antibodies directed to epitopes on influenza virus hemagglutinin. Arch. Virol. 1141-26. [DOI] [PubMed] [Google Scholar]
- 9.Corver, J., A. Ortiz, S. L. Allison, J. Schalich, F. X. Heinz, and J. Wilschut. 2000. Membrane fusion activity of tick-borne encephalitis virus and recombinant subviral particles in a liposomal model system. Virology 26937-46. [DOI] [PubMed] [Google Scholar]
- 10.Crill, W. D., N. B. Trainor, and G. J. Chang. 2007. A detailed mutagenesis study of flavivirus cross-reactive epitopes using West Nile virus-like particles. J. Gen. Virol. 881169-1174. [DOI] [PubMed] [Google Scholar]
- 11.Diamond, M. S., T. C. Pierson, and D. H. Fremont. 2008. The structural immunology of antibody protection against West Nile virus. Immunol. Rev. 225212-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferlenghi, I., M. Clarke, T. Ruttan, S. L. Allison, J. Schalich, F. X. Heinz, S. C. Harrison, F. A. Rey, and S. D. Fuller. 2001. Molecular organization of a recombinant subviral particle from tick-borne encephalitis virus. Mol. Cell 7593-602. [DOI] [PubMed] [Google Scholar]
- 13.Friguet, B., A. F. Chaffotte, L. Djavadi-Ohaniance, and M. E. Goldberg. 1985. Measurements of the true affinity constant in solution of antigen-antibody complexes by enzyme-linked immunosorbent assay. J. Immunol. Methods 77305-319. [DOI] [PubMed] [Google Scholar]
- 14.Fritz, R., K. Stiasny, and F. X. Heinz. 2008. Identification of specific histidines as pH sensors in flavivirus membrane fusion. J. Cell Biol. 183353-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goncalvez, A. P., R. H. Purcell, and C. J. Lai. 2004. Epitope determinants of a chimpanzee Fab antibody that efficiently cross-neutralizes dengue type 1 and type 2 viruses map to inside and in close proximity to fusion loop of the dengue type 2 virus envelope glycoprotein. J. Virol. 7812919-12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gromowski, G. D., and A. D. Barrett. 2007. Characterization of an antigenic site that contains a dominant, type-specific neutralization determinant on the envelope protein domain III (ED3) of dengue 2 virus. Virology 366349-360. [DOI] [PubMed] [Google Scholar]
- 17.Guirakhoo, F., F. X. Heinz, and C. Kunz. 1989. Epitope model of tick-borne encephalitis virus envelope glycoprotein E: analysis of structural properties, role of carbohydrate side chain, and conformational changes occurring at acidic pH. Virology 16990-99. [DOI] [PubMed] [Google Scholar]
- 18.Heinz, F. X., R. Berger, O. Majdic, W. Knapp, and C. Kunz. 1982. Monoclonal antibodies to the structural glycoprotein of tick-borne encephalitis virus. Infect. Immun. 37869-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heinz, F. X., and C. Kunz. 1981. Homogeneity of the structural glycoprotein from European isolates of tick-borne encephalitis virus: comparison with other flaviviruses. J. Gen. Virol. 57263-274. [DOI] [PubMed] [Google Scholar]
- 20.Heinz, F. X., C. Mandl, R. Berger, W. Tuma, and C. Kunz. 1984. Antibody-induced conformational changes result in enhanced avidity of antibodies to different antigenic sites on the tick-borne encephalitis virus glycoprotein. Virology 13325-34. [DOI] [PubMed] [Google Scholar]
- 21.Heinz, F. X., C. W. Mandl, H. Holzmann, C. Kunz, B. A. Harris, F. Rey, and S. C. Harrison. 1991. The flavivirus envelope protein E: isolation of a soluble form from tick-borne encephalitis virus and its crystallization. J. Virol. 655579-5583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heinz, F. X., K. Stiasny, G. Puschner-Auer, H. Holzmann, S. L. Allison, C. W. Mandl, and C. Kunz. 1994. Structural changes and functional control of the tick-borne encephalitis virus glycoprotein E by the heterodimeric association with protein prM. Virology 198109-117. [DOI] [PubMed] [Google Scholar]
- 23.Holzmann, H., K. Stiasny, M. Ecker, C. Kunz, and F. X. Heinz. 1997. Characterization of monoclonal antibody-escape mutants of tick-borne encephalitis virus with reduced neuroinvasiveness in mice. J. Gen. Virol. 7831-37. [DOI] [PubMed] [Google Scholar]
- 24.Holzmann, H., K. Stiasny, H. York, F. Dorner, C. Kunz, and F. X. Heinz. 1995. Tick-borne encephalitis virus envelope protein E-specific monoclonal antibodies for the study of low pH-induced conformational changes and immature virions. Arch. Virol. 140213-221. [DOI] [PubMed] [Google Scholar]
- 25.Kanai, R., K. Kar, K. Anthony, L. H. Gould, M. Ledizet, E. Fikrig, W. A. Marasco, R. A. Koski, and Y. Modis. 2006. Crystal structure of West Nile virus envelope glycoprotein reveals viral surface epitopes. J. Virol. 8011000-11008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaufmann, B., G. E. Nybakken, P. R. Chipman, W. Zhang, M. S. Diamond, D. H. Fremont, R. J. Kuhn, and M. G. Rossmann. 2006. West Nile virus in complex with the Fab fragment of a neutralizing monoclonal antibody. Proc. Natl. Acad. Sci. USA 10312400-12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuhn, R. J., W. Zhang, M. G. Rossmann, S. V. Pletnev, J. Corver, E. Lenches, C. T. Jones, S. Mukhopadhyay, P. R. Chipman, E. G. Strauss, T. S. Baker, and J. H. Strauss. 2002. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laemmli, U. K., and M. Favre. 1973. Maturation of the head of bacteriophage T4. I. DNA packaging events. J. Mol. Biol. 80575-599. [DOI] [PubMed] [Google Scholar]
- 29.Li, Y., H. Li, F. Yang, S. J. Smith-Gill, and R. A. Mariuzza. 2003. X-ray snapshots of the maturation of an antibody response to a protein antigen. Nat. Struct. Biol. 10482-488. [DOI] [PubMed] [Google Scholar]
- 30.Lindenbach, B. D., H. J. Thiel, and C. M. Rice. 2006. Flaviviridae: the viruses and their replication, p. 1101-1152. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 31.Lok, S. M., V. Kostyuchenko, G. E. Nybakken, H. A. Holdaway, A. J. Battisti, S. Sukupolvi-Petty, D. Sedlak, D. H. Fremont, P. R. Chipman, J. T. Roehrig, M. S. Diamond, R. J. Kuhn, and M. G. Rossmann. 2008. Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins. Nat. Struct. Mol. Biol. 15312-317. [DOI] [PubMed] [Google Scholar]
- 32.Mandl, C. W., F. Guirakhoo, H. Holzmann, F. X. Heinz, and C. Kunz. 1989. Antigenic structure of the flavivirus envelope protein E at the molecular level, using tick-borne encephalitis virus as a model. J. Virol. 63564-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mandl, C. W., F. X. Heinz, and C. Kunz. 1988. Sequence of the structural proteins of tick-borne encephalitis virus (western subtype) and comparative analysis with other flaviviruses. Virology 166197-205. [DOI] [PubMed] [Google Scholar]
- 34.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2003. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl. Acad. Sci. USA 1006986-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2004. Structure of the dengue virus envelope protein after membrane fusion. Nature 427313-319. [DOI] [PubMed] [Google Scholar]
- 36.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2005. Variable surface epitopes in the crystal structure of dengue virus type 3 envelope glycoprotein. J. Virol. 791223-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mukhopadhyay, S., B. S. Kim, P. R. Chipman, M. G. Rossmann, and R. J. Kuhn. 2003. Structure of West Nile virus. Science 302248. [DOI] [PubMed] [Google Scholar]
- 38.Nybakken, G. E., C. A. Nelson, B. R. Chen, M. S. Diamond, and D. H. Fremont. 2006. Crystal structure of the West Nile virus envelope glycoprotein. J. Virol. 8011467-11474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oliphant, T., M. Engle, G. E. Nybakken, C. Doane, S. Johnson, L. Huang, S. Gorlatov, E. Mehlhop, A. Marri, K. M. Chung, G. D. Ebel, L. D. Kramer, D. H. Fremont, and M. S. Diamond. 2005. Development of a humanized monoclonal antibody with therapeutic potential against West Nile virus. Nat. Med. 11522-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oliphant, T., G. E. Nybakken, M. Engle, Q. Xu, C. A. Nelson, S. Sukupolvi-Petty, A. Marri, B. E. Lachmi, U. Olshevsky, D. H. Fremont, T. C. Pierson, and M. S. Diamond. 2006. Antibody recognition and neutralization determinants on domains I and II of West Nile virus envelope protein. J. Virol. 8012149-12159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Page, G. S., A. G. Mosser, J. M. Hogle, D. J. Filman, R. R. Rueckert, and M. Chow. 1988. Three-dimensional structure of poliovirus serotype 1 neutralizing determinants. J. Virol. 621781-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pierson, T. C., and M. S. Diamond. 2008. Molecular mechanisms of antibody-mediated neutralisation of flavivirus infection. Expert Rev. Mol. Med. 10e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pierson, T. C., D. H. Fremont, R. J. Kuhn, and M. S. Diamond. 2008. Structural insights into the mechanisms of antibody-mediated neutralization of flavivirus infection: implications for vaccine development. Cell Host Microbe 4229-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rey, F. A., F. X. Heinz, C. Mandl, C. Kunz, and S. C. Harrison. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature 375291-298. [DOI] [PubMed] [Google Scholar]
- 45.Roehrig, J. T. 2003. Antigenic structure of flavivirus proteins. Adv. Virus Res. 59141-175. [DOI] [PubMed] [Google Scholar]
- 46.Roehrig, J. T., R. A. Bolin, and R. G. Kelly. 1998. Monoclonal antibody mapping of the envelope glycoprotein of the dengue 2 virus, Jamaica. Virology 246317-328. [DOI] [PubMed] [Google Scholar]
- 47.Saito, T., G. Taylor, W. G. Laver, Y. Kawaoka, and R. G. Webster. 1994. Antigenicity of the N8 influenza A virus neuraminidase: existence of an epitope at the subunit interface of the neuraminidase. J. Virol. 681790-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schaffner, G. and C. Weissmann. 1973. A rapid, sensitive, and specific method for the determination of protein in dilute solution. Anal. Biochem. 56502-514. [DOI] [PubMed] [Google Scholar]
- 49.Schalich, J., S. L. Allison, K. Stiasny, C. W. Mandl, C. Kunz, and F. X. Heinz. 1996. Recombinant subviral particles from tick-borne encephalitis virus are fusogenic and provide a model system for studying flavivirus envelope glycoprotein functions. J. Virol. 704549-4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stevens, F. J. 1987. Modification of an ELISA-based procedure for affinity determination: correction necessary for use with bivalent antibody. Mol. Immunol. 241055-1060. [DOI] [PubMed] [Google Scholar]
- 51.Stiasny, K., S. Kiermayr, H. Holzmann, and F. X. Heinz. 2006. Cryptic properties of a cluster of dominant flavivirus cross-reactive antigenic sites. J. Virol. 809557-9568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thiel, H. J., M. S. Collett, E. A. Gould, F. X. Heinz, M. Houghton, G. Meyers, R. H. Purcell, and C. M. Rice. 2005. Family Flaviviridae, p. 981-998. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA.
- 53.Van Regenmortel, M. H. 1999. The antigenicity of tobacco mosaic virus. Philos. Trans. R. Soc. Lond. B 354559-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang, W., P. R. Chipman, J. Corver, P. R. Johnson, Y. Zhang, S. Mukhopadhyay, T. S. Baker, J. H. Strauss, M. G. Rossmann, and R. J. Kuhn. 2003. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat. Struct. Biol. 10907-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang, Y., W. Zhang, S. Ogata, D. Clements, J. H. Strauss, T. S. Baker, R. J. Kuhn, and M. G. Rossmann. 2004. Conformational changes of the flavivirus E glycoprotein. Structure 121607-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.