Abstract
Human immunodeficiency virus type 1 (HIV-1) assembles poorly in murine cells, reflecting inefficient targeting of the Gag structural polyprotein to the plasma membrane. Virus particle production can be restored by replacing the cis-acting Rev response element (RRE) in Gag-Pol mRNAs with multiple copies of the CTE (4×CTE), suggesting a mechanistic link between HIV-1 RNA trafficking and productive Gag assembly. In this report, we demonstrate that Gag molecules generated from RRE-dependent transcripts are intrinsically defective for assembly in murine 3T3 cells. When controlled for the intracellular Gag level, modulations of the Gag matrix (MA) domain that enhance Gag membrane association (e.g., deletion of the MA globular head) substantially improve assembly for Gag derived from RRE- but not 4×CTE-dependent transcripts. Gag mutants carrying a leucine zipper replacement of the nucleocapsid (NC) domain remain largely assembly defective when derived from RRE-dependent transcripts, indicating that the defect does not reflect aberrant NC/RNA-driven Gag multimerization. We further demonstrate that single changes in uncharged amino acids implicated in Gag/MA myristoyl switch regulation, most notably replacing the leucine at position 21 with serine, improve assembly for Gag derived from RRE-dependent transcripts. In sum, we provide genetic evidence to suggest that HIV-1 RNA metabolism specifically modulates the activation of MA-dependent membrane targeting.
The nuclear export of full-length genomic RNAs (gRNAs) initiates the late stages of retrovirus replication, including virus particle assembly and egress (19). In the cytosol, gRNAs serve as the mRNAs encoding the Gag (and Gag-Pol [GP]) structural polyproteins. Although Gag is sufficient to drive assembly in the absence of other viral factors, gRNAs not only bind to Gag to ensure their packaging into viral cores, but may also play a structural role in promoting Gag self-association (16). As with all mRNAs, gRNA metabolism and transport are highly regulated processes, and the cellular mechanisms underlying the interplay between the Gag protein and gRNA during assembly are not well defined (11, 49).
In the nucleus, all primary retroviral RNA transcripts undergo pre-mRNA splicing, so that gRNAs, containing functional introns, require specialized machineries to ensure their stabilization and nuclear export (14, 22). Human immunodeficiency virus type 1 (HIV-1) encodes its own viral accessory factor, Rev, that regulates nuclear export by binding to a structured cis-acting RNA signal, the Rev response element (RRE), found only in full-length gRNAs and partially spliced viral transcripts (40). Rev contains a leucine-rich nuclear export signal (NES) that recruits the cellular factor CRM1 (also known as exportin 1) to direct nuclear export of a gRNA/Rev ribonucleoprotein (RNP) complex. Of retroviruses that do not encode Rev equivalents, the best understood is Mason-Pfizer monkey virus, which carries a distinct RNA export element, the constitutive transport element (CTE), in all viral transcripts, including gRNAs (5). The CTE directly recruits the cellular factor NXF1(TAP), thereby regulating nuclear export through the NXF1/NXT pathway (21). Whereas CRM1 regulates the nuclear export of proteins with leucine-rich NESs, 5S ribosomal RNAs, U snRNAs, and a subset of mRNAs associated with NES-dependent RNA binding proteins (14, 28), the NXF1/NXT pathway is responsible for the bulk of cellular mRNA nuclear export (24). Taken together, the RRE and CTE constitute discrete and transferable cis-acting RNA signals that ensure gRNA transport to the cytosol, each trafficking gRNAs through a distinct nuclear export pathway.
HIV-1 assembly is notably poor in murine cells, a defect we have previously linked to viral-RNA nuclear export (50). In murine cells modified to support low levels of HIV-1 infection, Gag precursors interact poorly with cellular membranes and do not undergo efficient proteolytic processing, and virus particle generation is weak (4, 8, 27, 32, 42). The defect is rescued by fusing infected murine cells with human cells, or in human-mouse hybrid cells, suggesting the lack of one or more human trans-acting factor(s) (4, 12, 31, 52). In the absence of human factors, the replacement of the RRE in subgenomic Gag and GP mRNA transcripts with four copies of the CTE (4×CTE) is sufficient to restore Gag membrane binding and virus particle production in murine 3T3 cells, demonstrating that viral-RNA nuclear export using the NXF1/NXT pathway provides the cellular cues necessary for assembly whereas the canonical RRE/Rev pathway does not (50). In contrast, both nuclear export pathways allow productive assembly in human cell lines (5, 50, 54).
Whether the murine assembly defect reflects a true species-specific defect in Gag assembly function remains controversial. Many experimental vector systems in murine cells yield low levels of Gag expression, and low intracellular Gag levels have been correlated with poor Gag plasma membrane targeting, even in some human cell lines (e.g., HOS and 293T cells) (23, 39). This defect may be explained, at least in part, by a requirement for a threshold concentration of intracellular Gag to activate membrane targeting through a “myristoyl switch” mechanism (23, 39, 51). Gag is cotranslationally modified with a hydrophobic myristoyl group at its N terminus, a modification required for efficient membrane targeting and virus particle production (6, 20). The myristoyl group can adopt an “exposed” conformation that promotes membrane binding or be sequestered, and thereby inactivated, by the globular-head domain of the Gag matrix (MA) domain (44, 51). Deletion of the MA globular-head domain, resulting in constitutive exposure of the myristoyl group, can significantly improve Gag membrane targeting and virion assembly at low intracellular Gag levels, demonstrating that the globular-head domain plays an autoinhibitory function in this context and suggesting that myristoyl exposure is driven by cooperative Gag-Gag interactions (39). Consistent with these ideas, similar deletions of the MA globular head that retain the native myristoylation signal (or, alternatively, the addition of the myristoylated membrane-targeting signal from the Src kinase to the N terminus of intact Gag) substantially improves HIV-1 assembly in murine cells (23). Additionally, replacements of HIV-1 MA with other retroviral MA regions (e.g., from murine leukemia virus) also improve assembly in murine cells (8, 9, 42).
In this study, we addressed the relationship between HIV-1 gRNA nuclear export, Gag expression levels, and assembly efficiency in murine 3T3 cells using surrogate gRNA transcripts routed through alternative RRE/Rev- or CTE-dependent nuclear export pathways and encoding wild-type Gag or previously characterized Gag mutants. We demonstrate that the presence of the 4×CTE nuclear export element in Gag-encoding transcripts substantially improves Gag assembly efficiency compared to the RRE/Rev nuclear export pathway and that this effect is independent of the capacity of the 4×CTE to stimulate Gag expression. Modulations in MA thought to activate or circumvent the necessity for a myristoyl switch, most notably a single amino acid change (L21S), provide for efficient assembly in either pathway and abolish the enhancement in assembly efficiency associated with the 4×CTE export element. The assembly of Gag mutants carrying a leucine zipper replacement of the nucleocapsid (NC) domain remains inefficient in the RRE/Rev pathway, suggesting that RNA-driven Gag multimerization does not underlie the defect. Together, these observations reinforce the notion that posttranscriptional RNA regulation is linked to protein function in the context of HIV-1 assembly and suggest that this cross talk specifically targets the activation of MA-dependent membrane targeting.
MATERIALS AND METHODS
Cell culture and plasmids.
Murine 3T3 cells and human HeLa cells were cultured in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum plus l-glutamine and penicillin/streptomycin. Expression plasmids for Gag/GP and the Gag mutants shown in Fig. 1 to 4 were generated by insertion of HIV-1HXB2 proviral DNA fragments (nucleotides 678 to 5097) encoding GP into pcDNA3.1-based (Invitrogen) plasmid vectors carrying the RRE or multimeric 4×CTE upstream of a bovine growth hormone poly(A) signal (50) using HindIII and EcoRI sites. DNAs encoding HIV-1HXB2 wild-type and protease-defective GP and mutants Δ8-126 (43), ZWT-p6, and ZIL-p6 (1) were the kind gifts of Heinrich Göttlinger (University of Massachusetts Medical School, Worcester, MA). For the MA mutation analysis (Fig. 5 and 6), HIV-1NL4-3 fragments (nucleotides 683 to 5748) encoding Gag/GP and Vif were inserted into the same RRE and 4×CTE vectors using NheI and EcoRI sites; the derivative L8A, W16A, L21S, and W36A Gag mutants were generated using overlapping PCR. DNA encoding the G2A mutant (17) was a kind gift of Eric Freed (NCI-Frederick, Frederick, MD). DNAs encoding HIV-1 Rev (pcRev) and firefly luciferase were also cloned into pcDNA3.1 (50).
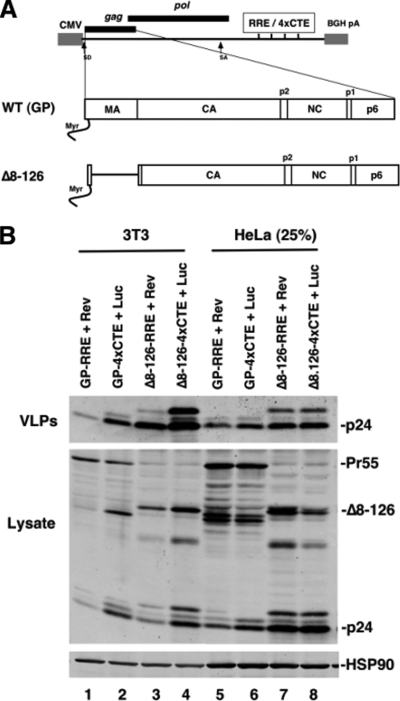
FIG. 1.
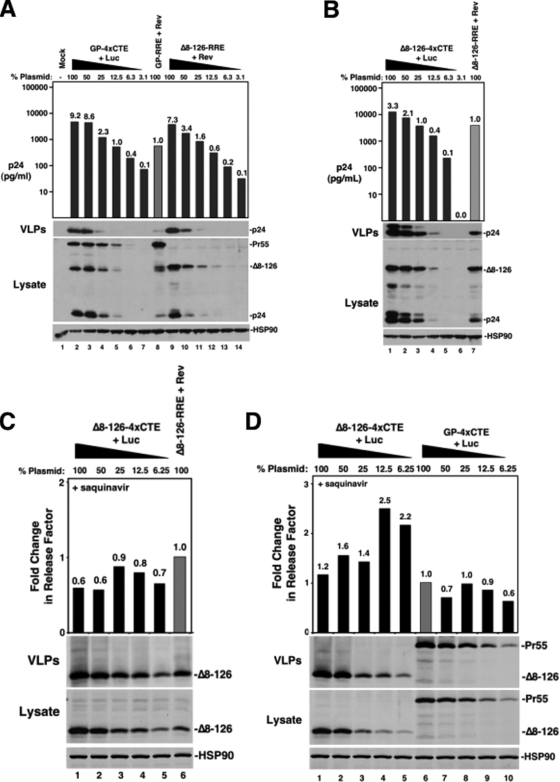
4×CTE-dependent RNA nuclear export and deletion of the MA globular head both increase VLP production in murine cells. (A) Depiction of cytomegalovirus major immediate-early-driven surrogate gRNA transcripts carrying either RRE or 4×CTE nuclear export elements and encoding either wild-type (WT) Gag (GP) or Gag carrying a deletion of the MA globular-head domain (Δ8-126). (B) 3T3 (lanes 1 to 4) or HeLa (lanes 5 to 8) cells were cotransfected with 1 μg or 250 ng (25%), respectively, Gag expression plasmids encoding the indicated transcripts plus plasmids encoding either Rev or luciferase (Luc) as indicated. Cell lysates and VLPs were collected 48 h posttransfection, resolved by SDS-PAGE, and immunoblotted for Gag and HSP90 (loading control).
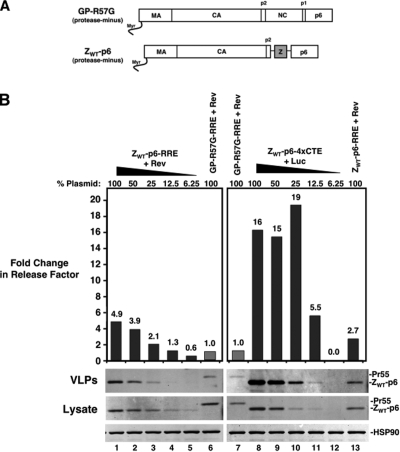
FIG. 4.
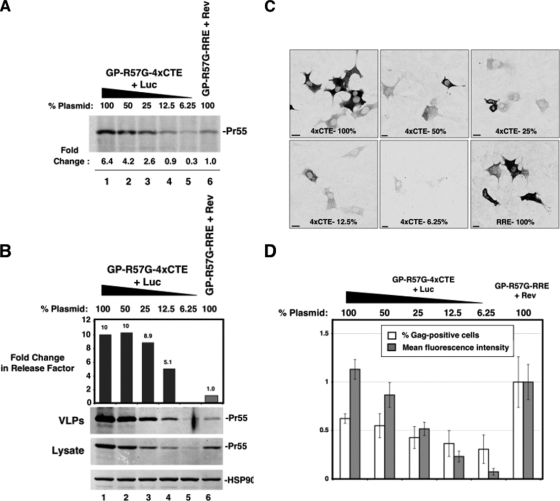
The 4×CTE affects assembly independently of NC-dependent Gag multimerization. (A) Depiction of protease-minus Gag (GP-R57G) and the ZWT-p6 mutant carrying a leucine zipper replacement of NC. (B) A twofold 4×CTE plasmid dilution series followed by quantitative immunoblotting similar to that described for Fig. 2C comparing RRE- and 4×CTE-dependent transcripts for the ZWT-p6 mutant. The calculated release factors were normalized to the GP-R57G-RRE plus Rev sample (lanes 6 and 7; gray bars). The change is indicated above each bar.
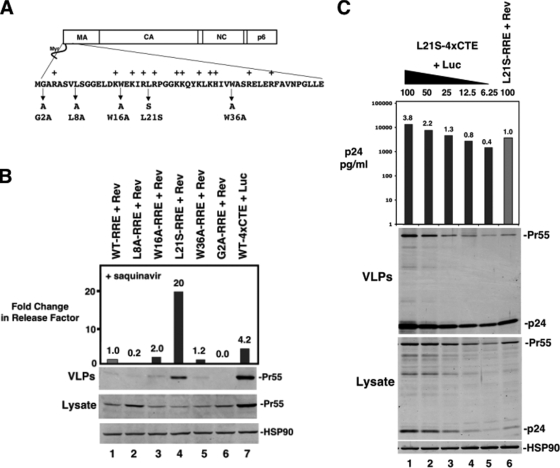
FIG. 5.
Single MA amino acid changes implicated in myristoyl switch regulation improve RRE/Rev-derived Gag assembly efficiency in murine cells. (A) Depiction of N-terminal MA single-amino-acid mutations generated for this analysis. Basic residues are indicated by “+.” (B) 3T3 cells were transfected with the indicated MA mutant and cultured in the presence of saquinavir, and the cells and supernatants were processed for quantitative assembly assay as described for Fig. 2C. Release factors were normalized to the wild-type (WT)-RRE plus Rev sample. The change in the release factor is indicated above each bar. (C) A twofold 4×CTE plasmid dilution series comparing the assembly efficiency of Gag (L21S) mutants derived from 4×CTE-dependent transcripts (lanes 1 to 5) to that derived from RRE/Rev-dependent transcripts (lane 6). As for Fig. 2A, VLP production was assessed both by immunoblotting and p24Gag ELISA. The change in p24Gag release is indicated above each bar.
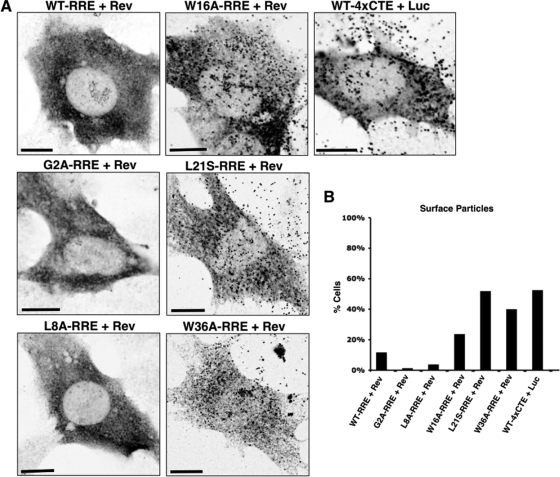
FIG. 6.
MA mutants rescue Gag membrane targeting and particle production. (A) 3T3 cells transfected with Gag or the indicated Gag mutant were fixed 24 h posttransfection prior to indirect immunofluorescence detection of Gag using an anti-p17Gag polyclonal antiserum. The images were collected using a laser scanning confocal microscope and represent a single optical slice near the cell-coverslip junction. Bars, 10 μm. (B) For the experiment in panel A, coverslips were assessed blindly and >100 transfected cells were scored for the detectable presence of Gag-positive surface punctae, consistent with VLP production. The bars represent average values from duplicate samples.
Assembly assays.
Plasmid transfection was carried out using FuGene 6 reagent (Roche) following the manufacturer's instructions. A standard transfection mixture contained 1 μg Gag plasmid plus 0.5 μg Rev plasmid (for RRE-containing transcripts) or 0.5 μg control firefly luciferase plasmid (for 4×CTE-containing transcripts) added to a 35-mm dish of cells plated at ∼30% confluence. Plasmid dilution series were normalized to 1.5 μg total plasmid DNA using the luciferase-encoding plasmid. The medium was changed 24 h posttransfection, and cells and supernatants were harvested at 48 h posttransfection. In some experiments (e.g., those shown in Fig. 2C and D and 5B), the protease inhibitor saquinavir (NIH AIDS Research and Reference Reagent Program) was added to a final concentration of 1 μM at 24 h posttransfection. Supernatants were filtered through 0.45-μm filters, and virus-like particles (VLPs) were collected by pelleting them through a 20% sucrose cushion using centrifugation at ∼21,000 × g for 2 h prior to resuspension in 1× dissociation buffer (62.5 mM Tris-HCl, pH 6.8, 10% glycerol, 2% sodium dodecyl sulfate [SDS], 5% β-mercaptoethanol). The cells were lysed in 1× RIPA buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 1% Triton X-100, 1% sodium deoxycholate), and the lysates were further disrupted by 10 passages through a 26.5-gauge needle prior to 1:1 dilution with 2× dissociation buffer. Proteins were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) using conventional procedures and transferred to nitrocellulose membranes. Gag was detected using a mouse monoclonal antibody recognizing p24Gag (24-2; diluted 1:1,000) (18) and secondary antibodies conjugated to horseradish peroxidase for enhanced chemiluminescence detection (Fig. 2A and B) or secondary antibodies conjugated to the infrared fluorophore IRDye800 (Li-Cor Biosciences) for quantitative immunoblotting (Fig. 2C and D and 3 to 5) using an Odyssey infrared scanner (Li-Cor Biosciences). HSP90 was detected in all experiments as a loading control using rabbit anti-HSP90 antiserum (Santa Cruz Biotechnology). p24Gag levels were determined by enzyme-linked immunosorbent assay (ELISA) (Perkin Elmer) according to the manufacturer's instructions.
FIG. 2.
The 4×CTE enhancement of assembly in murine cells requires an intact MA globular-head domain. (A) 3T3 cells were transfected with Gag expression plasmids or dilutions thereof, as indicated. Lysates and VLPs were harvested for immunoblot analysis as described for Fig. 1B. p24Gag released into the supernatant was quantified by ELISA (bars); the value above each bar represents the change for values normalized to the GP-RRE plus Rev sample (gray bar; lane 8). HSP90 served as a loading control for cell lysates. (B) Comparison of assembly of Δ8-126 Gag mutants encoded by transcripts undergoing either RRE/Rev- or 4×CTE-dependent nuclear export. (C) An experiment similar to that presented in panel B for the Δ8-126 mutant carried out in culture medium containing 1 μM saquinavir to prevent Gag processing. Protein levels were measured by quantitative immunoblotting. A release factor (bars) representing assembly efficiency was calculated as the ratio of Gag detected in VLPs to Gag detected in cell lysates. For comparison, all values were normalized to the Δ8-126-RRE plus Rev condition (gray bar; lane 6). The change is indicated above each bar. (D) Comparison of assembly efficiencies for the Δ8-126 mutant Gag and wild-type Gag encoded by transcripts undergoing 4×CTE-dependent nuclear export.
FIG. 3.
RRE/Rev-derived Gag is intrinsically defective for assembly in murine cells independent of the expression level. (A) 3T3 cells were metabolically radiolabeled with [35S]Met/Cys for 10 min 24 h posttransfection with the indicated amounts of protease-minus Gag (GP-R57G) expression plasmids. Gag was immunoprecipitated from cell lysates using an anti-p24Gag polyclonal serum and resolved by SDS-PAGE. Radiolabeled protein was quantified by phosphorimager, and the values were normalized to the GP-RRE plus Rev sample (lane 6). (B) Analysis of virus assembly efficiency by quantitative immunoblotting (as described for Fig. 2C) for 3T3 cells transfected in a fashion identical to that for panel A. HSP90 served as a loading control for cell lysates. The change in the release factor is indicated above each bar. (C) Visual comparison of intracellular Gag (GP-R57G) levels for 3T3 cell populations transfected in a fashion identical to that for panels A and B. Cells were seeded on glass coverslips and fixed 48 h posttransfection. Gag was detected by indirect immunofluorescence using an anti-p24Gag serum and visualized by confocal microscopy. Identical laser settings were used for each acquisition. Representative images are presented. Bars, 10 μm. (D) Comparison of intracellular Gag levels for transfected 3T3 cell populations analyzed by flow cytometry. Cells were transfected in a fashion identical to that for panels A to C prior to being trypsinized, fixed, permeabilized, and stained with a monoclonal anti-p24Gag serum conjugated to fluorophore RD1 at 48 h posttransfection. The percentage of cells expressing detectable levels of Gag (white bars) or the mean fluorescence intensity for each Gag-positive population (gray bars) for each twofold dilution of 4×CTE plasmid was normalized to the RRE plus Rev condition (bars on right). The mean values for three separate transfections are presented. The error bars represent ±1 standard deviation.
Metabolic labeling.
Twenty-four hours posttransfection, cells were washed and incubated in Cys−/Met− depletion medium containing 10% dialyzed serum (Invitrogen) for 20 min prior to the addition of 0.25 mCi/ml [35S]Cys/Met (Amersham) for 10 min at 37°C. The cells were washed and lysed in 1× RIPA buffer, and Gag was immunoprecipitated using a polyclonal rabbit antiserum specific for p24Gag (UP598; diluted 1:500) (48) prior to SDS-PAGE and visualization by autoradiography. Relative expression was calculated based on measurements made using a Storm phosphorimager (GE Healthcare).
Microscopy.
3T3 cells were plated on glass coverslips and transfected as described above using FuGene6. Twenty-four hours posttransfection, the cells were washed with phosphate-buffered saline and fixed in 4% paraformaldehyde (EM Sciences) for 15 min, permeabilized with 0.2% Triton X-100 for 15 min, and blocked/quenched in buffer NGB (50 mM NH4Cl, 1% goat serum, 1% bovine serum albumin) for 1 h. Gag was detected using mouse monoclonal anti-capsid (p24Gag) antiserum (24-2; diluted 1:1,000 in NGB) (18) (Fig. 3C) or rabbit polyclonal anti-matrix (p17Gag) serum (UP595; diluted 1:500 in NGB) (50) (Fig. 6A) and secondary goat anti-rabbit immunoglobulin G conjugated to Alexa Fluor 488 (Invitrogen). Laser scanning confocal imaging was performed on a DM IRE2 microscope (Leica), and the images were processed using LCS (Leica) and Openlab (Improvision) software packages.
Flow cytometry.
Cells were trypsinized, washed, and prepared for flow cytometry 48 h posttransfection using the Intrastain Fixation and Permeabilization Kit (Dako) and a mouse monoclonal anti-p24Gag antibody (KC57) conjugated to RD1 (Beckman Coulter). Flow cytometry was performed using a BD Biosciences FACS Canto II Flow Cytometry System.
RESULTS
HIV-1 particle assembly in murine 3T3 cells is improved by 4×CTE -dependent Gag mRNA nuclear export or deletion of the MA globular-head domain.
Murine blocks to HIV-1 replication include viral entry, postentry, transcriptional elongation, oversplicing, and particle assembly (2-4, 29, 30, 32, 52, 53, 56). To focus specifically on RNA export and assembly, we established a genetic approach based on the transfection of plasmids carrying subgenomic HIV-1 RNAs encoding Gag (and GP) (50). To provide efficient transcription, the HIV-1 U3 promoter region was replaced with the cytomegalovirus major immediate-early promoter. To prevent or reduce oversplicing, only the major 5′ splice donor and a subset of proximal splice acceptors were retained (Fig. 1A). Differential control of nuclear export was provided by including either the RRE (GP-RRE) directing nuclear export by the Rev/CRM1 pathway or multiple copies of the CTE (GP-4×CTE) (54) that recruits NXF1 for export using the NXF1/NXT mRNA export pathway. Efficient Gag production in the GP-RRE context requires cotransfection of a plasmid encoding Rev, while Gag production from GP-4×CTE is Rev independent. In order to improve Gag expression levels in murine cells, we also deleted the majority of the 5′ untranslated leader region upstream of the SL1 stem-loop. At the level of virus assembly, this deletion did not affect sensitivity to the alternative export elements compared to transcripts including the authentic 5′ leader (data not shown).
We first evaluated MA-dependent Gag membrane targeting in the context of alternative RNA nuclear export pathways in murine (3T3) and human (HeLa) cell lines. We selected a previously characterized Gag mutant carrying a deletion within the MA globular head, Δ8-126, that retains the N-terminal Gag myristoylation sequence and supports efficient VLP production (43). DNA fragments encoding either wild-type Gag or the Δ8-126 mutant were subcloned into RRE- or 4×CTE-containing expression plasmids (Fig. 1A) and cotransfected into 3T3 or HeLa cells with plasmids encoding Rev (RRE plus Rev) or luciferase (4×CTE plus Luc), respectively. Forty-eight hours posttransfection, the cells were lysed and VLPs were harvested from the supernatants prior to SDS-PAGE and Gag detection using an anti-p24Gag antiserum. As expected, wild-type Gag derived from RRE/Rev-dependent transcripts was not efficiently processed in 3T3 cells. VLP production was low in comparison to HeLa cells transfected with 25% (250 ng) total Gag plasmid, which was done to achieve comparable levels of Gag expression (Fig. 1B, lane 1). In contrast, Gag encoded by a 4×CTE-dependent transcript (GP-4×CTE) restored Gag processing and substantially improved VLP production, specifically from 3T3 cells (Fig. 1B, lane 2). Deletion of the MA globular head in the RRE context (Δ8-126-RRE plus Rev) similarly improved particle production from 3T3 cells (Fig. 1B, lane 3). Combining this mutation with the 4×CTE (Δ8-126-4×CTE plus Luc) further stimulated VLP production (Fig. 1B, compare lane 4 to lanes 2 and 3). In contrast, none of these modulations significantly affected assembly in HeLa cells at any level of expression tested (Fig. 1B, lanes 5 to 8, and data not shown). These results implied that 3T3 cells harbor genetically distinct assembly defects that are rescued by MA manipulation or by 4×CTE-dependent nuclear export.
The 4×CTE-dependent rescue of assembly requires an intact MA domain.
MA-dependent autoinhibition is at least partially responsible for Gag assembly defects under conditions of low Gag expression (23, 39). To address assembly efficiency (i.e., levels of VLP production relative to intracellular Gag levels) in the context of varying intracellular Gag levels, we performed a twofold vector dilution series comparing GP-4×CTE or Δ8-126-RRE/Rev to the GP-RRE/Rev context. For both GP-4×CTE and Δ8-126-RRE/Rev, similar levels of VLP production were achieved by transfecting only 125 to 250 ng (12.5 to 25% of total plasmid) relative to 1 μg (100%) of GP-RRE plasmid, despite considerably lower levels of Gag detected in cell lysates for either condition (Fig. 2A, compare lanes 5 and 11 to lane 8). As such, both the 4×CTE and deletion of the MA globular head substantially improve virus assembly at apparently lower levels of expression in 3T3 cells.
In contrast, a similar dilution series comparing Δ8-126-4×CTE to Δ8-126-RRE/Rev revealed that there was little difference in Gag expression levels when normalized for levels of VLP production, indicating that the presence of the 4×CTE no longer enhanced virus assembly efficiency in the context of the Δ8-126 mutation (Fig. 2B, compare lane 3 to lane 7). Accordingly, the improvement of VLP production from combining the Δ8-126 mutation with 4×CTE-dependent nuclear export (Fig. 1B, lane 4) could be attributed to an increase in intracellular Gag abundance induced by the 4×CTE (Fig. 2B, compare lane 1 to lane 7). Indeed, metabolic labeling of cells with [35S]cysteine/methionine for 10 minutes followed by immunoprecipitation with a Gag-specific antiserum confirmed that the 4×CTE increased the amount of Gag translated relative to the RRE/Rev context by three- to ninefold over four independent experiments (Fig. 3A and data not shown).
We repeated these plasmid titration-based analyses in the absence of Gag proteolytic processing by either culturing cells in medium containing 1 μM saquinavir, an inhibitor of the HIV-1 protease (Fig. 2C and D and 5B), or expressing protease-inactivated Gag mutants (Fig. 3B and 4B). This approach allowed us to quantify Gag levels after SDS-PAGE as a single protein species using fluorescence-based quantitative immunoblotting. For each condition, we could determine the assembly efficiency for a variety of intracellular Gag levels by calculating a “release factor,” i.e., the ratio of released Gag (VLPs) to cellular Gag (lysate). In the absence of Gag processing, the 4×CTE improved the efficiency of VLP production from 4- to 20-fold relative to the RRE plus Rev (Fig. 3B and data not shown), demonstrating that these assembly effects were not related to the presence or absence of a functional viral protease. Using this approach, we confirmed that the 4×CTE did not improve assembly efficiency for the Δ8-126 mutant at any intracellular Gag level tested (Fig. 2C), consistent with the data presented in Fig. 2B. Compared to wild-type Gag in the 4×CTE context, the Δ8-126 MA deletion resulted in a moderate (1.2- to 3.7-fold) improvement in assembly efficiency (Fig. 2D). Therefore, the 4×CTE-dependent rescue can be considered slightly suboptimal, while the Δ8-126 mutation confers a dominant rescue of virus particle production for Gag derived from either nuclear export pathway.
Taken together, these experiments demonstrate that the 4×CTE enhancement requires an intact MA globular-head domain for its activity. This result implies that these manipulations, which are accepted as affecting either Gag mRNA metabolism or Gag protein activity, are, in fact, linked to the same step/stage of the HIV-1 assembly pathway.
RRE/Rev-dependent nuclear export results in defective assembly in murine cells independently of Gag expression kinetics.
Although the above-mentioned results implied that the 4×CTE stimulates assembly independently of intracellular Gag levels, we sought a more definitive approach for addressing possible effects of Gag expression kinetics on assembly. As noted above, we performed a 35S metabolic-labeling experiment for a twofold dilution series expressing a protease-defective Gag mutant (designated GP-R57G; protease residue arginine-57 mutated to glycine). Similar levels of expression were achieved at ∼125 ng (∼12.5%) of GP-R57G-4×CTE plasmid compared to 1 μg (100%) of GP-R57G-RRE (Fig. 3A, compare lanes 3 and 4 to lane 6). Despite these differences, the 4×CTE improved assembly efficiency at lower levels of intracellular Gag concentration relative to GP-R57G-RRE/Rev (Fig. 3B, compare lanes 3 and 4 to lane 6).
Corresponding analyses by confocal microscopy (Fig. 3C) or using flow cytometry (Fig. 3D) confirmed that the above-mentioned results were obtained in the context of decreases in Gag abundance per cell for successive dilutions of 4×CTE plasmid. In particular, cells transfected with 100% RRE plasmid (plus Rev) were ∼4-fold and ∼2-fold brighter on average than cells expressing 12.5% or 25% 4×CTE plasmid, respectively (Fig. 3C and D). In sum, although the 4×CTE improves Gag expression relative to the RRE/Rev context, assembly efficiency is substantially improved by the 4×CTE at similar or lower levels of Gag expression and intracellular accumulation. Thus, the assembly efficiency of Gag produced from the RRE/Rev pathway is intrinsically low in murine 3T3 cells.
RNA-independent multimerization cannot rescue the murine assembly defect.
Our results implied an intersection between the functions of viral-RNA trafficking and Gag, possibly occurring at the level of Gag-RNA association and/or formation of Gag/RNA assembly intermediates. Gag binds to RNA predominantly through its NC region, and RNA can promote Gag-Gag self-association (16, 33). We therefore tested the relevance of differential RNA nuclear export trafficking in the context of protease-minus Gag oligomerization mutants in which NC was deleted and replaced with the efficient self-associating leucine zipper dimerization motif from the yeast GAL4 protein (ZWT-p6) (Fig. 4A). These mutants assemble efficiently in human cells and produce VLPs that package little to no viral or cellular RNA (1, 13, 55). Expression of the ZWT-p6 mutant in the RRE-dependent context improved assembly up to fivefold compared to GP-R57G-RRE (Fig. 4B, compare lane 1 or 13 to lane 6 or 7). However, unlike for the Δ8-126 MA mutation, the assembly efficiency of ZWT-p6 mutants was enhanced to a substantially greater extent by the 4×CTE at all levels of expression (Fig. 4B, compare lanes 9 and 10 to lane 1 or 13). Similar results were obtained for a Gag mutant carrying a zipper driving trimerization (ZIL-p6, data not shown). These data suggest that the 4×CTE does not affect particle assembly at the level of NC/RNA-dependent Gag multimerization. Furthermore, the forced multimerization of Gag molecules is insufficient to fully activate Gag membrane targeting in the murine system.
Alterations in single uncharged amino acids in MA improve HIV-1 assembly in murine cells.
Deletion of the MA globular head relieves sequestration of the N-terminal myristoyl group to constitutively activate Gag membrane targeting (39, 43). The N-terminal glycine residue is the target of myristoylation, and replacement of this glycine with alanine (G2A) severely reduces Gag membrane binding and virus particle production (6, 20). Additional mutations of residues within MA located between the myristoylation signal and the helical core of the globular head (e.g., V7R, L8A, and L8I) also reduce Gag membrane binding by preventing myristoyl exposure (17, 35, 36, 38, 45). Gottlinger and colleagues have reported second-site suppressors of an L8A mutant that include W16A, L21S, and W36A (38).
To test for a link between nuclear export and the myristoyl switch, we measured VLP production in 3T3 cells for the G2A, L8A, W16A, L21S, and W36A mutants in the RRE/Rev context (Fig. 5A). To calculate a release factor for a single 55-kDa Gag species, the protease inhibitor saquinavir was added to cultures 24 h after transfection. As expected, the G2A and L8A Gag mutants assembled poorly, even relative to wild-type Gag (wild type-RRE/Rev) (Fig. 5B, compare lanes 2 and 6 to lane 1). In contrast, the W16A and W36A mutations in Gag improved assembly moderately (one- to fivefold over four independent experiments) (Fig. 5B, compare lanes 3 and 5 to lane 1, and data not shown). Strikingly, the L21S mutant exhibited up to 20-fold enhancement of assembly efficiency relative to the wild type (Fig. 5B, compare lane 4 to lane 1). We also compared the L21S mutant in the 4×CTE context to L21S-RRE/Rev and found that, reminiscent of the Δ8-126 mutant, the two modes of assembly rescue were equivalent when controlled for similar intracellular Gag levels (Fig. 5C, compare lane 3 or 4 to lane 6). As such, both a single point mutation in MA (L21S) and 4×CTE-dependent nuclear export efficiently restore Gag assembly in murine cells.
To address the membrane targeting of Gag more directly, we examined the intracellular distribution of Gag derived from RRE/Rev-dependent transcripts by laser scanning confocal microscopy using a polyclonal antibody specific for MA. The wild-type, G2A, and L8A Gags were detected in a diffuse cytosolic distribution in the vast majority of cells, consistent with a lack of plasma membrane targeting (Fig. 6A and B). In contrast, the W16A, L21S, and W36A mutants each improved Gag accumulation at the plasma membrane (Fig. 6A). Additionally, the formation of surface punctae, which is consistent with VLP production, was readily detectable in a substantially higher proportion of transfected cells for each mutant (W16A, 21% Gag-positive cells; L21S, 46%; W36A, 35%), similar to the phenotype of the wild type-4×CTE context (46%) (Fig. 6A and B). Why the W36A mutant performed so well in this visual assay relative to the virus release assay is not clear, though we have noted aberrant Gag processing for this mutant, possibly indicating a secondary assembly defect downstream of membrane targeting (data not shown).
Taken together, single-amino-acid changes in MA implicated in Gag myristoyl switch regulation (or circumvention), most notably the L21S mutation, improve assembly efficiency for Gag derived from Rev/RRE-dependent transcripts. When controlled for intracellular Gag levels, the Δ8-126 and L21S mutants assemble equivalently irrespective of whether Gag is derived from mRNAs following either the RRE/Rev- or 4×CTE-dependent nuclear export pathway. Therefore, the inefficiency of HIV-1 assembly in murine cells likely reflects a cellular deficiency in the RRE/Rev-dependent nuclear export pathway that is specifically linked to the activation of Gag membrane targeting by the regulatory MA globular-head domain.
DISCUSSION
The unprocessed HIV-1 Gag precursor accumulates in the cytosol of murine cells, consistent with a defect in Gag trafficking/targeting to the plasma membrane. Membrane targeting and particle assembly are significantly improved by replacing the RRE in Gag mRNAs with the multimeric CTE (4×CTE) or, at the protein level, by mutations in MA that have been previously implicated in myristoyl switch regulation (e.g., Δ8-126 and L21S). Analyses by plasmid dilution and metabolic labeling reveal that the 4×CTE does improve Gag expression rates relative to transcripts from the RRE/Rev pathway (Fig. 3A). Despite this, we have demonstrated here that the 4×CTE also has the capacity to substantially improve assembly efficiency even at significantly lower levels of intracellular Gag concentration (Fig. 3B to D). Therefore, while likely relevant, low intracellular Gag levels are not the only cause of defective HIV-1 assembly in murine cells.
When controlled for intracellular Gag levels, the 4×CTE substantially improves assembly efficiency for wild-type Gag but has no further enhancing effect on Δ8-126 and L21S Gag mutants (Fig. 2 and 5). Thus, an intact globular head or, strikingly, a leucine and not a serine at MA position 21 is required for the ability of 4×CTE-dependent RNA nuclear export to improve Gag assembly competency in murine cells. These observations lead us to propose that 4×CTE-dependent RNA nuclear export and these gain-of-function mutants affect a common defective step in the assembly pathway. Because these mutations are implicated in activation or circumvention of the myristoyl switch, the data reinforce the view that MA exerts an autoinhibitory function in certain contexts (23) and suggest that HIV-1 RNA metabolism can be linked to myristoyl switch activation, at least in murine cells. Poor virus production and the diffuse cytosolic distribution of Gag derived from RRE/Rev-dependent transcripts clearly resemble the phenotypes of the G2A (myristoyl-minus) or L8A (myristoyl-sequestered) mutants (Fig. 5 and 6). In contrast, all mutants implicated in myristoyl switch activation or circumvention improve assembly efficiency (at least mildly) and can restore virus particle production at the plasma membrane (Fig. 5 and 6). While it is likely that deletion of the globular head results in constitutive exposure of the myristoyl group, it is not certain that the L21S mutant acts in precisely the same manner. Recent evidence suggests that myristoyl exposure can also be driven after the Gag-membrane interaction, promoted by phosphatidylinositol(4,5)bisphosphate [PI(4,5)P2], a phosphoinositide that resides at the inner leaflet of the plasma membrane (34, 46). A similar, but not necessarily equivalent, mutation (L21K) was shown to be unresponsive to PI(4,5)P2 in terms of myristoyl exposure but instead exhibited a twofold improvement in binding to PI(4,5)P2 independently of the myristoyl group (45). Alternatively, it is also conceivable that these gain-of-function mutations in MA could represent alterations in cis-acting elements operating at the level of the Gag mRNA, with consequences for assembly. Efforts are ongoing in our laboratory to address this issue.
Forced dimerization or trimerization of Gag by Z-p6 mutants can only partially restore assembly for the Rev/RRE context in murine cells, consistent with a multistep pathway for assembly wherein NC-independent multimerization provides only a “weak” assembly signal that is further enhanced by 4×CTE-induced activation of MA-dependent membrane binding (Fig. 4). The formation of Gag multimers may be initiated upstream of membrane association; in particular, (i) high intracellular Gag levels have been correlated with improvements in Gag membrane targeting and virus production, leading to the suggestion that the initiation of Gag membrane targeting is a cooperative process (39); (ii) forced dimerization of the MA subunit substantially improves MA-membrane binding (15), consistent with previous work demonstrating that the membrane binding capacity of MA is improved in the context of the full-length Gag molecule, provided that the NC domain is intact (47, 57); and (iii) analyses of MA using nuclear magnetic resonance have confirmed that low-order Gag multimerization (predominantly trimerization) acts as a trigger for myristoyl exposure (51). In contrast with the last study, a more recent analysis by nuclear magnetic resonance identified a role for PI(4,5)P2 in driving myristoyl exposure and stabilization (46), suggesting that activation of the myristoyl switch may occur predominantly after Gag is trafficked to the plasma membrane. Importantly, these models are not mutually exclusive; for example, Gag interactions in the cytosol may initiate low-level membrane binding, while PI(4,5)P2 may function as a plasma membrane-specific “receptor” necessary for higher-order assembly. Because the inclusion of a leucine zipper in place of NC gave only a modest rescue of assembly (Fig. 4), we suggest that the deficiency in the Rev/RRE nuclear export pathway affects an aspect of the Gag membrane-targeting mechanism that is separable from that triggered by cooperative Gag-Gag interactions.
The question remains, what specifically couples viral-RNA nuclear export to Gag membrane-targeting capacity? Because mouse-human heterokaryons or somatic cell hybrids can rescue the virus assembly defect, we favor the idea that one or more human factors are lacking or nonfunctional in the murine RRE/Rev-dependent pathway and that, in the permissive context, these factors regulate the activation of MA-dependent membrane targeting. In addition to the 4×CTE, other Gag transcripts rendered Rev independent due to codon optimization (23, 50) or the addition of the posttranscriptional regulatory element from hepatitis B virus (25) also result in improved HIV-1 Gag assembly in murine cells, further suggesting that the cellular defect is specific to the Rev/RRE-dependent nuclear export pathway. We previously hypothesized that nuclear “marking” of viral RNAs (i.e., differences in RNP composition) affects cytosolic RNA localization with relevance to intracellular gRNP/Gag regulation (50). In this context, we now show that the 4×CTE clearly improves cytoplasmic utilization of Gag mRNAs in 3T3 cells to achieve enhanced Gag expression (Fig. 3A); we speculate that this may be attributable to the recruitment of NXT1 multimers, which has previously been demonstrated to improve expression from CTE-containing transcripts (26). How gRNP composition specifically affects MA-dependent membrane targeting is not yet clear. However, Crist et al. recently showed that, in vitro, Gag “zipper” mutants similar to those examined here required additional cofactors for high-order assembly, including nucleic acids, phosphoinositides (IP5 or IP6), and/or alternative polyanions that likely act to neutralize inhibitory electrostatic repulsions attributed to the MA basic face (amino acids 15 to 32) (13).
In sum, HIV-1 assembly appears to require both a strong C-terminal interaction involving NC and cofactor-dependent regulation that relieves the MA autoinhibitory function to facilitate membrane binding. Multiple studies have suggested that MA/RNA interactions may contribute to assembly function (7, 10, 13, 37) and that specific charged residues within the polybasic face (e.g., lysine-18 and arginine-22) are implicated in the direct binding of viral RNA (41). In this context, we propose that gRNA trafficking ordinarily intersects (potentially via provision of a cofactor that may be gRNA itself) with MA function to facilitate membrane binding and efficient virus assembly. In murine cells, however, the RRE/Rev nuclear export pathway is uncoupled from MA function, resulting in marked deficiencies in the membrane association of Gag and the resulting inefficient assembly of viral particles.
Acknowledgments
We thank Eric Freed, Heinrich Göttlinger, and the NIH AIDS Research and Reference Reagent Program for reagents.
This work was supported by the United Kingdom Medical Research Council. N.M.S. is a Long-Term Fellow (ALTF 176-2007) of the European Molecular Biology Organization, C.M.S. is a Research Councils United Kingdom Academic Fellow, and M.H.M. is an Elizabeth Glaser Scientist.
Footnotes
Published ahead of print on 17 June 2009.
REFERENCES
- 1.Accola, M. A., B. Strack, and H. G. Gottlinger. 2000. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 745395-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atchison, R. E., J. Gosling, F. S. Monteclaro, C. Franci, L. Digilio, I. F. Charo, and M. A. Goldsmith. 1996. Multiple extracellular elements of CCR5 and HIV-1 entry: dissociation from response to chemokines. Science 2741924-1926. [DOI] [PubMed] [Google Scholar]
- 3.Baumann, J. G., D. Unutmaz, M. D. Miller, S. K. Breun, S. M. Grill, J. Mirro, D. R. Littman, A. Rein, and V. N. KewalRamani. 2004. Murine T cells potently restrict human immunodeficiency virus infection. J. Virol. 7812537-12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bieniasz, P. D., and B. R. Cullen. 2000. Multiple blocks to human immunodeficiency virus type 1 replication in rodent cells. J. Virol. 749868-9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bray, M., S. Prasad, J. W. Dubay, E. Hunter, K. T. Jeang, D. Rekosh, and M. L. Hammarskjold. 1994. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc. Natl. Acad. Sci. USA 911256-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bryant, M., and L. Ratner. 1990. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc. Natl. Acad. Sci. USA 87523-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burniston, M. T., A. Cimarelli, J. Colgan, S. P. Curtis, and J. Luban. 1999. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J. Virol. 738527-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, B. K., I. Rousso, S. Shim, and P. S. Kim. 2001. Efficient assembly of an HIV-1/MLV Gag-chimeric virus in murine cells. Proc. Natl. Acad. Sci. USA 9815239-15244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, P., W. Hubner, K. Riviere, Y. X. Liu, and B. K. Chen. 2006. Chimeric HIV-1 containing SIV matrix exhibit enhanced assembly in murine cells and replicate in a cell-type-dependent manner in human T cells. Virology 3491-12. [DOI] [PubMed] [Google Scholar]
- 10.Cimarelli, A., and J. Luban. 1999. Translation elongation factor 1-alpha interacts specifically with the human immunodeficiency virus type 1 Gag polyprotein. J. Virol. 735388-5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cochrane, A. W., M. T. McNally, and A. J. Mouland. 2006. The retrovirus RNA trafficking granule: from birth to maturity. Retrovirology 318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coskun, A. K., M. van Maanen, D. Janka, D. Stockton, P. Stankiewicz, S. Yatsenko, and R. E. Sutton. 2007. Isolation and characterization of mouse-human microcell hybrid cell clones permissive for infectious HIV particle release. Virology 362283-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crist, R. M., S. A. Datta, A. G. Stephen, F. Soheilian, J. Mirro, R. J. Fisher, K. Nagashima, and A. Rein. 2009. Assembly properties of human immunodeficiency virus type 1 Gag-leucine zipper chimeras: implications for retrovirus assembly. J. Virol. 832216-2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cullen, B. R. 2003. Nuclear mRNA export: insights from virology. Trends Biochem. Sci. 28419-424. [DOI] [PubMed] [Google Scholar]
- 15.Dalton, A. K., D. Ako-Adjei, P. S. Murray, D. Murray, and V. M. Vogt. 2007. Electrostatic interactions drive membrane association of the human immunodeficiency virus type 1 Gag MA domain. J. Virol. 816434-6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.D'Souza, V., and M. F. Summers. 2005. How retroviruses select their genomes. Nat. Rev. Microbiol. 3643-655. [DOI] [PubMed] [Google Scholar]
- 17.Freed, E. O., J. M. Orenstein, A. J. Buckler-White, and M. A. Martin. 1994. Single amino acid changes in the human immunodeficiency virus type 1 matrix protein block virus particle production. J. Virol. 685311-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaddis, N. C., E. Chertova, A. M. Sheehy, L. E. Henderson, and M. H. Malim. 2003. Comprehensive investigation of the molecular defect in vif-deficient human immunodeficiency virus type 1 virions. J. Virol. 775810-5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goff, S. 2001. Retroviridae: the retroviruses and their replication, p. 1871-1940. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 20.Gottlinger, H. G., J. G. Sodroski, and W. A. Haseltine. 1989. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 865781-5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gruter, P., C. Tabernero, C. von Kobbe, C. Schmitt, C. Saavedra, A. Bachi, M. Wilm, B. K. Felber, and E. Izaurralde. 1998. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol. Cell 1649-659. [DOI] [PubMed] [Google Scholar]
- 22.Harris, M. E., and T. J. Hope. 2000. RNA export: insights from viral models. Essays Biochem. 36115-127. [DOI] [PubMed] [Google Scholar]
- 23.Hatziioannou, T., J. Martin-Serrano, T. Zang, and P. D. Bieniasz. 2005. Matrix-induced inhibition of membrane binding contributes to human immunodeficiency virus type 1 particle assembly defects in murine cells. J. Virol. 7915586-15589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herold, A., L. Teixeira, and E. Izaurralde. 2003. Genome-wide analysis of nuclear mRNA export pathways in Drosophila. EMBO J. 222472-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin, J., T. Sturgeon, C. Chen, S. C. Watkins, O. A. Weisz, and R. C. Montelaro. 2007. Distinct intracellular trafficking of equine infectious anemia virus and human immunodeficiency virus type 1 Gag during viral assembly and budding revealed by bimolecular fluorescence complementation assays. J. Virol. 8111226-11235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin, L., B. W. Guzik, Y. C. Bor, D. Rekosh, and M. L. Hammarskjold. 2003. Tap and NXT promote translation of unspliced mRNA. Genes Dev. 173075-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koito, A., H. Shigekane, and S. Matsushita. 2003. Ability of small animal cells to support the postintegration phase of human immunodeficiency virus type-1 replication. Virology 305181-191. [DOI] [PubMed] [Google Scholar]
- 28.Lei, E. P., and P. A. Silver. 2002. Protein and RNA export from the nucleus. Dev. Cell 2261-272. [DOI] [PubMed] [Google Scholar]
- 29.Maddon, P. J., A. G. Dalgleish, J. S. McDougal, P. R. Clapham, R. A. Weiss, and R. Axel. 1986. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell 47333-348. [DOI] [PubMed] [Google Scholar]
- 30.Malim, M. H., D. F. McCarn, L. S. Tiley, and B. R. Cullen. 1991. Mutational definition of the human immunodeficiency virus type 1 Rev activation domain. J. Virol. 654248-4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mariani, R., B. A. Rasala, G. Rutter, K. Wiegers, S. M. Brandt, H. G. Krausslich, and N. R. Landau. 2001. Mouse-human heterokaryons support efficient human immunodeficiency virus type 1 assembly. J. Virol. 753141-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mariani, R., G. Rutter, M. E. Harris, T. J. Hope, H. G. Krausslich, and N. R. Landau. 2000. A block to human immunodeficiency virus type 1 assembly in murine cells. J. Virol. 743859-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muriaux, D., J. Mirro, D. Harvin, and A. Rein. 2001. RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. USA 985246-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ono, A., S. D. Ablan, S. J. Lockett, K. Nagashima, and E. O. Freed. 2004. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc. Natl. Acad. Sci. USA 10114889-14894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ono, A., and E. O. Freed. 1999. Binding of human immunodeficiency virus type 1 Gag to membrane: role of the matrix amino terminus. J. Virol. 734136-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ono, A., M. Huang, and E. O. Freed. 1997. Characterization of human immunodeficiency virus type 1 matrix revertants: effects on virus assembly, Gag processing, and Env incorporation into virions. J. Virol. 714409-4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ott, D. E., L. V. Coren, and T. D. Gagliardi. 2005. Redundant roles for nucleocapsid and matrix RNA-binding sequences in human immunodeficiency virus type 1 assembly. J. Virol. 7913839-13847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paillart, J. C., and H. G. Gottlinger. 1999. Opposing effects of human immunodeficiency virus type 1 matrix mutations support a myristyl switch model of Gag membrane targeting. J. Virol. 732604-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perez-Caballero, D., T. Hatziioannou, J. Martin-Serrano, and P. D. Bieniasz. 2004. Human immunodeficiency virus type 1 matrix inhibits and confers cooperativity on Gag precursor-membrane interactions. J. Virol. 789560-9563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pollard, V. W., and M. H. Malim. 1998. The HIV-1 Rev protein. Annu. Rev. Microbiol. 52491-532. [DOI] [PubMed] [Google Scholar]
- 41.Purohit, P., S. Dupont, M. Stevenson, and M. R. Green. 2001. Sequence-specific interaction between HIV-1 matrix protein and viral genomic RNA revealed by in vitro genetic selection. RNA 7576-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reed, M., R. Mariani, L. Sheppard, K. Pekrun, N. R. Landau, and N. W. Soong. 2002. Chimeric human immunodeficiency virus type 1 containing murine leukemia virus matrix assembles in murine cells. J. Virol. 76436-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reil, H., A. A. Bukovsky, H. R. Gelderblom, and H. G. Gottlinger. 1998. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J. 172699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Resh, M. D. 2004. A myristoyl switch regulates membrane binding of HIV-1 Gag. Proc. Natl. Acad. Sci. USA 101417-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saad, J. S., E. Loeliger, P. Luncsford, M. Liriano, J. Tai, A. Kim, J. Miller, A. Joshi, E. O. Freed, and M. F. Summers. 2007. Point mutations in the HIV-1 matrix protein turn off the myristyl switch. J. Mol. Biol. 366574-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saad, J. S., J. Miller, J. Tai, A. Kim, R. H. Ghanam, and M. F. Summers. 2006. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc. Natl. Acad. Sci. USA 10311364-11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sandefur, S., V. Varthakavi, and P. Spearman. 1998. The I domain is required for efficient plasma membrane binding of human immunodeficiency virus type 1 Pr55Gag. J. Virol. 722723-2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simon, J. H., R. A. Fouchier, T. E. Southerling, C. B. Guerra, C. K. Grant, and M. H. Malim. 1997. The Vif and Gag proteins of human immunodeficiency virus type 1 colocalize in infected human T cells. J. Virol. 715259-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Swanson, C. M., and M. H. Malim. 2006. Retrovirus RNA trafficking: from chromatin to invasive genomes. Traffic 71440-1450. [DOI] [PubMed] [Google Scholar]
- 50.Swanson, C. M., B. A. Puffer, K. M. Ahmad, R. W. Doms, and M. H. Malim. 2004. Retroviral mRNA nuclear export elements regulate protein function and virion assembly. EMBO J. 232632-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang, C., E. Loeliger, P. Luncsford, I. Kinde, D. Beckett, and M. F. Summers. 2004. Entropic switch regulates myristate exposure in the HIV-1 matrix protein. Proc. Natl. Acad. Sci. USA 101517-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trono, D., and D. Baltimore. 1990. A human cell factor is essential for HIV-1 Rev action. EMBO J. 94155-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wei, P., M. E. Garber, S. M. Fang, W. H. Fischer, and K. A. Jones. 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92451-462. [DOI] [PubMed] [Google Scholar]
- 54.Wodrich, H., A. Schambach, and H. G. Krausslich. 2000. Multiple copies of the Mason-Pfizer monkey virus constitutive RNA transport element lead to enhanced HIV-1 Gag expression in a context-dependent manner. Nucleic Acids Res. 28901-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang, Y., H. Qian, Z. Love, and E. Barklis. 1998. Analysis of the assembly function of the human immunodeficiency virus type 1 Gag protein nucleocapsid domain. J. Virol. 721782-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng, Y. H., H. F. Yu, and B. M. Peterlin. 2003. Human p32 protein relieves a post-transcriptional block to HIV replication in murine cells. Nat. Cell Biol. 5611-618. [DOI] [PubMed] [Google Scholar]
- 57.Zhou, W., and M. D. Resh. 1996. Differential membrane binding of the human immunodeficiency virus type 1 matrix protein. J. Virol. 708540-8548. [DOI] [PMC free article] [PubMed] [Google Scholar]