Abstract
Single-strand conformation polymorphism (SSCP) analysis is used by many laboratories to study the quasispecies distribution of the hepatitis C virus (HCV). Here we question the validity of this experimental approach, as conclusions are drawn from the analysis of the migration patterns of two ssDNA molecules and not from RNA. Using previously characterized mutants of the HCV 5′ untranslated regions, we show that contrary to what has been predicted, SSCP migration patterns of DNA amplicons with differences in their nucleotide sequences generated from the full 5′ UTR of HCV are not necessarily unique.
Single-strand conformation polymorphism (SSCP) analysis is a method used for detecting sequence differences of single-stranded DNA (ssDNA) by nondenaturing polyacrylamide gel electrophoresis (PAGE) (25). In general, the SSCP process involves PCR amplification of the target DNA, denaturation of the double-stranded PCR product with heat and formamide (or other denaturants), and sample resolution by nondenaturing PAGE (10, 23, 25). During electrophoresis, ssDNA fragments are expected to fold into a three-dimensional shape depending mainly on their primary sequence (10, 23, 25). Several authors have suggested that even if the difference in the sequence between the wild-type sample and a mutated fragment is just a single nucleotide, a unique and distinct electrophoretic mobility pattern will be adopted by each sequence (9, 12). Therefore, complex mixtures of DNA species of the same size can be separated by nondenaturing PAGE into bands of different mobility, due to a difference in their predominant semistable conformations. At present, no adequate theoretical model exists for predicting the three-dimensional structure assumed by an ssDNA fragment with a known nucleotide sequence under a given set of conditions (10, 23). Therefore, for each ssDNA fragment, the number of stable conformations which give rise to bands with different mobilities during SSCP electrophoresis must be determined experimentally under rigorously controlled conditions.
Numerous parameters have been empirically found to affect the sensitivity of SSCP analysis, including the type of mutation, size of the DNA fragment, G and C content, content of polyacrylamide or other gel matrix constituents, gel size and potential, gel temperature during electrophoresis, DNA concentration, electrophoresis run time, and buffer composition, including ionic strength and pH (1, 12, 13, 15, 21, 23). In consequence, SSCP optimization today remains a highly empirical experimental approach. Yet, in despite of these apparent experimental constraints, SSCP analysis has been widely used to detect mutations in oncogenes, tumor suppressor genes, and genes responsible for genetic diseases, among others (10-12, 15, 17, 23, 25, 31). SSCP remains a favored method not only to screen potential sequence variations but also to identify new mutations within genes. Moreover, its speed and simplicity in the detection of mutations within DNA molecules make it attractive for use in clinical diagnostic laboratories (3, 8, 17).
SSCP analysis is used by many laboratories today to study virus quasispecies distribution. Populations of RNA viruses in vivo consist of heterogeneous mixtures of genetically different but closely related variants, referred to as “quasispecies” (5, 14, 30). The heterogeneity of the genome of RNA viruses is a consequence of continuous and elevated replication rates in combination with error-prone replication catalyzed by viral RNA-dependent RNA polymerases, which lack proofreading activity. The ability to form quasispecies endows the virus with an extraordinary capacity to adapt to new environmental conditions, such as immune pressure or antiviral therapy (6, 24). The study of RNA virus quasispecies distribution by SSCP requires an additional step to the above experimental procedure, namely, that of reverse transcription. Moreover, in quasispecies studies of RNA viruses, conclusions are drawn from the analysis of the migration patterns of two ssDNA molecules. These facts prompted us to question the validity of using SSCP assays in the study of quasispecies distribution of the hepatitis C virus (HCV), an RNA virus, classified in the genus Hepacivirus within the family Flaviviridae (20).
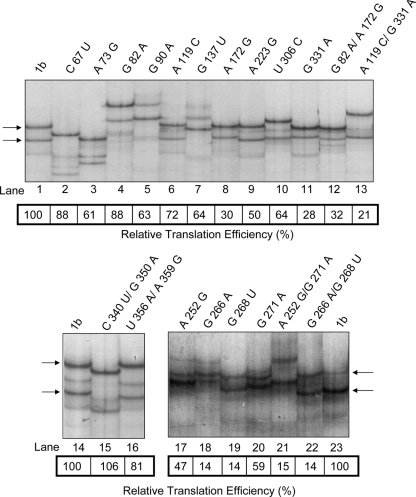
In a recent report, we described the isolation of a series of naturally occurring mutations present within the 5′ untranslated region (5′ UTR) of the HCV RNA (2). The HCV 5′ UTR corresponds to the most conserved region within the HCV RNA genome. The conservation of the 5′ UTR is not limited to the primary sequence but also applies to the overall RNA structure (7, 22). The HCV 5′ UTR harbors the internal ribosomal entry site (IRES), an RNA structure required for translation initiation of the viral polyprotein (7, 22). The ability of the different IRESs, isolated from clinical samples, to drive translation in rabbit reticulocyte lysate was evaluated in vitro using a dual luciferase (Renilla and firefly luciferase) bicistronic mRNA (2). The relative translational efficiency (RTE) was used in this study as an index of IRES activity, with the mean translation efficiency of the control HCV-1b IRES (wild type; nucleotides 13 to 383; GenBank no. AJ242654) being arbitrarily defined as 100%. To determine the influence of each of the identified mutations on viral IRES activity, substitutions were introduced into the HCV-1b IRES, and their individual effect on translation was evaluated in vitro (2). Structural analysis of the different RNA mutants revealed that the G266A/G268U double mutant conserved the structure of the wild-type IRES, yet its translational activity was abrogated (2). Based on this finding, we sought to establish whether, as predicted by others (9, 12), analysis of the SSCP pattern of the PCR amplification product of the different HCV IRES mutants (DNA plasmids) allowed their easy differentiation. We anticipated the SSCP patterns to be unique for each of the tested sequences (9, 12).
Figure 1 shows the SSCP patterns of the DNA amplicon and RTE for each RNA mutant (2). SSCP patterns for the 5′ UTR were established for 20 well-characterized HCV mutants following a previously described experimental protocol (33). Amplification of the HCV 5′ UTR was achieved using the sense primer 5′-TTG GGG GCG ACA CTC CAC CAT AGA TC-3′ and the antisense primer 5′-GTT ACG TTT GGT TTT TCT TTG AGG T-3′, generating a 370-bp amplicon. All amplicons were sequenced (Macrogen Corp, Rockville, MD). In these assays, the SSCP migration pattern of the wild-type HCV-1b IRES was used as a control (Fig. 1, lanes 1, 14, and 23). To lower the risk of false polymorphisms, which would lead to data misinterpretation, SSCP analysis was duplicated in an independent experiment using new DNA templates. PCR products were checked and used only if a single sharp product band was observed on a regular agarose gel. Data were considered only if the results of the control experiment were consistent with the initial findings. In agreement with what has been described by many (12, 13, 15, 23), SSCP patterns were shown to vary for the same sample in different assays (compare data for the wild-type HCV 5′ UTR in Fig. 1, lanes 1, 14, and 23). As expected, PCR products from most mutant 5′ UTRs yielded unique and distinct SSCP patterns (Fig. 1). Strikingly, mutants A119C (Fig. 1, lane 6), A223G (Fig. 1, lane 9), U356A/A359G (Fig. 1, lane 16), and G266A/G268U (Fig. 1, lane 22) exhibited SSCP patterns visually indistinguishable from the wild-type pattern (Fig. 1, lanes 1, 14, and 23). This observation was consistent internally; in most cases we were unable to compare the same sample in different SSCP runs due to elevated interassay variability. The presence of the HCV-1b control (Fig. 1, lanes 1, 14, and 23) turned out to be crucial in order to permit valid conclusions with respect to the significance of a particular SSCP pattern. No correlation between RTE and SSCP migration pattern could be established. For example, the patterns for mutants A119C (Fig. 1, lane 6) and A223G (Fig. 1, lane 9), or G82A (Fig. 1, lane 4) and G90A (Fig. 1, lane 5), were indistinguishable, yet their RTEs were not equivalent. Mutants A73G, G90A, A119C, and U306C (Fig. 1, lanes 3, 5, 6, and 10) exhibited comparable RTE values even though their SSCP patterns were not similar. An equivalent conclusion can be drawn from analyzing mutants C67U and G82A (Fig. 1, lanes 2 and 4).
FIG. 1.
SSCP analysis of the full HCV 5′ UTR. DNA from plasmids harboring the mutant HCV sequences (2) were used as PCR templates. SSCP assays were conducted as previously described (33). The RTEs of these mutant IRES elements (shown in boxes) were previously reported (2). Arrows indicate the SSCP migration pattern of the wild-type 1b HCV IRES in each gel.
Several studies on HCV compartmentalization or viral quasispecies distribution have drawn conclusions partially based on the SSCP analysis of the amplicons generated from the HCV 5′ UTR (4, 16, 18, 19, 27, 28, 33). However, data presented in Fig. 1 indicate that conclusions based exclusively on SSCP patterns of the PCR product of the full HCV 5′ UTR followed by sequencing of the major circulating viral populations might in fact underestimate the real number of virus quasispecies. A number of studies have described mutations within the HCV IRES capable of hindering translational activity without altering the overall RNA structure of this element (2, 26, 32). In the wild, these mutant viruses would clearly correspond to HCV quasispecies. Here, we report that the DNA product of one such mutant, G266A/G268U, exhibited an SSCP pattern that is indistinguishable from the pattern generated by the amplicon of the wild-type HCV IRES (Fig. 1, compare lanes 22 and 23). One possible explanation for the lack of sensitivity in the SSCP assay results presented herein might reside in the length of the analyzed amplicon (12, 13, 29). Yet, even accounting for this possible constraint and given the fact that most studies target the full 5′ UTR, we conclude that data generated using SSCP analysis of reverse transcription-PCR products must be regarded with caution. Contrary to what has been predicted by others, we provide evidence in this study that some mutated DNA amplicons generated by PCR may not be reliably distinguishable from wild-type HCV IRES PCR products by this widely used experimental approach.
Acknowledgments
We thank M. Rau for critical reading and editing of the manuscript.
This study was supported by FONDECYT no. 1080323 to A.S. and M.L.-L. J.V.-O. was supported by a MECESUP-USACH doctoral fellowship; M.I.B. was supported by a CONICYT and an IRD doctoral fellowship.
Footnotes
Published ahead of print on 24 June 2009.
REFERENCES
- 1.Balogh, K., A. Patocs, J. Majnik, K. Racz, and L. Hunyady. 2004. Genetic screening methods for the detection of mutations responsible for multiple endocrine neoplasia type 1. Mol. Genet. Metab. 8374-81. [DOI] [PubMed] [Google Scholar]
- 2.Barria, M. I., A. Gonzalez, J. Vera-Otarola, U. Leon, V. Vollrath, D. Marsac, O. Monasterio, T. Perez-Acle, A. Soza, and M. Lopez-Lastra. 2009. Analysis of natural variants of the hepatitis C virus internal ribosome entry site reveals that primary sequence plays a key role in cap-independent translation. Nucleic Acids Res. 37957-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Costabile, M., A. Quach, and A. Ferrante. 2006. Molecular approaches in the diagnosis of primary immunodeficiency diseases. Hum. Mutat. 271163-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Di Liberto, G., A. M. Roque-Afonso, R. Kara, D. Ducoulombier, G. Fallot, D. Samuel, and C. Feray. 2006. Clinical and therapeutic implications of hepatitis C virus compartmentalization. Gastroenterology 13176-84. [DOI] [PubMed] [Google Scholar]
- 5.Domingo, E., C. Escarmis, N. Sevilla, A. Moya, S. F. Elena, J. Quer, I. S. Novella, and J. J. Holland. 1996. Basic concepts in RNA virus evolution. FASEB J. 10859-864. [DOI] [PubMed] [Google Scholar]
- 6.Domingo, E., and J. J. Holland. 1997. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 51151-178. [DOI] [PubMed] [Google Scholar]
- 7.Fraser, C. S., and J. A. Doudna. 2007. Structural and mechanistic insights into hepatitis C viral translation initiation. Nat. Rev. Microbiol. 529-38. [DOI] [PubMed] [Google Scholar]
- 8.Gasser, R. B., and N. B. Chilton. 2001. Applications of single-strand conformation polymorphism (SSCP) to taxonomy, diagnosis, population genetics and molecular evolution of parasitic nematodes. Vet. Parasitol. 101201-213. [DOI] [PubMed] [Google Scholar]
- 9.Glavac, D., and M. Dean. 1993. Optimization of the single-strand conformation polymorphism (SSCP) technique for detection of point mutations. Hum. Mutat. 2404-414. [DOI] [PubMed] [Google Scholar]
- 10.Hayashi, K. 1992. PCR-SSCP: a method for detection of mutations. Genet. Anal. Technol. Appl. 973-79. [DOI] [PubMed] [Google Scholar]
- 11.Hayashi, K. 1991. PCR-SSCP: a simple and sensitive method for detection of mutations in the genomic DNA. PCR Methods Appl. 134-38. [DOI] [PubMed] [Google Scholar]
- 12.Hayashi, K., and D. W. Yandell. 1993. How sensitive is PCR-SSCP? Hum. Mutat. 2338-346. [DOI] [PubMed] [Google Scholar]
- 13.Highsmith, W. E., Jr., A. J. Nataraj, Q. Jin, J. M. O'Connor, S. H. El-Nabi, N. Kusukawa, and M. M. Garner. 1999. Use of DNA toolbox for the characterization of mutation scanning methods. II: evaluation of single-strand conformation polymorphism analysis. Electrophoresis 201195-1203. [DOI] [PubMed] [Google Scholar]
- 14.Holland, J. J., J. C. De La Torre, and D. A. Steinhauer. 1992. RNA virus populations as quasispecies. Curr. Top. Microbiol. Immunol. 1761-20. [DOI] [PubMed] [Google Scholar]
- 15.Humphries, S. E., V. Gudnason, R. Whittall, and I. N. Day. 1997. Single-strand conformation polymorphism analysis with high throughput modifications, and its use in mutation detection in familial hypercholesterolemia. International Federation of Clinical Chemistry Scientific Division: Committee on Molecular Biology Techniques. Clin. Chem. 43427-435. [PubMed] [Google Scholar]
- 16.Jang, S. J., L. F. Wang, M. Radkowski, J. Rakela, and T. Laskus. 1999. Differences between hepatitis C virus 5′ untranslated region quasispecies in serum and liver. J. Gen. Virol. 80711-716. [DOI] [PubMed] [Google Scholar]
- 17.Kakavas, V. K., P. Plageras, T. A. Vlachos, A. Papaioannou, and V. A. Noulas. 2008. PCR-SSCP: a method for the molecular analysis of genetic diseases. Mol. Biotechnol. 38155-163. [DOI] [PubMed] [Google Scholar]
- 18.Laskus, T., M. Radkowski, L. F. Wang, H. Vargas, and J. Rakela. 1998. Search for hepatitis C virus extrahepatic replication sites in patients with acquired immunodeficiency syndrome: specific detection of negative-strand viral RNA in various tissues. Hepatology 281398-1401. [DOI] [PubMed] [Google Scholar]
- 19.Laskus, T., M. Radkowski, J. Wilkinson, H. Vargas, and J. Rakela. 2002. The origin of hepatitis C virus reinfecting transplanted livers: serum-derived versus peripheral blood mononuclear cell-derived virus. J. Infect. Dis. 185417-421. [DOI] [PubMed] [Google Scholar]
- 20.Lauer, G. M., and B. D. Walker. 2001. Hepatitis C virus infection. N. Engl. J. Med. 34541-52. [DOI] [PubMed] [Google Scholar]
- 21.Liechti-Gallati, S., V. Schneider, D. Neeser, and R. Kraemer. 1999. Two buffer PAGE system-based SSCP/HD analysis: a general protocol for rapid and sensitive mutation screening in cystic fibrosis and any other human genetic disease. Eur. J. Hum. Genet. 7590-598. [DOI] [PubMed] [Google Scholar]
- 22.Lukavsky, P. J. 2008. Structure and function of HCV IRES domains. Virus Res. 139166-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nataraj, A. J., I. Olivos-Glander, N. Kusukawa, and W. E. Highsmith, Jr. 1999. Single-strand conformation polymorphism and heteroduplex analysis for gel-based mutation detection. Electrophoresis 201177-1185. [DOI] [PubMed] [Google Scholar]
- 24.Nijhuis, M., N. M. van Maarseveen, and C. A. Boucher. 2009. Antiviral resistance and impact on viral replication capacity: evolution of viruses under antiviral pressure occurs in three phases. Handb. Exp. Pharmacol. 2009299-320. [DOI] [PubMed] [Google Scholar]
- 25.Orita, M., H. Iwahana, H. Kanazawa, K. Hayashi, and T. Sekiya. 1989. Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc. Natl. Acad. Sci. USA 862766-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Psaridi, L., U. Georgopoulou, A. Varaklioti, and P. Mavromara. 1999. Mutational analysis of a conserved tetraloop in the 5′ untranslated region of hepatitis C virus identifies a novel RNA element essential for the internal ribosome entry site function. FEBS Lett. 45349-53. [DOI] [PubMed] [Google Scholar]
- 27.Roque-Afonso, A. M., D. Ducoulombier, G. Di Liberto, R. Kara, M. Gigou, E. Dussaix, D. Samuel, and C. Feray. 2005. Compartmentalization of hepatitis C virus genotypes between plasma and peripheral blood mononuclear cells. J. Virol. 796349-6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sampietro, M., S. Badalamenti, S. Salvadori, N. Corbetta, G. Graziani, G. Como, G. Fiorelli, and C. Ponticelli. 1995. High prevalence of a rare hepatitis C virus in patients treated in the same hemodialysis unit: evidence for nosocomial transmission of HCV. Kidney Int. 47911-917. [DOI] [PubMed] [Google Scholar]
- 29.Sheffield, V. C., J. S. Beck, A. E. Kwitek, D. W. Sandstrom, and E. M. Stone. 1993. The sensitivity of single-strand conformation polymorphism analysis for the detection of single base substitutions. Genomics 16325-332. [DOI] [PubMed] [Google Scholar]
- 30.Steinhauer, D. A., and J. J. Holland. 1987. Rapid evolution of RNA viruses. Annu. Rev. Microbiol. 41409-433. [DOI] [PubMed] [Google Scholar]
- 31.Sunnucks, P., A. C. Wilson, L. B. Beheregaray, K. Zenger, J. French, and A. C. Taylor. 2000. SSCP is not so difficult: the application and utility of single-stranded conformation polymorphism in evolutionary biology and molecular ecology. Mol. Ecol. 91699-1710. [DOI] [PubMed] [Google Scholar]
- 32.Tang, S., A. J. Collier, and R. M. Elliott. 1999. Alterations to both the primary and predicted secondary structure of stem-loop IIIc of the HCV-1b 5′ untranslated region (5′ UTR) lead to mutants severely defective in translation which cannot be complemented in trans by the wild-type 5′ UTR sequence. J. Virol. 732359-2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vera-Otarola, J., M. I. Barria, U. Leon, D. Marsac, P. Carvallo, A. Soza, and M. Lopez-Lastra. 2009. Hepatitis C virus quasispecies in plasma and peripheral blood mononuclear cells of treatment naive chronically infected patients. J. Viral Hepat. [Epub ahead of print.] [DOI] [PubMed]