Abstract
The original cotransfection replication assay identified eight human herpesvirus 8 (HHV8)-encoded proteins required for origin-dependent lytic DNA replication. Previously, we demonstrated that under conditions where K-Rta is overexpressed, a K-bZIP knockout bacmid displayed an aberrant subcellular localization pattern for the latency-associated nuclear protein (LANA). Additionally, these same studies demonstrated that K-bZIP interacts with LANA in the absence of K-Rta and that K-bZIP does not directly participate in, but may facilitate, the initiation of lytic DNA synthesis. We developed a modification of the transient cotransfection replication assay wherein both lytic (oriLyt) and latent (terminal repeat) DNA replication are evaluated simultaneously. We now show that LANA represses origin-dependent lytic DNA replication in a dose dependent manner when added to the cotransfection replication assay. This repression was overcome by increasing amounts of a K-bZIP expression plasmid in the cotransfection mixture or by dominant-negative inhibition of the interaction of LANA with K-bZIP by the overexpression of the K-bZIP-LANA binding domain. Chromatin immunoprecipitation assays show that LANA interacts with oriLyt in the absence of K-bZIP expression, suggesting that suppression of lytic replication by LANA is mediated by direct binding. The interaction of K-bZIP with oriLyt was dependent upon the expression of LANA; however, LANA interacted with oriLyt independently of K-bZIP expression. These data suggest that the interaction of LANA with K-bZIP modulates lytic and latent replication and that K-bZIP facilitates lytic DNA replication and modulates the switch from the latent phase of the virus.
Kaposi's sarcoma-associated herpesvirus or human herpesvirus 8 (KSHV or HHV8, respectively) is a gammaherpesvirus and the cause of Kaposi sarcoma, primary effusion lymphoma, and multicentric Castleman disease (6). The gene expression profile of HHV8 can be divided into two distinct infection phases, lytic and latent. During latent infection, there is no viral progeny produced and the HHV8 genome is maintained as multiple episomes in host cells. HHV8 viral DNA is replicated once per cell cycle and partitioned into daughter cells along with the host cell chromosomes (13, 31). Lytic replication is marked by an increase in gene expression and the production of infectious virus progeny.
Latently infected cells express only a small subset of genes thought to be critical for maintenance of the latent genome. During latent as well as lytic infection the latency-associated nuclear antigen (LANA) open reading frame 73 (ORF73) is the predominant viral antigen expressed (18). LANA facilitates latent viral DNA synthesis and tethers the HHV8 episome to the host chromosome, ensuring that the genome is distributed to daughter cells during each cell division. LANA is a 1,162-amino-acid (aa) peptide with a calculated molecular mass of 135 kDa although it typically migrates between 220 to 230 kDa on a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel (8, 17). LANA contains three distinct protein domains: an N-terminal basic domain of 337 aa, a middle 585-aa hydrophilic region, and a C-terminal basic 240-aa domain (29).
LANA is localized to the nucleus of HHV8 latently infected cells, and a nuclear localization sequence (NLS) between aa 24 to 30 was identified and is homologous to the NLS for Epstein-Barr virus EBNA1 (30). LANA can repress as well as activate transcription (11, 14, 15, 25, 28). LANA can autoactivate transcription from its own promoter, presumably to maintain the expression of latent proteins while suppressing other viral genes (15, 28). LANA maintains viral latency by regulating the immediate-early transcriptional regulator protein K-Rta and can repress the transcriptional activity of the K-Rta promoter, leading to a decrease in both HHV8 virus production and lytic cycle activation (22). Additionally, increased levels of K-Rta are observed when LANA protein expression is suppressed using small interfering RNA (10). These studies suggest that regulation of viral latency is by the functional interaction between LANA and K-Rta, which in turn appears to contribute to the switch between latent and lytic infection.
Recently, it was demonstrated that the gene product of K8, K-bZIP, interacts with LANA in transfected and infected cells (16). In the context of the viral genome, the absence of K-bZIP expression resulted in a significant increase in virus gene expression and production of virus under conditions where K-Rta was overexpressed (16). However, when tetradecanoyl phorbol acetate/n-butyrate was used to induce the virus lytic cycle, a marked decrease in gene expression occurred, and no virus production was observed (16). This result indicated that K-bZIP was not absolutely necessary for virus growth and that K-Rta was able to compensate for the lack of K-bZIP expression. Interestingly, immunofluorescence staining revealed that the subcellular localization of K-Rta was unchanged; however, a disruption of the nuclear punctate subcellular localization of LANA was observed in cells harboring recombinant bacterial artificial chromosome (BAC) with a deletion of K-bZIP (ΔK-bZIP). These data showed that K-bZIP influences LANA localization and suggested that it may regulate the activity of LANA with respect to controlling the switch to the lytic cycle.
In this report we define the protein binding domains for LANA and K-bZIP and investigate the significance of the interaction between these two proteins in the context of the regulation of lytic and latent DNA replication. Using a modification of the original transient cotransfection replication assay, we show that the interaction of K-bZIP with LANA modulates the switch from latent to lytic infection. We cotransfected plasmids containing HHV8 terminal repeat (TR) elements along with a plasmid containing oriLyt to evaluate both lytic and latent replication simultaneously. This lytic/latent replication assay was used to assess the contribution of K-bZIP and LANA to amplification of either replication element (5, 12, 13, 26). The presence of LANA alone in the cotransfection mixture repressed oriLyt amplification, and the interaction of K-bZIP with LANA was essential to modulate the switch from lytic to latent transient replication. This repression was relieved by transfection of a K-bZIP expression plasmid or a plasmid that expressed the K-bZIP peptide that interacts with LANA. Increasing concentrations of K-bZIP in the transfection mixture shifted amplification of the TR-containing plasmid to an increase in accumulation of replicated oriLyt. Lastly, we used a chromatin immunoprecipitation (ChIP) assay to show that LANA binds to a region of oriLyt that contains CCAAT/enhancer binding protein α (C/EBPα) transcription factor binding sites, the same region shown previously to interact with K-bZIP. Additionally, using LANA or K-bZIP knockout bacmids, we show that the interaction of K-bZIP with oriLyt is dependent upon LANA expression, whereas LANA interacted with oriLyt independent of K-bZIP expression. These results implicate K-bZIP as a regulator protein that does not directly participate in lytic DNA replication but instead modulates latent and lytic replication through an interaction with LANA and oriLyt.
MATERIALS AND METHODS
Cells and bacmids.
Vero and HEK293 cells were maintained in Dulbecco's modified Eagle medium supplemented with 10% bovine growth serum (HyClone, Logan, UT). The wild-type (wt) HHV8 bacmid and the LANA knockout bacmid were previously described (24) and were obtained from S.-J. Gao (University of Texas). The HHV8 recombinant bacmids that have a deletion of K-bZIP or of the ORF50 locus have been previously described (16, 33).
Vero cells containing the BAC constructs were maintained in Dulbecco's modified Eagle medium supplemented with 10% bovine growth serum and 250 mg/ml hygromycin.
Plasmids.
The set of plasmids that expressed the deletions of K-bZIP was previously described (1). The K-bZIP interaction domain expression plasmid, pZIP-ID, was generated by using primers that flanked the region of aa 101 to 134 and that had an in-frame ATG for proper protein translation and a 3′ FLAG epitope tag. The PCR product was ligated into phCMV-xi1 (Genlantis) with the forward primer cgacttaacagatctcgagctcaagcttcgaattcATGCTGAATGCAGAAACTAAATTCCACATCCCC and the reverse primercccgggcccgcggtaccgtcgactgcagaattcTCACTTATCGTCGTCATCCTTGTAATCGCCCTGTTTGGCCTTAG TGCATAAGCGTTC. The LANA-green fluorescent protein (GFP) and TR plasmid (pTR8) constructs were a generous gift from Kenneth Kaye (Harvard University).
Coimmunoprecipitation assay.
For cotransfection, HEK293 cells (2 × 106 cells/10-cm dish) were transfected with LANA and K-bZIP expression plasmids using TransIT LT1 (Mirrus). Forty-eight hours posttransfection, protein extracts were prepared using lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Trition X-100, and 0.1% NP-40), passed through a 23-gauge needle to shear the DNA, and centrifuged at 10,000 × g for 10 min to remove debris; lysate was precleared with mouse immunoglobulin G-agarose conjugate (Santa Cruz Biotechnology) at 4°C for 30 min, and then 50 μl of anti-hemagglutinin (HA) affinity agarose gel (Sigma) was added to the lysate. This mixture was rotated at 4°C overnight. The beads were then washed four times with 1 ml of Tris-buffered saline (Tris-HCl, pH 7.4, 150 mM NaCl), each time with rotation for 10 min at 4°C. Twenty microliters of the immunoprecipitated protein was separated through a 10% SDS-PAGE gel, which was subsequently transferred to an Immun-Blot polyvinylidene difluoride membrane (Bio-Rad). After an initial blocking step (15 min with Tris-buffered saline plus 5% nonfat milk), the blots were reacted with anti-HA, anti-LANA, or anti-K-bZIP antibodies overnight at 4°C, followed by washing and incubation with horseradish peroxidase-conjugated secondary antibody anti-immunoglobulin G (SantaCruz). Protein bands were visualized using a chemiluminescence substrate (Femto, Pierce).
ChIP assay.
For induced samples, a Vero cell line (4 × 107 cells) containing wt BAC36 was treated with tetradecanoyl phorbol acetate (25 ng/ml) for 5 days. For uninduced cells, cultures were grown to the same density, and both samples were treated as follows. The protocol for the ChIP assay was modified from the Active Motif ChIP-IT express kit (catalog number 53008). Cells were washed once with phosphate-buffered saline (PBS), fixed in a 1% formaldehyde-PBS solution for 10 min, and then washed twice in PBS. Three milliliters of PBS was added onto the cells, and the cells were scraped, spun down at 1,500 × g, resuspended in 750 μl of lysis buffer plus protease inhibitors, and incubated on ice for 30 min. This solution was then sonicated 10 times with a 10-s pulse. Sonicated samples were analyzed by agarose gel electrophoresis prior to further use. Prior to immunoprecipitation, 10 μl was removed for the input sample. Sheared samples (161 μl) were incubated for 15 h at 4°C with 25 μl of magnetic beads, 10 μl of ChIP buffer 1 (Active Motif), 3 μl of antibody (anti-K-LANA [ABI] or MAb84, an isotype control), and 1 μl protease inhibitor cocktail (Sigma). After incubation the magnetic beads were removed by being placed on a magnetic stand, and the supernatant was removed. Beads were washed once with 800 μl of ChIP buffer 1, followed by a two washes with 800 μl of ChIP buffer 2 (Active Motif), with rotation for 10 min at 4°C after the addition of each wash buffer. Beads were then resuspended with 50 μl of elution buffer AM2 and incubated for 15 min at room temperature. Fifty microliters of reverse cross-link buffer was added, and beads were quickly pelleted and transferred to a new tube. A total of 88 μl of ChIP buffer 2 and 2 μl of 5 M NaCl was added to input samples. Samples were then incubated at 94°C for 15 min. Two microliters of proteinase K solution was added, and samples were then incubated at 37°C for 1 h; finally, 2 μl of proteinase K stop solution was added. This solution was then used for PCR analysis.
The primers used for PCR amplification (forward, 5′-AATCCCCCATAATCCTCTGC-3′; reverse, 5′-GGAAAAATCAAAACAAAACTC-3′) corresponded to nucleotides 23326 to 23572 of HHV8 oriLyt. The control primers, 5′-ACGTCCGGAGAGTTGGAACTGTCA-3′ (forward) and 5′-GGGGTCCATGGGATGGGTTAGTCA-3′ (reverse), were complementary to the ORF45 region.
Transient lytic/latent replication assay.
Vero cells were transfected with plasmids containing HHV8 oriLyt (2) and TRs (4, 9, 26) as well as the complete set of core replication proteins, ORF6, ORF9, ORF40/41, ORF44, ORF56, ORF59, K-bZIP, and K-Rta, with or without a LANA expression plasmid. Cells were harvested 5 to 7 days posttransfection, and total cellular DNA was isolated and cleaved with EcoRI and DpnI. Cleaved DNA was separated using a 0.8% agarose gel and transferred to a nylon membrane and hybridized to a 32P-labeled pGEM probe.
RESULTS
A 33-aa region of K-bZIP mediates LANA binding.
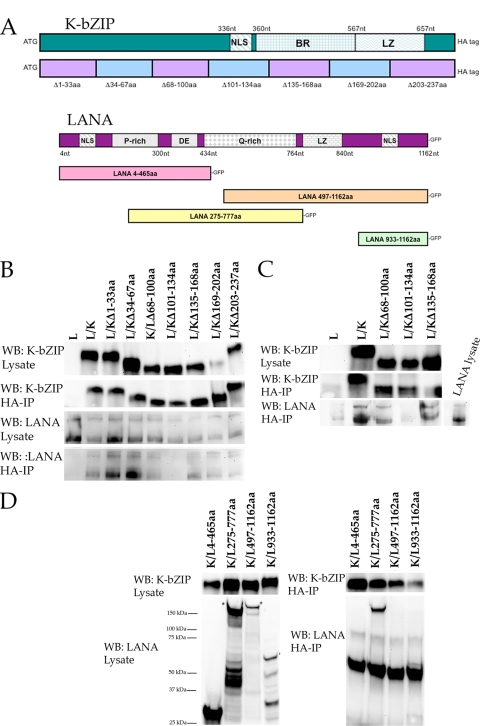
We previously demonstrated that K-bZIP interacts with LANA in transfected cells. To determine the region of the K-bZIP ORF mediating the binding with LANA, we generated a series of deletion mutants for K-bZIP and used the resulting expression plasmids in a series of cotransfection-immunoprecipitation experiments. We generated seven HA-tagged expression plasmids, each having a different 33-aa deletion within the K-bZIP ORF. Figure 1A is a schematic of the K-bZIP ORF showing a series of K-bZIP mutants used in cotransfection-coimmunoprecipitation assays. Plasmids expressing K-bZIP fragments were each transfected along with a full-length wt LANA expression plasmid that produced a LANA-enhanced GFP (EGFP) fusion protein. Protein extracts were prepared, and protein-protein complexes were immunoprecipitated using anti-HA antibody (SantaCruz). Protein-protein interactions were detected by Western blotting using anti-EGFP antibody (Santa Cruz). Western blots reveled that the K-bZIP mutant with a deletion of aa 101 to 134 (K-bZIP Δ101-134) failed to interact with wt LANA, whereas all other K-bZIP deletion mutants retained their ability to interact with LANA (Fig. 1B). As a control for expression we also evaluated protein lysates by Western blotting for both LANA and K-bZIP (Fig. 1B). All EGFP and HA protein fusion species were expressed at similar levels, and all HA-tagged protein species were efficiently immunoprecipitated (Fig. 1B). These results show that the region between amino acids 101 and 134 of the K-bZIP ORF contributes to the interaction with LANA.
FIG. 1.
Identification of K-bZIP and LANA interaction domains. (A) Schematic of HHV8 K-bZIP and LANA ORFs showing the relative locations of important protein domains and subclones or deletion mutants (K-bZIP) used in cotransfections and coimmunoprecipitations. BR denotes basic region and LZ denotes zipper region of K-bZIP. Full-length protein and subclones of K-bZIP contained an in-frame HA tag for the resulting protein product. For LANA subfragments, all protein products have an in-frame GFP tag. (B) LANA interacts with a domain located within aa 101 to 134 of the K-bZIP ORF. HEK293 cells were cotransfected with a plasmid expressing full-length LANA and various plasmids that express the full-length protein or a series of 7- to 33-aa deletions of the K-bZIP ORF. Protein extracts were immunoprecipitated using anti-HA antibody (HA-IP), and precipitated protein complexes were resolved through an SDS-PAGE; the gel was transferred to polyvinylidene difluoride, and blots were reacted with the specific antibodies listed at left (Western blotting [WB]). Lane L, LANA expression plasmid alone; lane L/K, cotransfection of LANA and full-length K-bZIP expression plasmids. Other lanes are identified by the amino acids deleted; e.g., lane L/KΔ1-33aa indicates the cotransfection of LANA and the K-bZIP Δ1-33 mutant. (C) A K-bZIP mutant lacking aa 101 to 134 fails to interact with LANA. HEK293 cells were transfected with either plasmids that express various deletion mutants of K-bZIP or full-length LANA. Protein extracts were prepared and mixed together prior to immunoprecipitation with anti-HA antibody (HA-IP). Antibodies used for Western blotting are shown at the left of the figure. Lane L, transfection of LANA expression plasmid; lane L/K, mixture of transfection of LANA and K-bZIP expression plasmids, followed by immunoprecipitation using anti-HA antibody. All other lanes represent the mixture of transfection of plasmids expressing LANA and K-bZIP deletion mutants, followed by immunoprecipitation using anti-HA antibody; K-bZIP mutants are identified by the deleted amino acid region, as described for panel B. (D) K-bZIP interacts with the Q-rich region of the LANA ORF. HEK293 cotransfection of a full-length K-bZIP expression plasmid and various plasmid expression regions of the LANA ORF. Protein extracts were immunoprecipitated using anti-HA antibody, resolved using an SDS-PAGE gel, blotted as described above, and reacted with antibodies shown to the left of the figures. Lanes are identified by cotransfections of the full-length K-bZIP (K) with and the indicated amino acid regions of the LANA (L) expression plasmids.
As shown in the schematic in Fig. 1, the region of aa 101 to 134 of K-bZIP contains the partial NLS for the protein. Hence, the fact that we did not observe any interaction of this deletion mutant of K-bZIP with LANA could be due to a lack of proper subcellular localization even though a high level of protein expression was observed by Western blotting. In order to confirm that the region of K-bZIP from aa 101 to 134 was indeed involved in LANA binding and to rule out that the lack of protein-protein interaction was due to the fact that this region of K-bZIP has been implicated in nuclear localization, we performed a lysate-mixing experiment. For this experiment protein lysates were prepared either from cells transfected with K-bZIP (deletion mutants or wt) or from full-length LANA. K-bZIP-transfected protein lysates were mixed with LANA prepared protein lysates, and immunoprecipitations were then performed. Under these conditions, proper subcellular localization is not required since there is no cotransfection, and protein interactions are assayed upon mixing of the lysates. Using this protocol, wt K-bZIP interacted with full-length LANA very efficiently (Fig. 1C). Also, the same protein interactions that were observed from the cotransfection immunoprecipitation assay were also observed using the protein lysate mixing protocol. However, no interaction was observed using the K-bZIP Δ101-134 deletion mutant (Fig. 1C). This result is consistent with the cotransfection data and strongly suggests that the region of interaction within the K-bZIP ORF is between amino acid residues 101 to 134.
LANA amino acids 465 to 777 interact with K-bZIP.
Figure 1 shows a schematic of the LANA ORF. LANA contains several protein domains that contribute to its function along with regions that are highly repetitive. To define the region of LANA that interacts with K-bZIP, we again performed the cotransfection immunoprecipitation assay using several GFP-tagged LANA expression plasmids, each having different amino acid truncations of the LANA ORF plus the full-length K-bZIP expression plasmid that produced an HA-tagged fusion protein. Protein extracts were prepared from cotransfected cells, and interacting proteins were immunoprecipitated using anti-HA antibody. Protein-protein interactions between K-bZIP and LANA were detected by reacting Western blots of the immunoprecipitated protein with an anti-GFP antibody. Protein expression in the lysate was evaluated for both K-bZIP (Fig. 1A, top panel) and LANA truncations (Fig. 1A, bottom panel). Immunoprecipitated protein from cotransfected cells revealed that only one of the LANA truncations interacted with wt K-bZIP (Fig. 1D). The LANA truncations that produced a protein product containing amino acids 4 to 465 did not interact with wt K-bZIP (Fig. 1D). LANA truncations that produced a protein from either aa 487 to 1162 or aa 933 to 1162 also failed to bind to K-bZIP (Fig. 1D). Based on this set of cotransfections and immunoprecipitations, we conclude that the interaction domain for LANA with K-bZIP is contained between amino acids 465 to 777.
LANA represses lytic DNA replication.
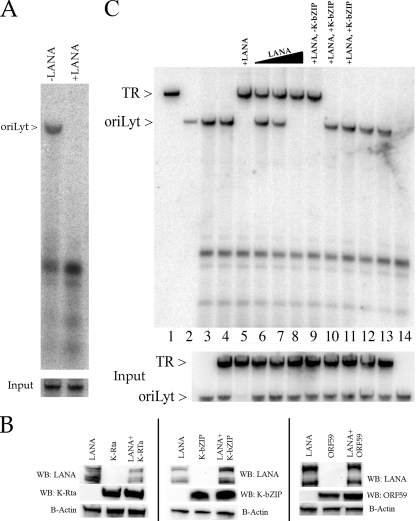
Since we established that K-bZIP, a protein involved in the lytic cycle, and LANA, a factor that has a clear role in latency but is present during the lytic phase of replication, interact in transfected/infected cells, we next wanted to investigate a possible role for these two proteins in lytic replication. The transient cotransfection replication assay involves the cotransfection of a set of plasmids that encode the viral proteins necessary to amplify the lytic origin of replication, oriLyt. For HHV8, oriLyt-dependent DNA replication requires the core replication proteins encoded by ORFs 6, 9, 40 to 41, 44, 56, and 59, which are common to all herpesviruses (1). In addition, the original assay identified ORF 50 (K-Rta) and K-bZIP as also necessary for efficient oriLyt amplification (1). We wanted to investigate the implications of adding a LANA expression plasmid to this transfection mixture. To this end, the cotransfection replication assay was performed as previously described except that a LANA expression plasmid was added to the transfection mixture. Amplification of HHV8-cloned oriLyt was detected by Southern blotting after cleavage of total cellular DNA with EcoRI and DpnI. As shown before, oriLyt efficiently replicated in cells cotransfected with the required replication protein encoding plasmids (Fig. 2A). However, detectable oriLyt amplification was eliminated when a LANA expression plasmid was added to the cotransfection mixture (Fig. 2A). This result suggested that LANA expression was capable of repressing oriLyt amplification.
FIG. 2.
Effects of LANA on lytic DNA replication. (A) LANA represses oriLyt-dependent DNA replication. Expression of LANA in the cotransfection replication assay suppressed amplification of oriLyt. Vero cells were cotransfected with plasmids that express the required replication proteins along with K-Rta and K-bZIP expression plasmids and a plasmid containing oriLyt. A LANA expression plasmid was either present (+LANA) or absent (−LANA) from the cotransfection mixture. The arrow to the left of the figure shows the presence of amplified (DpnI resistant) oriLyt. In the bottom panel is a Southern blot showing input oriLyt DNA. (B) Suppression of oriLyt amplification by LANA expression is not due to a decrease in protein production. Vero cells were transfected with a LANA expression plasmid and a K-Rta, K-bZIP, or ORF59 expression plasmid. Protein lysates were prepared, and protein expression was analyzed by Western blotting (WB). Blots were reacted with an antibody specific for LANA, K-Rta, K-bZIP, or ORF59. (C) K-bZIP modulates amplification of oriLyt and the TR element. Vero cells were cotransfected with the set of plasmids expressing the required replication proteins plus plasmids containing oriLyt and the TR. Southern blotting of total cellular DNA from cotransfections that contained HHV8 plasmids that encoded the required replication proteins along with oriLyt- and/or TR-containing plasmids hybridized with a pGEM 32P-labeled probe was carried out. Cotransfections also contained a LANA expression plasmid and plasmids containing K-bZIP and K-Rta ORFs, as indicated. All transfections contained 1 μg of each replication plasmid, 10 μg of oriLyt- or TR-containing plasmid, and 1 μg of K-bZIP and K-Rta expression plasmids except where otherwise stated. Lane 1, TR plasmid cleaved with EcoRI; lane 2, oriLyt plasmid cleaved with EcoRI; lane 3, cotransfection of required replication plasmids plus oriLyt; lane 4, cotransfection of required replication plasmids plus oriLyt and TR plasmids; lane 5, cotransfection of required replication plasmids plus LANA and TR plasmids; lanes 6, 7, and 8, cotransfection of required replication plasmids plus increasing amounts of a LANA expression plasmid (1, 2.5, and 5 μg, respectively), oriLyt, and TR; lane 9, cotransfection of required replication plasmids plus LANA, oriLyt, and TR and no K-bZIP expression plasmid; lanes 10 and 11, cotransfection of required replication plasmids plus LANA (5 μg), oriLyt, and TR plasmids and a K-bZIP expression plasmid (5 and 10 μg, respectively); lanes 12 and 13, cotransfection of required replication plasmids plus LANA (5 μg), oriLyt, and TR plasmids and plasmid expressing K-bZIP 101 to 134 (5 and 10 μg, respectively); lane 14, cotransfection of required replication plasmids plus LANA (5 μg) and oriLyt but not ORF59 expression plasmid. Input oriLyt and TR plasmids are shown in the bottom panel.
Overexpression of K-bZIP can reverse LANA-mediated suppression of oriLyt amplification.
The observed suppression of oriLyt amplification could be occurring for several reasons. One possibility is that LANA affects the level of protein expression, particularly the level of K-Rta, in the cotransfection mixture. Although this is unlikely since all expression plasmids used in the assay use the human cytomegalovirus immediate-early promoter for protein expression, we nevertheless tested this possibility by using Western blot analysis to measure protein accumulation for K-Rta, K-bZIP, and ORF59 in the presence of LANA expression. Cells were cotransfected with the LANA expression plasmid along with either K-Rta, K-bZIP, or ORF59 expression plasmids. Protein lysates were prepared and subjected to SDS-PAGE followed by Western blotting. Since all recombinant proteins were FLAG tagged, Western blots were reacted with anti-FLAG antibody (Sigma). As an internal control for protein loading, we also reacted the same blots with antibody-specific for β-actin (Santa Cruz). LANA expression had no effect on the levels of K-Rta, K-bZIP, or ORF59 protein accumulation as measured by Western blotting (Fig. 2B).
Since LANA had no effect on the level of protein accumulation in the cotransfection assay, we next sought to investigate the mechanism involved in the suppression of oriLyt amplification and to determine whether it involved the interaction of LANA with any of the proteins in the replication assay. We also wanted to examine latent and lytic replication simultaneously since, in the context of viral infection, both LANA and lytic replication factors are present during lytic reactivation. This evaluation would give some insight into the possible switch from lytic to latent origin amplification. The latent origin of HHV8, which is located within the leftward TR region of the genome, undergoes amplification in the presence of LANA (3, 4, 7, 19). To test the implications of LANA expression on both lytic and latent replication, we modified the original cotransfection replication assay to include the latent as well as the lytic origins of replication. In this modified assay cells were harvested at a slightly later time point than used in the transient lytic replication assay in order to maximize the amplification of the TR plasmid (latent origin). Hence, this system would allow us to monitor the effects of various viral proteins on both lytic and latent replication in the same cells.
This lytic/latent replication assay was used to evaluate the effect of increasing levels of LANA expression on lytic replication. We first evaluated the effects of adding the plasmid containing the TR of HHV8 to the transfection mixture. The addition of this plasmid to the cotransfection mixture had no apparent effect on lytic replication, as evidenced by the presence of the amplified oriLyt band (Fig. 2C, lane 4). As demonstrated above, the addition of the LANA expression plasmid to the transfection mixture resulted in the suppression of oriLyt amplification; however, due to LANA expression, the amplification of the TR plasmid can now be detected (Fig. 2C, lane 5). When lower concentrations of the LANA expression plasmid were added to the mixture, it was possible to obtain amplification of both the TR and oriLyt plasmids in the same cotransfection sample (Fig. 2C, lane 6). The presence of the LANA expression plasmid up to 2.5 μg still allowed for simultaneous replication of both latent and lytic cis-acting elements (Fig. 2C, lane 7). When an increasing amount of the LANA expression plasmid was added to the transfection mixture, the amplification of oriLyt was suppressed in a dose-dependent manner (Fig. 2C, lanes 6 to 8). Interestingly, the suppression of oriLyt had no effect on the accumulation of the latent origin plasmid, which remained constant (Fig. 2C, lanes 6 to 8, TR band). These experiments suggested that LANA expression could influence the regulation of lytic or latent DNA replication.
We next wanted to investigate if a protein that is known to interact with LANA, in this case K-bZIP, could influence the balance between lytic and latent replication in our system. We again performed the cotransfection replication assay, this time adding increasing concentrations of the K-bZIP expression plasmid to the mixture. When increasing amounts of the K-bZIP expression plasmid were added to the transfection mixture, lytic replication was restored, and amplification of the TR element was suppressed, as shown by the appearance of the oriLyt band and the disappearance of the TR band (Fig. 2C, lanes 10 and 11, respectively). This reversal of suppression of oriLyt amplification was also observed when increasing amounts of the plasmid expressing the interaction domain of K-bZIP were transfected with LANA (Fig. 2C, lanes 12 and 13). This result strongly suggested that the interaction of K-bZIP with LANA could modulate the switch from oriLyt to TR amplification in the replication assay.
K-Rta can compensate for K-bZIP in the transient oriLyt-dependent DNA replication assay.
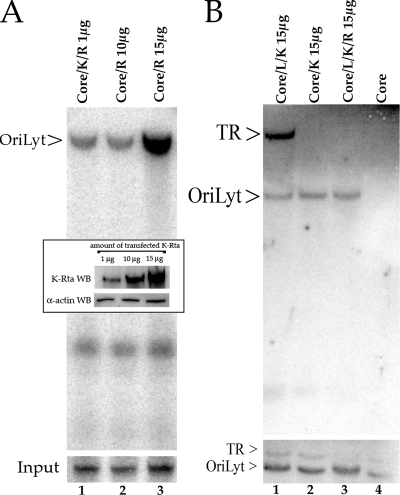
Deletion of the K-bZIP ORF from a recombinant HHV8 BAC displayed a high level of DNA replication and virus production upon infection with Ad50, a recombinant adenovirus that overexpresses K-Rta (16). These studies demonstrated that K-bZIP is not required for reactivation or lytic DNA replication under conditions where K-Rta is overexpressed. We wanted to reevaluate the requirement for K-bZIP in the transient cotransfection replication assay under conditions where increasing concentrations of a K-Rta expression plasmid are added to the transfection mixture, which is similar to the protocol used with the HHV8 ΔK-bZIP bacmid. Cells were cotransfected as before with the required core replication proteins, oriLyt, and a K-Rta expression plasmid, using concentrations from 5 to 15 μg of K-Rta plasmid DNA, which is 5 to 15 times more plasmid than used in the original assay. Under these conditions, oriLyt was efficiently amplified when cotransfection mixtures contained 10 or 15 μg of the K-Rta expression plasmid in the absence of K-bZIP expression (Fig. 3A, lanes 2 and 3). The inset figure shows the relative increase in K-Rta protein expression due to the increasing amounts of the K-Rta expression plasmid added to each cotransfection mixture. This result confirmed the HHV8 ΔK-bZIP bacmid data showing that K-bZIP does not directly participate in lytic DNA replication and that K-Rta can compensate for K-bZIP in the replication assay.
FIG. 3.
Evaluation of K-Rta on lytic and latent DNA replication. (A) Efficient oriLyt amplification in the absence of K-bZIP in the cotransfection/replication assay. Vero cells were cotransfected with the required core replication plasmids plus oriLyt in the presence or absence of a K-bZIP expression plasmid and increasing amounts of a K-Rta expression plasmid. Lane 1, cotransfection of core replication plasmids plus oriLyt, K-Rta (1 μg), and K-bZIP expression plasmids; lane 2, cotransfection of core replication plasmids plus oriLyt and K-Rta (10 μg); lane 3, cotransfection of core replication plasmids plus oriLyt and K-Rta (15 μg). The inset is a Western blot showing the relative K-Rta protein accumulation from cells cotransfected with core plasmids plus increasing amounts of a K-Rta expression plasmid. The lanes match the cotransfection performed as shown in panel A. (B) K-Rta expression does not affect TR amplification. Lane 1, cotransfection of plasmids encoding all the required core replication proteins plus oriLyt, TR, LANA expression plasmid, and 15 μg of the K-Rta expression plasmid; lane 2, cotransfection of plasmids encoding all the required core replication proteins plus oriLyt, TR, and 15 μg of the K-Rta expression plasmid; lane 3, cotransfection of plasmids encoding all the required core replication proteins plus oriLyt, TR, LANA expression plasmid, 15 μg of the K-Rta expression plasmid, and 1 μg of the K-bZIP expression plasmid; lane 4, cotransfection of plasmids encoding all the required core replication proteins plus oriLyt and TR plasmids. Core, required replication protein expression plasmids; K, K-bZIP; R, K-Rta; L, LANA.
To confirm that overexpression of K-Rta did not affect TR amplification in the presence or absence of K-bZIP, we again performed the lytic/latent cotransfection replication assay under conditions where we added a high concentration of the K-Rta expression plasmid to the cotransfection mixture. Addition of the K-Rta expression plasmid in the absence of K-bZIP and in the presence of TR did not result in amplification of TR in the replication assay (Fig. 3B, lane 2). In addition, as shown in previous experiments, the addition of K-bZIP to the cotransfection mixture did not produce a TR replication signal (Fig. 3B, lane 3). The control experiment, in which the K-Rta expression plasmid was omitted from the transfection mixture, failed to produce a detectable oriLyt band (Fig. 3B, lane 4). These experiments established that the overexpression of K-Rta in the cotransfection replication assay relieved the requirement for K-bZIP and did not influence amplification of TR in the presence or absence of K-bZIP.
LANA binds to oriLyt.
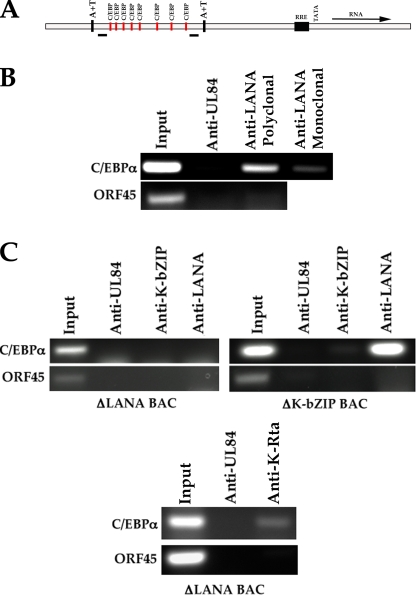
K-bZIP was shown to interact with oriLyt and binds to several C/EBPα sites located within oriLyt (32). The interaction of K-bZIP with oriLyt is proposed to occur via a “piggyback” formation, with the transcription factor C/EBPα making contact with oriLyt DNA. Although it was demonstrated that K-bZIP has no known intrinsic enzymatic or transactivation activity and is not required for DNA synthesis, its interaction with oriLyt does suggest some role in lytic DNA replication. Since the data presented here suggested that LANA and K-bZIP appeared to modulate oriLyt and TR amplification, we investigated the possible interaction of LANA with the HHV8 lytic origin. To identify any interaction, we performed a ChIP assay using DNA extracted from BAC36-infected Vero cells, followed by immunoprecipitation of protein-DNA complexes using LANA-specific antibodies. The immunoprecipitated DNA was evaluated using primers specific for the oriLyt region between nucleotides 23326 and 23572 of oriLyt, which contains the C/EBPα transcription factor binding sites known to be a substrate for K-bZIP (Fig. 4A). A positive PCR amplification product was detected using two different LANA-specific antibodies, indicating that LANA interacts with this region of oriLyt (Fig. 4B,). Control samples using an isotype-specific control antibody or primers designed to amplify a different HHV8 genomic locus failed to show any PCR signal (Fig. 4B).
FIG. 4.
Interaction of LANA with oriLyt. (A) Schematic of HHV8 oriLyt region containing C/EBPα transcription factor binding sites and location of flanking PCR amplification primers. RRE, Rta-responsive element. (B) LANA interacts with oriLyt. ChIP assay from HHV8 BAC36-infected cells using LANA-specific antibodies and primers flanking the C/EBPα binding motif sites within oriLyt. (C) The interaction of K-bZIP with oriLyt is dependent upon LANA expression. ChIP assays were performed using cells infected with the recombinant HHV8 bacmids that are defective for the expression of LANA or K-bZIP. The left and bottom panels show ChIP assays from LANA-knockout bacmid-infected cells. The right panel shows a ChIP assay from ΔK-bZIP bacmid-infected cells. ChIP assays were performed using anti-UL84-, anti-LANA-, anti-K-Rta-, or anti-K-bZIP-specific antibodies.
We also evaluated the ability of K-bZIP to interact with C/EBPα binding motifs within oriLyt in the absence of LANA expression. For these experiments we used the recombinant bacmid that is defective for LANA expression (24). This bacmid displays a lytic phenotype upon transfection into cells but is unable to establish a latent infection. Cells were transfected with the LANA knockout bacmid, and ChIP assays were performed as before. We failed to detect a positive PCR band, indicating that an interaction of K-bZIP with the C/EBPα binding motifs did not occur in the absence of LANA expression (Fig. 4C, left panel). Control ChIP experiments in which we used a nonspecific antibody (anti-UL84; Virusys) or the anti-LANA antibody did not show a specific PCR amplification product (Fig. 4C, left panel). Amplification of an unrelated region (ORF45) also failed to produce a PCR amplification product (Fig. 4C, left panel). Lastly, the absence of LANA expression did not affect the ability of K-Rta to bind to the C/EBPα binding motifs within oriLyt as demonstrated by a ChIP analysis from cells transfected with the LANA knockout virus (Fig. 4C, bottom panel). These results strongly suggested that the interaction of K-bZIP with oriLyt was dependent upon LANA binding to the C/EBPα binding motifs.
We also asked if LANA could interact with oriLyt C/EBPα binding motifs in the absence of K-bZIP expression. For these experiments we used the HHV8 ΔK-bZIP recombinant bacmid. We again performed the ChIP assay in BAC-infected cell lines. We were able to detect LANA binding to C/EBPα binding motifs, indicating that this interaction was not dependent upon the expression or binding of K-bZIP to oriLyt (Fig. 4C, right panel). Control ChIP experiments failed to show a positive PCR amplification product when anti-K-bZIP-specific antibody was used or amplification of the ORF45 locus (Fig. 4C, right panel).
This experiment demonstrated that LANA can bind to oriLyt in the region known to interact with K-bZIP and suggests that the interaction of K-bZIP with oriLyt is dependent upon LANA expression and interaction with oriLyt C/EBPα binding motifs. Further, LANA and K-bZIP appear to modulate DNA synthesis using a mechanism that involves direct suppression of oriLyt function.
DISCUSSION
The regulatory processes that determine the balance between latency and lytic reactivation are complex. Typically, each viral state is studied as a separate process. Here, we show the development of an in vitro system to study the regulatory and enzymatic mechanisms for both lytic and latent replication simultaneously. Clearly, our results indicated that viral proteins involved in latent replication regulate the initial events in lytic DNA synthesis. HHV8 LANA plays a major role in the establishment and maintenance of the latent viral genome. Although LANA is associated with the latent state of the genome, it is expressed throughout the infectious cycle and appears to remain at a constant protein level even upon viral induction to enter the lytic cycle (20). This seemingly would present the virus with the scenario of overcoming LANA's “push” toward latency even when the expression of the viral transactivator and switch protein K-Rta is expressed and subsequently drives the virus into the lytic phase. This control mechanism appears to involve the ability of LANA to repress the expression of, and interact with, K-Rta and suggests that a change in the regulation of K-Rta can lead to the expression of lytic phase-associated proteins, resulting in virus production. In addition, since K-Rta is a component of the virion (as is K-bZIP), it would appear to contribute to the observed initial burst of lytic replication followed by latent infection (20). In our earlier studies using an HHV8 bacmid deficient for K-bZIP expression, it was clear that K-Rta was sufficient to drive the expression of lytic genes, replicate viral DNA, and produce infections virus. The ΔK-bZIP bacmid displayed an enhanced growth phenotype when K-Rta was overexpressed, suggesting that K-bZIP had an inhibitory effect on virus growth. This modulation of the virus lytic cycle may involve the interaction of K-bZIP with LANA, given that ΔK-bZIP bacmid-infected cells showed an aberrant subcellular pattern for LANA. The fact that the overexpression of K-Rta was sufficient for reactivation in the absence of K-bZIP may indicate that the system involving LANA suppression can be overcome by a high concentration of K-Rta.
The interaction of K-bZIP and LANA involves the basic domain of the K-bZIP protein (aa 101 to 134), a region of the protein that also encodes the NLS (27). Although a K-bZIP subclone with this region deleted efficiently expressed protein, we performed mixing experiments to show that a mutated protein with this region deleted was unable to interact with LANA, whereas other mutated forms of K-bZIP still retained their ability to bind to LANA even when lysates were mixed together or when K-bZIP and LANA are expressed separately. The interaction domain for LANA with K-bZIP resides in the repeat region or Q-rich domain of the protein, which contains a high degree of glutamine amino acids.
We show here that LANA can directly suppress oriLyt-dependent DNA replication and that this suppression was not due to a general or specific decrease in gene expression of K-Rta or essential core replication protein expression. All gene expression of replication proteins and K-Rta was driven by the human cytomegalovirus immediate-early promoter, and protein levels were unaffected by the presence of LANA. This lack of an observed decrease in gene expression prompted us to investigate if the interaction of LANA with K-Rta and/or K-bZIP may influence DNA syntheses. We demonstrated that K-bZIP could influence the suppressive effects of LANA on lytic DNA replication. Further, like K-bZIP and K-Rta, LANA interacts with oriLyt, and the results of the experiments presented here suggest that this interaction may suppress lytic replication directly. Additionally, K-bZIP may serve to modulate this suppression via its binding to oriLyt and LANA. Our results strongly suggest that the expression and binding of LANA to oriLyt are essential for K-bZIP binding. This presents the possible scenario of another mechanism whereby LANA maintains the virus genome in the latent state along with suppression of K-Rta gene expression. In virus infection, K-bZIP enters the cell as part of the infectious virion, and this may serve to initially “orient” K-bZIP and LANA at oriLyt. Induction of the lytic cycle may displace the LANA-K-bZIP complex or allow for the binding of the protein involved in the origin recognition complex that subsequently results in lytic DNA synthesis. In this scenario, K-bZIP would not be necessary for lytic DNA replication but would act as a facilitator for DNA synthesis. In the BAC infection system, the overexpression of K-Rta may result in the displacement of LANA from oriLyt and subsequently lead to lytic DNA synthesis.
Much emphasis is placed on the association of LANA with K-Rta and the observation that LANA can regulate the expression of K-Rta in transient assays (21, 22). Previously, we demonstrated that LANA interacts with K-bZIP, the putative homolog to Epstein-Barr virus Zta. However it appears that K-bZIP has a distinctly different function in HHV8 lytic DNA replication. The evidence for the involvement of K-bZIP in the modulation of the effects of LANA suppression is based on the fact that we can achieve efficient oriLyt replication in the absence of K-bZIP when K-Rta is overexpressed in the transient cotransfection system. The overexpression of K-Rta appears to be sufficient to overcome the suppressive effects of LANA in both the transient system and in the context of the virus genome (16).
Recently, it was suggested that there may be cell-type-dependent differences with respect to the role and requirement for K-bZIP in initiation of lytic DNA synthesis (23). Using short hairpin RNA, it was shown that the expression of K-bZIP in primary effusion lymphoma cells was essential for lytic reactivation. The discrepancy between this system and the one we describe could be due to the fact that we used knockout viruses as a means to downregulate gene expression. Another explanation is that reactivation of lytic virus in the BAC system is more robust with a more efficient expression of K-Rta. Nevertheless, the results shown here add to the complex regulation of lytic and latent phases for HHV8.
Acknowledgments
We thank S.-J. Gao for the LANA knockout bacmid and Erle Robertson and Kenneth Kaye for LANA expression plasmids.
This work was supported by PHS grant CA11521179.
Footnotes
Published ahead of print on 24 June 2009.
REFERENCES
- 1.AuCoin, D. P., K. S. Colletti, S. A. Cei, I. Papouskova, M. Tarrant, and G. S. Pari. 2004. Amplification of the Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 lytic origin of DNA replication is dependent upon a cis-acting AT-rich region and an ORF50 response element and the trans-acting factors ORF50 (K-Rta) and K8 (K-bZIP). Virology 318542-555. [DOI] [PubMed] [Google Scholar]
- 2.AuCoin, D. P., K. S. Colletti, Y. Xu, S. A. Cei, and G. S. Pari. 2002. Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) contains two functional lytic origins of DNA replication. J. Virol. 767890-7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284641-644. [DOI] [PubMed] [Google Scholar]
- 4.Ballestas, M. E., and K. M. Kaye. 2001. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mediates episome persistence through cis-acting terminal repeat (TR) sequence and specifically binds TR DNA. J. Virol. 753250-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbera, A. J., J. V. Chodaparambil, B. Kelley-Clarke, K. Luger, and K. M. Kaye. 2006. Kaposi's sarcoma-associated herpesvirus LANA hitches a ride on the chromosome. Cell Cycle 51048-1052. [DOI] [PubMed] [Google Scholar]
- 6.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 2661865-1869. [DOI] [PubMed] [Google Scholar]
- 7.Cotter, M. A., II, C. Subramanian, and E. S. Robertson. 2001. The Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen binds to specific sequences at the left end of the viral genome through its carboxy-terminus. Virology 291241-259. [DOI] [PubMed] [Google Scholar]
- 8.Gao, S. J., L. Kingsley, D. R. Hoover, T. J. Spira, C. R. Rinaldo, A. Saah, J. Phair, R. Detels, P. Parry, Y. Chang, and P. S. Moore. 1996. Seroconversion to antibodies against Kaposi's sarcoma-associated herpesvirus-related latent nuclear antigens before the development of Kaposi's sarcoma. N. Engl. J. Med. 335233-241. [DOI] [PubMed] [Google Scholar]
- 9.Garber, A. C., J. Hu, and R. Renne. 2002. Latency-associated nuclear antigen (LANA) cooperatively binds to two sites within the terminal repeat, both sites contribute to the ability of LANA to suppress transcription and to facilitate DNA replication. J. Biol. Chem. 27727401-27411. [DOI] [PubMed] [Google Scholar]
- 10.Godfrey, A., J. Anderson, A. Papanastasiou, Y. Takeuchi, and C. Boshoff. 2005. Inhibiting primary effusion lymphoma by lentiviral vectors encoding short hairpin RNA. Blood 1052510-2518. [DOI] [PubMed] [Google Scholar]
- 11.Groves, A. K., M. A. Cotter, C. Subramanian, and E. S. Robertson. 2001. The latency-associated nuclear antigen encoded by Kaposi's sarcoma-associated herpesvirus activates two major essential Epstein-Barr virus latent promoters. J. Virol. 759446-9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grundhoff, A., and D. Ganem. 2003. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus permits replication of terminal repeat-containing plasmids. J. Virol. 772779-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu, J., A. C. Garber, and R. Renne. 2002. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus supports latent DNA replication in dividing cells. J. Virol. 7611677-11687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hyun, T. S., C. Subramanian, M. A. Cotter, 2nd, R. A. Thomas, and E. S. Robertson. 2001. Latency-associated nuclear antigen encoded by Kaposi's sarcoma-associated herpesvirus interacts with Tat and activates the long terminal repeat of human immunodeficiency virus type 1 in human cells. J. Virol. 758761-8771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeong, J. H., J. Orvis, J. W. Kim, C. P. McMurtrey, R. Renne, and D. P. Dittmer. 2004. Regulation and autoregulation of the promoter for the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Biol. Chem. 27916822-16831. [DOI] [PubMed] [Google Scholar]
- 16.Kato-Noah, T., Y. Xu, C. C. Rossetto, K. Colletti, I. Papouskova, and G. S. Pari. 2007. Overexpression of the Kaposi's sarcoma-associated herpesvirus transactivator K-Rta can complement a K-bZIP deletion bacmid and yields an enhanced growth phenotype. J. Virol. 8113519-13532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kellam, P., C. Boshoff, D. Whitby, S. Matthews, R. A. Weiss, and S. J. Talbot. 1997. Identification of a major latent nuclear antigen, LNA-1, in the human herpesvirus 8 genome. J. Hum. Virol. 119-29. [PubMed] [Google Scholar]
- 18.Komatsu, T., M. E. Ballestas, A. J. Barbera, and K. M. Kaye. 2002. The KSHV latency-associated nuclear antigen: a multifunctional protein. Front Biosci. 7d726-d730. [DOI] [PubMed] [Google Scholar]
- 19.Komatsu, T., A. J. Barbera, M. E. Ballestas, and K. M. Kaye. 2001. The Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen. Viral Immunol. 14311-317. [DOI] [PubMed] [Google Scholar]
- 20.Krishnan, H. H., P. P. Naranatt, M. S. Smith, L. Zeng, C. Bloomer, and B. Chandran. 2004. Concurrent expression of latent and a limited number of lytic genes with immune modulation and antiapoptotic function by Kaposi's sarcoma-associated herpesvirus early during infection of primary endothelial and fibroblast cells and subsequent decline of lytic gene expression. J. Virol. 783601-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lan, K., D. A. Kuppers, S. C. Verma, and E. S. Robertson. 2004. Kaposi's sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen inhibits lytic replication by targeting Rta: a potential mechanism for virus-mediated control of latency. J. Virol. 786585-6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lan, K., D. A. Kuppers, S. C. Verma, N. Sharma, M. Murakami, and E. S. Robertson. 2005. Induction of Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen by the lytic transactivator RTA: a novel mechanism for establishment of latency. J. Virol. 797453-7465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lefort, S., and L. Flamand. 2009. Kaposi's sarcoma-associated herpesvirus K-bZIP protein is necessary for lytic viral gene expression, DNA replication and virion production in primary effusion lymphoma cell lines. J. Virol. 835869-5880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, Q., F. Zhou, F. Ye, and S. J. Gao. 2008. Genetic disruption of KSHV major latent nuclear antigen LANA enhances viral lytic transcriptional program. Virology 379234-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim, C., Y. Gwack, S. Hwang, S. Kim, and J. Choe. 2001. The transcriptional activity of cAMP response element-binding protein-binding protein is modulated by the latency associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Biol. Chem. 27631016-31022. [DOI] [PubMed] [Google Scholar]
- 26.Lim, C., H. Sohn, D. Lee, Y. Gwack, and J. Choe. 2002. Functional dissection of latency-associated nuclear antigen 1 of Kaposi's sarcoma-associated herpesvirus involved in latent DNA replication and transcription of terminal repeats of the viral genome. J. Virol. 7610320-10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Portes-Sentis, S., E. Manet, G. Gourru, A. Sergeant, and H. Gruffat. 2001. Identification of a short amino acid sequence essential for efficient nuclear targeting of the Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 K8 protein. J. Gen. Virol. 82507-512. [DOI] [PubMed] [Google Scholar]
- 28.Renne, R., C. Barry, D. Dittmer, N. Compitello, P. O. Brown, and D. Ganem. 2001. Modulation of cellular and viral gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75458-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 9314862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwam, D. R., R. L. Luciano, S. S. Mahajan, L. Wong, and A. C. Wilson. 2000. Carboxy terminus of human herpesvirus 8 latency-associated nuclear antigen mediates dimerization, transcriptional repression, and targeting to nuclear bodies. J. Virol. 748532-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verma, S. C., K. Lan, and E. Robertson. 2007. Structure and function of latency-associated nuclear antigen. Curr. Top. Microbiol. Immunol. 312101-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu, F. Y., S. E. Wang, Q. Q. Tang, M. Fujimuro, C. J. Chiou, Q. Zheng, H. Chen, S. D. Hayward, M. D. Lane, and G. S. Hayward. 2003. Cell cycle arrest by Kaposi's sarcoma-associated herpesvirus replication-associated protein is mediated at both the transcriptional and posttranslational levels by binding to CCAAT/enhancer-binding protein α and p21CIP-1. J. Virol. 778893-8914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu, Y., D. P. AuCoin, A. R. Huete, S. A. Cei, L. J. Hanson, and G. S. Pari. 2005. A Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 ORF50 deletion mutant is defective for reactivation of latent virus and DNA replication. J. Virol. 793479-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]