Abstract
The influenza A virus M2 protein has important roles during virus entry and in the assembly of infectious virus particles. The cytoplasmic tail of the protein can be palmitoylated at a cysteine residue, but this residue is not conserved in a number of human influenza A virus isolates. Recombinant viruses encoding M2 proteins with a serine substituted for the cysteine at position 50 were generated in the A/WSN/33 (H1N1) and A/Udorn/72 (H3N2) genetic backgrounds. The recombinant viruses were not attenuated for replication in MDCK cells, Calu-3 cells, or in primary differentiated murine trachea epithelial cell cultures, indicating there was no significant contribution of M2 palmitoylation to virus replication in vitro. The A/WSN/33 M2C50S virus displayed a slightly reduced virulence after infection of mice, suggesting that there may be novel functions for M2 palmitoylation during in vivo infection.
Influenza A virus is a member of the Orthomyxoviridae and contains a segmented, negative-sense RNA genome that codes for 10 or 11 proteins, depending upon the virus strain (11). The integral membrane protein M2 is the viral ion channel protein that is required during virus entry (29) and for the production of infectious virus particles (4, 10, 12, 13). The sequences responsible for the latter map to the cytoplasmic tail of the protein and overlap with a number of sites for posttranslational modification, which include palmitoylation and phosphorylation (7, 26, 31). Palmitoylation occurs on the cysteine present at amino acid 50 and is not required for ion channel activity of the M2 protein from A/Udorn/72 (H3N2) (7). Palmitoylation of M2 appeared to be dispensable for the production of infectious virus particles using a reassortant virus consisting of seven segments from an H3N8 subtype virus (A/Equine/Miami/63) and the M segment from an H1N1 subtype virus (A/Puerto Rico/8/34) (2). No studies examining the role of M2 palmitoylation in the context of a naturally occurring influenza A virus strain have been published to date.
The significance of palmitoylation of the influenza A virus hemagglutinin (HA) protein can vary among virus strains. Palmitoylation of HA from an H7 and an H1 but not an H3 subtype is required for efficient membrane fusion (5, 24, 32), whereas palmitoylation of HA from an H3 but not an H1 subtype is required for virus assembly (5). An analysis of 3,532 sequences of influenza isolates from humans revealed that the M2 residue C50 is conserved in a strain-specific manner. A total of 2,602 of 2,610 H3N2 sequences code for a cysteine at this position; the cysteine, however, is conserved in only 330 of 1,051 H1N1 sequences (data not shown). A serine residue is substituted for cysteine in the majority of the H1N1 viruses that do not have a cytoplasmic palmitoylation site; the newly emerged 2009 H1N1 influenza A viruses, however, do have a cysteine at this position (3). The sequence alignment data are consistent with a strain-specific selective pressure to maintain the palmitoylation site on the M2 protein. Interestingly, other M2 cytoplasmic tail sequences display differential effects on infectious virus production, depending on the strain used (12).
To investigate the role of M2 palmitoylation in influenza A virus replication, we substituted a serine for the cysteine residue at position 50 (C50S) of the M2 protein in two influenza A virus strains, A/Udorn/72 (H3N2) (rUdorn) and A/WSN/33 (H1N1) (rWSN). The resultant viruses were tested for their ability to replicate in tissue culture cells, and the mouse-adapted virus was tested for virulence in a mouse model of infection. Neither mutant virus showed any defect in virus replication in tissue culture cells, in differentiated murine primary trachea epithelial cells (mTEC), or in the lungs of infected mice. The viruses lacking a palmitoylation site, however, did have a modest reduction in virulence, suggesting that M2 palmitoylation is dispensable for in vitro replication but contributes to virus virulence in vivo.
MATERIALS AND METHODS
Cells and viruses.
MDCK cells, human lung carcinoma cells (Calu-3), and human embryonic kidney cells (293T) were cultured in Dulbecco's modified Eagle medium (DMEM; Sigma) containing 10% fetal bovine serum (Atlanta Biologicals, Inc.), 100 U of penicillin/ml (Invitrogen), 100 μg of streptomycin/ml (Invitrogen), and 1 mM sodium pyruvate (Sigma). The cells were incubated at 37°C and 5% CO2.
Viruses.
Viruses were propagated on MDCK cells in DMEM containing 100 U of penicillin/ml, 100 μg of streptomycin/ml, 0.3% bovine serum albumin (BSA; Calbiochem or Sigma), and 4 μg/ml N-acetyl-trypsin (Sigma). The parental viruses used in this study were rUdorn (a recombinant virus derived from A/Udorn/72) and rWSN (a recombinant virus derived from A/WSN/33), which have been described previously (10, 13, 15, 29). The plasmids pHH21 M-Udorn and pHH21 M-WSN (15, 29) were altered from cysteine to serine at codon 50 of the M2 open reading frame by site-directed mutagenesis. Recombinant viruses were generated and plaque purified, and the sequence of the M segment of the viral RNAs was verified.
Infection of tissue culture cells.
Low-multiplicity growth curves on MDCK cells were carried out in triplicate by infecting confluent monolayers grown in 24-well plates at a multiplicity of infection (MOI) of 0.001 50% tissue culture infective dose (TCID50) per cell (three wells per virus at each time point). Cells were infected for 1 h at room temperature, the inoculum was aspirated, and 0.5 ml DMEM containing 4 μg/ml N-acetyl-trypsin, penicillin, streptomycin, and 0.3% BSA was added to each well. At the indicated times, the medium was removed and stored at −80°C. The amount of infectious virus present in each sample was determined by TCID50 assay as previously described (13, 20).
Infection of Calu-3 cells was carried out in manner similar to the experiments with MDCK cells, with the following exceptions. Calu-3 cells were grown on 0.33-cm Transwell inserts, and once a transepithelial resistance of approximately 400 Ω·cm2 was reached, the cells were infected as described above for MDCK cells except that N-acetyl-trypsin was omitted. At the indicated times, the medium in the apical chamber was removed, stored at −80°C, and replaced with 150 μl of fresh medium.
High-multiplicity infections of MDCK cells were carried out in T75 flasks (MOI = 3). Cells were infected at room temperature for 1 h in DMEM containing penicillin, streptomycin, and 4 μg/ml N-acetyl-trypsin. The inoculum was then removed, the cells were washed three times with phosphate-buffered saline (PBS), then 10 ml DMEM containing penicillin and streptomycin was added and the flasks were incubated at 37°C and 5% CO2 for 15 h. Supernatants were centrifuged at a low speed to remove cell debris and layered onto a 20% sucrose cushion. Virus particles were pelleted at 71,000 × g in a TH641 rotor (Sorval) for 1 h at 4°C. Supernatants were aspirated, and the pellets were resuspended in PBS. An aliquot was removed for determination of infectious titer, and 4× sodium dodecyl sulfate-polyacrylamide gel electrophoresis loading buffer was added to the remaining sample for subsequent Western blot analysis.
Infection of primary mouse trachea epithelial cells.
Primary mTEC cultures were generated as described previously (8, 17, 23). The cells were infected, and samples were harvested as described above for experiments with Calu-3 cells except that 3,300 TCID50 per well were used to infect the cells. If all of the cells in the culture were susceptible to influenza virus infection, this amount of virus would correspond to an approximate MOI of 0.01 (16).
Western blotting.
Samples were analyzed by Western blot analysis as described previously (13). Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride membrane (Immobilon-FL; Millipore). All antibodies were diluted in 5% skim milk in PBS-0.3% Tween 20. The antibodies used in this study were (i) 14c2 (anti-M2 monoclonal antibody) (35), (ii) anti-A/Udorn/72 virus goat serum (37), or (iii) a combination consisting of an anti-M1 monoclonal antibody (HB-64; American Type Culture Center), an anti-NP monoclonal antibody (HB-65; American Type Culture Center), and V-314-511-157 (a polyclonal goat anti-H0 A/PR/8/34 antibody; National Institute of Allergy and Infectious Diseases). Antibodies were detected with species-specific secondary antibodies conjugated to Alexa Fluor 647 (Invitrogen), and the blots were imaged using an FLA-5000 phosphorimager (Fujifilm).
Immunofluorescence microscopy.
MDCK cells were grown to confluence on tissue culture-treated glass coverslips. The medium was changed every other day, and infections were initiated on the fifth day after reaching confluence. Approximately 5 × 104 TCID50 rUdorn or rUdorn-M2C50S was used per coverslip. At 15 h postinfection (p.i.), the cells were placed on ice and incubated with goat anti-H3 sera raised against HA from A/Aichi/2/68 (V-314-591-157; National Institute of Allergy and Infectious Diseases) diluted in DMEM-5% fetal bovine serum for 1 h. After washing with ice-cold PBS, the cells were fixed with 3% paraformaldehyde-2% sucrose in PBS for 10 min. Samples were then washed three times with PBS-1% BSA-0.1% saponin (washing buffer) and incubated with donkey anti-goat immunoglobulin G conjugated with Alexa Fluor 555 (Invitrogen) and 4′,6-diamidino-2-phenylindole (DAPI; Roche Molecular Biochemical) diluted in washing buffer for 1 h at room temperature. Cells were washed three times with washing buffer and once with PBS before being mounted with ProLong Gold antifade reagent (Invitrogen).
Samples were imaged with a Leica confocal microscope (405-nm and 543-nm laser lines). For high-magnification images, a ×63 oil-immersion objective was used, and Z sections were taken at 0.5-μm intervals. Projections were made using the LSM Image Browser software (Zeiss). For quantitation of the extent of filament formation, images of 10 adjacent, nonoverlapping fields of view were taken in each of two independent experiments (×20 objective; Z sections taken at 1.9-μm intervals), and projections were made. Cells were then scored for whether they did or did not show filaments on their cell surfaces, and the percentage of infected cells with filaments in each field of view was calculated.
Animal experiments.
Six- to 8-week-old female BALB/c mice (Charles River) were infected with mouse-adapted rWSN or rWSN-M2C50S by the intranasal route as previously described (34). Virus doses for rWSN were 103 or 105 TCID50 per animal, and doses for rWSN-M2C50S were 103, 105, 106, or 107 TCID50 per animal. Ten animals were infected with each dose, with the exception of the experiments with 103 TCID50 per animal, which contained 5 animals per group. Animals were weighed prior to infection and on each subsequent day and scored for the day of death.
For experiments to examine virus load and cytokine production in the lungs, a total of seven animals from two independent experiments were infected with 105 TCID50 for each virus. On days 3 and 5 p.i., animals were euthanized and the lungs were removed and flash frozen. Lungs were then weighed and homogenized in an appropriate volume of DMEM-penicillin-streptomycin-0.3% BSA-4 μg/ml N-acetyl trypsin to make a 10% (wt/vol) homogenate. The amount of virus in each sample was determined by TCID50 assay.
Enzyme-linked immunosorbent assays.
Supernatant from homogenized lung tissue was used to measure macrophage inflammatory protein 1α (MIP-1α) (mouse CCL3/MIP-1 alpha DuoSet; R&D Systems) and interleukin-10 (IL-10) (OptEIA mouse IL-10 enzyme-linked immunosorbent assay set; BD Biosciences) using the manufacturers' protocols. Protein concentrations are expressed relative to grams of lung mass.
Statistical analyses.
Infectious virus production and body mass changes were analyzed using mixed analyses of variance with time and virus as the independent variables. Virus titers and concentrations of protein in the lungs were analyzed with two-way analyses of variance. Differences in the percentage of cells with filaments and the average day of death were calculated using t tests. If the data violated the assumptions of a normal distribution, then nonparametric tests were used. The mean day of death was determined by the Mann-Whitney rank sum test. Significant interactions were further evaluated using Tukey's test or the Dunn method for pairwise multiple comparisons. Mean differences were considered statistically significant if the P value was <0.05.
RESULTS
Growth of M2 palmitoylation mutant viruses in vitro.
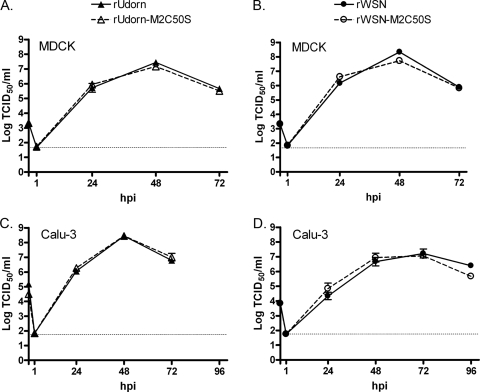
The effect of M2C50S mutations on the production of infectious virus was determined with low MOI growth curves on MDCK cells (Fig. 1A and B). Cultures infected with wild-type and mutant viruses developed virus-induced cytopathic effect (CPE) at a similar rate and to a similar extent (data not shown). Statistically significant differences in the amount of infectious virus produced by MDCK cells were not detected, indicating that palmitoylation of M2 is not required for the production of infectious H3N2 or H1N1 viruses in this cell type. To verify that the viruses did not simply revert to wild type, viral RNA was isolated from supernatants harvested at 48 h p.i. from duplicate wells of rUdorn-M2C50S- and rWSN-M2C50S-infected cells. Reverse transcription-PCR specific for the M segment was carried out, and the product was sequenced. All of the samples examined still contained the C50S mutation (data not shown), indicating that the lack of attenuation in vitro was not due to reversion of the C50S mutation.
FIG. 1.
Altering the palmitoylation site of M2 does not affect infectious virus production in tissue culture cells. MDCK cells (A and B) or polarized Calu-3 cells (C and D) were infected with the indicated viruses. The amount of infectious virus in cell supernatants harvested at the indicated times was determined by TCID50 assay. Data points indicate the average and error bars represent the standard error of the mean from triplicate samples. The horizontal dotted lines represent the limits of detection.
To determine whether there is a requirement for M2 palmitoylation in polarized respiratory epithelial cells, Calu-3 cells were infected. Calu-3 cells are derived from human lung tissue and can support infection with a number of respiratory viruses, including influenza A virus (30, 36) and severe acute-respiratory syndrome coronavirus (25). Cells were grown on Transwell inserts and were infected 4 to 5 days after confluence. At this time, the cells exhibited transepithelial resistance readings of >400 Ω·cm2, suggesting that they had formed a polarized monolayer (6). At the indicated times, the apical supernatants were harvested and replaced with fresh medium and the amount of infectious virus in each sample was determined on MDCK cells. As shown in Fig. 1C and D, there were strain-specific differences in the kinetics of virus replication on this cell type with peak infectious virus titers at 48 h p.i. for rUdorn and 72 h p.i. for rWSN. In addition, the amount of CPE produced by the replication of each strain was different. The rWSN-infected cells exhibited very little CPE, whereas rUdorn produced complete cell death in the monolayer by 48 h p.i. (data not shown). However, each of the M2C50S mutants exhibited CPE, replication kinetics, and peak titers that were similar to those of the parental control virus, indicating that C50 (and, therefore, M2 palmitoylation) is not required for replication in polarized Calu-3 cells.
Effect of M2C50S mutation on virus assembly.
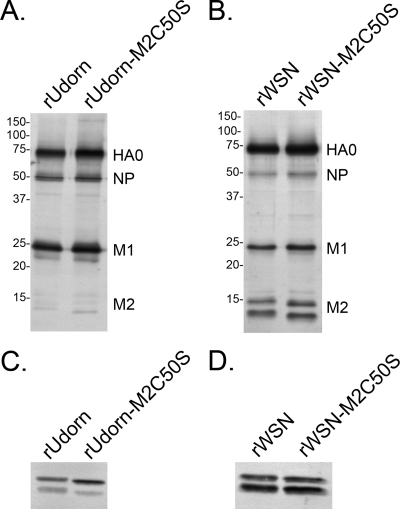
To determine whether palmitoylation of M2 affected the incorporation of proteins into virions, Western blot analysis for HA, NP, M1, and M2 proteins was carried out on virus particles concentrated by ultracentrifugation (Fig. 2A and B). For better visualization of M2 in virions, larger sample volumes (fivefold for Udorn and twofold for WSN) of the same virus preparations were used in the Western blot analyses (Fig. 2C and D). Although the two virus strains differed in the amount of M2 protein incorporated into virions, no defect in the amount of NP, M1, or M2 incorporation was detected for either of the M2C50S viruses, suggesting that palmitoylation of M2 is not required for virus assembly in MDCK cells. Consistent with published results (2, 7), introduction of the C50S mutation did not have an adverse effect on M2 protein expression level or oligomerization (data not shown). M2 and NP were both detected as multiple bands, which is likely due to cleavage by cell proteases as described previously (38).
FIG. 2.
Altering the palmitoylation site of M2 does not affect the protein composition of virions. Virus particles concentrated by ultracentrifugation through a 20% sucrose cushion were analyzed by Western blotting for HA, NP, M1 (A and B), and M2 incorporation (C and D). Molecular weight standards are shown to the left of each blot (A and B).
Effect of M2C50S mutation on filament formation.
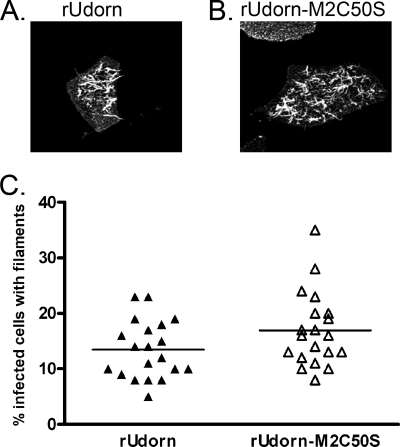
Most clinical isolates of influenza A virus and some laboratory strains (e.g., A/Udorn/72) form filamentous particles in addition to spherical particles (14, 21, 22) on a subset of infected MDCK cells. These long filaments can be visualized by immunofluorescence staining for HA. To determine whether M2 palmitoylation affects the formation of filaments on virus-infected cells, MDCK cells were infected with rUdorn or rUdorn-M2C50S. At 15 h p.i., the cells were immunostained for cell surface HA and nuclei counterstained with DAPI. The samples were then examined by confocal microscopy, and the relative number of infected cells that showed HA-positive filaments was determined (Fig. 3A and B). The filaments produced by cells infected with rUdorn-M2C50S were generally similar to those produced by cells infected with rUdorn. There were no obvious defects in either the number or the length of the filaments produced by rUdorn-M2C50S-infected cells. Quantitation of the percentage of infected cells with filaments in a total of 20 fields viewed with a ×20 objective taken in two independent experiments also revealed no statistically significant differences between infection with rUdorn and rUdorn-M2C50S (Fig. 3C). These data indicate that M2 palmitoylation is not required for the formation of filaments on infected MDCK cells.
FIG. 3.
Alteration of the M2 palmitoylation site does not affect filamentous virus particle formation. The ability of rUdorn or rUdorn-M2C50S to form filaments was examined by immunofluorescence assay for HA followed by confocal microscopy. Panels A and B show projections of z stacks taken at ×63 magnification. The percentage of infected cells with filaments was also determined for a total of 20 fields viewed with a ×20 objective (C).
Effect of M2C50S mutation of virus virulence.
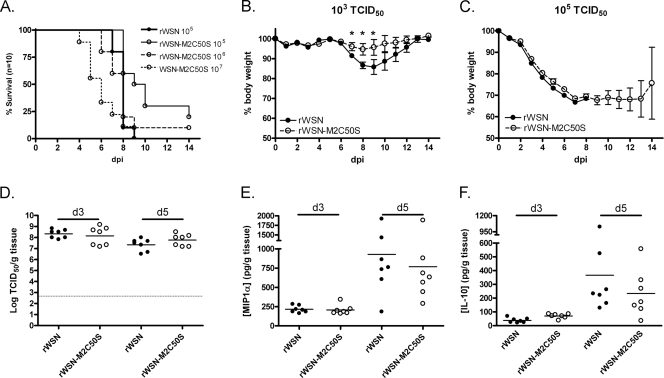
To determine whether M2 palmitoylation is required for virulence of influenza in vivo, female BALB/c mice were infected intranasally with the mouse-adapted rWSN or rWSN-M2C50S viruses as described previously (34). Mice were weighed daily, and the day of death was recorded (Fig. 4A to C). There was a significant delay in the time of death (P < 0.05) between mice infected with 105 TCID50 of rWSN (7.90 ± 0.18 days) and mice infected with 105 TCID50 rWSN M2C50S (10.30 ± 0.84 days). The survival curve for rWSN, however, was similar to that of mice infected with a 10-fold-higher dose of rWSN-M2C50S (Fig. 4A).
FIG. 4.
Altering the palmitoylation site of M2 attenuates virus virulence in vivo. Mice were infected intranasally with the indicated doses of rWSN or rWSN-M2C50S and monitored for 14 days for mortality (A) and weight loss (B and C). Each group contained 10 animals except for those infected with 103 TCID50, which contained 5 animals. Body mass p.i. was normalized to mass at the time of infection. Error bars indicate the standard error of the mean. The lungs from infected mice were homogenized, and the amount of infectious virus (D), MIP-1α (E), and IL-10 (F) per gram of tissue was determined. An asterisk indicates a P value of <0.05. The horizontal lines in the graphs represent mean titers.
When mice were infected with a low dose (i.e., 103 TCID50 per animal), all the mice survived, but mice infected with rWSN-M2C50S lost significantly less weight (P < 0.05) than mice infected with rWSN (Fig. 4B). When mice were infected with higher doses of virus, there was no difference in weight loss (Fig. 4C).
When mouse lungs were examined for the extent of virus replication, there was no statistical difference in the amount of virus in the lungs of rWSN- or rWSN-M2C50S-infected mice (Fig. 4D). Because the production of pro- and anti-inflammatory cytokines can have a significant effect on influenza A virus-induced disease in mice (19, 27, 28), concentrations of the anti-inflammatory cytokine IL-10 and the inflammatory chemokine MIP-1α (CCL3) were measured. Although concentrations of both cytokines were higher at 5 days p.i. than 3 days p.i., the levels were not statistically different between rWSN- and rWSN-M2C50S-infected mice (Fig. 4E and F). These data indicate that although the M2C50S mutation leads to a modest attenuation of influenza A virus virulence in the mouse model of infection, this attenuation cannot be attributed to statistically significant changes in virus load or to altered IL-10 or MIP-1α production in the lungs at the time points analyzed.
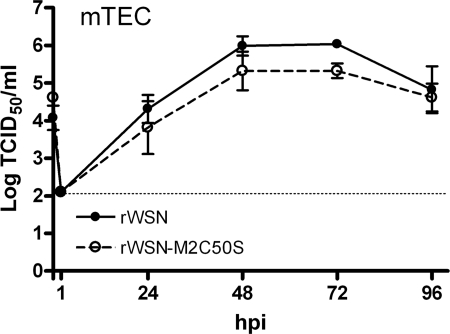
As a more stringent test of whether the M2C50S mutation affects virus replication in vitro, primary mTEC were prepared and infected with rWSN or rWSN-M2C50S. The rWSN virus strain was chosen for these studies, as rWSN is mouse adapted and replicates efficiently in these cultures (17). As shown in Fig. 5, there was no statistically significant difference in the amount of infectious rWSN-M2C50S produced by primary mTEC cultures compared to rWSN. These data suggest that the M2C50S mutation does not alter infectious virus production in mTEC cultures, consistent with the virus load data in infected mouse lungs.
FIG. 5.
Alteration of the M2 palmitoylation site does not affect virus replication in primary, differentiated mTEC. mTEC cultures were infected with the indicated viruses, and at the indicated times the apical supernatants were harvested. The amount of infectious virus in each sample was determined by TCID50 assay on MDCK cells. Data points indicate the average from two independent experiments each done in duplicate. Error bars indicate the standard error of the mean, and the dotted line indicates the limit of detection.
DISCUSSION
Together, these data indicate that although alteration of the palmitoylation site in M2 does not affect virus replication, assembly, or filament formation in tissue culture cells, it does have a minor effect on the virulence of the virus in vivo. There are several examples of mutations to the M2 cytoplasmic tail that result in viruses that have no obvious defects for in vitro replication but are attenuated in the mouse model of infection (33, 34); the mechanism(s) of attenuation, however, has yet to be identified. In the present study, the extended survival of mice infected with the mutant virus did not reflect any statistically significant change in the concentration of IL-10 or MIP-1α in the lungs, nor was there any change in virus load in the lungs or in mTEC cultures. Defining the precise mechanisms responsible for the subtle change in rWSN-M2C50S virulence awaits a more extensive analysis of virus replication in vivo. Because virus replication in differentiated mTEC cultures was not affected by mutation of the palmitoylation site in M2, it is interesting to speculate that the interaction of M2C50S viruses with other cell types in the lungs—dendritic cells, or macrophages, for example (9)—may be important in the attenuation observed with rWSN-M2C50S.
Interestingly, C50 is highly conserved in human H3N2 isolates but not human H1N1 isolates. Therefore, one might predict that this mutation would have a more severe effect on the in vivo virulence of an H3N2 virus than that seen with the H1N1 rWSN virus tested in this study. This hypothesis, however, could not be tested, because the H3N2 rUdorn strain used in this study is not mouse adapted and does not cause a significant infection in mouse models (1, 18). Incorporating the M2C50S mutations into a mouse-adapted H3 virus would allow for the determination of the importance of M2 palmitoylation in this antigenic subtype of influenza A viruses.
In contrast to the critically important role of palmitoylation of the HA protein for virus replication in vitro, palmitoylation of the M2 protein appears to be dispensable for replication in a number of different cell types and culture systems. The expression, oligomerization, and ion channel activity of Udorn M2C50S is comparable to that of wild-type M2 (7). The introduction of a C50S mutation into a reassortant virus consisting of the M segment of A/Equine/Miami/63 (H3N8) into the A/PR/8/34 genetic background increases infectious virus production after infection of MDCK cells (2), which contrasts with our data from infected MDCK cells, Calu-3 cells, and mTEC cultures. The reason for this discrepancy is not clear, but one obvious difference is that the present study focused on introducing mutations into a naturally occurring, wild-type influenza virus strain, while Castrucci et al. used a reassortant virus consisting of one equine influenza A virus segment in the genetic background of a mouse-adapted human influenza A virus. Interestingly and consistent with our results with infected mice, the virus that lacked M2 palmitoylation sites replicated in a manner identical to that of the wild-type virus in infected ferret nasal turbinates (2). Future studies into the mechanism(s) of in vivo attenuation of M2C50S and other more severely attenuated M2 cytoplasmic tail mutants should lead to a better understanding of influenza virus virulence and the pulmonary response to influenza virus infection.
Acknowledgments
We acknowledge and thank the members of the Pekosz Laboratory for their comments and suggestions and the Johns Hopkins University School of Medicine Institute for Basic Biomedical Sciences Microscope facility for use of the confocal microscope.
This study was supported by Public Health Service grants AI007417 (M.L.G.), AI061253 (A.P.), and AI053629 (A.P.) from the National Institutes of Allergy and Infectious Diseases. A.P. also acknowledges support from the Eliasberg Foundation and the Marjorie Gilbert Foundation.
Footnotes
Published ahead of print on 24 June 2009.
REFERENCES
- 1.Benton, K. A., J. A. Misplon, C. Y. Lo, R. R. Brutkiewicz, S. A. Prasad, and S. L. Epstein. 2001. Heterosubtypic immunity to influenza A virus in mice lacking IgA, all Ig, NKT cells, or gamma delta T cells. J. Immunol. 1667437-7445. [DOI] [PubMed] [Google Scholar]
- 2.Castrucci, M. R., M. Hughes, L. Calzoletti, I. Donatelli, K. Wells, A. Takada, and Y. Kawaoka. 1997. The cysteine residues of the M2 protein are not required for influenza A virus replication. Virology 238128-134. [DOI] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 2009. Swine influenza A (H1N1) infection in two children—Southern California, March-April 2009. MMWR Morb. Mortal. Wkly. Rep. 58400-402. [PubMed] [Google Scholar]
- 4.Chen, B. J., G. P. Leser, D. Jackson, and R. A. Lamb. 2008. The influenza virus M2 protein cytoplasmic tail interacts with the M1 protein and influences virus assembly at the site of virus budding. J. Virol. 8210059-10070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, B. J., M. Takeda, and R. A. Lamb. 2005. Influenza virus hemagglutinin (H3 subtype) requires palmitoylation of its cytoplasmic tail for assembly: M1 proteins of two subtypes differ in their ability to support assembly. J. Virol. 7913673-13684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foster, K. A., M. L. Avery, M. Yazdanian, and K. L. Audus. 2000. Characterization of the Calu-3 cell line as a tool to screen pulmonary drug delivery. Int. J. Pharm. 2081-11. [DOI] [PubMed] [Google Scholar]
- 7.Holsinger, L. J., M. A. Shaughnessy, A. Micko, L. H. Pinto, and R. A. Lamb. 1995. Analysis of the posttranslational modifications of the influenza virus M2 protein. J. Virol. 691219-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ibricevic, A., A. Pekosz, M. J. Walter, C. Newby, J. T. Battaile, E. G. Brown, M. J. Holtzman, and S. L. Brody. 2006. Influenza virus receptor specificity and cell tropism in mouse and human airway epithelial cells. J. Virol. 807469-7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ichinohe, T., H. K. Lee, Y. Ogura, R. Flavell, and A. Iwasaki. 2009. Inflammasome recognition of influenza virus is essential for adaptive immune responses. J. Exp. Med. 20679-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iwatsuki-Horimoto, K., T. Horimoto, T. Noda, M. Kiso, J. Maeda, S. Watanabe, Y. Muramoto, K. Fujii, and Y. Kawaoka. 2006. The cytoplasmic tail of the influenza A virus M2 protein plays a role in viral assembly. J. Virol. 805233-5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamb, R. A., and R. M. Krug. 2001. Orthomyxoviridae: the viruses and their replication, p. 1487-1532. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott, Williams & Wilkins, Philadelphia, PA.
- 12.McCown, M. F., and A. Pekosz. 2006. Distinct domains of the influenza A virus M2 protein cytoplasmic tail mediate binding to the M1 protein and facilitate infectious virus production. J. Virol. 808178-8189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCown, M. F., and A. Pekosz. 2005. The influenza A virus M2 cytoplasmic tail is required for infectious virus production and efficient genome packaging. J. Virol. 793595-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mosley, V. M., and R. W. G. Wyckoff. 1946. Electron micrography of the virus of influenza. Nature 157263. [DOI] [PubMed] [Google Scholar]
- 15.Neumann, G., T. Watanabe, H. Ito, S. Watanabe, H. Goto, P. Gao, M. Hughes, D. R. Perez, R. Donis, E. Hoffmann, G. Hobom, and Y. Kawaoka. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. USA 969345-9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newby, C. M., R. K. Rowe, and A. Pekosz. 2006. Influenza A virus infection of primary differentiated airway epithelial cell cultures derived from Syrian golden hamsters. Virology 35480-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newby, C. M., L. Sabin, and A. Pekosz. 2007. The RNA binding domain of influenza A virus NS1 protein affects secretion of tumor necrosis factor alpha, interleukin-6, and interferon in primary murine tracheal epithelial cells. J. Virol. 819469-9480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nguyen, H. H., Z. Moldoveanu, M. J. Novak, F. W. van Ginkel, E. Ban, H. Kiyono, J. R. McGhee, and J. Mestecky. 1999. Heterosubtypic immunity to lethal influenza A virus infection is associated with virus-specific CD8+ cytotoxic T lymphocyte responses induced in mucosa-associated tissues. Virology 25450-60. [DOI] [PubMed] [Google Scholar]
- 19.Perrone, L. A., J. K. Plowden, A. García-Sastre, J. M. Katz, and T. M. Tumpey. 2008. H5N1 and 1918 pandemic influenza virus infection results in early and excessive infiltration of macrophages and neutrophils in the lungs of mice. PLoS Pathog. 4e1000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reed, L. J., and H. Muench. 1938. A simple method of estimating 50 percent endpoints. Am. J. Hyg. 27493-499. [Google Scholar]
- 21.Roberts, P. C., and R. W. Compans. 1998. Host cell dependence of viral morphology. Proc. Natl. Acad. Sci. USA 955746-5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roberts, P. C., R. A. Lamb, and R. W. Compans. 1998. The M1 and M2 proteins of influenza A virus are important determinants in filamentous particle formation. Virology 240127-137. [DOI] [PubMed] [Google Scholar]
- 23.Rowe, R. K., S. L. Brody, and A. Pekosz. 2004. Differentiated cultures of primary hamster tracheal airway epithelial cells. In Vitro Cell Dev. Biol. Anim. 40303-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakai, T., R. Ohuchi, and M. Ohuchi. 2002. Fatty acids on the A/USSR/77 influenza virus hemagglutinin facilitate the transition from hemifusion to fusion pore formation. J. Virol. 764603-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaecher, S. R., E. Touchette, J. Schriewer, R. M. Buller, and A. Pekosz. 2007. Severe acute respiratory syndrome coronavirus gene 7 products contribute to virus-induced apoptosis. J. Virol. 8111054-11068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sugrue, R. J., R. B. Belshe, and A. J. Hay. 1990. Palmitoylation of the influenza A virus M2 protein. Virology 17951-56. [DOI] [PubMed] [Google Scholar]
- 27.Sun, J., R. Madan, C. L. Karp, and T. J. Braciale. 2009. Effector T cells control lung inflammation during acute influenza virus infection by producing IL-10. Nat. Med. 15277-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szretter, K. J., S. Gangappa, X. Lu, C. Smith, W.-J. Shieh, S. R. Zaki, S. Sambhara, T. M. Tumpey, and J. M. Katz. 2007. Role of host cytokine responses in the pathogenesis of avian H5N1 influenza viruses in mice. J. Virol. 812736-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takeda, M., A. Pekosz, K. Shuck, L. H. Pinto, and R. A. Lamb. 2002. Influenza A virus M2 ion channel activity is essential for efficient replication in tissue culture. J. Virol. 761391-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tumpey, T. M., C. F. Basler, P. V. Aguilar, H. Zeng, A. Solorzano, D. E. Swayne, N. J. Cox, J. M. Katz, J. K. Taubenberger, P. Palese, and A. Garcia-Sastre. 2005. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science 31077-80. [DOI] [PubMed] [Google Scholar]
- 31.Veit, M., H. D. Klenk, A. Kendal, and R. Rott. 1991. The M2 protein of influenza A virus is acylated. J. Gen. Virol. 721461-1465. [DOI] [PubMed] [Google Scholar]
- 32.Wagner, R., A. Herwig, N. Azzouz, and H. D. Klenk. 2005. Acylation-mediated membrane anchoring of avian influenza virus hemagglutinin is essential for fusion pore formation and virus infectivity. J. Virol. 796449-6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watanabe, T., S. Watanabe, J. H. Kim, M. Hatta, and Y. Kawaoka. 2008. Novel approach to the development of effective H5N1 influenza A virus vaccines: use of M2 cytoplasmic tail mutants. J. Virol. 822486-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu, W.-H., and A. Pekosz. 2008. Extending the cytoplasmic tail of the influenza A virus M2 protein leads to reduced virus replication in vivo but not in vitro. J. Virol. 821059-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zebedee, S. L., and R. A. Lamb. 1988. Influenza A virus M2 protein: monoclonal antibody restriction of virus growth and detection of M2 in virions. J. Virol. 622762-2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeng, H., C. Goldsmith, P. Thawatsupha, M. Chittaganpitch, S. Waicharoen, S. Zaki, T. M. Tumpey, and J. M. Katz. 2007. Highly pathogenic avian influenza H5N1 viruses elicit an attenuated type I interferon response in polarized human bronchial epithelial cells. J. Virol. 8112439-12449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang, J., A. Pekosz, and R. A. Lamb. 2000. Influenza virus assembly and lipid raft microdomains: a role for the cytoplasmic tails of the spike glycoproteins. J. Virol. 744634-4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhirnov, O. P., T. E. Konakova, W. Garten, and H. Klenk. 1999. Caspase-dependent N-terminal cleavage of influenza virus nucleocapsid protein in infected cells. J. Virol. 7310158-10163. [DOI] [PMC free article] [PubMed] [Google Scholar]