Abstract
Highly pathogenic influenza virus strains currently in circulation pose a significant risk of a global pandemic. Following the reported crystal structure of the endonuclease domain from the avian influenza virus polymerase PA subunit, here we report the results of a systematic X-ray crystallographic analysis of its complex with adenosine, uridine, and thymidine nucleoside monophosphates (NMPs). Electron density corresponding to the monophosphate moiety of each nucleotide was apparent in each NMP complex and bound to the catalytic metal. A hydrophobic site was found to contribute to nucleoside binding. The NMP complex structures should represent the conformation of the bound product after nuclease cleavage. Moreover, one solvent molecule was found to occupy an equivalent position to the second reported Mn2+ ion, where it mediates the interaction between bound NMPs and the N-terminal PA domain in the presence of the Mg2+ ion. The results presented here indicate a possible cleavage mechanism and identify a distinct nucleotide binding pocket. The identification of this binding pocket opens a new avenue for anti-influenza drug discovery, targeting the cap-dependent endonuclease, in response to the worldwide threat of influenza.
The recent emergence of highly pathogenic avian and swine influenza viruses poses a significant global threat to human health (8). A total of 421 human infections by avian influenza H5N1 viruses have been reported worldwide since 2003, with 257 fatalities (WHO, April 2009). The current global outbreak of swine influenza with the H1N1 subtype has resulted in more than 100 deaths since it emerged in March 2009 and has spread to almost 40 countries. While approved anti-influenza drugs are available, their effectiveness in the event of an influenza pandemic may be limited due to drug resistance of the influenza viruses. Elucidating the underlying mechanisms of the virus life cycle and identifying new targets to be exploited for the discovery of antiviral therapeutics are therefore paramount.
The influenza virus contains a segmented RNA genome with eight negative-sense segments encoding 11 proteins. The influenza virus polymerase is a heterotrimeric ∼250-kDa complex with the following three protein subunits: PA, PB1, and PB2. It plays central roles in the viral life cycle and is directly responsible for RNA synthesis for both viral replication and transcription. However, the mechanisms by which these two different RNA synthesis functions are regulated within the large polymerase complex remain unclear. There is still some controversy about the functions of the various subunits, which have been reviewed by Liu et al. previously (17). Briefly, PB1 contains conserved and well-characterized RNA-dependent RNA polymerase motifs (3), while PB2 is required for transcription (16) and methylated cap binding (7, 9). PA has been implicated in a diverse range of functions but has been confirmed to possess endonuclease activity (6, 23).
PA is a 80-kDa subunit and can be cleaved into two independent domains (10, 11), as follows: a smaller N-terminal domain with cap-dependent endonuclease activity (6, 23) and a larger C-terminal domain that mediates the interaction with PB1 (13, 20). The recent crystal structures of the N-terminal PA domain, termed PAN, confirmed its endonuclease activity (6, 23), although the mode of substrate binding, cleavage mechanism, and metal dependence by PAN remains unclear. Understanding such aspects should provide a handle for the discovery of specific drugs that block the cap-snatching step during influenza virus genome replication. To provide a structural basis for substrate binding by the cap-dependent endonuclease, we have determined the high-resolution crystal structures of complexes of PAN with three nucleoside monophosphates (NMPs). For these three NMP complexes, ribo-UMP (rUMP), rAMP, and TMP, we observe electron density near to the active site which can be readily interpreted as the phosphate moieties of the NMPs. An additional solvent molecule occupies a potential metal ion binding site and mediates the interaction between the phosphate moieties of the NMPs and residues Glu119 and Lys134 of PAN. Moreover, the location of their less-ordered nucleoside moieties indicates a relatively hydrophobic pocket (N site). His41 is also involved in both the N site and the ribose binding site (R site). In sharp contrast, unambiguous electron density indicates that rGMP and rCMP will not bind into this site, suggesting that this binding site has substrate specificity. The results reported here provide a more-detailed picture of PAN nuclease cleavage and provide a distinct binding pocket for anti-influenza drug discovery targeting the cap-dependent endonuclease.
MATERIALS AND METHODS
Cloning, expression, and purification.
PAN, covering residues 1 to 256, was cloned from an avian influenza A virus isolate [A/goose/Guangdong/1/96 (H5N1)], as described previously (23).
Crystallization and diffraction data collection.
Crystals of a form of the influenza virus PAN containing the N-terminal 256 residues were grown, as described previously (23). Crystals were soaked for 25 to 30 days at 16°C in a solution containing either rGMP, rUMP, rAMP, TMP, or rCMP at the concentration of 15 mM in 17.5% (wt/vol) PEG 3350, 100 mM MgAc, 100 mM MES (morpholineethanesulfonic acid) (pH 6.5). The soaking solution is enough for the cryoprotectant, and the crystals were flash-cooled by plunging them into liquid nitrogen. For soaking, all of these five nucleosides were added into soaking solution excessively, and precipitate was observed in the soaking solution. The nucleotide concentration used was 15 mM, indicating that the actual concentration of nucleotides is lower than that shown. To elucidate the difference between divalent metal ions, PAN crystals were presoaked in buffer containing 17.5% (wt/vol) PEG 3350, 10 mM EDTA, and 100 mM MES (pH 6.5) for 4 h to remove magnesium ions, then transferred to the buffer containing 17.5% (wt/vol) PEG 3350, 100 mM MnCl2, and 100 mM MES (pH 6.5), and soaked for another 4 h.
Diffraction data of PAN soaked with rGMP and Mn2+ were collected using an in-house Rigaku MicroMax-007 rotating anode X-ray generator, with an R-AXIS IV++ detector at a wavelength of 1.5418 Å. The remaining data sets were collected at BL17A of the Photon Factory (Japan) at a wavelength of 1.0000 Å. Data were processed and reduced with the HKL-2000 package (21). All data collection statistics are summarized in Table 1.
TABLE 1.
Data collection and refinement statistics
| Statistics | Value(s) forc: |
|||
|---|---|---|---|---|
| TMP | rAMP | rUMP | Mn2+ | |
| Data collectiond | ||||
| Space group | P1 | P1 | P1 | P1 |
| Wavelength (Å) | 1.0000 | 1.0000 | 1.0000 | 1.5418 |
| Resolution range (Å) | 50.0 (2.0)-1.9 | 50.0 (1.9)-1.8 | 50.0 (2.0)-1.9 | 50.0 (2.6)-2.5 |
| Total no. of reflections | 403,222 | 478,447 | 401,429 | 57,883 |
| No. of unique reflections | 54,857 | 63,099 | 54,981 | 19,014 |
| Completeness (%) | 94.2 (72.7) | 92.0 (64.1) | 96.1 (79.9) | 74.0 (51.0) |
| Average I/σ(I) | 11.2 (5.8) | 12.8 (4.2) | 13.4 (4.5) | 10.7 (2.1) |
| Rmerge (%)a | 6.4 (26.4) | 5.4 (27.4) | 6.2 (25.1) | 8.0 (30.3) |
| Refinement | ||||
| No. of reflections used (σ(F) > 0) | 52,062 | 59,908 | 52,165 | 18,020 |
| Rwork (%)b | 21.2 | 20.6 | 21.2 | 24.0 |
| Rfree (%)b | 27.6 | 26.2 | 27.4 | 28.8 |
| RMSD bond distance (Å) | 0.023 | 0.024 | 0.021 | 0.019 |
| RMSD bond angle (°) | 2.033 | 1.816 | 1.892 | 1.818 |
| Average B value (Å2) | 35.8 | 29.1 | 29.2 | 42.9 |
Rmerge = ΣhΣl(Iih − <Ih>)/ΣhΣI <Ih>, where <Ih> is the mean of multiple observations Iih of a given reflection, h.
Rwork = Σ{[Fp(obs)] − [Fp(calc)]}/Σ[Fp(obs)]. Rfree is an R factor for a selected subset (5%) of reflections that was not included in prior refinement calculations.
Numbers in parentheses are corresponding values for the highest-resolution shell.
The cell parameters for TMP, rAMP, rUMP, and Mn2+ are as follows: a, 51.1 Å; b, 59.6 Å; c, 66.9 Å; α, 96.3°; β, 96.6°; and γ, 109.4°.
Structure determination.
The program PHASER (18) was used for molecular replacement, with the structure of unliganded PAN as a search model (Protein Data Bank [PDB] accession no. 3EBJ) (23). Refinement was carried out with Refmac5 (19) and PHENIX (1). An initial positional and B-factor refinement of the molecular replacement solution led to an Rfree (4) value below 32%. The bound metal ions and NMPs were built under the guidance of Fo-Fc difference maps. Two peaks near Glu119 were clearly attributable to two catalytic metal ions when the metal used in the crystallization buffer was manganese.
After the bound NMPs and metal ions were built in the catalytic site and after the amino acid side chains surrounding these ions were manually adjusted, three cycles of refinement and automatic water molecule picking were performed. After addition of water molecules and automatic adjustment of the weight term by Refmac5, Rwork dropped below 23%, and Rfree dropped below 29%. Complete statistics for the data collection and refinement of PAN complexes are provided in Table 1.
Protein structure accession numbers.
Coordinates and structure factors for PAN have been deposited in the PDB with the following accession numbers: 3HW3 for the rUMP complex, 3HW4 for the TMP complex, 3HW5 for the rAMP complex, and 3HW6 for the Mn2+-bound structure.
RESULTS AND DISCUSSION
The NMP binding site in the catalytic center.
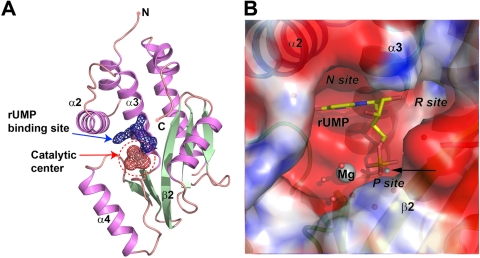
X-ray diffraction analysis of PAN crystals soaked with rUMP/rAMP/TMP and divalent cations under the conditions described in Materials and Methods revealed extra density around the catalytic site of the PAN endonuclease, suggesting that the affinity of PAN for nucleosides is therefore sufficiently high in the presence of Mg2+ (Fig. 1). This binding site is located in the top half of the catalytic center, where residues from α2, α4, and β2 contribute mainly to the interaction with bound rUMP (Fig. 1B). The phosphate moieties of each NMP were visible in the difference electron density maps, while the nucleoside moieties were not so clear, indicating that the nucleoside moieties are less ordered (Fig. 2). The refined temperature factors of these bound NMPs ranged between 50 Å2 and 60 Å2, depending on the particular complex. While PAN is reported to show higher nuclease activity in the presence of Mn2+ ions (6), the mode of binding by each NMP to PAN is expected to be the same in the presence of either Mn2+ or Mg2+. In the case of Mg2+-bound PAN, a solvent molecule occupies the position of the second Mn2+ ion. Crystallographic refinement was therefore pursued first for the models corresponding to complexes obtained in the presence of Mg2+. Furthermore, we also observed no electron density in the catalytic center binding site after soaking rUMP, TMP, or rAMP into crystals presoaked with EDTA, suggesting that the presence of divalent ions is crucial for substrate binding. Furthermore, comparison of PAN in both native and complexed forms gives a low root mean square deviation (RMSD) of less than 0.5 Å (calculated by LSQMAN [15], using all atoms of the PAN polypeptide), which indicates that substrate binding does not require any significant conformational change of PAN and that the binding site is enough to accommodate the bound NMP.
FIG. 1.
Overview of the binding site inside the catalytic center, which is revealed by the crystal structure of the PAN complex with rUMP. (A) Overall view of the PAN and rUMP complex. The rUMP binding site inside the catalytic center is represented by electron density and shown as blue mesh, while the bound Mg2+ and coordinated solvent molecules are covered by red mesh. Secondary structure elements of the PAN polypeptide are colored light pink (helices) and green (strands). (B) Electrostatic surface potential of the rUMP binding site. PAN is shown covered by an electrostatic surface, colored blue for positive charge and red for negative charge, while bound rUMP is shown as yellow sticks. The Mg2+ ions and solvent molecules are shown as spheres, and relevant secondary structure elements are labeled. The location of the second metal ion (corresponding to the Mn2+-bound structure) is indicated by a black arrow and small gray sphere.
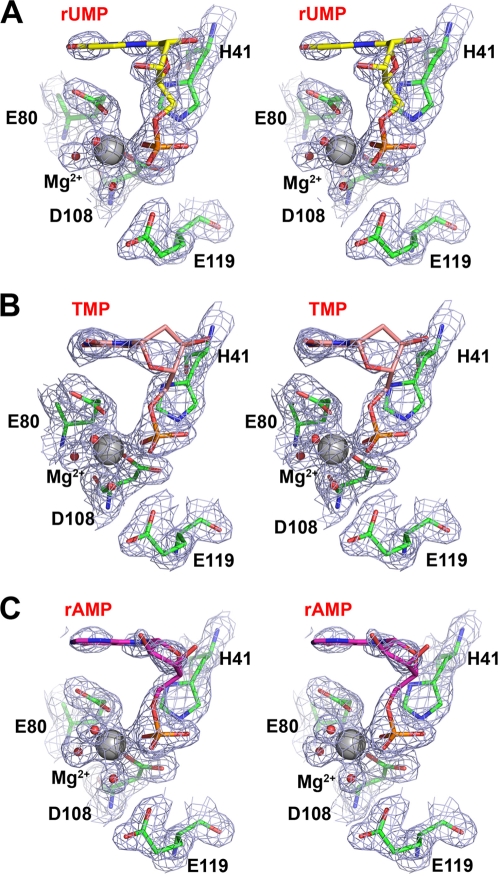
FIG. 2.
Electron density for bound rUMP (A), TMP (B), and rAMP (C). All NMPs are shown in stick representation, as follows: rUMP is colored yellow, TMP is colored pink, and rAMP is colored violet. Interacting residues are shown as green sticks. The Mg2+ ion and solvent molecules are shown as gray and red spheres, respectively. Electron density maps are shown as blue mesh and contoured at 1.5 σ.
Interaction between rUMP and PAN.
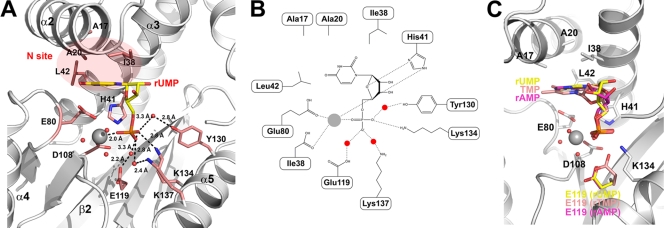
Residues from α2, α4, and β2 of PAN contribute mainly to NMP binding and form three subsites based on the binding with different moieties of the NMPs. We hereafter refer to the subsites as the P site (for the phosphate binding site), the R site (for the ribose binding site), and the N site (for the nucleoside binding site) (Fig. 1B). As the modes of interaction with PAN are similar for rUMP, rAMP, and TMP, we will discuss only the rUMP complex structure, unless otherwise stated.
The P site, which is responsible for phosphate binding, is formed mainly by the side chains of Glu119, Lys134, Lys137, and Tyr130. The O1P atom of the phosphate moiety, which occupies the position of the solvent molecule which was coordinated by Mg2+ and Asp80 in the crystal structure of native PAN (23), is stabilized by the ɛ-amino group of Lys137 from a water-mediated hydrogen bond, while the O2P atom interacts with the OH group of Tyr130 and the ɛ-amino group of Lys134 (Fig. 3A and B). The coordinated Mg2+ ion interacts with the O3P atom of the phosphate moiety, with a distance of 2.1 Å (Table 2). The side chain of Asp108 also bridges the Mg2+ ion and the water molecule, similar to the structure reported by Dias and colleagues (6). The ɛ-amino atom of His41 also contributes to stabilizing the phosphate moiety by forming bidentate hydrogen bonds with the O2P and O3P atoms, with the same distance of 3.4 Å.
FIG. 3.
Details of the interaction between PAN and bound rUMP. (A) Interaction between PAN and bound rUMP. The PAN polypeptide is shown in ribbon representation and colored white; relevant secondary structure elements are labeled. Bound rUMP and interacting residues are shown as yellow and pink sticks, respectively. The divalent Mg2+ ion and coordinating solvent molecules are shown as gray and red spheres, respectively. All possible interactions are shown as dashed lines, with related distances given. (B) Schematic showing the interaction between PAN and bound rUMP. Residues in the PAN polypeptide are labeled. Interactions are shown as dashed lines. (C) Superposition of bound rUMP, rAMP, and TMP. The PAN polypeptide is shown in ribbon representation and colored white; rUMP, TMP, and rAMP are colored yellow, pink, and violet, respectively. The residues involved in substrate binding but whose conformations do not change are shown as white sticks; Glu119 is colored using the same color scheme as that used for NMPs.
TABLE 2.
Hydrogen bonds between bound rUMP, the Mg2+ ion, and PAN atoms and residues
| rUMP atom | Hydrogen bond(s) between rUMP atoms anda: |
Distance (Å)b | |
|---|---|---|---|
| PAN residue | PAN atom | ||
| O4 | Leu16 (wat) | O | 2.4 (2.6) |
| Glu80 | Oε1 | 2.6 | |
| O2 | Leu42 | N | 2.9 |
| O4′ | His41 | Nδ | 2.9 |
| O1P | R137 (wat) | Nζ | 2.7 (2.4) |
| O2P | Ile120 | O | 3.3 |
| Tyr130 (wat) | OH (wat) | 2.8 (3.3) | |
| O3P | Mg2+ | 2.1 | |
wat, water-mediated hydrogen bond.
The value outside parentheses is the distance from rUMP atoms to water, while the value inside parentheses is the distance from water to PAN atoms.
While rUMP, rAMP, and TMP share a common mode of interaction with PAN (Fig. 2), a comparison of their complex structures indicates a different conformation of Glu119 in all three structures, in contrast to the other catalytic residues (Fig. 3C). In the crystal structures of PAN in complex with rUMP and rAMP, one of the two oxygen atoms in the side chain of Glu119 interacts with a free water molecule; the other oxygen interacts with a bound solvent molecule occupying an equivalent position to the second Mn2+ binding site. In the TMP complex structure, however, the side chain of Glu119 flips over and forms a bridge between the phosphate moiety and one of the water molecules coordinating the Mg2+ ion. One oxygen atom in its side chain will occupy the second Mn2+ binding site and directly interacts with the phosphate moiety, while the other one interacts with a solvent molecule coordinating the Mg2+ ion (Fig. 3C).
The R site, which contributes to stabilization of the ribose group of rUMP, is located in the middle of helix α4 of PAN and is formed mainly by the main chain atoms of Ala37-Cys39 and the side chain of His41. The two OH groups of the rUMP ribose moiety are directed toward the solvent and do not directly interact with the PAN polypeptide. At the opposite site of the ribose, the O4′ atom forms a 2.8-Å hydrogen bond with the Nδ atom of His41, suggesting that His41 also contributes to stabilization of the ribose of NMP.
Surprisingly, the nucleoside groups of rUMP, rAMP, and TMP show temperature factors above 50 Å2, compared with an average overall temperature factor of around 30 Å2. Only several atoms are well ordered in the electron density map in the N site, which is formed mainly by the side chains of Ala20, Leu38, Leu42, and Glu80. The N site accommodates the nucleobase of the bound ribonucleotide rUMP, and our structural observations suggest that the nucleobase is quite flexible in this site. Of the well-ordered atoms, the O2 atom of the uridine base interacts with the amide nitrogen of Leu42 and the Nδ atom of His41, with respective distances of 2.9 and 2.6 Å. The O4 atom of the uridine base interacts with the carbonyl atom of Leu16 from a water-mediated hydrogen bond and forms a 2.6-Å hydrogen bond with the side chain of Glu80. The side chains of Ala17, Ala20, Leu38, and Leu42 provide a hydrophobic platform against which the uridine group lies flat, almost perpendicular to the orientation of the phosphate and ribose moieties. As PAN is in the active form in the presence of Mg2+, this observation suggests that the crystal structures of PAN in complex with rAMP, rUMP, and TMP represent a scenario in which the products are bound to the protein after the nuclease cleavage reaction.
In contrast, it was evident that there was a lack of electron density in the catalytic center of PAN soaked with rGMP and rCMP at 15 mM for a period ranging from 10 days to 1 month (data not shown). The catalytic center in these two types of soaked crystals was identical to that in the native PAN, which would suggest that this binding site exerts some selectivity on the nucleotide binding. Notably, rGMP/rCMP usually generates strong secondary structures, while rAMP/rUMP is likely to form linear nucleic acid. We propose that this binding site recognizes nucleotides which are used mainly to form linear nucleic acid, which appears to be the heel of the 5′ cap of capped RNA and is consistent with the endonuclease function of PAN in the process of cap snatching. It is interesting to observe that one solvent molecule occupies the additional metal binding site which was found in the Mn2+-bound structure reported by Dias and colleagues but was not found in our previous Mg2+-bound PAN structure. This observation was also found in the structures of PAN in complex with rAMP and TMP. In both of the previously reported structures of native PAN (6, 23), these two different metal binding modes arose under different crystallization conditions. As these two PAN structures were derived from different influenza virus subtypes and strains, we used EDTA to remove the bound Mg2+ ions from our PAN crystals and then soaked them in buffers containing 100 mM Mg2+ and Mn2+, in order to examine the different metal binding characteristics of PAN.
Unsurprisingly, the clear electron density shows a Mn2+ binding mode similar to that in the structure observed by Dias and colleagues. This would suggest that these two different metal binding modes are the result of the artificial crystallization conditions. Together with the conformational change of Glu119 in different complexes, these observations suggest that the second Mn2+ binding site should be less stable and could be directly occupied by solvent or by an amino acid side chain in the absence of artificial buffer containing Mn2+ ions. Taken together and considering the much higher concentration of Mg2+ (1 to 5 mM) (2) than Mn2+ (μM) (5) under physiological conditions, it is more likely that Mg2+ is the natural cofactor of PAN, despite the higher nuclease activity in the presence of Mn2+.
Model for PAN with a nucleic acid substrate.
The results reported here provide a detailed high-resolution picture of the binding of NMPs to the cap-dependent endonuclease in the influenza A virus polymerase PA subunit. As previously reported, PAN shares high structural similarity with type II endonucleases (6, 23). Therefore, the comparison of our results with known structures with or without a substrate becomes more informative. The observation that rUMP/rAMP/TMP bind at the catalytic site with the divalent metal ion and hydrogen bonded to the polypeptide presents a picture of product binding and should be important for the conception of new, specific antiviral drugs. As the relevant interactions involve only divalent metal ions and the key catalytic residues of the PA subunit in influenza A virus, it is conceivable that the virus could not easily mutate to escape inhibition by compounds which block the endonuclease activity. Analogs of rUMP that bind to this site in this alternative mode with higher affinity, acting as competitive inhibitors of the enzyme, can be obtained. Indeed, some compounds with an inhibitory effect on influenza virus cap-dependent endonuclease have been reported (12, 22), although to date there is no evidence that they could directly act on PAN.
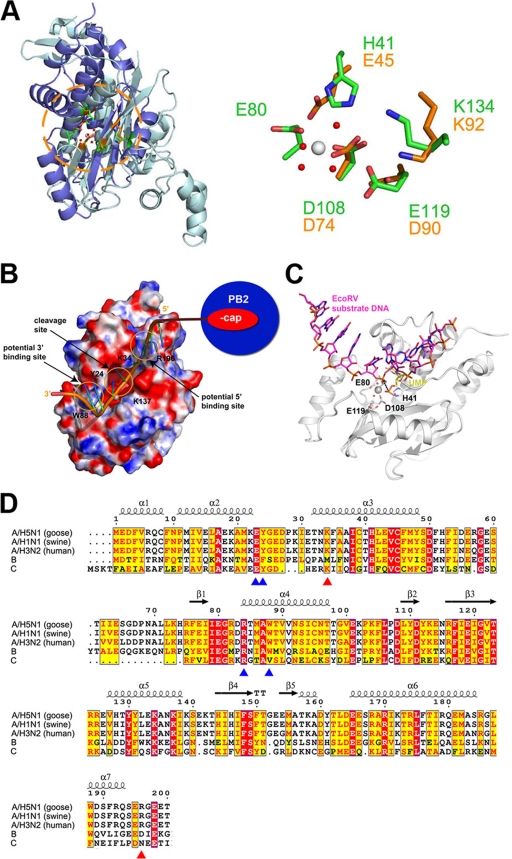
Based on our crystal structures of PAN in complex with NMPs reported here, we built a model for PAN in complex with a nucleic acid substrate based on the crystal structure of EcoRV endonuclease in complex with 5′-CGGGATATCCC DNA (PDB accession no. 1BGB) (14). The EcoRV endonuclease is a typical type II endonuclease and shares high structural similarity with PAN. The nucleic acid model was constructed by superimposing PAN onto EcoRV using all atoms of their key catalytic residues (Glu119 to Asp90, Asp108 to Asp74, His41 to Glu45), with an RMSD of 1.0 Å. Although the key catalytic residues of PAN and EcoRV superimpose well (Fig. 4A), the superimposed substrate DNA of EcoRV inside the catalytic center of PAN is distant from the bound rUMP. In further detail, the nucleotide of the superposed EcoRV substrate passes close to the divalent Mg2+ ion in the catalytic center of PAN. The phosphate group of the modeled substrate is about 4.5 Å from the equivalent part of bound rUMP, while the base and sugar groups have a 90° counterclockwise rotation (Fig. 4C). This observation suggests that the structures of PAN with NMPs at the catalytic center might represent the bound conformation of the products after the nuclease cleavage reaction. However, we cannot rule out the possibility that these observations are due to the limitations of molecular modeling, and further work is needed to confirm this.
FIG. 4.
Possible substrate binding model based on superposition with the crystal structure of EcoRV in complex with its substrate DNA. (A, left) Overview of superposition of PAN (blue) with EcoRV (cyan). The molecules are shown in ribbon representation. (A, right) Superposition of catalytic residues of PAN and EcoRV. The residues and relevant labels are colored using the same scheme; PAN is shown in green, while EcoRV is shown in gold. (B) Possible model of a capped RNA chain linked with PAN and the PB2 cap-binding domain. (C) Comparison of the modeled substrate and bound rUMP. PAN is shown in ribbon representation and colored white, and relevant residues are shown as sticks. The modeled substrate and bound rUMP are shown as magenta and yellow sticks, respectively. A nucleotide shift between the complex structure and substrate-bound model is also labeled. (D) Multiple sequence alignment of PAN for different influenza viruses, as follows: A/Guangdong/goose/1/1996 (avian influenza H5N1), A/California/04/2009 (swine influenza H1N1), A/Victoria/3/1975 (human influenza H3N2), B/Ann Arbor/1/1966 (B), and C/Johannesburg/1/1966 (C). Residue numbers and secondary structure elements were assigned according to PAN (avian influenza H5N1). The residues, which could be related to potential 5′- and 3′-end binding sites, are highlighted by red and blue triangles, respectively, at the bottom.
The modeled oligomeric substrate passes through the catalytic center of PAN, with its 5′ end close to a narrow cleft near the N terminus of PAN and its 3′ end close to the molecular surface of PAN. The 5′-end cleft is formed mainly by two basic residues, Lys34 and Arg196, which are strictly conserved in the influenza A virus (Fig. 4D). As Arg196 is immediately next to the linker from residues 197 to 256, connecting the N- and C-terminal domains of PA, it is possible that this linker could be directly involved in PA substrate binding, in addition to its other reported functions (10). The 3′ end of the substrate just crosses a gap formed mainly between Tyr24 and Trp88 face to face and accessorized by several other basic residues, which show relatively high conservation among the influenza A viruses. Although there is no direct biological evidence for this hypothesis, it is conceivable that these two sites might be involved in stabilizing the PAN substrate for the nuclease cleavage.
Conclusions.
The results presented here provide a detailed high-resolution picture of the binding of NMPs to the endonuclease domain of the avian influenza A virus polymerase PA subunit. The NMPs represent the cleavage products, with saturating conditions of the ligands used in order to maximize the occupancy and facilitate the interpretation of the electron density maps. Under these conditions, we were able to observe a distinct site inside the catalytic center which can selectively bind rUMP/rAMP/TMP (Fig. 2) but not rGMP or rCMP (data not shown). The NMPs do not form a strong secondary structure, suggesting that this selectivity might be related to its cap-dependent endonuclease activity with the linear nucleic acid after capping. Based on the superposition with the crystal structure of EcoRV in complex with its substrate, a model of PAN with a nucleic acid substrate was constructed to present the possible interaction between PAN and its substrate. The identification of a binding site in the catalytic center of PAN is particularly informative and should accelerate the discovery of new, specific antiviral drugs. It is conceivable that rUMP/rAMP/TMP analogues binding to this site with higher affinity, and which act as competitive inhibitors of the enzyme, can be employed as therapeutics to combat the current threat of highly pathogenic influenza viruses.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grants 30870486 and 3059943), the National Major Project of China (grants 2009zx/0004 and 2009zx09311-001), the Ministry of Science and Technology (MOST) 973 Project (grants 2006CB806503, 2006CB10901, and 2007CB914300), the MOST International Cooperation Project (grant 2006DFB32420), 863 Project (grants 2006AA020502 and 2006AA02A322), and the PSA II Project from MOST and KNAW (grant 2008AA000238).
Footnotes
Published ahead of print on 8 July 2009.
REFERENCES
- 1.Adams, P. D., R. W. Grosse-Kunstleve, L. W. Hung, T. R. Ioerger, A. J. McCoy, N. W. Moriarty, R. J. Read, J. C. Sacchettini, N. K. Sauter, and T. C. Terwilliger. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58:1948-1954. [DOI] [PubMed] [Google Scholar]
- 2.Alatossava, T., H. Jutte, A. Kuhn, and E. Kellenberger. 1985. Manipulation of intracellular magnesium content in polymyxin B nonapeptide-sensitized Escherichia coli by ionophore A23187. J. Bacteriol. 162:413-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biswas, S. K., and D. P. Nayak. 1994. Mutational analysis of the conserved motifs of influenza A virus polymerase basic protein 1. J. Virol. 68:1819-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brunger, A. T. 1997. Free R value: cross-validation in crystallography. Methods Enzymol. 277:366-396. [DOI] [PubMed] [Google Scholar]
- 5.Devasahayam, G., D. J. Burke, and T. W. Sturgill. 2007. Golgi manganese transport is required for rapamycin signaling in Saccharomyces cerevisiae. Genetics 177:231-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dias, A., D. Bouvier, T. Crepin, A. A. McCarthy, D. J. Hart, F. Baudin, S. Cusack, and R. W. Ruigrok. 2009. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 458:914-918. [DOI] [PubMed] [Google Scholar]
- 7.Fechter, P., L. Mingay, J. Sharps, A. Chambers, E. Fodor, and G. G. Brownlee. 2003. Two aromatic residues in the PB2 subunit of influenza A RNA polymerase are crucial for cap binding. J. Biol. Chem. 278:20381-20388. [DOI] [PubMed] [Google Scholar]
- 8.Gambotto, A., S. M. Barratt-Boyes, M. D. de Jong, G. Neumann, and Y. Kawaoka. 2008. Human infection with highly pathogenic H5N1 influenza virus. Lancet 371:1464-1475. [DOI] [PubMed] [Google Scholar]
- 9.Guilligay, D., F. Tarendeau, P. Resa-Infante, R. Coloma, T. Crepin, P. Sehr, J. Lewis, R. W. Ruigrok, J. Ortin, D. J. Hart, and S. Cusack. 2008. The structural basis for cap binding by influenza virus polymerase subunit PB2. Nat. Struct. Mol. Biol. 15:500-506. [DOI] [PubMed] [Google Scholar]
- 10.Guu, T. S., L. Dong, P. Wittung-Stafshede, and Y. J. Tao. 2008. Mapping the domain structure of the influenza A virus polymerase acidic protein (PA) and its interaction with the basic protein 1 (PB1) subunit. Virology 379:135-142. [DOI] [PubMed] [Google Scholar]
- 11.Hara, K., F. I. Schmidt, M. Crow, and G. G. Brownlee. 2006. Amino acid residues in the N-terminal region of the PA subunit of influenza A virus RNA polymerase play a critical role in protein stability, endonuclease activity, cap binding, and virion RNA promoter binding. J. Virol. 80:7789-7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hastings, J. C., H. Selnick, B. Wolanski, and J. E. Tomassini. 1996. Anti-influenza virus activities of 4-substituted 2,4-dioxobutanoic acid inhibitors. Antimicrob. Agents Chemother. 40:1304-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He, X., J. Zhou, M. Bartlam, R. Zhang, J. Ma, Z. Lou, X. Li, J. Li, A. Joachimiak, Z. Zeng, R. Ge, Z. Rao, and Y. Liu. 2008. Crystal structure of the polymerase PA(C)-PB1(N) complex from an avian influenza H5N1 virus. Nature 454:1123-1126. [DOI] [PubMed] [Google Scholar]
- 14.Horton, N. C., and J. J. Perona. 1998. Recognition of flanking DNA sequences by EcoRV endonuclease involves alternative patterns of water-mediated contacts. J. Biol. Chem. 273:21721-21729. [DOI] [PubMed] [Google Scholar]
- 15.Kleywegt, G. J., and T. A. Jones. 1997. Detecting folding motifs and similarities in protein structures. Methods Enzymol. 277:525-545. [DOI] [PubMed] [Google Scholar]
- 16.Li, M. L., P. Rao, and R. M. Krug. 2001. The active sites of the influenza cap-dependent endonuclease are on different polymerase subunits. EMBO J. 20:2078-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu, Y., Z. Lou, M. Bartlam, and Z. Rao. 2009. Structure-function studies of the influenza virus RNA polymerase PA subunit. Sci. China C 52:450-458. [DOI] [PubMed] [Google Scholar]
- 18.McCoy, A., R. Grosse-Kunstleve, P. Adams, M. Winn, L. Storoni, and R. Read. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40:658-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53:240-255. [DOI] [PubMed] [Google Scholar]
- 20.Obayashi, E., H. Yoshida, F. Kawai, N. Shibayama, A. Kawaguchi, K. Nagata, J. R. Tame, and S. Y. Park. 2008. The structural basis for an essential subunit interaction in influenza virus RNA polymerase. Nature 454:1127-1131. [DOI] [PubMed] [Google Scholar]
- 21.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode, p. 307-326. In C. W. Carter, Jr., and R. M. Sweet (ed.), Macromolecular crystallography, part A, vol. 276. Academic Press, San Diego, CA. [DOI] [PubMed] [Google Scholar]
- 22.Singh, S. B., and J. E. Tomassini. 2001. Synthesis of natural flutimide and analogous fully substituted pyrazine-2,6-diones, endonuclease inhibitors of influenza virus. J. Org. Chem. 66:5504-5516. [DOI] [PubMed] [Google Scholar]
- 23.Yuan, P., M. Bartlam, Z. Lou, S. Chen, J. Zhou, X. He, Z. Lv, R. Ge, X. Li, T. Deng, E. Fodor, Z. Rao, and Y. Liu. 2009. Crystal structure of an avian influenza polymerase PA(N) reveals an endonuclease active site. Nature 458:909-913. [DOI] [PubMed] [Google Scholar]