Abstract
Phosphoprotein ppUL44 of the human cytomegalovirus (HCMV) DNA polymerase plays an essential role in viral replication, conferring processivity to the DNA polymerase catalytic subunit pUL54 by tethering it to the DNA. Here, for the first time, we examine in living cells the function of the highly flexible loop of ppUL44 (UL44-FL; residues 162 to 174 [PHTRVKRNVKKAP174]), which has been proposed to be directly involved in ppUL44's interaction with DNA. In particular, we use a variety of approaches in transfected cells to characterize in detail the behavior of ppUL44Δloop, a mutant derivative in which three of the five basic residues within UL44-FL are replaced by nonbasic amino acids. Our results indicate that ppUL44Δloop is functional in dimerization and binding to pUL54 but strongly impaired in binding nuclear structures within the nucleus, as shown by its inability to form nuclear speckles, reduced nuclear accumulation, and increased intranuclear mobility compared to wild-type ppUL44. Moreover, analysis of cellular fractions after detergent and DNase treatment indicates that ppUL44Δloop is strongly reduced in DNA-binding ability, in similar fashion to ppUL44-L86A/L87A, a point mutant derivative impaired in dimerization. Finally, ppUL44Δloop fails to transcomplement HCMV oriLyt-dependent DNA replication in cells and also inhibits replication in the presence of wild-type ppUL44, possibly via formation of heterodimers defective for double-stranded DNA binding. UL44-FL thus emerges for the first time as an important determinant for HCMV replication in cells, with potential implications for the development of novel antiviral approaches by targeting HCMV replication.
The Betaherpesviridae subfamily member human cytomegalovirus (HCMV) is a major human pathogen, causing serious disease in newborns following congenital infection and in immunocompromised individuals (28, 42). Replication of its double-stranded DNA (dsDNA) genome occurs in the nuclei of infected cells via a rolling-circle process mediated by 11 virally encoded proteins (32, 33), including a viral DNA polymerase holoenzyme, comprising a catalytic subunit, pUL54, and a proposed processivity factor, ppUL44 (14). ppUL44 is readily detectable in virus-infected cells as a 52-kDa phosphoprotein of 433 amino acids with strong dsDNA-binding ability (30, 45). Defined as a “polymerase accessory protein” (PAP) whose function is highly conserved among herpesviruses, ppUL44 is an essential factor for viral replication in cultured cells and hence represents a potential therapeutic target to combat HCMV infection (39). It is a multifunctional protein capable of self-associating (5, 10), as well as interacting with a plethora of viral and host cell proteins, including the viral kinase pUL97 (29), the viral transactivating protein pUL84 (15), the viral uracil DNA glycosylase ppUL114 (37), and the host cell importin α/β (IMPα/β) heterodimer, which is responsible for its transport into the nucleus (4). The activities of ppUL44 as a processivity factor, including the ability to dimerize, as well as bind to, pUL54 and DNA, reside in the N-terminal portion (26, 45), whereas the C terminus is essential for phosphorylation-regulated, IMPα/β-dependent nuclear targeting of ppUL44 monomers and dimers (4-6). Once within the nucleus, ppUL44 is thought to tether the DNA polymerase holoenzyme to the DNA, thus increasing its processivity (14).
Recent studies have identified specific residues responsible for ppUL44 interaction with pUL54, as well as for the interaction with IMPα/β and homodimerization (4, 10, 27, 41). The crystal structure of ppUL44's N-terminal domain (Fig. 1A) reveals striking similarity to that of other processivity factors, such as proliferating cell nuclear antigen (PCNA) and its herpes simplex virus type 1 (HSV-1) homologue UL42 (10, 46). Unlike the PCNA trimeric ring, however, both ppUL44 and UL42, which bind to dsDNA as dimers and monomers, respectively, have an open structure, which is believed to be the basis for their ability to bind to dsDNA in the absence of clamp loaders and ATP (9, 10, 46). Both ppUL44 and UL42 share a very basic “back” face, which appears to be directly involved in DNA binding via electrostatic interactions (19, 22, 23, 38, 46). One striking difference between ppUL44 and UL42 is the presence on the former of an extremely basic flexible loop (UL44-FL, PHTRVKRNVKKAP174) protruding from the basic back face of the protein (Fig. 1A). Comparison of ppUL44 homologues from different betaherpesviruses, including human herpesvirus 6 (HHV-6) and 7 (HHV-7), showed that all possess similar sequences in the same position (44) (Fig. 1B), implying functional significance.
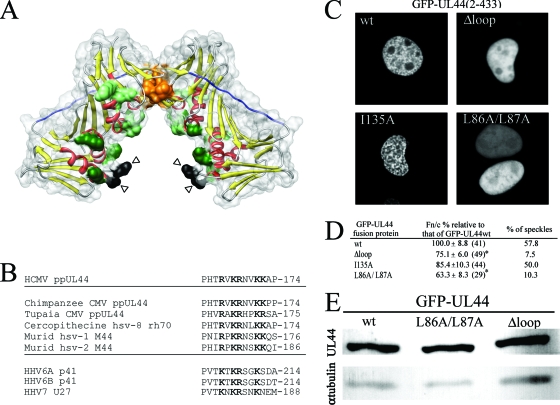
FIG. 1.
The highly conserved flexible loop (residues 162 to 174) within ppUL44 protrudes from ppUL44 basic face and is important for efficient nuclear accumulation and localization in nuclear speckles. (A) Schematic representation of ppUL44 N-terminal domain (residues 9 to 270, protein data bank accession no. 1T6L) generated using the Chimera software based on the published crystal structure (10, 35). Color: yellow, β-sheets; red, α-helices. Residues involved in ppUL44 dimerization (P85, L86, L87, L93, F121, and M123), as well as basic residues potentially involved in DNA binding (K21, R28, K32, K35, K128, K158, K224, and K237), are represented as spacefill in orange and green, respectively. Residues P162 and C175, in black, are indicated by arrowheads, while residues 163 to 174 are not visible in the electron density maps and could potentially extend in the cavity formed by ppUL44's basic face to directly contact DNA. Residues forming ppUL44 connector loop (128-142) are in blue. (B) Sequence alignment between HCMVUL44-FL and the corresponding region of several betaherpesvirus ppUL44 homologues. The single-letter amino acid code is used, with basic residues in boldface. (C) COS-7 cells were transfected to express the indicated GFP fusion proteins and imaged live 16 h after transfection using CLSM and a 40× water immersion objective lens. (D) Quantitative results for the Fn/c and speckle formation for GFP-UL44 fusion proteins. The data for the Fn/c ratios represent the mean Fn/c relative to each protein indicated as a percentage of the mean Fn/c relative to GFP-UL44wt ± the standard error of the mean, with the number of analyzed cells in parentheses. (E) HEK 293 cells expressing the indicated GFP-UL44 fusion proteins were lysed, separated by PAGE, and analyzed by Western blotting as described in Materials and Methods, using either the anti-GFP or the anti-α-tubulin MAbs.
A recent study revealed that substitution of UL44-FL basic residues with alanine residues strongly impairs the ability of a bacterially expressed N-terminal fragment of UL44 to bind 30-bp dsDNA oligonucleotides in vitro, suggesting that UL44-FL could be involved in dsDNA-binding during viral replication (22). However, the role of UL44-FL in mediating the binding of full-length UL44 to dsDNA in cells and its role in DNA replication have not been investigated. We use here a variety of approaches to delineate the role of UL44-FL in living cells, our data revealing that UL44-FL is not required for ppUL44 dimerization or binding to the catalytic subunit pUL54 but is crucial for HCMV oriLyt-dependent DNA replication, being required for the formation of nuclear aggregates, nuclear accumulation/retention, and DNA binding of ppUL44. Importantly, ppUL44Δloop exhibits a transdominant-negative phenotype, inhibiting HCMV oriLyt-dependent DNA replication in the presence of wild-type ppUL44, possibly via formation of heterodimers defective for dsDNA binding. This underlines ppUL44-FL as an important determinant for HCMV replication in a cellular context for the first time, with potential implications for the development of novel antiviral approaches.
MATERIALS AND METHODS
Generation of molecular graphics images.
A molecular graphic image of ppUL44 was generated by using the UCSF Chimera package (35) from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH P41 RR-01081).
Construction of expression plasmids.
Plasmid pDNR207-UL44Δloop encoding a point mutant derivative of ppUL44 in which three of the five basic residues within UL44-FL have been mutated (PHTRVKRNVKKAP174 → PHTgVngNVKKAP174), was generated by using a QuikChange mutagenesis kit (Stratagene) according to the manufacturer's recommendations, with appropriate oligonucleotide pairs and plasmid pDNR207-UL44 as a template (5). Plasmid pDNR207-UL44Δloop was used to perform LR Gateway system (Invitrogen) recombination reactions with the Gateway system compatible expression plasmids pDEST-FLAG, pEPI-DEST-GFP (36), and pBkCMV-DsRed2 (18), according to the manufacturer's recommendations, in order to generate mammalian expression vectors encoding N-terminally tagged fusion proteins. Plasmids pDEST-FLAG-UL44, pEGFPC1-H1E, pEPI-GFP-UL44ΔNLS, pEPI-GFP-UL54(1213-1242), pEPI-GFP-UL44wt, pEPI-GFP-UL44I135A, pEPI-GFP-UL44P85G, pEPI-GFP-UL44L86A/L87A, DsRed2-UL44wt, DsRed2-UL44I135A, and DsRed2-UL44L86A/L87A have been described (4, 5, 7, 16, 41).
Cell culture and transfection.
Daoy and COS-7 cells were maintained in Dulbecco modified Eagle medium supplemented with 5% (vol/vol) fetal bovine serum (FBS), 50 U of penicillin/ml, 50 U of streptomycin/ml, and 2 mM l-glutamine. Human embryonic kidney 293 (HEK 293) and human foreskin fibroblast (HFF) cells were cultured in Dulbecco modified Eagle medium supplemented with 10% (vol/vol) FBS, 50 U of penicillin/ml, 50 U of streptomycin/ml, and 2 mM l-glutamine.
For all experiments, cells were treated with trypsin, and 8 × 104 cells were used to seed respective plates 1 day before transfection, which was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's specifications. For live cell imaging by confocal laser scanning microscopy (CLSM), COS-7 cells were seeded onto coverslips in six-well plates. For Western blotting and nuclear matrix extraction experiments, HEK 293 cells were seeded in six-well plates, and for imaging of nuclear matrix-associated proteins, Daoy cells were seeded onto 2-mm glass-bottom Willcodishes (Willcowells). For HCMV oriLyt-dependent DNA replication assays, HFF cells were plated onto 6-cm-diameter dishes at a cell density of 105 per plate 24 h before transfection that was performed by using the calcium phosphate method (41).
CLSM/image analysis.
To determine the nuclear to cytoplasmic fluorescence ratio (Fn/c) values relative to GFP-UL44 fusion proteins when expressed in COS-7 cells, CLSM was used as previously described (4, 5, 7). Cells were analyzed by using an Olympus Fluoview 1000 equipped with a heated Planapo 60× water immersion lens (Nikon) in combination with a heated stage. The Fn/c ratios were calculated by using ImageJ 1.38 public domain software (National Institutes of Health) from single cell measurements for the nuclear (Fn) and cytoplasmic (Fc) fluorescence levels, subsequent to the subtraction of fluorescence due to autofluorescence and/or background.
FRAP analysis.
Fluorescence recovery after photobleaching (FRAP) analysis was performed in COS-7 cells transiently expressing GFP-UL44 wild-type and mutant derivatives by using an Olympus Fluoview 1000 (Olympus) microscope equipped with an Argon ion laser (40 mW) and a 100× oil immersion lens (Nikon) in combination with a heated stage. Three images were collected by using 3% total laser power with excitation at 488 nm (2× zoom, scanned at a rate of 8 μs/pixel) before photobleaching. The bleaching was performed by zooming 100-fold in a small area covering ca. 5% of the nucleus and by using 100% of the laser power (10 scans, at a rate of 8 μs/pixel). After bleaching, cells were immediately scanned, and fluorescence recovery was monitored by acquiring subsequent images at 20-s intervals for 6 min using detector and laser settings identical to those prior to photobleaching. Image analysis was performed as described above. The results were expressed as the fractional recovery of Fb/Fnb ratios (i.e., the fluorescence of the bleached area divided by the fluorescence of the nonbleached area) at several time points, divided by the prebleach Fb/Fnb value. FRAP data were fitted exponentially according to the formula y = a(1 − e−bx) as previously described (11, 24, 34) to determine the fractional recovery and t1/2 values.
Subcellular fractionation and Western blotting.
HEK 293 cells transfected to express the green fluorescent protein (GFP) fusion proteins of interest were harvested by gentle pipetting. After three washes in phosphate-buffered saline, cells were either resuspended in 100 μl of radioimmunoprecipitation assay buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate, 1 mM phenylmethylsulfonyl fluoride [PMSF], 10 μg of leupeptin/ml, 10 μg of aprotinin/ml), sonicated five times for 2 min on ice, and incubated for 1 h on ice to obtain whole-cell lysates or permeabilized for 15 min at room temperature using 100 μl of buffer A (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 5 mM MgCl2, 1% NP-40, 1 mM PMSF, 10 μg of leupeptin/ml, 10 μg of aprotinin/ml). Soluble proteins (S1) were isolated by centrifugation at 3,000 rpm for 10 min. Cell pellets were washed with 100 μl of buffer B (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 5 mM MgCl2, 1 mM PMSF, 10 μg of leupeptin/ml, 10 μg of aprotinin/ml), and pellets were separated from soluble proteins (S2) by centrifugation. DNA was digested by incubating pellets with buffer C (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 5 mM MgCl2, 100 U of DNase I [Fermentas]/ml, 1 mM PMSF, 10 μg of leupeptin/ml, 10 μg of aprotinin/ml) 1 h at room temperature. Digested DNA and DNA-bound proteins (S3) were separated from cellular pellets by centrifugation at 3,000 rpm for 10 min. Pellets were then washed with 100 μl of buffer B (S4). The nuclear matrix was separated from tightly DNA-bound proteins (S5) by treatment with buffer D (10 mM Tris-HCl [pH 7.4], 2 M NaCl, 5 mM MgCl2, 1 mM PMSF, 10 μg of leupeptin/ml, 10 μg of aprotinin/ml) and centrifugation at 3,000 rpm for 10 min. Matrix-associated proteins (S6) were solubilized with 150 μl of buffer E (90 mM Tris-HCl [pH 6.8], 100 mM dithiothreitol, 2% sodium dodecyl sulfate [SDS], 20% glycerol, 1 mM PMSF, 10 μg of leupeptin/ml, 10 μg of aprotinin/ml) and heating for 5 min at 95°C. Laemmli loading buffer was added to each fraction, the samples were boiled for 5 min at 95°C, and the mixtures were loaded onto 10% bis-Tris acrylamide gel prior to separation by polyacrylamide gel electrophoresis (PAGE). Electrophoretically separated proteins were then transferred to Hybond-P membranes (Amersham) as previously described (39). Membranes were blocked in blocking buffer F (5% [wt/vol] skin dry milk and 1× Tris-buffered saline) for 1 h at room temperature and washed three times with buffer G (0.05% Tween and 1× Tris-buffered saline). GFP fusion proteins were detected by incubating the membranes successively with either the anti-GFP (clone sc-9996; Santa Cruz Biotechnology; 1:400) or anti-α tubulin (clone B-5-1-2; Sigma; 1:1,000) monoclonal antibodies (MAbs) and horseradish peroxidase-coupled secondary antibody (Sigma; 1:500). Immunoblots were developed with the horseradish peroxidase substrate 4-Cl-1-naphthol (Bio-Rad) in the presence of H2O2. The intensity of bands relative to each fusion protein was quantified by using the ImageJ (National Institutes of Health) software.
Nuclear matrix preparation from intact cells.
Daoy cells grown on Willcodishes (Willcowells) were incubated for 10 min at 37°C in 5% CO2 with Hoechst 33342 (Invitrogen) at 1 μg/ml dissolved in Dulbecco modified Eagle medium containing 5% FBS. The subcellular localization of GFP fusion proteins was analyzed by using a Nikon Eclipse E600 inverted microscope (Nikon), equipped with a Nikon DXN1200 digital camera and a Nikon Plan Fluor ×40 objective lens (Nikon). Cells were then permeabilized with buffer A for 15 min at room temperature and washed twice with buffer B to remove the soluble proteins, and the subcellular localization of GFP fusion proteins was investigated by fluorescence microscopic analysis as described above. Cells were subsequently treated with buffer F (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 5 mM MgCl2, 40 U of DNase I [Fermentas]/ml, 1 mM PMSF, 10 μg of leupeptin/ml, 10 μg of aprotinin/ml) for 1 h at room temperature. DNA-bound proteins were then removed by incubating cells 10 min at room temperature with buffer D and washing them with buffer B, and the subcellular localization of the nuclear matrix-associated GFP fusion proteins was visualized as described above.
Immunoprecipitation of FLAG-tagged proteins.
At 48 h posttransfection, HEK 293 cells were harvested and washed with PBS. Cells were lysed in lysis buffer (50 mM HEPES [pH 7.4], 100 mM NaCl, 1% Nonidet-P 40, Complete Mini EDTA-free protease inhibitor cocktail tablets [Roche Applied Science]) for 10 min on ice and sonicated at low intensity for 10 s. After clarification, supernatants were incubated with 4 μg of the anti-FLAG at 4°C, with gentle rocking. The following day, 30 μl of protein A/G beads (Santa Cruz Biotechnology) was added, followed by incubation for 4 h. After washes with lysis buffer, the beads were resuspended in 2× Laemmli buffer, boiled at 95°C, and centrifuged at 16,000 × g for 5 min before analyzing the supernatant by SDS-10% PAGE and Western blotting using the anti-GFP (clone sc-9996; Santa Cruz Biotechnology; 1:10,000) and anti-FLAG (Sigma; 1:5,000) MAbs, followed by incubation with a peroxidase-coupled secondary antibody (Sigma; 1:10,000). The immunoblots were developed with ECL plus (Amersham).
oriLyt-dependent DNA replication transcomplementation assays.
To investigate the role of ppUL44 basic loop in HCMV DNA replication, HFF cells were transfected with the pSP50 plasmid, which contains the HCMV oriLyt DNA replication origin (8), a set of plasmids expressing the HCMV proteins essential for oriLyt-dependent DNA replication except for ppUL44 (32, 33) and either pEPI-GFP-UL44wt or pEPI-GFP-UL44Δloop plasmid. To test for the ability of ppUL44 mutants to interfere with the functionality of wild-type ppUL44, the GFP-UL44Δloop and the dimerization defective GFP-UL44L86A/L87A mutants were expressed in the presence of a set of plasmids encoding all of the HCMV proteins required for oriLyt-dependent DNA replication, including ppUL44, and of the pSP50 plasmid. Replication of pSP50 was detected by treatment of transfected cell DNA with DpnI, which cleaves only unreplicated, dam-methylated input DNA (32, 33), followed by Southern blotting as described previously (41).
RESULTS
The highly conserved ppUL44 flexible loop is crucial for ppUL44 localization in nuclear speckles and nuclear retention.
Based on the presence of a proline residue upstream of a stretch of basic amino acids, the N-terminal portion of UL44-FL (PHTRVKR168) had initially been proposed as a putative nuclear localization signal (NLS), which proved to be nonfunctional in terms of nuclear import (4, 25). Indeed, mutation of basic residues within UL44-FL does not prevent either binding to the IMPα/β heterodimer nor ppUL44 nuclear import. However, there is a difference in intranuclear distribution of ppUL44Δloop, a mutant derivative in which three of five UL44-FL basic residues were mutated, compared to wild-type ppUL44 (4). When fused to GFP, UL44Δloop showed diffuse nucleoplasmic localization rather than a speckled nuclear appearance, which has been described for PAPs from several herpesviruses and is believed to be the consequence of binding to dsDNA (1, 3-5, 7, 25, 44). Intriguingly, alanine substitution of residues L86 and L87, which are essential for dimerization of ppUL44, results in a similar loss of speckled pattern upon overexpression of the GFP-UL44L86A/L87A fusion protein in mammalian cells (41). Since ppUL44 dimerization defective mutants are also impaired for dsDNA in vitro (10), we hypothesized that the similar intranuclear localization of GFP-UL44Δloop and GFP-UL44L86A/L87A could be the result of impaired DNA binding. We therefore transiently expressed the aforementioned ppUL44 derivatives as GFP fusion proteins in COS-7 cells and analyzed their subcellular localization by CLSM (Fig. 1C). We also expressed GFP-UL44I135A, a point mutant derivative fully capable of dimerization and DNA binding but impaired in pUL54 binding as a control (27, 41). As expected, all proteins localized mainly in the nucleus of transfected cells. However, while GFP-UL44wt and GFP-UL44I135A often localized in distinctive intranuclear speckles (in >50% of cells [see Fig. 1C and D and reference 41]), both GFP-UL44L86A/L87A and GFP-UL44Δloop mutants did not (speckles were observed in ca. 10% of cells [see Fig. 1C and D]), a finding consistent with the idea that this results from impaired intranuclear binding. To verify this, we quantified the levels of nuclear accumulation for each protein. Both GFP-UL44L86A/L87A and GFP-UL44Δloop accumulated to significantly lower levels (P < 0.05) compared to GFP-UL44wt (Fn/c ca. 65 and 75%, respectively, of that of GFP-UL44wt [see Fig. 1D]), whereas the I135A substitution had no effect on ppUL44 nuclear accumulation (Fn/c ca. 90% of that of GFP-UL44wt). Thus, UL44-FL appears to play a key role in localizing ppUL44 in nuclear speckles and thereby increasing nuclear accumulation. Importantly, when we compared the level of expression of wild-type and mutant ppUL44 proteins by Western blot analysis, all proteins were expressed to comparable levels (Fig. 1E), indicating that the impaired ability to form nuclear speckles and accumulate into the nucleus observed for GFP-UL44L86A/L87A and GFP-UL44Δloop was not due to reduced expression levels.
UL44-FL is involved in ppUL44 intranuclear binding.
We reasoned that the reduced nuclear accumulation and speckle formation ability of GFP-UL44Δloop and GFP-UL44L86A/L87A could be due to reduced intranuclear binding (see above). To test this hypothesis, we analyzed their intranuclear mobility when transiently expressed in COS-7 cells using the FRAP technique. A small region within the nucleus was bleached and the recovery of the fluorescent signal monitored over time to calculate the fractional recovery (Fig. 2; see Movies S1 to S3 in the supplemental material). Using the laser power at 100% (10 scans, 8 μs/pixel), it proved possible to bleach a small area within the nucleus of cells expressing GFP-UL44 fusions (Fig. 2A). This was not possible in cells expressing GFP alone, where the treatment resulted in bleaching of fluorescence in the whole-cell (data not shown), which is consistent with the idea that ppUL44 binds to nuclear components such as dsDNA that markedly reduce its intranuclear mobility. Recovery of fluorescence on the bleached area of cells expressing GFP-UL44wt was three times slower than that of GFP-UL44L86A/L87A (t1/2 of ca. 40 s and 13 s, respectively; Fig. 2B and C, see also Movies S1 to S3 in the supplemental material), in keeping with ppUL44wt's ability to bind tightly to dsDNA. Similarly, GFP-UL44Δloop was also more mobile than GFP-UL44wt, with a t1/2 of ca. 20 s, implying that mutation of the UL44-FL reduces intranuclear binding and hence increases the intranuclear mobility of the protein. Consistent with the idea that ppUL44 binds to nuclear components, the fractional recovery of fluorescence in cells expressing GFP-UL44wt was ca. 0.6, indicating that ca. 40% of ppUL44 is immobile within the cell nucleus. In contrast, cells expressing GFP-UL44L86A/L87A exhibited significantly higher fractional recovery (ca. 0.8), supporting the idea that prevention of DNA binding through inhibition of ppUL44 dimerization increases its intranuclear mobility. Cells expressing GFP-UL44Δloop also exhibited significantly higher (ca. 0.8) fractional recovery, a finding consistent with impaired binding of the protein to nuclear components/DNA and further supporting the critical role of UL44-FL in intranuclear binding.
FIG. 2.
UL44-FL is required for ppUL44 intranuclear binding as determined by FRAP analysis. The intranuclear mobility of the indicated GFP fusions was analyzed by FRAP 16 h after transfection of COS-7 cells. (A) Visualization using CLSM of the return of intranuclear fluorescence after photobleaching in a specific area of the nucleus (black boxes) (see Materials and Methods). (B) Quantification of the recovery over time of specific nuclear fluorescence after photobleaching expressed in terms of the fractional recovery of the Fb/Fnb ratio (Fb [fluorescence of the bleached area above background] divided by Fnb [fluorescence of the unbleached area above background] at the indicated time points divided by the prebleach Fb/Fnb value). (C) Pooled data (average ± the standard error of the mean, n = 9) for the fractional recovery and half-time of the return of fluorescence (t1/2) for the wild type and the mutants, with significant differences denoted by the P values.
UL44-FL is required for intranuclear retention of ppUL44.
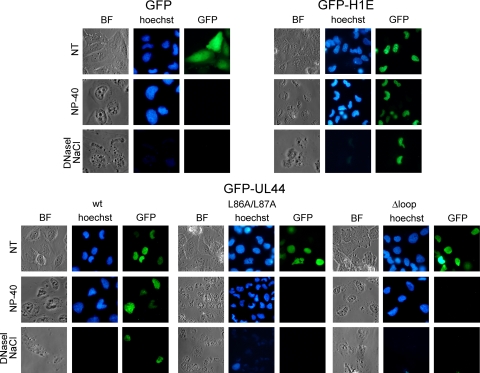
Our FRAP results strongly suggested that ppUL44 binds to intranuclear structures in living cells. We therefore decided to verify this hypothesis by imaging detergent-permeabilized cells. Since treatment of cells with detergents results in the release of nucleoplasmic proteins, we expressed GFP-UL44, GFP-UL44Δloop, and GFP-UL44L86/87A in Daoy cells and analyzed their subcellular localization before and after cell permeabilization with Nonidet P-40 (NP-40). As a negative control, we also analyzed the subcellular localization of GFP, which does not bind to nuclear components. As a further control, we expressed a fusion protein between GFP and the linker histone H1 variant H1E, which binds tightly to cellular chromatin, and it is therefore retained in the nucleus upon cell permeabilization (31).
As expected, before cell permeabilization GFP localized diffusely throughout the cytoplasm and the nucleus, whereas GFP-H1E strongly localized within the cell nucleus, occasionally colocalizing with cellular heterochromatin (Fig. 3). On the other hand, all GFP-UL44 mutant derivatives strongly localized within the cell nucleus, with GFP-UL44wt often colocalizing with cellular chromatin, and GFP-UL44Δloop and GFP-UL44L86A/L87A mainly localizing with a diffuse pattern (Fig. 3). Upon cell permeabilization, GFP alone was no longer detectable, indicating that GFP does not bind to nuclear components within the cell. As expected, GFP-H1E was mainly retained within the cell nucleus, as a consequence of its ability to strongly bind to cellular chromatin via its C-terminal tail (2). Importantly, a considerable fraction of GFP-UL44wt was still retained within the nucleus in a typical speckled pattern after cell permeabilization, whereas most of GFP-UL44L86A/L87A was lost, possibly as a consequence of its inability to efficiently bind to dsDNA (10). Similarly, cell permeabilization resulted in a strong decrease of the fluorescence relative to GFP-UL44Δloop (Fig. 3), a finding consistent with the involvement of UL44-FL in binding to dsDNA in vitro (22). These results clearly indicate that GFP-UL44, but not GFP-UL44L86A/L87A and GFPUL44Δloop, binds strongly to nuclear structures. Importantly, upon permeabilization, the colocalization of GFP-UL44 with cellular chromatin became clearer (see Fig. S1 in the supplemental material), in keeping with the idea that UL44 interacts with nuclear components. To verify that GFP-UL44 is able to bind to cellular dsDNA when transiently expressed in the absence of other viral proteins as suggested by previous studies (25), cells where further treated with DNase I before being washed with 2 M NaCl. A significant decrease in the signal relative to both GFP-H1E and GFP-UL44 was observed upon such treatment, suggesting that a portion of ppUL44 could bind to cellular dsDNA when expressed in the absence of other viral proteins.
FIG. 3.
GFP-UL44 associates with dsDNA and the nuclear matrix, in contrast to GFP-UL44L86A/L87A and GFP-UL44Δloop. The intracellular localization of the indicated GFP fusion proteins was investigated 24 h after transfection of Daoy cells, prior to treatment (NT), or after treatment with 1% NP-40 for 15 min at room temperature (NP-40) or after incubation with 40 U of DNase I/ml for 1 h at room temperature, followed by incubation with NaCl 2 M for 5 min (DNase I, NaCl). The bright-field (BF) and the green channel (GFP) are shown, with cellular dsDNA being shown in blue (Hoechst).
ppUL44-FL binds to dsDNA in cells.
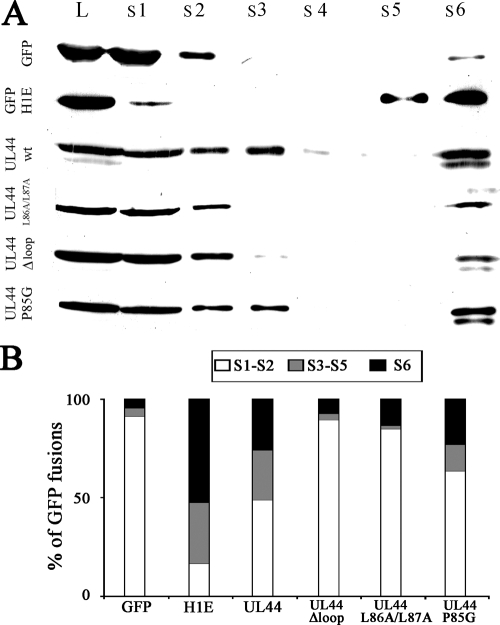
To verify the involvement of UL44-FL in DNA binding in cells, we transiently expressed several GFP-UL44 fusion proteins in HEK 293 cells and, at 48 h posttransfection, quantitatively analyzed the amount of proteins released after cell permeabilization and DNase I treatment by Western blotting. GFP and GFP-H1 were also expressed as controls. As expected, almost all of GFP (>90% [Fig. 4A and B]) was detectable in the S1 and S2 fractions, corresponding to completely soluble proteins. On the other hand, only ca. 20% of GFP-H1E was detectable in the soluble fractions (S1 and S2), and a considerable amount was detectable in the DNA-containing (S3 to S5, more than 30% of total) and matrix-associated (S6, ca. 50%) fractions. Importantly only ca. 50% of total GFP-UL44 could be detected in the soluble fractions, with a significant amount of protein (ca. 25% each) being found in the DNA- and nuclear matrix-containing fractions, confirming that ppUL44 can interact with cellular chromatin in the absence of viral DNA. As expected, the dimerization defective GFP-UL44L86A/L87A mutant derivative was also impaired in DNA binding, being detectable almost exclusively (ca. 90%) in the soluble fractions. Similar results were obtained for the GFP-UL44Δloop mutant derivative, which was mainly (ca. 85%) detectable in the soluble fractions and was almost completely absent (ca. 3% of total fusion protein) from the DNA-containing fractions. On the other hand, significant amounts of the control molecule GFP-UL44P85G, which is not impaired for dimerization and DNA binding, were detectable in both the DNA- and the matrix-associated fractions (ca. 15 and 25% of the total fusion protein, respectively). These results show that, when expressed in the absence of other viral proteins, ppUL44 can bind to nuclear components such as DNA and the nuclear matrix through a process that requires dimerization of the protein and its highly flexible loop.
FIG. 4.
ppUL44 dimerization and UL44-FL are required for in vivo DNA binding. (A) HEK 293 cells expressing the indicated fusion proteins were harvested 48 h after transfection, and cellular fraction collection and SDS-PAGE/Western blot analysis were performed as described in Materials and Methods. An anti-GFP MAb was used to detect the GFP fusion proteins extracted after incubation of the cells with the following buffers: S1, NP-40; S2, wash; S3, DNase I; S4, wash; S5, NaCl 2 M; S6, Laemmli buffer; and L, whole-cell lysates. (B) Images such as those in A were analyzed as described in Materials and Methods to calculate the relative amounts of the indicated GFP fusion protein present in the specified cellular fraction. The data represent the means of three independent experiments, where the amount of soluble (S1 and S2), DNA-bound (S3 to S5), and matrix-associated (S6) proteins are expressed as a percentage of the total. The data represent the means of three independent experiments.
UL44-FL is not required for ppUL44 dimerization and binding to pUL54.
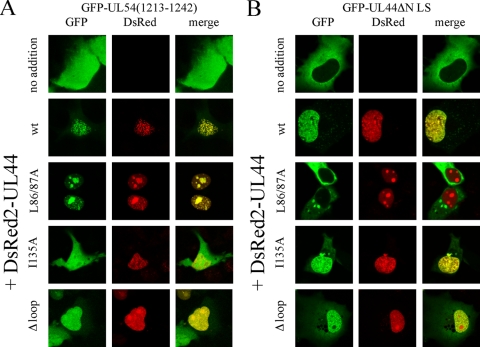
We next decided to analyze the ability of the ppUL44Δloop mutant derivative to bind to the catalytic subunit pUL54 (14) and to homodimerize (10), using our recently described live cell CLSM-based assays (5, 7, 41). We analyzed the subcellular localization of GFP-UL54(1213-1242) when expressed alone (Fig. 5A, upper panels) or in the presence of DsRed2-UL44wt and mutant derivatives thereof (Fig. 5A). As expected, GFP-UL54(1213-1242), a fusion protein containing the minimal binding site for ppUL44 but lacking the pUL54 NLS (7), localized both in the nucleus and in the cytoplasm of transfected cells with a diffuse pattern (Fig. 5A, upper panels). However, when GFP-UL54(1213-1242) was coexpressed with DsRed2-UL44wt and DsRed2-UL44L86A/L87A, the two proteins strongly colocalized within the cell nucleus (Fig. 5A), indicating functional interaction. A similar result was obtained upon coexpression of GFP-UL54(1213-1242) with DsRed2-UL44Δloop, indicating that the mutant derivative could still interact with pUL54. In contrast, no colocalization was observed when GFP-UL54(1213-1242) was coexpressed with DsRed2-UL44I135A, a fusion protein carrying a point mutation within the binding site for pUL54 that is sufficient to prevent the ppUL44-pUL54 interaction (41).
FIG. 5.
UL44-FL is not required for ppUL44 binding to pUL54 and homodimerization. COS-7 cells were transfected to express GFP-UL54(1213-1242) (A) or GFP-UL44ΔNLS (B), in the absence or the presence of the indicated DsRed2-UL44 fusions. Cells were imaged 24 to 36 h after transfection by CLSM. Merged images of the green (GFP) and red (DsRed2) channels are shown on the right, with yellow coloration indicative of colocalization.
The ability of DsRed2-UL44Δloop to dimerize was tested by investigating its ability to influence the subcellular localization of GFP-UL44ΔNLS, a mutant derivative unable to interact with IMPα/β and whose localization is therefore entirely restricted to the cytoplasm (4). As expected, GFP-UL44ΔNLS localized to the cytoplasm when expressed alone (Fig. 5B, upper panels), but coexpression with DsRed2-UL44wt and DsRed2-UL44I135A resulted in its marked relocalization to the cell nucleus as a consequence of the ability of ppUL44 to dimerize before being translocated into the nucleus (5). Coexpression of GFP-UL44ΔNLS with DsRed2-UL44Δloop, but not with DsRed2-UL44L86A/L87A, resulted in a similar relocalization (Fig. 5B). These results suggest that the basic residues within UL44-FL are essential neither for binding to pUL54 nor for ppUL44 homodimerization. They also suggest that the ppUL44Δloop mutant protein is not affected pleiotropically in multiple functions, and hence it is highly unlikely that its impaired DNA binding is the result of general misfolding. To verify this further, we analyzed the ability of FLAG-UL44Δloop to coimmunoprecipitate with GFP-UL54(1213-1242) compared to that of FLAG-UL44wt. Immunoprecipitation of both FLAG-UL44 and FLAG-UL44Δloop (see Fig. S2 in the supplemental material), using an anti-FLAG MAb, resulted in coimmunoprecipitation of GFP-UL54(1213-1242), further demonstrating that the UL44Δloop mutant derivative is not misfolded, with the effects of the mutation on intranuclear mobility/retention likely to be attributable to its impaired ability to bind to dsDNA.
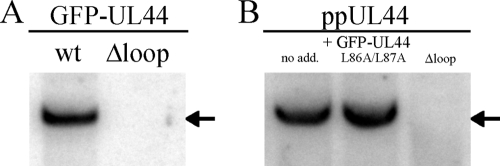
UL44-FL is required for HCMV oriLyt-dependent DNA replication in cells.
ppUL44 is believed to confer processivity to the DNA polymerase holoenzyme by tethering the catalytic subunit pUL54 on the dsDNA, meaning that mutations affecting its ability to bind to dsDNA could affect its activity as a processivity factor. We decided to investigate the ability of GFP-UL44Δloop to support HCMV oriLyt-dependent DNA replication using a cotransfection replication assay in HFF cells (41). Briefly, the pSP50 plasmid, bearing the HCMV oriLyt DNA replication origin (8), was cotransfected with a plasmid encoding wild-type or mutant GFP-UL44 (pEPI-GFP-UL44wt or pEPI-GFP-UL44Δloop), together with a set of plasmids encoding the remaining HCMV proteins essential for oriLyt-dependent DNA replication (32, 33). Replication of pSP50 was detected by treatment of transfected cells DNA with DpnI, which cleaves only unreplicated, dam-methylated input DNA (8). As expected, a DpnI-resistant replication product was detected in the presence of wild-type GFP-UL44 (Fig. 6A). On the other hand, oriLyt-mediated DNA replication was not detected in the presence of GFP-UL44Δloop (Fig. 6A), clearly indicating that the basic residues within UL44-FL are required for efficient HCMV oriLyt DNA replication in cells. Based on the fact that the GFP-UL44Δloop is capable of binding pUL54 (Fig. 5A and see Fig. S2 in the supplemental material), as well as homodimerizing (Fig. 5B), an interesting possibility was that the simultaneous expression of ppUL44wt and ppUL44Δloop could result in the formation of ppUL44wt-ppUL44Δloop heterodimers, which potentially may be impaired in dsDNA binding. We decided to test this possibility by testing the effect of overexpressing GFP-UL44Δloop to interfere with HCMV oriLyt-dependent DNA replication in HFF cells in the presence of wild type in our cotransfection replication assay. The dimerization-defective GFP-UL44L86A/L87A fusion protein was used as a control. The pSP50 plasmid and a set of plasmids encoding all of the HCMV proteins essential for oriLyt-dependent DNA replication, including ppUL44, were transfected in HFF cells in the absence or presence of GFP-UL44Δloop or GFP-UL44L86A/L87A. As expected, replicated pSP50 was detectable when cells were transfected to express to HCMV replicating proteins in the absence of the aforementioned ppUL44 mutants (Fig. 6B, left lane). Expression of GFP-UL44L86A/L87A, which is not capable of mediating oriLyt-dependent DNA replication but is defective for dimerization (41), did not affect pSP50 replication in the presence of ppUL44wt (Fig. 6B, middle lane). Importantly, the expression of GFP-UL44Δloop prevented the replication of pSP50 even in the presence of ppUL44wt (Fig. 6B, right lane), presumably due to the formation of DNA-binding-defective heterodimers.
FIG. 6.
UL44-FL is required for HCMV oriLyt-dependent DNA replication. (A) HFF cells were transiently transfected with the pSP50 plasmid, which contains the HCMV oriLyt DNA replication origin, a plasmid expressing GFP-UL44 (left lane) or GFP-UL44Δloop (right lane), as well as a set of plasmids expressing all other essential HCMV replication proteins. After DNA extraction and digestion with DpnI, undigested, replicated DNA (indicated by an arrow) was visualized by Southern blotting. (B) HFF cells were transiently transfected with the pSP50 plasmid, which contains the HCMV oriLyt DNA replication origin, a set of plasmids expressing all of the essential HCMV replication proteins, in the absence (left lane) or in the presence of the indicated GFP-UL44 mutant derivatives (middle and right lanes). After DNA extraction and digestion with DpnI, undigested, replicated DNA (indicated by an arrow) was visualized by Southern blotting.
DISCUSSION
ppUL44 is a multifunctional protein that is capable of interacting with dsDNA, through a process that is believed to involve electrostatic interactions between the dsDNA backbone and basic residues located both on ppUL44's basic face and within its highly flexible basic loop (UL44-FL) (4, 15, 29, 37, 45). Our results support this hypothesis, indicating a direct involvement of UL44-FL in DNA binding in living transfected cells and in a cell-based HCMV replication system (Fig. 6). Mutation of three of the five basic residues (PHTgVngNVKKAP174) does not affect ppUL44's ability to dimerize or to bind to the catalytic subunit pUL54 (Fig. 5 and see Fig. S2 in the supplemental material), but it is sufficient to impair nuclear accumulation of ppUL44, as well as to decrease its ability to form nuclear speckles (Fig. 1C and D). Since the level of nuclear accumulation of a protein is a product of its nuclear import and export rates, as well as of its ability to be retained within the nucleus upon nuclear entry (12, 40), and mutation of UL44-FL does not affect ppUL44 binding to the IMPα/β heterodimer, the reduced nuclear accumulation of GFP-UL44Δloop clearly implicates a role for UL44-FL in intranuclear binding.
Mutation of UL44-FL resulted in an increase in ppUL44 intranuclear mobility (Fig. 2) due to reduced binding to intranuclear components as supported by detergent extraction and/or fractionation experiments, where a significant amount of GFP-UL44 is retained in the nucleus associated with cellular chromatin after permeabilization of transfected Daoy cells (Fig. 3). In contrast, both GFP-UL44Δloop and GFP-UL44L86A/L87A are almost completely released upon cell permeabilization, as is GFP alone. This finding is also in keeping with the observation that HSV-1 UL42 nuclear localization is also resistant to detergent treatment in HSV-1-infected cells and that mutations affecting its binding to dsDNA in vitro also affect its resistance to detergent treatment (13, 21). Our quantitative analysis indicates that ca. 50% of GFP-UL44 is retained in the nucleus after detergent permeabilization of HEK 293 cells (Fig. 4). This is consistent with a fractional recovery of ca. 0.6 calculated for GFP-UL44 in COS-7 cells in FRAP experiments (see Fig. 2C), indicating that almost half of the protein is not rapidly moving within the nucleus. In contrast, up to 90% of GFP-UL44Δloop and GFP-UL44L86A/L87A are released by cell permeabilization (see Fig. 4), a finding consistent with their significantly higher fractional recoveries (ca. 0.8) determined in FRAP experiments and indicating that only a small fraction of the proteins is bound to intranuclear structures. A significant fraction of GFP-UL44wt, but not of GFP-UL44Δloop and GFP-UL44L86A/L87A, is released from permeabilized cells after DNase I treatment (see Fig. 4). Although we cannot exclude the possibility that UL44-FL's DNA-binding activity in live cells is indirect, i.e., through interaction with other nuclear proteins immobilized on DNA, our data strongly support the idea that UL44-FL is directly involved in the ppUL44-DNA interaction, as implicated by in vitro experiments (22). Our finding that ppUL44 can bind to cellular dsDNA when expressed even in the absence of viral DNA is not surprising, since ppUL44 appears to be able to bind to dsDNA without any sequence specificity (22).
Underlining the physiological significance of the results, GFP-UL44Δloop failed to support HCMV oriLyt-dependent DNA replication in cells (Fig. 6). Similarly, mutations preventing ppUL44 homodimerization in a cellular context also prevented ppUL44 from transcomplementing viral DNA replication in a transient-transfection assay (41), most likely due to the inability of these mutants to bind to dsDNA (see Fig. 3 and 4). Although the experiments here were not performed using the preferred experimental system of recombinant viruses, the results obtained are clearly consistent with a recent report showing that mutations impairing HSV-1 UL42 ability to bind to dsDNA also impair replication of a recombinant virus (20) and thus strong evidence for an important role for ppUL44-FL in HCMV replication. Importantly, the defect of ppUL44Δloop in mediating oriLyt-dependent DNA replication is not due to misfolding of the protein, as indicated by its ability to both bind to pUL54 and to heterodimerize with ppUL44wt (see Fig. 5AB and see Fig. S2 in the supplemental material). Hence, our results suggest that the inability of ppUL44Δloop to transcomplement oriLyt-dependent DNA replication depends directly on its reduced affinity for dsDNA (41; the present study). We cannot formally exclude the possibility that ppUL44 is impaired in binding to other viral or cellular proteins implicated in viral DNA replication despite still being able to bind to pUL54 (43), but its transdominant-negative phenotype in the replication assay is consistent with the defect of ppUL44Δloop in mediating HCMV DNA replication being at least partially due to its decreased ability to bind to dsDNA; the transdominant-negative effect can be explained by the formation of ppUL44Δloop/ppUL44wt heterodimers upon coexpression of the two proteins (see Fig. 5B and 6B). Formation of inactive protein complexes has been reported to be the molecular basis for the transdominant-negative phenotype of several viral multimeric proteins, including the human immunodeficiency virus type 1 protein Rev (17).
Whether UL44-FL is solely implicated in the binding of ppUL44 to dsDNA or also to cellular and viral proteins, interaction between herpesvirus PAPs and viral DNA represents a potential therapeutic target to hinder viral replication. In particular, UL44-FL appears as a potentially important determinant of ppUL44 biological activity in vivo and thus a target of great interest for the future.
Supplementary Material
Acknowledgments
This study was supported by the University of Bologna and the Italian Ministry of Education (60 and 40%), the AIDS Project of the Italian Ministry of Public Health, and the Australian National Health and Medical Research Council (fellowship 384109 and project grant 143710).
We thank Valerio Leoni (Bologna, Italy) for help with the Chimera software and Simone Avanzi (Bologna, Italy) for tissue culture.
Footnotes
Published ahead of print on 1 July 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Agulnick, A. D., J. R. Thompson, S. Iyengar, G. Pearson, D. Ablashi, and R. P. Ricciardi. 1993. Identification of a DNA-binding protein of human herpesvirus 6, a putative DNA polymerase stimulatory factor. J. Gen. Virol. 74(Pt. 6):1003-1009. [DOI] [PubMed] [Google Scholar]
- 2.Allan, J., T. Mitchell, N. Harborne, L. Bohm, and C. Crane-Robinson. 1986. Roles of H1 domains in determining higher order chromatin structure and H1 location. J. Mol. Biol. 187:591-601. [DOI] [PubMed] [Google Scholar]
- 3.Alvisi, G., S. Avanzi, D. Musiani, D. Camozzi, V. Leoni, J. D. Ly-Huynh, and A. Ripalti. 2008. Nuclear import of HSV-1 DNA polymerase processivity factor UL42 is mediated by a C-terminally located bipartite nuclear localization signal. Biochemistry 47:13764-13777. [DOI] [PubMed] [Google Scholar]
- 4.Alvisi, G., D. A. Jans, J. Guo, L. A. Pinna, and A. Ripalti. 2005. A protein kinase CK2 site flanking the nuclear targeting signal enhances nuclear transport of human cytomegalovirus ppUL44. Traffic 6:1002-1013. [DOI] [PubMed] [Google Scholar]
- 5.Alvisi, G., D. A. Jans, and A. Ripalti. 2006. Human cytomegalovirus (HCMV) DNA polymerase processivity factor ppUL44 dimerizes in the cytosol before translocation to the nucleus. Biochemistry 45:6866-6872. [DOI] [PubMed] [Google Scholar]
- 6.Alvisi, G., S. M. Rawlinson, R. Ghildyal, A. Ripalti, and D. A. Jans. 2008. Regulated nucleocytoplasmic trafficking of viral gene products: a therapeutic target? Biochim. Biophys. Acta 1784:213-227. [DOI] [PubMed] [Google Scholar]
- 7.Alvisi, G., A. Ripalti, A. Ngankeu, M. Giannandrea, S. G. Caraffi, M. M. Dias, and D. A. Jans. 2006. Human cytomegalovirus DNA polymerase catalytic subunit pUL54 possesses independently acting nuclear localization and ppUL44 binding motifs. Traffic 7:1322-1332. [DOI] [PubMed] [Google Scholar]
- 8.Anders, D. G., M. A. Kacica, G. Pari, and S. M. Punturieri. 1992. Boundaries and structure of human cytomegalovirus oriLyt, a complex origin for lytic-phase DNA replication. J. Virol. 66:3373-3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Appleton, B. A., J. Brooks, A. Loregian, D. J. Filman, D. M. Coen, and J. M. Hogle. 2006. Crystal structure of the cytomegalovirus DNA polymerase subunit UL44 in complex with the C terminus from the catalytic subunit. Differences in structure and function relative to unliganded UL44. J. Biol. Chem. 281:5224-5232. [DOI] [PubMed] [Google Scholar]
- 10.Appleton, B. A., A. Loregian, D. J. Filman, D. M. Coen, and J. M. Hogle. 2004. The cytomegalovirus DNA polymerase subunit UL44 forms a C clamp-shaped dimer. Mol. Cell 15:233-244. [DOI] [PubMed] [Google Scholar]
- 11.Axelrod, D., P. Ravdin, D. E. Koppel, J. Schlessinger, W. W. Webb, E. L. Elson, and T. R. Podleski. 1976. Lateral motion of fluorescently labeled acetylcholine receptors in membranes of developing muscle fibers. Proc. Natl. Acad. Sci. USA 73:4594-4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bauerle, M., D. Doenecke, and W. Albig. 2002. The requirement of H1 histones for a heterodimeric nuclear import receptor. J. Biol. Chem. 277:32480-32489. [DOI] [PubMed] [Google Scholar]
- 13.Chen, Y., C. M. Livingston, S. D. Carrington-Lawrence, P. Bai, and S. K. Weller. 2007. A mutation in the human herpes simplex virus type 1 UL52 zinc finger motif results in defective primase activity but can recruit viral polymerase and support viral replication efficiently. J. Virol. 81:8742-8751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ertl, P. F., and K. L. Powell. 1992. Physical and functional interaction of human cytomegalovirus DNA polymerase and its accessory protein (ICP36) expressed in insect cells. J. Virol. 66:4126-4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao, Y., K. Colletti, and G. S. Pari. 2008. Identification of human cytomegalovirus UL84 virus- and cell-encoded binding partners by using proteomics analysis. J. Virol. 82:96-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerlitz, G., I. Livnat, C. Ziv, O. Yarden, M. Bustin, and O. Reiner. 2007. Migration cues induce chromatin alterations. Traffic 8:1521-1529. [DOI] [PubMed] [Google Scholar]
- 17.Hope, T. J., N. P. Klein, M. E. Elder, and T. G. Parslow. 1992. trans-dominant inhibition of human immunodeficiency virus type 1 Rev occurs through formation of inactive protein complexes. J. Virol. 66:1849-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hubner, S., J. E. Eam, K. M. Wagstaff, and D. A. Jans. 2006. Quantitative analysis of localization and nuclear aggregate formation induced by GFP-lamin A mutant proteins in living HeLa cells. J. Cell Biochem. 98:810-826. [DOI] [PubMed] [Google Scholar]
- 19.Jiang, C., Y. T. Hwang, J. C. Randell, D. M. Coen, and C. B. Hwang. 2007. Mutations that decrease DNA binding of the processivity factor of the herpes simplex virus DNA polymerase reduce viral yield, alter the kinetics of viral DNA replication, and decrease the fidelity of DNA replication. J. Virol. 81:3495-3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang, C., Y. T. Hwang, G. Wang, J. C. Randell, D. M. Coen, and C. B. Hwang. 2007. Herpes simplex virus mutants with multiple substitutions affecting DNA binding of UL42 are impaired for viral replication and DNA synthesis. J. Virol. 81:12077-12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang, C., G. Komazin-Meredith, W. Tian, D. M. Coen, and C. B. Hwang. 2009. Mutations that increase DNA binding by the processivity factor of herpes simplex virus affect virus production and DNA replication fidelity. J. Virol. 83:7573-7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komazin-Meredith, G., R. J. Petrella, W. L. Santos, D. J. Filman, J. M. Hogle, G. L. Verdine, M. Karplus, and D. M. Coen. 2008. The human cytomegalovirus UL44 C clamp wraps around DNA. Structure 16:1214-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Komazin-Meredith, G., W. L. Santos, D. J. Filman, J. M. Hogle, G. L. Verdine, and D. M. Coen. 2008. The positively charged surface of herpes simplex virus UL42 mediates DNA binding. J. Biol. Chem. 283:6154-6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lang, I., M. Scholz, and R. Peters. 1986. Molecular mobility and nucleocytoplasmic flux in hepatoma cells. J. Cell Biol. 102:1183-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loh, L. C., V. D. Keeler, and J. D. Shanley. 1999. Sequence requirements for the nuclear localization of the murine cytomegalovirus M44 gene product pp50. Virology 259:43-59. [DOI] [PubMed] [Google Scholar]
- 26.Loregian, A., B. A. Appleton, J. M. Hogle, and D. M. Coen. 2004. Residues of human cytomegalovirus DNA polymerase catalytic subunit UL54 that are necessary and sufficient for interaction with the accessory protein UL44. J. Virol. 78:158-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loregian, A., B. A. Appleton, J. M. Hogle, and D. M. Coen. 2004. Specific residues in the connector loop of the human cytomegalovirus DNA polymerase accessory protein UL44 are crucial for interaction with the UL54 catalytic subunit. J. Virol. 78:9084-9092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malm, G., and M. L. Engman. 2007. Congenital cytomegalovirus infections. Semin. Fetal Neonatal Med. 12:154-159. [DOI] [PubMed] [Google Scholar]
- 29.Marschall, M., M. Freitag, P. Suchy, D. Romaker, R. Kupfer, M. Hanke, and T. Stamminger. 2003. The protein kinase pUL97 of human cytomegalovirus interacts with and phosphorylates the DNA polymerase processivity factor pUL44. Virology 311:60-71. [DOI] [PubMed] [Google Scholar]
- 30.Mocarski, E. S., L. Pereira, and N. Michael. 1985. Precise localization of genes on large animal virus genomes: use of lambda gt11 and monoclonal antibodies to map the gene for a cytomegalovirus protein family. Proc. Natl. Acad. Sci. USA 82:1266-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orrego, M., I. Ponte, A. Roque, N. Buschati, X. Mora, and P. Suau. 2007. Differential affinity of mammalian histone H1 somatic subtypes for DNA and chromatin. BMC Biol. 5:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pari, G. S., and D. G. Anders. 1993. Eleven loci encoding trans-acting factors are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA replication. J. Virol. 67:6979-6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pari, G. S., M. A. Kacica, and D. G. Anders. 1993. Open reading frames UL44, IRS1/TRS1, and UL36-38 are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA synthesis. J. Virol. 67:2575-2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Partikian, A., B. Olveczky, R. Swaminathan, Y. Li, and A. S. Verkman. 1998. Rapid diffusion of green fluorescent protein in the mitochondrial matrix. J. Cell Biol. 140:821-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pettersen, E. F., T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, and T. E. Ferrin. 2004. UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605-1612. [DOI] [PubMed] [Google Scholar]
- 36.Poon, I. K., C. Oro, M. M. Dias, J. Zhang, and D. A. Jans. 2005. Apoptin nuclear accumulation is modulated by a CRM1-recognized nuclear export signal that is active in normal but not in tumor cells. Cancer Res. 65:7059-7064. [DOI] [PubMed] [Google Scholar]
- 37.Prichard, M. N., H. Lawlor, G. M. Duke, C. Mo, Z. Wang, M. Dixon, G. Kemble, and E. R. Kern. 2005. Human cytomegalovirus uracil DNA glycosylase associates with ppUL44 and accelerates the accumulation of viral DNA. Virol. J. 2:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Randell, J. C., G. Komazin, C. Jiang, C. B. Hwang, and D. M. Coen. 2005. Effects of substitutions of arginine residues on the basic surface of herpes simplex virus UL42 support a role for DNA binding in processive DNA synthesis. J. Virol. 79:12025-12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ripalti, A., M. C. Boccuni, F. Campanini, and M. P. Landini. 1995. Cytomegalovirus-mediated induction of antisense mRNA expression to UL44 inhibits virus replication in an astrocytoma cell line: identification of an essential gene. J. Virol. 69:2047-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roth, D. M., I. Harper, C. W. Pouton, and D. A. Jans. Modulation of nucleocytoplasmic trafficking by retention in cytoplasm or nucleus. J. Cell Biochem., in press. [DOI] [PubMed]
- 41.Sinigalia, E., G. Alvisi, B. Mercorelli, D. M. Coen, G. S. Pari, D. A. Jans, A. Ripalti, G. Palu, and A. Loregian. 2008. Role of homodimerization of human cytomegalovirus DNA polymerase accessory protein UL44 in origin-dependent DNA replication in cells. J. Virol. 82:12574-12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steininger, C. 2007. Clinical relevance of cytomegalovirus infection in patients with disorders of the immune system. Clin. Microbiol. Infect. 13:953-963. [DOI] [PubMed] [Google Scholar]
- 43.Strang, B. L., E. Sinigalia, L. A. Silva, D. M. Coen, and A. Loregian. 2009. Analysis of the association of the human cytomegalovirus DNA polymerase subunit UL44 with the viral DNA replication factor UL84. J. Virol. 83:7581-7589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takeda, K., M. Haque, E. Nagoshi, M. Takemoto, T. Shimamoto, Y. Yoneda, and K. Yamanishi. 2000. Characterization of human herpesvirus 7 U27 gene product and identification of its nuclear localization signal. Virology 272:394-401. [DOI] [PubMed] [Google Scholar]
- 45.Weiland, K. L., N. L. Oien, F. Homa, and M. W. Wathen. 1994. Functional analysis of human cytomegalovirus polymerase accessory protein. Virus Res. 34:191-206. [DOI] [PubMed] [Google Scholar]
- 46.Zuccola, H. J., D. J. Filman, D. M. Coen, and J. M. Hogle. 2000. The crystal structure of an unusual processivity factor, herpes simplex virus UL42, bound to the C terminus of its cognate polymerase. Mol. Cell 5:267-278. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.