Abstract
The N terminus of the replicase nonstructural protein 2 (nsp2) of porcine reproductive and respiratory syndrome virus (PRRSV) contains a putative cysteine protease domain (PL2). Previously, we demonstrated that deletion of either the PL2 core domain (amino acids [aa] 47 to 180) or the immediate downstream region (aa 181 to 323) is lethal to the virus. In this study, the PL2 domain was found to encode an active enzyme that mediates efficient processing of nsp2-3 in CHO cells. The PL2 protease possessed both trans- and cis-cleavage activities, which were distinguished by individual point mutations in the protease domain. The minimal size required to maintain these two enzymatic activities included nsp2 aa 47 to 240 (Tyr47 to Cys240) and aa 47 to 323 (Tyr47 to Leu323), respectively. Introduction of targeted amino acid mutations in the protease domain confirmed the importance of the putative Cys55- His124 catalytic motif for nsp2/3 proteolysis in vitro, as were three additional conserved cysteine residues (Cys111, Cys142, and Cys147). The conserved aspartic acids (e.g., Asp89) were essential for the PL2 protease trans-cleavage activity. Reverse genetics revealed that the PL2 trans-cleavage activity played an important role in the PRRSV replication cycle in that mutations that impaired the PL2 protease trans function, but not the cis activity, were detrimental to viral viability. Lastly, the potential nsp2/3 cleavage site was probed. Mutations with the largest impact on in vitro cleavage were at or near the G1196|G1197 dipeptide.
Porcine reproductive and respiratory syndrome virus (PRRSV) is the causative agent of PRRS, a prevalent and troubling disease for the swine industry (4, 7, 8, 49, 50). PRRSV is a positive-strand RNA virus with a genome size of about 15.4 kb (30, 33) and, together with equine arteritis virus (EAV), murine lactate dehydrogenase-elevating virus, and simian hemorrhagic fever virus, forms the family Arteriviridae of the order Nidovirales (5). PRRSV has a genome organization similar to that of other members of its family and contains at least nine open reading frames (ORFs) (41). ORF1a and ORF1b encode the viral replicase polyproteins pp1a and pp1ab, while ORF2 to ORF7 code for viral structural proteins expressed from subgenomic RNAs that are most likely generated through a discontinuous transcription mechanism (35, 41). Replication of PRRSV first involves translation of polyproteins pp1a and pp1ab. The proteolytic maturation of these replicase precursors is thought to be mediated predominantly by virally encoded proteases specified by nonstructural protein 1 (nsp1), nsp2, and nsp4, which produce at least 14 mature nonstructural proteins most probably involved in viral RNA replication (15, 48, 53).
PRRSV nsp2 is a multidomain protein located immediately downstream of the first two nonstructural proteins (nsp1α and nsp1β) within ORF1a. It possesses a putative cysteine protease domain (PL2), a 500- to 700-amino-acid middle hypervariable region with unknown function, and a transmembrane domain, followed by a C-terminal tail of uncertain size (Fig. 1) (17, 18). The putative cysteine protease PL2 domain of PRRSV has a predicted core size of about 100 amino acids (aa) (nsp2 aa 47 to 147) and is believed to cleave the downstream nsp2 3 junction site, similar to its EAV PL2 counterpart (43, 45). The nsp2 PL2 protease core domain is well conserved not only among PRRSV strains but also among the members of the Arteriviridae family, especially for the putative catalytic dyad Cys55-His124 (numbered according to the nsp2 sequence of PRRSV strain VR-2332) (Fig. 1). The arterivirus PL2 protease shares features of both papain-like cysteine proteases and chymotrypsin-like cysteine proteases in that it possesses the signature Cys-His catalytic motif of viral papain-like proteases, as well as the marker of viral chymotrypsin-like cysteine proteases, in which the putative catalytic Cys residue is always followed by a Gly instead of an amino acid with a large side chain, as seen in papain-like proteases (10, 11, 42, 44, 45, 53). Bioinformatic analyses have also revealed an interesting relationship between arterivirus nsp2 PL2 proteases and mammalian ovarian tumor domain (OTU)-containing proteins (28). The OTU family was only recently recognized and represents a novel class of putative cysteine proteases that are homologous to the Ovarian tumor gene product of Drosophila, a protein important for oogenesis (28). More than 100 members have been predicted with origins from eukaryotes, bacteria, and viruses (Pfam:PF002338). However, only a limited number of them have been characterized. The sequence conservation among OTU members centered on the motifs containing the putative catalytic Cys and His residues, and hence, the arterivirus nsp2 proteases are thought to be new members of the OTU superfamily (28).
FIG. 1.
Schematic diagram of nsp2-3 and alignment of arterivirus nsp2 cysteine protease PL2 domains. The PRRSV genome with all identified ORFs is represented at the top. (A)n, poly(A) tail. ORF1A and ORF1B are posttranslationally cleaved (↓) by virally encoded papain-like proteases (PCP1α and PCP1β), a cysteine protease (PL2), and a poliovirus 3C-like serine protease (3CL). The following polymerase signature regions are indicated: RNA-dependent RNA polymerase (RdRp), cysteine and histidine rich (C/H), helicase (HEL), and nidovirus uridylate-specific endoribonuclease (NendoU). An enlargement of the PRRSV nsp2-3 protein is shown below the full genome. The nsp2 multidomain protein contains five different regions: hypervariable region I (HV-I; aa 1 to 46), the putative PL2 cysteine protease core domain (aa 47 to 147), the middle hypervariable region (HV-II; aa 147 to 847), predicted transmembrane domains (vertical bars; aa 876 to 898, 911 to 930, 963 to 979, and 989 to 1009), and a C-terminal tail. The identified PL1β cleavage site is represented by a gray triangle (10), and the site at which nsp3 is purportedly cleaved is illustrated by an open triangle. The proposed cleavage sites of nsp2 are shown by black triangles, and the positions are annotated on the basis of nsp2 amino acid position. nsp3 is predicted to be a highly hydrophobic protein with transmembrane domains annotated by vertical bars (aa 1203 to 1223, 1268 to 1290, 1302 to 1327, and 1339 to 1365) (21). At the bottom is a comparison of selected arterivirus PL2 cysteine protease domains generated with CLUSTALW (6). Conserved residues investigated in this study are highlighted by shaded boxes. Potential catalytic residues are shown in red, and completely conserved tryptophan residues, potentially contributing to the catalytic mechanism, are shown in blue. The nsp2s (accession numbers) are VR-2332 (AY150564), 16244B (AF046869), CC-1 (EF153486), CH-1a (AY032626), HB-2 (AY262352), JA142 (AY424271), MN184A (DQ176019), P129 (AF494042), MN-04-06_EU (AAW81311), SDPRRS 04-41 (ABO93301), SDPRRS 04-43 (ABO93305), EuroPRRSV (AY366525), LV (M96262), RPOA_SHFV (Q68722), RPOA_LDVC (Q06562), RPOA_LDVP (Q83017), Bucyrus (CAC42775), and CW96 (Q6V7J0).
Using a PRRSV reverse genetic system, we recently showed that deletion of either the region encoding the predicted PL2 protease core domain (aa 47 to 180) or the immediate downstream region (aa 181 to 323) is lethal to the virus (17), supporting the hypothesis that PRRSV nsp2 likely harbors an active enzyme that is required for nsp2/3 proteolysis. Another indirect link to the protease activity of the PL2 domain comes from a recent study that showed that the PRRSV nsp2 coding region had the ability to decrease the level of ubiquitin and ISG15 conjugates in transfected 293T cells (14).
In this study, the nsp2 PL2 domain of PRRSV strain VR-2332 was further characterized. The putative PL2 protease was shown to possess trans- and likely cis-cleavage activities with nsp2/3 proteolysis occurring near 1196G|G|G in CHO cells, and the highly divergent C-terminal extension region (Ser181 to Cys240) of the PL2 protease core domain (Tyr47 to Cys147) was found to be critical for PL2 catalytic activity. In addition, site-directed mutagenesis identified specific residues important for PL2 protease activity, including amino acids such as the putative Cys55-His124 catalytic motif. We also showed that the trans-cleavage activity of the PL2 protease could be distinguished from the cis-cleavage activity by point mutations. Finally, specific amino acids within the PL2 domain appeared to important for the replication cycle of PRRSV in infected MARC-145 cells.
MATERIALS AND METHODS
Antibodies.
Mouse monoclonal antibodies to the c-myc epitope (9E10; Developmental Studies Hybridoma Bank at the University of Iowa), rabbit polyclonal anti-c-myc antibodies (Abcam Inc., Cambridge, MA), mouse anti-hemagglutinin epitope (HA) antibodies (Covance Research Products, Denver, CO), mouse anti-FLAG epitope antibodies (M2; Sigma, St. Louis, MO), and horseradish peroxidase-conjugated anti-mouse immunoglobulin G or anti-rabbit immunoglobulin G secondary antibodies (SouthernBiotech, Inc., Birmingham, AL) were purchased. Rabbit polyclonal antibody V (Covance) was raised against the nsp2 aa 1078 to 1094 peptide (SEKPIAFAQLDEKKITA) of PRRSV strain VR-2332.
Plasmids.
Plasmid constructs were generated by standard recombinant DNA procedures. To create a HA-FLAG epitope-tagged vector, a linker containing HA-FLAG epitopes (YPYDVPDYAYPYDVPDYACTDYKDDDKDYKDDDDK) was inserted into the position between the XbaI and ApaI sites in plasmid vector pcDNA3.0 (Invitrogen) to generate pcDNA3/HA-FLAG (GenBank accession no. FJ524378). The construction of plasmid pV7-nsp2Δ324-433-GFP was reported previously (17). The green fluorescent protein (GFP)-encoding gene was replaced with three copies of the c-myc epitope (ASEQKLISEEDLEQKLISEEDLEQKLISEED) to generate plasmid pV7-nsp2Δ324-433-myc (GenBank accession no. FJ524377). The constructs expressing specific regions of nsp2 were generated by PCR amplification from plasmid pV7-nsp2Δ324-433-myc. Briefly, the region coding for nsp2-3 (PRRSV VR-2332 nucleotides [nt] 1368 to 5607) was amplified with primer pair Nsp2PL2-1U37/VR-5583L35, digested with BamHI and XbaI, and then cloned into pcDNA3/HA-FLAG to generate pNsp2-3. nsp2-3 is in frame with the HA-FLAG tag. Similarly, the region of the PRRSV VR-2332 genome including nt 2057 to 5618 was cloned to generate pNsp2-3Δ1-240. To generate pPL2(12-323) with a 10-aa c-myc tag at the C terminus, the nt 1368 to 2306 region of PRRSV VR-2332 ORF1a (nsp2 aa 12 to 323 region) was amplified with primer pair Nsp2CP2-1U37/Nsp2PL2-941L60, digested with HindIII and XhoI, and then cloned into pcDNA3. The PL2 truncation constructs, including pPL2(12-240), pPL2(12-180), pPL2(12-160), pPL2(47-323), pPL2(47-240), pPL2(47-180), and pPL2(47-160), were generated in a similar way. Plasmids pNsp2-3Δ181-323, pNsp2-3Δ241-323, and pNsp2-3Δ324-813 were generated from infectious cDNA clone plasmids pV7-nsp2Δ181-323, pV7-nsp2Δ241-323, and pV7-nsp2Δ324-813, respectively, by insertion of the corresponding nsp2-3 deletion fragment into the vector pcDNA3/HA-FLAG through PCR amplification. For these constructs, one c-myc epitope was added to the N terminus of nsp2. All of the constructs included a Kozak core sequence (GCCACCATGG) for optimal translation. The primers used in this study are listed in Table 1.
TABLE 1.
Primers used in construction and mutagenesis studiesa
| Primer use and name | VR-2332 genome position | Sequence |
|---|---|---|
| Construction of nsp2 and PL2 | ||
| HA-Flag Linker U100/ | HA-FLAG | 5′-CTGGCGGCCGCTCGAGCATGCATCTAGATACCCATACGAUTGTTCCAGATTACGCTTATCCTTACGACGTCCCAGACTATGCCTGTACAGATTACAAGGAT |
| /HA-Flag Linker L100 | HA-FLAG | 5′-TAGCATTTAGGTGACACTATAGAATAGGGCCCTTATCACTTGTCATCATCGTCCTTATAGTCCTTATCGTCGTCATCCTTGTAATCTGTACAGGCATAGT |
| Nsp2PL2-1U37/ | 1368-1386 | 5′-AGCTGGATCCGCCACCATGGTGGCGACTGCTACAGTC |
| Nsp2CP2-1U37/ | 1368-1386 | 5′-AGCTAAGCTTGCCACCATGGTGGCGACTGCTACAGTC |
| /Nsp2PL2-941L60 | 2290-2306 | 5′-ATGCTCGAGTTATCACAGATCCTCTTCTGAGATGAGTTTTTGTTCCAAGGACTTTTGAGT |
| /VR-5583L35 | 5582-5607 | 5′-CTATCTAGAAGACCCAAGCTGGGACGGGGTAAACA |
| VR-2058U43/ | 2055-2074 | 5′-CTCGGATCCGCCACCATGGGATGTTCCCAGAACAAAACCAACC |
| Nsp2-47U77/ | 1474-1499 | 5′-CCCGGATCCGCCACCATGGGAGAACAAAAACTCATCTCAGAAGAGGATCTGTACTCCCCGCCTGCCGAAGGGAATTG |
| /VR-1784L60 | 1786-1800 | 5′-ATGCTCGAGTTATCACAGATCCTCTTCTGAGATGAGTTTTTGTTCGGAACCAAGACCGCC |
| /VR-1859L60 | 1861-1875 | 5′-ATGCTCGAGTTATCACAGATCCTCTTCTGAGATGAGTTTTTGTTCGCTAGGCAGGTGCAT |
| /VR-2039L60 | 2041-2055 | 5′-ATGCTCGAGTTATCACAGATCCTCTTCTGAGATGAGTTTTTGTTCACAGCAGCAGTCCTC |
| VR-1475U47/ | 1474-1498 | 5′-AGCTAAGCTTGCCACCATGGTGTACTCTCCGCCTGCCGAAGGGAATT |
| pcDNA-726U25/ | 5′-CAACGGGACTTTCCAAAATGTCGTA | |
| /pcDNA-1066L22 | 5′-ACGGGGGAGGGGCAAACAACAG | |
| POK12-19U25/ | 5′-CTCCCCGCGCGTTGGCCGATTCATT | |
| /POK12-354L23 | 5′-AGCAGCCGGATCCGAGCTCTCCC | |
| Detection of ORF7 | ||
| pet71/ | 14887-14906 | 5′-GCGCATATGCCAAATAACAAGCGC |
| /P72 | 15343-15363 | 5′-CGCCCTAATTGAATAGGTGAC |
Restriction endonuclease sites are shown in boldface italics. Engineered start codons are shown in boldface. Forward primer names end with a slash, and reverse primer names begin with a slash. Mutagenesis primer sequences are available on request.
Site-directed mutagenesis.
Plasmids pPL2 and pNsp2-3 were subjected to site-directed mutagenesis by using a QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions. For mutagenesis of the infectious clone, the mutations were first introduced into shuttle plasmid pOK-I or pOK-II (17). After verification by sequencing, digested fragment I or II was transferred to the PRRSV strain VR-2332 infectious clone backbone (pVR-V7) as described previously (17).
Cell culture and transfection.
MARC-145 cells (ATCC CRL-11171) and CHO cells (Invitrogen) were maintained in minimum essential medium with Earle's balanced salts (catalog no. 56416C; Sigma-Aldrich Corp., St. Louis, MO) supplemented with 10% fetal bovine serum at 37°C with 5% CO2. Transient plasmid transfections of CHO cells were carried out as described here briefly. Solutions of Lipofectamine 2000 (10 μl; Invitrogen, Carlsbad, CA) and 8 to 10 μg plasmid diluted in 1 ml Opti-MEM (Invitrogen, Carlsbad, CA) were applied to CHO cell monolayers in 60-mm petri dishes. The transfection medium was removed after 4 h of incubation, and fresh culture medium (1× minimum essential medium with Earle's balanced salts and 10% fetal bovine serum) was added. At 48 h posttransfection, the cells were lysed with radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 7.4], 1% NP-40, 0.5% Na-deoxycholate, 150 mM NaCl, 1 mM EDTA, and a cocktail of protease inhibitors [catalog no. P8340; Sigma-Aldrich Corp., St. Louis, MO]). For transfection of the monocistronic construct, about 10 μg of each nsp2-3 mutant was transfected into CHO cells in 60-mm petri dishes. For cotransfection studies, 6 μg of each PL2 mutant was cotransfected with 5 μg of substrate plasmid pNsp2-3Δ1-240. For RNA transfection of MARC-145 cells, the full-length infectious clone plasmids were in vitro linearized, purified, transcribed, and transfected as described previously (17, 34).
Immunoprecipitation and Western blotting.
Transfected CHO cells were rinsed twice with cold phosphate-buffered saline (0.14 M NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.5 mM KH2PO4) and lysed with RIPA buffer on a shaker at 4°C for 30 min. The cell debris was removed by centrifugation at 13,000 rpm for 25 min. The supernatants were precleared with protein G PLUS (Santa Cruz Biotechnology, Santa Cruz, CA) or protein A agarose (Roche, Nutley, NJ) and then incubated with the proper antibodies, as well as protein G PLUS or protein A agarose, at 4°C overnight. Immunocomplexes were washed twice with cold RIPA buffer, once with 0.1% sodium dodecyl sulfate RIPA buffer, and once with phosphate-buffered saline. After boiling for 5 min in loading buffer (Invitrogen) with 5% β-mercaptoethanol, the proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then electrically transferred to a nitrocellulose membrane. For Western blot analysis, the membrane was blocked with 5% milk powder in PBST (20 mM NaPO4, 140 mM NaCl, 0.1% Tween 20) for 1 h and then incubated with the appropriate primary antibodies diluted in PBST-5% milk overnight at 4°C. After washing three times for 30 min with PBST, the blot was incubated with the appropriate secondary antibody in PBST for 1 h. The membrane was then washed and developed with the ECL Western blotting analysis system (Pierce Chemical, Rockford, IL).
Detection and titration of mutant viruses.
After transfection of full-length RNA transcripts into MARC-145 cells, the appearance of a virus-induced cytopathic effect (CPE), characterized by cell rounding and detachment, was monitored daily. For transfections that did not induce a CPE by 72 h, the cell supernatants were collected and blindly passaged onto fresh monolayers for a total of three passages. Total intracellular RNA was extracted with the RNeasy Mini Kit (Qiagen, Valencia, CA), and PRRSV was detected by reverse transcription (RT)-PCR with primers targeting ORF7, which encodes the viral nucleocapsid protein. For viable viruses, passage 1 mutant viruses were titrated by the endpoint dilution assay and expressed as 50% tissue culture infective doses (TCID50) (37). The parental infectious-clone-derived virus VR-V7 (17) served as a positive control.
RESULTS
nsp2 was efficiently processed from the nsp2/3 precursor.
In earlier reports, strain MN184 was shown to possess three discontinuous deletions compared to strain VR-2332 and the first of these deletions (aa 324 to 434) had no effect on growth rate when engineered into pVR-V7, the infectious clone of strain VR-2332 (17, 18). These findings led to the premise that the pV7-nsp2Δ324-434 deletion mutant was suitable for PL2 examination. In addition, the viability of PRRSV strain VR-2332 with an insertion of the GFP-encoding gene in place of nsp2 nonessential region aa 324 to 434 was demonstrated (17). To facilitate analysis of nsp2 processing in vitro and to bypass the need for the generation of nsp2-specific antibodies, the GFP-encoding gene was replaced with three consecutive c-myc epitopes to generate pV7-nsp2Δ324-434-myc, which generates viable virus with growth kinetics similar to those of the parent virus VR-V7 and will be described elsewhere (unpublished data). The c-myc tagged nsp2-3 fragment, corresponding to ORF1a aa 394 to 1806 (C394 to S1806), was then cloned into the vector pcDNA3/HA-FLAG to generate plasmid pNsp2-3 with the carboxyl terminus of nsp3 fused to the HA-FLAG tag (148.6 kDa; Fig. 2A).
FIG. 2.
The cysteine protease PL2 domain mediated efficient processing of nsp2 in CHO cells. (A) Schematic diagram of constructs used in examining PL2 cis processing. A c-myc epitope was inserted into the position where the nsp2 aa 324 to 434 region had been deleted, and a HA-FLAG tag was attached to the C terminus of nsp3. The plasmids were transfected into CHO cells. At 48 h posttransfection, the cells were lysed and immunoprecipitated with mouse monoclonal antibodies against c-myc (9E10) or HA. The immunoprecipitated (IP) proteins were separated by SDS-PAGE and then transferred to nitrocellulose sheets and reacted with selected antibodies in a Western blot (WB) assay. (B) Detection of nsp2-3 and the nsp2-3H124C mutant protein by rabbit anti-c-myc antibodies. (C) Mutation of the putative catalytic site His124 to cysteine abolished the processing of nsp2. The nsp2-3H124C mutant precursor, but not the wild-type nsp2-3 protein, was detected by anti-HA antibodies. (D) Detection of nsp3 by mouse anti-FLAG antibodies after immunoprecipitation with anti-HA antibodies. (E) Deletion of the nsp2 aa 1 to 240 region containing the PL2 domain blocked the processing of nsp2. nsp2 could be detected by both anti-c-myc and anti-HA antibodies. Molecular mass markers (kDa) are shown at the left of each panel. Use of MAb 9E10 for immunoprecipitation and anti-HA antibodies for Western blotting frequently detected nonspecific proteins (25 and 50 kDa, respectively), which are likely immunoglobulin light and heavy chains, respectively, that reacted with the secondary antibody.
To determine if nsp2 is processed from the nsp2-3 precursor (148.6 kDa), pNsp2-3 was transfected into CHO cells. At 48 h posttransfection, the cells were lysed and subjected to immunoprecipitation, followed by Western blot analysis. As shown in Fig. 2B, a major band of about 120 kDa of nsp2 was detected by anti-c-myc antibodies (Fig. 2B, lane 2) but not by anti-HA antibodies (Fig. 2C, lane 2), demonstrating that the nsp2-3 precursor was autoprocessed. To detect processed nsp3, the cell lysates were immunoprecipitated with the anti-HA antibodies and then subjected to Western blotting with anti-FLAG antibodies. A specific band of about 29 kDa that corresponded to the predicted size of nsp3-HA-FLAG (29 kDa) was detected (Fig. 2D, lane 1). These results demonstrated that nsp2 was efficiently processed in transfected CHO cells.
The PL2 domain mediated the processing of nsp2.
The cysteine protease PL2 domain is predicted (18, 45) to have a core size of about 100 aa (Y47 to C147) and is highly conserved among all arteriviruses (Fig. 1). Cys55-His124 is the putative catalytic dyad (45, 53). Two approaches were devised to determine if the PL2 domain mediates nsp2/3 processing. The first involved mutation of putative catalytic residue His124 to Cys (pNsp2-3H124C; 148.6 kDa). The H124C mutation produced a protein in transfected CHO cells (Fig. 2B, lane 1) that migrated more slowly than processed unmodified nsp2 (120 kDa; Fig. 2B, lane 2) and could be detected by both anti-c-myc (Fig. 2B, lane 1) and anti-HA antibodies (Fig. 2C, lane 1), providing evidence that the H124C mutation abolished nsp2 processing. The second approach involved deletion of nsp2 aa 1 to 240 containing the PL2 domain (pNsp2-3Δ1-240, 124 kDa; Fig. 2A). We hypothesized that if the PL2 domain mediated the processing of nsp2-3, deletion of the protease domain would leave nsp2-3Δ1-240 uncleaved. Anti-c-myc and anti-HA antibodies detected the intact nsp2-3Δ1-240 precursor (124 kDa) after transfection into CHO cells (Fig. 2E, lanes 1 and 3), demonstrating the absence of efficient nsp2-3 cleavage. Thus, results from two separate approaches were consistent with a mechanism by which the PL2 domain mediated nsp2/3 proteolysis.
We also detected an additional major nsp2-specific band with a molecular mass of around 70 to 80 kDa (Fig. 2B, lanes 1 and 2). This was likely a product cleaved by a cellular protease since it could be detected under conditions where the viral PL2 protease activity was blocked by an H124C mutation (Fig. 2B, lane 1) or after deletion of the PL2 domain (Fig. 2E, lane 1). The latter band possessed a smaller size (51 kDa) due to truncation of the N-terminal PL2 domain (24 kDa). Therefore, it was concluded that the major band corresponding in size to nsp2 was processed by the PRRSV PL2 protease, while the minor product (70 to 80 kDa) was probably cleaved by an unknown cellular protease or an undescribed second protease domain within nsp2.
The PL2 domain possessed trans-cleavage activity.
To test if the PL2 protease was active in trans, the fragment corresponding to nsp2 aa 12 to 323 (Cys12 to Leu323) was cloned into pcDNA3 to generate construct pPL2 (Fig. 3A). The rationale for including the downstream flanking sequence of the predicted PL2 core domain is that nsp2 aa 181 to 323 are critical for viral viability, as previously demonstrated in a PRRSV strain VR-2332 reverse genetic system (17). To facilitate detection of the encoded polypeptide, a c-myc epitope tag was added to the C terminus of the protein (Fig. 3A). To test the trans-cleavage activity of the PL2 domain, plasmid pPL2 was cotransfected along with pNsp2-3H124C into CHO cells, and transfection of a single plasmid, pPL2 or pNsp2-3H124C, served as a control. At 48 h posttransfection, the CHO cells were lysed and immunoprecipitated with anti-c-myc monoclonal antibody (9E10), followed by Western blotting with anti-c-myc polyclonal antibody (Fig. 3B and D) or anti-HA antibody (Fig. 3C and D). When the PL2 domain was provided in trans, the substrate nsp2-3H124C, previously shown not to self-cleave, underwent efficient proteolytic cleavage (Fig. 3B, lane 2, and C, lane 2), and processed nsp2 had a faster migration rate than the unprocessed nsp2-3 precursor (Fig. 3B, lane 3). In contrast, when the H124C mutation was introduced into the trans-provided PL2 domain (Fig. 3A), PL2H124C failed to cleave the substrate, as shown by detection of the nsp2-3 precursor with anti-HA antibodies (Fig. 3D, lane 3). Thus, we concluded that the PL2 protease possessed trans-cleavage activity.
FIG. 3.
The cysteine protease PL2 domain possessed trans-cleavage activity. (A) Diagrams of constructs used in trans-cleavage assays. Plasmid pNsp2-3H124C was cotransfected with either pPL2 or pPL2H124C into CHO cells. A plasmid encoding unmodified nsp2-3 or PL2 served as the control. CHO cells were lysed at 48 h posttransfection and immunoprecipitated (IP) by mouse monoclonal antibodies against the c-myc epitope (9E10). The immunoprecipitated proteins were separated by SDS-PAGE and analyzed by Western blotting (WB). (B) Analysis of protein expression by rabbit anti-c-myc antibodies. (C) Analysis of protein expression by anti-HA antibodies. (D) Mutation of His124 to Cys in the trans-provided PL2 domain restored detection of the nsp2-3H124C precursor by anti-HA antibodies.
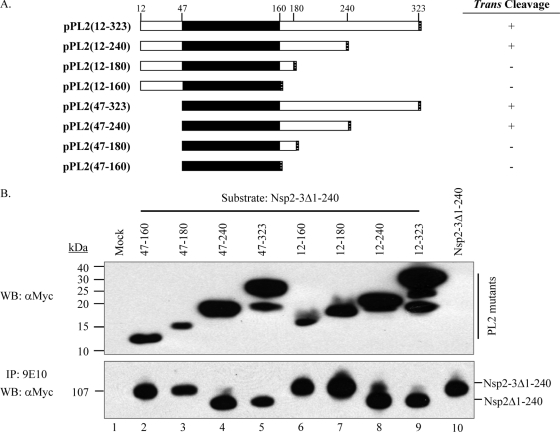
nsp2 aa 47 to 240 was required for PL2 trans-cleavage activity.
To identify the catalytic core for the PL2 trans-cleavage activity, a series of deletion mutants based on parental plasmid pPL2(12-323) were generated (Fig. 4A). Initially, pPL2(12-323) and three C-terminal truncation mutant constructs [pPL2(12-240), pPL2(12-180), and pPL2(12-160)] were cotransfected with the substrate plasmid pNsp2-3Δ1-240. Only PL2(12-240) and PL2(12-323) could effectively cleave the substrate nsp2-3Δ1-240 (Fig. 4B, lanes 8 and 9). Both the PL2(12-180) and PL2(12-160) truncated mutants lost the ability to cleave in trans (Fig. 4B, lanes 6 and 7). This suggested that the residues between 181 and 240 were required for trans proteolytic activity. Our previous work showed that nsp2 aa 12 to 35 are dispensable for nsp2 function (17). Alignment of nsp2 revealed that aa 12 to 46 are highly divergent among PRRSV strains (data not shown). Therefore, we made four N-terminal truncation mutant constructs: pPL2(47-323), pPL2(47-240), pPL2(47-180), and pPL2(47-160) (Fig. 4A). We found that nsp2-3Δ1-240 was efficiently processed when cotransfected with either pPL2(47-323) or pPL2(47-240) (Fig. 4B, lanes 4 and 5) but not when further truncations of the carboxyl terminus were tested [pPL2(47-180) and pPL2(47-160)] (Fig. 4B, lanes 2 and 3). Increasing the amount of mutant PL2 domain plasmid did not result in observable cleavage of the substrate (data not shown). We did not make further N-terminal truncations, as a C55A mutation inactivated the protease, as described below (see Fig. 6A, lane 1). The origin of the seemingly aberrant bands in lanes 5, 8, and 9 of the PL2 mutant protein Western blot assay (19 to 25 kDa) is unknown, but when SDS-PAGE was performed under nonreducing conditions, proteins of similar size were no longer detected (data not shown).
FIG. 4.
The nsp2 aa 47 to 240 region was the minimal size required for PL2 trans-cleavage activity. (A) Schematic representation of PL2 deletion mutants. The core PL2 region is indicated by the black bar. Each PL2 mutant construct was tagged with a c-myc epitope at the C terminus (gray hatched bar) and cotransfected with the substrate plasmid pNsp2-3Δ1-240. The transfected CHO cells were lysed and then subjected to immunoprecipitation (IP) and Western blot (WB) analysis. The trans-cleavage assay (Fig. 4B) results are summarized (+ or −) at the right. (B) The nsp2 aa 47 to 240 region was essential for PL2 trans-cleavage activity. The PL2 mutant proteins were analyzed directly by Western blotting with rabbit anti-c-myc antibodies, as shown at the top. The processing of nsp2-3Δ1-240 precursors was analyzed by Western blotting with rabbit anti-myc antibodies after immunoprecipitation with mouse monoclonal antibodies (9E10) against c-myc.
FIG. 6.
Separation of PL2 protease cis function from trans activity by site-directed mutagenesis. (A) To examine the cis condition, nsp2-3 mutants were transfected into CHO cells and protein processing was analyzed with rabbit anti-c-myc antibodies or mouse anti-HA antibodies after immunoprecipitation (IP) with mouse anti-c-myc monoclonal antibodies (9E10). (B) To examine the trans condition, PL2 mutant constructs were cotransfected with the substrate plasmid nsp2-3Δ1-240. Protein expression was analyzed by Western blotting (WB) with rabbit polyclonal antibodies against c-myc after immunoprecipitation with anti-c-myc monoclonal antibodies (9E10). (C) Mutations were introduced into the PRRSV strain VR-2332 infectious cDNA clone and tested for infectivity in MARC-145 cells. Viable viruses were titrated, and nonviable mutants were confirmed by RT-PCR against ORF7 after three blind passages. WT, wild type. The values on the left of panels A and B are molecular sizes in kDa.
PL2 downstream flanking sequence (aa 181 to 323) was critical for monocistronic nsp2/3 proteolysis.
The preceding experiment demonstrated that the downstream adjacent sequence (aa 181 to 240) of the PL2 core domain was critical for PL2 trans-cleavage activity. To test whether a similar region was important for nsp2 cleavage under monocistronic conditions, we initially generated two constructs, pNsp2-3Δ181-323 and pNsp2-3Δ241-323, both with a c-myc epitope tag fused to the N terminus (Fig. 5A). As shown in Fig. 5B, deletion of nsp2 aa 181 to 323 completely blocked nsp2/3 proteolytic processing of itself, as it was detected by both anti-c-myc and anti-HA antibodies (Fig. 5B, lane 4). Expression of mutant nsp2-3Δ241-323 was repeatedly unstable and could not be detected (data not shown). However, when a G1197P mutation was introduced to block the putative nsp2/3 cleavage site, the precursor nsp2-3Δ241-323G1197P (149.4 kDa) was detected by both anti-c-myc and HA antibodies (Fig. 5B, lane 7), an observation not yet understood. Plasmid pNsp2-3Δ181-323G1197P or pNsp2Δ241-323G1197P was then cotransfected with substrate plasmid pNsp2-3Δ1-240 (Fig. 5B, lane 3 or 6, respectively). In this experiment, nsp2-3Δ1-240 was efficiently cleaved in trans by nsp2Δ241-323G1197P (nsp2-3Δ1-240 protein size decreased; Fig. 5B, lane 6) but not by nsp2-3Δ181-323G1197P (Fig. 5B, lane 3), consistent with the result that nsp2 aa 241 to 323 were not essential for PL2 trans activity. Overall, we concluded that aa 181 to 323 played a crucial role in nsp2/3 cleavage under monocistronic conditions.
FIG. 5.
The nsp2 aa 181 to 323 region was essential for proteolysis under monocistronic conditions, and a G1197P mutation also prevented trans cleavage. (A) Diagram of nsp2 deletion constructs which were tagged with a c-myc epitope and a HA-FLAG tag. (B) Deletion of nsp2 aa 181 to 323 blocked processing of nsp2-3. The nsp2-3 precursor could still be detected by both anti-c-myc and HA antibodies after immunoprecipitation (IP) with mouse monoclonal antibodies (9E10) against c-myc when the nsp2 aa 181 to 323 region was deleted. The nsp2-3 precursor construct with an added mutation at the putative nsp2/3 cleavage site (G1197P) also did not permit trans cleavage. Deletion of nsp2 aa 241 to 323 resulted in an unstable product which could not be detected by anti-c-myc antibodies (data not shown), but a G1197P mutation allowed trans cleavage of nsp2-3Δ1-240 to occur. (C) nsp2 with a deletion of aa 324 to 813 was efficiently processed in CHO cells (lane 1) and showed a molecular weight shift compared to that of the precursor protein, in which a G1197P mutation blocked the cleavage (lane 2). WB, Western blotting.
The nsp2 aa 324 to 813 hypervariable region was not essential for proteolysis.
Previously, we demonstrated that PRRSV nsp2 tolerates deletions in the second hypervariable region (aa 324 to 813) of nsp2 (17). To evaluate the role of this hypervariable region in the processing of nsp2, we cloned the nsp2-3Δ324-813 fragment from infectious cDNA clone pVR-V7-nsp2Δ324-813 (17) and generated a new plasmid, pNsp2-3Δ324-813 (105 kDa; Fig. 5A). A c-myc epitope was attached to the N terminus of nsp2, and a HA-FLAG epitope was attached to the C terminus. At 48 h posttransfection of the plasmid into CHO cells, a band corresponding to processed nsp2Δ324-813 (76 kDa) was readily detected by anti-myc antibodies (Fig. 5C, lane 1), although a small amount (<10%) of intact nsp2-3 precursor could still be observed. When the putative cleavage site was changed via a G1197P mutation, the processing of the nsp2-3 substrate was blocked (Fig. 5C, lanes 2 and 5). Interestingly, two isoforms of nsp2-3Δ324-813G1197P were detected, seen as a smear of two bands, which could be due to posttranslational modifications or degradation. In conclusion, the nsp2 aa 324 to 813 hypervariable region was not essential for the PL2 protease to process its downstream substrate.
Separation of PL2 protease cis- and trans-cleavage activities by site-directed mutagenesis.
Next, the role of several conserved amino acids in the PL2 domain was investigated in order to determine their impact on protease activity. Alignment of arterivirus PL2 proteases revealed complete conservation at positions Cys55, Gly56, Trp86, Cys111, His124, Trp125, Cys142, and Cys147 (Fig. 1). Among these, Cys55 and His124 are the putative catalytic motif (36). Also, several aspartic acid residues that are commonly involved in the protease activity of virus-encoded trypsin-like serine or cysteine proteases were examined (11, 39, 40, 53). The aspartic acid residues at positions 85, 89, and 91 are well conserved among PRRSV isolates but not among other arteriviruses (Fig. 1). The conserved amino acids potentially contributing to the PL2 protease activity were tested by various amino acid substitutions in both the monocistronic construct pNsp2-3 (referred to as “cis condition”) (Fig. 6A) and in a cotransfection assay (referred to as “trans condition”) (Fig. 6B). After 48 h of transfection, the cells were lysed and the proteins were analyzed by immunoprecipitation with mouse anti-c-myc monoclonal antibody 9E10 and Western blotting with rabbit anti-c-myc polyclonal antibodies or mouse anti-HA antibodies. The experiments were repeated three times, and representative results are shown in Fig. 6A and B.
Consistent with the EAV PL2 domain mutagenesis findings (11, 39, 40, 53), mutation of the catalytic dyad (C55A and H124C) blocked nsp2/3 proteolysis in either the cis or the trans condition (Fig. 6A, lanes 1 and 12, and B, lanes 1 and 11), affirming their important role in catalysis. Mutagenesis results for the other three conserved cysteine residues (Cys111, Cys142, and Cys147) revealed differing phenotypes. The C111A mutation abolished the processing of nsp2-3 under both conditions (Fig. 6A, lane 11, and B, lane 10). In contrast, a C147A substitution appeared more detrimental to proteolysis in trans since the mutation only partially inhibited cleavage under the cis condition (Fig. 6A, lane 14) but completely blocked proteolysis in trans (Fig. 6B, lane 14). Replacement of Cys142 with an alanine residue severely affected the expression level of nsp2-3C142A, and we could not detect any protein expression (data not shown). When tested in the cotransfection assay, the PL2C142A mutant failed to cleave the substrate nsp2-3Δ1-240 (Fig. 6B, lane 13). An extended incubation time of up to 60 h or an increased transfection amount (10 μg) of plasmid pPL2C142A did not result in processing of the substrate (data not shown).
We also tested the importance of the residues following the Cys55-His124 dyad. As reported previously (45), the catalytic residue Cys55 is always followed by the small amino acid Gly56, not the bulky hydrophobic residue Trp, a typical feature of papain-like proteases. In EAV, a reversion of Gly to the canonical residue Trp completely inactivated EAV PL2 protease activity (45). Consistent with the EAV report, a G56W substitution also prevented the proteolysis of PRRSV nsp2 under both cis and trans conditions (Fig. 6A and B, lanes 3). Conversely, a G56A replacement did not affect the processing of nsp2 under the cis condition (Fig. 6A, lane 2) but partially impaired the PL2 protease activity in trans (Fig. 6B, lane 2). This suggested that an amino acid with a small side chain at position 56 is essential for optimal trans-cleavage activity of the PL2 protease. Next, we examined the role of the Trp125 residue that always follows the catalytic His124 residue in arteriviruses (Fig. 1). A W125G substitution abrogated proteolytic activity under both cis and trans conditions (Fig. 6A, lane 13, and B, lane 12). The same substitution for the conserved Trp86 residue did not affect the processing of nsp2 under cis conditions (Fig. 6A, lane 6) but, interestingly, partially impeded the protease activity under trans conditions (Fig. 6B, lane 5). These data suggested that Trp125 was much more important for PL2 protease activity than Trp86.
Aspartic acid is usually the key catalytic residue in chymotrypsin-like serine or cysteine proteases (11, 39, 40, 47, 53). The PL2 domain contains three aspartic acid residues that are highly conserved among PRRSV strains (Fig. 1), and it was reasonable to hypothesize that some or all might be required for the PL2 protease activity. To test this hypothesis, we made conserved substitutions at positions 85, 89, and 91 to either asparagine or glutamate. Most significantly, replacement of Asp89 with Asn resulted in almost total loss of the PL2 trans-cleavage activity since less than 10% of the substrate was processed (Fig. 6B, lane 6). The inefficient processing was not due to poor expression since the PL2D89N mutant had an expression level (Fig. 6B, lane 6) greater than that of control wild-type PL2 (Fig. 6B, lane 15). In contrast, the same mutation showed efficient processing under the monocistronic condition (>90% efficiency) (Fig. 6A, lane 7). A D89E mutation did not affect nsp2/3 processing under the cis condition (Fig. 6A, lane 8) but somewhat impaired the PL2 trans-cleavage activity (∼50% efficiency; Fig. 6B, lane 7), indicating a probable preference for an acidic amino acid at this position for catalytic activity. In addition, the mutational effect at this position was selective, since replacement of Asp91 with Asn or Glu did not impair efficient cleavage of the substrate under both conditions (Fig. 6A, lanes 9 and 10, and B, lanes 8 and 9). Furthermore, mutation of Asp85 to Asn was not as detrimental, resulting in incomplete cleavage only under trans conditions (∼75% efficiency; Fig. 6B, lane 4).
Overall, we observed a differential mutational effect at several positions (e.g., D85N, W86G, D89N, and D89E) that allowed efficient cleavage under the cis, but not the trans, condition. The proteolysis of nsp2/3 under the cis condition involved two possible mechanisms (cis and trans cleavages), in contrast to only one mechanism (trans cleavage) under the trans condition. The fact that defined mutations impaired trans cleavage under the trans condition suggested that the same mutation would most likely have affected PL2 trans cleavage under the cis condition. The stark difference in processing extents due to the D89N mutation is therefore a reflection of different mechanisms involved in the two different conditions and strongly indicated that cis activity is involved in nsp2-nsp3 processing (and does not rule out the possibility that trans cleavage also takes place under the cis condition) since the D89N mutation severely crippled the PL2 trans-cleavage activity (Fig. 6B, lane 6). Another strong piece of evidence supporting this conclusion is the mutational effect of C147A. The mutation of Cys147 to alanine totally abolished the PL2 trans-cleavage activity, as shown in a cotransfection assay (Fig. 6B, lane 14). However, when it was tested in the monocistronic construct, the nsp2-3 precursor was still processed, although the proteolysis was incomplete (Fig. 6A, lane 14). Therefore, we concluded that the PL2 protease possessed cis- and trans-cleavage activities that could be distinguished by point mutations in the protease domain.
Mutational analysis of the PL2 protease domain in a PRRSV reverse genetic system.
The experimental data presented above suggested that the PL2 protease trans-cleavage activity was more sensitive to mutations than was cis cleavage. To test whether the PL2 protease trans-cleavage activity played an important role in the viral replication cycle, similar mutations were introduced into PRRSV strain VR-2332 infectious cDNA clone pVR-V7 by site-directed mutagenesis. Plasmids containing the full-length mutated genome were linearized and transcribed in vitro as described previously (17). The full-length RNA transcripts were transfected into MARC-145 cells, which efficiently support PRRSV entry and replication. Virus-induced CPE, characterized by cell rounding, clustering, and detachment, was monitored for daily. For mutants that did not result in visible CPE, extracellular or intracellular RNA was examined by PRRSV ORF7 RT-PCR after three subsequent passages of cell supernatants to new culture monolayers and 5 to 6 days of incubation. Mutations that blocked cleavage of nsp2 under both cis and trans conditions (C55A, G56W, C111A, H124C, W125G, C142A, and C147A) were lethal to the virus since viable virus was not produced (Fig. 6C) and we failed to detect viral RNA after passage 3 (data not shown). Mutations that partially blocked the processing of nsp2 under the trans condition but did not alter cleavage under the cis condition were also detrimental to the virus (G56A, D85N, W86G, D89N, and D89E). Mutations that did not impair efficient cleavage of nsp2 in CHO cells did not affect viral viability (D85E, D91N, and D91E). The V7-D91N and V7-D91E mutants readily induced typical CPE similar to that caused by parental virus VR-V7 5 to 6 days after transfection. The V7-D85E mutant virus induced CPE at passage 1 but not posttransfection. Overall, these results support the hypothesis that a functional and efficient PL2 protease trans-cleavage activity is critical for PRRSV replication in cell culture.
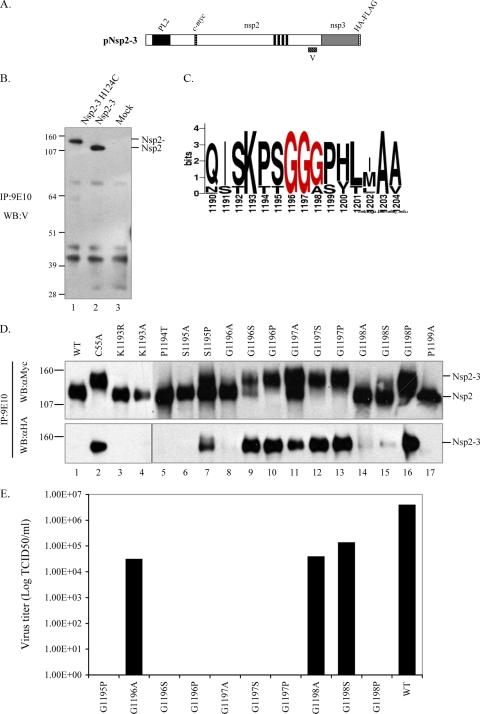
Mutational analysis of the putative nsp2/3 cleavage site.
In the experiments described above, one predominant product was detected following cleavage of the nsp2-3 substrate, suggesting that a single cleavage site was recognized by the PRRSV PL2 protease. Previously, two investigators proposed different cleavage sites for nsp2/3 proteolysis (981G|G and 1196G|G|G) (2, 53). In order to differentiate the relative cleavage position, we synthesized a rabbit peptide antibody (V) against the PRRSV strain VR-2332 nsp2 aa 1078 to 1094 region and used this antibody to probe the cleavage site of the nsp2-3 protein (Fig. 7). The V antibody recognized both nsp2 and the nsp2-3 precursor (Fig. 7B), suggesting that cleavage occurred downstream of nsp2 aa 1078 to 1094. This experiment ruled out 981G|G as the in vitro cleavage site.
FIG. 7.
Site-directed mutagenesis of the putative cleavage site. (A) Schematic of the plasmid used for cleavage site mutagenesis studies. (B) Rabbit peptide antibody (V) against the PRRSV VR-2332 strain nsp2 aa 1078 to 1094 region was prepared in order to differentiate the relative cleavage position. After immunoprecipitation (IP) with mouse anti-c-myc monoclonal antibodies (9E10), Western blotting (WB) with anti-V antibodies recognized both nsp2 and the nsp2-3 precursor, suggesting that the cleavage site was downstream of nsp2 aa 1078 to 1094. (C) Alignment of the region surrounding the putative cleavage site 11996G|G|G by WebLogo 3 (9). The bit score represents the strength of conservation. (D) Mutant protein nsp2-3 processing was analyzed by Western blotting with anti-c-myc or HA antibodies after immunoprecipitation with mouse anti-c-myc antibodies. (E) Mutations were introduced into PRRSV strain VR-2332 infectious cDNA clone pVR-V7 and tested in MARC-145 cells. Viruses were titrated in MARC-145 cells. The nonviable mutants were also confirmed by RT-PCR against ORF7, which codes for viral nucleocapsid protein, after three blind passages (data not shown). WT, wild type. The values on the left of panels B and D are molecular sizes in kDa.
Alignment of the region surrounding residues 1196G|G from 56 PRRSV strains revealed that the 1196G|G dipeptide is highly conserved (Fig. 7C). A series of amino acid substitutions in pNsp2-3 at positions Lys1193 to Pro1199 was completed. As shown in Fig. 7D, mutations at K1193 and P1194 and other alanine substitutions were well tolerated, except at position Gly1197 (Fig. 7D, lane 11). When nonconservative proline substitutions were introduced, the S1195P mutation only partially affected processing (Fig. 7D, lane 7). In contrast, the G1196P, G1197P, or G1198P substitution completely abolished cleavage of the substrate (Fig. 7D, lanes 10, 13, and 16), suggesting that residues at positions 1196 to 1198 were important for cleavage. Since a mutation to proline could potentially alter the local tertiary structure of the protein, a change to serine was tested at all three of these positions. The results revealed that Gly1196 was sensitive to the Ser substitution, with only partial cleavage observed (Fig. 7D, lane 9), and the G1197S mutation totally blocked proteolysis (Fig. 7D, lane 12). In contrast, a G1198S mutation did not inhibit efficient processing (Fig. 7D, lane 15). Thus, the G1196|G1197 dipeptide was most susceptible to mutations that resulted in impaired nsp2/3 cleavage site processing.
The consequences of the mutations at 1196G|G|G were tested in the PRRSV reverse genetic system (17). Virus titers were determined with MARC-145 cells. Mutations that totally abolished nsp2 processing in vitro (G1196P, G1197S, G1197P, and G1198P) were lethal to the virus, as neither titers nor transcripts (data not shown) could be detected at passage 3 (Fig. 7E). Mutations that did not impair efficient cleavage of nsp2 in vitro (G1196A, G1198A, and G1198S) did not affect viral viability (Fig. 7E), and CPE was readily observed after 4 to 5 days posttransfection (data not shown). Mutations that resulted in partial processing in vitro, such as G1196S and G1197A, were detrimental to the virus (Fig. 7E). Viable virus for these mutants could not be recovered after three continuous blind passages on MARC-145 cells, and no transcripts for the viral nucleocapsid gene were detected by RT-PCR at passage 3 (data not shown). Therefore, efficient processing of nsp2 appeared essential for productive replication of infectious virus progeny in MARC-145 cells.
DISCUSSION
We previously reported that deletion of the nsp2 PL2 domain is lethal to PRRSV (17). In this study, we further characterized the PL2 domain of PRRSV strain VR-2332 and provided evidence that the domain encoded a cis- and trans-active enzyme that mediated nsp2/3 proteolysis in vitro, with cleavage most likely occurring near the Gly1196|Gly1197 dipeptide. The PL2 core downstream flanking sequence (aa 181 to 323) was required for PL2 protease catalysis under either trans- or cis-cleavage conditions. Through site-directed mutagenesis, the putative catalytic motif [Cys55 (Gly) and His124 (Trp)] and the conserved cysteine residues (Cys111, Cys142, and Cys147) were confirmed and determined to be essential to PL2 protease activity. In addition, the conserved aspartic acid residues (Asp85 and Asp89) were shown to play an important role in PL2 trans-cleavage activity. Most significant was our finding that cis- and trans-cleavage activities could be distinguished by point mutations and the PL2 protease trans-cleavage activity, in particular, appeared to play an important role in PRRSV replication in cell culture.
Several results from our PRRSV PL2 domain studies were consistent with EAV PL2 protease characterization done previously (43, 45). The PL2 proteases from both EAV and PRRSV possessed trans-cleavage activity and were responsible for processing the nsp2/3 junction site, with a G|G dipeptide substrate preference. The putative Cys55-His124 catalytic motif was critical for the PL2 protease activity in both viruses. However, we also uncovered differences between the PL2 proteases of EAV and PRRSV. Although the EAV and PRRSV nsp2s are similar in domain organization in that a large middle region separates the N-terminal protease domain and its downstream substrate site, the middle region (aa 324 to 813) of PRRSV nsp2 was not critical for downstream processing (Fig. 5C) and deletion of aa 324 to 726 did not prevent virus viability (17). This is in direct contrast to EAV, where deletion of the EAV nsp2 middle region completely blocked PL2 cleavage in vitro (45). Second, the PRRSV and EAV PL2 proteases displayed different susceptibilities to protease domain point mutations. For instance, the C147A mutation only partially impaired nsp2/3 processing when tested in the monocistronic construct pNsp2-3 (Fig. 6A, lane 14). However, the same mutation at the corresponding cysteine in EAV totally abolished nsp2 proteolysis under similar conditions (45). Additionally, several mutations (e.g., W86G, D89N, D89E, and C147A) in the PRRSV PL2 protease domain differentially affected the cis- and trans-cleavage activities. Interestingly, this phenomenon is not without precedence in positive-stranded RNA viruses. Certain mutations (e.g., D38E, Y88L, and Y89L) in the 2A protease of poliovirus also caused specific loss of trans- but not cis-cleavage function (52).
PRRSV PL2 harbors features of both papain-like cysteine proteases and chymotrypsin-like serine proteases (45). The PL2 enzyme possesses a catalytic-motif (Cys-His) resemblance to papain-like cysteine proteases. Yet PRRSV PL2 contains the marker [Cys55 (Gly56)] of chymotrypsin-like cysteine proteases [His-Asp-Cys (Gly)], in which the catalytic Cys residue is always followed by Gly instead of Trp, which is seen in papain-like proteases [Cys (Trp)-His]. In PRRSV PL2, we showed that Gly and Trp were not interchangeable. This signifies a difference between PRRSV PL2 and papain-like proteases. In light of the recognition of arterivirus PL2 proteases as new members of the OTU superfamily (14, 28), we also analyzed the residues surrounding the catalytic Cys and His residues in OTU-containing proteases. The position following the active cysteine is less conserved and is often occupied by residues such as Leu, Phe, and Gln and sometimes Met, Val, and Cys (Pfam: PF02338). It will be interesting to investigate whether Gly56 of PRRSV PL2 can be mutated to these residues (L, F, Q, M, V, and C) in order to more firmly establish its relationship to other OTU family members. Since a mutation from Gly to Ala in the PRRSV PL2 domain somewhat impaired the trans-cleavage activity of PRRSV PL2 (Fig. 6B, lane 2), our prediction is that replacement with such residues with large side chains may not allow efficient trans cleavage. On the other hand, the highly conserved Trp125 residue following the catalytic His124 residue in the PRRSV PL2 protease also appeared to be important for cleavage (Fig. 6A, lane 13, and B, lane 12), since a W-to-G mutation abolished nsp2/3 proteolysis. The mutation at this position appears selective since a similar mutation at position 86 (W86G) did not affect nsp2/3 processing under the cis condition (Fig. 6A). Interestingly, the residue next to the putative catalytic His in members of the OTU superfamily is always occupied by a similar aromatic amino acid (e.g., F or Y) (PF02338) instead of an aliphatic acid (e.g., A, V, L, or G), as seen in papain-like (PF00112) and chymotrypsin-like (PF00548) cysteine proteases. This may reflect an evolutionary convergence within OTU domain-containing family members.
Aspartate, an active amino acid, is often involved in enzymatic hydrolysis and constitutes an important component of the catalytic triad (His-Asp-Ser/Cys) of virally encoded chymotrypsin-like serine or cysteine proteases (36, 39, 40, 47, 53). Papain-like cysteine proteases do not normally employ aspartic acid as a catalytic component (39, 40, 47, 53). Our mutagenesis analysis also revealed a critical role for aspartic acid, particularly Asp89, in PRRSV PL2 protease activity. The D89N mutation resulted in an almost total loss of trans activity in a cotransfection assay. The same mutation is not without consequence when tested in a monocistronic construct, impairing nsp2/3 processing but to a much lesser degree. When Asp89 was replaced with Glu, the activity of PL2 was partially restored, indicating that the electrical charge is important. Although Asp89 is highly conserved among PRRSV strains, a comparative analysis/alignment did not reveal any significant conservation at this position among OTU family members (PF02338). Considering differential mutational effects and the distinct mechanisms employed by the PL2 enzyme under trans and cis conditions in order to access the substrate (e.g., polyprotein folding to oblige enzyme-substrate interaction in cis versus normal enzyme-substrate binding in trans) (39, 40, 47), one explanation is that the Asp89 residue likely plays a structural role in substrate binding which may have a much stricter requirement under the trans condition. However, we could not rule out the possibility that the negatively charged Asp89 residue serves as an active site to stabilize the His124 residue that deprotonates the putative catalytic residue Cys55 during catalysis. Recently solved structures of several members from the OTU family revealed that these OTU-containing cysteine proteases employ Cys, Asp, and His as the catalytic triad (12, 19, 29, 32). The catalytic triad core may show a similar configuration; however, the position of the active aspartate is variable in the primary sequence. For example, the active residue Asp70 of the A20 protein is upstream of the catalytic Cys103 residue (19). For Otu1, in contrast, Asp226 is just 2 aa C terminal from the catalytic His224 residue (12). Resolving the crystal structure of the PRRSV PL2 domain would certainly provide insights into the mechanism of the enzyme-substrate interaction and reveal how aspartic acid and other residues contribute to the PL2 enzymatic activity.
We introduced directed mutations into the PRRSV full-length cDNA clone to test the contribution of the PL2 trans function to viral replication. Some mutations that impaired trans-cleavage activity but not cis-cleavage activity were surprisingly detrimental to the virus. Similar results were also found in poliovirus 2A mutants lacking trans function only (51). Several possible explanations may account for the defective phenotype. First, the trans-cleavage activity of the PL2 protease may be required for cleavage of other sites in the viral polyproteins during replicase maturation, in addition to processing of the current nsp2/3 junction site. Second, the trans-cleavage activity of the PL2 protease may play an active role in suppressing host innate immunity. Inevitably, almost all of the proteases from positive-stranded RNA viruses assume multiple roles and are actively involved in antagonizing cellular proteins required for host antiviral responses. For instance, the hepatitis C virus NS3/4A protease ablates alpha/beta interferon signaling by targeting the Toll-like receptor 3 adaptor protein TRIF and the mitochondrial antiviral signaling protein (22, 23, 25). The 2A, Lpro, and 3C proteases from picornaviruses cleave translation initiation factor eIF4FG, poly(A) binding protein, TATA box binding protein, and other cellular factors, thus effectively halting cap-dependent translation of host mRNAs (16, 20). The papain-like protease of coronaviruses has multiple functions, including deubiquitinating the ubiquitin-conjugated proteins and inducing degradation of the interferon regulatory factor IRF-3 in addition to proteolytic processing (24, 38a). As reported previously, PRRSV is a suppressor of interferon signaling and strongly inhibits the production of alpha and beta interferons, as well as interleukin-1, interleukin-6, and tumor necrosis factor alpha, during viral infection either in pigs or the supporting cell line MARC-145 (1, 31). Recently, it was reported that PRRSV particularly interferes with the RIG-1 signaling pathway and suppresses the activation of IRF-3 (26). By analogy with the proteases from these RNA viruses, it is reasonable to believe that ablation of PRRSV PL2 trans-cleavage activity may impair its ability to block intracellular innate immunity, which renders the virus susceptible to host antiviral responses. In line with this hypothesis, PRRSV nsp2 was recently shown to be able to decrease the level of ubiquitin and ISG15 conjugates (14), suggesting that the PRRSV nsp2 PL2 protease may be a deubiquitinating enzyme which uses its trans-cleavage activity to target ubiquitin conjugates. Thus, this may in part explain why the induction of interferon and some inflammatory cytokines is inhibited during viral infection. Future studies may be directed toward studying the deubiquitinating activity and substrate specificity of the PRRSV PL2 protease and examining its contribution to viral infection. Lastly, it is also possible that PL2 acts as a multifunctional domain that interacts with host or viral factors during viral replication, as occurs for the 2A protein of poliovirus virus, which binds to the 5′ untranslated region of poliovirus RNA (27). Thus, mutation of the conserved sites in the PL2 domain may affect PL2 function at the protein or RNA level, therefore impairing viral replication. Overall, one or two of the factors may not be sufficient to cripple PRRSV replication; however, a combination of these effects would most likely be lethal to the virus.
The third important feature concerns the potential function of the PL2 flanking sequence (nsp2 aa 180 to 322). Deletion of nsp2 aa 181 to 323 abolished both the cis and trans activities of the PL2 protease, consistent with the previous data showing that deletion of either nsp2 aa 181 to 323 or aa 241 to 323 is lethal to PRRSV (17). Alignment of the nsp2 aa 181 to 323 region shows it to be highly divergent, with less than 10% amino acid identity between North American and European PRRSV strains, but relatively conserved within PRRSV genotypes (13, 18, 38). Although it is not clear how the C terminus contributes to the PL2 protease activity, it is conceivable that several functions could be related to this domain: (i) involvement in enzyme-substrate molecule interactions, (ii) maintenance of overall PL2 enzyme folding, and/or (iii) an undefined nonproteolytic activity. It is noteworthy that the nsp2 aa 181 to 240 region is rich in cysteine and histidine residues in both PRRSV genotypes and may play a role in inter- or intramolecular interactions or mediate the formation of a zinc binding structure. Such interactions could be critical for protease catalysis or to maintain a favorable structure for substrate binding, as observed for other virally encoded cysteine or serine proteases (39, 40, 53).
Replicase polyprotein maturation constitutes a very critical part of the replication cycle of positive-stranded RNA viruses and occurs in a highly delicate and orchestrated order. The mature subunits have been shown to be largely involved in the assembly of viral replication complexes. Correct identification of the cleavage site is a prerequisite for studying the function of a protein. Our site-directed mutagenesis study yielded results consistent with the previous prediction that the arterivirus PL2 proteases prefer a G|G dipeptide as a substrate (39, 40, 53). However, the arterivirus nsp2 proteases were recently shown to have potential deubiquitinating activities (14), which may have important implications for substrate recognition. Characterized viral deubiquitinating enzymes have been shown to cleave after a G|G dipeptide located in the C terminus of ubiquitin, ISG15, Nedd8, or SUMO (46). A related nidovirus protease, coronavirus severe acute respiratory syndrome virus PLP2, has been shown to be a deubiquitinating enzyme and cleaves at a downstream LXGG motif (3, 24). Therefore, it is highly likely that PRRSV PL2 has similar substrate recognition properties. Our mutagenesis showed that the dipeptide Gly1196-Gly1197 was most sensitive to mutations, as even a conservative G1197A substitution significantly impaired cleavage efficiency. A structurally less dramatic G1196S substitution partially blocked cleavage, while a G1197S mutation completely abolished proteolytic processing. The mutational effect at these two positions is selective, since neither a G1198A nor a G1198S mutation affected the efficient proteolysis of nsp2/3. This was expected because Gly1198 is not completely conserved among PRRSV strains and is often replaced with an alanine residue. It appears that the G1196|G1197 dipeptide is spatially and structurally preferred in that the replacement of Gly1196 with serine did not produce protease cleavage at or near the Gly1197|Gly1198 dipeptide. Therefore, substrate recognition by the PL2 protease may be not only sequence specific but also structurally tuned. This might explain why the PL2 protease did not, in vitro, recognize other dipeptides (G647|G648, G981|G982, G828|G829|G830) that are conserved among PRRSV strains. Certainly, we cannot rule out the possibility that the PL2 protease may recognize and cleave these additional sites in PRRSV-susceptible macrophages and MARC-145 cells, perhaps with the help of host or viral cofactors.
In summary, we have identified the PRRSV nsp2 PL2 domain as an active protease which possessed both trans- and cis-cleavage activities. Importantly, the trans- and cis-cleavage activities of the PL2 protease could be distinguished by point mutations and appeared to play an important role in the PRRSV replication cycle. The assays established here will help to test the potential deubiquitinating activity of the PRRSV PL2 protease and its role in PRRSV infection in the future. Overall, these findings further extend our understanding of arterivirus PL2 proteases and establish the PL2 protease as a new therapeutic target for the development of antiviral drugs against PRRSV infection.
Acknowledgments
This research was supported by USDA NRI-CSREES (2006-01598).
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Footnotes
Published ahead of print on 8 July 2009.
REFERENCES
- 1.Albina, E., C. Carrat, and B. Charley. 1998. Interferon-alpha response to swine arterivirus (PoAV), the porcine reproductive and respiratory syndrome virus. J. Interferon Cytokine Res. 18:485-490. [DOI] [PubMed] [Google Scholar]
- 2.Allende, R., T. L. Lewis, Z. Lu, D. L. Rock, G. F. Kutish, A. Ali, A. R. Doster, and F. A. Osorio. 1999. North American and European porcine reproductive and respiratory syndrome viruses differ in non-structural protein coding regions. J. Gen. Virol. 80(Pt. 2):307-315. [DOI] [PubMed] [Google Scholar]
- 3.Barretto, N., D. Jukneliene, K. Ratia, Z. Chen, A. D. Mesecar, and S. C. Baker. 2005. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 79:15189-15198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benfield, D. A., E. Nelson, J. E. Collins, L. Harris, S. M. Goyal, D. Robison, W. T. Christianson, R. B. Morrison, D. Gorcyca, and D. Chladek. 1992. Characterization of swine infertility and respiratory syndrome (SIRS) virus (isolate ATCC VR-2332). J. Vet. Diagn. Investig. 4:127-133. [DOI] [PubMed] [Google Scholar]
- 5.Cavanagh, D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 142:629-633. [PubMed] [Google Scholar]
- 6.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christianson, W. T., J. E. Collins, D. A. Benfield, L. Harris, D. E. Gorcyca, D. W. Chladek, R. B. Morrison, and H. S. Joo. 1992. Experimental reproduction of swine infertility and respiratory syndrome in pregnant sows. Am. J. Vet. Res. 53:485-488. [PubMed] [Google Scholar]
- 8.Collins, J. E., D. A. Benfield, W. T. Christianson, L. Harris, J. C. Hennings, D. P. Shaw, S. M. Goyal, S. McCullough, R. B. Morrison, H. S. Joo, et al. 1992. Isolation of swine infertility and respiratory syndrome virus (isolate ATCC VR-2332) in North America and experimental reproduction of the disease in gnotobiotic pigs. J. Vet. Diagn. Investig. 4:117-126. [DOI] [PubMed] [Google Scholar]
- 9.Crooks, G. E., G. Hon, J. M. Chandonia, and S. E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.den Boon, J. A., K. S. Faaberg, J. J. Meulenberg, A. L. Wassenaar, P. G. Plagemann, A. E. Gorbalenya, and E. J. Snijder. 1995. Processing and evolution of the N-terminal region of the arterivirus replicase ORF1a protein: identification of two papainlike cysteine proteases. J. Virol. 69:4500-4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dougherty, W. G., and B. L. Semler. 1993. Expression of virus-encoded proteinases: functional and structural similarities with cellular enzymes. Microbiol. Rev. 57:781-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edelmann, M. J., A. Iphofer, M. Akutsu, M. Altun, K. di Gleria, H. B. Kramer, E. Fiebiger, S. Dhe-Paganon, and B. M. Kessler. 2009. Structural basis and specificity of human otubain 1-mediated deubiquitination. Biochem. J. 418:379-390. [DOI] [PubMed] [Google Scholar]
- 13.Fang, Y., D. Y. Kim, S. Ropp, P. Steen, J. Christopher-Hennings, E. A. Nelson, and R. R. Rowland. 2004. Heterogeneity in Nsp2 of European-like porcine reproductive and respiratory syndrome viruses isolated in the United States. Virus Res. 100:229-235. [DOI] [PubMed] [Google Scholar]
- 14.Frias-Staheli, N., N. V. Giannakopoulos, M. Kikkert, S. L. Taylor, A. Bridgen, J. Paragas, J. A. Richt, R. R. Rowland, C. S. Schmaljohn, D. J. Lenschow, E. J. Snijder, A. Garcia-Sastre, and H. W. Virgin III. 2007. Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell Host Microbe 2:404-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorbalenya, A. E., L. Enjuanes, J. Ziebuhr, and E. J. Snijder. 2006. Nidovirales: evolving the largest RNA virus genome. Virus Res. 117:17-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haghighat, A., Y. Svitkin, I. Novoa, E. Kuechler, T. Skern, and N. Sonenberg. 1996. The eIF4G-eIF4E complex is the target for direct cleavage by the rhinovirus 2A proteinase. J. Virol. 70:8444-8450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han, J., G. Liu, Y. Wang, and K. S. Faaberg. 2007. Identification of nonessential regions of the nsp2 replicase protein of porcine reproductive and respiratory syndrome virus strain VR-2332 for replication in cell culture. J. Virol. 81:9878-9890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han, J., Y. Wang, and K. S. Faaberg. 2006. Complete genome analysis of RFLP 184 isolates of porcine reproductive and respiratory syndrome virus. Virus Res. 122:175-182. [DOI] [PubMed] [Google Scholar]
- 19.Komander, D., and D. Barford. 2008. Structure of the A20 OTU domain and mechanistic insights into deubiquitination. Biochem. J. 409:77-85. [DOI] [PubMed] [Google Scholar]
- 20.Kräusslich, H. G., M. J. Nicklin, H. Toyoda, D. Etchison, and E. Wimmer. 1987. Poliovirus proteinase 2A induces cleavage of eucaryotic initiation factor 4F polypeptide p220. J. Virol. 61:2711-2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 22.Li, K., E. Foy, J. C. Ferreon, M. Nakamura, A. C. Ferreon, M. Ikeda, S. C. Ray, M. Gale, Jr., and S. M. Lemon. 2005. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc. Natl. Acad. Sci. USA 102:2992-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, X. D., L. Sun, R. B. Seth, G. Pineda, and Z. J. Chen. 2005. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc. Natl. Acad. Sci. USA 102:17717-17722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindner, H. A., N. Fotouhi-Ardakani, V. Lytvyn, P. Lachance, T. Sulea, and R. Menard. 2005. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J. Virol. 79:15199-15208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loo, Y. M., D. M. Owen, K. Li, A. K. Erickson, C. L. Johnson, P. M. Fish, D. S. Carney, T. Wang, H. Ishida, M. Yoneyama, T. Fujita, T. Saito, W. M. Lee, C. H. Hagedorn, D. T. Lau, S. A. Weinman, S. M. Lemon, and M. Gale, Jr. 2006. Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proc. Natl. Acad. Sci. USA 103:6001-6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo, R., S. Xiao, Y. Jiang, H. Jin, D. Wang, M. Liu, H. Chen, and L. Fang. 2008. Porcine reproductive and respiratory syndrome virus (PRRSV) suppresses interferon-beta production by interfering with the RIG-I signaling pathway. Mol. Immunol. 45:2839-2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Macadam, A. J., G. Ferguson, T. Fleming, D. M. Stone, J. W. Almond, and P. D. Minor. 1994. Role for poliovirus protease 2A in cap independent translation. EMBO J. 13:924-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Makarova, K. S., L. Aravind, and E. V. Koonin. 2000. A novel superfamily of predicted cysteine proteases from eukaryotes, viruses and Chlamydia pneumoniae. Trends Biochem. Sci. 25:50-52. [DOI] [PubMed] [Google Scholar]
- 29.Messick, T. E., N. S. Russell, A. J. Iwata, K. L. Sarachan, R. Shiekhattar, J. R. Shanks, F. E. Reyes-Turcu, K. D. Wilkinson, and R. Marmorstein. 2008. Structural basis for ubiquitin recognition by the Otu1 ovarian tumor domain protein. J. Biol. Chem. 283:11038-11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meulenberg, J. J., A. Petersen den Besten, E. de Kluyver, A. van Nieuwstadt, G. Wensvoort, and R. J. Moormann. 1997. Molecular characterization of Lelystad virus. Vet. Microbiol. 55:197-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller, L. C., W. W. Laegreid, J. L. Bono, C. G. Chitko-McKown, and J. M. Fox. 2004. Interferon type I response in porcine reproductive and respiratory syndrome virus-infected MARC-145 cells. Arch. Virol. 149:2453-2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nanao, M. H., S. O. Tcherniuk, J. Chroboczek, O. Dideberg, A. Dessen, and M. Y. Balakirev. 2004. Crystal structure of human otubain 2. EMBO Rep. 5:783-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nelsen, C. J., M. P. Murtaugh, and K. S. Faaberg. 1999. Porcine reproductive and respiratory syndrome virus comparison: divergent evolution on two continents. J. Virol. 73:270-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nielsen, H. S., G. Liu, J. Nielsen, M. B. Oleksiewicz, A. Botner, T. Storgaard, and K. S. Faaberg. 2003. Generation of an infectious clone of VR-2332, a highly virulent North American-type isolate of porcine reproductive and respiratory syndrome virus. J. Virol. 77:3702-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pasternak, A. O., W. J. Spaan, and E. J. Snijder. 2006. Nidovirus transcription: how to make sense? J. Gen. Virol. 87:1403-1421. [DOI] [PubMed] [Google Scholar]
- 36.Ratia, K., K. S. Saikatendu, B. D. Santarsiero, N. Barretto, S. C. Baker, R. C. Stevens, and A. D. Mesecar. 2006. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc. Natl. Acad. Sci. USA 103:5717-5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent end-points. Am. J. Hyg. 27:493-497. [Google Scholar]
- 38.Ropp, S. L., C. E. Wees, Y. Fang, E. A. Nelson, K. D. Rossow, M. Bien, B. Arndt, S. Preszler, P. Steen, J. Christopher-Hennings, J. E. Collins, D. A. Benfield, and K. S. Faaberg. 2004. Characterization of emerging European-like porcine reproductive and respiratory syndrome virus isolates in the United States. J. Virol. 78:3684-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38a.Roth-Cross, J. K., L. Martínez-Sobrido, E. P. Scott, A. García-Sastre, and S. R. Weiss. 2007. Inhibition of the alpha/beta interferon response by mouse hepatitis virus at multiple levels. J. Virol. 81:7189-7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ryan, M. D., and M. Flint. 1997. Virus-encoded proteinases of the picornavirus super-group. J. Gen. Virol. 78:699-723. [DOI] [PubMed] [Google Scholar]
- 40.Ryan, M. D., S. Monaghan, and M. Flint. 1998. Virus-encoded proteinases of the Flaviviridae. J. Gen. Virol. 79:947-959. [DOI] [PubMed] [Google Scholar]
- 41.Snijder, E. J., and J. J. Meulenberg. 1998. The molecular biology of arteriviruses. J. Gen. Virol. 79:961-979. [DOI] [PubMed] [Google Scholar]
- 42.Snijder, E. J., A. L. Wassenaar, and W. J. Spaan. 1993. Proteolytic processing of the N-terminal region of the equine arteritis virus replicase. Adv. Exp. Med. Biol. 342:227-232. [DOI] [PubMed] [Google Scholar]
- 43.Snijder, E. J., A. L. Wassenaar, and W. J. Spaan. 1994. Proteolytic processing of the replicase ORF1a protein of equine arteritis virus. J. Virol. 68:5755-5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Snijder, E. J., A. L. Wassenaar, and W. J. Spaan. 1992. The 5′ end of the equine arteritis virus replicase gene encodes a papainlike cysteine protease. J. Virol. 66:7040-7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Snijder, E. J., A. L. Wassenaar, W. J. Spaan, and A. E. Gorbalenya. 1995. The arterivirus Nsp2 protease. An unusual cysteine protease with primary structure similarities to both papain-like and chymotrypsin-like proteases. J. Biol. Chem. 270:16671-16676. [DOI] [PubMed] [Google Scholar]
- 46.Sulea, T., H. A. Lindner, and R. Menard. 2006. Structural aspects of recently discovered viral deubiquitinating activities. Biol. Chem. 387:853-862. [DOI] [PubMed] [Google Scholar]
- 47.ten Dam, E., M. Flint, and M. D. Ryan. 1999. Virus-encoded proteinases of the Togaviridae. J. Gen. Virol. 80:1879-1888. [DOI] [PubMed] [Google Scholar]
- 48.van Aken, D., J. Zevenhoven-Dobbe, A. E. Gorbalenya, and E. J. Snijder. 2006. Proteolytic maturation of replicase polyprotein pp1a by the nsp4 main proteinase is essential for equine arteritis virus replication and includes internal cleavage of nsp7. J. Gen. Virol. 87:3473-3482. [DOI] [PubMed] [Google Scholar]
- 49.Wensvoort, G., E. P. de Kluyver, J. M. Pol, F. Wagenaar, R. J. Moormann, M. M. Hulst, R. Bloemraad, A. den Besten, T. Zetstra, and C. Terpstra. 1992. Lelystad virus, the cause of porcine epidemic abortion and respiratory syndrome: a review of mystery swine disease research at Lelystad. Vet. Microbiol. 33:185-193. [DOI] [PubMed] [Google Scholar]
- 50.Wensvoort, G., C. Terpstra, J. M. Pol, E. A. ter Laak, M. Bloemraad, E. P. de Kluyver, C. Kragten, L. van Buiten, A. den Besten, F. Wagenaar, et al. 1991. Mystery swine disease in The Netherlands: the isolation of Lelystad virus. Vet. Q. 13:121-130. [DOI] [PubMed] [Google Scholar]
- 51.Yu, S. F., P. Benton, M. Bovee, J. Sessions, and R. E. Lloyd. 1995. Defective RNA replication by poliovirus mutants deficient in 2A protease cleavage activity. J. Virol. 69:247-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu, S. F., and R. E. Lloyd. 1991. Identification of essential amino acid residues in the functional activity of poliovirus 2A protease. Virology 182:615-625. [DOI] [PubMed] [Google Scholar]
- 53.Ziebuhr, J., E. J. Snijder, and A. E. Gorbalenya. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81:853-879. [DOI] [PubMed] [Google Scholar]