Abstract
Human parvovirus B19 (B19V) is a member of the genus Erythrovirus in the family Parvoviridae. In vitro, autonomous B19V replication is limited to human erythroid progenitor cells and in a small number of erythropoietin-dependent human megakaryoblastoid and erythroid leukemic cell lines. Here we report that the failure of B19V DNA replication in nonpermissive 293 cells can be overcome by adenovirus infection. More specifically, the replication of B19V DNA in the 293 cells and the production of infectious progeny virus were made possible by the presence of the adenovirus E2a, E4orf6, and VA RNA genes that emerged during the transfection of the pHelper plasmid. Using this replication system, we identified the terminal resolution site and the nonstructural protein 1 (NS1) binding site on the right terminal palindrome of the viral genome, which is composed of a minimal origin of replication spanning 67 nucleotides. Plasmids or DNA fragments containing an NS1 expression cassette and this minimal origin were able to replicate in both pHelper-transfected 293 cells and B19V-semipermissive UT7/Epo-S1 cells. Our results have important implications for our understanding of native B19V infection.
Human parvovirus B19 (B19V), a member of the family Parvoviridae, causes a variety of diseases in humans. These include erythema infectiosum in children; acute or chronic arthropathy, commonly seen in women; transient aplastic crisis in patients with chronic hemolytic anemia (e.g., sickle cell disease patients); pure red-cell aplasia due to persistent infection in immunocompromised patients; and hydrops fetalis in pregnant women (52). Recently, B19V infection also has been associated with chronic and acute myocarditis (36). Cytotoxicity from viral infection, e.g., in primary erythroid progenitor cells (EPCs), is often the direct cause of disease outcomes such as transient aplastic crisis and pure red-cell aplasia (5, 52).
B19V contains a small, linear, single-stranded DNA genome of 5.6 kb, which harbors two identical inverted terminal repeats (ITRs) that serve as origins of DNA replication (12). The genome encodes a single nonstructural protein (NS1) with a large open reading frame (ORF) in its left half and two capsid proteins (VP1 and VP2) from another ORF in the right half. It also encodes at least two other smaller proteins (7.5 kDa and 11 kDa) whose functions remain poorly defined. B19V shows an extreme tropism for the erythroid progenitor CFU erythroid cells (CFU-E) and burst-forming unit erythroid cells (BFU-E) of the human bone marrow (29, 40). A few semipermissive cell lines that support B19V infection with limited efficiency have been established. These include the megakaryoblastoid cell line UT7/Epo-S1 (23) and the erythroid leukemic cell line KU812Ep6 (22), both of which are dependent on erythropoietin (Epo) for growth. The blood-group P antigen (which serves as the cellular receptor for B19V), integrin α5β1, and KU80 (recently reported as coreceptors involved in B19V infection) (25, 47) are believed to be the main regulators of the B19V tropism. Additionally, the B19V tropism has been shown to be limited intracellularly to human blast cell lines, but the mechanism responsible for this restriction has not been clearly established (17, 29, 39). After B19V enters cells, the single-stranded B19V genome has to be converted to a duplex replicative form (RF) that is capable of being transcribed. However, such a form has not been detected in some cell types for which B19V exhibits a tropism (e.g., M-07 cells), and although it has been observed in others (e.g., UT7/Epo cells), in those cases it is limited to a small subpopulation (17). The tropism could theoretically also be a consequence of limited capsid protein expression due to a block in the production of full-length transcripts by internal polyadenylation (4). However, we have recently shown that a block in the generation of full-length transcripts can be overcome by replication of the viral genome (19).
Among the Parvoviridae, only members of the genera Erythrovirus and Dependovirus contain identical ITRs at both ends of their genome. B19V has an ITR of 383 nucleotides, and of these nucleotides, the terminal 365 can be folded into hairpins that are expressed in two alternative orientations, flip or flop (Fig. 1) (54). The genomes of members in other genera exhibit distinct terminal structures, most of which are smaller than the B19V hairpin (12). The mechanism whereby the B19V genome replicates is not yet understood. One model that has been proposed is the “rolling-hairpin” model of parvovirus replication (13); it is based on observations of the terminal hairpin structures on a single-stranded B19V genome under an electron microscope (2) and on in vitro studies involving priming DNA synthesis using Escherichia coli DNA polymerase (11).
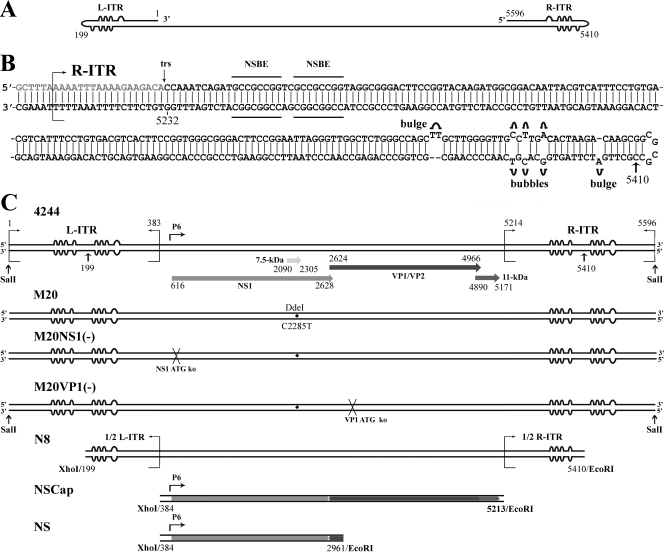
FIG. 1.
Structure of the B19V genome. (A) Schematic diagram of the minus strand of the B19V genome. Identical ITRs are present at each end of the genome, and these are depicted with unpaired or mismatched bases in the palindromes represented by “bulges” or “bubbles,” respectively. (B) Structure of the B19V R-ITR. The palindrome at 365 nt is depicted in the “flip” orientation, and the putative trs and NS1 binding elements (NSBE) thought to comprise the NS1 binding site are indicated. Nucleotide numbers are according to GenBank accession no. AY386330. The ITR is an almost-perfect duplex structure; the only exceptions are the three unpaired bases at two sites (bulges) and three mismatched bases at three sites (bubbles). (C) Schematic diagrams of B19V DNAs. B19V DNA 4244 is the duplex RF of the B19V genome, which has the capability to replicate and produce progeny virus in B19V-permissive cells (54). The bubbles within each ITR reflect potential interstrand folding. The position of the P6 promoter, as well as the ORFs for the five B19V proteins, is also indicated. B19V DNA M20 is likewise an infectious DNA and is nearly identical to 4244 RF DNA; it contains the single nucleotide substitution C2285T to distinguish it from the native 4244 RF DNA by the digestion of DdeI as shown (54). In B19V DNAs M20NS(−) and M20VP1(−), the start codons for NS1 and VP1 are knocked out, respectively. In B19V DNA N8, only half of the L-ITR (nt 199 to 383) and half of the R-ITR (nt 5214 to 5410) sequences are included. The palindromic structure is not present in N8, and the bubbles, which are shown only as a symbol of half of the ITR, will thus not be folded in strand. The B19V DNAs of NSCap and NS do not contain any ITR sequences and encode different subsets of the B19V proteins. The ORFs preserved in the NSCap and NS DNAs are depicted in the respective diagrams, and the nucleotide numbers of each DNA are indicated beneath the diagram. The restriction enzymes to excise these B19V DNAs from the corresponding B19V plasmids described in Materials and Methods are indicated at their ends.
The replication of the Dependovirus adeno-associated virus type 2 (AAV2) is characterized by a strand displacement mechanism that uses a hairpinned ITR as a primer (3). The virus-encoded large nonstructural proteins have the site-specific DNA helicase and endonuclease activities required for terminal resolution and reinitiation (38), and the hairpinned ITR provides a free 3′-OH for DNA synthesis (Fig. 2Aa). However, in vitro, Rep-dependent initiation of replication has been demonstrated to occur even in the absence of a hairpin structure (20, 26, 43). In the presence of a hairpin structure, site- and strand-specific cleavage at the terminal resolution site (trs) creates a base-paired 3′ nucleotide (3′ primer) that supports the assembly of a new replication fork (Fig. 2Ab), while the large Rep protein is covalently attached to the 5′ end of the DNA at the nicking site, where it serves as a DNA helicase (Fig. 2Ac). While the hairpin end, which can be as small as four base pairs (46), unfolds as a consequence of this helicase activity, the 3′ primer leads extension to the end of the template (Fig. 2Ad). When the hairpin is present, newly synthesized DNA can be refolded into a hairpin structure as a 3′ primer and resumes hairpin-primed strand displacement synthesis (Fig. 2Ae and Af). In B19V, the ITRs are likewise identical, and replication initiated at the origins may be the same as in AAV2 (45).
FIG. 2.
Models of parvovirus DNA replication. (A) Rolling-hairpin model of Dependovirus replication (13, 45). (a) The single-stranded DNA genome uses the hairpin as a primer for the synthesis of a duplex DNA molecule that is covalently closed at one end by the hairpin structure. (b) The ITR is cleaved at the nicking site by the NS1 protein (filled circle). (c, d) The hairpinned ITR is subsequently repaired, and this results in an open-ended duplex replication intermediate. (e, f) The repaired ITR is then denatured and reannealed, in a process termed reinitiation, to form a double-hairpinned intermediate that initiates a round of strand displacement synthesis. L, L-ITR; R, R-ITR. (B) Putative model of hairpin-independent B19V DNA replication in the current study. (a) A representative NSCap-based DNA fragment that contains the Ori, and a 500-nt extension, is shown, with each strand labeled as positive (+) or negative (−); no hairpin of more than 3 bp is present at the right end. (b) The Ori at the right end is nicked at the trs by NS1 (filled circle), which binds to the NS1 binding site (NSBS). (c) DNA is synthesized and primed from the free nucleotide (3′ OH) that is created by the nicking, with the complementary strand (negative [−]) serving as the template. (d) DNA synthesis goes to the 3′ end. (e) Following complementary-strand synthesis, the 3′ primer (possibly the last synthesized nucleotide) is used to initiate strand-displacement synthesis, through an unknown mechanism in which NS1 likely plays a key role. (f) The final replicated DNA product is composed of the prokaryotic template strand (+) and the newly synthesized complementary strand (−) which is resistant to DpnI digestion. dam, dam methylation.
In ex vivo cultures of expanded primary EPCs that closely resemble the BFU-E/CFU-E of human bone marrow (16), infection with B19V resulted in a high level of capsid expression and a 100- to 1,000-fold increase in B19V genomes (50). However, the low level of progeny virus production in this system, which was the most physiologically relevant one available, strongly suggested that B19V replication in primary EPCs does not recapitulate B19V replication in the BFU-E/CFU-E of human bone marrow in vivo and that some steps in B19V replication are restricted in in vitro cell culture systems.
B19V DNA replication has not been investigated in detail, due to the lack of an infectious clone. However, a full-length B19V clone was recently constructed; it has been shown to be infectious in B19V-semipermissive UT7/Epo-S1 cells but to produce only a low level of progeny virus (53). The use of this B19V infectious clone in the current study has enabled us to examine the replication of the B19V genome in cells by transfection. Our study confirms that B19V permissiveness is restricted by the replication of its genome. Specifically, the B19V-nonpermissive 293 cells supported B19V DNA replication only when infected with adenovirus or transfected with the necessary adenovirus genes, whereas the replication of B19V DNA in semipermissive UT7/Epo-S1 cells was facilitated by adenovirus infection. Our results suggest that the replication of B19V DNA is unique in that specific cellular factors or microenvironments may be required for it to occur.
MATERIALS AND METHODS
Cell lines and primary cells.
293 (ATCC CRL-10852), HepG2 (ATCC HB-8065), and A549 cells (ATCC CCL-185) were maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal calf serum (FCS) at 37°C in 5% CO2. Immortalized human microvascular endothelial cells (HMEC-1) were obtained from the CDC (Atlanta, GA) and cultured in DMEM with 10% FCS, hydrocortisone (1 μg/ml), and mouse epidermal growth factor (10 ng/ml) (Invitrogen). UT7/Epo-S1 cells were cultured in DMEM with 10% FCS and 2 units/ml of Epo (Epogen; Amgen, Thousand Oaks, CA) at 37°C in 5% CO2. The process of generating of primary human CD36+ EPCs has been described elsewhere (19, 50); large numbers of CD36+ EPCs, at day 8 of ex vivo culture, were used for B19V infection.
Transfection.
293, HepG2, A549, and HMEC-1 cells grown in 60-mm dishes were transfected with 2 μg of excised B19V DNA fragments or plasmids, as described in Results and indicated in each figure; the Lipofectamine and Plus reagents (Invitrogen) were used as previously described (35). For some of the transfection experiments, 293 cells were cotransfected with 2 μg of pHelper plasmid (Stratagene), which contains the adenovirus 5 (Ad5) E2a, E4orf6, and VA genes.
UT7/Epo-S1 cells were transfected with 2 μg of DNA per 2 × 106 cells with Nucleofector (Amaxa, MD) using a universal reagent (DNAproject, WA) and the program X005. After transfection, the cells were maintained at 2 × 105 cells/ml.
Virus and infection.
293 cells transfected with the B19V DNA together with pHelper plasmid, or UT7/Epo-S1 cells transfected with B19V DNA, were harvested 3 days after transfection. After the cells were frozen and thawed three times, the supernatant was collected. A total of 100 μl of the supernatant was incubated with 2 × 105 CD36+ EPCs in a volume of 250 μl DMEM with a slow rotation (15 rpm) at 4°C for 2 h. The infected cells were washed twice with DMEM and were cultured at a concentration of 2 × 105 cells/ml at 37°C with 5% CO2. mRNA was directly isolated from the cells 3 days later using the TurboCapture 8 mRNA kit (Qiagen) according to the manufacturer's instructions.
In some experiments, the 293 and UT7/Epo-S1 cells were infected with Ad5 at a multiplicity of infection of 5, as previously described (35).
Plasmids.
The infectious B19V clones (pB19-4244 and pB19-M20), the NS1 knockout mutant pB19-M20NS(−), the VP1 knockout mutant pB19-M20VP1(−), and pB19-N8, which has half of the left ITR (L-ITR; nucleotides [nt] 199 to 383) and half of the right ITR (R-ITR; nt 5214 to 5410), were described previously (53, 54). pB19-M20, pB19-4244, pB19-M20NS(−), and pB19-M20VP1(−) were digested with SalI, whereas pB19-N8 was digested with XhoI/EcoRI, to recover the excised B19V DNA fragments that were used for transfection. All five of these B19V DNA fragments are schematically depicted in Fig. 1C. Plasmids expressing Ad5 E2a, E4orf6, and VA RNA were gifts from Tom Shenk (Princeton University). The nucleotide numbering referred to here for the B19V genome is that of the J35 isolate of the B19V genome (GenBank accession no. AY386330). The nucleotide numbering of the lambda (λ) DNA refers to GenBank accession no. J02459.
(i) NSCap-based plasmids.
p1/2TRNSCap, pNSCap1/2TR, and pNSCap were generated by inserting the B19V sequences corresponding to nt 187 to 5213, nt 384 to 5410, and nt 384 to 5213 into pBluescript(+) (Stratagene), respectively. p1/2TRNSCap has only half of the B19V L-ITR (nt 199 to 383) at one end, pNSCap1/2TR has only half of the B19V R-ITR (nt 5214 to 5410) at one end, and pNSCap has no ITR. pNSCap1/2TRtrsKO1 was constructed by mutating a CAC within the putative trs (located at nt 5212 to 5214) of the parent plasmid pNSCap1/2TR to TGA; pNSCap1/2TRtrsKO2 was constructed by mutating the same CAC sequence to CGC. The XhoI/EcoRI-digested B19V DNA fragments from these five plasmids are schematically depicted in Fig. 5A. pNSCap5262, pNSCap5270, and pNSCap5280 were constructed by inserting the fragments nt 384 to 5262, nt 384 to 5270, and nt 384 to 5280 into pBluescript(+), respectively. pNSCap5280m18, pNSCap5280mNSBS1, and pNSCap5280mNSBS2 were constructed by mutating the sequences from nt 5263 to 5280, nt 5243 to 5260, and nt 5243 to 5254, respectively, to those shown in Fig. 5B. pNSCap5280J130, pNSCap5280J316, and pNSCap5280J500 were generated by inserting λ DNA sequences (nt 23301 to 23430, nt 23301 to 23616, and nt 23301 to 23800, respectively) after nt 5280 of the B19V sequence in the parent plasmid pNSCap5280. The XhoI/EcoRI-digested B19V DNA fragments of these four plasmids are schematically depicted in Fig. 7.
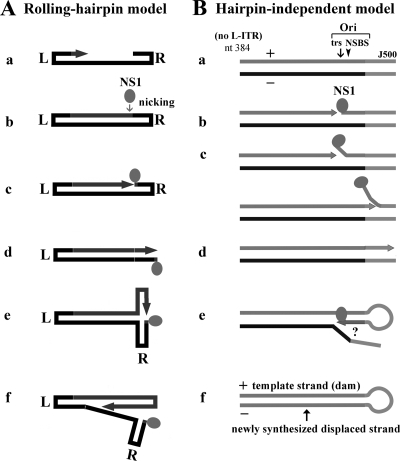
FIG. 5.
Identification of the trs and the NS1 binding site within the B19V R-ITR. Hirt DNA samples were extracted from intact cells. Southern blotting and testing for DNA replication, as described in the legend to Fig. 3, were carried out on additional excised B19V DNA fragments. (A) NSCap and four derivatives were tested to identify the trs. In one derivative, half of the L-ITR (1/2TRNSCap) is included; in a second, half of the R-ITR (NSCap1/2TR) is included. In both the third and fourth NSCap1/2TR-derived constructs, nucleotides of the putative trs are mutated. (B) Six additional mutants affecting nucleotides in the R-ITR were tested to identify the NS1 binding elements (NSBE) of the NS1 binding site. Three constructs are simply truncations. In the others, which harbor mutations in addition to being truncated at nt 5280, the mutated nucleotides are indicated as small characters and underlined.
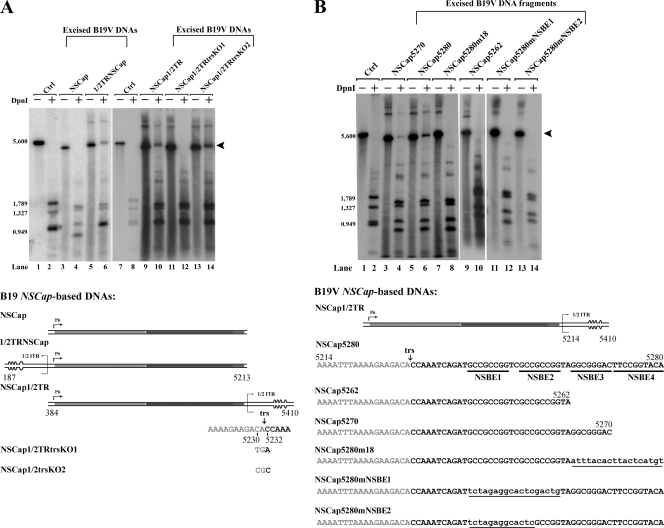
FIG. 7.
Hairpin-independent replication of B19V DNA. Hirt DNA samples were extracted from intact cells. Southern blotting and testing for DNA replication, as described in the legend to Fig. 3, were carried out on excised B19V NSCap5280 DNA and three variants, in which heterologous prokaryotic fragments of various lengths were fused to the parent DNA, as illustrated to the right. The arrowhead indicates the DpnI-resistant DNA band in DpnI-digested samples.
(ii) NS-based plasmids.
The parent pNS1 construct, which contains only the NS1 expression cassette, was constructed by inserting the sequence from nt 384 to 2961 into pBluescript(+). pNS1Ori5120-5280 was generated by inserting nt 5120 to 5280 after the NS1 expression cassette. pNS1Ori5120-5280J500 was made by inserting the λ DNA sequence (nt 23301 to 23800) after nt 5280 of the B19V sequence. The B19V sequence (nt 5120 to 5280) and the λ DNA sequence (nt 23301 to 23800) were inserted in the reverse orientation at the 3′ end of the NS1 expression cassette to generate pNS1J500Ori(−)5280-5120. pNS1J700Ori5120-5280, pNS1J700Ori5175-5280, pNS1J700Ori5201-5280, pNS1J700Ori, and pNS1J700Δtrs were constructed by inserting the λ DNA sequence (nt 23301 to 24000) between the NS1 cassette and the B19V sequences, starting from nt 5120, 5175, 5201, 5214, or 5232 and extending to nt 5280, respectively. All nine NS-based plasmids are schematically depicted in Fig. 6.
FIG. 6.
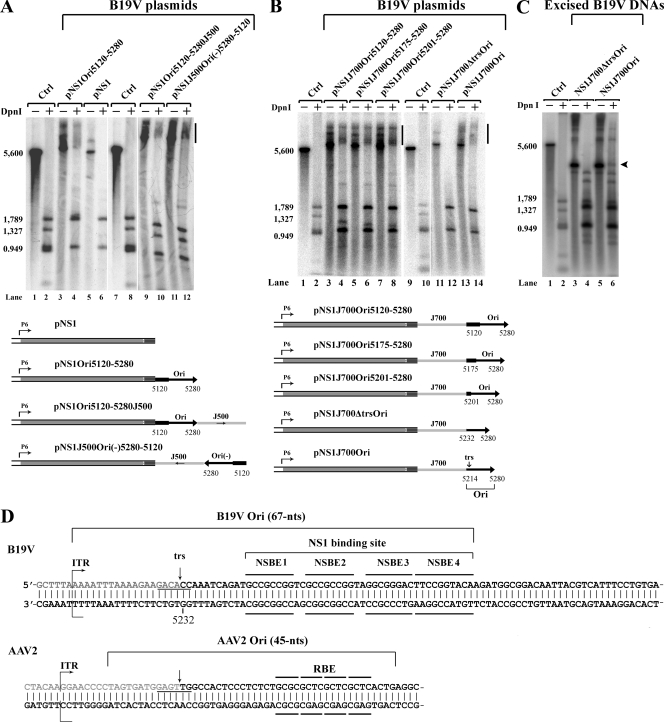
Identification of the B19V Ori. Hirt DNA samples were extracted from intact cells. Southern blotting and testing for DNA replication, as described in the legend to Fig. 3, was carried out on B19V DNA plasmids (A, B) and excised B19V DNA fragments (C). (A, B) The B19V plasmids schematically depicted at the bottoms of the panels were tested. The locations, positions, and lengths of the putative Ori are indicated, as are the lengths and orientations of the prokaryotic sequences that were inserted as described in Materials and Methods. Bars indicate smears of the DpnI-resistant DNA band in DpnI-digested samples. (C) Two B19V DNA fragments were excised from plasmids pNS1J700Ori and pNS1J700ΔtrsOri through XhoI/EcoRI digestion, respectively, and tested for DpnI-resistant DNA. The arrowhead indicates the DpnI-resistant DNA band in DpnI-digested samples. (D) Structure comparison of the Ori between B19V and AAV2. The nucleotide sequences of the region spanning the B19V and AAV2 Ori, as indicated, are shown, and the identified trs and NS1 binding elements (NSBE) that comprise the NS1 binding site are indicated. Ctrl, control; RBE, Rep binding element.
Isolation of low-molecular-weight (Hirt) DNA and Southern blot analysis.
Two days after transfection, the cells were collected and washed twice with phosphate-buffered saline (PBS). In some experiments, pure nuclei were isolated using a nucleus isolation kit (Sigma). The washed cells or the isolated nuclei were lysed in Hirt solution (10 mM Tris, 10 mM EDTA [pH 7.5], 0.6% sodium dodecyl sulfate) for 10 min. NaCl was then added to a concentration of 5 M, and the lysates were incubated overnight at 4°C. After centrifugation at 15,000 rpm for 15 min at 4°C, the supernatant was collected and mixed with 3 volumes of QG buffer (Qiagen). DNA was collected by binding to a DNA miniprep spin column (Zymo, Orange, CA) and eluted with Tris-EDTA buffer. The Hirt DNA samples were digested with EcoRI (control) or DpnI at a final concentration of 1.5 units/μl overnight at 37°C, whereas the Hirt DNA samples extracted from B19V plasmid-transfected cells were digested with DpnI at the same concentration as above and mock digestion was used as a control (Dpn−). EcoRI/mock-digested and DpnI-digested samples were run on a 1% agarose gel. Southern blotting was performed essentially as described previously (33), and the blots were hybridized with the B19V NSCap probe (nt 199 to 5410). They were then either exposed to X-ray film or processed using a GE phosphorimager and Quant TL software (GE Healthcare) for quantification.
Immunofluorescence.
293, HepG2, A549, and HMEC-1 cells were harvested 2 days after transfection and fixed on multiple-spot slides (Thermo) in cold acetone for 20 min. For the suspension cultures, UT7/Epo-S1 cells were cytocentrifuged at 1,500 rpm for 5 min and fixed in a mixture of acetone and methanol (1:1) at −20°C for 15 min. The slides were air dried, washed twice in PBS, and then incubated with an antiserum against glutathione S-transferase-fused amino acids 1 to 181 of the B19V NS1 at a dilution of 1:100 in PBS with 10% FCS for 1 h at 37°C. This was followed by incubation with a fluorescein isothiocyanate-conjugated secondary antibody. Evans blue dye was used for counterstaining. The slides were observed under a Nikon confocal microscope.
Real-time RT-PCR.
A multiplex real-time reverse transcription-PCR (RT-PCR) system was developed to detect B19V-specific mRNAs, with β-actin mRNA serving as an internal control. Isolated mRNA from infected cells was reverse transcribed to cDNA using random hexamers (Promega) and Moloney murine leukemia virus RT (Invitrogen), following the manufacturer's instructions but scaling the reaction volume to 50 μl. Then, the cDNA copy number was determined by real-time PCR as described previously (19), using TaqMan universal PCR master mix (Takara).
The forward and reverse primers and probes used in the real-time PCR for cDNAs converted from the respective B19V mRNAs were as follows: for VP2-encoding mRNA, 5′-GACCAGTTCAGGAGAATCAT-3′ (nt 2256 to 2275), 5′-TTCTGAGGCGTTGTAAGC-3′ (nt 3292 to 3275), and 5′-Cy5-TCGTGAACT/GTGCAGCTGCCCCTGTG-BHQ-3′ (splice donor site D2/acceptor site A2-1); for 11-kDa-encoding mRNA, 5′-GAAGCCTTCTACACACCTTTGG-3′ (nt 2323 to 2344), 5′-TGGCTGTCCACAATTCTTCAGG-3′ (nt 4946 to 4925), and 5′-FAM-AGACCAGTTTCGTGAACT/CTACAGATGCA-BHQ-3′ (D2/A2-2); for D1/A1-1 mRNA, 5′-TCTTGCTGTTTTTTGTGAGCTAACTAA-3′ (nt 557 to 583), 5′-TGATACTGGTGTCTGTGACAATTGG-3′ (nt 2143 to 2119), and 5′-FAM-AG/ATGCCCT CCACCCAGACCTCCA-BHQ-3′ (D1/A1-1). All the splice donor (D1 and D2) and acceptor (A1-1, A2-1, and A2-2) sites were described previously (19).
cDNA copies of β-actin mRNA were quantified using the forward and reverse primers and probe as follows: 5′-GGCACCCAGCACAATGAAG-3′, 5′-GCCGATCCACACGGAGTACT-3′, and 5′-JOE-TCAAGATCATTGCTCCTCCTGAGCGC-BHQ1-3′ (21). Standard curves for β-actin and three B19V cDNAs were generated from serial dilutions of a β-actin cDNA plasmid and three B19V cDNA plasmids, respectively (50). The copy numbers of all three of these B19V mRNAs are presented as copies per μl of the cDNA reaction mixture and were normalized with respect to the copy number of the β-actin mRNA produced in the same reaction.
RESULTS
Replication of B19V DNA in 293 cells is facilitated by the presence of an adenovirus gene set.
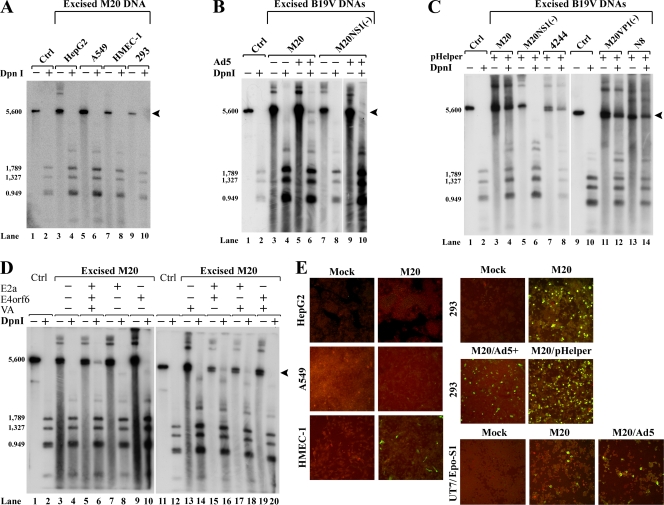
Using the RF of B19V DNA, which was excised from the pB19-M20 infectious clone and designated M20 (Fig. 1C), we were able to directly examine the replication of viral DNA in nonpermissive cells. We first transfected the M20 DNA into HepG2, A549, HMEC-1, and 293 cells. Hirt preparations of viral DNA were digested with either EcoRI (DpnI−; resulting in a 5.6-kb fragment as a control for DpnI digestion) or DpnI (DpnI+) and were then subjected to Southern blotting. The aim was to identify newly replicated DNA, which is resistant to DpnI digestion due to its cleavage at the dam site only when both adenine residues are methylated (49) (both hemimethylated and unmethylated DNA products are resistant to DpnI digestion). This analysis revealed, to our surprise, that none of these transfected cell lines harbor nascent M20 DNA (the absence of DpnI-resistant bands in Fig. 3A, lanes 4, 6, 8, and 10). Since NS1 is the only nonstructural protein known to be essential for B19V replication (53), we next attempted to detect NS1 expression in these transfected cells, using a rat antiserum against 181 amino acids at the NS1 N terminus (the antiserum generation is described in reference 41; A. Y. Chen, et al., unpublished data). Interestingly, in the M20-transfected HepG2 and A549 cells, we were unable to detect NS1 expression by immunofluorescence (Fig. 3E), in spite of the fact that the M20 DNA was present in the nuclei (Fig. 3A, lanes 3 and 5). These findings suggested that P6 promoter-based transcription is limited in these cell types. Moreover, although NS1 was expressed in both HMEC-1 and 293 cells (Fig. 3E), the transfection of the M20 DNA did not lead to the generation of DpnI-resistant bands (Fig. 3A, lanes 8 and 10), indicating that the replication of the B19V RF DNA is not supported in these cells. Notably, when 293 cells infected with Ad5 were tested, a weak but significant DpnI-resistant band was detected (Fig. 3B, lane 6). Furthermore, the transfection of the NS1 knockout RF DNA [M20NS1(−) (Fig. 1C)], which was used as a control, did not result in a DpnI-resistant band, regardless of whether or not the cells had been infected with Ad5 (Fig. 3B, lanes 8 and 10). These results confirm that the DpnI-resistant band produced in 293 cells infected with Ad5 represents newly synthesized B19V DNA.
FIG. 3.
Adenovirus infection and transfection with adenovirus gene products support B19V DNA replication in 293 cells. (A to D) Various cell types were transfected with excised B19V DNAs. Hirt DNA samples were extracted either from isolated nuclei (A) or from intact cells (B to D) and were used to test for DNA replication by Southern blotting using the B19V NSCap probe, with DpnI digestion (DpnI+) versus EcoRI digestion (DpnI−), revealing the levels of newly replicated and total input B19V DNA, respectively. In each experiment, DpnI− and DpnI+ M20 DNA (6 ng) were run in lanes 1 and 2, respectively, to serve as controls for DpnI digestion and as DNA size markers. Lanes 9 and 10 in panel C and lanes 11 and 12 in panel D are the same controls but for the respective blots. Arrowheads indicate DpnI-resistant DNA bands in DpnI-digested samples. (A) Infectious M20 DNA was transfected into HepG2, A549, HMEC-1, and 293 cells. (B) Both M20 and noninfectious M20NS1(−) DNA were transfected into 293 cells with or without Ad5 infection (Ad5+ or Ad5−). (C) A variety of infectious B19V DNAs [M20, M20VP1(−), or 4244] or noninfectious B19V DNAs [M20NS1(−) and N8] were transfected into 293 cells, either alone (pHelper−) or with pHelper (pHelper+), as indicated. (D) M20 was transfected into 293 cells along with different combinations of three adenovirus genes (E2a, E4orf6, and VA RNA) as indicated. (E) M20 DNA was transfected into HepG2, A549, HMEC-1, 293, or UT7/Epo-S1 cells, which were subsequently examined for NS1 expression. In the case of the 293 cells, cotransfection with pHelper was also tested, as was transfection into cells infected with Ad5 (Ad5+), as indicated. UT7/Epo-S1 cells also were tested in the presence or absence of Ad5 infection as indicated. NS1 expression was detected by immunofluorescence, using a polyclonal antibody against NS1 and a fluorescein isothiocyanate-conjugated secondary antibody (green); the cells were counterstained with Evans blue (red). The confocal images were taken at ×20 magnification with an Eclipse C1 Plus confocal microscope (Nikon). Ctrl, control.
Adenovirus is a native helper virus for AAV2 replication, and only five adenovirus genes, E1a, E1b55K, E2a, E4orf6, and VA RNA, are required for this helper function (18). We tested whether these five adenovirus genes are sufficient to support B19V DNA replication. When pHelper, a plasmid that contains the Ad5 E2a, E4orf6, and VA RNA genes, was cotransfected with M20 DNA, the latter replicated efficiently, as revealed by the presence of a strong DpnI-resistant band by Southern blotting (Fig. 3C, lane 4). In contrast, when pHelper was cotransfected with NS1 knockout RF DNA [M20NS(−)], DpnI-resistant DNA was not produced (Fig. 3C, lane 6), indicating that NS1 and three Ad5 helper genes from pHelper together confer the ability to synthesize B19V DNA in 293 cells, which themselves express the Ad5 E1a and E1b genes. The transfection of other B19V DNAs containing full-length ITRs [i.e., M20VP1(−) and 4244 RF DNA] also led to the production of DpnI-resistant bands at levels equivalent to those produced from M20 RF DNA (Fig. 3C, lanes 8 and 12). Surprisingly, the transfection of B19V N8 DNA, which lacks half of each ITR (Fig. 1C), produced significant quantities of DpnI-resistant DNA (Fig. 3C, lane 14), indicating that B19V DNA replication does not require the full-length ITRs in pHelper-transfected 293 cells.
We next examined the abilities of individual Ad5 genes to support the replication of M20 RF DNA. As shown in Fig. 3D, the transfection of the E2a, E4orf6, or VA RNA gene alone did not appear to significantly enhance DNA replication. Likewise, the transfection of combinations of two of the genes led to only little (Fig. 3D, lanes 16 and 18) or no (Fig. 3D, lane 20) replication. However, when all three genes were cotransfected, the replication of M20 DNA reached a higher level (Fig. 3D, lane 6). Although we detected less replicated DNA relative to total DNA when we performed cotransfections with the three individual genes (Fig. 3C, lane 6) than with pHelper (Fig. 3C, lane 4), this difference is likely explained by the relatively low efficiency of transfection with three independent helper plasmids versus with one. Our results thus suggested that at least the three Ad5 genes in pHelper (E2a, E4orf6, and VA RNA) are required for replication and that their products function in concert with the E1a and E1b gene products that are already expressed in 293 cells.
NS1 protein was confirmed to be expressed at similar levels under each of the above-described transfection conditions, with the exception of the cells into which the NS1 knockout DNA [M20NS1(−)] was transfected (data not shown). Representative immunofluorescence images showing staining with the anti-NS1 antiserum are shown for 293 cells under various conditions (Fig. 3E).
Taken together, our results demonstrate that the B19V genome resembles that of the Dependovirus genome in that it is supported by the products of adenovirus genes, i.e., Ad5 E1a, E1b, E2a, E4orf6, and VA RNA.
Progeny virus production in pHelper-transfected 293 cells versus the semipermissive UT7/Epo-S1 cells.
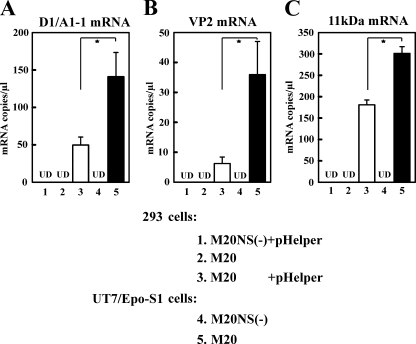
We next set out to determine whether B19V DNA replication in pHelper-transfected 293 cells produces infectious progeny virus. To this end, a lysate generated from M20/pHelper-cotransfected 293 cells was used to infect B19V-permissive CD36+ EPCs. A lysate generated from UT7/Epo-S1 cells transfected with M20 DNA served as a control. The presence of productive virus was established based on the infectivity of the CD36+ EPCs and was quantified by real-time RT-PCR analysis of spliced viral mRNAs produced in these cells. The infectivity of progeny virus obtained from the lysates of M20/pHelper-cotransfected 293 cells was comparable to that generated in the lysates of M20-transfected UT7/Epo-S1 cells; however, infectivity was significantly higher when cell lysates from M20-transfected UT7/Epo-S1 cells were tested (Fig. 4A to C, bar 5 versus bar 3). More specifically, in the 293 cells cotransfected with M20 and pHelper, the 11-kDa-encoding mRNA, mRNAs spliced from the D1 donor to A1-1 acceptor sites, and the VP2-encoding mRNA were expressed at levels of approximately 2 × 105, 5 × 104, and 1 × 104 copies per ml, respectively. In the absence of pHelper, the transfection of 293 cells with the M20 DNA did not result in detectable infectivity (Fig. 4A to C, bar 2). Similarly, the cotransfection of pHelper with M20NS1(−) DNA failed to generate productive virus (Fig. 4A to C, bar 1), as did the transfection of M20NS1(−) DNA into UT7/Epo-S1 cells (Fig. 4A to C, bar 4). Collectively, these results indicated that the infectivity of the cell lysate generated from M20/pHelper-cotransfected 293 cells resulted specifically from the support function of the set of Ad5 gene products in pHelper-transfected 293 cells.
FIG. 4.
Cotransfection of B19V RF M20 DNA into 293 cells with pHelper generates B19V progeny virus at a level comparable to that produced in UT7/Epo-S1 cells. 293 and UT7/Epo-S1 cells transfected with B19V DNAs were tested for the efficiency of virus production, based on the ability of lysates to stimulate virus mRNA production in CD36+ EPCs. Transfections were carried out on 2 × 106 cells per sample, using B19V infectious RF M20 DNA or noninfectious M20NS(−) DNA; transfections in 293 cells were carried out with or without the cotransfection of pHelper. Cell lysates prepared at 3 days posttransfection were used to infect CD36+ EPCs, and the infectivity of these cell lysates was quantified by a real-time RT-PCR strategy that detected specifically D1/A1-1 (A), VP2 (B), and 11-kDa-encoding mRNAs (C) in the CD36+ EPC lysates. In each sample, the absolute number of mRNA copies was normalized to the level of β-actin mRNA (104 copies per μl). Results shown represent the averages and standard deviations of data from at least three independent experiments. UD stands for undetectable. A single star indicates a P value of <0.05, as assessed based on the Student's t test.
Identification of the trs and the NS1 binding site on the B19V R-ITR.
The high transfection efficiency of the pHelper-transfected 293 cell system provided us, for the first time, with a physiologically relevant system in which to investigate which cis-elements are required for B19V DNA replication. Our analysis focused on the B19V R-ITR. Given that transfecting B19V N8 DNA, which lacks half of the L-ITR and half of the R-ITR, generated the DpnI-resistant DNA band (Fig. 3C, lane 14), we worked with various excised B19V DNAs that contain only the NSCap gene, NSCap plus half of the L-ITR (1/2TRNSCap), or NSCap plus half of the R-ITR (NSCap1/2TR). Both of the half-ITR-containing B19V DNAs generated DpnI-resistant bands (Fig. 5A, lanes 6 and 10) and thus were competent to replicate, but the NSCap DNA was not (Fig. 5A, lane 4). However, the relative levels of replication in response to transfection with each half-ITR-containing B19V DNA were significantly lower than those in response to transfection with B19V DNAs that include complete replication elements at both ends, i.e., M20, N8, M20VP1(−), and 4244 DNAs (Fig. 3C, lanes 4, 8, 12, and 14). For this reason, we decided to dissect the replication elements on the R-ITR next.
The trs on the B19V R-ITR had previously been predicted (12). To confirm the functionality of this trs experimentally, we first mutated its sequence (nt 5230 to 5232) within the NSCap1/2TR DNA, as shown in the diagram in Fig. 5A. The mutation of all three nucleotides of this site (CAC→TGA) abolished the DpnI-resistant band completely (Fig. 5A, lane 12). Notably, the mutation of only the nucleotide at which cleavage occurs (A→G) did not affect replication, as indicated by the presence of the DpnI-resistant band (Fig. 5A, lane 14). Thus, our results confirmed the putative trs at nt 5230 to 5232 within the R-ITR and suggested that the cleaved nucleotide per se can vary in this trs.
Next, we carried out a series of mutations and deletions of the sequences beyond the trs on the B19V DNA of NSCap1/2TR. It had previously been postulated that the NS1 binding site comprises two copies of the GC-rich elements (5′GCCGCCGG3′) (12). Surprisingly, transfecting the NSCap5262 DNA, which contains both the trs and this putative NS1 binding site, did not support replication (Fig. 5B, lane 10), suggesting that this putative NS1 binding site is not sufficient to confer the ability to replicate. We found that the NS1 binding site extends beyond the two GC-rich elements present in the NSCap5262 DNA; transfecting NSCap5270 DNA, which contains additional sequences encompassing another GC-rich element (5′GGCGGGAC3′), led to DNA replication, as evidenced by the DpnI-resistant band (Fig. 5B, lane 4). However, even this DNA was less effective than NSCap1/2TR, whereas the NSCap5280 DNA supported DNA replication at a level comparable to that observed when NSCap1/2TR was used (Fig. 5, compare the ratio for lane 6 to lane 5 in panel B with that for lane 10 to lane 9 in panel A). The mutation of the B19V sequence between nt 5262 and 5280 (NSCap5280m18), and also mutations affecting the two GC-rich elements (NSCap5280mNSBE1 and NSCap5280mNSBE2), abolished DNA replication from the R-ITR (Fig. 5B, lanes 8, 12, and 14, respectively). In all cases of 293 cell transfection, NS1 was conformed to be consistently expressed (data not shown).
These results reveal that the NS1 binding site is required for DNA replication from an origin on the R-ITR. Two GC-rich elements (5′GCCGCCGG3′) and one degenerative GC-rich element (5′GGCGGGAC3′) comprise the minimal NS1 binding site, and a fourth motif (5′TTCCGGTACA3′) is required to support maximal replication.
Identification of the B19V Ori and the discovery of hairpin-independent replication.
Next, we used a plasmid transfection strategy to identify the B19V minimal origin of replication (Ori). The transfection of the pNS1 plasmid, in which the capsid-encoding region and both ITRs are deleted, did not result in the production of DpnI-resistant DNA (Fig. 6A, lane 6), although NS1 expression was detected by immunofluorescence (data not shown). However, when any plasmid bearing a short fragment encompassing nt 5120 to 5280 was ligated to pNS1 [pNS1Ori5120-5280, pNS1Ori5120-5280J500 and pNS1J500Ori(−)5280-5120], replication occurred regardless of the position and orientation of that fragment, as evidenced by a smear of DpnI-resistant DNA bands (Fig. 6A, lanes 4, 10, and 12, respectively). This suggests that the B19V DNA fragment of nt 5120 to 5280 contains the B19V Ori. Since the input DNAs were circular supercoiled plasmids that contain only one B19V Ori on the right side, the sizes of the replicated DNA varied, accounting for the smearing of DpnI-resistant DNA on Southern blots (Fig. 6A and B). By truncating the sequence of this DNA in pNS1J700Ori, we were able to delimit the Ori to a DNA fragment encompassing nt 5214 to 5280 (Fig. 6B, lane 14). As expected, when the fragment was further shortened, deleting the trs, replication was completely abolished (Fig. 6B, lane 12). To confirm that replication resulted from the transfection of the Ori-containing plasmids, we transfected an excised DNA fragment (NS1J700Ori) containing the NS1 expression cassette and only the Ori. The transfection of this DNA produced a DpnI-resistant DNA band (Fig. 6C, lane 6), whereas the transfection of the control excised DNA without the trs (NS1J700ΔtrsOri) did not (Fig. 6C, lane 4). This supports the idea that we have identified the B19V Ori, which is composed of only 67 nt (nt 5214 to 5280) and essentially contains only the trs and the NS1 binding site (Fig. 6D).
Among parvoviruses, only Erythrovirus and Dependovirus have identical ITRs at each end (12), which suggests that they might share a common mechanism of replication. The results from the in vitro replication assays with AAV2 have suggested that when the Rep78/Rep68 proteins are involved in replication, only the Ori (containing the trs and the Rep binding element) is required for replication in vitro and the hairpin structure is not essential (20, 26, 43). Since we observed that a B19V DNA fragment containing the Ori in the absence of other ITR DNA was sufficient to produce DpnI-resistant DNA, we fused heterologous prokaryotic DNAs of various lengths to the Ori. The transfection of these B19V DNA chimeras fully supported DNA replication from this Ori (Fig. 7, lanes 6, 8, and 10, respectively). Since there is no cryptic replication origin in the NSCap DNA (Fig. 5, lane 4), these results strongly suggest that when NS1 is involved in replication, an Ori comprising only the trs and an NS1 binding site in cis is sufficient to turn the direction of replication for second-strand replacement synthesis and that a hairpin structure is not involved in this process (Fig. 2Be). Even when NSCap5280J500, which carries a 500-nt extension of heterologous prokaryotic DNA, was transfected, newly synthesized DNA was resistant to DpnI digestion (Fig. 7, lane 10). NS1 expression was confirmed in all of these transfections into 293 cells (data not shown).
Our findings suggest that NS1, rather than the base-paired hairpin, controls the direction of B19V DNA synthesis in our transfection system.
B19V DNA replication in UT7/Epo-S1 cells is facilitated by Ad5 infection.
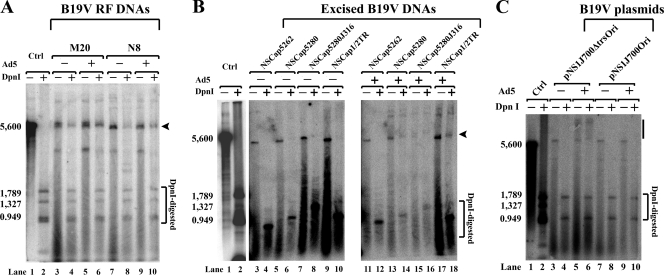
Having demonstrated the ability of adenovirus genes to facilitate the replication of B19V DNA in a nonpermissive cell line, we also studied the ability of adenovirus to support B19V DNA replication in the semipermissive cell line UT7/Epo-S1. The B19V infectious M20 RF DNA and replication-competent N8 DNA were each transfected into UT7/Epo-S1 cells with or without Ad5 infection, respectively. In cells infected with Ad5, there was a slight increase in the intensity of the DpnI-resistant DNA bands relative to these seen in uninfected cells (Fig. 8A, compare lane 6 with 4 and lane 10 with 8, respectively), suggesting that Ad5 enhances B19V DNA replication in UT7/Epo-S1 cells.
FIG. 8.
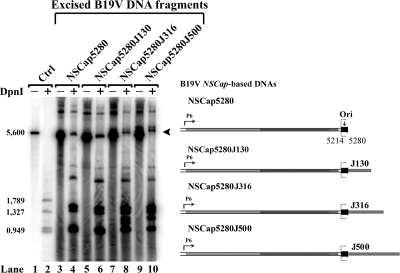
Adenovirus facilitates B19V DNA replication in UT7/Epo-S1 cells. Hirt DNA samples were extracted from intact cells. Southern blotting and testing for DNA replication, as described in the legend to Fig. 3, were carried out on excised B19V DNA fragments (A, B) and B19V DNA plasmids (C) transfected into the B19V-semipermissive UT7/Epo-S1 cells. As indicated, six B19V DNAs (A, B) and two B19V plasmids (C) were tested in either Ad5-infected (+) or uninfected (−) cells. Arrowheads in panels A and B and the bar in panel C identify the DpnI-resistant DNA band in DpnI-digested samples.
We next tested the physiological relevance of the identified B19V Ori in UT7/Epo-S1 cells. Representative B19V DNAs that had been used to transfect 293 cells were now transfected into UT7/Epo-S1 cells in the presence or absence of Ad5. The transfection of NSCap5262, which has an incomplete NS1 binding site, failed to support replication in either the infected or uninfected cells, as illustrated by the absence of DpnI-resistant bands (Fig. 8B, lanes 4 and 12). As in the pHelper-transfected 293 cells, the Ori enabled replication by a mechanism that is independent of a hairpin structure but was very weak (Fig. 8B, lanes 6, 8, and 10). Replication was supported when a heterologous prokaryotic DNA extension of 316 nt (NSCap5280J316) was present (Fig. 8B, lane 8). However, in all cases, the DpnI-resistant bands were more intense when the DNA was transfected into Ad5-infected UT7/Epo-S1 cells (Fig. 8B, lanes 14, 16, and 18). Moreover, we observed that replication was supported in the Ori-containing pNS1J700Ori plasmid in the presence of Ad5 (Fig. 8C, compare lane 6 with 4), whereas it was not supported in the control plasmid without an intact Ori (pNS1J700ΔtrsOri) (Fig. 8C, lanes 8 and 10). The fact that the input DNA in Fig. 8C was a circular supercoiled plasmid means that the sizes of the replicated DNA varied, which accounts for the smearing of DpnI-resistant DNA. The transfection of the excised NS1J700Ori (containing the Ori) into Ad5-infected UT7/Epo-S1 cells also produced a clear DpnI-resistant DNA band (data not shown). NS1 was confirmed to be consistently expressed in all transfections of UT7/Epo-S1 cells; representative immunofluorescence images of M20-transfected UT7/Epo-S1 cells with or without Ad5 infection are shown in Fig. 3E.
Taken together, these results from the transfection of UT7/Epo-S1 cells indicate that the replication of B19V DNA in UT7/Epo-S1 cells is facilitated by Ad5 infection. Furthermore, they reveal that the B19V Ori supports the replication of both excised B19V DNA fragments and B19V DNA-containing plasmids in UT7/Epo-S1 cells. This suggests that replication in UT7/Epo-S1 cells, at least with respect to NS1-mediated nicking and the self-priming of second-strand displacement synthesis, resembles that in pHelper-transfected 293 cells.
DISCUSSION
B19V DNA replication is restricted in B19V-nonpermissive cells.
In this study, we show that B19V DNA replication is limited in nonpermissive cells but that these cells become replication competent in response to either infection with adenovirus or transfection with a subset of adenovirus genes. We also have shown that in semipermissive cells, B19V DNA replication can be enhanced by adenovirus infection.
The replication of parvoviruses depends on the rapid growth of infected cells during the S phase. In the case of AAV2, an in vitro-reconstituted replication system has been developed, and it requires the following group of factors provided by the host cell: replication factor C, proliferating cell nuclear antigen (PCNA), polymerase δ, and the minichromosome maintenance complex (27, 28). In the case of the in vitro replication of the minute virus of mice (MVM), an overlapping set of factors—replication factor C, polymerase δ, PCNA, replication protein A, and parvovirus initiation factor—has been shown to be involved (9). It is generally accepted that all of the host-cell factors required for parvovirus DNA replication exist in all mammalian cells, and even in insect cells (42), and these cellular factors are likely also used for B19V DNA replication.
B19V infection, however, is primarily restricted to highly differentiating and proliferating EPCs (29, 40). It occurs only in particular conditions that exist in EPCs, a small number of Epo-dependent myeloid cell lines such as UT7/Epo-S1 (22, 23), and—as we have shown here—nonpermissive cells with help from adenovirus genes. We hypothesize that the replication of B19V DNA either is inhibited in the cellular microenvironments of nonpermissive cell types (some of which support the replication of other parvovirus DNA) or requires special cellular factors that are abundantly expressed in EPCs and UT7/Epo-S1 cells. We have previously shown that the replication of the B19V genome is essential to generating sufficient capsid-encoding mRNAs to produce viable progeny virus after virus successfully enters the host cells (19). Thus, aside from the cellular receptors required for virus entry into cells, genome replication is a key determinant of B19V tropism.
Control of transcription from the P6 promoter.
Our results also support the previous observation that the B19V P6 promoter is cell type specific, transcribing actively in EPCs and myeloid cell lines (15, 44) but poorly in epithelial cell types other than 293 cells (15, 31). Previous studies suggested that HepG2 and HMEC-1 cells could be infected by B19V (14, 32). However, our data indicate that this infectivity is unlikely. In HepG2 cells transfected with infectious B19V RF DNA, NS1 expression was extremely low and DNA replication was not detectable. Although NS1 was expressed at a moderate level in transfected HMEC-1 cells, replicated B19V DNA was again not detectable. Our results suggest that both cell lines are likely nonpermissive to B19V infection. We have also attempted to transfect the B19V infectious DNA into other B19V-nonpermissive cells, e.g., HeLa cells, by adenovirus infection. Although we observed the rescue of B19V DNA replication in these cell lines (data not shown), this rescue was accompanied by a significant increase in NS1 expression in the presence of adenovirus, suggesting that the transactivation of the P6 promoter, as previously reported (31), may contribute to the rescue in this case. Nevertheless, the rescued replicated B19V DNA in HeLa cells could be partly due to the helper function of adenovirus, because no replicated DNA was detected in HeLa cells in the absence of adenovirus infection (data not shown). We did not observe a significant difference in the expression of NS1 in 293 cells and UT7/Epo-S1 cells, regardless of whether adenovirus gene products were present (Fig. 3E).
Identification of the trs and Ori.
The Ori identified in B19V is only 67 nt long and is composed of the NS1 binding site and the trs. As such, it is quite similar to the Ori of the Dependovirus AAV2 (Fig. 6D), which is 45 nt long and contains the Rep binding site and the trs (26, 37). It is shorter than the 125-nt OriR of MVM (10). The B19V Ori has a unique nicking site (5′GACA/CCA3′) and thus differs from the MVM nicking site (5′CTWW/TCA3′) (10), the goose parvovirus (GPV) trs (5′TGAG/TTG3′), and the AAV2 trs (5′GAGT/TGG3′) (37). Thus, each parvovirus appears to be designated to be nicked by the large nonstructural protein encoded by its own genome. At least three GC-rich elements are essential for B19V NS1 binding and subsequent nicking at the trs. Interestingly, a fourth binding motif (5′TTCCGGTACA3′) is absolutely required for maximal replication and could be involved in stabilizing the replication fork initiated by the NS1/Ori complex. However, all four of these motifs are clustered together at the NS1 binding site, as is the case for the Rep binding site in Dependovirus AAV2 (Fig. 6D) and GPV (34, 37, 45). Moreover, the B19V ITR with its Ori resembles the ITR of GPV in that both have an arrow-like hairpin structure (12). Our identification of the B19V Ori is important in that it serves as a starting point for future studies that will clarify which cellular factors interact with this Ori and are essential for B19V DNA replication.
Hairpin-independent replication.
In transfecting the excised B19V NS/NSCap-based B19V DNA fragments that contain the Ori, we always observed replicated DNA when NS1 was expressed. Since the left Ori is not present on these excised B19V DNA fragments, the replicated bands that were resistant to DpnI digestion presumably were hemimethylated DNAs (Fig. 2Bf) (49). In AAV2, the hairpinned ITR serves as the 3′ primer that provides a free 3′-OH for DNA synthesis, yet Rep-dependent initiation of replication can occur in the absence of a hairpin structure in vitro, in spite of the fact that the trs and Rep binding elements are still required (20, 26, 43). It has been reported that a hairpin with minimally 4 bp can be folded to prime DNA synthesis in AAV2 (46). In the case of these NS- and NSCap-based B19V DNA fragments or plasmids, hairpin structures of more than 3 bp were not present. Therefore, the B19V DNA fragment with the Ori is replicated in a hairpin-independent but NS1-dependent manner. We propose that, after NS1-mediated nicking takes place, NS1 can unwind blunt-ended B19V DNA by an unknown mechanism and redirect the last base-paired 3′ nucleotide to serve as the 3′ primer for strand displacement synthesis (Fig. 2Bd and Be), which results in the synthesis of hemimethylated DNAs (Fig. 2Bf). Perhaps, a “template-switching” nature of the unidirectional, NS1-initiated fork is involved in this hairpin-independent replication. The template-switching mechanism of DNA replication has been confirmed at the Ori of the porcine circovirus (7, 8) but less well documented in parvoviral DNA replication. For NS-based plasmids that only contain one Ori at their right end, replication may simply be initiated by NS1-mediated nicking, followed by strand extension with the last base-paired 3′ nucleotide serving as the 3′ primer. Alternatively, NS1 may redirect this 3′ primer to undergo strand displacement synthesis, which could result in the smearing of replicated DNA. In fact, in vitro, AAV2 Rep can unwind blunt-ended DNA substrates that contain a Rep binding site (55). In hairpin-independent replication with long 3′ ends after the Ori, e.g., 500 nt (Fig. 2Ba), we observed that the position of the DpnI-resistant bands was located slightly above that of the input DNA fragments (Fig. 7, lanes 6, 8, and 10). This might be because the replicated DNA contains a loop region of 75 to 250 nt of single-stranded DNA sequence, thus altering the conformation of these newly synthesized DNAs. In MVM, NS1 appears to be directly involved in refolding the resulting extended DNA at the right end to form a rabbit-ear structure (48). The mechanism whereby the B19V NS1 unwinds blunt-ended DNA and redirects the 3′ primer to strand displacement synthesis warrants further investigation in an in vitro replication system with lysates from B19V-permissive CD36+ EPCs.
While a hairpin structure is present in B19V RF DNA M20, M20VP1(−), and 4244 (Fig. 3C), the replication of these B19V DNAs was likely a consequence of NS1-mediated nicking, unwinding, and refolding of the hairpin structure, and thus, replication could proceed by a mechanism similar to that thought to take place for AAV2 replication, as illustrated in Fig. 2A.
Regulation of B19V DNA replication.
We have shown, based on progeny virus production, that transfection with pHelper facilitates the second step of B19V DNA replication (strand displacement synthesis) and the generation of single-stranded DNA in the normally nonpermissive 293 cell line. Although the transfected cells remained limited in the volume of virus progeny generated, the fact remains that pHelper made it possible for replication to occur at all. The helper function that the set of five Ad5 genes (three from pHelper and the others present endogenously in 293 cells) provides to B19V replication is certainly poor in comparison to its support of AAV2 replication and virus production (18). In the case of the semipermissive UT7/Epo-S1 cell line, we did not observe a significant increase in infectivity when comparing the results of transfecting B19V M20 DNA in the presence of adenovirus to those of transfecting cells that were not subjected to Ad5 infection (data not shown). Highly differentiating/proliferating EPCs supported B19V replication and amplified the copies of the B19V genome up to 1,000-fold (50) but without producing more progeny virus (W. Guan et al., unpublished data). These highly differentiating/proliferating cells never reproduced the replication efficiency and virus production seen in the BFU-E/CFU-E cells of the bone marrow of infected humans, indicating that B19V virus replication is controlled at multiple steps, e.g., the generation of single-stranded DNA and its packaging. B19V infection of humans normally produces viral titers as high as 1012 genomic copies per ml plasma (19, 50), indicating that B19V replication is stimulated exceptionally in the bone marrow. This is consistent with reports, based on in vitro B19V infection of primary EPCs in 1% O2 of hypoxia, that the hypoxic conditions in bone marrow contribute to the support of B19V replication (30).
The most important steps during B19V infection of humans are its initial propagation and its transport to human bone marrow, both of which remain poorly understood. The transmission of B19V is thought to occur via the respiratory tract (1), through blood transfusions (24, 51), and vertically from mother to fetus (6), but only transmission through the respiratory tract has been experimentally proven (1). Our observation that the replication of B19V DNA in nonpermissive cells can be supported by the presence of either adenovirus or certain of its gene products strongly supports the possibility that B19V infection may take place as an outcome of the coinfection of other viruses—such as adenovirus—within tissues of the respiratory tracts.
Acknowledgments
This work was supported by PHS grant RO1 AI070723 from NIAID and grant P20 RR016443 from the NCRR COBRE program.
We thank David Pintel for critical readings of the manuscript.
Footnotes
Published ahead of print on 8 July 2009.
REFERENCES
- 1.Anderson, M. J., P. G. Higgins, L. R. Davis, J. S. Willman, S. E. Jones, I. M. Kidd, J. R. Pattison, and D. A. Tyrrell. 1985. Experimental parvoviral infection in humans. J. Infect. Dis. 152:257-265. [DOI] [PubMed] [Google Scholar]
- 2.Astell, C. R., W. Luo, J. Brunstein, and J. Amand. 1997. B19 parvovirus: biochemical and molecular features, p. 16-41. In L. J. Anderson and N. Young (ed.), Human parvovirus B19. Karger, Basel, Switzerland.
- 3.Berns, K. I., and C. R. Parrish. 2007. Parvoviridae, p. 2437-2477. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed. Lippincott Williams, New York, NY.
- 4.Bloom, M. E., and N. Young. 2001. Parvoviruses, p. 2361-2379. In D. M. Knipe (ed.), Fields virology. Lippincott Williams and Wilkins, Philadelphia, PA.
- 5.Brown, K. E., and N. Young. 1997. Human parvovirus B19: pathogenesis of disease, p. 105-119. In L. J. Anderson and N. Young (ed.), Human parvovirus B19, vol. 20. Karger, Basel, Switzerland. [Google Scholar]
- 6.Brown, T., A. Anand, L. D. Ritchie, J. P. Clewley, and T. M. Reid. 1984. Intrauterine parvovirus infection associated with hydrops fetalis. Lancet ii:1033-1034. [DOI] [PubMed] [Google Scholar]
- 7.Cheung, A. K. 2004. Detection of template strand switching during initiation and termination of DNA replication of porcine circovirus. J. Virol. 78:4268-4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung, A. K. 2004. Palindrome regeneration by template strand-switching mechanism at the origin of DNA replication of porcine circovirus via the rolling-circle melting-pot replication model. J. Virol. 78:9016-9029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christensen, J., and P. Tattersall. 2002. Parvovirus initiator protein NS1 and RPA coordinate replication fork progression in a reconstituted DNA replication system. J. Virol. 76:6518-6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cotmore, S. F., J. Christensen, and P. Tattersall. 2000. Two widely spaced initiator binding sites create an HMG1-dependent parvovirus rolling-hairpin replication origin. J. Virol. 74:1332-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cotmore, S. F., and P. Tattersall. 1984. Characterization and molecular cloning of a human parvovirus genome. Science 226:1161-1165. [DOI] [PubMed] [Google Scholar]
- 12.Cotmore, S. F., and P. Tattersall. 2005. Structure and organization of the viral genome, p. 73-94. In J. Kerr, S. F. Cotmore, M. E. Bloom, R. M. Linden, and C. R. Parrish (ed.), Parvoviruses. Hodder Arnold, London, United Kingdom.
- 13.Cotmore, S. F., and P. Tattersall. 2005. A rolling-hairpin strategy: basic mechanisms of DNA replication in the parvoviruses, p. 171-181. In J. Kerr, S. F. Cotmore, M. E. Bloom, R. M. Linden, and C. R. Parrish (ed.), Parvoviruses. Hodder Arnold, London, United Kingdom.
- 14.Duechting, A., C. Tschope, H. Kaiser, T. Lamkemeyer, N. Tanaka, S. Aberle, F. Lang, J. Torresi, R. Kandolf, and C. T. Bock. 2008. Human parvovirus B19 NS1 protein modulates inflammatory signaling by activation of STAT3/PIAS3 in human endothelial cells. J. Virol. 82:7942-7952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ekman, A., K. Hokynar, L. Kakkola, K. Kantola, L. Hedman, H. Bonden, M. Gessner, C. Aberham, P. Norja, S. Miettinen, K. Hedman, and M. Soderlund-Venermo. 2007. Biological and immunological relations among human parvovirus B19 genotypes 1 to 3. J. Virol. 81:6927-6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freyssinier, J. M., C. Lecoq-Lafon, S. Amsellem, F. Picard, R. Ducrocq, P. Mayeux, C. Lacombe, and S. Fichelson. 1999. Purification, amplification and characterization of a population of human erythroid progenitors. Br. J. Haematol. 106:912-922. [DOI] [PubMed] [Google Scholar]
- 17.Gallinella, G., E. Manaresi, E. Zuffi, S. Venturoli, L. Bonsi, G. P. Bagnara, M. Musiani, and M. Zerbini. 2000. Different patterns of restriction to B19 parvovirus replication in human blast cell lines. Virology 278:361-367. [DOI] [PubMed] [Google Scholar]
- 18.Geoffroy, M. C., and A. Salvetti. 2005. Helper functions required for wild type and recombinant adeno-associated virus growth. Curr. Gene Ther. 5:265-271. [DOI] [PubMed] [Google Scholar]
- 19.Guan, W., F. Cheng, Y. Yoto, S. Kleiboeker, S. Wong, N. Zhi, D. J. Pintel, and J. Qiu. 2008. Block to the production of full-length B19 virus transcripts by internal polyadenylation is overcome by replication of the viral genome. J. Virol. 82:9951-9963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hong, G., P. Ward, and K. I. Berns. 1992. In vitro replication of adeno-associated virus DNA. Proc. Natl. Acad. Sci. USA 89:4673-4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kammula, U. S., K. H. Lee, A. I. Riker, E. Wang, G. A. Ohnmacht, S. A. Rosenberg, and F. M. Marincola. 1999. Functional analysis of antigen-specific T lymphocytes by serial measurement of gene expression in peripheral blood mononuclear cells and tumor specimens. J. Immunol. 163:6867-6875. [PubMed] [Google Scholar]
- 22.Miyagawa, E., T. Yoshida, H. Takahashi, K. Yamaguchi, T. Nagano, Y. Kiriyama, K. Okochi, and H. Sato. 1999. Infection of the erythroid cell line KU812Ep6 with human parvovirus B19 and its application to titration of B19 infectivity. J. Virol. Methods 83:45-54. [DOI] [PubMed] [Google Scholar]
- 23.Morita, E., A. Nakashima, H. Asao, H. Sato, and K. Sugamura. 2003. Human parvovirus B19 nonstructural protein (NS1) induces cell cycle arrest at G1 phase. J. Virol. 77:2915-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mortimer, P. P., N. L. Luban, J. F. Kelleher, and B. J. Cohen. 1983. Transmission of serum parvovirus-like virus by clotting-factor concentrates. Lancet ii:482-484. [DOI] [PubMed] [Google Scholar]
- 25.Munakata, Y., T. Saito-Ito, K. Kumura-Ishii, J. Huang, T. Kodera, T. Ishii, Y. Hirabayashi, Y. Koyanagi, and T. Sasaki. 2005. Ku80 autoantigen as a cellular coreceptor for human parvovirus B19 infection. Blood 106:3449-3456. [DOI] [PubMed] [Google Scholar]
- 26.Musatov, S., J. Roberts, D. Pfaff, and M. Kaplitt. 2002. A cis-acting element that directs circular adeno-associated virus replication and packaging. J. Virol. 76:12792-12802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nash, K., W. Chen, W. F. McDonald, X. Zhou, and N. Muzyczka. 2007. Purification of host cell enzymes involved in adeno-associated virus DNA replication. J. Virol. 81:5777-5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nash, K., W. Chen, and N. Muzyczka. 2008. Complete in vitro reconstitution of adeno-associated virus DNA replication requires the minichromosome maintenance complex proteins. J. Virol. 82:1458-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ozawa, K., G. Kurtzman, and N. Young. 1986. Replication of the B19 parvovirus in human bone marrow cell cultures. Science 233:883-886. [DOI] [PubMed] [Google Scholar]
- 30.Pillet, S., G. N. Le, T. Hofer, F. NguyenKhac, M. Koken, J. T. Aubin, S. Fichelson, M. Gassmann, and F. Morinet. 2004. Hypoxia enhances human B19 erythrovirus gene expression in primary erythroid cells. Virology 327:1-7. [DOI] [PubMed] [Google Scholar]
- 31.Ponnazhagan, S., M. J. Woody, X. S. Wang, S. Z. Zhou, and A. Srivastava. 1995. Transcriptional transactivation of parvovirus B19 promoters in nonpermissive human cells by adenovirus type 2. J. Virol. 69:8096-8101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poole, B. D., Y. V. Karetnyi, and S. J. Naides. 2004. Parvovirus B19-induced apoptosis of hepatocytes. J. Virol. 78:7775-7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qiu, J., F. Cheng, L. R. Burger, and D. Pintel. 2006. The transcription profile of Aleutian mink disease virus (AMDV) in CRFK cells is generated by alternative processing of pre-mRNAs produced from a single promoter. J. Virol. 80:654-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qiu, J., F. Cheng, Y. Yoto, Z. Zadori, and D. Pintel. 2005. The expression strategy of goose parvovirus exhibits features of both the Dependovirus and Parvovirus genera. J. Virol. 79:11035-11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qiu, J., and D. J. Pintel. 2002. The adeno-associated virus type 2 Rep protein regulates RNA processing via interaction with the transcription template. Mol. Cell. Biol. 22:3639-3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schowengerdt, K. O., J. Ni, S. W. Denfield, R. J. Gajarski, N. E. Bowles, G. Rosenthal, D. L. Kearney, J. K. Price, B. B. Rogers, G. M. Schauer, R. E. Chinnock, and J. A. Towbin. 1997. Association of parvovirus B19 genome in children with myocarditis and cardiac allograft rejection: diagnosis using the polymerase chain reaction. Circulation 96:3549-3554. [DOI] [PubMed] [Google Scholar]
- 37.Smith, D. H., P. Ward, and R. M. Linden. 1999. Comparative characterization of Rep proteins from the helper-dependent adeno-associated virus type 2 and the autonomous goose parvovirus. J. Virol. 73:2930-2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Snyder, R. O., R. J. Samulski, and N. Muzyczka. 1990. In vitro resolution of covalently joined AAV chromosome ends. Cell 60:105-113. [DOI] [PubMed] [Google Scholar]
- 39.Srivastava, A., E. Bruno, R. Briddell, R. Cooper, C. Srivastava, K. van Besien, and R. Hoffman. 1990. Parvovirus B19-induced perturbation of human megakaryocytopoiesis in vitro. Blood 76:1997-2004. [PubMed] [Google Scholar]
- 40.Srivastava, A., and L. Lu. 1988. Replication of B19 parvovirus in highly enriched hematopoietic progenitor cells from normal human bone marrow. J. Virol. 62:3059-3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun, Y., A. Y. Chen, F. Cheng, W. Guan, F. B. Johnson, and J. Qiu. 2009. Molecular characterization of infectious clones of the minute virus of canines reveals unique features of bocaviruses. J. Virol. 83:3956-3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Urabe, M., C. Ding, and R. M. Kotin. 2002. Insect cells as a factory to produce adeno-associated virus type 2 vectors. Hum. Gene Ther. 13:1935-1943. [DOI] [PubMed] [Google Scholar]
- 43.Urcelay, E., P. Ward, S. M. Wiener, B. Safer, and R. M. Kotin. 1995. Asymmetric replication in vitro from a human sequence element is dependent on adeno-associated virus Rep protein. J. Virol. 69:2038-2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, X. S., M. C. Yoder, S. Z. Zhou, and A. Srivastava. 1995. Parvovirus B19 promoter at map unit 6 confers autonomous replication competence and erythroid specificity to adeno-associated virus 2 in primary human hematopoietic progenitor cells. Proc. Natl. Acad. Sci. USA 92:12416-12420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ward, P. 2005. Replication of adeno-associated virus DNA, p. 189-211. In J. Kerr, S. F. Cotmore, M. E. Bloom, M. E. Linden, and C. R. Parrish (ed.), Parvoviruses. Hodder Arnold, London, United Kingdom.
- 46.Ward, P., and K. I. Berns. 1995. Minimum origin requirements for linear duplex AAV DNA replication in vitro. Virology 209:692-695. [DOI] [PubMed] [Google Scholar]
- 47.Weigel-Kelley, K. A., M. C. Yoder, and A. Srivastava. 2003. Alpha5beta1 integrin as a cellular coreceptor for human parvovirus B19: requirement of functional activation of beta1 integrin for viral entry. Blood 102:3927-3933. [DOI] [PubMed] [Google Scholar]
- 48.Willwand, K., E. Mumtsidu, G. Kuntz-Simon, and J. Rommelaere. 1998. Initiation of DNA replication at palindromic telomeres is mediated by a duplex-to-hairpin transition induced by the minute virus of mice nonstructural protein NS1. J. Biol. Chem. 273:1165-1174. [DOI] [PubMed] [Google Scholar]
- 49.Wobbe, C. R., F. Dean, L. Weissbach, and J. Hurwitz. 1985. In vitro replication of duplex circular DNA containing the simian virus 40 DNA origin site. Proc. Natl. Acad. Sci. USA 82:5710-5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wong, S., N. Zhi, C. Filippone, K. Keyvanfar, S. Kajigaya, K. E. Brown, and N. S. Young. 2008. Ex vivo-generated CD36+ erythroid progenitors are highly permissive to human parvovirus B19 replication. J. Virol. 82:2470-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yee, T. T., C. A. Lee, and K. J. Pasi. 1995. Life-threatening human parvovirus B19 infection in immunocompetent haemophilia. Lancet 345:794-795. [DOI] [PubMed] [Google Scholar]
- 52.Young, N. S., and K. E. Brown. 2004. Mechanism of disease: parvovirus B19. N. Engl. J. Med. 350:586-597. [DOI] [PubMed] [Google Scholar]
- 53.Zhi, N., I. P. Mills, J. Lu, S. Wong, C. Filippone, and K. E. Brown. 2006. Molecular and functional analyses of a human parvovirus B19 infectious clone demonstrates essential roles for NS1, VP1, and the 11-kilodalton protein in virus replication and infectivity. J. Virol. 80:5941-5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhi, N., Z. Zadori, K. E. Brown, and P. Tijssen. 2004. Construction and sequencing of an infectious clone of the human parvovirus B19. Virology 318:142-152. [DOI] [PubMed] [Google Scholar]
- 55.Zhou, X., I. Zolotukhin, D. S. Im, and N. Muzyczka. 1999. Biochemical characterization of adeno-associated virus Rep68 DNA helicase and ATPase activities. J. Virol. 73:1580-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]