Abstract
Lytic infection with the two human gammaherpesviruses, Kaposi's sarcoma-associated herpesvirus (KSHV) and Epstein-Barr virus (EBV), leads to significant depletion of the cellular transcriptome. This host shutoff phenotype is driven by the conserved herpesviral alkaline exonuclease, termed SOX in KSHV and BGLF5 in EBV, which in gammaherpesviruses has evolved the genetically separable ability to target cellular mRNA. We now show that host shutoff is also a prominent consequence of murine gammaherpesvirus 68 (MHV68) infection, which is widely used as a model system to study pathogenesis of these viruses in vivo. The effector of MHV68-induced host shutoff is its SOX homolog, here termed muSOX. There is remarkable functional conservation of muSOX host shutoff activities with those of KSHV SOX, including the recently described ability of SOX to induce mRNA hyperadenylation in the nucleus as well as cause nuclear relocalization of the poly(A) binding protein. SOX and muSOX localize to both the nucleus and cytoplasm of infected cells. Using spatially restricted variants of these proteins, we go on to demonstrate that all known host shutoff-related activities of SOX and muSOX are orchestrated exclusively from the cytoplasm. These results have important mechanistic implications for how SOX and muSOX target nascent cellular transcripts in the nucleus. Furthermore, our findings establish MHV68 as a new, genetically tractable model to study host shutoff.
Efficient viral replication and immune evasion often require manipulation of cellular gene expression either in a targeted or in a global manner. Widespread inhibition of host gene expression, termed host shutoff, is a conserved feature of a diverse set of viruses, including picornaviruses, coronaviruses, orthomyxoviruses, and herpesviruses (3, 13, 23, 32, 37). Host shutoff likely confers a selective advantage to these viruses, perhaps by facilitating redirection of cellular resources toward the virus or by dampening immune stimulatory signals that could restrict viral replication. Within the herpesvirus family, both alphaherpesviruses (alpha-HVs) and gammaherpesviruses (gamma-HVs) induce a prominent host shutoff phenotype (13). Interestingly, while both subfamilies potently decrease the population of cellular mRNA, the viral host shutoff factors and their mechanisms of action are distinct. Herpes simplex viruses (HSV), of the alpha subfamily, encode an RNase termed vhs that degrades cytoplasmic mRNA (6, 7, 26, 45, 53) as well as a second factor, ICP27, which inhibits splicing of cellular messages later in infection (16, 17, 36, 42). Kaposi's sarcoma-associated herpesvirus (KSHV) and Epstein-Barr virus (EBV), two oncogenic human viruses of the gamma subfamily, target cellular mRNA via the activity of the viral alkaline exonuclease (DNase), termed SOX in KSHV and BGLF5 in EBV (12, 41). In contrast to vhs, SOX and BGLF5 lack in vitro RNase activity and are presumed to instead promote host shutoff by activating cellular mRNA turnover pathways. Thus, different members of the herpesvirus family have independently evolved distinct mechanisms to deplete the cellular transcriptome. Such an example of convergent evolution underscores the likely importance of host shutoff for overall viral fitness.
The alkaline exonuclease gene is widely conserved across all herpesviruses, where it participates in the resolution of complex branched structures that arise during rolling circle replication of the viral DNA genome (14, 33, 38, 39). In KSHV and EBV, this protein has also evolved a genetically separable host shutoff function (11, 54). While cellular mRNA turnover pathways targeted by SOX or BGLF5 remain unknown, SOX has recently been shown to induce two prominent phenotypes directly related to host shutoff. First, upon SOX expression, nascent cellular mRNAs undergo poly(A) tail extension (hyperadenylation), and as a consequence they are likely retained in the nucleus (28). Although hyperadenylation has not been characterized in human cells, in yeast it is thought to be a quality control mechanism arising as a consequence of aberrant mRNA 3′-end processing or nuclear export defects (19, 22, 30). The second SOX-induced phenotype is a striking relocalization of cytoplasmic poly(A) binding protein (PABPC) into the nucleus. PABPC has prominent roles in cytoplasmic mRNA stability and translation, and its nuclear relocalization by SOX during KSHV infection is coincident with turnover of reporter mRNAs in the cytoplasm (28). Thus, SOX may prevent export of nascent nuclear messages and deplete preexisting cytoplasmic mRNAs via distinct polyadenylation-related mechanisms. SOX and BGLF5 localize predominantly to the nucleus but are also detectable to a lesser extent in the cytoplasm of transfected cells. The nuclear population is anticipated to participate in the viral genome processing function, but it has been proposed that the cytoplasmic fraction of SOX and BGLF5 contributes to mRNA turnover in that locale (11, 41). It has yet to be elucidated whether the recently characterized PABPC import and hyperadenylation activities are similarly carried out by cytoplasmic SOX. This point is particularly relevant for understanding the means by which SOX induces nuclear hyperadenylation, as direct modulation of the RNA 3′-end processing machinery by SOX should require its presence in the nucleus.
While both KSHV and EBV are associated with significant human disease (10, 40), they are difficult to manipulate, and the potential for in vivo studies is limited due to their strict species specificity. A third gamma-HV, murine gamma-HV 68 (MHV68), possesses a significantly more tractable genetic system and has proven to be a robust tool for the study of gamma-HV pathogenesis in both cell culture and murine models (47). MHV68 is therefore a potentially valuable model in which to assess the role of host shutoff in gamma-HV replication and maintenance.
Here we demonstrate that MHV68 infection induces a global block to cellular gene expression that initiates early in infection and is caused by the MHV68 SOX homolog, termed muSOX. All known activities of SOX and BGLF5 are mimicked by muSOX, underscoring a high level of functional conservation of this phenotype within the gamma-HV subfamily. Using spatially restricted variants of SOX and muSOX, we go on to show that, remarkably, all host shutoff-related activities, including nuclear mRNA hyperadenylation, are orchestrated exclusively by the cytoplasmic pool of SOX and muSOX. These results have important implications for the mechanisms underlying gamma-induced host shutoff, as well as establish MHV68 as a new model to study the contributions of global inhibition of cellular gene expression toward viral replication and pathogenesis.
MATERIALS AND METHODS
Plasmids.
Primers and restriction sites used for plasmid construction are shown in Table 1. A hemagglutinin (HA) tag was introduced onto the N terminus of muSOX by using PCR methods and cloned into the EcoRI/NotI sites of pCDEF3, creating pCDEF3-HAmuSOX. The pCDEF3-SOX-NLS mutant (NLSmut) construct has been previously described (11). An HA tag was introduced onto the N terminus of the wild-type (WT) nuclear retention signal (NRS) of hnRNP-C1 (amino acids [aa] 88 to 165) or mutant NRS (aa 98 to 146) (kindly provided by Jens Lykke-Andersen) by standard PCR methods. To ensure that the mutant NRS was the same size as the WT, an additional 29 aa from green fluorescent protein (GFP) were incorporated to compensate for the deleted NRS sequence. These PCR products were cloned into the N terminus of muSOX using KpnI/EcoRI sites to generate pCDEF3-HA-NRS-muSOX and pCDEF3-HA-ΔNRS-muSOX. An N-terminally tagged HSV-1 alkaline exonuclease (AE) nuclear localization signal (NLS) mutant with alanines at amino acid residues 35 and 36 within the predicted NLS was generated by overlap extension PCR and cloned into the EcoRI/NotI sites of pCDEF3 to generate pCDEF3-HA-AE-NLSmut.
TABLE 1.
Primers used for plasmid construction
| Construct | Primer name | Primer sequence |
|---|---|---|
| ΔmuSOX BAC | ORF36-DsRed | GGAAGGGTCGATTATTCTGGATTTTTTTGAAGCCTCCTCCGAGAACGTCATC |
| ΔmuSOX BAC | DsRed-ORF37 | CGCCACCACCTGTTCCTGCCAGATAGTATTGTGTATGAAATAAAGTCCAGATATAAG |
| ΔmuSOX, ΔmuSOX MR BACs | BamHI-ORF36-fwd | ATTAGGATCCGACAGCACTGTACAAGGAGGC |
| ΔmuSOX, ΔmuSOX MR BACs | ORF38-NotI-rev | TTAAGCGGCCGCGTGTTTGCCTCGTTCATATTCAATG |
| HA-NRS-muSOX | KpnI-HA-NRS-fwd | GGGGTACCATGGCTTACCCATACGATGTACCTGACTATGCGCCAAAAGTGAACCGAGGAAAA |
| HA-NRS-muSOX | EcoRI-NRS-rev | CGGAATTCTCCACTCTTAGAATTGAAGCC |
| HA-ΔNRS-muSOX | KpnI- HA-ΔNRS-fwd | GGGGTACCATGGCTTACCCATACGATGTACCTGACTATGCGAAACGATCTGCAGCGGAG |
| HA-ΔNRS-muSOX | EcoRI- HA-ΔNRS-rev | CGGAATTCTTTCGAGGGCACTACAGCCCG |
| GFP-29aa (for ΔNRS) | EcoRI-GFP-29aa-fwd | GGGAATTCCAGGATGATGGCACGCTGCCC |
| GFP-29aa (for ΔNRS) | EcoRI-GFP-29aa-rev | CGGAATTCATTGATCCTAGCAGAAGCACA |
| HA-UL12 | EcoRI-UL12-fwd | GGAATTCATGGCTTACCCATACGATGTACCTGACTATGCGATGGAGTCCACGGTAGGC |
| HA-UL12 | NotI-UL12-rev | ATAGTTTAGCGGCCGCTCAGCGAGACGACCTCCC |
| HA-UL12-NLSmut | UL12-NLSmut-fwd | CGTGGCCCCGACAGCCCCCCCGCGGCCCCCCGCCCTAACAGTCTTCCG |
| HA-UL12-NLSmut | UL12-NLSmut-rev | CGGAAGACTGTTAGGGCGGGGGGCCGCGGGGGGGCTGTCGGGGCCACG |
| HA-muSOX | EcoRI-HA-ORF37 fwd | CGGAATTCATGGCTTACCCATACGATGTACCTGACTATGCGATGGAAGGGTCGATTATTC |
| HA-muSOX | NotI-ORF37 rev | ATAGTTTAGCGGCCGCTTAGGGGGTTATGGGTTTTCT |
Cells, transfections, and viruses.
293T, NIH 3T3, NIH 3T12, and human foreskin fibroblast (HFF) cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS). BHK-21 cells (clone 15) were propagated in RPMI 1640 medium (Invitrogen) supplemented with 5% FBS. 293T cells were transfected with Effectene (Qiagen), following the manufacturer's protocol. BHK and 3T3 cells were transfected with SuperFect (Qiagen). KSHV stocks were prepared from induced BCBL-1 cells as described previously (2). KSHV infections of HFFs were carried out in the presence of 8 μg/ml Polybrene, and lytic reactivation was induced by superinfection with an adenoviral vector expressing RTA (Ad-RTA) as described previously (2). Bacterial artificial chromosome (BAC)-derived MHV68 was generated by transfecting 2 μg of BAC DNA per well of a six-well plate of NIH 3T3 using SuperFect (Qiagen) and then propagated in NIH 3T12 cells, and the titer was determined by using a plaque assay on NIH 3T3 cells. To harvest the virus, cells were passed through a Dounce homogenizer and cellular debris was removed by pelleting and passing the supernatant through a 0.45-μm filter.
The GFP-expressing MHV68 BAC infectious clone has been described previously elsewhere (1). Mutants were generated by allelic exchange as described previously (46). To generate the ΔmuSOX BAC, a targeting region consisting of a DsRed coding sequence flanked upstream by the terminal 979 nucleotides (nt) of the open reading frame 36 (ORF36) coding region (including the first 33 nt of the ORF37 coding region) and downstream by 756 nt of the 3′ end of the ORF37 coding region, followed by 178 nt of the ORF38 coding region, was generated by splicing by overlap extension PCR. The product was ligated into pGS284 between the BglII and NotI sites and electroporated into the S17λpir strain of Escherichia coli. The targeting vector for the rescue BAC was generated by ligating the WT region of the BAC into pGS284. Targeting vector-containing cells were cross-streaked with BAC-containing GS500 cells, and successful recombinants were identified by colony PCR. BAC DNA was isolated from positive clones using the Qiagen large-construct kit (Qiagen). BAC variants were verified by diagnostic restriction digestion with EcoRI and PvuII and sequencing of the region surrounding the recombination site.
qPCR.
RNA was isolated from transfected cells using RNA-Bee reagent (Tel-Test) or the Zymo Mini RNA II isolation kit (Zymo Research). RNA samples were DNase-treated and reverse transcribed using AMV RT (Promega) and an oligo(dT) or 18S-specific primer. GFP mRNA copies were quantified by quantitative PCR (qPCR) on the resulting cDNA as described previously (28). GFP cDNA copies were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA copies using rodent GAPDH endogenous controls or to 18S cDNA copies using ribosomal control reagents (Applied Biosystems).
Plasmid DNA was analyzed for degradation by transfecting 293T cells with pCDEF3-GFP and pCDEF3 or pCDEF3-HAmuSOX. At 24 h posttransfection, cells were washed in phosphate-buffered saline and treated with micrococcal nuclease for 30 min at 37°C (15 U/μl in 50 mM NaCl, 10 mM Tris, pH 7.5, 5 mM MgCl2, and 1 mM CaCl2). The reaction was stopped by adding EDTA to 5 mM. DNA was isolated by lysing the cells in 20 mM Tris, pH 8.0, 50 mM EDTA, 200 mM NaCl, and 1.2% sodium dodecyl sulfate (SDS), followed by phenol-chloroform extraction. Samples were diluted to 0.15 μg/μl and quantitated by qPCR as described above.
DNase assays.
Proteins analyzed for DNase activity were in vitro transcribed and translated (IVT) using the rabbit reticulocyte lysate system (Promega). From each reaction mixture, 8 μl of protein was incubated with 200 ng of NotI-linearized pCDEF3 plasmid DNA in 42 μl of degradation assay buffer (0.1 M MgCl2, 0.5 M Tris, pH 9.0, 100 μg/ml bovine serum albumin, 5 mM β-mercaptoethanol) at 37°C for the indicated time period and then phenol-chloroform extracted. Pellets were resuspended in 20 μl of water, resolved on a 1% agarose gel, and visualized by ethidium bromide staining. One-third of each IVT reaction was also resolved by SDS-polyacrylamide gel electrophoresis (PAGE), and gels were fixed, dried, and visualized by autoradiography to verify equivalent protein expression.
Cell extracts, Western blots, and Northern blots.
For metabolic labeling experiments, infected 3T3 cells were incubated in methionine- and cysteine-free Dulbecco's modified Eagle's medium containing 10% FBS for 1 h and then pulse labeled in the same medium containing 0.1 mCi/ml 35S-labeled EasyTag Express protein labeling mix (PerkinElmer) for 30 min. For this and other experiments, cell lysates were prepared in RIPA buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM EDTA, 1% Nonidet P-40, 0.1% SDS) containing protease inhibitor cocktail (Roche) and quantified by the Bradford assay (Bio-Rad). Equivalent masses of each sample were resolved by SDS-PAGE and either dried and exposed to film or transferred to a polyvinylidene difluoride membrane and Western blotted with either HA 12CA5 monoclonal antibodies (MAbs) (1:4,000; Abcam), SOX MAbs (1:1,000), or muSOX polyclonal antibodies (pAbs) (1:250) and either horseradish peroxidase-conjugated goat anti-rabbit or goat anti-mouse secondary antibodies (1:5,000; Southern Biotechnology Associates). Rabbit pAbs were raised against a maltose binding protein (MBP)-tagged full-length muSOX by standard methods (18). Serum was then affinity purified over MBP-muSOX and MBP columns. To generate SOX MAb, MBP-tagged SOX was purified with amylose resin and subsequently cleaved with FactorXa (New England Biolabs) to remove the MBP moiety. BALB/c mice were injected intraperitoneally with 10 μg of SOX in Ribi adjuvant at 8 weeks, 4 weeks, and 4 days prior to the fusion of splenocytes with P3X63-Ag8.653 myeloma cells. Hybridomas were selected in hypoxanthine-aminopterin-thymidine medium for 2 weeks, followed by enzyme-linked immunosorbent assay screening using SOX-transfected 293T cell lysates or vector-transfected lysates. Positive wells were rescreened using immunofluorescence assays (IFA) and Western blotting, followed by two rounds of cloning and retesting by enzyme-linked immunosorbent assay, IFA, and Western blotting. The 5H5 hybridoma supernatants were collected once per week. The MAb was purified by protein G chromatography (GE Healthcare), and purity was confirmed by SDS-PAGE, followed by Coomassie blue staining.
For Northern blotting, total RNA was harvested using RNA-Bee (Tel-Test) and analyzed by agarose-formaldehyde gel electrophoresis. RNAs were transferred to a 0.45-μm nylon membrane and probed with 32P-labeled DNA probes generated using the Rediprime II random prime labeling system (GE Healthcare). RNase H digests were performed by combining 5 μg of RNA with 500 pmol of oligo(dT)15 primer in a 25.8-μl reaction volume and then incubating at 65°C for 8 min. Samples were then incubated at 37°C for 30 min in the presence of 1 U RNase H (New England Biolabs), 1× RNase H buffer, and 40 U RNasin (Promega); reactions were terminated by the addition of 1 μl of 0.5 M EDTA, pH 8.0, and the RNA was ethanol precipitated.
IFA and oligo(dT) in situ hybridization.
293T cells grown on coverslips were fixed in 4% formaldehyde and processed for IFA as described previously (31). In situ samples were processed as described previously (http://www.singerlab.org/protocols) using 2 ng/μl of Alexa Fluor 546-labeled oligo(dT)15 (Molecular Probes). After oligonucleotide hybridization, samples were incubated with either anti-SOX or anti-HA (Abcam) MAbs at a 1:100 or 1:500 dilution, respectively, in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.1% Triton X-100 for 3 h at 37°C and subsequently with Alexa Fluor 488-labeled goat anti-mouse secondary antibodies (Molecular Probes) and mounted with 4′,6-diamidino-2-phenylindole (DAPI)-containing VectaShield mounting medium (Vector Labs, Inc.). IFA not involving in situ hybridization was done as described previously (31) using anti-SOX MAbs (1:100 dilution), rabbit polyclonal anti-PABPC pAbs (1:25 dilution) (Cell Signaling Technology), or rabbit anti-muSOX pAbs (1:25 dilution) and Alexa Fluor 488- or 546-labeled goat anti-rabbit or anti-mouse secondary antibodies (1:1,500 dilution) (Invitrogen).
RESULTS
MHV68 lytic replication induces a shutoff of host gene expression.
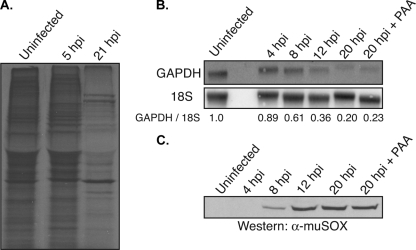
Lytic infection with KSHV and EBV leads to a profound reduction in host gene expression driven by enhanced cellular mRNA turnover (12, 41), suggesting that host shutoff may be a phenotype conserved among gamma-HVs. We therefore sought to determine whether infection with the related murine herpesvirus 68 (MHV68) resulted in a similar reduction in host gene expression. MHV68 represents an attractive model system to explore gamma-HV biology, as unlike the human gamma-HVs, it possesses highly tractable genetics (1). Global efficiency of cellular protein synthesis was monitored by [35S]Cys/Met metabolic labeling of 3T3 cells either mock infected or infected with MHV68 (Fig. 1A). At 21 h postinfection, there was a clear reduction in overall cellular protein synthesis, indicative of host shutoff. Proteins accumulating after infection are presumably viral, as many of these are not present in the uninfected sample. Both KSHV and EBV promote host shutoff by degrading mRNA (12, 41); therefore, we assayed the levels of select host transcripts during a time course of MHV68 infection by Northern blotting (Fig. 1B). By 12 h postinfection, the level of GAPDH mRNA decreased threefold, and by 20 h, the magnitude of its reduction increased to fivefold relative to that of mock-infected samples (Fig. 1B). Similar results were observed for β-actin mRNA (data not shown). To determine whether MHV68-induced host shutoff manifested as a result of early or late viral gene expression, we also infected cells in the presence of phosphonoacetic acid (PAA), which blocks viral genome replication, thereby prohibiting the expression of late viral genes (35). PAA treatment had no effect on host shutoff, indicating that this phenotype is caused by an early viral gene(s) (Fig. 1B).
FIG. 1.
MHV68 promotes host shutoff. 3T3 cells were either mock infected or infected with MHV68 at a multiplicity of infection of 10. (A) At the indicated times postinfection, cells were pulse-labeled with [35S]-labeled Cys/Met and lysates were resolved by SDS-PAGE. The gel was then dried and visualized by autoradiography. (B) RNA was harvested from uninfected cells or cells infected for the indicated times and Northern blotted with GAPDH and 18S (loading control) probes. Quantification (normalized to 18S levels) is shown, with the level of GAPDH mRNA in the uninfected sample set to 1.0. PAA indicates the sample infected and cultured in the presence of 200 μg/ml PAA. (C) Lysates were harvested from either uninfected cells or cells infected with MHV68 for the indicated times and Western blotted with anti-muSOX antibodies. All figures are representative of three independent experiments. hpi, hours postinfection; α-muSOX, anti-muSOX.
In other gamma-HVs, the host shutoff function maps to the alkaline exonuclease homolog (named SOX in KSHV and BGLF5 in EBV). This protein, termed muSOX and encoded by ORF37 in MHV68, represents the most likely candidate for the MHV68 host shutoff factor. We therefore generated and affinity purified pAbs against muSOX to monitor its expression during infection. Western blotting showed that muSOX expression initiates approximately 8 h postinfection and peaks by 12 h postinfection (Fig. 1C). In agreement with viral microarray data (5, 34), muSOX is an early gene, as its expression is uninhibited by PAA (Fig. 1C). Thus, muSOX expression is contemporaneous with the reduction in cellular mRNAs.
muSOX induces mRNA degradation.
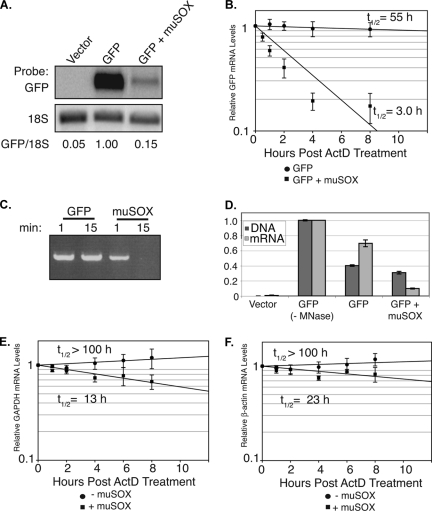
We hypothesized that, similar to SOX and BGLF5, muSOX could promote mRNA turnover. To test this hypothesis directly, we measured by Northern blotting the mRNA levels of a GFP reporter in 293T cells cotransfected with empty vector or a muSOX expression plasmid. As predicted, Northern blotting revealed a significant muSOX-induced reduction in reporter mRNA levels (Fig. 2A). RNA half-life measurements performed in the presence of actinomycin D (ActD) subsequently confirmed that the decreased GFP mRNA levels in muSOX-expressing cells were a result of enhanced mRNA degradation, as the message half-life decreased from 55 h to 3 h in the presence of muSOX (Fig. 2B). Given that muSOX possesses DNA exonuclease activity like its homologs (Fig. 2C), we wanted to ensure that these results were not an indirect effect of muSOX degrading GFP plasmid DNA. We therefore simultaneously measured the levels of the GFP reporter plasmid DNA and mRNA in the presence or absence of muSOX by qPCR (Fig. 2D). Cells were treated with micrococcal nuclease just prior to lysis to remove any extracellular nucleic acid. Importantly, muSOX expression resulted in a significant decrease in GFP mRNA but not DNA levels (Fig. 2D). This result is in agreement with prior observations, indicating that the analogous DNase activity of SOX does not contribute measurably to its host shutoff function (12).
FIG. 2.
muSOX enhances mRNA degradation. (A) 293T cells were transfected with the indicated plasmids for 24 h. Total RNA was then isolated and analyzed by Northern blotting with GFP and 18S probes. Quantification (normalized to 18S levels) is shown, with the level of GFP mRNA in the absence of muSOX set to 1.0. (B) Cells were transfected as described above, and the GFP mRNA half-life was calculated by qPCR at different time points post-ActD (2 μg/ml) treatment. (C) IVT muSOX or GFP (as a negative control) was incubated for 1 or 15 min with linear plasmid DNA in degradation assay buffer at 37°C. The DNA was then extracted, resolved by agarose gel electrophoresis, and visualized by ethidium bromide staining. (D) 293T cells were transfected with empty vector or with pCDEF3-GFP alone or together with pCDEF3-muSOX. With the exception of the control (−MNase), all samples were treated with micrococcal nuclease prior to cell lysis to remove extracellular nucleic acids. Samples were then divided in half and harvested for either total cellular DNA or RNA. GFP DNA and mRNA levels were then calculated via qPCR. (E and F) Cells were transfected as described for panel A. The endogenous GAPDH (E) or β-actin (F) mRNA half-life was then calculated via quantification of Northern blots (normalized to 18S) at different time points post-ActD treatment. Errors bars show the standard error between samples. All graphs represent a compilation of at least three independent experiments. t1/2, half-life.
To verify that muSOX-induced mRNA turnover was not just an artifact specific to the exogenously expressed GFP reporter, we also measured the half-life of endogenous GAPDH and β-actin mRNAs in cells transfected with muSOX or empty vector (Fig. 2E and F). Indeed, both messages were degraded four- to eightfold more rapidly in muSOX-expressing cells. We anticipate this to be a conservative estimation of the rate of endogenous mRNA turnover by muSOX, because the ∼20% to 30% of untransfected cells present in the muSOX samples will inflate the estimated mRNA half-life. We conclude that muSOX expression induces host shutoff via enhanced mRNA degradation and that it is a true functional homolog of SOX and BGLF5.
muSOX is necessary for viral replication and host shutoff.
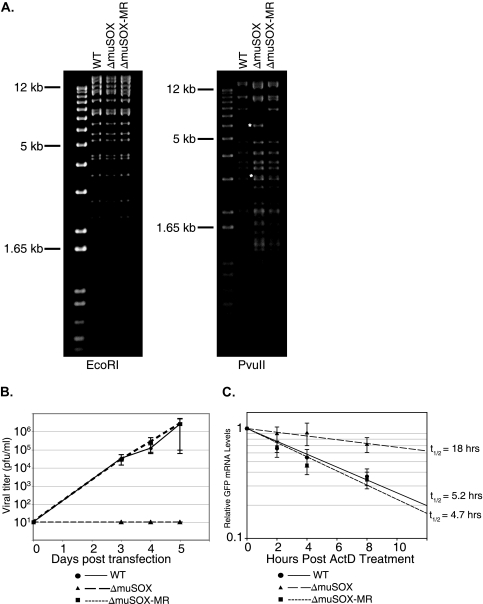
We next examined the contribution of muSOX toward MHV68-induced host shutoff. To this end, we sought to generate an MHV68 muSOX deletion mutant virus (MHV68ΔmuSOX) and the corresponding marker rescue (MR) virus (MHV68ΔmuSOX-MR) using an MHV68 BAC-based infectious clone (Fig. 3A) (1). We were unable to recover any virus following transfection of the deletion mutant BAC genome into 3T3 cells, whereas the WT and the MR BAC generated high viral titers (Fig. 3B). This result was not unexpected, as the muSOX homologs HSV-1 UL12 (AE) and EBV BGLF5 have been shown to be essential due to their roles in viral genome processing during DNA replication (9, 15, 33). Indeed, we observe in vitro DNase activity for muSOX, as has been reported for other herpesviral AE proteins (Fig. 2C). We presume that this genome processing activity by muSOX is similarly required for MHV68 replication, although host shutoff activity may also contribute to efficient virus production.
FIG. 3.
muSOX is necessary for viral replication and host shutoff during infection. (A) EcoRI and PvuII digests of WT MHV68, MHV68ΔmuSOX, and MHV68ΔmuSOX-MR. In the PvuII digest, asterisks represent cleavage products generated as a result of introducing an additional PvuII site upon deletion of the muSOX gene. (B) Growth curve comparing viral titer following transfection of 3T3 cells with WT, MHV68ΔmuSOX, and MHV68ΔmuSOX-MR BAC DNA. Limit of detection is 10 PFU/ml. Data are a compilation of three independent replicates with the WT and deletion BACs and two independent replicates with the MR BAC. (C) The indicated BAC DNAs were transfected into BHK cells in the presence of 200 μg/ml PAA. BAC-derived GFP mRNA abundance was determined by qPCR at different time points posttreatment with 5 μg/ml ActD. Errors bars show the standard error between samples from three independent experiments. t1/2, half-life.
Failure of the MHV68ΔmuSOX BAC to produce virus, coupled with low transfection efficiency of the BAC DNA, prevented measurement of endogenous cellular mRNA stability in the presence of this mutant. We therefore opted to compare the half-life of the BAC-derived GFP message in BHK-21 cells transfected with WT, MHV68ΔmuSOX, and MHV68ΔmuSOX-MR BAC DNAs. To eliminate any complicating effects of viral DNA replication, experiments were conducted in the presence of PAA. Cells were treated with ActD at 18 h posttransfection, and RNA was isolated at the indicated time points thereafter. Significantly, there was a marked increase in the half-life of the GFP transcript in cells transfected with the ΔmuSOX BAC relative to those in WT and MR BAC-transfected cells (Fig. 3C). Collectively, these data indicate that muSOX plays a prominent role in MHV68-induced host shutoff.
SOX homologs localize to both the nucleus and cytoplasm.
All herpesvirus AE homologs examined to date localize predominantly or exclusively to the nucleus, in agreement with their roles in processing and packaging newly replicated viral DNA (14, 33, 38, 39). However, several lines of evidence suggest that cytoplasmic localization of the gamma-HV SOX homologs plays an important role in host shutoff. First, KSHV SOX and EBV BGLF5 localize both to the nucleus and cytoplasm of cells, whereas HSV-1 AE, which lacks host shutoff activity, is constrained to the nucleus (12, 41). Second, a SOX NLS mutant retains WT mRNA turnover activity in transfected cells (11). Finally, mRNA half-life studies demonstrated that cytoplasmic messages are destabilized in SOX-expressing cells (28).
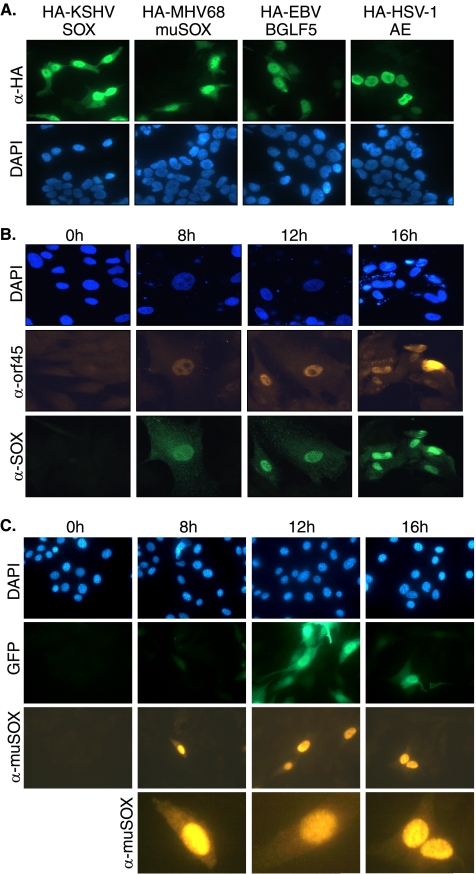
We therefore predicted that MHV68 muSOX would also exhibit partial cytoplasmic localization, which might be integral to its host shutoff function. Indeed, similar to its KSHV and EBV homologs, HA-tagged muSOX was detected in both the nucleus and cytoplasm of transfected 293T cells, whereas HSV-1 AE remained exclusively nuclear (Fig. 4A). The absence of antibodies against any gamma-HV SOX homologs that function in IFA had previously prevented confirmation of this partial cytoplasmic SOX localization during viral infection. Given that plasmid-based overexpression does not always faithfully mimic endogenous protein localization, we generated both MAbs against KSHV SOX and affinity-purified pAbs against MHV68 muSOX to monitor their localization throughout the course of infection. The localization and kinetics of expression of SOX and muSOX were examined by IFA in HFF and 3T3 cells lytically infected with KSHV and MHV68, respectively (Fig. 4B and C). Expression of both SOX and muSOX was detected beginning at 8 h postinfection, in agreement with previous results (Fig. 1C) (12), and expression was maintained throughout the lytic cycle. Importantly, SOX and muSOX localized to both the nucleus and the cytoplasm during viral infection, although, in agreement with the localization of the overexpressed proteins, the majority of SOX and muSOX exists in the nucleus (Fig. 4B and C; a magnified view of representative muSOX-expressing cells is shown in the bottom panels of Fig. 4C).
FIG. 4.
SOX homologs of gamma-HVs localize to both the nucleus and cytoplasm. (A) 293T cells were transfected with empty vector or HA-tagged SOX (KSHV), muSOX (MHV68), BGLF5 (EBV), and AE (HSV-1). Twenty-four hours posttransfection, cells were subjected to IFA with anti-HA antibodies. (B) HFF cells were either mock infected or infected with KSHV and lytically reactivated with an adenoviral vector expressing RTA. At the indicated times, samples were subjected to IFA with anti-ORF45 (KSHV lytic marker) (center panels) and anti-SOX (bottom panels) antibodies. (C) 3T3 cells were either mock infected or infected with BAC-derived MHV68. At the indicated times, samples were subjected to IFA with anti-muSOX antibodies. GFP is encoded by the BAC and serves as a marker of infection. Bottom panels show a magnified view of representative muSOX-expressing cells at each time point. All samples were costained with DAPI to visualize nuclei. α, anti.
Restriction of muSOX to the nucleus abolishes its shutoff activity.
Having shown that both SOX and muSOX exhibit partial cytoplasmic localization during lytic infection, we next sought to determine whether this cytoplasmic fraction was necessary for host shutoff. A SOX nuclear localization signal mutant (SOX-NLSmut) had previously been shown to be predominantly cytoplasmic and to retain mRNA turnover activity (11). We initially attempted to generate a similar NLS mutant in muSOX to explore the level of phenotypic conservation between these homologs. However, we found that unlike the monopartite SOX NLS, muSOX possesses a bipartite NLS (aa 314 to 318 and aa 409 to 412), the latter component of which resides in a highly conserved region of the protein that cannot be mutated in either SOX or muSOX without destroying both DNase and host shutoff activity (unpublished observations).
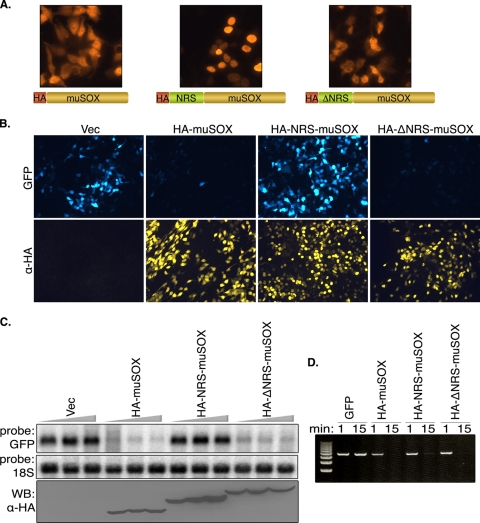
We therefore pursued the converse approach of restricting muSOX to the nucleus by fusing it to an NRS derived from the heterogeneous nuclear ribonucleoprotein C1 (HA-NRS-muSOX) (44). As a control, we fused muSOX to a mutant NRS peptide of identical length that is incapable of nuclear restriction (HA-ΔNRS-muSOX). IFA staining confirmed that the HA-NRS-muSOX fusion was constrained to the nucleus, whereas the WT HA-muSOX and the HA-ΔNRS-muSOX fusion were present both in the nucleus and the cytoplasm (Fig. 5A). We then measured host shutoff by testing the ability of these fusions to block expression of GFP. We observed no shutoff of GFP expression in cells coexpressing HA-NRS-muSOX, whereas GFP protein expression was strongly inhibited in cells coexpressing HA-muSOX or HA-ΔNRS-muSOX (Fig. 5B). Northern blot analysis of the GFP message confirmed that the inability of HA-NRS-muSOX to block GFP protein expression was due to its failure to promote mRNA turnover, even at high concentrations (Fig. 5C). Western blotting of the muSOX variants shows that transfecting increased amounts of HA-muSOX or HA-ΔNRS-muSOX does not lead to a significant increase in their protein levels, presumably because they are inducing degradation of their own transcripts (Fig. 5C). This is not the case with HA-NRS-muSOX, whose protein levels continually increase as more plasmid DNA is transfected, further supporting the observation that this protein is defective for RNA turnover. Notably, the inability of HA-NRS-muSOX to promote RNA turnover is not due to gross misfolding of the protein, since this fusion retains enzymatic activity in DNase assays (Fig. 5D). As this was not the case for NRS-tagged SOX from KSHV, which lost all activity upon fusion to any large tag, we were unable to perform similar experiments with SOX (data not shown). Thus, we conclude that the nuclear fraction of muSOX does not contribute appreciably to mRNA turnover, whereas the cytoplasmic fraction is critical for this function.
FIG. 5.
Host shutoff is orchestrated from the cytoplasm. (A) 293T cells were transfected with the indicated plasmid expressing HA-tagged muSOX or muSOX fused to a WT or mutant NRS, as diagramed. Localization of each muSOX protein was visualized by IFA with anti-HA antibodies. (B) The host shutoff activity of each muSOX variant was assessed by cotransfection of a GFP reporter with either empty vector or the indicated muSOX-expressing plasmid. At 24 h posttransfection, cells were subjected to IFA with anti-HA antibodies. (C) 293T cells were transfected with increasing amounts of the indicated muSOX constructs (100 to 300 ng). Total RNA was harvested 24 h posttransfection and Northern blotted with GFP and 18S probes. Protein levels were assessed by Western blotting with anti-HA antibodies. (D) Linearized pCDEF3 plasmid was incubated with aliquots of the indicated IVT protein for 1 or 15 min in degradation assay buffer at 37°C. The DNA was then extracted, resolved by agarose gel electrophoresis, and visualized by ethidium bromide staining. α, anti; Vec, vector.
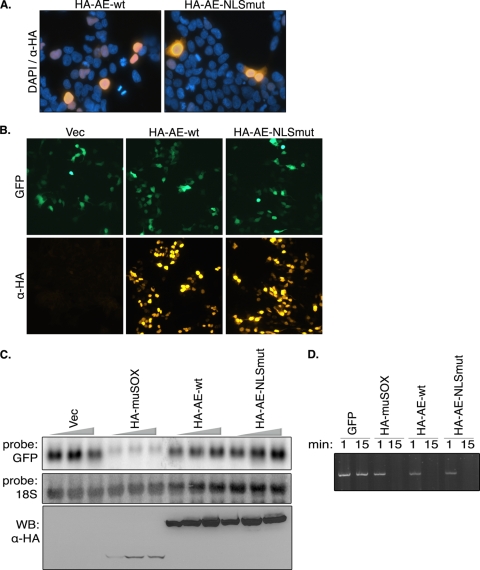
Given that cytoplasmic localization is critical for host shutoff activity, we considered the possibility that HSV-1 AE fails to promote mRNA turnover simply because it is not present in the cytoplasm. We therefore sought to create an HSV-1 AE mutant that localized to the cytoplasm, similar to its gamma-HV homologs. An HSV-1 AE NLS has previously been mapped to the N terminus of the protein (aa 1 to 126) (38). Two point mutations (in bold) were introduced into the predicted AE NLS (33PPKRPR38 to 33PPAAPR38) to generate HA-AE-NLSmut, and the protein localization was monitored by IFA in 293T cells (Fig. 6A). The HA-AE-NLSmut protein exhibited partial cytoplasmic localization, very similar to WT SOX. Despite its presence in the cytoplasm, this mutant was unable to promote host shutoff, as measured by GFP protein and mRNA accumulation (Fig. 6B and C). HA-AE-NLSmut retained WT enzymatic activity in DNase assays (Fig. 6D), indicating that failure of this mutant to induce host shutoff was not likely due to gross misfolding of the protein. We therefore conclude that cytoplasmic localization of the ΑΕ homologs, while critical for activity, is not the sole determinant of host shutoff and that the gamma-HV SOX proteins have indeed evolved additional functions not present in HSV-1 AE.
FIG. 6.
Partial relocalization of HSV-1 AE into the cytoplasm is not sufficient to induce mRNA turnover. (A) 293T cells were transfected with plasmids expressing either WT HSV-1 AE or AE-NLSmut for 24 h and then subjected to IFA with anti-HA antibodies. DAPI staining was used to visualize nuclei. (B) 293T cells were transfected with a GFP reporter plasmid alone or together with the indicated AE construct for 24 h and then subjected to IFA with anti-HA (α-HA) antibodies. (C) 293T cells were transfected with increasing quantities of the indicated plasmids (100 to 300 ng). Total RNA was harvested 24 h posttransfection and Northern blotted with GFP and 18S probes. AE and muSOX levels were assessed by Western blotting with anti-HA antibodies. (D) The linearized pCDEF3 plasmid was incubated with aliquots of the indicated IVT protein for 1 or 15 min in degradation assay buffer at 37°C. The DNA was then extracted, resolved by agarose gel electrophoresis, and visualized by ethidium bromide staining. Vec, vector.
Cytoplasmic fraction of muSOX and SOX drives mRNA hyperadenylation and PABPC nuclear import.
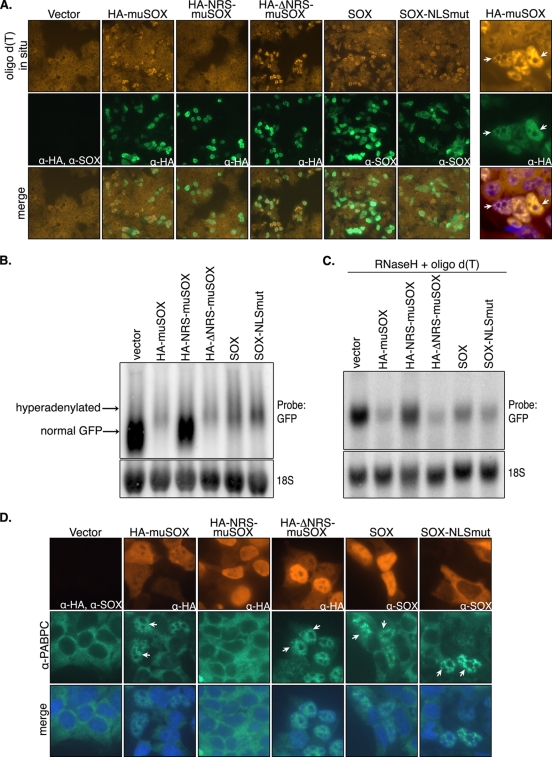
It has recently been shown that SOX induces aberrant hyperadenylation of nascent cellular messages within the nucleus (28). The mechanism by which SOX causes such mRNA 3′-end processing defects has not been elucidated, yet this activity is dependent on its host shutoff function (28). We observed that muSOX similarly promotes mRNA hyperadenylation, as measured both by oligo(dT) staining to detect accumulation of endogenous nuclear poly(A) RNA and by Northern blotting to show an increased mRNA length (Fig. 7A and B). Similar to our previous observations with KSHV SOX (28), the increased poly(A) RNA signal in muSOX-expressing cells occurred only in the nucleus, whereas the cytoplasm was depleted of poly(A) RNA (Fig. 7A, right). To confirm that the GFP mRNA size difference observed in the presence of muSOX was due to hyperadenylation, we removed the mRNA poly(A) tails by hybridizing the RNA to oligo(dT) and digesting with RNase H. Indeed, after deadenylation, the size of the GFP mRNA was identical in the presence or absence of muSOX (Fig. 7C).
FIG. 7.
The cytoplasmic fractions of muSOX and SOX drive mRNA hyperadenylation and PABPC nuclear import. (A) 293T cells were transfected with the indicated plasmids for 24 h and then subjected to oligo(dT) in situ hybridization (top panels), followed by staining with anti-SOX MAbs (SOX samples), anti-HA antibodies (muSOX samples), or both anti-HA and anti-SOX antibodies (vector sample) (center panels). The bottom panels show a merge of the in situ and IFA staining. The far right panels show a magnified view of a field of muSOX-transfected cells; the bottom right panel of this column shows a merge of the in situ signal and DAPI-stained nuclei. Arrows identify select muSOX-expressing cells. (B) 293T cells were transfected with a GFP reporter plasmid alone or together with the indicated muSOX- or SOX-expressing plasmid. At 24 h posttransfection, samples were treated with 5 ng/ml leptomycin B for 6 h to stabilize hyperadenylated RNAs, which are detected as the slower-migrating population (28). Total RNA was then harvested and Northern blotted with GFP and 18S probes. (C) A fraction of each RNA sample from that shown in panel B was incubated with oligo(dT) to bind poly(A) tails and then subjected to RNase H digestion to deadenylate the mRNAs prior to agarose-formaldehyde gel electrophoresis and Northern blotting. (D) 293T cells were transfected with empty vector or with a plasmid expressing the indicated muSOX fusion protein for 24 h and then subjected to double-label IFA with polyclonal anti-PABPC antibodies (center panels) and anti-HA or anti-SOX antibodies (top panels). The bottom panel shows a merge of the PABPC-stained and DAPI-stained nuclei. Arrows point to representative cells exhibiting PABPC nuclear import. α, anti.
Given that hyperadenylation occurs within the nucleus, we reasoned that direct modulation of the cellular mRNA processing machinery would necessarily involve the nuclear fraction of muSOX or SOX. Conversely, if the nuclear population of SOX were dispensable for hyperadenylation, this would be indicative of SOX indirectly altering mRNA 3′-end processing in the nucleus via one or more of its cytoplasmic activities. We therefore assessed the ability of the muSOX NRS fusions, as well as that of the SOX NLS mutant, to promote mRNA hyperadenylation. Surprisingly, we observed that nucleus-restricted HA-NRS-muSOX was deficient for nuclear hyperadenylation, as measured by both oligo(dT) in situ hybridization and Northern blotting (Fig. 7A and B). In contrast, the cytoplasmic NLS mutant of SOX and the control HA-ΔNRS-muSOX retained WT nuclear hyperadenylation activity (Fig. 7A and B). RNase H-mediated poly(A) tail removal confirmed that the increased GFP mRNA size in the presence of HA-ΔNRS-muSOX, SOX, and SOX-NLSmut was due to hyperadenylation (Fig. 7C). These data indicate that SOX and muSOX do not directly target nuclear mRNA processing machinery, but instead must indirectly alter its regulation to promote hyperadenylation in the nucleus.
An additional prominent activity of SOX is to promote the relocalization of PABPC from the cytoplasm into the nucleus of cells in a host shutoff-dependent manner (28). We first confirmed by IFA that muSOX expression also induces nuclear relocalization of endogenous PABPC (Fig. 7D). We then explored whether PABPC relocalization was dependent on the cytoplasmic or nuclear activities of muSOX. Again, we observed that HA-NRS-muSOX was unable to alter PABPC localization, whereas HA-ΔNRS-muSOX and SOX-NLSmut induced PABPC nuclear import as efficiently as WT SOX and muSOX (Fig. 7D). In summary, our data support the conclusion that all known host shutoff-related activities of the gamma-HV proteins are orchestrated exclusively from the cytoplasm of cells.
DISCUSSION
The ability to severely restrict cellular gene expression is a conserved phenotype within the gamma-HV subfamily and therefore presumably plays an important role in viral replication and/or fitness. Within the alpha-HV, vhs host shutoff activity influences viral pathogenesis and contributes to the establishment of HSV latency and reactivation in mice (29, 49-51). Host shutoff is likely to similarly participate in generalized immune evasion during gamma-HV infection, perhaps as a complement to viral proteins with specific immune evasion functions, such as inhibition of major histocompatibility complex class I presentation (4, 21, 48). Analysis of the role of host shutoff during human gamma-HV infection has been hampered by difficulties in generating and propagating KSHV and EBV mutants. Our demonstration that host shutoff is phenotypically mimicked in MHV68, which is far more genetically tractable, indicates that this virus presents a robust model to dissect the contribution of host shutoff toward gamma-HV replication and maintenance both in cultured cells and in vivo. However, in addition to its role in cellular mRNA turnover, muSOX is also likely essential for viral DNA processing, as has recently been described in detail for the EBV homolog BGLF5 (9). We predict that our failure to recover virus from a muSOX deletion mutant BAC is due in large part to an inability of this virus to process and package newly replicated viral DNA. Thus, specific examination of the host shutoff function of muSOX during infection will require generation of single-function mutants that retain the DNA processing activity. Such mutants have successfully been isolated for SOX and BGLF5 (11, 54), and efforts are currently under way to identify similar single-function muSOX mutants.
It is notable that GFP mRNA expressed from a pCDEF3 plasmid in the absence of muSOX had a 55-h half-life, but when expressed from the ΔmuSOX BAC its half-life was only 18 h (compare Fig. 2B and 3C). We anticipate that the difference in the half-life of the GFP message in these two experiments may be due to a combination of factors. First, these are not identical transcripts; the mRNA in the muSOX transfections contains extra sequences in the coding region that control GFP protein stability, whereas the GFP mRNA derived from the BAC lacks these sequences. Furthermore, there are extensive sequence differences within the 5′ untranscribed regions of these two transcripts, and they are transcribed from different promoters: pCDEF3 contains an EF-1a promoter, and the BAC-derived GFP is expressed from a cytomegalovirus promoter. Collectively, these differences may affect the overall stability of the mRNAs. Alternatively, the difference in these GFP mRNA half-lives may indicate that there are additional viral host shutoff factors contributing to instability of the BAC-derived GFP. Future experiments will address whether either of these possibilities is correct.
Homologs of SOX are found throughout the herpesvirus family, yet only in gamma-HVs have these proteins evolved the ability to induce cellular mRNA turnover. A notable difference between these proteins in alpha-HV versus gamma-HVs is their subcellular localization; gamma-HV SOX proteins are present in both the nucleus and the cytoplasm, whereas alpha-HV AE is exclusively nuclear. However, while cytoplasmic localization is clearly necessary for the host shutoff function of these proteins, partial redirection of HSV-1 AE to the cytoplasm is not sufficient to induce mRNA turnover. Thus, gamma-HV SOX homologs have evolved additional activities required for host shutoff. The fact that SOX lacks apparent RNase activity and can function in the absence of other viral factors suggests that it must interface with one or more cytoplasmic cellular factors not targeted by HSV-1 AE to trigger mRNA turnover. Thus far, however, no cellular proteins have been identified that physically interact with either SOX or muSOX during infection. Although PABPC is specifically targeted for relocalization by the gamma-HV SOX proteins and not by HSV-1 AE, we have not detected an interaction between SOX and PABPC (unpublished observations). PABPC localization must therefore be regulated by another SOX or muSOX cytoplasmic target.
While cytoplasmic localization is clearly important for host shutoff, one possibility is that the SOX homologs are nuclear-cytoplasmic shuttling proteins whose shuttling activity is also required to promote mRNA turnover. However, we have observed that while treatment with the CRM1-mediated nuclear protein export inhibitor leptomycin B increases the concentration of SOX in the nucleus (28), the elevated nuclear population of SOX does not decrease upon drug removal (unpublished observations). Thus, while we cannot yet formally rule out shuttling activity, this observation suggests that once SOX enters the nucleus it is very inefficiently (if at all) transported back into the cytoplasm. Presumably, one or more cellular factors must retain a portion of SOX in the cytoplasm, thereby facilitating host shutoff.
SOX-induced host shutoff is tightly linked to its ability to promote hyperadenylation of nascent messages in the nucleus (28). mRNA polyadenylation is normally carried out by cellular poly(A) polymerase II (PAPII) in concert with nuclear poly(A) binding protein (PABPN) (24, 25, 52). PABPN is proposed to participate in tail formation and length control by facilitating interactions between PAPII and the cleavage and polyadenylation specificity factor complex over a defined tail length (27). Similar to cellular mRNAs, SOX-induced hyperadenylation requires both PAPII and PABPN, and the resulting aberrantly processed mRNAs fail to be exported for translation (28). In yeast, hyperadenylated mRNAs accumulate at their site of transcription in mutants defective in either 3′-end processing or export (19, 22, 30). Hyperadenylation may therefore be viewed as a quality control signal to retain improperly processed messages in the nucleus and, additionally, may occur on mRNAs whose nuclear biogenesis and export do not take place in a timely manner. Because mRNA processing and export are tightly linked processes (8), it is difficult to dissect which of these events is targeted primarily by SOX and muSOX to promote hyperadenylation. However, our results demonstrate that hyperadenylation is orchestrated by SOX and muSOX from the cytoplasm. Given that hyperadenylation is a nuclear event, we conclude that these viral host shutoff factors do not directly target the cellular polyadenylation machinery or any other nuclear RNA processing factors. Instead, their ability to extend the poly(A) tail length must be indirect, perhaps mediated by one or more cellular targets within the cytoplasm that either directly affect RNA processing or participate in RNA export. It is possible that PABPC may contribute to this phenotype, as it associates with nuclear pre-mRNAs and poly(A) polymerase-γ (20) and has recently been found to be a component of the mRNA 3′-end processing machinery (43). Like hyperadenylation, PABPC import is similarly mediated by the cytoplasmic fraction of SOX and muSOX. However, siRNA-mediated knockdown of the predominant PABPC1 isoform does not prevent SOX-induced hyperadenylation (28), perhaps indicating that other factors participate in this phenotype as well. Identification of these factors, and how they orchestrate mRNA turnover in the nucleus and cytoplasm, will undoubtedly shed light on mechanisms of global repression of gene expression, either during viral infection or in response to other physiological stimuli. Additionally, we anticipate that SOX and its homologs will be powerful tools to help dissect RNA metabolism in metazoans.
Acknowledgments
We thank all members of the Glaunsinger laboratory for helpful discussions and critical readings of the manuscript.
This research was supported by a Howard Temin Career Development Award (5K01CA117982) and a W. M. Keck Foundation Distinguished Young Investigator Award to B.A.G. and by a Chancellor Diversity Fellowship to S.C.
Footnotes
Published ahead of print on 8 July 2009.
REFERENCES
- 1.Adler, H., M. Messerle, M. Wagner, and U. H. Koszinowski. 2000. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J. Virol. 74:6964-6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bechtel, J. T., Y. Liang, J. Hvidding, and D. Ganem. 2003. Host range of Kaposi's sarcoma-associated herpesvirus in cultured cells. J. Virol. 77:6474-6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, Z., and R. M. Krug. 2000. Selective nuclear export of viral mRNAs in influenza-virus-infected cells. Trends Microbiol. 8:376-383. [DOI] [PubMed] [Google Scholar]
- 4.Coscoy, L., and D. Ganem. 2000. Kaposi's sarcoma-associated herpesvirus encodes two proteins that block cell surface display of MHC class I chains by enhancing their endocytosis. Proc. Natl. Acad. Sci. USA 97:8051-8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ebrahimi, B., B. M. Dutia, K. L. Roberts, J. J. Garcia-Ramirez, P. Dickinson, J. P. Stewart, P. Ghazal, D. J. Roy, and A. A. Nash. 2003. Transcriptome profile of murine gammaherpesvirus-68 lytic infection. J. Gen. Virol. 84:99-109. [DOI] [PubMed] [Google Scholar]
- 6.Elgadi, M. M., C. E. Hayes, and J. R. Smiley. 1999. The herpes simplex virus vhs protein induces endoribonucleolytic cleavage of target RNAs in cell extracts. J. Virol. 73:7153-7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Everly, D. N., Jr., P. Feng, I. S. Mian, and G. S. Read. 2002. mRNA degradation by the virion host shutoff (Vhs) protein of herpes simplex virus: genetic and biochemical evidence that Vhs is a nuclease. J. Virol. 76:8560-8571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fasken, M. B., and A. H. Corbett. 2005. Process or perish: quality control in mRNA biogenesis. Nat. Struct. Mol. Biol. 12:482-488. [DOI] [PubMed] [Google Scholar]
- 9.Feederle, R., H. Bannert, H. Lips, N. Muller-Lantzsch, and H. J. Delecluse. 2009. The Epstein-Barr virus alkaline exonuclease BGLF5 serves pleiotropic functions in virus replication. J. Virol. 83:4952-4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ganem, D. 2007. Kaposi's sarcoma-associated herpesvirus, p. 2847-2888. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 11.Glaunsinger, B., L. Chavez, and D. Ganem. 2005. The exonuclease and host shutoff functions of the SOX protein of Kaposi's sarcoma-associated herpesvirus are genetically separable. J. Virol. 79:7396-7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glaunsinger, B., and D. Ganem. 2004. Lytic KSHV infection inhibits host gene expression by accelerating global mRNA turnover. Mol. Cell 13:713-723. [DOI] [PubMed] [Google Scholar]
- 13.Glaunsinger, B. A., and D. E. Ganem. 2006. Messenger RNA turnover and its regulation in herpesviral infection. Adv. Virus Res. 66:337-394. [DOI] [PubMed] [Google Scholar]
- 14.Goldstein, J. N., and S. K. Weller. 1998. In vitro processing of herpes simplex virus type 1 DNA replication intermediates by the viral alkaline nuclease, UL12. J. Virol. 72:8772-8781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldstein, J. N., and S. K. Weller. 1998. The exonuclease activity of HSV-1 UL12 is required for in vivo function. Virology 244:442-457. [DOI] [PubMed] [Google Scholar]
- 16.Hardwicke, M. A., and R. M. Sandri-Goldin. 1994. The herpes simplex virus regulatory protein ICP27 contributes to the decrease in cellular mRNA levels during infection. J. Virol. 68:4797-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardy, W. R., and R. M. Sandri-Goldin. 1994. Herpes simplex virus inhibits host cell splicing, and regulatory protein ICP27 is required for this effect. J. Virol. 68:7790-7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harlow, E., and D. Lane. 1999. Using antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 19.Hilleren, P., and R. Parker. 2001. Defects in the mRNA export factors Rat7p, Gle1p, Mex67p, and Rat8p cause hyperadenylation during 3′-end formation of nascent transcripts. RNA 7:753-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hosoda, N., F. Lejeune, and L. E. Maquat. 2006. Evidence that poly(A) binding protein C1 binds nuclear pre-mRNA poly(A) tails. Mol. Cell. Biol. 26:3085-3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishido, S., C. Wang, B. S. Lee, G. B. Cohen, and J. U. Jung. 2000. Downregulation of major histocompatibility complex class I molecules by Kaposi's sarcoma-associated herpesvirus K3 and K5 proteins. J. Virol. 74:5300-5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jensen, T. H., K. Patricio, T. McCarthy, and M. Rosbash. 2001. A block to mRNA nuclear export in S. cerevisiae leads to hyperadenylation of transcripts that accumulate at the site of transcription. Mol. Cell 7:887-898. [DOI] [PubMed] [Google Scholar]
- 23.Kamitani, W., K. Narayanan, C. Huang, K. Lokugamage, T. Ikegami, N. Ito, H. Kubo, and S. Makino. 2006. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc. Natl. Acad. Sci. USA 103:12885-12890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keller, R. W., U. Kuhn, M. Aragon, L. Bornikova, E. Wahle, and D. G. Bear. 2000. The nuclear poly(A) binding protein, PABP2, forms an oligomeric particle covering the length of the poly(A) tail. J. Mol. Biol. 297:569-583. [DOI] [PubMed] [Google Scholar]
- 25.Kerwitz, Y., U. Kuhn, H. Lilie, A. Knoth, T. Scheuermann, H. Friedrich, E. Schwarz, and E. Wahle. 2003. Stimulation of poly(A) polymerase through a direct interaction with the nuclear poly(A) binding protein allosterically regulated by RNA. EMBO J. 22:3705-3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krikorian, C. R., and G. S. Read. 1991. In vitro mRNA degradation system to study the virion host shutoff function of herpes simplex virus. J. Virol. 65:112-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Küehn, U., M. Güendel, A. Knoth, Y. Kerwitz, S. Rüedel, and E. Wahle. 8 June 2009. Poly(A) tail length is controlled by the nuclear poly(A) binding protein regulating the interaction between poly(A) polymerase and the cleavage and polyadenylation specificity factor. J. Biol. Chem. [Epub ahead of print.] [DOI] [PMC free article] [PubMed]
- 28.Lee, Y., and B. Glaunsinger. 2009. Aberrant herpesvirus-induced polyadenylation correlates with cellular messenger RNA destruction. PLoS Biol. 7:e1000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leib, D. A., T. E. Harrison, K. M. Laslo, M. A. Machalek, N. J. Moorman, and H. W. Virgin. 1999. Interferons regulate the phenotype of wild-type and mutant herpes simplex viruses in vivo. J. Exp. Med. 189:663-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Libri, D., K. Dower, J. Boulay, R. Thomsen, M. Rosbash, and T. H. Jensen. 2002. Interactions between mRNA export commitment, 3′-end quality control, and nuclear degradation. Mol. Cell. Biol. 22:8254-8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lukac, D. M., R. Renne, J. R. Kirshner, and D. Ganem. 1998. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology 252:304-312. [DOI] [PubMed] [Google Scholar]
- 32.Lyles, D. S. 2000. Cytopathogenesis and inhibition of host gene expression by RNA viruses. Microbiol. Mol. Biol. Rev. 64:709-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez, R., R. T. Sarisky, P. C. Weber, and S. K. Weller. 1996. Herpes simplex virus type 1 alkaline nuclease is required for efficient processing of viral DNA replication intermediates. J. Virol. 70:2075-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martinez-Guzman, D., T. Rickabaugh, T. T. Wu, H. Brown, S. Cole, M. J. Song, L. Tong, and R. Sun. 2003. Transcription program of murine gammaherpesvirus 68. J. Virol. 77:10488-10503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Overby, L. R., E. E. Robishaw, J. B. Schleicher, A. Rueter, N. L. Shipkowitz, and J. C. Mao. 1974. Inhibition of herpes simplex virus replication by phosphonoacetic acid. Antimicrob. Agents Chemother. 6:360-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phelan, A., M. Carmo-Fonseca, J. McLaughlan, A. I. Lamond, and J. B. Clements. 1993. A herpes simplex virus type 1 immediate-early gene product, IE63, regulates small nuclear ribonucleoprotein distribution. Proc. Natl. Acad. Sci. USA 90:9056-9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Racaniello, V. R. 2007. Picornaviridae: the viruses and their replication, p. 795-838. In D. M Knipe and P. M. Howley (ed.), Fields virology, 5th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 38.Reuven, N. B., S. Antoku, and S. K. Weller. 2004. The UL12.5 gene product of herpes simplex virus type 1 exhibits nuclease and strand exchange activities but does not localize to the nucleus. J. Virol. 78:4599-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reuven, N. B., A. E. Staire, R. S. Myers, and S. K. Weller. 2003. The herpes simplex virus type 1 alkaline nuclease and single-stranded DNA binding protein mediate strand exchange in vitro. J. Virol. 77:7425-7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rickinson, A. B., and E. Kieff. 2007. Epstein-Barr virus, p. 2655-2700. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 41.Rowe, M., B. Glaunsinger, D. van Leeuwen, J. Zuo, D. Sweetman, D. Ganem, J. Middeldorp, E. J. Wiertz, and M. E. Ressing. 2007. Host shutoff during productive Epstein-Barr virus infection is mediated by BGLF5 and may contribute to immune evasion. Proc. Natl. Acad. Sci. USA 104:3366-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sandri-Goldin, R. M., M. K. Hibbard, and M. A. Hardwicke. 1995. The C-terminal repressor region of herpes simplex virus type 1 ICP27 is required for the redistribution of small nuclear ribonucleoprotein particles and splicing factor SC35; however, these alterations are not sufficient to inhibit host cell splicing. J. Virol. 69:6063-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi, Y., D. C. Di Giammartino, D. Taylor, A. Sarkeshik, W. J. Rice, J. R. Yates III, J. Frank, and J. L. Manley. 2009. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol. Cell 33:365-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh, G., S. Jakob, M. G. Kleedehn, and J. Lykke-Andersen. 2007. Communication with the exon-junction complex and activation of nonsense-mediated decay by human Upf proteins occur in the cytoplasm. Mol. Cell 27:780-792. [DOI] [PubMed] [Google Scholar]
- 45.Smiley, J. R., M. M. Elgadi, and H. A. Saffran. 2001. Herpes simplex virus vhs protein. Methods Enzymol. 342:440-451. [DOI] [PubMed] [Google Scholar]
- 46.Smith, G. A., and L. W. Enquist. 1999. Construction and transposon mutagenesis in Escherichia coli of a full-length infectious clone of pseudorabies virus, an alphaherpesvirus. J. Virol. 73:6405-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Speck, S. H., and H. W. Virgin. 1999. Host and viral genetics of chronic infection: a mouse model of gamma-herpesvirus pathogenesis. Curr. Opin. Microbiol. 2:403-409. [DOI] [PubMed] [Google Scholar]
- 48.Stevenson, P. G., S. Efstathiou, P. C. Doherty, and P. J. Lehner. 2000. Inhibition of MHC class I-restricted antigen presentation by gamma 2-herpesviruses. Proc. Natl. Acad. Sci. USA 97:8455-8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strelow, L., T. Smith, and D. Leib. 1997. The virion host shutoff function of herpes simplex virus type 1 plays a role in corneal invasion and functions independently of the cell cycle. Virology 231:28-34. [DOI] [PubMed] [Google Scholar]
- 50.Strelow, L. I., and D. A. Leib. 1996. Analysis of conserved domains of UL41 of herpes simplex virus type 1 in virion host shutoff and pathogenesis. J. Virol. 70:5665-5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strelow, L. I., and D. A. Leib. 1995. Role of the virion host shutoff (vhs) of herpes simplex virus type 1 in latency and pathogenesis. J. Virol. 69:6779-6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wahle, E. 1995. Poly(A) tail length control is caused by termination of processive synthesis. J. Biol. Chem. 270:2800-2808. [DOI] [PubMed] [Google Scholar]
- 53.Zelus, B. D., R. S. Stewart, and J. Ross. 1996. The virion host shutoff protein of herpes simplex virus type 1: messenger ribonucleolytic activity in vitro. J. Virol. 70:2411-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zuo, J., W. Thomas, D. van Leeuwen, J. M. Middeldorp, E. J. Wiertz, M. E. Ressing, and M. Rowe. 2008. The DNase of gammaherpesviruses impairs recognition by virus-specific CD8+ T cells through an additional host shutoff function. J. Virol. 82:2385-2393. [DOI] [PMC free article] [PubMed] [Google Scholar]