Abstract
The parainfluenza virus simian virus 5 (SV5) is a poor inducer of innate immune responses. In contrast, the naturally occurring SV5 variant Wake Forest parainfluenza virus (WF-PIV) activates the synthesis of proinflammatory cytokines and beta interferon (IFN-β). Comparison of SV5 and WF-PIV genome sequences revealed nine nucleotide differences within the viral genomic promoter, including two substitutions (U5C and A14G) in the most highly conserved 3′-end promoter element. To test the consequences of these promoter variations, a recombinant SV5 mutant [Le-(U5C, A14G)] was engineered to harbor the two WF-PIV genomic promoter substitutions in an otherwise wild-type (WT) SV5 background. Human lung epithelial cells infected with the Le-(U5C, A14G) mutant had higher rates of viral protein synthesis and levels of mRNA than cells infected with WT SV5, but levels of genomic RNA were not changed. Unlike WT SV5, the Le-(U5C, A14G) mutant was a potent inducer of interleukin-6 and IFN-β synthesis, despite expressing a functional V protein antagonist. Cytokine responses to Le-(U5C, A14G) infection were reduced either by small interfering RNA-mediated knockdown of retinoic acid-inducible gene I (RIG-I) or after infection of cells that were engineered to express the reovirus sigma3 double-stranded RNA-binding protein. Le-(U5C, A14G) induced cytopathic effects not seen with WT SV5, and the extent of cell killing correlated with elevated levels of viral F protein and cell-cell fusion. Our results support a model whereby the SV5 promoter has evolved to function at an attenuated level in order to limit (i) synthesis of aberrant RNAs which induce RIG-I-mediated responses and (ii) overproduction of mRNA for potentially toxic gene products, such as the F protein. Control of genomic promoter activity may be particularly important for viruses such as SV5, that express a V protein targeting mda-5 but do not encode antagonists such as the paramyxovirus C proteins, that specifically target RIG-I.
Innate host cell responses to viral infection are important factors that play major roles in viral pathogenesis, adaptive immunity, and tropism for specific tissues or cells (2). These antiviral responses include the stimulation of type I interferon (IFN) pathways and the secretion of proinflammatory cytokines, such as interleukin-6 (IL-6) and IL-8 (21, 41). Paramyxoviruses, such as simian virus 5 (SV5), can prevent activation of host antiviral responses by directly inhibiting the pathways that lead to activation (4) or by attenuating the expression of viral products that are the activators of a host cell response (9). Here we provide evidence for the role of the viral genomic promoter in enabling SV5 to avoid activation of innate responses.
Members of the paramyxovirus family of negative-strand RNA viruses employ a diverse range of mechanisms to circumvent host cell antiviral responses (reviewed in references 7 and 15). Many of these mechanisms for counteracting cytokine responses have been attributed to products of the paramyxovirus P/V (or P/V/C) gene, which, depending on the particular virus, encodes the phosphoprotein P subunit of the RNA-dependent RNA polymerase, the accessory V protein, and in some cases the family of C proteins (8, 26, 14, 33). For SV5, accurate transcription of the P/V gene results in an mRNA coding for the V protein. The P mRNA is identical to the V mRNA except for the addition of two nontemplated G residues that are inserted by the viral polymerase at a precise location in the P/V transcript (44). The P and V proteins have unique C-terminal regions, with the V protein encoding a highly conserved cysteine-rich domain that is required for many V-associated functions (8, 17, 38).
A major function of the SV5 V protein is the inhibition of IFN signaling, which occurs through V-mediated targeting of signal transducer and activator of transcription 1 (STAT1) for ubiquitylation and degradation (1, 8, 45). The SV5 V protein also blocks activation of the IFN-β promoter during virus infection (17) or following transfection of double-stranded RNA (dsRNA) (4, 38). The paramyxovirus V protein inhibits IFN-β induction by targeting the IFN-inducible RNA helicase encoded by the melanoma differentiation-associated gene 5 (mda-5) (4, 5). Inhibition of mda-5 activation requires only the V protein cysteine-rich C-terminal domain (4, 38), which prevents mda-5 from binding dsRNA and self-association (5). In contrast, the alternative RNA helicase retinoic acid-inducible gene I (RIG-I) does not appear to be inhibited by V protein (4, 5). V protein also appears to act as a decoy substrate for kinases that activate IFN regulatory factor 3 (IRF-3) (29) and IRF-7 (36). Similarly, the SV5 V protein is reported to block IL-6 induction (27), although virus-infected cells still respond to tumor necrosis factor alpha-mediated induction of IL-6 (50).
In addition to the V protein, some paramyxoviruses, such as Sendai virus (SeV) and measles virus, encode a group of C proteins that are expressed from the P/V/C gene (26). The SeV C protein can prevent IFN-β induction (42) by a mechanism that is thought to involve the inhibition of RIG-I activation (42). Thus, paramyxoviruses that express both V and C proteins have the capacity to block both mda-5 via the V protein and RIG-I via the C proteins. Rubulaviruses, such as SV5 and mumps virus, express a V protein but do not encode C proteins (26). This raises the important question of how SV5 can establish a robust, highly productive infection of epithelial cells without activation of RIG-I.
Wild-type (WT) SV5 is a poor inducer of host cell responses, including the activation of proinflammatory cytokines and IFN-β (6, 17, 46). In contrast, we have shown previously that the naturally occurring SV5 variant Wake Forest isolate of parainfluenza virus (WF-PIV) has a delay in viral RNA synthesis but is a potent inducer of IL-8 and IFN-β responses (50). WF-PIV is closely related to the CPI strain of SV5 and was isolated in our lab from persistently infected Vero cells (50). Sequence analysis has identified many nucleotide differences between WF-PIV and WT SV5 (50). Exchanging the P/V gene from WF-PIV or a second variant, CPI-, into an otherwise WT SV5 background results in a chimeric virus that overexpresses viral gene products and activates cytokine secretion and apoptosis (10, 46, 50). These results illustrate that chimeras can have phenotypes that are very different from those of the two parental viruses (10) and suggest that the trans-acting paramyxovirus gene products function in concert to coordinate viral gene expression and limit host cell responses. Here we show that the sequence of the WF-PIV genomic promoter differs substantially from that of WT SV5 in a key regulatory domain, and we analyze the consequences of variability in these cis-acting sequences for gene expression and antiviral responses.
The paramyxovirus genome contains a promoter at its 3′ end that directs both mRNA transcription and genome replication (26) and is transcribed into a short noncoding leader RNA. For many paramyxoviruses, such as SV5, SeV, and human parainfluenza virus type 3, the genomic promoter contains two 18-base promoter elements (PrE) that are essential for viral RNA synthesis (19, 43, 31, 32). PrE-I is located at the ultimate 3′ end of the genome and is separated from a second internal PrE, PrE-II, by a sequence-independent spacer region (Fig. 1). The WF-PIV genomic promoter differs from that of WT SV5 at nine positions, but unexpectedly two of the WF-PIV changes are in the highly conserved PrE-I. We introduced these two nucleotide changes into the SV5 genomic promoter and created a mutant virus which overexpressed viral mRNA and protein. Despite expressing a functional V protein, this promoter mutant virus unexpectedly also activated host cell cytokine responses through pathways that were dependent on RIG-I and dsRNA. Our results support a model whereby the SV5 genomic promoter plays a role in limiting production of aberrant viral RNA that can trigger RIG-I-mediated antiviral pathways. Control of genomic promoter activity may be particularly important for viruses such as SV5, that express a V protein targeting mda-5 but do not encode antagonists such as the C proteins, that specifically target RIG-I (42).
FIG. 1.
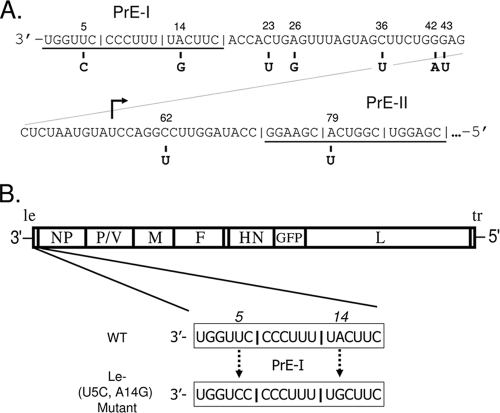
Schematic diagram of SV5 genomic promoter and viruses used in this study. (A) Genomic promoter. The 3′-to-5′ sequence of the 90-base SV5 genomic promoter is shown, with the positions of nucleotides that differ between WT SV5 (top) and the WF-PIV variant of SV5 (bottom). The three hexamers that compose PrE-I and PrE-II are denoted by underlining, and the start site for transcription of the NP gene is shown by an arrow. (B) The genome structure of SV5 is shown, with the addition of the GFP gene between HN and L, as described previously (16). The sequences at positions 5 and 14 in the PrE-I region that differ between WT SV5-GFP and the Le-(U5C, A14G) mutant are indicated. le, leader; tr, trailer.
MATERIALS AND METHODS
Cells, viruses, and growth analysis.
Monolayer cultures of A549 and HeLa cells were grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% heat-inactivated fetal bovine serum. A549 cells that constitutively express reovirus type 3 Dearing sigma3 protein were generated by transfection with pCXN-S4T3D (25), followed by selection in DMEM containing 0.5 mg/ml G418. Individual colonies were picked, expanded in media containing G418, and screened by Western blotting with polyclonal rabbit serum against sigma3. The sigma3 plasmid and antiserum were kindly provided by T. Kobayashi and T. Dermody (Vanderbilt University School of Medicine).
WT SV5 expressing green fluorescent protein (SV5-GFP) was recovered, as described previously (46), from a cDNA plasmid (16) kindly provided by Robert Lamb (Northwestern University) and Biao He (Penn State University). SV5-GFP stocks were grown in MDBK cells. To construct the SV5 leader mutant Le-(U5C, A14G), PCR was used to introduce a T-to-C change in position 5 and an A-to-G change in position 14 of the leader region of the infectious clone for WT SV5-GFP. The Le-(U5C, A14G) mutant was recovered as described previously (46), with minor modifications.
Virus stock was generated in Vero cells by low-multiplicity of infection (MOI) infection, as described previously (13), in order to prevent generation of defective particles. To assay virus growth, ∼106 A549 cells in six-well dishes were infected at an MOI of 0.05 (multistep growth) or 10 (single-step growth) for 1 h and replaced with DMEM containing 2% fetal bovine serum. Supernatant was collected at various times, and plaque assays were performed with CV-1 cells as described previously (46).
Western blotting and metabolic labeling.
For Western blotting, six-well dishes of cells were infected as described in the figure legends. At each time point, cells were washed with phosphate-buffered saline and lysed in 1% sodium dodecyl sulfate (SDS). The cell lysate protein concentration was determined by a bicinchoninic acid assay (Pierce Chemicals), and equivalent amounts of protein were analyzed by Western blotting with rabbit antiserum to cellular STAT-1 (Santa Cruz Biotechnology), monoclonal anti-V5 (Invitrogen) for V protein, or rabbit sera specific for the individual SV5 NP, P, and M proteins (9). As a loading control, lysates were also probed with mouse anti-β-actin antibodies (A5316; Sigma). Blots were visualized by horseradish peroxidase-conjugated secondary antibodies and enhanced chemiluminescence (Pierce Chemicals).
To examine rates of protein synthesis, cells were mock infected or infected at a high MOI. At the times postinfection (p.i.) indicated in the figure legends, cells were starved for 25 min with DMEM lacking cysteine and methionine and then radiolabeled for 20 min with 100 μCi/ml Tran[35S]-label or [35S]cysteine for detecting V protein. For chase experiments, cells were radiolabeled and then incubated in nonradioactive media for 3 h and lysed in 1% SDS, and equal amounts of protein were immunoprecipitated using rabbit polyclonal antisera to SV5 NP, P, M, and F proteins as described previously (9).
Analysis of viral RNA and small interfering RNA (siRNA) experiments.
The WF-PIV genomic promoter was cloned using 5′ rapid amplification of cDNA ends, according to the manufacturer's protocols (Invitrogen), on purified nucleocapsid-derived RNA isolated from infected cells. Details and primer sequences are available upon request. The resulting PCR-amplified fragment was cloned into a pGEM5 vector and sequenced.
An RNase protection assay (RPA) was carried out using an Ambion RPAIII kit according to the manufacturer's protocols and as previously described (9). To detect mRNA, RNA was collected from mock-infected or high-MOI-infected samples by use of Trizol (Invitrogen). To assay levels of viral genomes, cells were collected in phosphate-buffered saline containing 50 mM EDTA and cell lysates were treated with micrococcal nuclease to digest RNAs not encapsidated by NP (24, 35). RNA was isolated by Trizol extraction.
Riboprobes were generated by in vitro transcription of a linearized plasmid encoding negative-sense M (for mRNA) or positive-sense leader-NP junction sequences (for genomes) in the presence of [α-32P]CTP, as detailed previously (9). Hybridized RNA samples were treated with RNase A/T1, resolved on 9 M urea-6% polyacrylamide gels, visualized by autoradiography, and quantified by phosphorimaging.
For knockdown of RIG-I, A549 cells in 24-well plates were transfected using Transit siQuest (Mirus, Inc.) with siRNA specific for RIG-I (Dharmacon M-0125-01; 100 nM final concentration) or nontarget control RNA (Dharmacon D-001206-13; 100 nM final concentration) according to the manufacturer's instructions and as previously described (13). Forty-eight hours posttransfection, cells were infected as described above.
ELISAs and cell viability assays.
Immunoreactive IL-6 (BD Opt EIA; BD Biosciences) or IFN-β (PBL Biomedical Laboratories) in extracellular medium was quantified by a dual-antibody sandwich enzyme-linked immunosorbent assay (ELISA) according to the manufacturer's protocols. To allow comparison between experiments, cytokine levels were determined for the number of cells at the time of infection and values were normalized to the level for 106 cells. Cell viability was measured by using a CellTiter 96 AQueaous One Solution cell proliferation assay (Promega) according to the manufacturer's instructions.
Microscopy.
Phase microscopy and fluorescent microscopy were carried out as described previously (50). Analysis of IRF-3 nuclear translocation was performed as detailed elsewhere (13). Images were captured using a QImaging digital camera and processed using Q-capture software. Exposure times were set manually to be constant between samples.
RESULTS
Substitutions in the genomic PrE-I region lead to elevated viral protein and RNA synthesis.
cDNA clones for the WF-PIV 3′ genomic promoter region were generated from antigenomic RNA using 5′ rapid amplification of cDNA ends-reverse transcription-PCR on nucleocapsid-associated RNA. As shown in Fig. 1A, the WT SV5 genomic promoter consists of the 18-base 3′-end PrE-I, the 18-base internal PrE-II, and a 54-nucleotide spacer region (32). Compared to the sequence of WT SV5, the sequence of the WF-PIV genomic promoter contained nine nucleotide differences, with most of the variation clustering in the spacer region. One nucleotide difference was in the internal PrE-II domain at position 79, but results with a minigenome system have shown that substitutions in this position do not affect SV5 RNA synthesis (32). Likewise, while the 54-base region between the 3′-end PrE-I and the internal PrE-II showed six nucleotide differences, we have shown previously that this region plays a very minor role in RNA synthesis (24). The WF-PIV PrE-I region contained two nucleotide differences: a U-to-C change at position 5 and an A-to-G change at position 14. These PrE-I differences had the most potential significance, because PrE-I is among the most highly conserved regions in the genomes of related paramyxoviruses (31).
The above-described sequencing results raised the hypothesis that nucleotide differences in the highly conserved PrE-I region would alter viral RNA synthesis. To test this hypothesis, a recombinant SV5 [Le-(U5C, A14G)] was engineered to harbor only the U5C and A14G substitutions in PrE-I, with the remainder of the genome being derived from WT SV5. As shown in Fig. 1B, the Le-(U5C, A14G) mutant also contained a GFP gene inserted between the HN and L genes, as described previously (16). For these studies, WT SV5-GFP (16) was used as the proper control for analysis of the Le-(U5C, A14G) mutant.
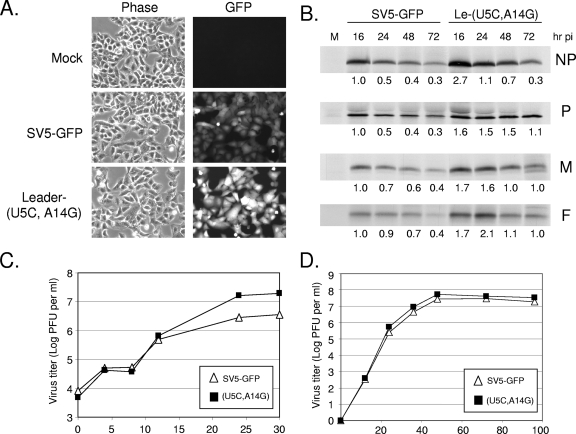
Cells infected with the Le-(U5C, A14G) mutant were found to overexpress viral protein. This is evident in the micrographs in Fig. 2A, where GFP expression was enhanced in cells 24 h p.i. with the Le-(U5C, A14G) mutant compared to that seen with WT SV5-GFP. Western blotting analysis revealed similar results for other viral proteins (data not shown). To determine whether increased levels of viral protein correlated with elevated rates of translation, infected cells were radiolabeled with Tran[35S]-label at various times p.i., and proteins were analyzed by immunoprecipitation. As shown in Fig. 2B, the rate of protein synthesis at 16 h p.i. for the Le-(U5C, A14G) mutant was 1.6- to 2.7-fold higher than that for SV5-GFP, and this was seen for proteins encoded in both the 3′-proximal (e.g., NP) and the 3′-distal (e.g., F) regions of the genome. The rate of viral protein synthesis for both SV5-GFP and the Le-(U5C, A14G) mutant decreased at later times p.i. to ∼30 to 40% of that seen at 16 h p.i. However, translation for the mutant was always higher than that seen for SV5-GFP at all time points and for all genes examined.
FIG. 2.
The Le-(U5C, A14G) mutant overexpresses viral protein compared to WT SV5. (A) Microscopy. A549 cells were mock infected or infected at an MOI of 10 with SV5-GFP or the Le-(U5C, A14G) mutant. Cells were examined by microscopy at 24 h p.i. (B) Rate of viral protein synthesis. A549 cells were mock infected (lane M) or infected at an MOI of 10 with SV5-GFP or the Le-(U5C, A14G) mutant. At the indicated times p.i., cells were radiolabeled for 20 min with Tran[35S]-label. Cells were lysed, and equal amounts of protein were immunoprecipitated with antibodies to the indicated proteins before analysis by SDS-polyacrylamide gel electrophoresis and autoradiography. Numbers beneath the autoradiograms indicate the expression levels relative to those for WT-infected cells at 16 h p.i. (C and D) Virus growth kinetics. A549 cells were infected at an MOI of 10 (C) or 0.05 (D), and virus in the media at the indicated times p.i. was quantitated by plaque assay. Data are representative of three independent experiments.
The Le-(U5C, A14G) mutant grew to a titer ∼1 log higher than that for SV5-GFP under single-step growth conditions (Fig. 2C), but growth of the mutant virus was very similar to that of SV5-GFP by multistep growth analysis (Fig. 2D). As we have reported previously (13), the enhanced gene expression and WT growth properties for the Le-(U5C, A14G) mutant are not consistent with defective viral particles, and the PFU-to-HA ratios were very similar between WT (10,766 ± 1,335) and mutant (8,723 ± 1,897) viruses.
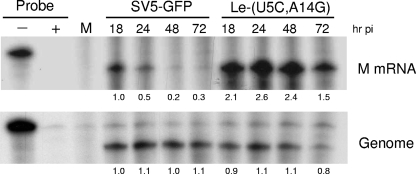
Levels of viral protein expression for negative-strand RNA viruses are thought to be regulated primarily at the level of transcription (26). To determine whether levels of viral RNA were also altered for the Le-(U5C, A14G) mutant, RPAs were carried out using riboprobes specific for viral genomic RNA or for M mRNA as a representative viral gene. As shown in Fig. 3, M mRNA levels in cells infected with SV5-GFP were initially high at 18 h p.i., but levels decreased to the level of detection by 48 h p.i. This result is consistent with the decrease in translation rates shown in Fig. 2B. In contrast, cells infected with the Le-(U5C, A14G) mutant maintained levels of mRNA that were more than twofold higher than those for WT SV5 at all times p.i. In contrast to mRNA, levels of genomic RNA were very similar for cells infected with WT and mutant viruses. Genomic RNA remained relatively constant out to 72 h p.i., with the exception of slightly lower levels for the Le-(U5C, A14G) mutant at 72 h p.i., which may reflect the cytopathic effect (see below). Taken together, these results indicate that the Le-(U5C, A14G) mutant overexpresses viral protein, which correlates with an increase in the ratio of viral mRNA to genomic RNA compared to the ratio for WT SV5.
FIG. 3.
Accumulation of Le-(U5C, A14G) mutant mRNA and genomic RNA. A549 cells were mock infected (lane M) or infected at an MOI of 10 with SV5-GFP or the Le-(U5C, A14G) mutant. At the indicated times p.i., RNA was harvested as described in Materials and Methods and analyzed by an RPA using 32P-labeled riboprobes specific for M mRNA or the leader-NP junction for genomic RNA. − and + indicate control samples where riboprobe alone was incubated without or with nuclease, respectively. The − lanes represent 1/50 of the input probe for each sample. The number for each lane indicates the change in signal (n-fold) relative to the level at 18 h p.i., with the level for SV5-GFP set at 1.0.
The Le-(U5C, A14G) mutant induces cytokine responses, despite expressing a functional V protein.
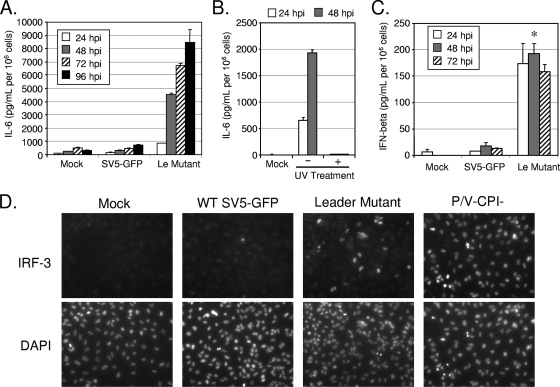
The above-described finding of elevated viral mRNA synthesis with the leader mutant raised the hypothesis that the Le-(U5C, A14G) virus would induce antiviral responses. To test this hypothesis, A549 cells were mock infected or infected at a high MOI with SV5-GFP or the Le-(U5C, A14G) mutant, and levels of secreted IL-6 and IFN-β were examined by ELISA. As shown in Fig. 4A, cells infected with the Le-(U5C, A14G) mutant showed a time-dependent increase in IL-6 secretion, which was not seen with SV5-GFP infection. IL-6 induction was eliminated by treatment of the leader mutant virus with UV light (Fig. 4B). Likewise, Le-(U5C, A14G) infection resulted in IFN-β secretion, but unlike with IL-6, the levels of secreted IFN-β were relatively low and did not increase further after 24 h p.i. (Fig. 4C). This IFN induction did not affect low multicycle growth (Fig. 2), consistent with previous work showing that multicycle growth of SV5 expressing a WT V protein is relatively insensitive to low levels of IFN (47).
FIG. 4.
Induction of proinflammatory cytokines and IFN by the Le-(U5C, A14G) mutant but not by WT SV5. (A, B, and C) Induction of cytokines. A549 cells were mock infected or infected at an MOI of 10 with SV5-GFP or the Le-(U5C, A14G) mutant, and at the indicated times p.i., levels of secreted IL-6 (A and B) or IFN-β (C) were determined by ELISA. Results are the expressed as mean values from triplicate samples, with error bars representing standard deviations, and are normalized to the level for 106 cells. *, P value of <0.005 compared to corresponding samples from control cells. For panel B, the Le-(U5C, A14G) mutant virus was treated with (+) or without (−) UV light prior to infection. (D) IRF-3 nuclear translocation. A549 cells were infected at an MOI of 10 with the indicated viruses. At 24 h p.i., cells were stained for IRF-3 using a monoclonal antibody and for the nucleus using DAPI (4′,6-diamidino-2-phenylindole).
IFN-β synthesis requires the activation and translocation of IRF-3 to the nucleus to initiate transcription of the IFN-β gene (18). To determine the extent of IRF-3 activation by the leader mutant, A549 cells were infected at a high MOI with WT SV5-GFP, the Le-(U5C, A14G) mutant, or SV5-P/V-CPI-, a mutant that we have previously shown to be a potent inducer of IFN-β (9). As shown in Fig. 4D, the diffuse cytoplasmic IRF-3 location in WT SV5-GFP-infected cells resulted in a low labeling intensity that was indistinguishable from that with mock-infected cells, as described previously (9). In the case of the positive-control P/V-CPI- infection, nearly all cells showed intense labeling resulting from translocation of IRF-3 to the nucleus. This is consistent with previous results showing this mutant virus inducing ∼4,000 pg/ml of IFN-β at 24 h p.i. (9). Most of the cells infected with the Le-(U5C, A14G) mutant showed diffuse cytoplasmic IRF-3 staining, although a few cells in the population showed bright nuclear localization. These data are consistent with the results in Fig. 4C showing a low-level induction of IFN-β by the Le-(U5C, A14G) mutant.
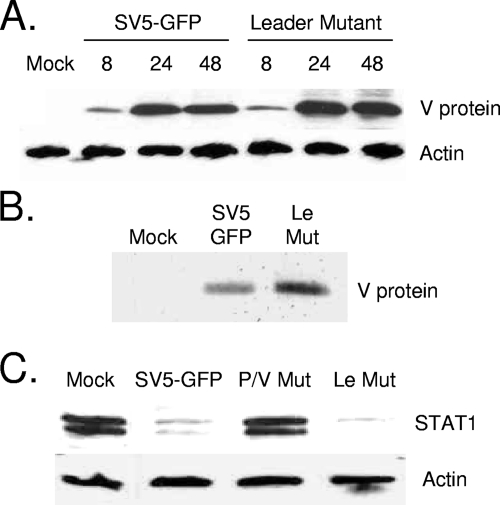
The SV5 V protein has been shown to block induction of both IL-6 and IFN-β (17, 27) and can target STAT1 for degradation (8). The backbone of the Le-(U5C, A14G) mutant encodes a WT V protein. As shown in Fig. 5A, the kinetics of V protein accumulation were slightly higher for cells infected with the Le-(U5C, A14G) mutant than for cells infected with SV5-GFP. Consistent with this, immunoprecipitation of lysates from radiolabeled cells showed that the rate of V protein synthesis was elevated for the Le-(U5C, A14G) mutant compared to that for SV5-GFP (Fig. 5B). To confirm that V protein functioned in the context of the Le-(U5C, A14G) mutant infection, A549 cells were infected at an MOI of 10 with the viruses indicated in Fig. 5C, and levels of STAT1 were analyzed by Western blotting at 8 h p.i. This time point was chosen in order to detect any small differences in the STAT1 levels that may not be evident at later times p.i. WT SV5-GFP showed reduced STAT1 levels compared to those for mock-infected cells. STAT1 levels were not decreased following infection with the P/V mutant P/V-CPI-, which is defective in targeting STAT1 degradation (46). Cells infected with the Le-(U5C, A14G) mutant showed a loss of STAT1 equivalent to that seen for WT SV5-GFP. These data indicate that the V protein expression levels are similar to that of WT SV5 and that V protein encoded by the Le-(U5C, A14G) mutant retains function in blocking antiviral signaling.
FIG. 5.
The Le-(U5C, A14G) mutant (Mut) encodes a functional V protein. (A) Time course of V protein expression. A549 cells were infected at a high MOI, and at the indicated times p.i., cell lysates were analyzed by Western blotting for levels of V protein and cellular actin. (B) Cells infected with the indicated viruses were labeled at 24 h p.i. for 30 min with [35S]cysteine before immunoprecipitation with anti-V protein antibody. (C) STAT1 levels. A549 cells were infected at an MOI of 10 with the indicated viruses. Cell lysates were prepared at 8 h p.i., and levels of STAT1 and actin were determined by Western blotting.
Role of RIG-I and dsRNA in cytokine induction by the Le-(U5C, A14G) mutant.
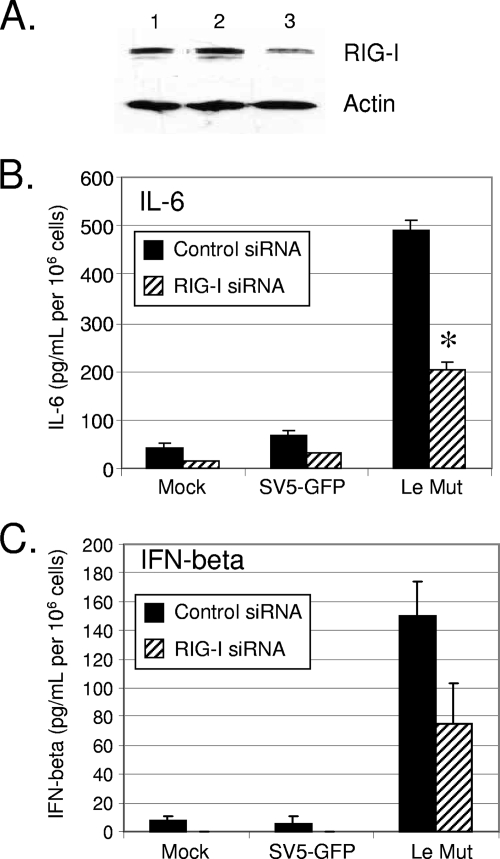
The V protein has been shown to bind to and inhibit the RNA helicase mda-5, but not RIG-I (1, 4). The above-described findings that the Le-(U5C,A14G) mutant induced cytokine secretion in the presence of WT V protein raised the hypothesis that RIG-I contributed to IL-6 and IFN induction. siRNA experiments were carried out to determine whether knockdown of RIG-I levels affected the induction of cytokines by Le-(U5C,A14G) infection. Western blotting showed that levels of RIG-I at 48 h posttransfection were reduced by >50% in A549 cells treated with siRNAs specific for RIG-I (Fig. 6A, lane 3) but not in control samples (Fig. 6A, lane 2). At 48 h after siRNA treatment, cells were mock infected or infected with SV5-GFP or Le-(U5C, A14G). Twenty-four hours later, media were collected and cytokine levels determined by ELISA. As shown in Fig. 6B and C, there was a substantial decrease in levels of IL-6 and IFN-β released by 24 h from infected cells in which RIG-I levels were reduced, indicating a role for RIG-I in activation of cytokines by the Le-(U5C, A14G) mutant. Consistent with other published data regarding A549 cells (3), the level of RIG-I knockdown was only ∼60%. We propose that if knockdown were more complete, the levels of cytokine induction would be further decreased.
FIG. 6.
Role of RIG-I in cytokine induction by the Le-(U5C, A14G) mutant (Mut). (A) RIG-I knockdown. A549 cells were left untreated (lane 1) or transfected with control siRNA (lane 2) or siRNA targeting RIG-I (lane 3). Forty-eight hours posttransfection, levels of RIG-I and actin were assayed by Western blotting. (B and C) Cytokine secretion. A549 cells were transfected with control siRNA or siRNA specific for RIG-I. At 48 h posttransfection, cells were mock infected or infected at an MOI of 10 with SV5-GFP or the Le-(U5C, A14G) mutant, and levels of secreted IL-6 (B) or IFN-β (C) were measured by ELISA at 24 h p.i. Results are expressed as mean values from triplicate samples, with error bars representing standard deviations, and are normalized to the level for 106 cells. *, P value of <0.005 compared to corresponding samples from control cells.
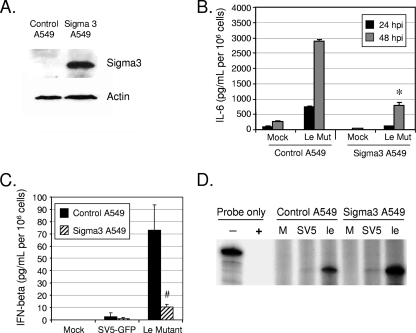
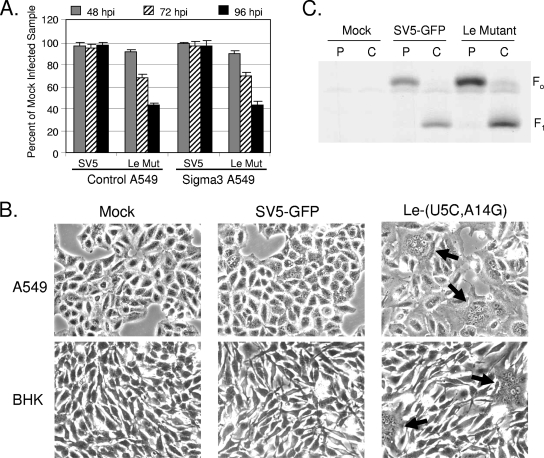
RIG-I can respond to the presence of dsRNA (23). To determine the role of dsRNA in cytokine induction by the Le-(U5C, A14G) mutant, an A549 cell line was engineered to express the reovirus sigma3 protein, which is known to specifically bind dsRNA (Fig. 7A). A549 control and sigma3-expressing cells were mock infected or infected at a high MOI with the Le-(U5C, A14G) mutant, and levels of cytokines were assayed at various times p.i. As shown in Fig. 7B and C, there was a significant decrease in IL-6 and IFN-β secretion by the Le-(U5C, A14G) mutant in sigma3-expresssing A549 cells compared to the level in control A549 cells. In both the RIG-I knockdown experiment (Fig. 6) and the sigma3-expressing cells (Fig. 7), cytokine induction by WT SV5 was lower than that in the control cells. This is consistent with the published proposal (49) that while SV5 is a poor inducer of cytokines, this is not an absolute, and that infection with WT virus does induce a relatively low level of antiviral response.
FIG. 7.
Role of dsRNA in cytokine induction by the Le-(U5C, A14G) mutant (Mut). (A) A549 cells expressing reovirus sigma3 protein. Control A549 cells or cells that stably express the reovirus sigma3 protein were analyzed by Western blotting for levels of sigma3. (B and C) Cytokine secretion. Control or sigma3-expressing A549 cells were mock infected or infected at an MOI of 10 with the Le-(U5C, A14G) mutant, and levels of secreted IL-6 were measured by ELISA at the indicated times p.i. For panel C, IFN-β levels were determined at 24 h p.i. Results are expressed as mean values from triplicate samples, with error bars representing standard deviations. * and #, P values of <0.005 and <0.05, respectively, compared to corresponding samples from control cells. (D) Viral M mRNA levels in sigma3-expressing cells. Control or sigma3-expressing A549 cells were mock infected or infected at an MOI of 10 with the indicated viruses. RNA harvested at 48 h p.i. was analyzed by an RPA with a probe specific for M mRNA. − and + indicate control samples where riboprobe alone was incubated without or with nuclease, respectively. The − lane represents 1/50 of the input probe for each sample. M, mock; SV5, SV5-GFP; le, Le-(U5C, A14G) mutant.
Reduced cytokine secretion from sigma3-expressing cells could be due to a reduction in levels of viral mRNA. To test this possibility, levels of M mRNA were determined by RPA at 48 h p.i., a time when there was a large increase in cytokine responses. As shown in Fig. 7D, mRNA levels for WT SV5-GFP were at the limit of detection, as demonstrated in Fig. 3 above), and were not altered by sigma3 expression. Importantly, levels of M mRNA for the Le-(U5C, A14G) mutant were not reduced in the sigma3-expressing cells but instead were ∼3-fold higher than those for the control A549 cells. Together, these results indicate that the Le-(U5C, A14G) mutant induces proinflammatory cytokines through a mechanism that involves production of dsRNA and activation of RIG-I signaling.
The Le-(U5C, A14G) mutant induces cytopathic effects that correlate with increased levels of F protein and cell-cell fusion.
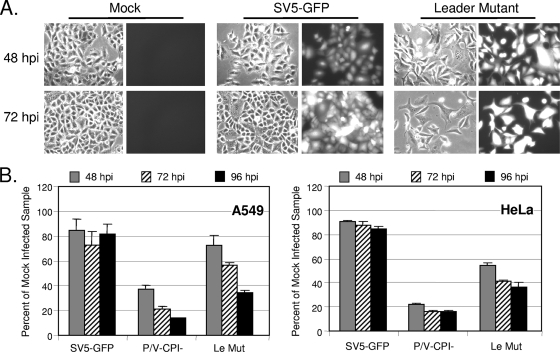
Cells infected with the Le-(U5C, A14G) mutant showed cytopathic effects. This is evident in the microscopy pictures in Fig. 8A, where WT SV5-GFP established a largely noncytopathic infection even out to 72 h p.i. In contrast, cultures infected with the Le-(U5C, A14G) mutant showed fewer cells, cell rounding, and syncytia starting as early as 48 h p.i. By use of a cell viability assay (Fig. 8B), high-MOI infection with the Le-(U5C, A14G) mutant resulted in a time-dependent loss of viability in both A549 and HeLa cells that was not seen with SV5-GFP infection. The loss of viability in cultures infected with the Le-(U5C, A14G) mutant was slower and less extensive than that for the highly cytopathic infection with the positive-control P/V-CPI- mutant (46).
FIG. 8.
Cytopathic effect induced by the Le-(U5C, A14G) mutant (Mut) but not WT SV5-GFP. (A) A549 cells were infected at an MOI of 10 with SV5-GFP or the Le-(U5C, A14G) mutant, and microscopy pictures were taken at 48 and 72 h p.i. (B) Cell viability. A549 or HeLa cells were mock infected or infected with the indicated viruses at an MOI of 10. Cell viability was determined at 48, 72, and 96 h p.i. using a cell viability assay. Data are from quadruplicate samples and are expressed as percentages of the values obtained with mock-infected cells. Error bars represent the standard deviations.
Le-(U5C, A14G)-infected cells had high levels of cleaved caspase-3 (data not shown), indicating that apoptotic pathways were activated. To determine whether production of dsRNA by the leader mutant contributed to cytopathic effects, control or sigma3-expressing A549s were mock infected or infected with SV5-GFP or Le-(U5C, A14G) and the viability of the population of cells was monitored at 48, 72, and 96 h p.i. As shown in Fig. 9A, the Le-(U5C, A14G) mutant induced nearly identical losses of cell viability in both control and sigma3-expressing cells. Similarly, cell killing by the Le-(U5C, A14G) mutant was not affected by RIG-I knockdown (not shown).
FIG. 9.
The Le-(U5C, A14G) mutant (Mut) induces cytopathic effects that correlate with increased levels of F protein and cell-cell fusion. (A) Cell viability. Control or sigma3-expressing A549 cells were mock infected or infected with the indicated viruses at an MOI of 10. Cell viability was determined at 48, 72, and 96 h p.i. using a cell viability assay as described in the legend for Fig. 8. Error bars represent the standard deviations. (B) Microscopy pictures. A549 or BHK cells were mock infected or infected with the indicated viruses at an MOI of 10. Microscopy pictures were taken at 48 h p.i. Arrows indicate syncytium pockets. (C) Rate of F protein expression and cleavage. A549 cells that were mock infected or infected with the indicated viruses were pulse-labeled for 20 min with Tran[35S]-label. Cells were either lysed immediately (pulse lanes, P) or lysed after incubation for 3 h in nonradioactive media (chase lanes, C), immunoprecipitated with anti-F antiserum, and analyzed by SDS-polyacrylamide gel electrophoresis and autoradiography.
During the course of these studies, it was observed that cells infected with the Le-(U5C, A14G) mutant had increased levels of cell-cell fusion. As shown in Fig. 9B, both A549 and BHK cells showed relatively little fusion after infection with SV5-GFP, but very large pockets of syncytia were evident in cultures infected with the Le-(U5C, A14G) mutant. Increased cell-cell fusion correlated with increased levels of the SV5 F protein. This is shown in the pulse-chase radiolabeling experiment with results illustrated in Fig. 9C, where cells infected with the Le-(U5C, A14G) mutant synthesized ∼2- to 3-fold-higher levels of F protein than cells infected with WT SV5-GFP. Likewise, after a 3-h chase period, elevated levels of F were transported through the Golgi body, as assayed by the cleavage of F0 into F1. Together, these data support the contention that cells infected with the Le-(U5C, A14G) mutant have higher levels of cleaved F protein as a result of the global increase in viral gene expression and that this leads to increased cell-cell fusion and cell death.
DISCUSSION
WT SV5 is a poor inducer of host cell antiviral responses, whereas the naturally occurring SV5 variant WF-PIV induces proinflammatory cytokines, IFN-β, and low levels of cell killing (6, 17, 46). The present study was initiated by our finding that SV5 and WF-PIV differ in the sequences of their 3′-end genomic promoters. Based on the finding that the 18-base PrE-I is the most highly conserved stretch of nucleotides in the rubulavirus genome (31), we have tested the hypothesis that SV5 harboring these PrE-I changes would display an altered RNA synthesis profile. Introducing the WF-PIV substitutions into the SV5 genomic promoter enhanced viral mRNA and protein ∼2-fold but had no apparent effect on genome replication levels. However, our most striking finding was that the leader mutant virus was a strong activator of cytokine secretion and cell killing, despite expressing a functional V protein antagonist. As detailed below, our data support the proposal that the paramyxovirus genomic promoter can be an important determinant of host cell antiviral responses and cell killing.
A priori, it was not clear what effect incorporating the WF-PIV substitutions into the highly conserved region of the WT SV5 promoter would have on RNA synthesis. For many negative-strand viruses, changes to the leader sequence have been shown to decrease viral RNA synthesis. For example, with the HPIV3 minigenome system, it has been reported that changes made in various positions of the first 16 bases of the leader RNA result in decreased reporter gene expression (19). Likewise, single point mutations made for each of the 3′-terminal 22 nucleotides of the rinderpest virus genome all resulted in lower levels of CAT expression in a minigenome system (30). We have taken a different approach, which is based on infectious virus and the use of naturally occurring variability in the genomic promoter.
Unexpectedly, the Le-(U5C, A14G) mutant accumulated elevated levels of viral mRNA, but the accumulation of viral genomes was not significantly changed over that seen with WT SV5. Thus, enhanced mRNA levels appeared to reflect elevated transcription, which was not an indirect effect of having an increased number of genomic RNA templates. This result contrasts with results for our previously described P/V mutant, which directs ∼10-fold-higher levels of both mRNA and genomic RNA (9, 46). Current models for viral determinants of transcription versus replication include the kinetics of NP accumulation and the binding of NP to the nascent leader RNA (26). However, results with our leader mutant are consistent with previous proposals that additional factors, such as cis-acting sequences, can dictate the balance between transcription and replication (12). A detailed mutational analysis of the SV5 PrE-I region using our established minigenome system (32, 40) should reveal the cis-acting determinants for transcription versus replication.
The most striking finding from our work is that two nucleotide substitutions in the genomic promoter convert WT SV5 into a virus that activates host cell responses. Our results with siRNA knockdown and expression of the reovirus sigma3 protein indicate that cytokine responses are activated through a cellular pathway involving RIG-I and dsRNA. RIG-I is activated by RNA containing a 5′ triphosphate (20), but recent evidence has shown that RIG-I also has specificity for short dsRNAs (23). Thus, our results are consistent with a model whereby changes in the SV5 genomic promoter lead to aberrant synthesis of short RNAs that have at least partial double-stranded properties and/or contain an accessible 5′ triphosphate. There are three possible sources of viral RNA with these properties that could activate RIG-I. One source could be the free 55-base leader RNA which is uncapped, has a 5′ triphosphate, and also may contain dsRNA structure. Previous data from transfection experiments have shown that the measles virus leader RNA can activate RIG-I (37). Second, elevated viral transcription could yield mRNAs with incomplete capping, which would yield 5′-triphosphate mRNAs with dsRNA structure. Finally, it is possible that changes to the genomic promoter lead to increased levels of aborted genome replication products that are inefficiently encapsidated by NP. The most straightforward hypothesis for RIG-I activation by the Le-(U5C, A14G) mutant involves elevated transcription from the viral genome to produce higher levels of free leader RNA, but further studies are needed to test this hypothesis.
It has been reported that the cellular protein La binds to the leader RNA of rinderpest virus (39) and of respiratory syncytial virus (RSV) (3). In the case of RSV, it is proposed that La binding to leader RNA masks recognition by RIG-I (3), although other reports have indicated a role for RIG-I in early responses to RSV infection (28). If La protein is also involved in SV5 infections, it is possible that elevated transcription from the Le-(U5C, A14G) mutant results in inefficient binding by La protein either due to an excess of leader RNA or due to alterations to La binding sites on the leader RNA. Work is in progress to determine whether SV5 produces a free leader RNA in infected cells and what type of role La protein has in RIG-I activation by the Le-(U5C, A14G) mutant.
Our data indicate that the Le-(U5C, A14G) mutant produces levels of dsRNA and RIG-I activation that are not normally seen in the case of WT SV5-infected cells. Consistent with this model, cytokines are induced in cells that are coinfected with both WT SV5-GFP and the Le-(U5C, A14G) mutant (not shown). Using immunofluorescence with an anti-dsRNA antibody, we have been unable to detect increased levels of dsRNA in cells infected with the Le-(U5C, A14G) mutant (not shown). This is consistent with the work of Weber et al. (48), which did not detect dsRNA during infections with negative-strand RNA viruses. Thus, the inhibition of host cell responses in cells expressing the dsRNA-binding protein likely reflects a low level of synthesis of dsRNA from the Le-(U5C, A14G) mutant, which can be sequestered by sigma3. An online RNAfold program (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi) predicts that the 55-base Le-(U5C, A14G) leader RNA folds into a different secondary structure than does WT leader RNA (not shown), although the free energy of folding is not significantly different. Thus, the differential structure of free leader RNA or interactions with RNA binding proteins could form the basis for differences in host cell response to WT SV5 and Le-(U5C, A14G) mutant infections.
There are a number of cellular pattern recognition receptors other than RIG-I that recognize dsRNA to induce host cell responses, including mda-5 and protein kinase R (PKR). The Le-(U5C, A14G) mutant expresses a functional WT V protein with kinetics that match that of the parental WT SV5 virus, making it reasonable to assume that mda-5 is not involved in responses to the leader mutant. Le-(U5C, A14G) infection does not significantly activate PKR or induce a shutdown of translation (data not shown), two properties of a previously described SV5 P/V mutant (13). This differential effect of Le-(U5C, A14G) on activation of RIG-I pathways but not PKR could reflect the relative sensitivities of these two cytoplasmic receptors to levels or lengths of dsRNA, with RIG-I being more easily activated than PKR. Similarly, the Le-(U5C, A14G) mutant induces apoptotic markers, but cell killing by the mutant was not affected by expression of sigma3 (Fig. 8A). Together, these results are consistent with a model whereby cytokine responses to the leader mutant are induced by a RIG-I-dependent pathway that is activated by dsRNA but whereby apoptosis is either independent of dsRNA or more sensitive to very low levels of dsRNA that have escaped sequestering by the dsRNA-binding protein sigma3.
The Le-(U5C, A14G) mutant induced a cytopathic effect that was not seen in cells infected with WT SV5, and infected cells showed increased levels of cell-cell fusion. The SV5 F protein is constitutively cleaved into an active form due to the presence of five arginine residues at the cleavage site between F1 and F2 (34). The mechanisms that control paramyxovirus fusion activity are not completely understood, but F expression level has been shown to be an important parameter (11). Consistent with this, the leader mutant showed ∼2- to 3-fold-higher levels of F protein synthesis as part of the global increase in gene expression across the Le-(U5C, A14G) genome. We propose that the SV5 genomic promoter has evolved to function at a level that is lower than maximal in order to prevent overexpression of potentially toxic gene products, such as the F protein. Our proposal for the genomic promoter is consistent with previous data whereby changes in mRNA start frequency for the SeV F gene lead to increased fusion (22).
In summary, our data support a model whereby the viral genomic promoter plays two important roles in allowing SV5 to limit antiviral responses and cell killing through (i) controlling the production of aberrant viral RNA that can trigger RIG-I-mediated antiviral pathways and also (ii) attenuating viral mRNA transcription across the genome in order to avoid overproduction of potentially toxic gene products, such as the F protein. Controlling production of dsRNA from the genomic promoter may be particularly important for viruses, such as SV5, that express a V protein targeting mda-5 but do not encode antagonists, such as the SeV C proteins, that specifically target RIG-I (42). Likewise, attenuating promoter activity could be an important property of viruses, such as SV5, which can establish highly productive long-term infections with only minimal cytopathic effects.
Acknowledgments
We thank members of the Parks lab and Doug Lyles for helpful comments on the manuscript. We are grateful to Biao He and Robert A. Lamb (Northwestern University) for the original gift of the SV5 infectious clone and Takeshi Kobayashi and Terry Dermody (Vanderbilt University) for the kind gift of sigma3 plasmid and antiserum.
This work was supported by NIH grant AI-42023.
Footnotes
Published ahead of print on 8 July 2009.
REFERENCES
- 1.Andrejeva, J., K. S. Childs, D. F. Young, R. S. Carlos, N. Stock, S. Goodbourn, and R. E. Randall. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. USA 101:17264-17269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biron, C. A., and G. C. Sen. 2007. Innate responses to viral infections, p. 249-278. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 3.Bitko, V., A. Musiyenko, M. A. Bayfield, R. J. Maraia, and S. Barik. 2008. Cellular La protein shields nonsegmented negative-strand RNA viral leader RNA from RIG-I and enhances virus growth by diverse mechanisms. J. Virol. 82:7977-7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Childs, K. S., N. Stock, C. Ross, J. Andrejeva, L. Hilton, M. Skinner, R. Randall, and S. Goodbourn. 2007. mda-5, but not RIG-I, is a common target for paramyxovirus V proteins. Virology 359:190-200. [DOI] [PubMed] [Google Scholar]
- 5.Childs, K. S., J. Andrejeva, R. E. Randall, and S. Goodbourn. 2009. Mechanism of mda-5 inhibition by paramyxovirus V proteins. J. Virol. 83:1465-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choppin, P. W. 1964. Multiplication of myxovirus (SV5) with minimal cytopathic effects and without interference. Virology 23:224-233. [DOI] [PubMed] [Google Scholar]
- 7.Conzelmann, K.-K. 2005. Transcriptional activation of alpha/beta interferon genes: interference by nonsegmented negative-strand RNA viruses. J. Virol. 79:5241-5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. The V protein of simian virus 5 inhibits interferon signalling by targeting STAT1 for proteasome-mediated degradation. J. Virol. 73:9928-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dillon, P. J., and G. D. Parks. 2007. Role for the phosphoprotein P subunit of the paramyxovirus polymerase in limiting induction of host cell antiviral responses. J. Virol. 81:11116-11127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dillon, P. J., E. K. Wansley, V. A. Young, M. A. Alexander-Miller, and G. D. Parks. 2006. Exchanges of P/V genes between two noncytopathic SV5 variants results in a recombinant virus that kills cells through death pathways that are sensitive to caspase inhibitors. J. Gen. Virol. 87:3643-3648. [DOI] [PubMed] [Google Scholar]
- 11.Dutch, R. E., S. B. Joshi, and R. A. Lamb. 1998. Membrane fusion promoted by increasing surface densities of the paramyxovirus F and HN proteins: comparison of fusion reactions mediated by simian virus 5 F, human parainfluenza virus type 3 F, and influenza virus HA. J. Virol. 72:7745-7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fearns, R., M. E. Peeples, and P. L. Collins. 1997. Increased expression of the N protein of RSV stimulates minigenome replication but does not alter the balance between the synthesis of mRNA and antigenome. Virology 236:188-201. [DOI] [PubMed] [Google Scholar]
- 13.Gainey, M. D., P. J. Dillon, K. M. Clark, M. J. Manuse, and G. D. Parks. 2008. Paramyxovirus-induced shutoff of host and viral protein synthesis: role of the P and V proteins in limiting PKR activation. J. Virol. 82:828-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcin, D., P. Latorre, and D. Kolakofsky. 1999. Sendai virus C proteins counteract the interferon-mediated induction of an antiviral state. J. Virol. 73:6559-6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signaling, immune modulation, antiviral responses and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 16.He, B., R. G. Paterson, C. D. Ward, and R. A. Lamb. 1997. Recovery of infectious SV5 from cloned DNA and expression of a foreign gene. Virology 237:249-260. [DOI] [PubMed] [Google Scholar]
- 17.He, B., R. G. Paterson, N. Stock, J. E. Durbin, R. K. Durbin, S. Goodbourn, R. E. Randall, and R. A. Lamb. 2002. Recovery of paramyxovirus simian virus 5 with a V protein lacking the conserved cysteine-rich domain: the multifunctional V protein blocks both interferon-beta induction and interferon signaling. Virology 303:15-32. [DOI] [PubMed] [Google Scholar]
- 18.Hiscott, J., P. Pitha, P. Genin, H. Nguyen, C. Heylbroeck, Y. Mamane, M. Algarte, and R. Lin. 1999. Triggering the interferon response: the role of IRF-3 transcription factor. J. Interferon Cytokine Res. 19:1-13. [DOI] [PubMed] [Google Scholar]
- 19.Hoffman, M. A., and A. K. Banerjee. 2000. Precise mapping of the replication and transcription promoters of human parainfluenza virus type 3. Virology 269:201-211. [DOI] [PubMed] [Google Scholar]
- 20.Hornung, V., J. Ellengast, S. Kim, K. Brzozka, A. Jung, H. Kato, H. Poeck, S. Akira, K.-K. Conzelmann, M. Schlee, S. Endres, and G. Hartmann. 2006. 5′-triphosphate RNA is the ligand for RIG-I. Science 314:994-997. [DOI] [PubMed] [Google Scholar]
- 21.Horvath, C. M. 2000. STAT proteins and transcriptional responses to extracellular signals. Trends Biochem. Sci. 25:496-502. [DOI] [PubMed] [Google Scholar]
- 22.Kato, A., K. Kiyotani, M. K. Hasan, T. Shioda, Y. Sakai, T. Yoshida, and Y. Nagai. 1999. Sendai virus gene start signals are not equivalent in reinitiation capacity: moderation at the fusion protein gene. J. Virol. 73:9237-9346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kato, H., O. Takeuchi, E. Mikamo-Satoh, R. Hirai, T. Kawai, K. Matsushita, A. Hiiragi, T. S. Dermody, T. Fujita, and S. Akira. 2008. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 205:1601-1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keller, M. A., and G. D. Parks. 2003. Positive- and negative-acting signals combine to determine differential RNA replication from the paramyxovirus simian virus 5 genomic and antigenomic promoters. Virology 306:347-358. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi, T., A. A. R. Antar, K. W. Boehme, P. Danthi, E. A. Eby, K. M. Gugliemi, G. H. Holm, E. M. Johnson, M. S. Maginnis, S. Naik, W. B. Skelton, J. D. Wetzel, G. J. Wilson, J. D. Chapell, and T. S. Dermody. 2007. A plasmid-based reverse genetics system for animal double-stranded RNA viruses. Cell Host Microbe 1:147-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamb, R. A., and G. D. Parks. 2007. Paramyxoviridae: the viruses and their replication, p. 1449-1496. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 27.Lin, Y., M. Sun, S. M. Fuentes, C. D. Keim, T. Rothermel, and B. He. 2007. Inhibition of interleukin-6 expression by the V protein of parainfluenza virus 5. Virology 368:262-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, P., M. Jamaluddin, K. Li, R. P. Garofalo, A. Casola, and A. R. Brasier. 2007. Retinoic acid-inducible gene I mediates early antiviral response and Toll-like receptor 3 expression in respiratory syncytial virus-infected airway epithelial cells. J. Virol. 81:1401-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu, L. L., M. Puri, C. M. Horvath, and G. C. Sen. 2008. Select paramyxoviral V proteins inhibit IRF3 activation by acting as alternative substrates for inhibitor of kB kinase epsilon (IKKe)/TBK1. J. Biol. Chem. 283:14269-14276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mioulet, V., T. Barrett, and M. D. Baron. 2001. Scanning mutagenesis identifies critical residues in the rinderpest virus genome promoter. J. Gen. Virol. 82:2905-2911. [DOI] [PubMed] [Google Scholar]
- 31.Murphy, S., K. Y. Ito, and G. D. Parks. 1998. A functional antigenomic promoter for the paramyxovirus simian virus 5 requires proper spacing between an essential internal segment and the 3′ terminus. J. Virol. 72:10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murphy, S. K., and G. D. Parks. 1999. RNA replication for the paramyxovirus simian virus 5 requires an internal repeated (CGNNNN) sequence motif. J. Virol. 73:805-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parisien, J. P., J. F. Lau, J. J. Rodriguez, B. M. Sullivan, A. Moscona, G. D. Parks, R. A. Lamb, and C. M. Horvath. 2001. The V protein of human parainfluenza virus 2 antagonizes type I interferon responses by destabilizing signal transducer and activator of transcription 2. Virology 283:230-239. [DOI] [PubMed] [Google Scholar]
- 34.Paterson, R. G., M. A. Shaughnessy, and R. A. Lamb. 1989. Analysis of the relationship between cleavability of a paramyxovirus fusion protein and length of the connecting peptide. J. Virol. 63:1293-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peeples, M. E., and P. L. Collins. 2000. Mutations in the 5′ trailer region of a respiratory syncytial virus minigenome which limit RNA replication to one step. J. Virol. 74:146-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pfaller, C. K., and K.-K. Conzelmann. 2008. Measles virus V protein is a decoy substrate for IκB kinase α and prevents Toll-like receptor 7/9-mediated interferon induction. J. Virol. 82:12365-12373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plumet, S., F. Herschke, J. M. Bourhis, H. Valentin, S. Longhi, and D. Gerlier. 2007. Cytosolic 5′ triphosphate ended viral leader transcript of measles virus as activator of the RIG-I mediated interferon response. PLoS One 2:e279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poole, E., B. He, R. A. Lamb, R. E. Randall, and S. Goodbourn. 2002. The V proteins of simian virus 5 and other paramyxoviruses inhibit induction of interferon-beta. Virology 303:33-46. [DOI] [PubMed] [Google Scholar]
- 39.Raha, T., R. Pudi, S. Das, and M. S. Shaila. 2004. Leader RNA of Rinderpest virus binds specifically with cellular La protein: a possible role in virus replication. Virus Res. 104:101-109. [DOI] [PubMed] [Google Scholar]
- 40.Rassa, J. C., and G. D. Parks. 1999. Highly diverse intergenic regions of the paramyxovirus simian virus 5 cooperate with the gene end U tract in viral transcription termination and can influence reinitiation at a downstream gene. J. Virol. 73:3904-3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sen, G. C. 2000. Novel functions of interferon-induced proteins. Semin. Cancer Biol. 10:93-101. [DOI] [PubMed] [Google Scholar]
- 42.Strähle, L., J.-B. Marq, A. Brini, S. Hausmann, D. Kolakofsky, and D. Garcin. 2007. Activation of the beta interferon promoter by unnatural Sendai virus infection requires RIG-I and is inhibited by viral C proteins. J. Virol. 81:12227-12237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tapparel, C., D. Maurice, and L. Roux. 1998. The activity of Sendai virus genomic and antigenomic promoters requires a second element past the leader template regions: a motif (GNNNNN)3 is essential for replication. J. Virol. 72:3117-3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomas, S. M., R. A. Lamb, and R. G. Paterson. 1988. Two mRNAs that differ by two nontemplated nucleotides encode the amino co-terminal proteins P and V of paramyxovirus SV5. Cell 54:891-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ulane, C. M., A. Kentsis, C. D. Cruz, J.-P. Parisien, K. L. Schneider, and C. M. Horvath. 2005. Composition and assembly of STAT-targeting ubiquitin ligase complexes: paramyxovirus V protein carboxyl terminus is an oligomerization domain. J. Virol. 79:10180-10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wansley, E. K., and G. D. Parks. 2002. Naturally occurring substitutions in the P/V gene convert the noncytopathic paramyxovirus simian virus 5 into a virus that induces alpha/beta interferon synthesis and cell death. J. Virol. 76:10109-10121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wansley, E. K., P. J. Dillon, M. D. Gainey, J. Tam, S. D. Cramer, and G. D. Parks. 2005. Growth sensitivity of a recombinant simian virus 5 P/V mutant to type I interferon differs between tumor cell lines and normal primary cells. Virology 335:131-144. [DOI] [PubMed] [Google Scholar]
- 48.Weber, F., V. Wagner, S. B. Rasmussen, R. Hartmann, and S. R. Paludan. 2006. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 80:5059-5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Young, D. F., L. Andrejeva, A. Livingstone, S. Goodbourn, R. A. Lamb, P. L. Collins, R. M. Elliott, and R. E. Randall. 2003. Virus replication in engineered human cells that do not respond to interferons. J. Virol. 77:2174-2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Young, V. A., P. J. Dillon, and G. D. Parks. 2006. Variants of the paramyxovirus simian virus 5 with accelerated or delayed viral gene expression activate proinflammatory cytokine synthesis. Virology 350:90-102. [DOI] [PubMed] [Google Scholar]