Abstract
Coxsackieviruses are significant human pathogens, and the neonatal central nervous system (CNS) is a major target for infection. Despite the extreme susceptibility of newborn infants to coxsackievirus infection and viral tropism for the CNS, few studies have been aimed at determining the long-term consequences of infection on the developing CNS. We previously described a neonatal mouse model of coxsackievirus B3 (CVB3) infection and determined that proliferating stem cells in the CNS were preferentially targeted. Here, we describe later stages of infection, the ensuing inflammatory response, and subsequent lesions which remain in the adult CNS of surviving animals. High levels of type I interferons and chemokines (in particular MCP-5, IP10, and RANTES) were upregulated following infection and remained at high levels up to day 10 postinfection (p.i). Chronic inflammation and lesions were observed in the hippocampus and cortex of surviving mice for up to 9 months p.i. CVB3 RNA was detected in the CNS up to 3 months p.i at high abundance (∼106 genomes/mouse brain), and viral genomic material remained detectable in culture after two rounds of in vitro passage. These data suggest that CVB3 may persist in the CNS as a low-level, noncytolytic infection, causing ongoing inflammatory lesions. Thus, the effects of a relatively common infection during the neonatal period may be long lasting, and the prognosis for newborn infants recovering from acute infection should be reexplored.
Early damaging events on the central nervous system (CNS) by infection can result not only in severe physical and intellectual disability but also in less obvious neurological deficits. For example, children who were thought to have fully recovered from neonatal CNS virus infections exhibited some deficiency in scholastic performance (12). Thus, the enduring harmful effects of childhood infections on the CNS may be greatly underappreciated. Picornaviruses including polioviruses, coxsackieviruses, and other unclassified enteroviruses frequently infect the CNS (60). Although these infections often are considered acute and self-limiting, evidence is emerging that these viruses—or at least the viral RNAs—may persist for months or years after the initial infection. For example, ∼50 years after the primary infection, a large percentage (∼30%) of polio victims are now experiencing new symptoms (postpolio syndrome), which some investigators have correlated with the presence of viral RNA in the CNS (43). Worldwide distribution of enterovirus infection is revealed by the detection of enterovirus-specific antibodies in the serum of approximately 75% of individuals within developed countries. For example, in 1996, approximately 10 to 15 million diagnosed cases of enterovirus infection occurred in the United States alone (49). Few studies have been done to determine if enteroviruses, or their close relatives, have the ability to persist and cause long-term damage in the CNS (10, 56) or whether previous infection of neurons may indirectly lead to the degeneration of aging motor neurons.
Coxsackievirus, a member of the enterovirus genus, is a fairly frequent childhood infection and may cause severe morbidity and mortality in humans, predominantly in the very young. Infants infected with coxsackievirus have been shown to be extremely susceptible to meningitis and encephalitis. Severe demyelinating diseases may occur following infection, including acute disseminated encephalomyelitis (18) and acute transverse myelitis (27). Also, a number of delayed neuropathologies have been associated with previous coxsackievirus infection, including schizophrenia (47, 52), encephalitis lethargica (16), and amyotrophic lateral sclerosis (62, 63). If human neurotropic viruses persist, they could provide a chronic inflammatory stimulus, leading to regional cytokine induction and activation of autoreactive T cells through molecular mimicry and bystander activation (32, 45). This may be especially true for viruses, such as coxsackievirus, which have the ability to infect stem cells (24) and neurons (1). Recently, we have shown that coxsackievirus B3 (CVB3) targets proliferating cells in regions of the neonatal CNS supporting neurogenesis (24). Nonetheless, infected migratory neuronal progenitor cells were able to differentiate into mature neurons. Many neurons eventually underwent caspase-3-mediated apoptosis at later stages of disease (22).
Intriguingly, viral RNA was detected in the CNS of surviving pups in the absence of infectious virus for up to 30 days postinfection (p.i.). The detection of CVB3 RNA in target tissues may have great significance for CVB3-mediated disease, given that the long-term presence of replication-restricted CVB3 RNA in the heart (generated using transgenic techniques) has been directly associated with dilated cardiomyopathy in a previous study by Wessely et al. (59). We were therefore interested in expanding this notable observation in the CNS by significantly increasing the number of animals examined, more precisely quantifying the amounts of viral RNA, and determining how long viral RNA might persist in the CNS. In addition, we thoroughly assessed the nature and degree of neuropathology in surviving animals harboring CVB3 RNA. These studies may help predict the lasting neurological sequelae of a previous viral infection on the developing host.
MATERIALS AND METHODS
Isolation and production of recombinant coxsackieviruses.
The generation of recombinant coxsackieviruses expressing enhanced green fluorescent protein (eGFP) or lymphocytic choriomeningitis virus (LCMV) T-cell epitopes has been described previously (50). Briefly, the CVB3 infectious clone (pH 3; obtained from Kirk Knowlton at University of California at San Diego) was engineered to contain a unique SfiI site which facilitates the insertion of any foreign sequence into the CVB3 genome. All virus stocks were grown on HeLa RW cells maintained in Dulbecco's modified Eagle's medium (Gibco-BRL, Gaithersburg, MD), supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 100 U of penicillin, and 100 μg of streptomycin per ml. Virus titrations were carried out as described previously (23).
Mice and viral inoculations.
Mouse experimentation conformed to the requirements of the Scripps Research Institute Animal Research Committee and the National Institutes of Health. BALB/c mice were obtained from the Scripps Research Institute animal facilities. Breeding pairs were checked every day, and 1-day-old or 3-day-old pups were infected intracranially (i.c.) with 104 PFU of recombinant CVB3 virus expressing LCMV nucleoprotein residues 118 to 126 (rCVB3-LCMVNP118-126) or with 2 × 106 to 2 × 107 PFU of CVB3 expressing eGFP (eGFP-CVB3). Two different recombinant coxsackieviruses were employed in order to minimize the possibility that any findings of persistent viral materials were an artifact of the particular virus that was used. Viral doses for both recombinant viruses and time of administration (1 day or 3 days postbirth) were optimized to inoculate the greatest amount of infectious virus leading to greater than 50% mouse survival. Since rCVB3 replication kinetics depends upon the size insert, rCVB3-LCMVNP118-126 required less inoculum to reach a similar level of mouse survival. The procedure for i.c. inoculation of 1-day-old pups has been described previously (22). Young pups (1 to 5 days p.i.) were sacrificed by hypothermia/CO2, followed by immediate decapitation. Older pups (10 days and beyond) were sacrificed by halothane treatment, followed by cervical dislocation. The brains were fixed by immersion in 10% neutral-buffered formalin for >4 h, paraffin-embedded, and stained with hematoxylin and eosin.
Viral RNA extraction and nested RT-PCR.
All work involving PCR was carried out in a designated PCR-clean area. Viral RNA was isolated from brain tissue using TRIzol reagent (Gibco-BRL, Rockville, MD), as described by the manufacturer. Reverse transcription-PCR (RT-PCR) was done utilizing first-round primers (CVB3 first-round forward primer, 5′-GCGGCCTGCCTATGGGGAAACC-3′; CVB3 first-round reverse primer, 5′-CTCTCAATTGTCACCATAAGCAGCCAGTA-3′) which amplified the 5′ untranslated leader of CVB3. Second-round (nested) primers (CVB3 second-round forward primer, 5′-GCTAGTTGGTAATCCTCCGGCCCCTGAATG-3′; CVB3 second-round reverse primer, 5′-AATAAAATGAAACACGGACACCCAAAGTAG-3′) recognized sequences internal to the product generated by the first-round primers. For nested (second round) PCR, 2 μl of the first-round product was added to the second-round reaction mixture. PCR amplification products were detected by electrophoresis on a 1.2% agarose gel. The first-round forward primer was designed to bind 370 nucleotides upstream from the 5′ termini of CVB3 (Woodruff strain).
Quantitative real-time RT-PCR.
5′ Untranslated region (UTR) CVB3-specific primers (forward primer, 5′-CACACTCCGATCAACAGTCA-3′; reverse primer, 5′-GAACGCTTTCTCCTTCAACC-3′) and a FAM/TAMRA (6-carboxyfluorescein/6-carboxytetramethylrhodamine)-labeled probe (5′-CGTGGCACACCAGCCATGTTT-3′) were designed for the TaqMan method of quantitative real-time RT-PCR. The first-round forward primer was designed to bind 120 nucleotides downstream from the 5′ terminus of CVB3. Reverse transcription reaction mixtures included 5 μg of total RNA and were carried out using SuperScript III reverse transcriptase (Invitrogen Inc., Carlsbad, CA) following the procedure described by the manufacturer. Separate RT reaction mixtures included either the reverse primer or forward primer to quantify the number of positive- and negative-sense viral genomes, respectively. The RT reaction mixture also included 1 μl of RNaseOUT (Invitrogen Inc.). Prior to PCR amplification, completed RT reaction mixtures were treated with 1 μl of RNase H (Invitrogen Inc.) to remove RNA complementary to the cDNA. PCR amplification was done using Platinum quantitative PCR SuperMix-UDG ready-to-use cocktail (Invitrogen Inc.) containing all components except the amplification primers and FAM/TAMRA-labeled probe, as described by the manufacturer. Quantitative analysis of viral RNA was carried out using a Stratagene Mx3000P real-time PCR system in 96-well optical reaction plates heated to 50°C for 2 min to digest dUTP-containing contaminants, followed by 95°C for 2 min to deactivate uracil N-glycosylase and activate Platinum Taq DNA polymerase. Forty cycles of denaturation at 95°C for 15 s and annealing and extension at 60°C for 30 s were carried out. All samples were evaluated in triplicate amplification reactions. To normalize the amount of RNA in each sample, separate RT reactions were generated using random hexamers instead of CVB3-specific primers and amplified for 18S rRNA. 18S ribosomal products were amplified in triplicate with 18S forward (5′-CGGCTACCACATCCAAGGAA-3′) and reverse (5′-GCTGGAATTACCGCGGCT-3′) primers and including a FAM/TAMRA-labeled probe (5′-TGCTGGCACCAGACTTGCCCTC-3′). Two separate standards using both diluted pH 3 CVB3 plasmid and in vitro transcribed pH 3 CVB3 RNA generated the standard curve. The standard curve was based on threshold cycle (CT) values (cycle number at which relative fluorescence crossed the threshold), and CT values from unknown samples (normalized for 18S rRNA signal) were compared to the standard curve to determine viral RNA copy numbers.
RPA.
mRNA levels for chemokines, cytokines, and type I interferon (IFN) response genes in the CNS were determined by multiprobe RNase protection assays (RPAs), as described in detail previously (51). The probe sets used included chemokine 1 and 2 probe sets (6, 9), cytokine probe sets ML11 and ML26 (31, 46), and an IFN-regulated probe set (7). The levels of mRNA were quantified using ImageJ software (NIH imaging software; http://rsb.info.nih.gov/ij).
Histochemical and immunofluorescence staining.
Paraffin-embedded sections were stained by hematoxylin and eosin (H&E). In addition, sections were stained with Oil Red O, which identifies lipids and lipoproteins, and counterstained with hematoxylin. The procedure for immunofluorescence staining has been described previously (22). Briefly, paraffin-embedded sections (3-μm thickness) were deparaffinized with three washes in xylene and serial washes in 100%, 90%, and 70% ethanol, followed by a final wash in phosphate-buffered saline. Sections were blocked with 10% normal goat serum for 30 min and incubated overnight with primary antibody at 4°C. Alternatively, sections stained with a mouse primary monoclonal antibody were treated with a Mouse on Mouse kit (Vector Labs), as described previously (24). All antibody dilutions were made in 2% normal goat serum. The detection of CD3 and Iba1 required high-temperature antigen unmasking in 0.01 M citrate buffer (pH 6.0). Sections were incubated with a rabbit primary monoclonal antibody against Iba1 at 1:100 (Wako Chemicals USA Inc., Richmond, VA) and CD3 at 1:50 (Biocare Medical Inc., Walnut Creek, CA) at 4°C overnight. A goat secondary antibody [goat biotinylated anti-rabbit immunoglobulin G(H+L)] at 1:500, (Vector Laboratories Inc., Burlingame, CA) was diluted in 2% normal goat serum and incubated on sections for 30 min. After being stained with secondary antibodies, all sections were washed twice with phosphate-buffered saline and incubated for 30 min with a streptavidin-rhodamine red complex (Jackson ImmunoResearch Laboratories Inc., West Grove, PA) diluted 1:500 in 2% normal goat serum. Specificity controls for immunostaining included sections stained in the absence of primary antibody or in the presence of rabbit immunoglobulin G control antibody at 0.1 μg/ml (Vector Laboratories, Inc). Sections were overlaid with Vectashield mounting medium (Vector Laboratories, Inc.) containing DAPI (4′, 6-diamidino-2-phenylindole) to detect DNA/nuclei (blue). The sections were observed by fluorescence microscopy (Zeiss Axiovert 200 inverted microscope). Green, red, and blue channel images were merged using AxioVision software. Confocal microscopy was done using a Rainbow Radiance 2100 laser scanning confocal system attached to a Nikon TE2000-U inverted microscope (Bio-Rad-Zeiss) located at the Core Microscopy Facility at the Scripps Research Institute. Images were acquired using Laser Sharp 2000 software, and further analysis was done with 3D Imaris Isosurface analysis software (Bitplane Inc, Saint Paul, MN) and ImageJ software.
RESULTS
Acute CVB3 infection in the CNS is followed by viral RNA persistence.
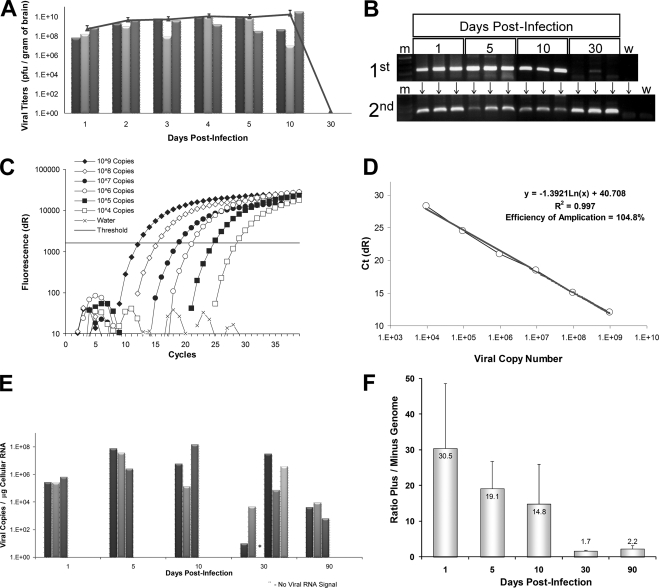
To determine the kinetics of virus clearance from the CNS and to investigate the possibility of persistence, 21 1-day-old BALB/c pups were infected with rCVB3- LCMVNP118-126 (104 PFU i.c.). Virus titers were determined at seven time points, ending at 30 days p.i.; a recombinant virus was used because the probability of lethal disease is greatly reduced, and the window of survival is larger than for wild-type virus (data not shown), thus facilitating our investigation of this long-term outcome. In parallel, the presence of viral RNA was determined using nested RT-PCR at four time points. High levels of infectious virus were detected as early as 24 h after infection and remained high to at least day 10 p.i. (Fig. 1A). However, infectious virus could no longer be detected at 30 days p.i. Despite the lack of infectious virus at this time point, viral RNA was detected after one round of RT-PCR in the brain tissue of two out of three mice and in all three mice after nested RT-PCR (Fig. 1B). The presence of viral RNA in the absence of infectious virus suggests that CVB3 may establish a persistent infection in the CNS at the viral RNA level, similar to what has been observed in other target tissues (34, 36, 48).
FIG. 1.
CVB3 RNA, but not infectious virus, persists in the neonatal CNS. Neonatal mice were infected with rCVB3-LCMVNP118-126, and the brains were harvested (three mice per time point) at the indicated time points p.i. (A) Viral titers were determined over the course of 30 days, as described in Materials and Methods. Viral titers increased over the course of 10 days. By 30 days p.i., infectious virus could no longer be detected. Each bar indicates virus titer results from an individual mouse, and the single line represents the average viral titer for three animals over time (error bars represent standard deviation for three mice per group). (B) RNA from the brains of mice was utilized for nested RT-PCR. Two sets of CVB3-specific nested primers (5′ UTR region) were used for amplification. The first- and second-round virus-specific PCR-amplified products (269-bp band) were separated and visualized by gel electrophoresis on a 1.2% agarose gel. Viral RNA was detected after the first round of PCR in all samples harboring infectious virus. Viral RNA was also detected in two of three samples isolated at 30 days p.i. at a time when no infectious virus was observed. After a second round of PCR, viral RNA was detected in one additional sample isolated at 30 days p.i. A DNA molecular weight marker lane (m) was included. Water control reactions (w lanes) were run in parallel for both the first and second rounds of PCR. (C) DNA (CVB3 plasmid) and RNA (in vitro transcribed viral RNA) standards were diluted to between 10° and 109 viral copies and amplified utilizing a Stratagene Mx3000P real-time PCR system with 5′ UTR CVB3-specific primers and a FAM-labeled probe designed for the TaqMan method of quantitative real-time RT-PCR. Fluorescence units (dR) were plotted versus PCR cycle number, and the threshold value was calculated automatically using Stratagene software. (D) The efficiency of amplification (104.8%) and R2 value (0.997) were determined by plotting CT values versus viral copy number using Stratagene software. Both values were found to be within acceptable ranges. (E) The number of viral RNA copies in the brains of infected animals was calculated by real-time RT-PCR and graphed over time. Viral RNA levels increased over time and remained high through 10 days p.i. By day 30 p.i., two out of three animals continued to have detectable, although greatly reduced, levels of viral RNA in the brain. Each bar indicates viral RNA results from an individual mouse. Asterisks indicate the lack of detectable viral RNA in a mouse found to be positive by nested RT-PCR. (F) The ratio of values for positive- and negative-sense-strand viral RNA was calculated for all time points determined in panel C. High ratio values (ratio of 30.5/1) were observed during early (day 1) infection, which may be expected during the high levels of viral replication and positive-sense stranded viral RNA production occurring initially. However after 5 and 10 days p.i., the ratios (ratio of 19.1/1 and 14.8/1, respectively) dropped with time, indicating lower levels of positive-sense-strand and higher levels of negative-sense-strand viral RNA generated later during infection. By 30 days p.i. (a time when no infectious virus is observed), the ratio decreased closer to a 1/1 ratio (ratio of 1.7) of positive- to negative-sense-strand viral RNA and stayed near this ratio (ratio of 2.2/1) up to 90 days p.i. Error bars represent the standard deviation for three mice per group. Statistical significance (P = 0.0146) was determined by (nonparametric) one-way analysis of variance.
Viral RNA in the CNS quantified by real-time RT-PCR.
Next, the copy number of viral RNA present during acute and persistent infection was determined using quantitative real-time RT-PCR. 5′ UTR CVB3-specific primers and a FAM/TAMRA-labeled probe were designed for the TaqMan method of quantitative real-time RT-PCR. Both plasmid DNA (serial dilutions of CVB3 infectious clone plasmid) and RNA standards (in vitro transcribed RNA from the CVB3 plasmid) were generated and compared by real-time RT-PCR in separate triplicate reactions. The results for the DNA standards amplification are shown in Fig. 1C. The detection limit of real-time RT-PCR was found to be approximately 100 copies of diluted DNA (Fig. 1C) or RNA standard. After the standard curve for viral copies was generated using Stratagene Mx3000P software, both DNA (Fig. 1D) and RNA standards revealed similar R2 values (0.997 and 0.996, respectively). An R2 value of 1.000 indicates that all data points lie perfectly on the graphed data set line. Both DNA and RNA standards also generated efficiency of amplification values within acceptable ranges (104.8% and 99.0%, respectively). An efficiency of amplification equal to 100% indicates that the amount of template doubles with each cycle of PCR.
High levels of viral RNA persist in the CNS.
Using the DNA standard curve, we calculated the number of viral copies/μg of cellular RNA for each sample after determining the detection CT for each sample (automatically calculated by the Stratagene Mx3000P software package). In addition, input cellular RNA was normalized for each sample in separate triplicate reactions by including primers plus a FAM-labeled probe for 18S rRNA. Quantification of viral RNA by real-time RT-PCR (Fig. 1E) closely paralleled results determined by plaque assay. Replicate samples within any time point showed substantial correlation, perhaps reflecting the high degree of accuracy for both the plaque assay and real-time RT-PCR. The highest amount of positive-sense-strand viral RNA was seen in the CNS of animals on days 5 and 10 p.i. Additional animals analyzed for day 30 and day 90 p.i. also demonstrated detectable levels of viral RNA (Fig. 1E). In total, viral RNA was detected in five of six mice harvested at 30 days p.i. Furthermore, all mice harvested at day 90 p.i. showed detectable levels of viral RNA despite the lack of infectious virus. Analysis by real-time RT-PCR revealed the presence of ∼104 copies of positive-sense-strand viral RNA per μg of cellular RNA in the CNS of one animal from day 90 p.i. (mouse 2). Assuming a yield of 5 μg of cellular RNA per mg of brain tissue and an average brain weight of 200 mg for a young (30-day-old) mouse, we estimate that up to 10 million copies of persisting CVB3 RNA may be isolated from brain tissue. These results were suggestive of relatively high viral copy numbers in the CNS despite the apparent lack of infectious virus (by viral plaque assay) at these later time points. We cloned and sequenced representative amplified viral products using the real-time RT-PCR primers described above. By standard T/A cloning methodology (TOPO PCR Cloning kit; Invitrogen Inc.), viral sequences were obtained from infected mice at days 1, 30, and 90, yet sequence analysis revealed only a synonymous nucleotide change (U to C) in a single day 90 viral clone (data not shown).
Similar amounts of plus- and minus-strand viral RNA during CVB3 persistence.
Quantification of negative-sense-strand viral RNA also was determined for each sample. The ratio of positive- to negative-sense-strand viral RNA was calculated and graphed over time (Fig. 1F). Intriguingly, the ratio was high early after infection (day 1 p.i.; ratio of 30.5/1) and slowly decreased over the course of infection. By day 30 p.i., a ratio of positive- to negative-sense-strand viral RNA closer to 1/1 was observed (ratio of 1.7/1) in the CNS. The ratio of positive- to negative-sense-strand viral RNA remained relatively low (ratio of 2.2/1) up to day 90 p.i. In a previous publication, a 1/1 ratio has been found in other tissues persistently infected with CVB3 and may indicate a latent double-stranded viral RNA complex (54).
Rapid upregulation of chemokines following CVB3 infection in the CNS.
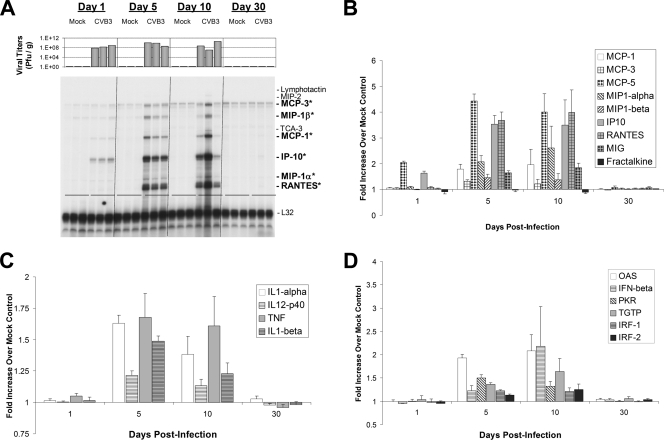
We determined RNA expression levels of chemokines, cytokines, and type I IFN response genes in the CNS of infected animals by utilizing RPAs. A representative RPA autoradiographic film for chemokine RNA expression levels (chemokine probe set 1) is shown in Fig. 2A. Additional chemokines (chemokine probe set 2) were quantitatively evaluated, and the data are graphically represented in Fig. 2B. Each lane in Fig. 2A represents chemokine RNA expression levels from an individual mouse, and the expected size band for each chemokine is depicted on the right side of the autoradiographic film. In addition, viral titers determined for each mouse are represented above each lane. Within 24 h p.i., IFN-γ-inducible protein 10 (IP10/CXCL10) RNA expression levels increased dramatically in the CNS for three infected animals compared to three mock-infected control samples (Fig. 2A). Similar kinetics were observed for MCP-5 (macrophage/monocyte chemotactic protein-5) RNA expression levels analyzed using chemokine probe set 2 (Fig. 2B). By 5 days p.i., high RNA levels of many chemokines (RANTES/CCL5, MIG/CXCL9, macrophage inflammatory protein-1α [MIP-1α]/CCL3, MIP-1β/CCL4, IP10, MCP-1/CCL2, MCP-3, and MCP-5) were preferentially upregulated in CVB3-infected animals. The expression levels of these chemokine genes remained high for at least 10 days p.i. By day 30 p.i., expression of activated chemokines dropped to undetectable levels or levels similar to those of mock-infected animals. The relative increase in chemokine RNA levels compared to mock-infected animals was quantified using ImageJ software and is graphically illustrated in Fig. 2B. The highest relative increase for all chemokines examined was seen for MCP-5, IP10, and RANTES (between a 3.5- and 4.5-fold increase) at days 5 and 10 p.i.
FIG. 2.
Quantitative analysis of chemokines, cytokines, and type I IFN response gene expression levels in the neonatal CNS over time. One-day-old BALB/c pups (three mice per time point) described in the legend of Fig. 1 were evaluated for chemokine gene expression by RNase protection assay on days 1, 5, 10, and 30 p.i. (A) Each sample lane of the autoradiographic film represents chemokine gene expression levels from a single mouse. Three mock-infected and three infected animals were analyzed per time point. Viral titers (PFU/g) for each mouse are depicted above each sample lane. Nine chemokines were analyzed for gene expression levels (lymphotactin, MIP-2, MCP-3, MIP-1β, TCA-3, MCP-1, IP-10, MIP-1α, and RANTES). Six of nine chemokines (MCP-3, MIP-1β, MCP-1, IP-10, MIP-1α, and RANTES, indicated by boldface with asterisks) were upregulated specifically within infected animals as late as 10 days p.i. However by day 30 p.i., chemokine gene expression levels returned to levels observed in mock-infected animals. The ribosomal gene L32 was probed to demonstrate equal loading of cellular RNA per lane. Band intensities evaluated for chemokine (B), cytokine (C), and type I IFN (D) gene expression by RPAs were calculated using ImageJ software (NIH public domain software), and arbitrary units were determined after compensating for RNA levels utilizing the housekeeping ribosomal gene, L32. Expression levels (in triplicate) were graphed as a relative increase over mock-infected controls. Standard error bars are shown for each sample. (B) As illustrated in panel A, chemokines were induced to high levels relative to mock-infected animals at days 5 and 10 p.i In particular, high levels of MCP-5 and IP10 were observed as early as 24 h p.i. (C) Inflammatory cytokines, including TNF, were induced at days 5 and 10 p.i. (D) Many type I IFN response genes, including IFN-β, were induced 5 and 10 days following infection. By 30 days p.i., chemokine, cytokine, and type I IFN response gene expression returned to levels seen in mock-infected control mice. Error bars represent the standard deviation for each group.
In addition to the chemokines described above, a number of chemokine genes showed no change in RNA levels compared to levels in mock-infected control animals. These included lymphotactin, MIP-2, TCA-3 and -4, SDF1α, KC, and eotaxin. In contrast, fractalkine RNA levels increased steadily in both infected and mock-infected animals. Similar results describing the developmental upregulation of fractalkine expression levels in the neonatal CNS have been published previously (37).
Cytokine induction in the CNS following CVB3 infection.
Similar experiments were carried out using two cytokine probe sets and revealed the upregulation of a number of cytokine mRNAs (tumor necrosis factor [TNF], interleukin-1α [IL-1α], IL-1β, and IL-12 p40) in response to CNS infection (Fig. 2C). The temporal pattern of cytokine mRNA upregulation was similar to that observed for chemokines. At 5 and 10 days p.i., a small increase in TNF mRNA levels was observed in each of three animals (average of 1.5-fold) compared to mock-infected control animals. In contrast, little or no IFN-γ mRNA expression was observed at any time points following infection (data not shown). A slight upregulation of IL-12 p40 mRNA was also observed at 5 and 10 days pi. IL-12, a heterodimeric protein composed of p40 and p35 subunits, plays a central role in cell-mediated immunity. The p40 subunit is also a component of IL-23 and by itself may be a chemoattractant for macrophages (28). The preferential upregulation of the p40 subunit mRNA has been observed in a number of autoimmunity models (4, 17). The lack of IL-12 p35 mRNA upregulation (data not shown) suggests that IL-12 p40 mRNA was expressed by microglia or infiltrating monocytes/macrophages (13).
In addition, IL-1α and IL-1β mRNAs were upregulated in CVB3-infected animals at 5 and 10 days p.i. IL-1-α and IL-1-β are known to be primarily released from stimulated macrophages and monocytes, and these molecules play key roles in the inflammatory response. The preferential induction of TNF, IL-1α, IL-1β, and the IL-12 p40 subunit mRNAs, along with high levels of MIG, RANTES, and IP10 gene expression, suggests that activated microglia or macrophages play a primary role during the initial response to CVB3 infection in the CNS. As with the chemokine gene expression pattern, cytokines activated in response to CVB3 infection fell to normal or undetectable levels by 30 days p.i. Also, no changes were observed in the CNS for other cytokine genes, including lymphotoxin-α, IL-2 through IL-7, IL-10, and IL-13 (data not shown).
Induction of type I IFN response genes following CVB3 infection.
Type I IFNs (IFN-α and IFN-β) and their downstream response genes are critical components of innate immunity. By RPA analysis, relatively high expression levels of IFN-β mRNA were detected in the neonatal CNS at 5 and 10 days p.i. compared to levels in mock-infected control animals (Fig. 2D). As might be expected, downstream targets of IFN-β were upregulated specifically in CVB3-infected animals, including IFN response factors 1 and 2 (IRF-1 and IRF-2), double-stranded RNA-activated protein kinase R (PKR), 2′-5′ oligoadenylate synthetase (OAS), and TGTP1 antiviral GTPase. In contrast, little or no increase in the levels of IFN-α1, IFN-κ, IFN-τ, and the major histocompatibility complex class II transactivator was seen at any time point after infection (data not shown). Similar to what was observed for fractalkine, P58IPK (a cellular PKR inhibitor) expression levels increased during CNS development equally in both mock-infected and infected animals. P58IPK has been shown previously to be recruited by influenza virus to inhibit PKR function during viral infection although it does so by inhibiting protein-protein interactions rather than by influencing mRNA expression levels (40). RPA analysis indicated the lack of IFN-β gene upregulation or expression of downstream mediator genes (PKR, OAS, IRF-1, or IRF-2) at a time when viral RNA remained in the CNS (day 30 p.i.). These data indicate that remaining viral material detected at later time points following infection may not efficiently activate the type I IFN response. Alternatively, CVB3 may alter the type I IFN response during viral RNA persistence.
Acute infection leads to reactive microgliosis in the neonatal CNS.
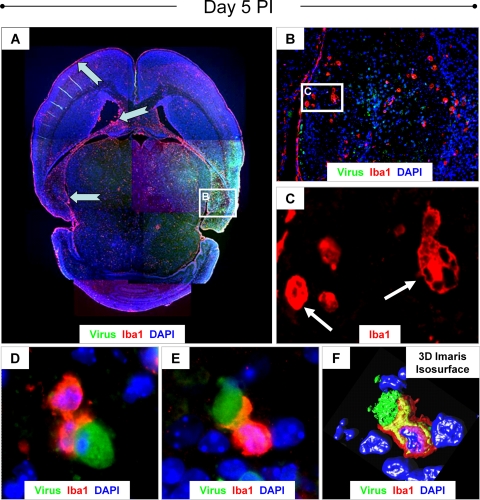
We evaluated the degree of microglial activation during acute infection with eGFP-CVB3 (day 5 p.i.). At this time point, high levels of virus protein expression (Fig. 3A) and neuropathology were observed within the hippocampus and cortex. A number of Iba1+ microglia/macrophages (red) were observed within these regions, as well as in the subventricular zones, the lateral ventricles, and the meninges (Fig. 3A). Numerous Iba1+ cells of distinctive morphology were identified within infected regions of the hippocampus (Fig. 3B). Higher magnification of Fig. 3B revealed gitter cells or compound granular corpuscles (Fig. 3C), indicative of microglial activation. Engulfment or phagocytosis of infected cells (green) was readily observed by Iba1+ cells (Fig. 3D and E) and confirmed by confocal microscopy plus 3D Imaris Isosurface reconstruction (Fig. 3F).
FIG. 3.
Induction of reactive microgliosis during acute infection. One-day-old pups were infected with eGFP-CVB3 (2 × 106 PFU i.c.) or mock infected. Brain sections from pups harvested on day 5 p.i. were immunostained for GFP (viral protein expression; green) and Iba1 (marker for activated macrophages and microglia; red) and counterstained with DAPI (nuclear stain; blue). (A) Fluorescence images of overlapping fields at a magnification of ×5 were taken and assembled into a composite image. Iba1 staining revealed high numbers of activated microglia/macrophages in the meninges, the lateral ventricles and subventricular zones, the hippocampus, and regions of the cortex (notched cyan arrows). Many of these regions correspond to areas of the CNS targeted by CVB3. (B) Higher magnification (×20) of the image in panel A revealed many gitter cells or compound granular corpuscles in the hippocampus, indicative of microglial activation. (C) White arrows point to high-magnification regions of the dentate gyrus which further demonstrate the distinctive morphology of gitter cells (shown at a magnification of ×100.8 plus a further ∼×2 digital magnification). (D) In many examples, activated microglia/macrophages were observed engulfing CVB3-infected target cells (shown at a magnification of ×100.8 plus a further ∼×2 digital magnification). The engulfment of infected target cells by activated microglia/macrophages (magnification of ×100.8 plus a further ∼×4 digital magnification) was confirmed by confocal microscopy (E) and 3D Imaris Isosurface reconstruction (F). The fluorescent data are representative images from two pups.
Chronic inflammation and lesions in the CNS of surviving mice previously infected with CVB3.
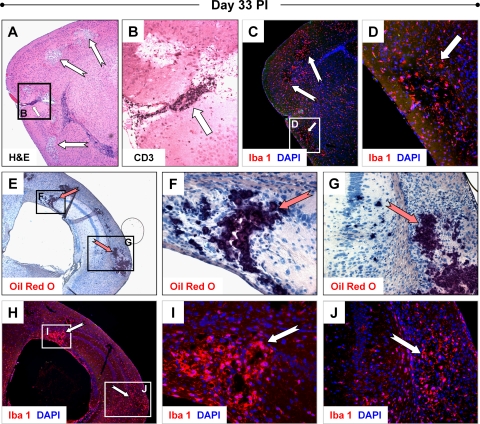
Reactive microgliosis continued for extended periods of time in pups surviving initial infection. In one litter of 3-day-old pups infected with eGFP-CVB3 (2 × 106 PFU i.c.), all four animals exhibited ataxia and showed signs of hydrocephalus. Overt signs of hydrocephalus included a rounded, enlarged cranium, cerebral spinal fluid exudate upon dissection of the cranium, and enlarged ventricles identified by H&E-stained sections of brain. We therefore examined these animals for clinical motor disturbance and determined that these animals displayed flaccid tail tone and hind limb weakness with mild paresis. When lifted by the base of their tails, these mice exhibited a significant hind limb clasping response and a diminished righting response. These moribund animals were euthanized at 33 days p.i. No viral titers were detected in these mice although by real-time RT-PCR all animals displayed relatively high levels of CVB3 RNA. No viral protein could be detected in these mice by immunohistochemistry, possibly due to the relatively low sensitivity of this technique. Representative histopathology of one animal from this litter is shown in Fig. 4. By H&E staining, lesions and perivascular cuffs adjacent to lesions were readily observed in regions of the cortex (Fig. 4A). Perivascular cuffs were populated with high numbers of CD3+ T cells (Fig. 4B). In addition, widespread Iba1 staining revealed reactive microgliosis (Fig. 4C) although the strongest Iba1 signal was seen within lesions (Fig. 4D). Furthermore, the majority of Iba1+ cells was morphologically represented by pleomorphic or activated forms of microglia. No obvious histopathology was observed in H&E-stained or Luxol fast blue/periodic acid-Schiff-stained sections of spinal cord (data not shown), suggesting that the hind limb dysfunction observed following infection may be associated with the histopathology seen in the brain.
FIG. 4.
Chronic inflammation and CNS pathology during persistent infection. Three-day-old pups were infected with rCVB3 (2 × 106 PFU i.c.). Brains were harvested 33 days later for histological analysis and the presence of persistent viral RNA by real-time RT-PCR. All four animals from one litter (one of which is represented below) suffered from hind limb dysfunction, showed signs of hydrocephalus, and exhibited similar histopathology and immunofluorescence staining, as described below. (A) Lesions (notched white arrows) associated with inflammatory cells and perivascular cuffing (small white arrow) were observed in cortex (magnification, ×5). (B) Higher magnification (×20) of the images in panel A stained with an antibody against CD3 identified the vast majority of cells within the perivascular cuff as T cells. CD3+ T cells were also observed within the lesions and the surrounding parenchyma. (C) Iba1 staining of parallel sections revealed the presence of numerous activated microglia/macrophages (magnification, ×5). (D) Higher magnification (×20) of the images from panel C demonstrated the elevated presence of Iba1+ cells within lesions (white arrow). (E) Lipid-laden microglia or macrophages, indicative of phagocytic gitter cells, were detected by Oil Red O staining (magnification, ×5). Sections stained with Oil Red O (dark red) and counterstained with hematoxylin revealed dark staining regions (notched red arrows) which overlapped precisely with lesions in the cortex and hippocampus. Higher magnification (×40 and ×20, respectively) revealed staining in damaged pyramidal cells and surrounding areas of the hippocampus (F) and within inflammatory regions seen in the cortex (G). (H) Parallel sections of the images from panel E stained with an antibody against Iba1 (red) showed high levels of expression in a subset of pyramidal cells of the hippocampus and in damaged regions of the cortex (magnification, ×5). (I and J) Higher magnification (×40 and ×20, respectively) illustrated the presence of numerous activated microglia or macrophages (notched white arrows) in regions corresponding to Oil Red O staining.
The presence of lipid-laden, activated microglia was determined by Oil Red O staining, a fat-soluble dye used for staining lipids and lipoproteins present in regions of neurodegeneration (3). Oil Red O staining was observed extensively in the damaged regions of the cortex and hippocampus (Fig. 4E). Higher magnification revealed Oil Red O staining in pyramidal neurons (Fig. 4F) and within regions of the cortex where inflammation was apparent (Fig. 4G). Parallel sections demonstrated the presence of Iba1+ cells in identical regions showing Oil Red O staining (Fig. 4H). Higher magnification showed the greater numbers of Iba+ cells preferentially found within Oil Red O-stained lesions (Fig. 4I and J). These results suggest that microglia or macrophages present within damaged regions of the cortex and hippocampus contained large amounts of lipid or lipoprotein debris. No infectious virus was detected in these mice, and no viral protein was observed by immunohistochemistry. However, when analyzed by real-time RT-PCR, all brains contained relatively high levels of CVB3 RNA (see Fig. 6A).
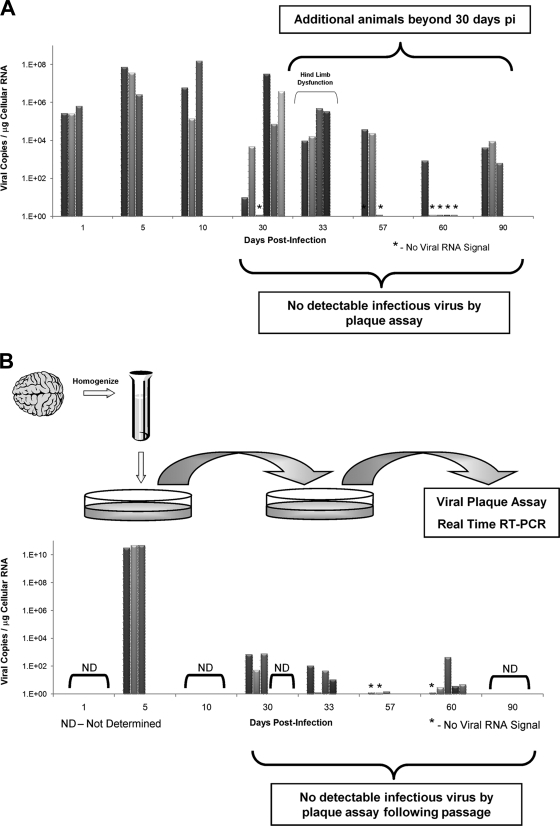
FIG. 6.
Viral RNA persistence in the CNS and in vitro passage of noncytolytic virus in tissue culture. Additional animals were infected at either 1 or 3 days postbirth with rCVB3 (2 × 106 to 107 PFU i.c.), and the brains were harvested up to day 90 p.i. None of the additional mice harvested beyond day 30 p.i. had detectable levels of infectious virus, as determined by viral plaque assay. (A) In contrast, 15 of 21 animals analyzed at or after 30 days p.i. contained viral RNA, as detected by real-time RT-PCR. Of interest, one litter of pups euthanized after 33 days and suffering from hind limb dysfunction appeared to have relatively high copy numbers of viral RNA in the brain. The histopathology of one animal from this litter is depicted in Fig. 4. (B) The brain homogenates from 3 acutely infected pups and 15 infected animals (day 30 p.i. and beyond) displaying no infectious virus by plaque assay were passaged on HeLa cells. Supernatants from treated HeLa cells were isolated and plated onto fresh HeLa cells. After 1 h, supernatants were removed, and the HeLa cells were washed and placed in complete medium. After 24 h, supernatants only (no cell lysates) from the second-passage HeLa cells were analyzed by viral plaque assay and real-time RT-PCR for viral RNA. As expected, second-passage supernatants from acutely infected pups gave rise to high levels of infectious virus (as determined by virus plaque assay) and viral RNA copies. Surprisingly, low levels of viral RNA, in the absence of infectious virus, were observed in a high proportion (12 of 15) of HeLa cell supernatants from infected animals.
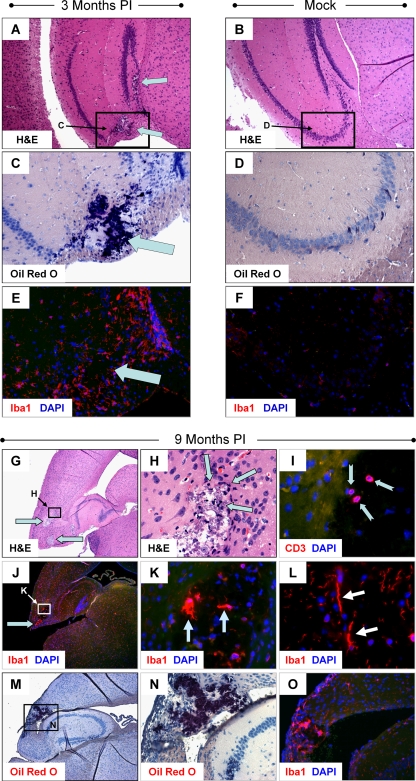
Remarkably, histopathology and reactive microgliosis were still ongoing in the hippocampus 3 months after infection (in three mice, all of which also were positive for viral RNA by RT-PCR) and in the cortex of one mouse harvested 9 months after infection (RNA for this sample not available for real-time RT-PCR analysis). Lesions were observed in the dentate gyrus and CA3 regions of the hippocampus at 3 months p.i. (Fig. 5A) . Furthermore, Oil Red O staining could be seen in identical regions of the hippocampus (Fig. 5C). A dramatic increase in Iba1+ microglia was observed and was shown to localize near lesion regions (Fig. 5E). No lesions, Oil Red O staining, or signs of microgliosis were observed in mock-infected control mice (Fig. 5A, D, and F, respectively).
FIG. 5.
Signs of CNS pathology and chronic inflammation remain up to 9 months p.i. Three-day-old pups were infected with rCVB3 (2 × 106 to 107 PFU i.c.). Brains were harvested 3 months and 9 months later for histological analysis. (A) Lesions were observed in the dentate gyrus and CA3 region of the hippocampus in three mice harvested 90 days p.i. (cyan arrows; representative sample shown). Oil Red O staining (C; cyan arrow) and microgliosis/Iba+ cells (E; cyan arrow) were seen in parallel sections. No lesions (B), Oil Red O staining (D), or microgliosis (F) was observed in the hippocampus of mock-infected control mice (representative of three mice). (G) Lesions in the cortex (magnification, ×5) were observed as late as 9 months after initial infection (magnification, ×5; cyan arrows). (H) Furthermore, inflammatory cells were identified within or adjacent to these lesions (magnification, ×63; cyan arrows). (I) The continued presence of CD3+ T cells, although reduced in number, was observed within the lesions (magnification, ×63; notched cyan arrows). (J) Similarly, Iba1+ cells continued to be found in high numbers within the cortex (cyan arrow) and hippocampal lesions (magnification, ×5). (K) Higher magnification (magnification, ×63) of image in panel J demonstrated pleomorphic Iba1+ microglia within the cortex lesions. (L) The presence of rod-shaped Iba1+ cells near lesions indicated chronically activated microglia (magnification, ×63). (M) Parallel sections of images in panel J stained with Oil Red O revealed dark-staining regions overlapping with lesions in the cortex and Iba1+ cells (magnification, ×5). (N) Higher magnification (magnification, ×63) showed Oil Red O-stained lesions in the cortex adjacent to the hippocampus. (O) Iba1+ cells were found in high numbers within regions outlined by Oil Red O staining (magnification, ×63).
At 9 months p.i., lesions in the retrosplenial cortex remained clearly visible (Fig. 5G), and higher magnification revealed inflammatory cells encircling the lesions (Fig. 5H). At this later time point, CD3+ cells (i.e., T cells), although few in number, remained visible near the lesion (Fig. 5I). In addition, pleomorphic Iba1+ microglia were found in higher numbers within or near lesions (Fig. 5J and K). In some cases, chronically activated microglia (rod-shaped form) were observed nearby (Fig. 5L). Similar to results described in the legends of Fig. 4E and Fig. 5C, Oil Red O staining was present within the lesion (Fig. 5M). Higher magnification of parallel sections showed the presence pleomorphic Iba1+ microglia within the Oil Red O staining region (Fig. 5N and O).
Viral RNA detected in second-round cultures of passaged brain homogenates from infected animals.
Analyses of additional mice infected with rCVB3 indicated that viral persistence in the CNS of surviving animals may be a frequent event. In all, 15 of 21 animals harvested at 30 days p.i. and beyond displayed detectable levels of viral RNA, as determined by real-time RT-PCR (Fig. 6A). Furthermore, no animals harvested at these late time points showed signs of infectious virus, as determined by a standard viral plaque assay (data not shown). We determined if genome replicative activity could be detected in vitro by cocultivating brain homogenates from RNA-positive animals with HeLa cells, similar to experiments published previously for cardiac tissue persistently infected with CVB3 (35). By real-time RT-PCR, we detected the presence of viral RNA in the supernatants of second-round HeLa cells passaged with brain homogenates from 12 out of 15 RNA-positive animals (Fig. 6B). Significantly lower viral copy numbers were detected in supernatants of HeLa cells passaged with brain homogenates from mice at day 30 to 60 p.i. than in acutely infected brain homogenates (day 5 p.i.). Also, no cytolytic virus was detected by a standard viral plaque assay from the second-passage HeLa cell supernatants of animals at day 30 to 60 p.i. These results suggest that an attenuated variant of CVB3 may remain in the CNS of surviving animals, similar, perhaps, to the virus that was recently identified in the heart by Kim et al. (35). Further analysis of the molecular structure of persisting CVB3, both in vivo and in culture, will be necessary to determine whether deletions or nucleotide changes in the viral genome are responsible for the ability of CVB3 to persist for extended periods of time in the adult CNS following acute infection early in life.
DISCUSSION
Enteroviruses, which include coxsackieviruses, are among the most widespread and medically important human pathogens and remain a frequent cause of CNS disease (44). Indeed, enterovirus infections are the most common causes of encephalitis and meningitis in the United States (11). Evidence in a clinical setting suggests that type B coxsackieviruses (CVB) can cause acute demyelinating diseases, such as acute disseminated myelitis and acute transverse myelitis (18, 27). Intriguingly, persistent CVB infections have been associated with myocarditis (29, 61), diabetes (32), chronic inflammatory myopathy (53), and, most recently, Sjogren's disease (55). Viral RNA persistence is thought to correlate with chronic disease, which suggests that the long-term presence of viral materials may be a potential mechanism for disease induction. The lack of infectious virus during viral RNA persistence has been cited by others to suggest that CVB3-associated diseases may occur through autoimmune mechanisms. However, an alternative model is that CVB3 may slowly replicate or remain in a latent state with periodic reactivation, similar to other latent viruses, such as herpes virus (23). Alternatively, residual amounts of protein generated by the persisting viral genome may be toxic to cells harboring CVB3 (57, 59) or, conversely, may contribute to a chronic inflammatory response in the CNS (60).
Previously, we have shown that CVB3 targets proliferating neuronal progenitor cells in the neonatal CNS (24) and that acute CVB3 infection induced profound pathology in specific regions of the developing CNS of mice (including the hippocampus, olfactory bulb, and entorhinal cortex). Here, we analyzed the ability of low-dose CVB3 to persist in the CNS following infection early in the life of the host. We report that many pups given lower doses of CVB3 or infected at a later age survive the initial infection and that viral RNA persistence commonly ensued. Utilizing quantitative real-time RT-PCR and extensive histopathological analysis of harvested brain tissue, we demonstrated that CVB3 can persist in the CNS, at least at the level of viral RNA. Furthermore, the presence of viral RNA in surviving animals was associated with chronic immunopathology in specific regions of the CNS. Finally, brain homogenates were passaged on HeLa cells, and the continued presence of viral RNA was detected in second-passage supernatants harvested and analyzed by quantitative real-time RT-PCR. These results suggest that CVB3 infection of the CNS may be characterized by sporadic reactivation or chronic, low-level replication of attenuated virus.
How does CVB, an inherently lytic virus, persist in vivo, and what mechanisms might regulate CVB persistence? Two recent publications suggest that the presence of CVB in the heart relies upon the generation of noncytolytic variants with 5′ terminal mutations and deletions (35, 38). Yet it is not clear from this study if attenuated, noncytolytic variants are found in human patients and how these variants may out-compete wild-type virus, assuming that CVB grows without major impediments in cardiomyocytes, as has been shown in vitro (2, 30, 39). Furthermore, recent data suggest that engineered deletions of the 5′ UTR of an infectious CVB3 clone (similar, although not identical, to those identified by Kim et al.) failed to give rise to infectious progeny or detectable levels of viral protein expression (33).
Data from other CVB models suggest that, contrary to the mechanism proposed by the above studies, virus persistence is facilitated not by genetic alterations in the virus that give rise to replication-defective forms but by the genetic stability of double-stranded complexes of wild-type viral RNA (54). Data from a number of laboratories which have identified a 1/1 ratio of positive- to negative-strand viral RNA genome during persistence in the heart would fit well with the model proposed by Tam and Messner (54). We determined the ratio of positive- to negative-strand CVB3 RNA genome in the brain and observed a reduction in this ratio over time. By day 30 p.i., the ratio approached 1/1 (ratio of 1.7/1), which suggests that CVB3 RNA might persist in the CNS in a double-stranded conformation. The ratio remained close to 1/1 in samples isolated from mice at 90 days p.i. Along with our previous data which support the view that proliferating neuronal progenitor cells are the target cells for active CVB3 replication, the data suggest that virus may remain in a double-stranded latent conformation in nondividing, differentiated cells of the CNS. Cells harboring persistent CVB3 RNA may represent the progeny of infected stem cells that survived initial infection during their differentiation process and progressed into mature neurons. However, the passage of brain homogenates (described in the legend of Fig. 6) may indicate the presence of limited amounts of noncytolytic virus which may replicate sporadically or at a low level, thereby not greatly affecting ratios of positive- to negative-strand CVB3 RNA genome.
The persistence of potentially labile viral RNA within a cellular environment rich with degradative enzymes might be a difficult phenomenon to consider. Viral RNA may be retained in the absence of infectious virus by at least two mechanisms. Viral RNA may remain in a latent state which sporadically reactivates in response to physiological stimuli (23). Alternatively, viral RNA may denote the presence of slowly replicating attenuated viral variants; the lack of infectious virus may be a consequence of the inability of attenuated viral variants to induce cytopathic effects necessary for detection by standard viral plaque assays. These two mechanisms might not be mutually exclusive. For example, attenuated virus may replicate sporadically in vivo, which may allow sluggish viral variants to partially circumvent the innate and adaptive immune response. Furthermore, a cellular environment inhospitable to viral replication (cellular quiescence and activation of cellular molecules with antiviral activity) may assist attenuated viral variants to outcompete rapidly growing wild-type virus.
We determined if virus material in brain homogenates from infected mice showing no infectious virus by a standard plaque assay could be passaged in HeLa cells. We hypothesized that the detection of viral RNA signal (by real-time RT-PCR) from HeLa cells cultured with brain homogenate would disappear during serial passage, and any remaining viral RNA detected within the supernatants of second-round HeLa cell cultures might indicate the presence of replicative virus (although perhaps not cytolytic) (35). Surprisingly, viral RNA was readily detected by real-time RT-PCR in second-round HeLa cell cultures passaged with brain homogenates from 12 of 15 infected animals showing no infectious virus by plaque assay. In contrast, no plaque titers (by standard viral plaque assay) were detected for these samples. Consequently, the lack of detectable infectious virus in brain homogenates (as identified by viral plaque assay) may be primarily due to the generation of noncytopathic viral variants in vivo. These conclusions would parallel a recent publication describing attenuated, noncytopathic CVB3 with 5′ deletions in persistently infected cardiac tissue (35).
For persistent or latent viruses, the ability to circumvent the immune system is vital. RNA viruses would be expected to induce type I IFNs and associated IFN response genes following the appearance of double-stranded RNA during viral replication. Likewise, mice deficient in type I IFN responses are highly susceptible to CVB3 infection and disease (20, 25, 58), suggesting that these immune molecules partially control CVB3 replication. Nevertheless, picornaviruses have mechanisms to evade the immune response during persistence/latency (14, 19, 21). Since CVB3 establishes persistence in many of its target organs, we thought it important to determine whether chemokines and cytokines were upregulated during acute and persistent infection. The rationale for infecting mice with rCVB3-LCMVNP118-126 was to eventually track and follow adoptively transferred memory T cells from LCMV NP118-126 T-cell receptor transgenic (SMARTA) mice and determine viral protein expression levels during CVB3 persistence. These studies are ongoing. However, both eGFP-CVB3 and rCVB3-LCMVNP118-126 were able to persist at the RNA level and induce similar immunopathologies in the CNS of mice surviving neonatal infection. Viral doses for both recombinant viruses were optimized to inoculate the largest amount of infectious virus leading to greater than 50% mouse survival. Since CVB3 replication kinetics depends upon the size insert, rCVB3-LCMVNP118-126 required less inoculum to reach a similar level of mouse survival.
A number of chemokine genes were induced, some within 24 h (CXCL10 and MCP-5), following infection in the neonatal CNS. Many of these upregulated chemokine (CCL5, CXCL9, and CXCL10) genes have been described in other models of viral infection in the CNS (8), and these immune molecules may play a direct role in chronic inflammation and disease. It remains to be determined which cell types are contributing to early chemokine production although other investigators have described astrocytes and activated microglia as the main chemokine producers during the early phase of a generalized infection in the CNS. Activation of a subset of cytokines was also observed after CVB3 infection. IL-12 p40 mRNA was upregulated in the absence of IL-12 p35 mRNA, which suggests that IL-23 may preferentially be upregulated in the CNS. IL-23 may, in turn, activate microglia or recruit macrophages into the CNS (17). Alternatively, the presence of IL-23 may reflect the degree of microgliosis and susceptibility to autoimmunity (5). The upregulation of the IL-1α and IL-1β genes also indicates the activation of microglia or macrophages in the CNS in response to CVB3 infection. As expected, a number of type I IFN response genes (IFN-β, PKR, OAS, TGTP1, and antiviral GTPase) were upregulated in the CNS following infection although others (IFN-α) were not. It remains to be determined if CVB3 infection influences the preferential activation of a subset of IFN response genes, perhaps for its own advantage. We failed to observe the upregulation of a type I IFN response and other immune regulatory genes during the persistent stage of infection. The lack of upregulated immune molecules at later time points at a time when viral RNA is present (30 days p.i.) may be explained by at least three possible scenarios. (i) CVB3 may remain in a latent state with little or no apparent virus replication or viral protein expression. (ii) An extremely low level of chemokine, cytokine, or type I IFN response gene induction may be produced but may be beyond detection limits of our RPA analysis. (iii) Viral modulation of the innate immune response may diminish the activation of chemokines, cytokines, or type I IFN response genes. Although one might expect a direct correlation of chemokine/cytokine gene expression levels with viral load, many in vivo variables may make such a close correlation uncertain. For example, slight differences in viral replication rates in certain regions of the CNS (hippocampus versus the cortex) may lead to a slightly altered immune response and subsequent chemokine/cytokine induction when individual animals are compared. Stochastic processes may also be in play during the activation and recruitment of immune cells responding to infection.
CVB3 may influence normal CNS development by inducing apoptosis in both infected stem cells and neurons (22, 24). Potential disruption of normal neurogenic activity might lead to developmental defects and CNS deficiencies, such as cognitive dysfunction (10). An extended immunopathology in the CNS following CVB3 infection may adversely affect normal CNS development and function. In our present study, reactive microgliosis was seen in the developing CNS during acute and persistent infection. In addition, signs of inflammation including activated microglia and infiltrating T cells were readily visualized within lesions in the hippocampus and cortex as late as 9 months after initial infection. Although older pups inoculated with a greater amount rCVB3 were not analyzed for chemokine and cytokine mRNA levels, the presence of inflammatory cells in the CNS suggests that immune molecules might be detected in older pups given a higher dose of virus. Furthermore, greater levels of persistent viral RNA were generally observed in these high-dose pups, in parallel with more profound signs of disease in the form of hind limb dysfunction. The hind limb dysfunction may possibly be a consequence of the inflammatory response or, alternatively, of acute or persistent infection and apoptosis of targeted neurons.
Defective or attenuated viral variants with 5′ or 3′ deletions have been identified for other persistent RNA viruses (41, 42). However, one might expect a low level of either attenuated or wild-type virus to induce detectable levels of type I IFN response molecules and other chemokines/cytokines during a chronic infection, something we have not seen. Therefore, it remains to be determined whether the viral RNA detected by real-time RT-PCR during second-passage brain homogenates represents active virus replication of attenuated variants within the CNS. For example, picornaviral RNA is considered infectious; cultivation of brain homogenates with HeLa cells may allow for the engulfment/transfection of latent CVB3 RNA into an activated cell population, which may stimulate attenuated viral replication. Further genomic analysis of second-passage virus isolates may help determine if 5′ or 3′ deletions occur in the viral genome and allow attenuated CVB3 to persist in the CNS. Also, future studies are necessary to determine which cell types are contributing to CVB3 persistence in the CNS. Virus might persist in a subpopulation of quiescent stem cells which remain, albeit at low frequency, in the adult CNS. Although stem cells might be clear candidates, differentiated neurons or other CNS cell types (astrocytes, microglia, and oligodendrocytes) may also harbor persistent viral RNA. The relatively quiescent state of differentiated cells may help the virus to remain in a restricted, nonreplicative state, similar to what has been described for poliovirus (15, 26). Regardless of the site of persistence, the presence of viral material in the CNS for extended periods of time may contribute to chronic immunopathology and long-term clinical consequences in the surviving host.
Acknowledgments
We are grateful to Annette Lord for excellent secretarial support.
This work was supported by National Institutes of Health (NIH) awards R01 NS054108-02 (to R.F.), R01 AI-42314 (to J.L.W.), MH62331, and NS036979 (to I.L.C.); Advanced Postdoctoral Fellowships from the National Multiple Sclerosis Society (FA 1551-A-1 to R.F. and FA-1552-A-1 to S.J.C); and NIH award F32 AI-065095 (to C.T.C.).
We have no conflicts of interest that are relevant to the content of this study.
This is manuscript number 18737-MIND from the Scripps Research Institute.
Footnotes
Published ahead of print on 1 July 2009.
REFERENCES
- 1.Ahn, J., J. Choi, C. H. Joo, I. Seo, D. Kim, S. Y. Yoon, Y. K. Kim, and H. Lee. 2004. Susceptibility of mouse primary cortical neuronal cells to coxsackievirus B. J. Gen. Virol. 85:1555-1564. [DOI] [PubMed] [Google Scholar]
- 2.Ahn, J., C. H. Joo, I. Seo, D. Kim, Y. K. Kim, and H. Lee. 2005. All CVB serotypes and clinical isolates induce irreversible cytopathic effects in primary cardiomyocytes. J. Med. Virol. 75:290-294. [DOI] [PubMed] [Google Scholar]
- 3.Akassoglou, K., L. Probert, G. Kontogeorgos, and G. Kollias. 1997. Astrocyte-specific but not neuron-specific transmembrane TNF triggers inflammation and degeneration in the central nervous system of transgenic mice. J. Immunol. 158:438-445. [PubMed] [Google Scholar]
- 4.Alleva, D. G., E. B. Johnson, J. Wilson, D. I. Beller, and P. J. Conlon. 2001. SJL and NOD macrophages are uniquely characterized by genetically programmed, elevated expression of the IL-12(p40) gene, suggesting a conserved pathway for the induction of organ-specific autoimmunity. J. Leukoc. Biol. 69:440-448. [PubMed] [Google Scholar]
- 5.Andersson, A., R. Kokkola, J. Wefer, H. Erlandsson-Harris, and R. A. Harris. 2004. Differential macrophage expression of IL-12 and IL-23 upon innate immune activation defines rat autoimmune susceptibility. J. Leukoc. Biol. 76:1118-1124. [DOI] [PubMed] [Google Scholar]
- 6.Asensio, V. C., and I. L. Campbell. 1997. Chemokine gene expression in the brains of mice with lymphocytic choriomeningitis. J. Virol. 71:7832-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asensio, V. C., J. Maier, R. Milner, K. Boztug, C. Kincaid, M. Moulard, C. Phillipson, K. Lindsley, T. Krucker, H. S. Fox, and I. L. Campbell. 2001. Interferon-independent, human immunodeficiency virus type 1 gp120-mediated induction of CXCL10/IP-10 gene expression by astrocytes in vivo and in vitro. J. Virol. 75:7067-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bergmann, C. C., T. E. Lane, and S. A. Stohlman. 2006. Coronavirus infection of the central nervous system: host-virus stand-off. Nat. Rev. Microbiol. 4:121-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boztug, K., M. J. Carson, N. Pham-Mitchell, V. C. Asensio, J. DeMartino, and I. L. Campbell. 2002. Leukocyte infiltration, but not neurodegeneration, in the CNS of transgenic mice with astrocyte production of the CXC chemokine ligand 10. J. Immunol. 169:1505-1515. [DOI] [PubMed] [Google Scholar]
- 10.Buenz, E. J., M. Rodriguez, and C. L. Howe. 2006. Disrupted spatial memory is a consequence of picornavirus infection. Neurobiol. Dis. 24:266-273. [DOI] [PubMed] [Google Scholar]
- 11.Centers for Disease Control and Prevention. 2003. Outbreaks of aseptic meningitis associated with echoviruses 9 and 30 and preliminary surveillance reports on enterovirus activity—United States, 2003. MMWR Morb. Mortal. Wkly. Rep. 52:761-764. [PubMed] [Google Scholar]
- 12.Chamberlain, R. N., P. N. Christie, K. S. Holt, R. M. Huntley, R. Pollard, and M. C. Roche. 1983. A study of school children who had identified virus infections of the central nervous system during infancy. Child Care Health Dev. 9:29-47. [DOI] [PubMed] [Google Scholar]
- 13.Constantinescu, C. S., K. Frei, M. Wysocka, G. Trinchieri, U. Malipiero, A. Rostami, and A. Fontana. 1996. Astrocytes and microglia produce interleukin-12 p40. Ann. N. Y. Acad. Sci. 795:328-333. [DOI] [PubMed] [Google Scholar]
- 14.Cornell, C. T., W. B. Kiosses, S. Harkins, and J. L. Whitton. 2006. Inhibition of protein trafficking by coxsackievirus B3: multiple viral proteins target a single organelle. J. Virol. 80:6637-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Couderc, T., S. Girard, I. Pelletier, A. S. Gosselin, J. Destombes, D. Thiesson, F. Colbere-Garapin, and B. Blondel. 2001. Inhibition of poliovirus RNA synthesis as a molecular mechanism contributing to viral persistence in the mouse central nervous system. Dev. Biol. (Basel) 105:225-230. [PubMed] [Google Scholar]
- 16.Cree, B. C., G. L. Bernardini, A. P. Hays, and G. Lowe. 2003. A fatal case of coxsackievirus B4 meningoencephalitis. Arch. Neurol. 60:107-112. [DOI] [PubMed] [Google Scholar]
- 17.Cua, D. J., J. Sherlock, Y. Chen, C. A. Murphy, B. Joyce, B. Seymour, L. Lucian, W. To, S. Kwan, T. Churakova, S. Zurawski, M. Wiekowski, S. A. Lira, D. Gorman, R. A. Kastelein, and J. D. Sedgwick. 2003. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature 421:744-748. [DOI] [PubMed] [Google Scholar]
- 18.David, P., D. Baleriaux, W. O. Bank, D. Amrom, T. D. De, C. Babusiaux, C. Matos, S. C. Van, C. Lloret-Pastor, and H. B. Szliwowski. 1993. MRI of acute disseminated encephalomyelitis after coxsackie B infection. J. Neuroradiol. 20:258-265. [PubMed] [Google Scholar]
- 19.Deitz, S. B., D. A. Dodd, S. Cooper, P. Parham, and K. Kirkegaard. 2000. MHC I-dependent antigen presentation is inhibited by poliovirus protein 3A. Proc. Natl. Acad. Sci. USA 97:13790-13795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deonarain, R., D. Cerullo, K. Fuse, P. P. Liu, and E. N. Fish. 2004. Protective role for interferon-beta in coxsackievirus B3 infection. Circulation 110:3540-3543. [DOI] [PubMed] [Google Scholar]
- 21.Dodd, D. A., T. H. Giddings, Jr., and K. Kirkegaard. 2001. Poliovirus 3A protein limits interleukin-6 (IL-6), IL-8, and beta interferon secretion during viral infection. J. Virol. 75:8158-8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feuer, R., I. Mena, R. R. Pagarigan, S. Harkins, D. E. Hassett, and J. L. Whitton. 2003. Coxsackievirus B3 and the neonatal CNS: the roles of stem cells, developing neurons, and apoptosis in infection, viral dissemination, and disease. Am. J. Pathol. 163:1379-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feuer, R., I. Mena, R. R. Pagarigan, M. K. Slifka, and J. L. Whitton. 2002. Cell cycle status affects coxsackievirus replication, persistence, and reactivation in vitro. J. Virol. 76:4430-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feuer, R., R. R. Pagarigan, S. Harkins, F. Liu, I. P. Hunziker, and J. L. Whitton. 2005. Coxsackievirus targets proliferating neuronal progenitor cells in the neonatal CNS. J. Neurosci. 25:2434-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flodstrom, M., A. Maday, D. Balakrishna, M. M. Cleary, A. Yoshimura, and N. Sarvetnick. 2002. Target cell defense prevents the development of diabetes after viral infection. Nat. Immunol. 3:373-382. [DOI] [PubMed] [Google Scholar]
- 26.Girard, S., A. S. Gosselin, I. Pelletier, F. Colbere-Garapin, T. Couderc, and B. Blondel. 2002. Restriction of poliovirus RNA replication in persistently infected nerve cells. J. Gen. Virol. 83:1087-1093. [DOI] [PubMed] [Google Scholar]
- 27.Graber, D., C. Fossoud, E. Grouteau, C. Gayet-Mengelle, and J. P. Carriere. 1994. Acute transverse myelitis and coxsackie A9 virus infection. Pediatr. Infect. Dis.J. 13:77. [DOI] [PubMed] [Google Scholar]
- 28.Ha, S. J., C. H. Lee, S. B. Lee, C. M. Kim, K. L. Jang, H. S. Shin, and Y. C. Sung. 1999. A novel function of IL-12p40 as a chemotactic molecule for macrophages. J. Immunol. 163:2902-2908. [PubMed] [Google Scholar]
- 29.Henke, A., S. A. Huber, A. Stelzner, and J. L. Whitton. 1995. The role of CD8+ T lymphocytes in coxsackievirus B3-induced myocarditis. J. Virol. 69:6720-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herzum, M., V. Ruppert, B. Kuytz, H. Jomaa, I. Nakamura, and B. Maisch. 1994. Coxsackievirus B3 infection leads to cell death of cardiac myocytes. J. Mol. Cell Cardiol. 26:907-913. [DOI] [PubMed] [Google Scholar]
- 31.Hobbs, M. V., W. O. Weigle, D. J. Noonan, B. E. Torbett, R. J. McEvilly, R. J. Koch, G. J. Cardenas, and D. N. Ernst. 1993. Patterns of cytokine gene expression by CD4+ T cells from young and old mice. J. Immunol. 150:3602-3614. [PubMed] [Google Scholar]
- 32.Horwitz, M. S., L. M. Bradley, J. Harbertson, T. Krahl, J. Lee, and N. Sarvetnick. 1998. Diabetes induced by coxsackie virus: initiation by bystander damage and not molecular mimicry. Nat. Med. 4:781-785. [DOI] [PubMed] [Google Scholar]
- 33.Hunziker, I. P., C. T. Cornell, and J. L. Whitton. 2007. Deletions within the 5′ UTR of coxsackievirus B3: consequences for virus translation and replication. Virology 360:120-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim, K. S., G. Hufnagel, N. M. Chapman, and S. Tracy. 2001. The group B coxsackieviruses and myocarditis. Rev. Med. Virol. 11:355-368. [DOI] [PubMed] [Google Scholar]
- 35.Kim, K. S., S. Tracy, W. Tapprich, J. Bailey, C. K. Lee, K. Kim, W. H. Barry, and N. M. Chapman. 2005. 5′-Terminal deletions occur in coxsackievirus B3 during replication in murine hearts and cardiac myocyte cultures and correlate with encapsidation of negative-strand viral RNA. J. Virol. 79:7024-7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klingel, K., C. Hohenadl, A. Canu, M. Albrecht, M. Seemann, G. Mall, and R. Kandolf. 1992. Ongoing enterovirus-induced myocarditis is associated with persistent heart muscle infection: quantitative analysis of virus replication, tissue damage, and inflammation. Proc. Natl. Acad. Sci. USA 89:314-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Labrada, L., X. H. Liang, W. Zheng, C. Johnston, and B. Levine. 2002. Age-dependent resistance to lethal alphavirus encephalitis in mice: analysis of gene expression in the central nervous system and identification of a novel interferon-inducible protective gene, mouse ISG12. J. Virol. 76:11688-11703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee, C. K., K. Kono, E. Haas, K. S. Kim, K. M. Drescher, N. M. Chapman, and S. Tracy. 2005. Characterization of an infectious cDNA copy of the genome of a naturally occurring, avirulent coxsackievirus B3 clinical isolate. J. Gen. Virol. 86:197-210. [DOI] [PubMed] [Google Scholar]
- 39.Luo, H., J. Zhang, C. Cheung, A. Suarez, B. M. McManus, and D. Yang. 2003. Proteasome inhibition reduces coxsackievirus B3 replication in murine cardiomyocytes. Am. J. Pathol. 163:381-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Melville, M. W., S. L. Tan, M. Wambach, J. Song, R. I. Morimoto, and M. G. Katze. 1999. The cellular inhibitor of the PKR protein kinase, P58(IPK), is an influenza virus-activated co-chaperone that modulates heat shock protein 70 activity. J. Biol. Chem. 274:3797-3803. [DOI] [PubMed] [Google Scholar]
- 41.Meyer, B. J., and C. Schmaljohn. 2000. Accumulation of terminally deleted RNAs may play a role in Seoul virus persistence. J. Virol. 74:1321-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meyer, B. J., and P. J. Southern. 1997. A novel type of defective viral genome suggests a unique strategy to establish and maintain persistent lymphocytic choriomeningitis virus infections. J. Virol. 71:6757-6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muir, P., F. Nicholson, M. K. Sharief, E. J. Thompson, N. J. Cairns, P. Lantos, G. T. Spencer, H. J. Kaminski, and J. E. Banatvala. 1995. Evidence for persistent enterovirus infection of the central nervous system in patients with previous paralytic poliomyelitis. Ann. N. Y. Acad. Sci. 753:219-232. [DOI] [PubMed] [Google Scholar]
- 44.Muir, P., and A. M. van Loon. 1997. Enterovirus infections of the central nervous system. Intervirology 40:153-166. [DOI] [PubMed] [Google Scholar]
- 45.Oldstone, M. B. 1998. Molecular mimicry and immune-mediated diseases. FASEB J. 12:1255-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pagenstecher, A., S. Lassmann, M. J. Carson, C. L. Kincaid, A. K. Stalder, and I. L. Campbell. 2000. Astrocyte-targeted expression of IL-12 induces active cellular immune responses in the central nervous system and modulates experimental allergic encephalomyelitis. J. Immunol. 164:4481-4492. [DOI] [PubMed] [Google Scholar]
- 47.Rantakallio, P., P. Jones, J. Moring, and L. von Wendt. 1997. Association between central nervous system infections during childhood and adult onset schizophrenia and other psychoses: a 28-year follow-up. Int. J. Epidemiol. 26:837-843. [DOI] [PubMed] [Google Scholar]
- 48.Reetoo, K. N., S. A. Osman, S. J. Illavia, C. L. Cameron-Wilson, J. E. Banatvala, and P. Muir. 2000. Quantitative analysis of viral RNA kinetics in coxsackievirus B3-induced murine myocarditis: biphasic pattern of clearance following acute infection, with persistence of residual viral RNA throughout and beyond the inflammatory phase of disease. J. Gen. Virol. 81:2755-2762. [DOI] [PubMed] [Google Scholar]
- 49.Sawyer, M. H. 2002. Enterovirus infections: diagnosis and treatment. Semin. Pediatr. Infect. Dis. 13:40-47. [DOI] [PubMed] [Google Scholar]
- 50.Slifka, M. K., R. R. Pagarigan, I. Mena, R. Feuer, and J. L. Whitton. 2001. Using recombinant coxsackievirus B3 to evaluate the induction and protective efficacy of CD8+ T cells in controlling picornaviral infection. J. Virol. 75:2377-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stalder, A. K., and I. L. Campbell. 1994. Simultaneous analysis of multiple cytokine receptor mRNAs by RNase protection assay in LPS-induced endotoxemia. Lymphokine Cytokine Res. 13:107-112. [PubMed] [Google Scholar]
- 52.Suvisaari, J., N. Mautemps, J. Haukka, T. Hovi, and J. Lonnqvist. 2003. Childhood central nervous system viral infections and adult schizophrenia. Am. J. Psychiatry 160:1183-1185. [DOI] [PubMed] [Google Scholar]
- 53.Tam, P. E., D. R. Fontana, and R. P. Messner. 2003. Coxsackievirus B1-induced chronic inflammatory myopathy: differences in induction of autoantibodies to muscle and nuclear antigens by cloned myopathic and amyopathic viruses. J. Lab. Clin. Med. 142:196-204. [DOI] [PubMed] [Google Scholar]
- 54.Tam, P. E., and R. P. Messner. 1999. Molecular mechanisms of coxsackievirus persistence in chronic inflammatory myopathy: viral RNA persists through formation of a double-stranded complex without associated genomic mutations or evolution. J. Virol. 73:10113-10121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Triantafyllopoulou, A., N. Tapinos, and H. M. Moutsopoulos. 2004. Evidence for coxsackievirus infection in primary Sjogren's syndrome. Arthritis Rheum. 50:2897-2902. [DOI] [PubMed] [Google Scholar]
- 56.Verboon-Maciolek, M. A., F. Groenendaal, F. Cowan, P. Govaert, A. M. van Loon, and L. S. de Vries. 2006. White matter damage in neonatal enterovirus meningoencephalitis. Neurology 66:1267-1269. [DOI] [PubMed] [Google Scholar]
- 57.Wessely, R., A. Henke, R. Zell, R. Kandolf, and K. U. Knowlton. 1998. Low-level expression of a mutant coxsackieviral cDNA induces a myocytopathic effect in culture: an approach to the study of enteroviral persistence in cardiac myocytes. Circulation 98:450-457. [DOI] [PubMed] [Google Scholar]
- 58.Wessely, R., K. Klingel, K. U. Knowlton, and R. Kandolf. 2001. Cardioselective infection with coxsackievirus B3 requires intact type I interferon signaling: implications for mortality and early viral replication. Circulation 103:756-761. [DOI] [PubMed] [Google Scholar]
- 59.Wessely, R., K. Klingel, L. F. Santana, N. Dalton, M. Hongo, L. W. Jonathan, R. Kandolf, and K. U. Knowlton. 1998. Transgenic expression of replication-restricted enteroviral genomes in heart muscle induces defective excitation-contraction coupling and dilated cardiomyopathy. J. Clin. Investig. 102:1444-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Whitton, J. L., C. T. Cornell, and R. Feuer. 2005. Host and virus determinants of picornavirus pathogenesis and tropism. Nat. Rev. Microbiol. 3:765-776. [DOI] [PubMed] [Google Scholar]
- 61.Whitton, J. L., and R. Feuer. 2004. Myocarditis, microbes, and autoimmunity. Autoimmunity 37:375-386. [DOI] [PubMed] [Google Scholar]
- 62.Woodall, C. J., and D. I. Graham. 2004. Evidence for neuronal localisation of enteroviral sequences in motor neurone disease/amyotrophic lateral sclerosis by in situ hybridization. Eur. J. Histochem. 48:129-134. [DOI] [PubMed] [Google Scholar]
- 63.Woodall, C. J., M. H. Riding, D. I. Graham, and G. B. Clements. 1994. Sequences specific for enterovirus detected in spinal cord from patients with motor neurone disease. BMJ. 308:1541-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]