Abstract
The requirement for multiple mutations for protease inhibitor (PI) resistance necessitates a better understanding of the molecular basis of resistance development. The novel bioinformatics resistance determination approach presented here elaborates on genetic profiles observed in clinical human immunodeficiency virus type 1 (HIV-1) isolates. Synthetic protease sequences were cloned in a wild-type HIV-1 background to generate a large number of close variants, covering 69 mutation clusters between multi-PI-resistant viruses and their corresponding genetically closely related, but PI-susceptible, counterparts. The vast number of mutants generated facilitates a profound and broad analysis of the influence of the background on the effect of individual PI resistance-associated mutations (PI-RAMs) on PI susceptibility. Within a set of viruses, all PI-RAMs that differed between susceptible and resistant viruses were varied while maintaining the background sequence from the resistant virus. The PI darunavir was used to evaluate PI susceptibility. Single sets allowed delineation of the impact of individual mutations on PI susceptibility, as well as the influence of PI-RAMs on one another. Comparing across sets, it could be inferred how the background influenced the interaction between two mutations, in some cases even changing antagonistic relationships into synergistic ones or vice versa. The approach elaborates on patient data and demonstrates how the specific mutational background greatly influences the impact of individual mutations on PI susceptibility in clinical patterns.
The clinical use of protease inhibitors (PIs) for the treatment of human immunodeficiency virus (HIV) infection has led to a remarkable decline in HIV-1-related morbidity and mortality, and PIs are now a cornerstone of highly active antiretroviral therapy (14). However, the clinical benefit of PIs is limited by several factors, including long-term safety and tolerability, resistance development, and drug-drug interactions.
The combination of extremely high levels of virus production and a high mutation rate is resulting in a growing resistance to anti-HIV drugs, making these less effective over time (1). In addition, an increasing proportion of primary infections involve the transmission of resistant viruses, including strains with reduced susceptibility to approved PIs (17). Therefore, patients need to be monitored for development of drug resistance, and treatment regimens have to be adapted accordingly. Most currently approved PIs are based on similar chemical structures, and therefore extensive cross-resistance can occur (7).
In order to investigate the molecular basis of resistance development, we used the PI darunavir (DRV) as a model. DRV, previously known as TMC114, was approved in 2006 for the treatment of highly experienced patients and in 2008 for treatment of naïve patients. DRV has a high in vitro and in vivo potency against wild-type (WT) HIV, and this activity is maintained against HIV variants that are highly cross-resistant to other licensed PIs (2, 15). Moreover, there appears to be a very high genetic barrier to the development of resistance to DRV (3). A diminished virological response to DRV was only observed at week 24 (POWER studies [4]), when at least three specific baseline protease mutations (of V11I, V32I, L33F, I47V, I50V, I54L/M, G73S, L76V, I84V, and L89V) occurred in a background containing multiple protease mutations (median of at least 10 International AIDS Society-USA [IAS-USA] PI resistance-associated mutations [PI-RAMs] [11]).
Mutations can interact as part of higher-order networks in complex and frequently overlapping patterns (7, 16, 18). In such patterns, the effect of an individual protease mutation on drug susceptibility depends on the presence of other mutations, PI-RAMs as well as background mutations. Many of the background mutations act synergistically with PI-RAMs and increase resistance to specific drugs. In addition, some of these mutations favor the development of other drug resistance mutations, thus lowering the genetic barrier to the development of PI resistance. In contrast, some mutations in the mutational background antagonize the effects of an individual PI-RAM. As resistance mutations are usually associated with reduced viral fitness, it may be that certain background mutations could (partly) compensate for this (12).
In order to design drugs with high genetic barriers to resistance, a full understanding of the molecular basis of resistance development is needed. This includes the complex interplay between resistance mutations that can be studied only by exploring genetically close variants. Because of the high variability of HIV, it is difficult to find the genetically related variants required for such a study in patient databases, even if they contain sequences from thousands of virus isolates. Traditional approaches utilizing site-directed mutagenesis to create close variants by modifying the protease amino acids in existing viruses are feasible only on a small scale. The advent of mature gene assembly technologies makes the large-scale generation of closely related variants practicable. Here we describe a novel approach, bioinformatics resistance determination (BIRD), in which we created PI resistance sets between viral genotypes observed in patient samples. By varying a specific set of mutations in an invariable genetic background, the complex interactions between these mutations could be carefully dissected. Our studies illustrate how some mutations do not influence other mutations, while other changes act synergistically or antagonistically toward a specific RAM. Moreover, by comparing sets, we show how a specific background can alter the interplay between mutations.
MATERIALS AND METHODS
Identification from patient data of pairs of a resistant virus and a susceptible virus.
Using phenotypic characteristics, two groups of viruses were selected from an in-house database containing matched phenotypic and genotypic profiles of HIV-1 clinical isolates: 2,286 viruses that were susceptible to DRV (fold change in 50% inhibitory concentration [FC] of <10) and 718 viruses with a reduced susceptibility to DRV (FC of ≥40). For each of the 718 latter viruses, a clinical isolate with a closely related genotype but a divergent phenotype was selected among the 2,286 susceptible viruses by minimizing the following genotypic distance metric D: D = 0.1 × Mixsusc + 0.01 × Mutsusc + ∑WPI-RAM, where Mixsusc is the number of mutation mixes in the susceptible virus, Mutsusc is the number of mutations in the susceptible virus, and WPI-RAM is the weight of PI-RAM present in susceptible or in resistant virus, but not in both. The weights of the PI-RAMs were based on a 2004 linear model for DRV resistance. When a selected DRV-susceptible virus contained a mutation mix, this virus was considered to be two viruses, hence matching a given resistant virus with two susceptible viruses.
Using the above-described approach, 1,015 pairs of a susceptible virus and a genetically closely related resistant virus were identified. Then, due to restricted resources, the number of pairs had to be further reduced to 69 (Table 1). The following restrictions were applied to reach this goal. First, susceptible and resistant viruses had to be different in at least two mutations considered to be important: V32I, L33F, L33I, M46I, M46L, I47V, I50V, I54L, I54M, I54V, L76V, V82A, V82F, I84V, and L90M. Second, the shift in antiviral activity for any given mutation in the pair had to be consistent with the shift observed for this mutation in other pairs, hence excluding errors in measuring phenotypes and reducing the potential influence of mutations at loci outside the protease gene. Then, viruses that originated from clinical samples of DRV clinical studies were chosen. Next, pairs with a difference of three to five PI-RAMs between the susceptible and the resistant viruses were chosen. In a last step, the selection was further reduced by a manual selection of the pairs that contained the most interesting mutational patterns.
TABLE 1.
Overview of the 69 sets studied and the PI-RAMs varied within each of these sets
| Setc | Mutationa |
FCb |
No. of viruses | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L10 | K20 | V32 | L33 | M36 | M46 | I47 | G48 | I50 | F53 | I54 | A71 | G73 | L76 | V77 | V82 | I84 | N88 | L90 | Source | Target | ||
| 1 | V | V | V | 7.95 | 25.84 | 8 | ||||||||||||||||
| 2 | F | V | I | M | 0.79 | 5.40 | 16 | |||||||||||||||
| 3 | R | L | L | A | 7.20 | — | 16 | |||||||||||||||
| 4 | L | V | V | M | 0.69 | 4.33 | 16 | |||||||||||||||
| 5 | V | L | V | V | M | 0.48 | 2.48 | 32 | ||||||||||||||
| 6 | I | I | V | V | 1.99 | — | 16 | |||||||||||||||
| 7 | F | V | L | V | M | 5.85 | 71.42 | 32 | ||||||||||||||
| 8 | L | V | V | 0.99 | 32.48 | 8 | ||||||||||||||||
| 9 | L | M | V | I | 1.45 | 6.43 | 16 | |||||||||||||||
| 10 | I | V | I | 1.65 | 9.60 | 8 | ||||||||||||||||
| 11 | V | V | V | C | 2.08 | 9.04 | 16 | |||||||||||||||
| 12 | T | L | V | S | 20.28 | — | 16 | |||||||||||||||
| 13 | L | V | D | M | 0.79 | — | 16 | |||||||||||||||
| 14 | I | S | M | 1.73 | 1.40 | 8 | ||||||||||||||||
| 15 | I | L | I | M | 0.85 | 33.06 | 16 | |||||||||||||||
| 16 | F | I | M | 4.94 | 11.83 | 8 | ||||||||||||||||
| 17 | M | V | M | 1.79 | 7.84 | 8 | ||||||||||||||||
| 18 | I | I | V | 0.73 | 5.25 | 8 | ||||||||||||||||
| 19 | V | M | T | M | 1.59 | — | 16 | |||||||||||||||
| 20 | L | V | M | 2.36 | 27.93 | 8 | ||||||||||||||||
| 21 | R | I | V | A | 2.04 | 39.32 | 16 | |||||||||||||||
| 22 | I | V | V | M | 5.43 | — | 16 | |||||||||||||||
| 23 | T | I | L | V | S | 15.87 | — | 32 | ||||||||||||||
| 24 | R | I | V | M | D | 1.52 | 85.80 | 32 | ||||||||||||||
| 25 | I | I | V | A | 4.70 | 70.60 | 16 | |||||||||||||||
| 26 | I | V | L | 7.62 | 10.96 | 8 | ||||||||||||||||
| 27 | I | V | L | 0.55 | 16.49 | 8 | ||||||||||||||||
| 28 | I | V | M | 3.78 | 27.53 | 8 | ||||||||||||||||
| 29 | I | M | M | 2.51 | 20.36 | 8 | ||||||||||||||||
| 30 | A | V | A | 3.29 | — | 8 | ||||||||||||||||
| 31 | L | I | L | M | 3.61 | 28.50 | 16 | |||||||||||||||
| 32 | I | L | S | A | 17.44 | 24.96 | 16 | |||||||||||||||
| 33 | I | F | L | I | 0.98 | 46.88 | 16 | |||||||||||||||
| 34 | I | V | V | M | 0.45 | 7.58 | 16 | |||||||||||||||
| 35 | I | S | V | M | 9.48 | — | 16 | |||||||||||||||
| 36 | I | V | M | 0.69 | — | 8 | ||||||||||||||||
| 37 | R | I | V | 13.71 | 16.01 | 8 | ||||||||||||||||
| 38 | L | V | A | 14.17 | 41.10 | 8 | ||||||||||||||||
| 39 | V | L | V | T | A | 57.07 | 68.58 | 32 | ||||||||||||||
| 40 | I | I | I | A | 7.52 | 37.56 | 16 | |||||||||||||||
| 41 | I | V | V | 1.13 | — | 8 | ||||||||||||||||
| 42 | I | R | L | V | 0.24 | 4.47 | 16 | |||||||||||||||
| 43 | I | I | L | 1.59 | 18.05 | 8 | ||||||||||||||||
| 44 | R | V | L | A | 3.41 | — | 16 | |||||||||||||||
| 45 | M | S | V | 3.67 | 29.81 | 8 | ||||||||||||||||
| 46 | I | I | A | M | 20.83 | 22.74 | 16 | |||||||||||||||
| 47 | I | T | T | V | 0.49 | — | 16 | |||||||||||||||
| 48 | I | V | V | V | M | 5.98 | — | 32 | ||||||||||||||
| 49 | V | V | M | 1.45 | — | 8 | ||||||||||||||||
| 50 | L | V | V | M | 1.67 | 23.26 | 16 | |||||||||||||||
| 51 | R | I | A | V | M | 19.11 | 54.13 | 32 | ||||||||||||||
| 52 | I | I | F | M | 4.12 | 18.63 | 16 | |||||||||||||||
| 53 | I | I | M | 1.82 | 54.92 | 8 | ||||||||||||||||
| 54 | I | I | V | S | 1.90 | 35.04 | 16 | |||||||||||||||
| 55 | R | I | V | C | 24.05 | 118.80 | 16 | |||||||||||||||
| 56 | V | L | V | 13.50 | — | 8 | ||||||||||||||||
| 57 | V | V | T | V | 4.68 | 384.47 | 16 | |||||||||||||||
| 58 | V | L | V | V | 31.35 | 43.85 | 16 | |||||||||||||||
| 59 | T | I | I | M | 0.79 | 42.06 | 16 | |||||||||||||||
| 60 | I | S | A | V | M | 9.70 | 79.09 | 32 | ||||||||||||||
| 61 | V | I | L | L | F | 0.75 | 206.95 | 32 | ||||||||||||||
| 62 | I | I | V | A | M | 6.49 | 18.75 | 32 | ||||||||||||||
| 63 | R | I | I | V | A | 20.07 | 30.21 | 32 | ||||||||||||||
| 64 | I | V | T | A | 5.59 | 29.10 | 16 | |||||||||||||||
| 65 | R | I | I | V | 1.06 | 12.49 | 16 | |||||||||||||||
| 66 | I | V | M | 59.99 | — | 8 | ||||||||||||||||
| 67 | T | F | I | L | V | 15.82 | 107.08 | 32 | ||||||||||||||
| 68 | V | I | V | M | 1.64 | — | 16 | |||||||||||||||
| 69 | I | L | V | 0.94 | — | 8 | ||||||||||||||||
Based on the IAS-USA 2004 guidelines (10), primary PI mutations are indicated in bold and DRV-RAMs are indicated in bold with underlining.
The DRV susceptibilities of source virus (with none of the indicated PI-RAMs) and target virus (with all of the indicated PI-RAMs) virus are shown as the change in 50% inhibitory concentrations compared to the WT 50% inhibitory concentrations (dashes indicate target viruses that were not available).
For background sequences of the 69 sets, see supplemental Table S1.
Creation of synthetic HIV-1 protease sequences.
As the designed variations were confined to the protease sequence, one large 2.3-kb fragment containing the WT (HIV-1 HXB2) 3′ end of Gag and the entire protease and reverse transcriptase sequences was first synthesized and cloned, and then the protease variants were synthesized as smaller fragments (0.7 kb) that were each subcloned into the 2.3-kb fragment. BamHI and EcoRI sites were added to the 2.3-kb fragment for directional cloning into the pBluescript II KS(+) vector (Stratagene, La Jolla, CA) to create the base construct. A BamHI site was added and a BsrGI site was retained (from within the original 2.3-kb fragment) in each of the protease variant fragments for directional cloning into the 2.3-kb construct. All sequences were padded to a specified length.
The same process was followed to synthesize the original 2.3-kb fragment and all of the small fragments encompassing the protease variants (5, 6). Briefly, padded sequences were parsed into contiguous segments of equal length on both the forward and reverse strands. Each segment was then chemically synthesized as an oligonucleotide using GeneWriter (Centocor, San Diego, CA) technology and purified by reversed-phase high-pressure liquid chromatography (Dionex, Sunnyvale, CA). The purified oligonucleotides were assembled into the full-length 2.3-kb fragment using proprietary gene assembly technology (GeneAssembler; Centocor).
The 2.3-kb fragment was amplified, digested (enzymes from New England Biolabs, Ipswich, MA), and cloned into pBluescript II KS(+). To facilitate the assembly and cloning of the numerous protease variant fragments, the variants were grouped into combinatorial subset libraries ranging in size from 8 to 32 members. For each combinatorial subset library, the complete set of oligonucleotides for all members was then simultaneously assembled and cloned as described above into the pBluescript II KS(+) base construct. Sequencing of the 0.7-kb protease library region was used to identify all members of a combinatorial subset library. Competent cells of Escherichia coli strain DH5α were used for all cloning steps (Invitrogen, Carlsbad, CA). The entire 2.3-kb fragment of each variant was sequence confirmed using plasmid DNA as sequencing template, prepared with a Qiagen BioRobot 3000 (Qiagen, Hilden, Germany). Sequencing was done with an ABI 3730x1 DNA sequencer using standard BigDye version 3 chemistry (Applied Biosystems, Foster City, CA).
Production of recombinant viruses and antiviral assay.
The 2.3-kb inserts, cloned into pBluescript II KS(+) and containing the synthetic protease and WT Gag- and reverse transcriptase-coding sequences, were reamplified with a nested PCR. The resulting amplicons were homologously recombined into a proviral clone by cotransfection of the inserts into MT4 cells with a proviral clone lacking the 3′ end of Gag, protease, and reverse transcriptase, as previously described (8). Transfected cells were incubated, and when cytopathic effect occurred, the recombinant viruses were harvested and titers were determined.
The in vitro susceptibilities of the synthetic-gene-derived recombinant viruses to DRV were determined in a cell-based HIV-1 replication assay with enhanced green fluorescent protein (EGFP)-based fluorescent readout (9). Briefly, to determine the viral growth in the presence of antiviral compounds, MT4 cells equipped with long terminal repeat-EGFP at 500,000 cells/ml were inoculated with different dilutions of compound. The plates were incubated at 37°C with 5% CO2 for 3 to 4 days, and the fluorescence of the wells was read at different time points postinfection. The results were expressed as 50% inhibitory concentrations, defined as the concentration of compound achieving 50% inhibition of the virus-induced EGFP signals compared to the untreated virus-infected control cells. A change in susceptibility was calculated by dividing the 50% inhibitory concentration for the tested virus by the 50% inhibitory concentration for the WT virus (HIV-1/IIIB) tested in parallel, expressed in FC. Susceptibility measurements were performed in duplicate. A third measurement was performed when the two measured values were too divergent.
RESULTS
Identification from patient data of pairs of a resistant virus and a susceptible virus.
A Tibotec database containing matched phenotypic and genotypic profiles of HIV-1 clinical isolates was analyzed for viruses with a reduced susceptibility to DRV (FC of ≥40) (Fig. 1). For each such virus, a corresponding clinical isolate with a closely related genotype but a divergent phenotype was identified. While the susceptibilities of these corresponding clinical isolates to DRV were much higher (FC of <10), their protease-coding sequence was very similar to that of the more resistant virus, differing by only a few PI-RAMs. The genotypic distance metric used to select the virus pairs aimed at the selection of sets with the cleanest and most interesting synergies, i.e., sets that delivered an unexpectedly high FC for the PI-RAMs involved. As the two viruses did not originate from the same patient, mutational pathways in a traditional directional sense were not studied.
FIG. 1.
Selection of susceptible (susceptible to DRV; FC of <10) and resistant (reduced sensitivity for DRV; FC of ≥40) clinical isolates with closely related genotypes, differing in only a few PI-RAMs. The color coding of the genotype table is based on the IAS-USA 2004 guidelines (10): primary PI mutations are indicated in orange, and DRV-RAMs are indicated in purple.
Thus, 1,015 pairs of a PI-susceptible virus and a genetically closely related resistant virus were identified. Sixty-nine of these virus pairs were chosen to include the most important DRV-RAMs. The following PI-RAMs (based on the IAS-USA 2004 guidelines [10]) varied within the resulting 69 sets: L10F/I/V, K20I/R/T, V32I, L33F/I, M36I/L, M46I/L, I47A/V, G48V, I50V, F53L, I54L/M/V, A71T/V, G73C/S/T, L76V, V77I, V82A/F, I84V, N88D, and L90M (Table 1).
Production of recombinant viruses.
For each virus pair, a set of synthetic protease sequences carrying all combinations of PI-RAMs different between the resistant and susceptible viruses was designed. Since it was presumed that background plays an important role in retaining viability in the highly PI-resistant viruses, the background protease sequence from the resistant virus was retained to increase the probability that the viruses would still be able to replicate in vitro. Thus, the original susceptible viruses were used only to identify interesting PI-RAMs, but their background protease sequence was not included in the set of synthetic proteases. Rather, a sequence containing only the background of the resistant virus but lacking the studied PI-RAMs was included instead. The protease sequences were constructed using the gene assembly technology of Egea Biosciences (Centocor). All synthetic protease sequences were cloned in the same fragment containing WT Gag and reverse transcriptase sequences.
This exercise resulted in the design of 1,104 synthetic protease sequences that were used to generate recombinant viruses. Up to 88% of these protease sequences resulted in the production of viable virus, and 85% of the recombinant viruses had a virus titer sufficient to generate a measurable EGFP signal after 3 days of incubation in a cellular antiviral assay. The majority of the produced viruses contained the expected mutations after synthesis of the protease and virus generation. Signals of slower-growing but viable viruses (3.4%) were not measured. Generally, the most mutated viruses were the most difficult ones to generate and test for DRV susceptibility; hence, the FC values for some of the target viruses could not be established (Table 1). In some cases, removal of a mutation resulted in the loss of viral growth, but this was noticed more than once only for L33F and A71V. In a few cases, one of the intended mutations (in particular I84V) reverted or mutational mixtures appeared. Because of such artifacts, a few intermediate mutants were lacking.
Barring nonviable or slower-growing virus, the resulting set of viruses contained the PI-susceptible virus with the background sequence (source), the PI-resistant virus bearing the cluster of PI-RAMs (target), and all intermediate viruses resulting from all possible combinations of the PI-RAMs between source and target. Consequently, the cumulative effect of mutations on PI susceptibility could be studied starting from the source virus and ending with the target virus.
Impact of individual protease mutations on PI susceptibility.
By comparing the PI susceptibility of a recombinant virus containing a certain PI-RAM with the PI susceptibility of the virus that lacked this mutation (but was otherwise genetically identical), the impact of an individual mutation on susceptibility could be assessed in several contexts. It was found that among the total set of 1,104 recombinant viruses, mutations V32I, I50V, I54L, I54M, L76V, and V82F had the highest impact on HIV-1 susceptibility to DRV (Table 2), with a median change in susceptibility greater than threefold. Although the addition of a certain mutation had a rather consistent effect on DRV susceptibility, the variability of the impact (interquartile range) across sets points to the considerable influence of the complex PI-resistant background on this effect.
TABLE 2.
Impact of individual mutations on HIV-1 susceptibility to DRV in a background of multiple PI-RAMs, measured across the total set of 1,104 viruses
| Mutation | ΔFC DRVa |
Countb | No. of sets | |
|---|---|---|---|---|
| Median | Interquartile range | |||
| V32 → V32I | 5.06 | 3.25-7.80 | 109 | 20 |
| V82 → V82F | 4.91 | 3.91-5.99 | 16 | 1 |
| L76 → L76V | 4.27 | 2.57-8.05 | 49 | 11 |
| I54V → I54L | 4.17 | 2.76-5.11 | 3 | 1 |
| I50 → I50V | 3.16 | 2.12-5.48 | 21 | 3 |
| I54 → I54M | 3.11 | 1.97-5.26 | 31 | 7 |
| I54V → I54M | 3.11 | 2.82-3.59 | 6 | 1 |
| I54 → I54L | 2.97 | 2.11-4.20 | 51 | 10 |
| G73 → G73T | 2.36 | 1.37-2.93 | 6 | 2 |
| L33 → L33F | 2.33 | 2.04-3.29 | 26 | 4 |
| I84 → I84V | 2.18 | 1.20-2.85 | 76 | 12 |
| L10 → L10F | 1.95 | 1.21-2.95 | 15 | 1 |
| G73 → G73C | 1.82 | 1.40-2.39 | 10 | 2 |
| I47 → I47V | 1.74 | 1.26-3.67 | 76 | 13 |
| K20 → K20T | 1.72 | 1.19-2.01 | 31 | 4 |
| G73 → G73S | 1.67 | 1.02-2.33 | 50 | 8 |
| M36 → M36L | 1.65 | 1.36-2.44 | 18 | 3 |
| F53 → F53L | 1.51 | 0.99-2.02 | 80 | 8 |
| L90 → L90M | 1.48 | 1.11-2.10 | 157 | 25 |
| F53Y → F53L | 1.39 | 1.11-1.75 | 6 | 1 |
| M46 → M46I | 1.38 | 1.09-1.85 | 94 | 15 |
| G73S → G73T | 1.34 | 0.82-1.60 | 12 | 2 |
| L10 → L10V | 1.27 | 1.00-1.79 | 39 | 5 |
| K20 → K20R | 1.25 | 1.02-1.6 | 72 | 9 |
| M36 → M36I | 1.20 | 0.91-1.85 | 80 | 9 |
| M46 → M46L | 1.16 | 0.90-1.62 | 24 | 4 |
| A71 → A71V | 1.13 | 0.78-1.67 | 99 | 17 |
| L10 → L10I | 1.12 | 0.86-1.52 | 7 | 2 |
| G48 → G48V | 1.12 | 0.91-1.16 | 3 | 1 |
| K20R → K20I | 1.12 | 0.66-1.72 | 4 | 1 |
| V82I → V82A | 1.01 | 0.87-1.05 | 3 | 2 |
| A71 → A71T | 1.00 | 0.79-1.30 | 5 | 2 |
| V77 → V77I | 0.98 | 0.79-1.21 | 31 | 6 |
| N88 → N88D | 0.98 | 0.81-1.43 | 15 | 2 |
| L10F → L10I | 0.97 | 0.65-1.45 | 9 | 1 |
| K20M → K20R | 0.95 | - | 1 | 1 |
| I54 → I54V | 0.92 | 0.58-1.12 | 51 | 10 |
| V82 → V82A | 0.84 | 0.63-1.25 | 109 | 12 |
| L33 → L33I | 0.73 | 0.60-1.07 | 26 | 5 |
| K20V → K20I | 0.71 | 0.66-0.76 | 2 | 1 |
ΔFC, difference in FC caused by the presence of the indicated mutation (values of ≥3.0 are in bold).
Count, number of experiments in which the mutation was varied.
Mutual influence of PI-RAMs.
The influence of the background on the impact on PI susceptibility to a given PI-RAM was inferred by analyzing different sets of viruses. This concept is illustrated with data on the susceptibility to DRV of one set of source and target viruses, differing from each other in the presence of mutations M46I, I54M and L90M (set 29) (Fig. 2), along with its intermediates. The design of all intermediate genotypes between the source and target viruses resulted in a set containing eight viruses with a constant background of seven other PI-RAMs. The interplay between two individual mutations is represented by a tetragon. When the tetragon resembles a parallelogram, the mutations are not synergistic or antagonistic toward each other. In this case, tetragon sides that reflect addition of the same mutation have an identical ΔFC, with ΔFC being the change in FC caused by presence of a mutation. Distortion of the parallelogram indicates mutual influence of the mutations. Thus, it was seen that the effect of mutation M46I is independent of the presence of mutation I54M or L90M. However, I54M and L90M do influence each other's effect on DRV susceptibility, with I54M having a 2- to 2.5-fold-greater impact when introduced after L90M and vice versa, indicating synergy. In this set, I54M had a higher individual impact on DRV susceptibility than the M46I and L90M mutation, in accordance with the impact of individual mutations studied over all sets (Table 2).
FIG. 2.
Mutual influence of L90M, I54M, and M46I on DRV susceptibility (set 29). The source virus contained the PI-RAMs in the background shown in the table beneath the graph and no mutations at positions M46, I54, and L90. Compared to the source virus, the genotypes of the other viruses in the graph contained additional PI-RAMs as indicated. Color coding of the genotype table is based on the IAS-USA 2004 guidelines (10): primary PI mutations are indicated in orange, and DRV-RAMs are indicated in purple.
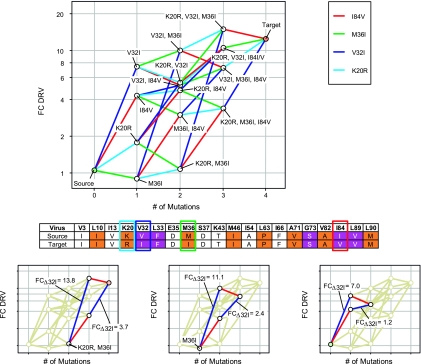
The complex interactions between different PI-RAMS were further explored in a set of viruses with variations of the PI-RAMs K20R, V32I, M36I, and I84V in a background of nine other PI-RAMs (set 65) (Fig. 3), resulting in 14 intermediate genotypes between the source and target viruses (Table 3). The influence of the K20R or the M36I mutation on the HIV-1 susceptibility to DRV was minimal (Fig. 3). The V32I mutation had the highest impact, as its presence caused a 2- to 10-fold decrease in susceptibility to DRV (Fig. 3). The high impact of the V32I mutation was consistent over the whole set, independent of the presence of K20R and M36I. The impact of I84V on HIV-1 susceptibility to DRV was smaller but still considerable. Interestingly, I84V had no additional effect when V32I was already present in a background of L10I, L33F, M46I, L63P, A71V, G73S, V82A, L89V, and L90M, whereas the individual impact of V32I was decreased when introduced after I84V, indicating antagonism (Fig. 3, bottom). As shown in Table 4, the individual impact of V32I was 4.7 times lower when I84V was present, irrespective of the presence of K20R or M36I as underscored by the small standard deviation (0.9) for this value.
FIG. 3.
Mutual influence of K20R, V32I, M36I, and I84V on DRV susceptibility (set 65). (Top) The source virus contained the PI-RAMs in the background shown in the table beneath the graph and no mutations at positions K20, V32, M36, and I84. Compared to the source virus, the genotypes of the other viruses in the graph contained additional PI-RAMs as indicated. (Bottom) Tetragons representing the interplay between the mutations V32I and I84V. Tetragons resembling a parallelogram indicate a lack of mutual influence; distorted tetragons point to synergistic or antagonistic influences between two mutations. Color coding of the genotype table is based on the IAS-USA 2004 guidelines (10): primary PI mutations are indicated in orange, and DRV-RAMs are indicated in purple.
TABLE 3.
Design of the intermediate genotypes between selected source and target viruses to study the impact of the K20R, V32I, M36I, and I84V mutations (set 65)
| Virus | Studied mutation(s) | Mutation in intermediate genotypea |
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| V3 | L10 | I13 | K20 | V32 | L33 | E35 | M36 | S37 | K43 | M46 | I54 | L63 | I66 | A71 | G73 | V82 | I84 | L89 | L90 | ||
| Source | None | I | I | V | F | D | D | T | I | A | P | F | V | S | A | V | M | ||||
| Virus 1 | V32I | I | I | V | I | F | D | D | T | I | A | P | F | V | S | A | V | M | |||
| Virus 2 | M36I | I | I | V | F | D | I | D | T | I | A | P | F | V | S | A | V | M | |||
| Virus 3 | I84V | I | I | V | F | D | D | T | I | A | P | F | V | S | A | V | V | M | |||
| Virus 4 | K20R | I | I | V | R | F | D | D | T | I | A | P | F | V | S | A | V | M | |||
| Virus 5 | V32I, M36I | I | I | V | I | F | D | I | D | T | I | A | P | F | V | S | A | V | M | ||
| Virus 6 | K20R, I84V | I | I | V | R | F | D | D | T | I | A | P | F | V | S | A | V | V | M | ||
| Virus 7 | K20R, M36I | I | I | V | R | F | D | I | D | T | I | A | P | F | V | S | A | V | M | ||
| Virus 8 | K20R, V32I | I | I | V | R | I | F | D | D | T | I | A | P | F | V | S | A | V | M | ||
| Virus 9 | V32I, I84V | I | I | V | I | F | D | D | T | I | A | P | F | V | S | A | V | V | M | ||
| Virus 10 | M36I, I84V | I | I | V | F | D | I | D | T | I | A | P | F | V | S | A | V | V | M | ||
| Virus 11 | V32I, M36I, I84V | I | I | V | I | F | D | I | D | T | I | A | P | F | V | S | A | V | V | M | |
| Virus 12 | K20R, V32I, I84I/V | I | I | V | R | I | F | D | D | T | I | A | P | F | V | S | A | I/Vb | V | M | |
| Virus 13 | K20R, V32I, M36I | I | I | V | R | I | F | D | I | D | T | I | A | P | F | V | S | A | V | M | |
| Virus 14 | K20R, M36I, I84V | I | I | V | R | F | D | I | D | T | I | A | P | F | V | S | A | V | V | M | |
| Target | K20R, V32I, M36I, I84V | I | I | V | R | I | F | D | I | D | T | I | A | P | F | V | S | A | V | V | M |
Based on the IAS-USA 2004 guidelines (10), primary PI mutations are indicated in bold and DRV-RAMs are indicated in bold with underlining.
Virus 12 contained a mixture of viruses bearing the I84V mutation and viruses with WT I84 and was therefore not used to assess the influence of I84V.
TABLE 4.
Mutual influence of I84V and V32I on DRV susceptibility (set 65)a
| Background | ΔFCV32I |
Change in ΔFCV32I | |
|---|---|---|---|
| −I84 | +I84V | ||
| +K20R, +M36I | 13.8 | 3.7 | 3.7 |
| −K20, +M36I | 11.1 | 2.4 | 4.6 |
| −K20, −M36 | 7.0 | 1.2 | 5.8 |
The FC caused by addition of V32I to the genotype (ΔFCV32I) was measured in the presence and absence of I84V, K20R, and/or M36I. K20R and M36I are indicated as genetic background because they did not influence the V32I FC value. The individual impact of V32I on DRV susceptibility was 4.7 ± 0.9 times lower when I84V was present.
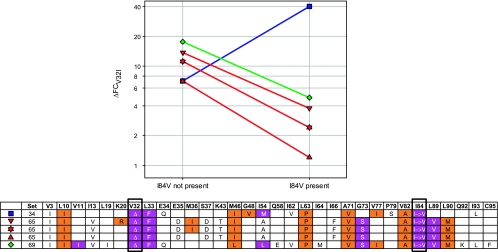
Influence of genetic background.
To evaluate the impact of genetic background, interactions between PI-RAMs were evaluated between sets. This is illustrated by studying the impact of background mutations on the influence of I84V on the FC in DRV susceptibility caused by V32I addition to the genotype (ΔFCV32I) (Fig. 4). ΔFCV32I values are shown in the absence and presence of I84V for all sets where the mutual influence of both mutations could be evaluated (i.e., sets 34, 65, and 69; set 68 was incomplete). Lines connect ΔFCV32I values for identical genotypes except for position 84. When these lines run parallel for two different sets, the genetic background does not influence the interaction between the mutations.
FIG. 4.
Effect of I84V on the change in DRV FC value caused by the addition of V32I to the genotype (ΔFCV32I) in different genetic backgrounds. The ΔFCV32I was measured in the presence and absence of I84V and other PI-RAMs as indicated. Color coding of the genotype table is based on the IAS-USA 2004 guidelines (10): primary PI mutations are indicated in orange, and DRV-RAMs are indicated in purple. Some combinations of mutations are not present in the graph, due to lacking intermediate viruses in the underlying sets.
As described above, the individual impact of V32I on DRV FC in set 65 was lower when I84V was present. This was also true for set 69. Moreover, in both sets the genetic background did not influence the interaction between I84V and V32I. Within set 65, the interaction was independent of the presence of K20R and/or M36I, and despite the even greater difference in genetic background between sets 65 and 69, no impact on this interaction was evident, as indicated by the parallel lines in Fig. 4.
In set 34 the relationship between I84V and V32I was completely opposite. Here, the individual impact of V32I was twofold higher when I84V was present.
This further illustrates how the specific genetic background greatly influences the impact of a certain mutation on the PI susceptibility of a virus, even changing an antagonistic relationship into a synergistic one.
DISCUSSION
The study of complex interactions between antiviral drug resistance-associated mutations is seriously hampered by the difficulty of discerning the influence of an isolated mutation in a cluster of mutations. Studies using genetic variants identified and catalogued in a database of clinical virus isolates are restricted by the content of the database, where individual mutations of interest are residing in a wide variety of genetic backgrounds. Traditional approaches that include deriving variants through serial site-directed mutagenesis based on isolated DNA from HIV strains of interest or creating desired variants via an entirely site-directed-mutagenesis-based method (from a single starting DNA), are limited by availability of variant DNAs or by the time and the effort required for the requisite serial rounds of mutagenesis.
The BIRD approach capitalizes on the advent of mature gene assembly technologies that allow the creation of large numbers of genetically close variants. Via a bioinformatics-based strategy with a database containing matched phenotypic and genotypic profiles of HIV-1 clinical isolates, 69 pairs of viruses with reduced in vitro susceptibility to DRV and the corresponding genetically closely related but DRV-susceptible viruses were identified. Next, a set of all viruses intermediate between the susceptible virus and the more resistant virus were created. By choosing the susceptible virus such that the PI-RAM difference with the resistant virus was as low as possible, the number of possible combinations of mutations and hence the number of set members was minimized. The 69 selected pairs of susceptible and resistant viruses gave rise to 1,104 possible intermediate synthetic genes and resulting viruses. Loss of data from several variant sets due to reduced viral viability, emergence of mixed bases at certain positions, or other artifacts could be tolerated because the number of viral sets generated was so large.
Pairs of source and target viruses were entirely based on patient data. By retaining the background sequence from the target in the synthesized protease variants, the variants have mutational patterns more similar to clinically relevant ones than could be achieved with site-directed mutagenesis-based strategies using WT HIV-1. Additionally, retaining the background kept its influence constant and consequently facilitated the analysis. As it was presumed that the genetic background plays an important role in retaining viability in the highly PI-resistant viruses, the background sequence from the resistant virus was chosen to be kept in each set.
Synthetic mutant protease sequences were recombined in a WT HIV-1 background. Therefore, the possible influence of mutations outside the protease (e.g., Gag) on reduced susceptibility was not assessed. In our set, there was a tendency toward a higher DRV susceptibility of the recombined resistant viruses compared with the original clinical isolates, but further research is needed to confirm possible influences of mutations outside protease.
The current study was not designed to investigate the impact of individual mutations on viral fitness, although viral fitness is an important factor in the success of antiviral therapy (13). The observation that removal of certain mutations (in particular L33F and A71V) resulted in the loss of viral growth in certain sets demonstrates that a BIRD approach would prove valuable in this respect. A clinical data set specifically focused on the relationship between genotype and viral fitness would be needed to devise such a study.
The ability to generate vast numbers of close variants that differ by one point mutation at a time enables a detailed dissection of the isolated influence of individual mutations on drug susceptibility. It also allows analysis of how the genetic background can influence this mutational effect. Considering the total set of recombinant viruses, V32I, I50V, I54L, I54M, L76V, and V82F generally had the highest impact on HIV-1 susceptibility to DRV. For all these mutations except V82F, this was in line with later observations in the clinic (4). However, the exact impact of individual mutations differed from set to set. In set 29, where three amino acid positions were varied, M46I had no impact on the susceptibility effect of I54M or L90M, while there was synergy between the latter mutations. In set 65, mutations at positions K20, V32, M36, and I84 were studied. Of particular interest was the interaction between I84V and V32I, with I84V having no additional effect when V32I was already present, whereas the individual impact of V32I was decreased when introduced after I84V. The same effect was seen in set 69, where the mutual influence of V32I, F53L, and I84V was studied. However, in set 34, varying V32I, G48V, I84V, and L90M, the relationship between I84V and V32I was reversed, and the individual impact of V32I was higher when I84V was present. Together, these findings illustrate how the effect of individual HIV-1 PI mutations on drug susceptibility is highly influenced by complex interactions with the genetic background. The findings also illustrate how difficult it is to draw a comprehensive overall picture without analyzing huge numbers of close variants in different genetic backgrounds. Our strategy makes such analysis feasible.
In the current study, mutational pathways were not studied in a traditional directional sense. Rather, susceptible and resistant viruses originated from different patients, and the intermediate viruses represented artificial steps between source and target viruses. Although it is not known if all intermediate viruses really exist in vivo, knowing the complex interplay between resistance mutations helps to select valuable candidates for drug development. In the future, BIRD analysis will be repeated with each pair of susceptible and resistant viruses originating from the same patient, allowing the study of real mutational pathways leading to resistance development. Information resulting from such an approach should allow for more accurate monitoring of resistance progression in patients and ultimately for a more directed follow-on drug discovery process.
When testing some first-generation PIs on the BIRD collection of viruses (data not shown), all these viruses proved to be resistant. For high-genetic-barrier drugs, several mutations are needed to confer resistance, and these mutations interact with each other. For lower-genetic-barrier drugs, fewer mutations are needed to confer resistance, and hence interactions between mutations may be less influential. Actually, to assess other drugs, specific panels of viruses for these drugs should be developed.
The BIRD approach could be used to examine complex interactions between individual mutations beyond mutations within the HIV-1 protease-coding sequence only. With a different setup, it should be possible to study the interplay between mutations in several HIV-1 genes. Likewise, the same approach could be used to study the isolated impact of mutations on phenotype, and the complex interplays between them, in any biological system.
Supplementary Material
Footnotes
Published ahead of print on 8 July 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Clavel, F., and A. J. Hance. 2004. HIV drug resistance. N. Engl. J. Med. 350:1023-1035. [DOI] [PubMed] [Google Scholar]
- 2.de Béthune, M.-P., and K. Hertogs. 2006. Screening and selecting for optimized antiretroviral drugs: rising to the challenge of drug resistance. Curr. Med. Res. Opin. 22:2603-2612. [DOI] [PubMed] [Google Scholar]
- 3.De Meyer, S., H. Azijn, D. L. N. G. Surleraux, D. Jochmans, A. Tahri, R. Pauwels, P. Wigerinck, and M.-P. de Béthune. 2005. TMC114, a novel human immunodeficiency virus type 1 protease inhibitor active against protease inhibitor-resistant viruses, including a broad range of clinical isolates. Antimicrob. Agents Chemother. 49:2314-2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Meyer, S., T. Vangeneugden, B. Van Baelen, E. De Paepe, H. Van Marck, G. Picchio, E. Lefebvre, and M.-P. de Béthune. 2008. Resistance profile of darunavir: combined 24-week results from the POWER trials. AIDS Res. Hum. Retroviruses 24:379-388. [DOI] [PubMed] [Google Scholar]
- 5.Evans, G. A. September 1998. Method for the complete chemical synthesis and assembly of genes and genomes. U.S. patent 6,521,427.
- 6.Evans, G. A. August 2001. Method for assembly of a polynucleotide encoding a target polypeptide. U.S. patent 6,670,127.
- 7.Hertogs, K., S. Bloor, S. D. Kemp, C. Van den Eynde, T. M. Alcorn, R. Pauwels, M. van Houtte, S. Staszewski, V. Miller, and B. A. Larder. 2000. Phenotypic and genotypic analysis of clinical HIV-1 isolates reveals extensive protease inhibitor cross-resistance: a survey of over 6000 samples. AIDS 14:1203-1210. [DOI] [PubMed] [Google Scholar]
- 8.Hertogs, K., M.-P. de Béthune, V. Miller, T. Ivens, P. Schel, A. Van Cauwenberge, C. Van den Eynde, V. van Gerwen, H. Azijn, M. van Houtte, F. Peeters, S. Staszewski, M. Conant, S. Bloor, S. Kemp, B. Larder, and R. Pauwels. 1998. A rapid method for simultaneous detection of phenotypic resistance to inhibitors of protease and reverse transcriptase in recombinant human immunodeficiency virus type 1 isolates from patients treated with antiretroviral drugs. Antimicrob. Agents Chemother. 42:269-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jochmans, D., J. Deval, B. Kesteleyn, H. Van Marck, E. Bettens, I. De Baere, P. Dehertogh, T. Ivens, M. Van Ginderen, B. Van Schoubroeck, M. Ehteshami, P. Wigerinck, M. Götte, and K. Hertogs. 2006. Indolopyridones inhibit human immunodeficiency virus reverse transcriptase with a novel mechanism of action. J. Virol. 80:12283-12292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson, V. A., F. Brun-Vezinet, B. Clotet, B. Conway, R. T. D'Aquila, L. M. Demeter, D. R. Kuritzkes, D. Pillay, J. M. Schapiro, A. Telenti, and D. D. Richman. 2004. Update of the drug resistance mutations in HIV-1: 2004. Top. HIV Med. 12:119-124. [PubMed] [Google Scholar]
- 11.Johnson, V. A., F. Brun-Vezinet, B. Clotet, B. Conway, D. R. Kuritzkes, D. Pillay, J. M. Schapiro, A. Telenti, and D. D. Richman. 2005. Update of the drug resistance mutations in HIV-1: fall 2005. Top. HIV Med. 13:125-131. [PubMed] [Google Scholar]
- 12.Maisnier-Patin, S., and D. I. Andersson. 2004. Adaptation to the deleterious effects of antimicrobial drug resistance mutations by compensatory evolution. Res. Microbiol. 155:360-369. [DOI] [PubMed] [Google Scholar]
- 13.Quinones-Mateu, M. E., D. M. Moore-Dudley, O. Jegede, J. Weber, and E. J. Arts. 2008. Viral drug resistance and fitness. Adv. Pharmacol. 56:257-296. [DOI] [PubMed] [Google Scholar]
- 14.Simon, V., D. D. Ho, and Q. Abdool Karim. 2006. HIV/AIDS epidemiology, pathogenesis, prevention, and treatment. Lancet 368:489-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Surleraux, D. L. N. G., A. Tahri, W. G. Verschueren, G. M. E. Pille, H. A. de Kock, T. H. M. Jonckers, A. Peeters, S. De Meyer, H. Azijn, R. Pauwels, M.-P. de Bethune, N. M. King, M. Prabu-Jeyabalan, C. A. Schiffer, and P. B. T. P. Wigerinck. 2005. Discovery and selection of TMC114, a next generation HIV-1 protease inhibitor. J. Med. Chem. 48:1813-1822. [DOI] [PubMed] [Google Scholar]
- 16.Svicher, V., F. Ceccherini-Silberstein, F. Erba, M. Santoro, C. Gori, M. C. Bellocchi, S. Giannella, M. P. Trotta, A. d. A. Monforte, A. Antinori, and C. F. Perno. 2005. Novel human immunodeficiency virus type 1 protease mutations potentially involved in resistance to protease inhibitors. Antimicrob. Agents Chemother. 49:2015-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wensing, A. M., and C. A. Boucher. 2003. Worldwide transmission of drug-resistant HIV. AIDS Rev. 5:140-155. [PubMed] [Google Scholar]
- 18.Wu, T. D., C. A. Schiffer, M. J. Gonzales, J. Taylor, R. Kantor, S. Chou, D. Israelski, A. R. Zolopa, W. J. Fessel, and R. W. Shafer. 2003. Mutation patterns and structural correlates in human immunodeficiency virus type 1 protease following different protease inhibitor treatments. J. Virol. 77:4836-4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.