Abstract
We report on a proteomic analysis of ex vivo human immunodeficiency virus (HIV) type 1 infection in human primary CD4 cells by shotgun liquid chromatography-tandem mass spectrometry analysis, revealing two distinct proteomic profiles at two phases of virus replication. Relative to mock-infected cells, 168 signature proteins exhibited abundance changes at the first sign of Gag p24 production (8 h postinfection [p.i.]) or the peak of virus replication (24 h p.i.); interestingly, most of the changes were exclusive to only one phase of virus replication. Based on characterization by functional ontology and known human-HIV protein interactions, we observed the enrichment for protein abundance increases pertaining to protein synthesis and nucleasomal reorganization amid an otherwise placid cellular proteome at the first sign of HIV replication. In contrast, we observed indications of decreased protein turnover, concomitant with heightened DNA repair activities and preludes to apoptosis, in the presence of robust virus replication. We also observed hints of disruptions in protein and small molecule trafficking. Our label-free proteomic strategy allowed us to perform multiplexed comparisons—we buttressed our detection specificity with the use of a reverse transcriptase inhibitor as a counterscreen, enabling highlighting of cellular protein abundance changes unique to robust virus replication as opposed to viral entry. In conjunction with complementary high-throughput screens for cellular partners of HIV, we put forth a model pinpointing specific rerouting of cellular biosynthetic, energetic, and trafficking pathways as HIV replication accelerates in human primary CD4 cells.
Human immunodeficiency virus type 1 (HIV-1) infections have been associated with functional defects in CD4 T cells, marked by unresponsiveness to T-cell signaling, as well as an increased propensity to cell activation and apoptosis (21). Processes culminating in CD4 T-cell compromise have been the subject of intense research. The viral envelope, Tat, Nef, and Vpr proteins have been shown to induce CD4 cell apoptosis and eventual death (4, 11, 36, 58). The virus life cycle also calls for numerous interfaces of host-virus interactions: at the point of entry, the viral envelope Env protein docks to the CD4 receptor and coreceptors on the target cells. Release of the viral nucleocapsid in the cytoplasm requires direct interactions with the host actin polymerization machinery (32, 55). Devoid of any replication enzymes, the virus depends on the host RNA polymerase II machinery for transcription from the HIV long terminal repeat (LTR). Integration of viral genetic materials into host chromosomes, a hallmark of the retroviral life cycle, calls for direct interactions of the host karyopherin machinery with the viral integrase, Vpr, Vif, and Rev proteins (12, 33, 35). Following productive transcription, viral gene products make use of the host ubiquitination system for virion assembly at and eventual egress from the plasma membrane (20, 30). While current anti-HIV-1 therapy employs cocktails of drugs that target viral proteins, cellular cofactors of infection present attractive targets for novel therapeutics, given the lower probability of spontaneous mutations.
To this end, recent discoveries of pro- and antiviral cellular factors by cDNA (39, 49, 50) and small interfering RNA (siRNA) screens (5, 28, 59) have uncovered novel and often seemingly unexpected cellular functions that affect HIV replication. Prominent cellular processes revealed include nuclear transport and DNA repair, likely to be associated with retroviral integration and trafficking of viral products across the nuclear envelope. In addition, the identification of proviral factors in the mitochondria along with cellular processes involved in energy production and mitochondrial integrity point to the siphoning of specific energy sources (e.g., NADH) away from the host by the replicating virus. The advent of microarray technologies has provided an alternative avenue for characterizing multifaceted host-HIV interactions. Earlier work done with HIV-infected CD4 T-cell lines has implicated cellular functions, including cholesterol biosynthesis, cell cycle arrest, apoptosis, and cell signaling, based on transcriptomic changes in the presence of HIV replication (8, 10, 19, 51, 54). Changes associated with apoptosis induction have recently been reported to occur early during ex vivo HIV infection of primary CD4 T cells (23), which is accompanied by notable induction of p53 transcription.
Recent proteomic characterizations of abundance changes in HIV-infected (6, 42) and -transfected (9) cells offer yet another way to characterize the interplay between HIV and the inopportune host; arguably, proteins are the effectors of most cellular functions and may not be governed by transcriptomic changes. In the first proteomic analyses of HIV-1 infection in the CEMx174 cell line, we reported on protein abundance changes that correlated with G1/G0 cell cycle arrest and alterations in Ran-mediated nuclear transport (6). A two-dimensional (2D) fluorescence difference gel electrophoresis analysis of HIV infection in the Jurkat cell line (42) also revealed changes in the protein abundance of host factors involved in nuclear transport, carbohydrate metabolism, and cell cycle progression in virus-producing CD4 cells. These analyses demonstrated the value of mass spectrometry (MS) as a mainstay for the characterization of complex proteomes. In particular, the widespread application of liquid chromatography (LC) and MS in complex proteome analyses (53) underscores the versatility of the approach, for both the quantification and the identification of proteins.
In this study, we profiled cellular proteome changes following ex vivo HIV-1 infection of primary CD4 cells isolated from five individual donors. To maximize the utilization of limiting analytes from primary CD4 cells, we adopted a label-free LC-MS approach (2, 14, 38, 48). We further enhanced the specificity of our findings with respect to productive HIV replication in a multiplexed comparison of protein abundance changes over a virus replication time course (i.e., 8 h postinfection [p.i.] versus 24 h p.i.), as well as in the presence and absence of reverse transcription (i.e., 24 h p.i. with or without efavirenz). In all, we report on the temporal effects of HIV replication on cellular protein profiles, demarcating two phases of HIV replication in primary human CD4 cells.
MATERIALS AND METHODS
Isolation and ex vivo infection of primary CD4 cells.
Peripheral blood mononuclear cells were isolated by Ficoll-Paque gradient separation (GE Lifesciences) from whole blood collected from six HIV-negative human donors; samples from five donors were used for LC-MS analysis, and the rest were used for Western blotting verification. Isolated peripheral blood mononuclear cells were negatively selected for with the CD4 Isolation Kit II in accordance with the manufacturer's protocol (Miltenyi Biotec). Isolated cells were stimulated with autologous serum for 24 h in the presence of 400 pg/ml recombinant interleukin-2 (IL-2; Calbiochem). Surface CD4 expression was verified in aliquots of isolated cells by flow cytometry on an FC500 flow cytometer (Beckman-Coulter) via fluorescein isothiocyanate (FITC)-conjugated anti-CD4 labeling (Miltenyi Biotec). HIV-1LAI (3, 52) virus inoculum was prepared as described previously (6). Enriched CD4+ CD3+ cells were exposed to virus at a multiplicity of infection of 2 50% tissue culture infective doses/cell for 2 h at 37°C, and excess virus was subsequently removed with phosphate-buffered saline as previously described (6). Mock-infected cells were treated in a like manner, with virus inoculums substituted for with medium collected from cells cultured in the absence of virus. All cells were then cultured in 12% fetal bovine serum- and recombinant-IL-2-supplemented RPMI 1640 medium for up to 28 h as described later.
Assessment of intracellular Gag p24 expression and cell viability by flow cytometry.
Primary CD4 cells were infected as described above and harvested at the times indicated following HIV and mock infections, with or without efavirenz treatment. To assess infectivity, 5 × 105 cells from each condition were fixed and stained with the Fix and Perm kit (Caltag) and anti-p24 KC57-FITC antibody (Beckman-Coulter) as described previously (19, 51). To assess cell viability, 5 × 105 cells were stained with annexin V-FLUOS and propidium iodide (PI) on ice for 15 min in accordance with the manufacturer's instructions (Roche Applied Sciences). Biparametric flow cytometric analysis was performed with intact cells gated for on the basis of forward and side scattering. Flow cytometric data analysis was carried out in WinMDI v2.9.
Dose response to a nonnucleoside reverse transcriptase inhibitor (NNRTI).
Efavirenz powder was dissolved in dimethyl sulfoxide and added to cultured primary CD4 cells following HIV-1 infection, achieving final concentrations ranging from 0 to 100 μM. Cells were monitored for cytotoxic effects by light microscopy based on trypan blue exclusion, by flow cytometry based on PI and annexin V incorporation, and for HIV-1 replication based on intracellular Gag p24 expression. Where indicated for the time course experiment, cells were cultured in the presence of efavirenz following the 2-h exposure to viral inoculums (i.e., h 0 of the time course).
Protein extraction and digestion.
HIV- and mock-infected cells were harvested at the times indicated and lysed as described previously (6). Protein concentrations were measured by bicinchoninic acid assay (Pierce). Proteins were denatured and digested with trypsin (1:50, wt/wt) as described previously (6).
LC-MS data acquisition, feature extraction, and alignment.
Tryptic peptides (250 ng in 20 μl 100 mM ammonium bicarbonate buffer) were loaded onto a BioBasic C18 Gold capillary column (2.1 mm [inside diameter] by 30 cm) with packed 1.9-μm particles (ThermoFisher Scientific). An 85-min high-performance LC gradient was run with 5% to 35% solvent B (0.1% formic acid in acetonitrile) against solvent A (0.1% formic acid in water) with a flow rate of 200 nl/min at the electrospray tip upon injection into an LTQ-Orbitrap (ThermoFisher Scientific). RAW files were imported into the Elucidator system v.3.2 (Rosetta Biosoftware) for alignment, feature extraction, and deconvolution as described previously (38).
Accurate mass and time (AMT) tag databases and VIPER analysis.
Alignment of Elucidator-extracted LC-MS features against the T-cell line CEMx174 AMT tag database (6) was done by using VIPER and following previously described procedures (34). The monoisotopic mass of peptide isotope groups from PeakTeller were matched to the monoisotopic mass of AMT tags. Proteins were annotated according to human IPI v.3.54.
Protein identification and differential analysis.
LC-MS features were first annotated on the basis of data-dependent analysis (DDA) by tandem MS (MS/MS) where possible. SEQUEST search results (SageN Research) were processed by PeptideTeller and ProteinTeller (37) within the Elucidator system; ProteinTeller results were used for annotation, with a predicted error rate of <3%. Remaining LC-MS features were annotated on the basis of alignment with AMT tag databases; matches with both SLiC and ΔSLiC (1) of ≥0.95 were selected for annotation as detailed in Results.
At a ProteinTeller-predicted error rate of <3%, 4,841 (5.7%) of the 84,913 extracted LC-MS features with charge states between 1 and 6 were annotated, corresponding to 4,213 peptides and 877 proteins. The 84,913 monoisotopic features were aligned against AMT tags from our previous study of HIV-infected CEMx174 cells (6) by using the VIPER software; at SLiC and ΔSLiC scores of ≥0.95, 6,773 (5.3%) features were annotated, corresponding to the annotation of 5,323 peptides and 2,680 proteins (Table 2; see Table S1 in the supplemental material). Of the 1,155 features annotated by both approaches, 952 (82.4%) were annotated with the same peptide sequences.
TABLE 2.
Cellular pathways enriched for differential expression at 8 h and 24 h p.i. reported by Ingenuity Pathway Analysisa
| Signature protein group and cellular pathway | P value | No. of proteins |
|---|---|---|
| Exclusive to 8 h p.i. | ||
| Glycerolipid metabolism | 0.045 | 1 |
| Bile acid biosynthesis | 0.045 | 1 |
| Exclusive to 24 h p.i. | ||
| 14-3-3-mediated signaling | 2.15 × 10−5 | 9 |
| IGF-1 signaling | 6.65 × 10−5 | 7 |
| G2/M DNA damage checkpoint regulation | 8.97 × 10−4 | 4 |
| Phosphatidylinositol 3-kinase/AKT signaling | 2.26 × 10−2 | 7 |
Cellular processes enriched for differential expression upon HIV-1 infection in primary CD4 cells. Ingenuity Pathway Analysis processes are deemed enriched for differential expression at 8 h and 24 h p.i. based on a t-test P value of <0.05.
Peptide intensities were used as proxies for label-free peptide abundance measurements (38). Ratios of peptide and protein abundance in infected samples, relative to time-matched, mock-infected samples, were computed on an error-weighted basis. Differential analysis by hierarchical clustering was performed within the Rosetta Elucidator system. Conversion from IPI index numbers to Human Genome Organization Gene Nomenclature Committee gene symbols and long descriptions was performed with ProteinCenter v2.0 (Proxeon Biosystems).
As an indicator of run-to-run variability, ion intensities of trypsin autolysis peptides served as an internal control, as trypsin was added to all protein extracts at a constant ratio of 1:50, and the same amount of total protein digests from every sample was loaded for LC-MS/MS analysis. Based on the error model built into the Rosetta Elucidator system, the unitless abundance value of a given protein was derived from the error-weighted summed abundance of individual peptides; likewise, peptide abundance was determined on the basis of the error-weighted summed abundance of the constituent feature ion intensities. Abundance values from each of the five biological replicates were collected individually from the 25 LC-MS/MS runs; the representative feature, peptide, and protein abundance values were, in turn, derived on the basis of an error-weighted average from the five biological replicates. For the differential analysis of any two conditions (e.g., mock infected versus HIV infected), the total error calculated by the error model was used to derive a P value. The P value corresponded to the probability of falsely rejecting the null hypothesis that there was, in fact, no change in abundance.
Western blotting validation.
Proteins were extracted from 106 primary CD4 cells with M-PER reagent (Pierce) and resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were probed for with primary antibodies against the cellular proteins BANF1, TRIM28 (Abcam), and Ran (Santa Cruz Biotechnology), and the viral Gag p24 antigen (16, 46), as well as horseradish peroxidase-conjugated secondary antibodies. To enrich HIV virions for Western blotting, cell culture medium supernatants were labeled with anti-CD26 antibody and subsequently enriched for with the μMACS streptavidin kit (Miltenyi Biotec); proteins bound on magnetic beads were dissolved with M-PER, resolved by 4 to 20% SDS-PAGE, transferred to polyvinylidene difluoride membranes, and visualized by chemiluminescence with ECL Plus (GE Lifesciences) on a Storm PhosphorImager (Molecular Dynamics).
RESULTS
HIV-1 infection kinetics and primary CD4 cell viability in the early and late phases of virus replication.
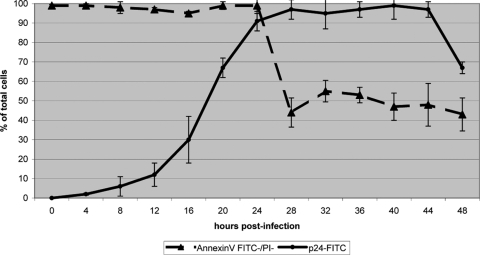
We based our proteomic analysis on the label-free quantification by MS (Fig. 1). To capture proteomic changes representative of virus replication in primary CD4 cells, cells were isolated from the blood of five healthy donors and subsequently cultured in autologous serum for 24 h, followed by infection with HIV-1LAI at 2 50% tissue culture infective doses/cell for 2 h. Cells (5 × 105) were harvested at the times indicated (Fig. 2), and virus replication was determined on the basis of intracellular staining of the HIV-1 Gag p24 antigen, while cell viability was determined on the basis of the staining of phosphatidylserine and genomic DNA by annexin V and PI, respectively. Intracellular Gag p24 was first detected at 8 h p.i. by flow cytometry, reaching >91% by 24 h p.i. (Fig. 2). As cell viability appeared to decrease rapidly at the time of robust virus production in our earlier cell line-based analysis (6), we opted to measure apoptosis by biparametric flow cytometry via annexin V-FITC and PI staining. Cell viability and integrity were maintained in almost all cells up to and including 24 h p.i. (Fig. 2; see Fig. S1 in the supplemental material). While virus replication continued to increase beyond 24 h p.i., 44% of the cultured cells were stained positively with annexin V at 28 h p.i.; this proportion of apoptotic cells continued to increase thereafter (Fig. 2). To capture events representative of the early and late phases of the initial HIV replication cycle in naïve primary CD4 cells, we sampled cells at 8 h and 24 h p.i. for our differential proteomic analyses in order to minimize confounding arising from compromised cell viability and integrity.
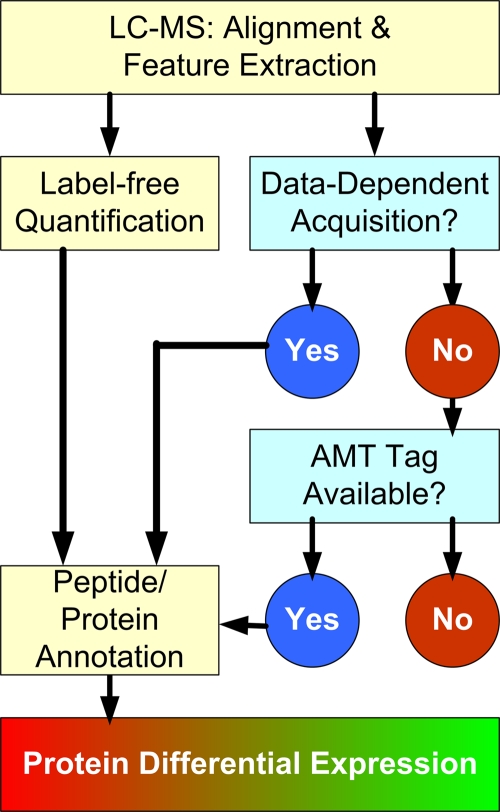
FIG. 1.
Workflow of label-free quantitative LC-MS proteomics with two-step peptide identification. Twenty five LC-MS LTQ-Orbitrap runs were acquired and processed by the Elucidator system. Following retention time alignment of spectral intensity maps and feature extraction, peptide and protein annotations were first performed on the basis of MS/MS DDA. Separately, features were also aligned to a previously published human AMT tag database for additional annotations. Subsequently, peptide and protein annotations were imported back into the Elucidator system to enable protein level analysis.
FIG. 2.
HIV-1 replication and cell viability. Flow cytometric measurements of virus replication based on the staining of intracellular Gag p24 with FITC-conjugated antibodies and cell viability based on negative staining with both annexin V-FITC and PI as functions of time. Error bars represent 1 standard deviation based on five measurements, corresponding to the five individual donors.
Effects of efavirenz on HIV-1 infection.
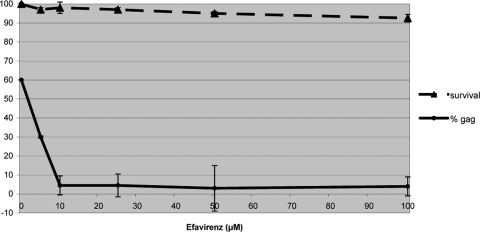
To help determine if the observed protein abundance changes were specific to HIV-1 replication, we posited that treatment with the NNRTI efavirenz would block reverse transcriptase activity and thereby ablate proteomic changes associated with robust HIV-1 replication following successful reverse transcription. To establish the optimal dosage, cells were subjected to up to100 μM efavirenz, corresponding to typical concentrations reported in the blood of AIDS patients undergoing antiretroviral therapies (22, 57) (Fig. 3). Reduction of intracellular HIV Gag p24 detection was noticeable with as little as 10 μM efavirenz. At the same time, cell survival, as assessed by the incorporation of annexin V and PI, showed little effect upon exposure to efavirenz, even at 100 μM. Consequently, we selected the minimum effective dosage of 10 μM for all subsequent analyses.
FIG. 3.
Effects of efavirenz. Dose response of HIV-infected cells to efavirenz at 24 h p.i. Percentages of cells expressing intense Gag p24-FITC staining and of cells negatively staining for both annexin V and PI are plotted as functions of increasing concentrations of efavirenz. Error bars represent 1 standard deviation based on three technical replicate measurements.
Identification of proteins by DDA and AMT tag annotations: a comparison.
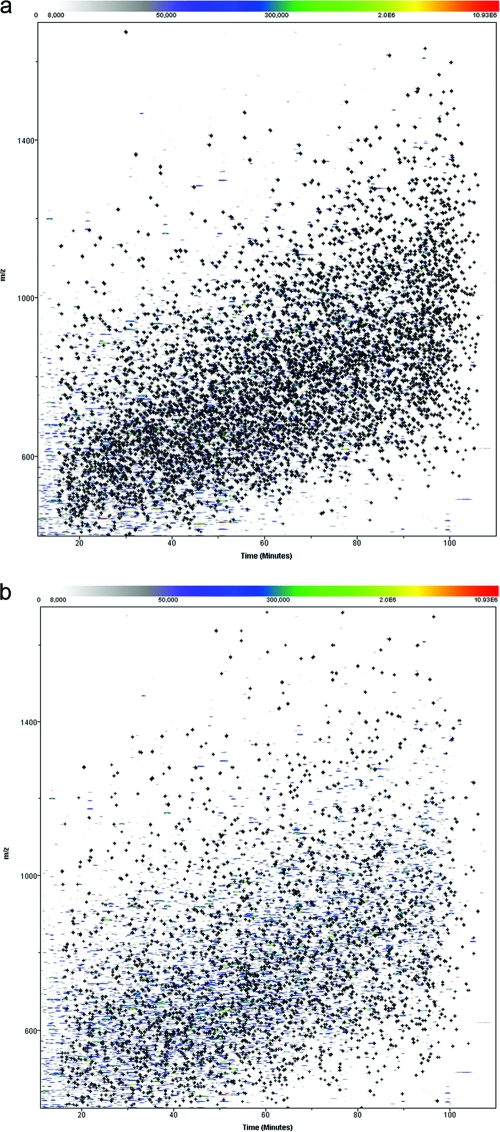
LC-MS features were extracted following alignment of 25 LC-MS/MS runs and charge state deconvolution with the PeakTeller algorithm (38) (Fig. 4). We followed a hybrid approach for the peptide and protein annotation of monoisotopic LC-MS features (Fig. 1): first relying on ProteinTeller-processed SEQUEST results from DDA generated in our present study and then based on matches to our existing human lymphocyte AMT tags (6) accumulated from our past studies; the AMT tag approach takes advantage of the accurate mass and LC retention time measurements of individual peptides as unique identifying features for future references. In all, 952 out of 1,155 features were attributed to the same 808 peptides by both the DDA and AMT tag approaches, corresponding to 82.4% agreement (Table 1; see Table S1 in the supplemental material). The annotated features were distributed throughout, relative to mass and LC elution time (Fig. 5a and b), showing little bias in annotation by either approach.
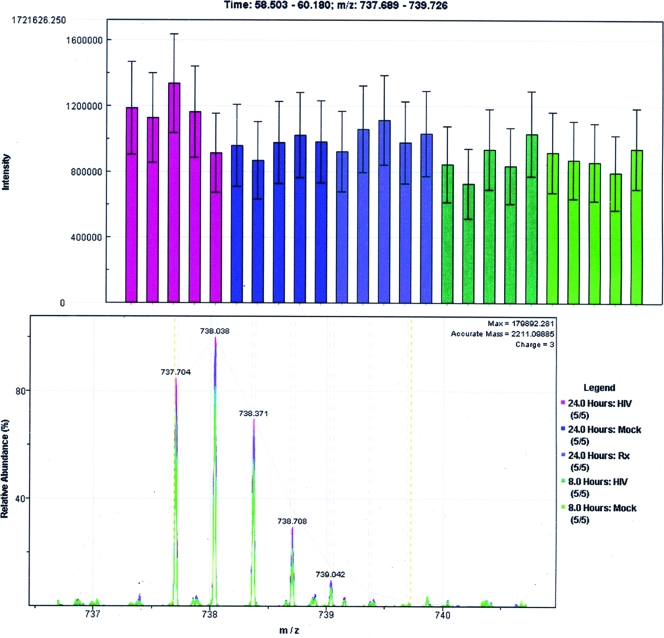
FIG. 4.
Quantification of trypsin autolysis product. Isotope group ion intensities of the trypsin autolysis product LGEHNIDVLEGNEQFINAAK are presented. (Top) Histogram of corresponding ion currents from the 25 LC-MS/MS runs; error bars correspond to 1 standard deviation. Although pairwise t tests were performed, with the average value from the 24 h p.i. mock-infected samples serving as a common reference, no significant difference (P < 0.05) was detected among the 25 runs. (Bottom) Overlaid mass spectra for the same peptide, colored based on treatment conditions as shown at the bottom right, showing all peaks in the isotope group.
TABLE 1.
LC-MS features annotated by DDA and AMT tag approaches
| Parameter | No. of features | No. of peptides | No. of proteins |
|---|---|---|---|
| Monoisotopic features annotated by DDA and/or AMT tags | |||
| DDA | 4,841 | 4,213 | 877 |
| AMT | 6,773 | 5,323 | 2,680 |
| Agreementa | 952 | 808 | 418 |
| LC-MS features annotated by AMT tags alone | |||
| AMT (no DDA available) | 5,618 | 4,714 | 2,514 |
| High confidenceb | 2,220 | 2,220 | 495 |
1,155 monoisotopic features were annotated by both DDA and AMT tags; 952 of the 1,155 received identical peptide annotations via both methods.
AMT tag annotations were deemed of high confidence if the corresponding proteins were at least identified by DDA (ProteinTeller-predicted error of <3%) or represented by a minimum of three unique peptides.
FIG. 5.
Peptide and protein annotation by DDA and AMT tags. Distribution of 84,913 features detected by PeakTeller is shown as m/z versus retention time. Marked by the crosses are (a) 4,841 and (b) 6,773 features annotated by DDA and AMT tag matching, respectively.
For the remainder of our analyses, we prioritized our annotation strategies as follows (Fig. 1): (i) peptide annotations were first accepted on the basis of a ProteinTeller-predicted error rate of <3%; (ii) for the 5,618 features annotated solely by AMT tags and therefore lacking ProteinTeller estimates (Table 1), annotations were accepted only if the presence of the corresponding protein identification was also supported by DDA (at a ProteinTeller error rate of <3%) or were supported by at least three unique peptide sequences. In total, 2,220 monoisotopic features (39.5%) satisfied at least one of these requirements. Adhering to this hybrid annotation strategy, a total of 7,061 features were annotated; 1,146 cellular proteins and 5,996 peptides were identified, along with HIV Gag-Pol and trypsin (Table 1; see Tables S1 and S2 in the supplemental material).
Label-free quantification of protein abundance changes in early and late phases of HIV replication.
In all, 178 cellular proteins of the 1,146 identified exhibited differential expression, with a threshold set at a ≥1.50-fold change relative to time-matched, mock-infected cells; we also chose a P value of <0.01, which corresponded to the likelihood of false discovery of differential expression. In contrast, trypsin autolysis products that were added to all samples in equimolar amounts did not meet the threshold for differential expression as defined above (Fig. 4). Three major patterns of differential abundance emerged: 114 and 24 proteins exhibited abundance increases and decreases exclusively at 24 h p.i., respectively; 24 proteins exhibited abundance increases exclusively at 8 h p.i., while another 1 exhibited decreased abundance (Fig. 6a; see Table S2 in the supplemental material). The label-free workflow also allowed us to perform multiplexed comparisons. To this end, four proteins (CHCHD3, RMB17, glucose-6-phosphate dehydrogenase [G6PD], and TRIM28) exhibited increases at 8 h p.i. and in the presence of efavirenz at 24 h p.i.; ribophorin (RPN1), an endoplasmic reticulum (ER) marker, decreased in abundance at both time points. An additional 10 of the 178 cellular proteins also exhibited differential expression at 24 h p.i.; however, we could not attest to their specificity to HIV replication, as the administration of efavirenz showed no ablation of the abundance changes at 24 h p.i. In all, 168 proteins showed differential abundance over the course of HIV infection in primary CD4 cells; notably, we observed a strong temporal effect as 162 proteins showed exclusivity in differential abundance at a single time point.
FIG. 6.
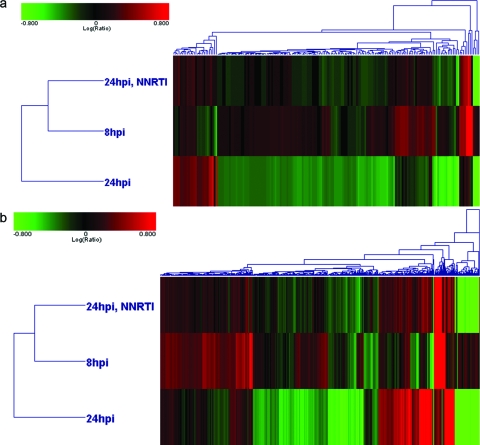
Shotgun analysis of differential expression regardless of protein identification availability. Hierarchical clustering of (a) protein and (b) peptide isotope groups, with red and green corresponding to increased and decreased abundance, respectively, relative to that of time-matched, mock-infected primary CD4 cells. Features exhibiting differential expression (≥1.50-fold change, P < 0.01) were included in the analysis. The diagrams are visual representations of agglomerative clusters based on Euclidean distance correlation of protein and isotope group ratios.
To determine if annotation by DDA might have been biased toward the labeling of high-abundance proteins, we also assessed the expression patterns of isotope group ratios independent of the availability of peptide and protein annotations (Fig. 6b). In accord with our observations made with annotated proteins (Fig. 6a), 2D hierarchical clustering showed that the expression pattern of isotope group abundance at 8 h p.i. more closely resembled that of efavirenz-treated cells than that of untreated cells at 24 h p.i.; this pattern was maintained irrespective of annotation status. By virtue of being an inhibitor of reverse transcription, efavirenz had, in effect, held in check many of the abundance changes otherwise observed as HIV replication progressed from 8 h to 24 h p.i.
Functional analysis of signature proteins—impact on cellular functions and host-virus interactions.
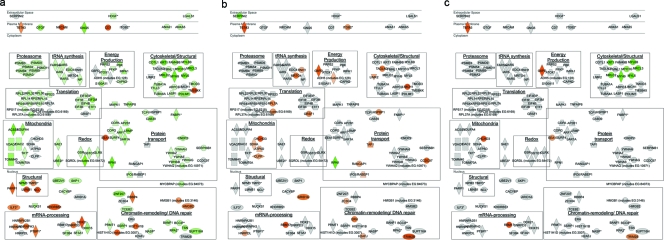
Proteins exhibiting abundance changes were characterized on the basis of known cellular functions and interactions with HIV proteins. While abundance changes of constituent proteins might affect the activities of any corresponding cellular processes, given the inherent redundancies in the eukaryotic machinery, the enrichment for abundance changes in selected cellular pathways would reflect a higher likelihood of perturbation in those processes. To this end, interrogation of our list of 168 signature proteins was performed against the Ingenuity Pathway Analysis knowledgebase (Table 2). At 8 h p.i., we observed an enrichment for abundance changes impacting key energetic processes, as exemplified by the upregulation of alcohol dehydrogenase (ADH5) involved in fatty acid oxidation, as well as by increased flux through glycolysis aided by the increased abundance G6PD (Table 2). In contrast, abundance changes at 24 h p.i. were enriched in mitogen-activated protein kinase signaling and the generation of cell proliferative signals. The addition of efavirenz blocked all of the abundance decreases in these cell survival signaling pathways, which also showed no change at 8 h p.i.; these observations coincided with our flow cytometric results, showing an abrupt increase in annexin V staining between 24 h and 28 h p.i. (Fig. 2; see Fig. S1 in the supplemental material). Characterization of protein abundance changes based on subcellular localization also revealed distinct changes at 24 h p.i., relative to both 8 h p.i. and the presence of efavirenz at 24 h p.i. (see Fig. 8a to c).
FIG. 8.
Subcellular and functional distributions of signature proteins. Shown are signature proteins (n = 168) on the basis of subcellular distribution as registered in the Ingenuity Pathway Analysis database. Individual proteins are annotated with Human Genome Organization Gene Nomenclature Committee gene symbols; the red and green shading corresponds to up- and downregulation, respectively. Gray indicates no differential regulation detected. Shown here are differential expression results from (a) 24 h p.i., (b) 8 h p.i., and (c) 24 h p.i. in the presence of 10 μM efavirenz. Proteins corresponding to similar functions are grouped in boxes as labeled; functions are deduced from Ingenuity Pathway Analysis and Gene Ontology annotations, as well as a literature review.
We also interrogated our lists of signature proteins for known interaction partners of HIV proteins (Table 3). We reasoned that abundance changes of these interaction partners would reveal potential impacts on different stages of the viral life cycle, spanning from exposure to the viral envelope, through transcriptional induction of and by Tat, all the way to the production of late viral gene products. Of the 1,433 known HIV-human interactions registered in the NIAID database (17), 37 were present among our list of signature proteins. At 8 h p.i., of the three signature proteins with known interactions with HIV proteins, all had been reported to interact with Tat; moreover, the integrin protein ITGB2 has also been shown to bind to the viral Env gp120 protein as part of the uncoating process. All of these proteins exhibited increases in abundance relative to those in mock-infected cells. In contrast, most of the 34 known interaction partners at 24 h p.i. exhibited decreases in abundance; additionally, both early and late (i.e., post Tat transactivation) HIV products were represented. Notably, we observed the downregulation of six tyrosine/tryptophan monooxygenase proteins and four proteasomal subunits at 24 h p.i. The decreased abundance of constituents from complexes involved in oxidation-reduction and protein degradation might themselves reflect disruptions in those processes, as well as the activities imparted by their known interactions with viral proteins.
TABLE 3.
Differentially expressed proteins with known interactions with HIV proteinsa
| Signature protein group and HIV-1 protein(s) | Cellular protein(s) | Direction of abundance change relative to mock infection |
|---|---|---|
| Exclusive to 8 h p.i. | ||
| Tat | H2AFJ | ↑ |
| Tat | TAP1 | ↑ |
| Tat, Env | ITGB2 | ↑ |
| Exclusive to 24 h p.i. | ||
| Tat, Nef, Env, Gag | AP2B1 | ↓ |
| Nef | ARF1 | ↓ |
| Gag, integrase, matrix | BANF1 | ↓ |
| Env | CAPN2 | ↓ |
| Tat | HIST1H3F | ↓ |
| Rev | ILF3 | ↑ |
| Tat, Rev, Vpr, integrase, matrix | IPO5 | ↓ |
| Rev | KHDRBS1 | ↑ |
| Env | LGALS1 | ↓ |
| Tat, Vpr | LMNB1 | ↑ |
| Env, Rev, Nef, Tat, Vpr, Vif, matrix, Gag, reverse transcriptase | MAPK1 | ↓ |
| Env, Nef, Tat, Vpr, matrix, integrase | PARP1 | ↑ |
| Rev, Tat | PTBP1 | ↑ |
| Vpr, reverse transcriptase | RPA2 | ↓ |
| Vpr | SF3B4 | ↓ |
| Tat | SNRPD3 | ↓ |
| Tat, Vif | TCEB2 | ↓ |
| Env, Rev, Tat | TUBA1A, TUBA4A | ↓ |
| p6 | UBE2I | ↓ |
| Vpr | VDAC1 | ↓ |
| Env, Vpr | YWHAB, YWHAE, YWHAG, YWHAH, YWHAQ, YWHAZ | ↓ |
| Tat, Vif, integrase | PSMA3, PSMA4, PSMB3, PSMB9, PSMD9 | ↓ |
Signature proteins exclusive to 8 h p.i. and 24 h p.i. with known interactions with HIV-1 proteins (17).
Western blotting for validation of selected signature proteins.
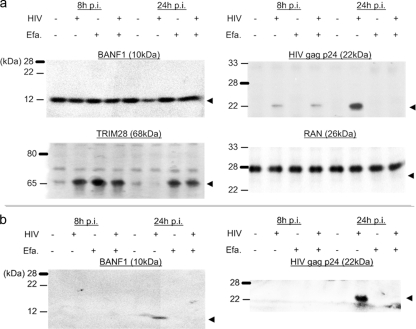
We selectively verified our quantitative LC-MS results via orthogonal validation by Western blotting. We probed for the protein abundance of the RNA-binding protein TRIM28, which exhibited an almost threefold increase at 8 h p.i. on the basis of our LC-MS analysis (see Table S2 in the supplemental material). With primary CD4 cells from a sixth donor not used for the LC-MS/MS analyses described forthwith, our Western blotting results (Fig. 7) were in line with our MS analysis. As an example of signatures unique to 24 h p.i., we blotted for the abundance of BANF1, a cellular protein known to be associated with HIV virions (7, 18, 43). We speculated that robust virus production would have exported these virion-associated proteins out of the infected cells, thereby appearing as downregulated protein abundance measurements on the basis of our approach. As confirmed by Western blotting, the abundance of BANF1 was lower at 24 h p.i. in infected cells without efavirenz treatment (Fig. 7a); the highly abundant Ran GTPase showed no change in abundance and served as a loading control. On the other hand, our blots showed that BANF1 was present in anti-CD26-enriched virion lysates from infected cell culture medium (Fig. 7b). Our observations with BANF1 served as a reminder that our LC-MS/MS analysis reflected solely the intracellular abundance of proteins, which might not necessarily reflect gross protein abundance changes stemming from the dynamics of protein synthesis and/or steady-state maintenance of proteins; robust virion production at 24 h p.i. might have further exacerbated this phenomenon.
FIG. 7.
Western blot validation of protein abundance. Protein extracts from (a) cell and (b) anti-CD26-enriched virion lysates collected under the conditions indicated were resolved by SDS-4 to 20% PAGE. Western blotting was performed with antiserum against the cellular proteins TRIM28 and BANF1, as well as against the viral Gag p24 antigen. Abundance of the Ran GTPase was probed for as a loading control. Efa., treatment with 10 μM efavirenz.
DISCUSSION
In this report, we profiled cellular proteome changes following ex vivo HIV-1 infection of primary CD4 cells isolated from five individual donors. To maximize the utilization of analytes for our profiling efforts, we adopted a label-free LC-MS approach (38); by eliminating the need for performing pairwise comparisons, as would otherwise be required of stable-isotope labeling and the 2D fluorescence difference gel electrophoresis approaches, we gained the ability to perform one-to-multiple comparisons. To help ascertain the specificity of our present findings with respect to productive HIV replication, we report on a multiplexed comparison of protein abundance changes between the start and peak of virus replication (i.e., 8 h p.i. versus 24 h p.i.); we also compared the cellular proteomic profiles in the presence and absence of reverse transcription (i.e., 24 h p.i. with or without efavirenz). We reasoned that efavirenz, an NNRTI common to anti-HIV “cocktails” (22, 57), could serve as a counterscreen for changes attributable to productive viral replication at 24 h p.i. In all, the profiles of protein abundance changes pointed to distinct impact on selected cellular functions as virus replication progressed from 8 h to 24 h p.i. Notably, early infection was accompanied by the induction of biosynthetic processes, in contrast to the disruption of protein synthesis and degradation machineries when virus production peaked at 24 h p.i. From a technical perspective, our present study benefited from the versatility of the label-free, quantitative LC-MS/MS approach. Furthermore, the temporal effects of HIV replication on cellular protein profiles echoed the observations made in earlier high-throughput studies, attesting to the complementarity of these screening approaches in our efforts to decipher complex virus-host relationships.
We have presented the first proteomic analysis of ex vivo HIV-1 infection in human primary CD4 cells, revealing 168 cellular proteins that changed in abundance relative to that in mock-infected human primary CD4 cells. Most of the abundance changes occurred at 24 h p.i., the time point at which intracellular virus production reached its maximum before apoptosis became noticeable by annexin V staining and flow cytometry (Fig. 2). Furthermore, 164 of the 168 signature proteins were exclusive to one of the two sampled time points, strongly suggesting that the intracellular proteome was highly dynamic as viral replication progressed. Our label-free LC-MS approach allowed us to use efavirenz as a counterscreen in a multiplexed comparison manner, otherwise not practical given the limited amount of primary cells we had on hand. We attempted to identify abundance changes specific to late, post-reverse transcription events in the viral life cycle; interestingly, treatment with 10 μM efavirenz resulted in the ablation of abundance changes in a majority of the signature proteins exclusive to 24 h p.i. The proteomic profile at 8 h p.i. was more similar to that of efavirenz-treated, HIV-1-infected cells at 24 h p.i. than to that of cells not treated with the reverse transcriptase inhibitor (Fig. 6). We speculate that the fruitful completion of viral gene expression and subsequent production of viral gene products, otherwise not available at 8 h p.i. or with reverse transcriptase activities inhibited, were contributors to the strong temporal effect we observed.
Induction of protein synthesis and energy production upon viral entry; the dynamics of subcellular structural integrity.
At the first sign of viral production at 8 h p.i., 24 of the 26 signature proteins increased in abundance relative to that in mock-infected primary CD4 cells (Fig. 8b; see Table S2 in the supplemental material). We detected the increase in the abundance of selected tRNA synthetase and ribosomal proteins, indicative of an overall induction of protein synthesis in infected cells. This could promote the translation of cellular proteins, which would be a logical progression from increased transcription permitted by the opening up of host chromosomes in the increased presence of histones and ribonucleoproteins; increased protein synthesis would, in turn, be matched by the increased abundance of the chaperone proteins CLP and calponin for proper protein folding (Fig. 8b). Coincidentally, there also appeared to be an increase in ATP production and flux through the glycolysis pathway, as reflected by the increased abundance of alcohol dehydrogenase and G6PD in the cytoplasm; increased abundance of ATP synthase components in the mitochondria rendered further support to the notion that energy production was needed to sustain increased biosynthesis. The increased abundance of selected cytoskeletal proteins such as transgelin and actin-associated factors could be attributed to the surge in translation activities.
It bears noting that our LC-MS analysis was performed with equal amounts of total proteins from individual conditions; as such, our analysis was not designed to detect global shifts in cellular or total protein abundance. In fact, increased transcriptional and translational activities could have impacted cellular and viral proteins alike: we observed the upregulation of GRSF1, an mRNA-binding protein that has recently been shown to aid in the nuclear localization of spliced HIV transcripts (24); the increased availability could also conceivably promote the translation of selected cellular gene products (27). The stability of viral transcripts could also benefit from the increased availability of ribonucleoproteins mentioned above. Furthermore, the increases in abundance of cytoskeletal proteins, including integrin and transgelin, were in line with their known roles in the uncoating of the internalized viral nucleocapsid (29, 44) and thus would facilitate the progression of the virus replication cycle. The promotion of cellular and viral protein biosynthesis need not be mutually exclusively; in fact, an overall state of cell proliferation as would be promoted by the decrease in the proapoptotic signal of Serpini 2—one of two cellular proteins with significant downregulation at 8 h p.i.—could very well promote the initiation of the viral gene expression program and the survival of the host cells. The decreased abundance of the ER marker RPN1 could be indicative of ER stress, as part of the toll exacted on CD4 cells that were otherwise exhibiting signs of a surge in protein biosynthesis.
Robust virus production coincided with compromised cellular integrity, energy deprivation, and tempering of overall biosynthesis.
In contrast to 8 h p.i., robust virus production at 24 h p.i. was marked by notable downregulation of cellular protein abundance in biosynthetic pathways. We observed decreases in the abundance of ribosomal proteins, as well as translation initiation factors and tRNA synthetases, strongly indicating an overall hampering of intracellular protein synthesis (Fig. 8a). Also in contrast to the earlier time point, we observed a concomitant decrease in the abundance of cytoskeletal components, including actin- and tubulin-associated factors, indicative of compromised cellular integrity. Furthermore, the decreased abundance of such structural components could be a reflection of the accumulation of cell fusion and cytopathic effects, given the ability of HIV-1LAI to form syncytia in peripheral blood T cells (31, 45, 47). Interestingly, this pathway has also been ascribed a possible role in macrophage cytopathology in a surface-enhanced laser desorption ionization chip-based proteomic analysis, with monocyte-derived macrophages showing increased secretion of actin and profilin following HIV-1ADA infection along with reorganization of cytoskeletal elements within the macrophages (26). Notwithstanding differences specific to T cells and macrophages in terms of virus dissemination, our findings have hinted at potential contributions of the cytoskeletal machinery to robust virus replication.
The decrease in protein synthesis we observed at 24 h p.i. also coincided with a decrease in the abundance of 26S proteasome components. We speculate that the decrease in protein abundance was more likely attributable to the tempering of biosynthetic pathways rather than to protein degradation. Functional analysis based on the Ingenuity knowledgebase revealed an enrichment of abundance changes in antiapoptotic 14-3-3 signaling proteins (Table 2). Further indications of apoptosis could be deduced from the increased abundance of poly(ADP)-ribose polymerase, a common marker of early stages of apoptosis, in the nucleus. On the other hand, signs of apoptosis induction were contrasted by the prosurvival signals, as could be seen in the decreased abundance of anion channels in the mitochondria, VDAC1 and VDAC2, as well as the concerted increases in abundance of high-mobility group proteins. Our observations were the direct opposite of those reported by Ringrose et al. for infected Jurkat cells (42); nonetheless, as seen in the Jurkat cell model, an “apoptosis tug-of-war” appeared to be under way. Since our flow cytometric analyses revealed a marked surge in annexin V-staining cells between 24 h and 28 h p.i. (Fig. 2; see Fig. S1 in the supplemental material), HIV-infected cells could conceivably gradually shift from a proliferative state, as observed earlier at 8 h p.i., to one conducive to cell death. In spite of the absence of apoptosis detectable by flow cytometry at 24 h p.i., signs of exhaustion could be gleaned from compromised mitochondrial structural integrity—decreased abundance of components of the inner and outer mitochondrial membranes (e.g., TOMM4 and TOMM70A), along with glutathione metabolic enzymes (e.g., GSR and GSTO1) (Fig. 8a), would likely deprive the infected cells of energy needed for biosynthesis. In particular, the decreased abundance of GSR would, in turn, reduce the availability of glutathione, which would otherwise suppress late stages of HIV replication and virion production (40).
Robust virus production was accompanied by induction of DNA repair and nucleasomal modification machineries.
The decreases in protein abundance in cell proliferation/survival and energetic pathways at 24 h p.i. were contrasted by concomitant increases in the abundance of cellular factors responsible for DNA repair and nucleasome assembly (Fig. 8a). The increases in the abundance of histone 1 components, hnRNPs, as well as mRNA-splicing factors, were in accord with the induction of DNA repair and VDJ recombination (15); such a response would be in line with DNA repair of chromosomal DNA following integration of the viral DNA. Additional signs of dynamic changes in the nucleus could be deduced from the increased abundance of thymopoietin, a nuclear lamina-associated protein that has been shown to induce and suppress transcriptional activation from the HIV LTR (25). On the other hand, increased abundance of thymopoietin would also be in line with its role in nuclear envelope repair, presumably in response to the nuclear herniation caused by HIV Vpr (13). In this regard, the maintenance of an intact nucleus would be in agreement with the signs of heightened transcriptional activities observed at 24 h p.i. based on the predominance of protein abundance increases in the nucleus and the fact that the HIV LTR is a potent regulator of transcription. Furthermore, in spite of possible confounding from dying cells, our flow cytometric analyses indicated that robust virus replication continued well beyond 24 h p.i. (Fig. 2)—such production could only be sustained by a constant supply of viral gene products. Nevertheless, normal cellular activities were undeniably disrupted at 24 h p.i. The present study and the two cell line-based studies (6, 42) all showed signs of cell cycle arrest; such disruptions could be related to the decrease in the abundance of CDK and cyclin-related proteins observed in all three studies. In addition, similar to the CEMx174 cell line study, we observed the downregulation of importin 5. Downregulation of this nuclear importer has been shown to impede cell proliferation (41), presumably because of perturbations in the trafficking of cell cycle regulators across the nuclear envelope via the RNA-dependent nuclear transport pathway.
Selected signature proteins had known interactions with early and late HIV proteins.
Interrogation of our lists of signature proteins against the NIAID database of human-HIV protein interactions also revealed a temporal effect: several signature proteins at 8 h p.i. (Table 3) had known interactions with Tat, an early viral gene product, as well as with Env, presumably from incoming viral particles. In contrast, signature proteins at 24 h p.i. had more known interactions with a wider range of viral proteins. Notwithstanding the fact HIV-1 is a relatively “young” virus, we speculate that retroviruses have likely evolved to make use of different cellular factors as their abundance levels alter over the various stages of the viral life cycle—the two distinct proteomic profiles at 8 h and 24 h p.i. we obtained attested to the dynamics of cellular protein abundance levels over less than one virus replication cycle. Abundance increases in the three cellular proteins with known interactions with Tat at 8 h p.i. (Table 3) would serve to promote the nuclear localization and activation of Tat (29, 44), which in turn marks the start of the full viral gene expression program. Our observation that the RNA-binding protein TRIM28 increased in abundance at 8 h p.i., as well as in efavirenz-treated cells at 24 h p.i., is also in line with such a view. Though not registered in the human-HIV interaction database, TRIM28 has been attributed to the stabilization of the tRNA primer during the retroviral life cycle (56); it is conceivable that similar mechanisms for promoting virus production would also apply to HIV-infected CD4 cells.
Perturbations of nuclear trafficking, DNA repair, and cell survival were also noted in complementary high-throughput screens.
The temporal effects on the host cellular protein profile were in accord with the transcriptomic profile reported recently by Imbeault et al. (23). The observation that the proapoptotic factor p53 was upregulated in HIV-1-infected primary CD4 cells would be in line with the prelude to apoptosis seemingly evident in our present study at 24 h p.i. However, none of the proteomic analyses to date have identified the upregulation of p53 at the protein level; the fact that the proapoptotic activity of p53 is regulated via phosphorylation could pose a technical problem for mass spectrometric analysis, as proteins with posttranslational modifications are refractive to routine MS/MS analysis and amino acid sequence search algorithms. In addition, the relatively low infectivity in that microarray study could introduce a large background from uninfected cells, which could, in turn, obscure the detection of differential expression. Indeed, the proportion of 168 signature proteins out of 1,146 quantified in our present study was much larger than the report of 336 signature transcripts out of an array of >12,000 human transcripts. In addition, the absence of cell-activating reagents such as IL-2 or phytohemagglutinin in the transcriptomic study could have further delayed the detection of impacts on cellular gene expression. Nonetheless, given that protein abundance changes spawning from translational changes should, by and large, correlate with perturbations in cellular transcription, it is not surprising that cellular pathways including DNA repair and RNA metabolism were implicated by both transcriptomic and proteomic profiling of HIV-1-infected primary CD4 cells.
We also noted with interest the overlap with cellular cofactors of HIV reported in recent cDNA and siRNA screens. In their screen for cofactors implicated in the postentry phase of the HIV life cycle, König et al. reported the RNA splicing factor RBM17 and the actin reorganization factor TAGLN2 as proviral factors (28); both of these proteins were upregulated at 8 h p.i. but not at 24 h p.i. The identification of RAN-mediated nuclear import as another cellular pathway impacting HIV replication by both König et al. and Brass et al. (5, 28) was also in agreement with the downregulation of importin 5 we observed at 24 h p.i., as well as with the dysregulated karyopherin-mediated transport reported in our CEMx174 cell line study. Other cellular processes mentioned in the siRNA screens, such as DNA repair and mitochondrial functions, also mirrored our observations in those processes. However, just as the overlap of individual genes/proteins was minimal among the screens, the agreement between these high-throughput approaches only holds up when categorizing genes/proteins by their corresponding cellular functions instead of by individual gene/protein names. We submit that no high-throughput screen would be all encompassing; in our present study, we addressed this inherent deficiency via the systematic characterization of protein lists by cellular functions, as well as by subcellular localization and known interactions with HIV proteins. Furthermore, as our observation with BANF1 has shown, we have to be cognizant of the specifics of experimental designs; our LC-MS/MS of whole-cell protein extracts would not have captured virion-incorporated cellular proteins.
In summary, our LC-MS/MS analysis of HIV-1-infected primary CD4 cells provided snapshots of the intracellular proteome at two distinct stages of the HIV-1 life cycle. The application of the label-free approach was optimal for the multiplexed quantitative comparisons given the limited supply of primary CD4 cells; the use of efavirenz treatment as a counterscreen helped increase our identification of cellular processes specific to robust virus production in primary CD4 cells. In spite of potential confounding from viral transformation and/or tumorigenesis, aspects of our observations in primary CD4 cells were in agreement with prior cell line-based reports, including the dynamic changes in the host ribosomal machinery, the exhaustion of host resources, the compromise of mitochondrial and nuclear integrity, and the presaging of apoptosis. Such overlaps increased our confidence in the interpretation of data from primary cells, given their inherent propensity to exhibit greater biological variability. In addition, we identified many changes in protein abundance that were not previously identified; in that regard, our present report adds extra information to ongoing efforts in the search for cellular cofactors as targets for novel therapeutics. By drawing on data from different high-throughput screening and profiling approaches, as well as by leveraging our knowledge about the retroviral life cycle, we will continue to add to the arsenal in our fight against the HIV epidemic.
Supplementary Material
Acknowledgments
We thank Marcus J. Korth and Robert E. Palermo for insightful comments and discussions. We also thank Leo E. Bonilla for assistance in planning for the study and Matthew E. Monroe for assistance with the VIPER software. We thank the Thermo Fisher Scientific BRIMS (Biomarker Research in Mass Spectrometry) Center for the use of MS instruments. The HIV-1 Gag p24 monoclonal (24-2) reagent was obtained from Michael H. Malim through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH.
E.Y.C. was supported by an STD/AIDS research training grant (NIH T32 AI07140). This work was supported by Public Health Service grants R24 RR016354 and P51 RR000166 from the National Center for Research Resources and P30 DA015625 from the National Institute on Drug Abuse.
Footnotes
Published ahead of print on 8 July 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Anderson, K., M. Monroe, and D. Daly. 2006. Estimating probabilities of peptide database identifications to LC-FTICR-MS observations. Proteome Sci. 4:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Askenazi, M., J. Sutton, R. Sadygov, T. Richmond, X. Shi, R. Gerszten, L. Bonilla, and A. Zumvalt. 2005. The Biomarker SIEVE: a computational framework for label-free MS-based proteomic biomarker discovery. Human Proteome Organization, Munich, Germany. http://www.thermo.com/eThermo/CMA/PDFs/Various/File_30421.pdf.
- 3.Barré-Sinoussi, F., J. C. Chermann, F. Rey, M. T. Nugeyre, S. Chamaret, J. Gruest, C. Dauguet, C. Axler-Blin, F. Vezinet-Brun, C. Rouzioux, W. Rozenbaum, and L. Montagnier. 1983. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science 220:868-871. [DOI] [PubMed] [Google Scholar]
- 4.Bartz, S. R., and M. Emerman. 1999. Human immunodeficiency virus type 1 Tat induces apoptosis and increases sensitivity to apoptotic signals by up-regulating FLICE/caspase-8. J. Virol. 73:1956-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brass, A. L., D. M. Dykxhoorn, Y. Benita, N. Yan, A. X. Engelman, J. Ramnik, J. Lieberman, and S. J. Elledge. 2008. Identification of host proteins required for HIV infection through a functional genomic screen. Science 319:921-926. [DOI] [PubMed] [Google Scholar]
- 6.Chan, E. Y., W. J. Qian, D. L. Diamond, T. Liu, M. A. Gritsenko, M. E. Monroe, D. G. Camp II, R. D. Smith, and M. G. Katze. 2007. Quantitative analysis of human immunodeficiency virus type 1-infected CD4+ cell proteome: dysregulated cell cycle progression and nuclear transport coincide with robust virus production. J. Virol. 81:7571-7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, H., and A. Engelman. 1998. The barrier-to-autointegration protein is a host factor for HIV type 1 integration. Proc Natl. Acad. Sci. USA 95:15270-15274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chun, T. W., J. S. Justement, R. A. Lempicki, J. Yang, G. Dennis, Jr., C. W. Hallahan, C. Sanford, P. Pandya, S. Liu, M. McLaughlin, L. A. Ehler, S. Moir, and A. S. Fauci. 2003. Gene expression and viral production in latently infected, resting CD4+ T cells in viremic versus aviremic HIV-infected individuals. Proc Natl. Acad. Sci. USA 100:1908-1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coiras, M., E. Camafeita, T. Urena, J. A. Lopez, F. Caballero, B. Fernandez, M. R. Lopez-Huertas, M. Perez-Olmeda, and J. Alcami. 2006. Modifications in the human T cell proteome induced by intracellular HIV-1 Tat protein expression. Proteomics 6(Suppl. 1):S63-S73. [DOI] [PubMed] [Google Scholar]
- 10.Corbeil, J., D. Sheeter, D. Genini, S. Rought, L. Leoni, P. Du, M. Ferguson, D. R. Masys, J. B. Welsh, J. L. Fink, R. Sasik, D. Huang, J. Drenkow, D. D. Richman, and T. Gingeras. 2001. Temporal gene regulation during HIV-1 infection of human CD4+ T cells. Genome Res. 11:1198-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cottrez, F., F. Manca, A. G. Dalgleish, F. Arenzana-Seisdedos, A. Capron, and H. Groux. 1997. Priming of human CD4+ antigen-specific T cells to undergo apoptosis by HIV-infected monocytes. A two-step mechanism involving the gp120 molecule. J. Clin. Investig. 99:257-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cullen, B. R. 2002. Using retroviruses to study the nuclear export of mRNA. Results Probl. Cell Differ. 35:151-168. [DOI] [PubMed] [Google Scholar]
- 13.de Noronha, C. M. C., M. P. Sherman, H. W. Lin, M. V. Cavrois, R. D. Moir, R. D. Goldman, and W. C. Greene. 2001. Dynamic disruptions in nuclear envelope architecture and integrity induced by HIV-1 Vpr. Science 294:1105-1108. [DOI] [PubMed] [Google Scholar]
- 14.Fang, R., D. A. Elias, M. E. Monroe, Y. Shen, M. Mcintosh, P. Wang, C. D. Goddard, S. J. Callister, R. J. Moore, Y. A. Gorby, J. N. Adkins, J. K. Fredrickson, M. S. Lipton, and R. D. Smith. 2006. Differential label-free quantitative proteomic analysis of Shewanella oneidensis cultured under aerobic and suboxic conditions by accurate mass and time tag approach. Mol. Cell. Proteomics 5:714-725. [DOI] [PubMed] [Google Scholar]
- 15.Fornace, A. J., I. Alamo, and M. C. Hollander. 1988. DNA damage-inducible transcripts in mammalian cells. Proc Natl. Acad. Sci. USA 85:8800-8804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fouchier, R. A., B. E. Meyer, J. H. Simon, U. Fischer, and M. H. Malim. 1997. HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J. 16:4531-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu, W., B. E. Sanders-Beer, K. S. Katz, D. R. Maglott, K. D. Pruitt, and R. G. Ptak. 2009. Human immunodeficiency virus type 1, human protein interaction database at NCBI. Nucleic Acids Res. 37:D417-D422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furukawa, K. 1999. LAP2 binding protein 1 (L2BP1/BAF) is a candidate mediator of LAP2-chromatin interaction. J. Cell Sci. 112:2485-2492. [DOI] [PubMed] [Google Scholar]
- 19.Geiss, G. K., R. E. Bumgarner, M. C. An, M. B. Agy, A. B. van 't Wout, E. Hammersmark, V. S. Carter, D. Upchurch, J. I. Mullins, and M. G. Katze. 2000. Large-scale monitoring of host cell gene expression during HIV-1 infection using cDNA microarrays. Virology 266:8-16. [DOI] [PubMed] [Google Scholar]
- 20.Goff, S. P. 2007. Host factors exploited by retroviruses. Nat. Rev. Microbiol. 5:253-263. [DOI] [PubMed] [Google Scholar]
- 21.Gougeon, M. L., H. Lecoeur, A. Dulioust, M. G. Enouf, M. Crouvoiser, C. Goujard, T. Debord, and L. Montagnier. 1996. Programmed cell death in peripheral lymphocytes from HIV-infected persons: increased susceptibility to apoptosis of CD4 and CD8 T cells correlates with lymphocyte activation and with disease progression. J. Immunol. 156:3509-3520. [PubMed] [Google Scholar]
- 22.Hammer, S. M., J. J. Eron, Jr., P. Reiss, R. T. Schooley, M. A. Thompson, S. Walmsley, P. Cahn, M. A. Fischl, J. M. Gatell, M. S. Hirsch, D. M. Jacobsen, J. S. G. Montaner, D. D. Richman, P. G. Yeni, and P. A. Volberding. 2008. Antiretroviral treatment of adult HIV infection: 2008 recommendations of the International AIDS Society—USA panel. JAMA 300:555-570. [DOI] [PubMed] [Google Scholar]
- 23.Imbeault, M., M. Ouellet, and M. Tremblay. 2009. Microarray study reveals that HIV-1 induces rapid type-I interferon-dependent p53 mRNA up-regulation in human primary CD4+ T cells. Retrovirology 6:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jablonski, J. A., and M. Caputi. 2009. Role of cellular RNA processing factors in human immunodeficiency virus type 1 mRNA metabolism, replication, and infectivity. J. Virol. 83:981-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacque, J. M., and M. Stevenson. 2006. The inner-nuclear-envelope protein emerin regulates HIV-1 infectivity. Nature 441:641-645. [DOI] [PubMed] [Google Scholar]
- 26.Kadiu, I., M. Ricardo-Dukelow, P. Ciborowski, and H. E. Gendelman. 2007. Cytoskeletal protein transformation in HIV-1-infected macrophage giant cells. J. Immunol. 178:6404-6415. [DOI] [PubMed] [Google Scholar]
- 27.Kash, J. C., D. M. Cunningham, M. W. Smit, Y. Park, D. Fritz, J. Wilusz, and M. G. Katze. 2002. Selective translation of eukaryotic mRNAs: functional molecular analysis of GRSF-1, a positive regulator of influenza virus protein synthesis. J. Virol. 76:10417-10426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.König, R., Y. Zhou, D. Elleder, T. L. Diamond, G. M. C. Bonamy, J. T. Irelan, C. Y. Chiang, B. P. Tu, P. D. De Jesus, C. E. Lilley, S. Seidel, A. M. Opaluch, J. S. Caldwell, M. D. Weitzman, K. L. Kuhen, S. Bandyopadhyay, T. Ideker, A. P. Orth, L. J. Miraglia, F. D. Bushman, J. A. Young, and S. K. Chanda. 2008. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell 135:49-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lafrenie, R. M., L. M. Wahl, J. S. Epstein, I. K. Hewlett, K. M. Yamada, and S. Dhawan. 1996. HIV-1-Tat protein promotes chemotaxis and invasive behavior by monocytes. J. Immunol. 157:974-977. [PubMed] [Google Scholar]
- 30.Lama, J., and V. Planelles. 2007. Host factors influencing susceptibility to HIV infection and AIDS progression. Retrovirology 4:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lifson, J. D., M. B. Feinberg, G. R. Reyes, L. Rabin, B. Banapour, S. Chakrabarti, B. Moss, F. Wong-Staal, K. S. Steimer, and E. G. Engleman. 1986. Induction of CD4-dependent cell fusion by the HTLV-III/LAV envelope glycoprotein. Nature 323:725-728. [DOI] [PubMed] [Google Scholar]
- 32.Liu, B., R. Dai, C. J. Tian, L. Dawson, R. Gorelick, and X. F. Yu. 1999. Interaction of the human immunodeficiency virus type 1 nucleocapsid with actin. J. Virol. 73:2901-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luban, J. 2008. HIV-1 infection: going nuclear with TNPO3/transportin-SR2 and integrase. Curr. Biol. 18:R710-R713. [DOI] [PubMed] [Google Scholar]
- 34.Monroe, M. E., N. Tolic, N. Jaitly, J. L. Shaw, J. N. Adkins, and R. D. Smith. 2007. VIPER: an advanced software package to support high-throughput LC-MS peptide identification. Bioinformatics 23:2021-2023. [DOI] [PubMed] [Google Scholar]
- 35.Moore, M. J., and M. Rosbash. 2001. Cell biology: TAPping into mRNA export. Science 294:1841-1842. [DOI] [PubMed] [Google Scholar]
- 36.Muthumani, K., A. Y. Choo, D. S. Hwang, M. A. Chattergoon, N. N. Dayes, D. Zhang, M. D. Lee, U. Duvvuri, and D. B. Weiner. 2003. Mechanism of HIV-1 viral protein R-induced apoptosis. Biochem. Biophys. Res. Commun. 304:583-592. [DOI] [PubMed] [Google Scholar]
- 37.Nesvizhskii, A. I., A. Keller, E. Kolker, and R. Aebersold. 2003. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75:4646-4658. [DOI] [PubMed] [Google Scholar]
- 38.Neubert, H., T. P. Bonnert, K. Rumpel, B. T. Hunt, E. S. Henle, and I. T. James. 2008. Label-free detection of differential protein expression by LC/MALDI mass spectrometry. J. Proteome Res. 7:2270-2279. [DOI] [PubMed] [Google Scholar]
- 39.Nguyen, D. G., H. Yin, Y. Zhou, K. C. Wolff, K. L. Kuhen, and J. S. Caldwell. 2007. Identification of novel therapeutic targets for HIV infection through functional genomic cDNA screening. Virology 362:16-25. [DOI] [PubMed] [Google Scholar]
- 40.Palamara, A. T., C. F. Perno, S. Aquaro, M. C. Bue, L. Dini, and E. Garaci. 1996. Glutathione inhibits HIV replication by acting at late stages of the virus life cycle. AIDS Res. Hum. Retrovir. 12:1537-1541. [DOI] [PubMed] [Google Scholar]
- 41.Quensel, C., B. Friedrich, T. Sommer, E. Hartmann, and M. Kohler. 2004. In vivo analysis of importin α proteins reveals cellular proliferation inhibition and substrate specificity. Mol. Cell. Biol. 24:10246-10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ringrose, J., R. E. Jeeninga, B. Berkhout, and D. Speijer. 2008. Proteomic studies reveal coordinated changes in T-cell expression patterns upon HIV-1 infection. J. Virol. 82:4320-4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roy, B. B., J. Hu, X. Guo, R. S. Russell, F. Guo, L. Kleiman, and C. Liang. 2006. Association of RNA helicase A with human immunodeficiency virus type 1 particles. J. Biol. Chem. 281:12625-12635. [DOI] [PubMed] [Google Scholar]
- 44.Shrikant, P., D. J. Benos, L. P. Tang, and E. N. Benveniste. 1996. HIV glycoprotein 120 enhances intercellular adhesion molecule-1 gene expression in glial cells. Involvement of Janus kinase/signal transducer and activator of transcription and protein kinase C signaling pathways. J. Immunol. 156:1307-1314. [PubMed] [Google Scholar]
- 45.Shutt, D. C., J. T. Stapleton, R. C. Kennedy, and D. R. Soll. 1995. HIV-induced syncytia in peripheral blood cell cultures crawl by extending giant pseudopods. Cell. Immunol. 166:261-274. [DOI] [PubMed] [Google Scholar]
- 46.Simon, J. H., R. A. Fouchier, T. E. Southerling, C. B. Guerra, C. K. Grant, and M. H. Malim. 1997. The Vif and Gag proteins of human immunodeficiency virus type 1 colocalize in infected human T cells. J. Virol. 71:5259-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sodroski, J., W. C. Goh, C. Rosen, K. Campbell, and W. A. Haseltine. 1986. Role of the HTLV-III/LAV envelope in syncytium formation and cytopathicity. Nature 322:470-474. [DOI] [PubMed] [Google Scholar]
- 48.Sutton, J., T. Richmond, X. Shi, M. Athana, C. Ptak, R. Gerszten, and L. Bonilla. 2008. Performance characteristics of an FT MS-based workflow for label-free differential MS analysis of human plasma: standards, reproducibility, targeted feature investigation, and application to a model of controlled myocardial infarction. Proteomics Clin. Appl. 2:862-881. [DOI] [PubMed] [Google Scholar]
- 49.Valente, S. T., and S. P. Goff. 2009. Somatic cell genetic analyses to identify HIV-1 host restriction factors. Methods Mol. Biol. 485:235-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Valente, S. T., G. M. Gilmartin, C. Mott, B. Falkard, and S. P. Goff. 2009. Inhibition of HIV-1 replication by eIF3f. Proc Natl. Acad. Sci. USA 106:4071-4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van 't Wout, A. B., G. K. Lehrman, S. A. Mikheeva, G. C. O'Keeffe, M. G. Katze, R. E. Bumgarner, G. K. Geiss, and J. I. Mullins. 2003. Cellular gene expression upon human immunodeficiency virus type 1 infection of CD4+-T-cell lines. J. Virol. 77:1392-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wain-Hobson, S., J. P. Vartanian, M. Henry, N. Chenciner, R. Cheynier, S. Delassus, L. P. Martins, M. Sala, M. T. Nugeyre, D. Guétard, D. Klatzmann, J.-C. Gluckmann, W. Rosenbaum, F. Barré-Sinoussi, and L. Montagnier. 1991. LAV revisited: origins of the early HIV-1 isolates from Institut Pasteur. Science 252:961-965. [DOI] [PubMed] [Google Scholar]
- 53.Washburn, M. P., D. Wolters, and J. R. Yates. 2001. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19:242-247. [DOI] [PubMed] [Google Scholar]
- 54.Wen, W., S. Chen, Y. Cao, Y. Zhu, and Y. Yamamoto. 2005. HIV-1 infection initiates changes in the expression of a wide array of genes in U937 promonocytes and HUT78 T cells. Virus Res. 113:26-35. [DOI] [PubMed] [Google Scholar]
- 55.Wilk, T., B. Gowen, and S. D. Fuller. 1999. Actin associates with the nucleocapsid Domain of the human immunodeficiency virus Gag polyprotein. J. Virol. 73:1931-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wolf, D., and S. P. Goff. 2007. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell 131:46-57. [DOI] [PubMed] [Google Scholar]
- 57.Young, S. D., S. F. Britcher, L. O. Tran, L. S. Payne, W. C. Lumma, T. A. Lyle, J. R. Huff, P. S. Anderson, D. B. Olsen, and S. S. Carroll. 1995. L-743, 726 (DMP-266): a novel, highly potent nonnucleoside inhibitor of the human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 39:2602-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zauli, G., D. Gibellini, P. Secchiero, H. Dutartre, D. Olive, S. Capitani, and Y. Collette. 1999. Human immunodeficiency virus type 1 Nef protein sensitizes CD4+ T lymphoid cells to apoptosis via functional upregulation of the CD95/CD95 ligand pathway. Blood 93:1000-1010. [PubMed] [Google Scholar]
- 59.Zhou, H., M. Xu, Q. Huang, A. T. Gates, X. D. Zhang, J. C. Castle, E. Stec, M. Ferrer, B. Strulovici, D. J. Hazuda, and A. S. Espeseth. 2008. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe 4:495-504. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.