Abstract
Sterile alpha and HEAT/Armadillo motif (SARM) is a highly conserved Toll/interleukin-1 receptor (TIR)-containing adaptor protein that is believed to negatively regulate signaling of the pathogen recognition receptors Toll-like receptor 3 (TLR3) and TLR4. To test its physiological function in the context of a microbial infection, we generated SARM−/− mice and evaluated the impact of this deficiency on the pathogenesis of West Nile virus (WNV), a neurotropic flavivirus that requires TLR signaling to restrict infection. Although SARM was preferentially expressed in cells of the central nervous system (CNS), studies with primary macrophages, neurons, or astrocytes showed no difference in viral growth kinetics. In contrast, viral replication was increased specifically in the brainstem of SARM−/− mice, and this was associated with enhanced mortality after inoculation with a virulent WNV strain. A deficiency of SARM was also linked to reduced levels of tumor necrosis factor alpha (TNF-α), decreased microglia activation, and increased neuronal death in the brainstem after WNV infection. Thus, SARM appears to be unique among the TIR adaptor molecules, since it functions to restrict viral infection and neuronal injury in a brain region-specific manner, possibly by modulating the activation of resident CNS inflammatory cells.
Understanding the outcome of a host-pathogen encounter requires identification of the molecular mechanisms by which microbes are recognized and inflammatory signals are generated. Pattern recognition receptors (PRR) detect conserved microbial structural elements identified as pathogen-associated molecular patterns. PRR that recognize single- and double-stranded genomes of RNA viruses include Toll-like receptors (TLR) on the cell surface or within endosomes, which signal through the adaptor molecules MyD88 and TRIF, and the RIG-I and MDA5 cytoplasmic helicases, which signal through the adaptor protein IPS-1 (also known as MAVS, Cardif, and VISA) (reviewed in references 18 and 41). Recognition of viral RNA by these sensors results in the downstream activation and nuclear translocation of master transcriptional regulators, including IRF-3, IRF-7, and NF-κB, which induce gene transcription of proinflammatory cytokines, including type I interferon (IFN-α and -β) and tumor necrosis factor alpha (TNF-α) (34). PRR signaling and the immune responses that ensue are regulated in part through the expression and localization of cell type-specific adaptor molecules.
Sterile alpha and HEAT/Armadillo motif (SARM) is the fifth identified Toll/interleukin-1 receptor (TIR)-containing adaptor protein and is highly conserved across many species (23). Diverse functions have been attributed to SARM. The results of in vitro studies have suggested that human SARM specifically inhibits the function of TRIF, an adaptor that mediates the MyD88-independent signaling of TLR3 and TLR4 (3). When overexpressed in 293T cells, human SARM associates with TRIF via its TIR and sterile-alpha motif (SAM) domains to block the induction of proinflammatory genes (3). In contrast, studies of mouse macrophages lacking SARM have demonstrated that cytokine production is unaltered after TLR stimulation (20). In vivo, its function is also uncertain. In Caenorhabditis elegans, SARM protects against infection in a TLR-independent manner by promoting the induction of antimicrobial peptides (5). In mice, SARM appeared proapoptotic in hippocampal neurons under metabolic stress, suggesting that it has tissue-specific signaling functions (20). Thus, SARM likely has diverse molecular targets in animals, and its role in modulating immunity remains to be fully elucidated.
West Nile virus (WNV) is a single-stranded, positive-polarity RNA virus of the Flaviviridae family. In humans, WNV infection is commonly asymptomatic, although in a subset of individuals it can cause a febrile illness and progress to encephalitis (15). The elderly and immunocompromised are at greatest risk for developing severe neurological disease, suggesting a role for the host immune response in the control of infection. The mouse model of WNV recapitulates many features of human disease and results in infection of myeloid cells and neurons in peripheral and central nervous system (CNS) tissues, respectively (reviewed in reference 31). Studies of mice also have highlighted the importance of the IFN-α/-β and TNF-α signaling pathways in controlling lethal WNV infection and minimizing cell death of CNS neurons (19, 30, 36). Recent studies have also demonstrated an essential protective role for TLR3 and IRF-3 in controlling WNV pathogenesis in the brain (6, 7). Given the importance of TLR-mediated immunity in modulating WNV pathogenesis, we reasoned that this model would be well suited to define the role of SARM in immunity in vivo. To test this hypothesis, we generated SARM−/− mice and evaluated the impact of this deficiency on WNV pathogenesis. Little if any direct antiviral effect was observed on WNV replication ex vivo in several cell types. However, a deficiency of SARM enhanced WNV pathogenesis specifically in the brainstem and was associated with altered TNF-α production, microglia activation, and neuronal cell death.
MATERIALS AND METHODS
Generation of SARM−/− mice.
A targeting vector was designed to replace a 7.3-kb fragment containing exons 1 and 2 of the SARM gene (encoding the start site and first 363 amino acids) with an MC1-neor gene flanked by loxP sites (Fig. 1A). The construct was electroporated into SSC#10 (129X1/SvJ) embryonic stem cells by the Siteman core ES facility (Washington University School of Medicine), and 225 clones were expanded and screened. One targeted clone was identified and confirmed by Southern blotting using multiple restriction enzyme digests and external and internal probes. The clone was injected into C57BL/6 blastocysts, and the resulting chimeras were bred to C57BL/6 mice expressing a Cre transgene under the cytomegalovirus promoter to delete the MC1-neor gene. Germ line transmission was assessed by coat color, and the presence of the SARM mutation by PCR. The SARM deletion was subsequently backcrossed onto C57BL/6 mice, which was facilitated by genome-wide screening of polymorphic microsatellite markers at 10-centimorgan intervals at each generation (performed by the Rheumatology Speed Congenics Core Laboratory at Washington University School of Medicine).
FIG. 1.
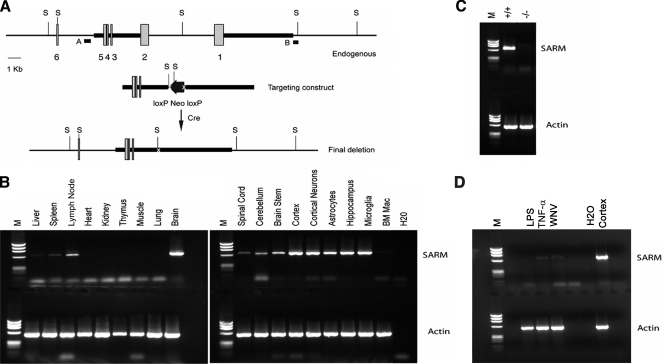
Expression and generation of SARM knockout mice. (A) Schematic of the SARM locus and targeting cassette. Exons 1 through 6 of SARM are represented by gray boxes, and the fragments included in the targeting construct by a heavy black line. The locations of external probes are shown, as are the restriction sites used for screening. S, SphI. (B) A representative image of the RT-PCR analysis of SARM mRNA from whole organs, specific brain regions, and primary cells derived from wild-type C57BL/6 mice is shown. The SARM transcript, at approximately 650 bp, is indicated; a 519-bp fragment of β-actin was amplified in parallel as a control. BM Mac, bone marrow-derived macrophages. (C) A representative image of the RT-PCR analysis of SARM−/− and wild-type mouse mRNA is shown. SARM and β-actin mRNA were amplified from equivalent amounts of mRNA derived from wild-type, SARM+/−, and SARM−/− astrocytes. (D) A representative image of RT-PCR analysis of SARM mRNA in macrophages activated with LPS and TNF-α or infected with WNV is shown. Cerebral cortex mRNA was used as a positive control. M, molecular size ladder.
RNA and RT-PCR.
Total RNA was prepared from cells and tissues by using an RNeasy kit (Qiagen). cDNA was synthesized by using oligo(dT), a 650-bp fragment of SARM amplified with 5′-AGGTTCTTTAGGGAGCTCACAGAG-3′ and 5′-TTGATGCCGTTGAAGGTGAGTACAGCCTG-3′, and a 519-bp fragment of actin amplified with 5′-AGCCATGTACGTAGCCATCCA-3′ and 5′-GTGGTACCACCAGACAACACT-3′. The PCR conditions were 94°C for 4 min, 35 cycles of 94°C for 30 s, 63°C for 30 s, and 72°C for 30 s, with a final 5 min at 72°C.
Virus propagation and titration.
The lineage 1 New York WNV strain (WNV-NY) (3000.0259) was isolated in 2000 (10) and passaged once in C6/36 Aedes albopictus cells to generate a stock that was used in most experiments. The lineage 2 Madagascar WNV strain (DakAnMg798) (WNV-MAD) was isolated in 1978 (1) and amplified once in Vero cells. BHK21-15 and Vero cells were used to measure the viral titer of infected cells or tissues by plaque assay (9). Viral burden was also measured by analyzing viral RNA levels using quantitative real-time PCR (RT-PCR) (9).
Mouse experiments and tissue preparation.
Eight- to 12-week-old SARM−/− or age-matched wild-type control mice were bred in parallel and used for all in vivo studies. Peripheral infection was performed by footpad inoculation of 102 PFU of WNV-NY diluted in 50 μl of Hanks balanced salt solution (HBSS) with 1% heat-inactivated fetal bovine serum (FBS). Intracranial inoculation was performed using 101 PFU of WNV-MAD diluted in 10 μl of HBSS with 1% heat-inactivated FBS. On specified days, mice were euthanized and perfused with 20 ml of iced phosphate-buffered saline (PBS), and organs were removed, weighed, and stored at −80°C until further processing. Alternatively, groups of mice were monitored for survival for 21 days postinfection.
Serological analysis.
WNV-specific immunoglobulin M (IgM) and IgG levels were determined by enzyme-linked immunosorbent assay (ELISA) as previously described (9). Soluble WNV envelope (E) protein (24) was absorbed to Maxi-Sorp microtiter plates (Nunc) overnight at 4°C, followed by blocking at 37°C for 1 h with PBS containing 1% bovine serum albumin, 3% horse serum, 0.05% Tween 20, and 0.025% NaN3. Fourfold serial dilutions of heat-inactivated serum samples were added to the plates and incubated overnight at 4°C. Plates were washed, incubated with biotin-conjugated goat anti-mouse IgM or IgG (Southern Biotech) followed by streptavidin-horseradish peroxidase (Invitrogen), and developed with tetramethylbenzidine substrate (Dako). Antibody titers represent the serum dilution yielding a value for the optical density at 450 nm equivalent to three standard deviations above the background of the assay.
Immunohistochemistry of brain tissues.
Tissues were prepared essentially as described previously (32). Briefly, following extensive perfusion with chilled PBS, brains were harvested, fixed in 4% paraformaldehyde overnight at 4°C, and then cryoprotected in 30% sucrose solutions over three days at 4°C. Brain tissues were embedded in optimal cutting temperature (O.C.T.) compound (Tissue Tek) and flash frozen prior to sectioning on a cryostat. Terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) staining was performed by using a NeuroTACS II in situ apoptosis detection kit (Trevigen) or an in situ cell death detection kit (Roche) according to the manufacturer's protocols, with some modifications (32). For fluorescence staining and confocal microscopic imaging, frozen sections were rehydrated with PBS and permeabilized and blocked with 0.1% Triton X-100 and 10% normal goat serum (Sigma) for 1 h at 37°C in a humidified chamber. Sections were incubated overnight at 4°C with 1 μg/ml anti-CD11b (BD Biosciences) or anti-microtubule-associated protein 2 (anti-MAP-2; Chemicon) antibody, followed by detection with 5 μg/ml Alexa Fluor 488-conjugated secondary antibody (Molecular Probes) for 1 h at room temperature. Nuclei were counterstained with ToPro-3 (Molecular Probes). Slides were mounted with ProLong Gold antifade reagent (Molecular Probes). Sections were visualized on a Zeiss LSM 510 META confocal laser scanning microscope.
Quantification of brain leukocytes.
Whole brains were harvested from wild-type and SARM−/− mice on day 8 after subcutaneous infection with WNV-NY or intracranial infection with WNV-MAD. Following perfusion, brains were homogenized through a 70-μm cell strainer, digested with a collagenase solution (500 μg/ml collagenase D, 0.1 μg/ml Nα-p-tosyl-l-lysine chloromethyl ketone [TLCK] trypsin inhibitor, 10 μg/ml DNase I, 10 mM HEPES in HBSS) for 1 h at room temperature. Cells were separated by centrifugation on a discontinuous 70-to-37-to-30% Percoll gradient. For detection of resident microglia and infiltrating macrophages, cells were incubated with fluorescein isothiocyanate-conjugated anti-major histocompatibility complex class II, phycoerythrin-conjugated anti-CD45, and allophycocyanin-conjugated anti-CD11b antibodies (BD Biosciences). For detection of WNV-specific CD8+ T cells, leukocytes were restimulated ex vivo with a Db-restricted immunodominant NS4B WNV peptide (27) and incubated with fluorescein isothiocyanate-conjugated anti-CD3 or allophycocyanin-conjugated anti-CD8 (BD Biosciences) at 4°C for 30 min. Subsequently, cells were washed, fixed in 1% paraformaldehyde, permeabilized, and incubated additionally with phycoerythrin-conjugated anti-IFN-γ or -TNF-α prior to analysis by flow cytometry.
Primary cell cultures.
Cortical and hippocampal neurons were generated as described previously (21). All experiments with neuron cultures were performed on cells propagated for 4 days. Bone marrow-derived macrophages and myeloid dendritic cells were isolated and maintained as described previously (33). Murine embryonic fibroblasts were generated from embryonic-day-14 wild-type or SARM−/− congenic mice according to established protocols and were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% FBS. Astrocytes were prepared essentially as described previously for rats, with some adaptations (47). Briefly, forebrains were dissected from 1- to 2-day-old mice, gently dissociated by trituration in DMEM containing 10% FBS, and filtered through a 70-μm cell strainer. Dissociated cells were seeded into poly-d-lysine-laminin-coated 75-cm2 flasks and cultured for 8 to 10 days until confluent. Flasks were shaken at 150 RPM for 1 h to remove nonadherent microglia, and fresh DMEM containing 3% FBS was added. This procedure was repeated for three consecutive days, and on the last day, astrocyte monolayers were recovered after trypsin treatment and seeded into poly-d-lysine-laminin-coated culture plates.
Quantification of cytokines and chemokines.
Cell culture supernatants and clarified tissue homogenates were analyzed by using an ELISA kit (R&D Systems) and Bioplex cytokine bead array (Bio-Rad) according to the manufacturers' protocols. Quantitative RT-PCR was used to measure levels of gene expression as previously described (21, 37). Briefly, RNA was isolated by using an RNeasy kit (Qiagen), followed by DNase I treatment (Invitrogen) and cDNA generation using a high-capacity cDNA reverse transcription kit (Applied Biosystems). Gene expression was measured by using Sybr green PCR master mix (Applied Biosystems) and primers for TNF-α and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (21); the latter was used for normalization of cycle threshold values to account for the amount of input tissue.
Statistical analysis.
All data were analyzed with Prism software (Graphpad Software, Inc.). For survival analysis, Kaplan-Meier survival curves were analyzed by using the log-rank test. For viral burden analysis, significance was determined by using the Mann-Whitney test. An unpaired Student's t test was used to determine significant differences in cytokine production and TUNEL assay results.
RESULTS
SARM expression and generation of null mice.
Recent studies with human cells and transgenic mice have suggested that SARM is a negative regulator of TRIF-dependent TLR3 signaling and a mediator of stress-induced toxicity in hippocampal neurons (3, 20). However, it remains uncertain how SARM contributes to the immune response during a pathogen challenge, when multiple PRR are stimulated concurrently. The results of previous experiments had suggested that SARM was distinct from other TLR adaptors in its preferential expression in neurons (20). To confirm this, we examined the expression of murine SARM in various tissues by using RT-PCR. Relatively high levels of SARM mRNA were observed in the brain, whereas lower levels were observed in other tissues (Fig. 1B). Consistent with this, SARM mRNA was detected in primary neurons, microglia, and astrocytes but was undetectable or at very low levels in primary lymphocytes and bone marrow-derived macrophages (Fig. 1B and data not shown). Activated macrophages that were pretreated with lipopolysaccharide (LPS) or TNF-α or infected with WNV also expressed little, if any, SARM mRNA (Fig. 1D). We were unable to confirm protein expression of SARM in tissue sections, as only one anti-mouse SARM antibody is available commercially and it fails to recognize SARM reliably in fixed or frozen tissue sections (K. Szretter and M. Diamond, unpublished observations).
To elucidate the function of SARM in the innate immune response, we generated SARM−/− mice by a homologous recombination strategy (Fig. 1A). Homozygous SARM−/−, heterozygotic, and wild-type mice were born at expected Mendelian ratios and confirmed by Southern blotting and RT-PCR (Fig. 1C and data not shown). Flow cytometric analyses of SARM−/− mice revealed normal hematopoietic cell development, as well as typical B- and T-cell maturation profiles (data not shown).
SARM restricts WNV pathogenesis.
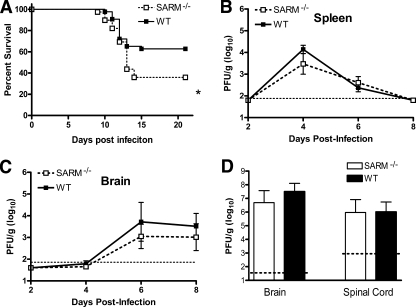
Given that SARM is primarily expressed in the CNS, we evaluated its function in vivo in the context of infection by WNV, a neurotropic RNA flavivirus. We used WNV as the model pathogen for two reasons: (i) it infects and injures neurons in diverse brain regions, including the hippocampus, brainstem, and cerebral cortex (35); and (ii) TLR3 restricts WNV infection in neurons in vitro and in vivo (7). As SARM is reported to control infection in some model systems and negatively regulate TLR3 signaling in others, we anticipated that challenging SARM-deficient mice might result in a phenotype that would clarify its function. SARM−/− and wild-type mice were infected subcutaneously with 102 PFU of a pathogenic North American WNV strain (WNV-NY) and monitored for disease. SARM−/− mice were more vulnerable to WNV-NY infection (36% survival rate) than age-matched wild-type mice (63% survival rate) (Fig. 2A) (P < 0.03).
FIG. 2.
Survival and viral burden analysis for wild-type and SARM−/− mice following WNV challenge. (A) Age-matched 8- to 12-week-old wild-type or SARM−/− mice were inoculated with 102 PFU of WNV-NY by footpad injection and monitored for 21 days. Survival differences were statistically significant (39 SARM−/− and 43 wild-type mice, P < 0.03). (B, C) Viral burden was determined by plaque assay in the spleen (B) and brain (C) on days 2, 4, 6, and 8 after subcutaneous WNV-NY infection. (D) Eight- to 12-week-old mice were inoculated with 101 PFU of WNV-MAD by intracranial injection, and viral burdens in brain and spinal cord were determined by plaque assay on day 6 after infection. Viral burden data are expressed as log10 PFU per gram ± the standard error of the mean for 6 to 10 mice per time point. The dotted lines denote the limit of detection of the viral plaque assay.
To better understand how a deficiency of SARM caused excess mortality following WNV infection, wild-type and SARM−/− mice were infected and their viral burdens in the spleen, brain, and spinal cord were measured by viral plaque assay at days 2, 4, 6, and 8 (Fig. 2B and C and data not shown); these tissues were previously defined as targets for WNV replication during its distinct phases of pathogenesis (9). Notably, no statistically significant difference (P > 0.1) in viral burdens was observed between wild-type and SARM−/− mice in any of these tissues following subcutaneous infection. To confirm the absence of a virologic phenotype in the CNS, we inoculated 101 PFU of an attenuated lineage 2 WNV strain (WNV-MAD) by an intracranial route and assessed viral replication. The WNV-MAD strain was used because its attenuation prolongs survival (1) and allows the resolution of differences in viral replication in the highly permissive neurons of the brain (19). Similar to the viral burden observed after peripheral infection of WNV-NY, no discernible difference in viral burden was observed on day 6 after intracranial infection in SARM−/− mice (Fig. 2D). Moreover, no significant difference in rates of mortality was observed between wild-type and SARM−/− mice after infection with the attenuated WNV-MAD by an intracranial route (data not shown).
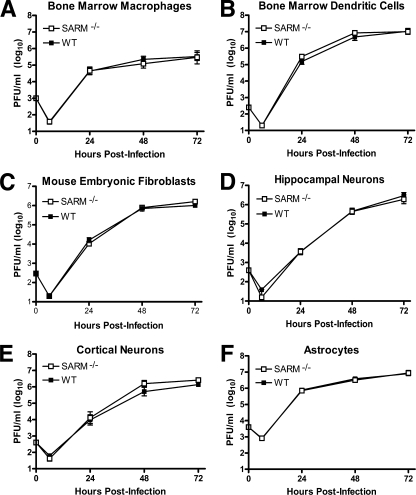
Consistent with the lack of an apparent virologic phenotype, multistep growth curve analysis with primary macrophages, myeloid dendritic cells, fibroblasts, cortical and hippocampal neurons, and astrocytes from wild-type and SARM−/− mice showed no difference in WNV-NY infection (Fig. 3A to F). Based on this, we hypothesized that although SARM contributes to survival after infection with a virulent WNV isolate, unlike TLR3 or other PRR signaling molecules (6-8, 42), it did not directly modulate viral replication.
FIG. 3.
SARM does not have a direct antiviral effect against WNV in primary cells. Primary cells derived from SARM−/− and wild-type mice (described in Materials and Methods) were infected with a multiplicity of infection of 0.001, and virus replication was measured at 6, 24, 48, and 72 h postinfection by viral plaque assay. Multistep growth curves showed no difference in viral yields between SARM−/− and wild-type (A) macrophages, (B) dendritic cells, (C) fibroblasts, (D) hippocampal neurons, (E) cortical neurons, or (F) astrocytes. Viral burden data are expressed as log10 PFU per ml ± the standard error of the mean for triplicate samples from two independent experiments.
WNV-specific B- and T-cell responses are normal in SARM−/− mice.
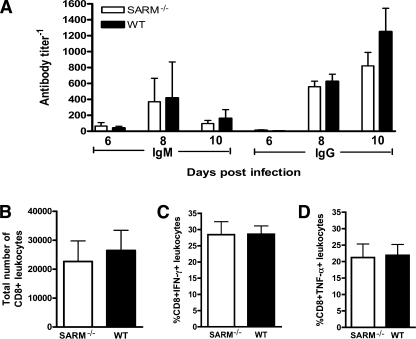
Intact adaptive B- and CD8+ T-cell immune responses are required to restrict CNS infection, neuronal injury, and progression to lethal WNV disease (31). To evaluate whether the increased mortality observed in WNV-infected SARM−/− mice was associated with a defect in adaptive immunity, we examined the B- and T-cell responses during WNV infection. No difference (P > 0.2) in WNV-specific IgG and IgM antibody titers was observed between wild-type and SARM−/− mice on days 6, 8, and 10 after subcutaneous infection (Fig. 4A). To test whether a deficiency of SARM affected CD8+ T-cell priming and migration, we measured the number of infiltrating T cells in the brain on day 8 after subcutaneous WNV infection. Wild-type and SARM−/− mice showed no difference in the total number of CD8+ T cells in the brain (Fig. 4B) or the number of CD8+ T cells expressing IFN-γ or TNF-α after ex vivo restimulation of brain leukocytes with a Db-restricted immunodominant WNV NS4B peptide (Fig. 4C and D). Thus, SARM is not required for efficient priming of WNV-specific adaptive immunity.
FIG. 4.
Lymphocyte responses after WNV-NY infection in SARM−/− mice are intact. (A) Wild-type and SARM−/− mice were inoculated with 102 PFU of WNV-NY by footpad injection, and serum samples collected on days 6, 8, and 10 after subcutaneous WNV-NY infection were assayed for WNV E-specific IgM and IgG. Titers are expressed as the reciprocal serum dilution that was three standard deviations above background. Differences were not statistically significant (P > 0.2). (B to D) Wild-type and SARM−/− mice were inoculated with 102 PFU of WNV-NY by footpad injection, and brains were harvested on day 8. Leukocytes were isolated by Percoll gradient centrifugation; stimulated ex vivo with NS4B WNV peptide; stained for CD3, CD8, and intracellular IFN-γ or TNF-α; and analyzed by flow cytometry. (B) The total number of brain CD8+ T cells was determined by the percentage of CD3+ CD8+ cells multiplied by the total cell count. The percentage of CD3+ CD8+ T cells positive for intracellular IFN-γ (C) or TNF-α (D) is indicated. Differences were not statistically significant (P > 0.9). Error bars show standard errors of the means.
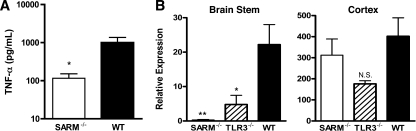
TNF-α responses are blunted in the CNS of SARM−/− mice.
Given that SARM has been postulated to modulate the expression of proinflammatory cytokines via the TLR3 pathway and our recent observation that a deficiency of TNF-α-TNF-R1 interactions is associated with greater lethality of WNV infection in C57BL/6 mice (36), we assessed the levels of TNF-α in whole-brain homogenates from wild-type, TLR3−/−, and SARM−/− mice by using a commercial ELISA (Fig. 5A and data not shown). SARM−/− mice had a 10-fold-lower concentration (P < 0.02) of TNF-α protein in whole-brain homogenates at day 6; although TLR3−/− mice showed a similar trend (5-fold) toward reduced TNF-α levels compared to the levels in wild-type mice, this did not attain statistical significance (P > 0.2). However, based on analysis of TNF-α mRNA levels in distinct brain regions, the reductions in SARM−/− and TLR3−/− mice correlated primarily with differences in gene expression in the brainstem (Fig. 5B) (P ≤ 0.02) and were not observed in the cortex (Fig. 5C) (P ≥ 0.4). In contrast, other inflammatory cytokine (e.g., interleukin-6) and chemokine (e.g., CXCL10) mRNA levels that are elevated in CNS cells after stimulation with the TLR3 synthetic ligand poly(I:C) (43) did not differ significantly between SARM−/−, TLR3−/−, and wild-type mice regardless of the brain region (data not shown).
FIG. 5.
A deficiency of SARM affects the TNF-α response during WNV infection. Wild-type, TLR3−/−, or SARM−/− mice were inoculated with 101 PFU of WNV-MAD by intracranial injection, and brains were harvested on day 6. (A) Whole-brain homogenates were assayed for TNF-α by ELISA. Data represent the average results for 8 to 10 mice from at least two independent experiments. (B) Brainstem and cortex regions of brain were harvested and analyzed by quantitative RT-PCR to measure TNF-α mRNA levels. Data represent the average results for 10 mice from at least two independent experiments. Error bars show standard errors of the means. *, P < 0.02; **, P < 0.005; N.S., not significant.
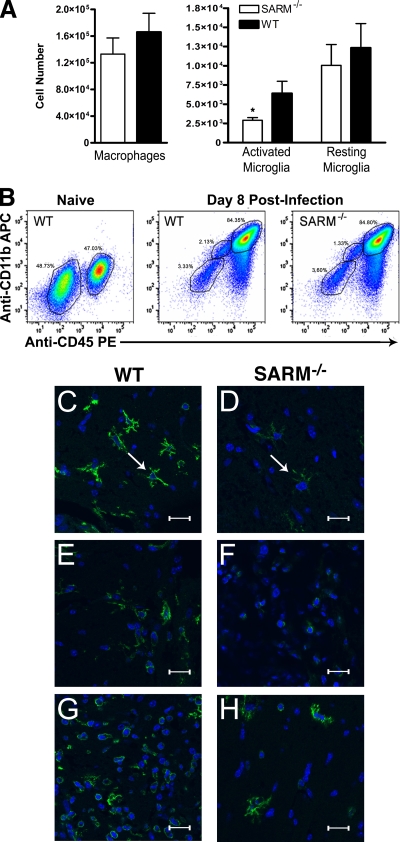
Effect of SARM on myeloid cell accumulation and activation in the CNS.
As prior experiments with TNF-R1−/− and TLR3−/− mice showed defects in the accumulation and activation of macrophages and microglia in the brain after WNV infection (36) or poly(I:C) injection (43), respectively, we evaluated the numbers and phenotypes of CD45+ CD11b+ cell populations in wild-type and SARM−/− mice on day 8 after infection. Based on the classification of CNS myeloid cells of Ford et al. (11), SARM−/− mice had significantly fewer CD11bhigh CD45low, activated microglia (P < 0.04), whereas the numbers of CD11bhigh CD45high, activated macrophages and CD11blow CD45low, resting microglia were not substantially different from those in WNV-infected wild-type mice (Fig. 6A and B). Immunohistochemistry confirmed these findings, as fewer ramified, activated microglia cells were observed by confocal microscopy, especially in the brainstem (Fig. 6C and D) and, to a lesser degree, in the cerebral cortex (Fig. 6E and F) of SARM−/− mice. In comparison, the activation state of myeloid cells in the cerebellum in WNV-infected wild-type and SARM−/− mice was similar (Fig. 6G and H). Given that myeloid cells express little, if any, SARM mRNA (Fig. 1B), we speculate that the differential activation of brain myeloid cells in SARM−/− mice may result from altered cell-extrinsic signals, such as the blunted levels of TNF-α in the brain.
FIG. 6.
SARM promotes microglia activation during WNV infection. Wild-type or SARM−/− mice were inoculated with 101 PFU of WNV-MAD by intracranial injection, and brains were harvested on day 8. Leukocytes were isolated by Percoll gradient centrifugation, stained for CD11b and CD45, and analyzed by flow cytometry. (A) Expression profiles for activated macrophages (CD11bhigh CD45high), activated microglia (CD11bhigh CD45low), and resting microglia (CD11blow CD45low) were evaluated. Error bars show standard errors of the means. (B) Representative flow cytometry profiles of CD11b and CD45 staining of brain leukocytes from naïve wild-type mice and from wild-type and SARM−/− mice 8 days after WNV infection are shown. (C to H) Representative confocal microscopic images of CD11b (green) and ToPro-3 nuclear staining (blue) of microglia and macrophages in brainstem (C, D), cortex (E, F), and cerebellum (G, H) are shown (*, P < 0.04). White arrows indicate ramified microglia. The scale bar represents ∼15 μm.
Effect of SARM on neuronal death after WNV infection.
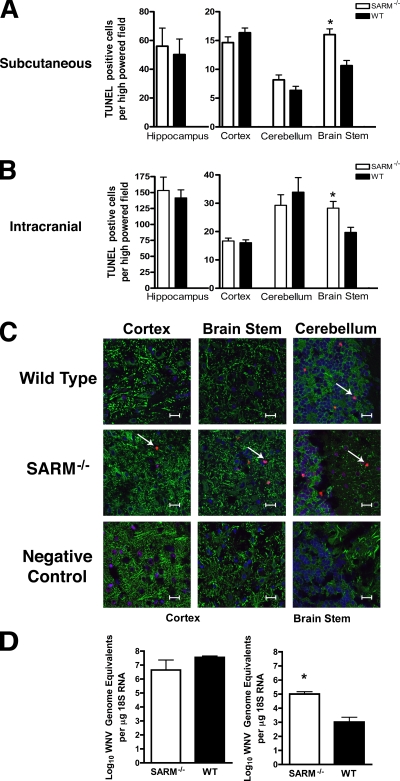
Microglia can modulate neuronal survival (13), and pathological studies of WNV infection in humans and animals have described microglial nodules apposed to infected and injured neurons (29, 45). As SARM−/− mice showed decreased microglia activation in the brain after WNV infection, we hypothesized that the decreased survival phenotype could be due to increased neuronal death or decreased clearance of infected dying neurons, which could cause bystander-cell injury. To assess this, brain sections from wild-type and SARM−/− mice were prepared at day 10 after subcutaneous WNV-NY inoculation (Fig. 7A) or day 8 after intracranial WNV-MAD infection (Fig. 7B) and the numbers of TUNEL-positive cells were evaluated in different brain regions by immunohistochemical analysis. For both routes of infection, SARM−/− mice showed higher numbers of TUNEL-positive neurons within the brainstem (Fig 7A [P < 0.001] and B [P < 0.005]), whereas there was little difference observed in the cortex, hippocampus, or cerebellum region. Confocal microscopy of frozen sections confirmed that TUNEL-positive cells costained to a great extent with the cytoplasmic neuronal marker MAP-2 and generally corroborated the immunohistochemistry data (Fig. 7C). Although no difference in infection of primary neuron cultures or overall viral titers in the brains from wild-type and SARM−/− mice was observed (Fig. 2 and 3), the neuronal death phenotype in the brainstem could be modulated by regional differences in WNV replication. Initially, to evaluate this, plaque assays were performed on brainstem homogenates from WNV-infected wild-type and SARM−/− mice. Because of the small size of the tissue, the amount of recoverable virus was at or below the limit of detection (data not shown). To overcome this, we analyzed viral RNA in infected mice by quantitative RT-PCR (Fig. 7D). WNV RNA levels were significantly elevated in the brainstem of SARM−/− mice compared to the results for wild-type mice (P < 0.01). In comparison, no statistically significant difference was observed in viral RNA in the cortex (P > 0.7). Overall, our data suggest that a deficiency of SARM results in increased neuronal death after WNV infection in a brain region-specific manner. This may be due to a cell-intrinsic effect on SARM−/− brainstem neurons that facilitates increased viral replication or, possibly, a cell-extrinsic effect in which a decrease in the number of activated microglia in the brainstem of SARM−/− mice attenuates the clearance of virus from infected neurons.
FIG. 7.
SARM protects against neuronal death during WNV infection. (A, B) Wild-type or SARM−/− mice were inoculated with either 102 PFU of WNV-NY by footpad injection (A) or 101 PFU of WNV-MAD by intracranial injection (B). Brains were harvested on day 10 or 8, respectively, and were analyzed for neuronal death by using an immunohistochemical apoptosis assay. The number of TUNEL-positive cells was quantified from 10 high-power fields per brain region per mouse for three to five independent mice. Note, fewer TUNEL-positive cells are observed after footpad than after intracranial injection. (C) Representative confocal microscopy images of MAP-2 (green; neuronal cell bodies and dendrites), terminal deoxynucleotidyl transferase (red; TUNEL, nuclear), and ToPro-3 (blue; nuclear) fluorescence staining of neurons in different brain regions in uninfected (negative control) or WNV-MAD-infected wild-type and SARM−/− mice. Arrows indicate TUNEL-positive cells. The scale bar represents ∼15 μm. Data are representative of results for three independent mice. (D) Cerebral cortex and brainstem regions of the brain were harvested and analyzed by quantitative RT-PCR for levels of WNV RNA. Data were normalized for tissue 18S RNA levels and represent the average results for 10 mice from at least two independent experiments. Error bars show standard errors of the means.
DISCUSSION
The gene for SARM (also known as Myd88-5) is a member of the Myd88 gene family and is highly conserved from insects to mammals. Unlike other TIR domain-containing proteins, SARM comprises an N-terminal HEAT/Armadillo and SAM domain that has been linked to regulation of protein-protein interaction and intracellular signaling (17, 22). The overexpression of human SARM in 293T cells showed decreased activation of NF-κB or IFN-β-regulated reporter genes after triggering by the TLR ligands LPS and poly(I:C) (3). Additionally, small interfering RNA knockdown of human SARM in 293T cells resulted in enhanced TRIF but not Myd88-dependent gene induction (3). The results of these experiments suggested that SARM negatively regulates TRIF signaling. Recent studies have questioned this function because mouse macrophages from SARM−/− mice showed no distinct phenotype in response to microbial products or TLR agonists compared to the phenotype of wild-type cells (20). Moreover, in mice, SARM was expressed preferentially in neurons and colocalized with mitochondria and Jun N-terminal protein kinase 3. As SARM−/− hippocampal neurons were protected from death after deprivation of oxygen and glucose, SARM was suggested to mediate stress-induced neuronal toxicity. Our in vivo studies with a pathogenic encephalitic flavivirus establish a novel neuroprotective function, as SARM−/− mice showed greater mortality after WNV infection, which was associated with decreased microglia activation and increased neuronal death in the brainstem.
The cell and tissue specificity of TLR signaling is modulated through the differential expression and recruitment of highly conserved adaptor proteins, which regulate the downstream activation of master transcription factors (26). Recent work has highlighted that innate immune signaling molecules play distinct roles in diverse cell types to restrict viruses (6, 16). CNS-resident cells in particular may utilize unique innate signaling programs, as neurons are a critical nonrenewing cell population. Studies to date in different model systems have reported that SARM may act as a negative regulator of TLR stimulation in vitro (2, 3, 5), have no effect on TLR responses in primary cells (20), and modulate neuronal survival (20). These data suggest that SARM likely has distinct functions in different cell types. Given its preferential expression in the CNS and putative function as a negative regulator, it seemed plausible that SARM could act to rapidly control the inflammatory response to minimize immunopathological sequelae. However, Kim et al. found that SARM−/− mouse cells had no apparent defect in response to TLR stimuli (20); our own unpublished studies with primary macrophages and dendritic cells from independently generated SARM−/− mice corroborate these findings. These contrasting results could be explained by the difference in levels of expression or species-specific functions of SARM. For example, the C. elegans ortholog of SARM, TIR-1, promotes antifungal immune function and neuronal development, whereas in Drosophila melanogaster (Ect-4) and horseshoe crabs (CrSARM), SARM appears to negatively regulate their respective TLR homologues (2, 3, 5, 28). An analogous disparity in innate immune signaling was observed when comparing transgenic overexpression data for the human LGP2 gene in 293T cells (46) with the results of functional studies of primary cells of LGP2−/− mice (44). Clearly, further studies must be performed to establish the nature of the phylogenetic differences in TLR adaptor functions of SARM proteins.
Mice lacking SARM were more susceptible to lethal WNV disease. In vivo experiments with SARM−/− mice showed blunted TNF-α responses within the CNS, decreased microglia activation, and greater brainstem-specific neuronal cell death. This phenotype was also associated with region-specific differences in WNV replication, with higher levels of viral RNA observed in the brainstem of SARM−/− mice. The data on neuronal death contrasts with the results of a recent study with independently generated knockout mice in which a lack of SARM protected against metabolic stress-induced death in hippocampal neurons (20). The disparity in results likely reflects fundamental mechanistic differences between metabolic and virus-induced cell death pathways in distinct neuron populations.
The extent of microbial infection in specific brain regions and magnitude of TNF-α production affect whether TNF-α plays a neurotoxic or neuroprotectve role in the CNS (reviewed in reference 40). A recent study demonstrated that CD8+ T cells and myeloid cells require systemic TNF-R1-dependent signals to cross the blood-brain barrier, accumulate in the brain parenchyma, and control WNV infection in the CNS (36). SARM appears to regulate the induction of TNF-α locally within the brain during pathogenic viral challenge. Interestingly, we also observed decreased TNF-α gene induction in the brainstem of TLR3−/− mice. The generation of SARM−/− × TLR3−/− and SARM−/− × TRIF−/− mice may address whether the SARM-induced signal for TNF-α production in the brain after WNV infection occurs through TLR3.
Diminished TNF-α production within the brain correlated with decreased local activation of microglia. Microglia are myeloid-lineage CNS-specific immune cells that are among the first responders to brain injury and function to secrete proinflammatory cytokines, enhance antigen presentation, destroy pathogens before they injure neurons, and clear dying cells (14). Activated microglia become juxtaposed to WNV-infected neurons in human and animal brain tissue pathological specimens, where they may have protective or pathogenic roles (4, 25, 43). Interestingly, a recent study using an attenuated lineage 2 Sarafend WNV strain suggested (36) that increases in the total number of activated microglia during infection may have a pathogenic role (12). In contrast, using a more-virulent lineage 1 WNV-NY strain, we observed a localized decrease of activated microglia in the brainstem of SARM−/− mice that was associated with increased neuronal death and mortality.
Though several factors contribute to microglial stimulation, TNF-α directly regulates activation. Mice lacking TNF-α receptors showed decreased microglia activation and correspondingly higher levels of neuronal injury in the hippocampus after treatment with a dopaminergic neurotoxin (38, 39). Our data are thus consistent with a model in which SARM regulates local production of TNF-α, which in turn modulates microglia activation, viral replication, and neuronal survival. Although SARM modulates microglia activation regionally, the cellular basis of this phenotype also remains uncertain. It could reflect a cell-autonomous regulatory effect within microglia or, given the expression pattern of SARM in the CNS, a complex circuit in which SARM regulates the production of cytokines (e.g., TNF-α) in neurons or astrocytes, resulting in cell-extrinsic effects on microglia. As an alternative model, region-specific differences in local TNF-α production could attenuate the flux of nonresident inflammatory monocyte-derived microglia across the blood-brain barrier (12); indeed, our prior studies have shown that TNF-α modulates the trafficking of monocytes into the brain, possibly by affecting early adhesion steps (36). Clearly, the specific signaling pathways that SARM uses to regulate microglia activation in the brainstem remain uncertain and an avenue for future studies.
In summary, the results of our experiments suggest that SARM modulates the host response to WNV infection in a tissue- and cell-specific manner. Thus, in contrast to its role in human cells, murine SARM in the CNS appears to positively regulate TNF-α induction, which affects microglia activation and accumulation and protects critical neuron subsets from virus-induced pathology. These studies illuminate the host and tissue-specific complexity of TLR adaptor molecules and enhance our understanding of the balance between an effective innate response and immunopathology and injury to neurons in the CNS.
Acknowledgments
This work was supported by the NIH (U54 AI057160 [Midwest Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research]), a predoctoral fellowship from the Howard Hughes Medical Institute (M.A.S.), a W. M. Keck Postdoctoral Fellowship in Molecular Medicine, and a Ruth L. Kirschstein Postdoctoral NRSA (K.J.S.).
We thank Michael White for blastocyst injections and the Speed Congenics Laboratory at Washington University School of Medicine for high-density microsatellite mapping.
Footnotes
Published ahead of print on 8 July 2009.
REFERENCES
- 1.Beasley, D. W., C. T. Davis, M. Whiteman, B. Granwehr, R. M. Kinney, and A. D. Barrett. 2004. Molecular determinants of virulence of West Nile virus in North America. Arch. Virol. Suppl. 2004:35-41. [DOI] [PubMed] [Google Scholar]
- 2.Belinda, L. W., W. X. Wei, B. T. Hanh, L. X. Lei, H. Bow, and D. J. Ling. 2008. SARM: a novel Toll-like receptor adaptor, is functionally conserved from arthropod to human. Mol. Immunol. 45:1732-1742. [DOI] [PubMed] [Google Scholar]
- 3.Carty, M., R. Goodbody, M. Schroder, J. Stack, P. N. Moynagh, and A. G. Bowie. 2006. The human adaptor SARM negatively regulates adaptor protein TRIF-dependent Toll-like receptor signaling. Nat. Immunol. 7:1074-1081. [DOI] [PubMed] [Google Scholar]
- 4.Cheeran, M. C., S. Hu, W. S. Sheng, A. Rashid, P. K. Peterson, and J. R. Lokensgard. 2005. Differential responses of human brain cells to West Nile virus infection. J. Neurovirol. 11:512-524. [DOI] [PubMed] [Google Scholar]
- 5.Couillault, C., N. Pujol, J. Reboul, L. Sabatier, J. F. Guichou, Y. Kohara, and J. J. Ewbank. 2004. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat. Immunol. 5:488-494. [DOI] [PubMed] [Google Scholar]
- 6.Daffis, S., M. A. Samuel, B. C. Keller, M. Gale, Jr., and M. S. Diamond. 2007. Cell-specific IRF-3 responses protect against West Nile virus infection by interferon-dependent and -independent mechanisms. PLoS Pathog. 3:e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daffis, S., M. A. Samuel, M. S. Suthar, M. Gale, Jr., and M. S. Diamond. 2008. Toll-like receptor 3 has a protective role against West Nile virus infection. J. Virol. 82:10349-10358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daffis, S., M. A. Samuel, M. S. Suthar, B. C. Keller, M. Gale, Jr., and M. S. Diamond. 2008. Interferon regulatory factor IRF-7 induces the antiviral alpha interferon response and protects against lethal West Nile virus infection. J. Virol. 82:8465-8475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diamond, M. S., B. Shrestha, A. Marri, D. Mahan, and M. Engle. 2003. B cells and antibody play critical roles in the immediate defense of disseminated infection by West Nile encephalitis virus. J. Virol. 77:2578-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ebel, G. D., J. Carricaburu, D. Young, K. A. Bernard, and L. D. Kramer. 2004. Genetic and phenotypic variation of West Nile virus in New York, 2000-2003. Am. J. Trop. Med. Hyg. 71:493-500. [PubMed] [Google Scholar]
- 11.Ford, A. L., E. Foulcher, F. A. Lemckert, and J. D. Sedgwick. 1996. Microglia induce CD4 T lymphocyte final effector function and death. J. Exp. Med. 184:1737-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Getts, D. R., R. L. Terry, M. T. Getts, M. Muller, S. Rana, B. Shrestha, J. Radford, N. Van Rooijen, I. L. Campbell, and N. J. King. 2008. Ly6c+ “inflammatory monocytes” are microglial precursors recruited in a pathogenic manner in West Nile virus encephalitis. J. Exp. Med. 205:2319-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glezer, I., A. R. Simard, and S. Rivest. 2007. Neuroprotective role of the innate immune system by microglia. Neuroscience 147:867-883. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez-Scarano, F., and G. Baltuch. 1999. Microglia as mediators of inflammatory and degenerative diseases. Annu. Rev. Neurosci. 22:219-240. [DOI] [PubMed] [Google Scholar]
- 15.Hayes, E. B., J. J. Sejvar, S. R. Zaki, R. S. Lanciotti, A. V. Bode, and G. L. Campbell. 2005. Virology, pathology, and clinical manifestations of West Nile virus disease. Emerg. Infect. Dis. 11:1174-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishii, K. J., S. Koyama, A. Nakagawa, C. Coban, and S. Akira. 2008. Host innate immune receptors and beyond: making sense of microbial infections. Cell Host Microbe 3:352-363. [DOI] [PubMed] [Google Scholar]
- 17.Jault, C., L. Pichon, and J. Chluba. 2004. Toll-like receptor gene family and TIR-domain adapters in Danio rerio. Mol. Immunol. 40:759-771. [DOI] [PubMed] [Google Scholar]
- 18.Kawai, T., and S. Akira. 2006. Innate immune recognition of viral infection. Nat. Immunol. 7:131-137. [DOI] [PubMed] [Google Scholar]
- 19.Keller, B. C., B. L. Fredericksen, M. A. Samuel, R. E. Mock, P. W. Mason, M. S. Diamond, and M. Gale, Jr. 2006. Resistance to alpha/beta interferon is a determinant of West Nile virus replication fitness and virulence. J. Virol. 80:9424-9434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, Y., P. Zhou, L. Qian, J. Z. Chuang, J. Lee, C. Li, C. Iadecola, C. Nathan, and A. Ding. 2007. MyD88-5 links mitochondria, microtubules, and JNK3 in neurons and regulates neuronal survival. J. Exp. Med. 204:2063-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klein, R. S., E. Lin, B. Zhang, A. D. Luster, J. Tollett, M. A. Samuel, M. Engle, and M. S. Diamond. 2005. Neuronal CXCL10 directs CD8+ T-cell recruitment and control of West Nile virus encephalitis. J. Virol. 79:11457-11466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mink, M., and K. Csiszar. 2005. SARM1: a candidate gene in the onset of hereditary infectious/inflammatory diseases. Clin. Immunol. 115:333-334. [DOI] [PubMed] [Google Scholar]
- 23.Mink, M., B. Fogelgren, K. Olszewski, P. Maroy, and K. Csiszar. 2001. A novel human gene (SARM) at chromosome 17q11 encodes a protein with a SAM motif and structural similarity to Armadillo/beta-catenin that is conserved in mouse, Drosophila, and Caenorhabditis elegans. Genomics 74:234-244. [DOI] [PubMed] [Google Scholar]
- 24.Oliphant, T., M. Engle, G. E. Nybakken, C. Doane, S. Johnson, L. Huang, S. Gorlatov, E. Mehlhop, A. Marri, K. M. Chung, G. D. Ebel, L. D. Kramer, D. H. Fremont, and M. S. Diamond. 2005. Development of a humanized monoclonal antibody with therapeutic potential against West Nile virus. Nat. Med. 11:522-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Omalu, B. I., A. A. Shakir, G. Wang, W. I. Lipkin, and C. A. Wiley. 2003. Fatal fulminant pan-meningo-polioencephalitis due to West Nile virus. Brain Pathol. 13:465-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Neill, L. A., and A. G. Bowie. 2007. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat. Rev. Immunol. 7:353-364. [DOI] [PubMed] [Google Scholar]
- 27.Purtha, W. E., N. Myers, V. Mitaksov, E. Sitati, J. Connolly, D. H. Fremont, T. H. Hansen, and M. S. Diamond. 2007. Antigen-specific cytotoxic T lymphocytes protect against lethal West Nile virus encephalitis. Eur. J. Immunol. 37:1845-1854. [DOI] [PubMed] [Google Scholar]
- 28.Raha, D., Q. D. Nguyen, and A. Garen. 1990. Molecular and developmental analyses of the protein encoded by the Drosophila gene ectodermal. Dev. Genet. 11:310-317. [DOI] [PubMed] [Google Scholar]
- 29.Sampson, B. A., and V. Armbrustmacher. 2001. West Nile encephalitis: the neuropathology of four fatalities. Ann. N. Y. Acad. Sci. 951:172-178. [PubMed] [Google Scholar]
- 30.Samuel, M. A., and M. S. Diamond. 2005. Alpha/beta interferon protects against lethal West Nile virus infection by restricting cellular tropism and enhancing neuronal survival. J. Virol. 79:13350-13361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Samuel, M. A., and M. S. Diamond. 2006. Pathogenesis of West Nile Virus infection: a balance between virulence, innate and adaptive immunity, and viral evasion. J. Virol. 80:9349-9360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samuel, M. A., J. D. Morrey, and M. S. Diamond. 2007. Caspase 3-dependent cell death of neurons contributes to the pathogenesis of West Nile virus encephalitis. J. Virol. 81:2614-2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samuel, M. A., K. Whitby, B. C. Keller, A. Marri, W. Barchet, B. R. Williams, R. H. Silverman, M. Gale, Jr., and M. S. Diamond. 2006. PKR and RNase L contribute to protection against lethal West Nile Virus infection by controlling early viral spread in the periphery and replication in neurons. J. Virol. 80:7009-7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sato, M., H. Suemori, N. Hata, M. Asagiri, K. Ogasawara, K. Nakao, T. Nakaya, M. Katsuki, S. Noguchi, N. Tanaka, and T. Taniguchi. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity 13:539-548. [DOI] [PubMed] [Google Scholar]
- 35.Shrestha, B., D. Gottlieb, and M. S. Diamond. 2003. Infection and injury of neurons by West Nile encephalitis virus. J. Virol. 77:13203-13213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shrestha, B., B. Zhang, W. E. Purtha, R. S. Klein, and M. S. Diamond. 2008. Tumor necrosis factor alpha protects against lethal West Nile virus infection by promoting trafficking of mononuclear leukocytes into the central nervous system. J. Virol. 82:8956-8964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sitati, E., E. E. McCandless, R. S. Klein, and M. S. Diamond. 2007. CD40-CD40 ligand interactions promote trafficking of CD8+ T cells into the brain and protection against West Nile virus encephalitis. J. Virol. 81:9801-9811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sriram, K., J. M. Matheson, S. A. Benkovic, D. B. Miller, M. I. Luster, and J. P. O'Callaghan. 2006. Deficiency of TNF receptors suppresses microglial activation and alters the susceptibility of brain regions to MPTP-induced neurotoxicity: role of TNF-alpha. FASEB J. 20:670-682. [DOI] [PubMed] [Google Scholar]
- 39.Sriram, K., J. M. Matheson, S. A. Benkovic, D. B. Miller, M. I. Luster, and J. P. O'Callaghan. 2002. Mice deficient in TNF receptors are protected against dopaminergic neurotoxicity: implications for Parkinson's disease. FASEB J. 16:1474-1476. [DOI] [PubMed] [Google Scholar]
- 40.Sriram, K., and J. P. O'Callaghan. 2007. Divergent roles for tumor necrosis factor-alpha in the brain. J. Neuroimmune Pharmacol. 2:140-153. [DOI] [PubMed] [Google Scholar]
- 41.Thompson, A. J., and S. A. Locarnini. 2007. Toll-like receptors, RIG-I-like RNA helicases and the antiviral innate immune response. Immunol. Cell Biol. 85:435-445. [DOI] [PubMed] [Google Scholar]
- 42.Town, T., F. Bai, T. Wang, A. T. Kaplan, F. Qian, R. R. Montgomery, J. F. Anderson, R. A. Flavell, and E. Fikrig. 2009. Toll-like receptor 7 mitigates lethal West Nile encephalitis via interleukin 23-dependent immune cell infiltration and homing. Immunity 30:242-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Town, T., D. Jeng, L. Alexopoulou, J. Tan, and R. A. Flavell. 2006. Microglia recognize double-stranded RNA via TLR3. J. Immunol. 176:3804-3812. [DOI] [PubMed] [Google Scholar]
- 44.Venkataraman, T., M. Valdes, R. Elsby, S. Kakuta, G. Caceres, S. Saijo, Y. Iwakura, and G. N. Barber. 2007. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J. Immunol. 178:6444-6455. [DOI] [PubMed] [Google Scholar]
- 45.Xiao, S. Y., H. Guzman, H. Zhang, A. P. Travassos da Rosa, and R. B. Tesh. 2001. West Nile virus infection in the golden hamster (Mesocricetus auratus): a model for West Nile encephalitis. Emerg. Infect. Dis. 7:714-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoneyama, M., M. Kikuchi, K. Matsumoto, T. Imaizumi, M. Miyagishi, K. Taira, E. Foy, Y. M. Loo, M. Gale, Jr., S. Akira, S. Yonehara, A. Kato, and T. Fujita. 2005. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 175:2851-2858. [DOI] [PubMed] [Google Scholar]
- 47.Zhang, B., L. Yang, Y. Konishi, N. Maeda, M. Sakanaka, and J. Tanaka. 2002. Suppressive effects of phosphodiesterase type IV inhibitors on rat cultured microglial cells: comparison with other types of cAMP-elevating agents. Neuropharmacology 42:262-269. [DOI] [PubMed] [Google Scholar]