Abstract
Hepatitis C virus (HCV) RNA genome replicates within the ribonucleoprotein (RNP) complex in the modified membranous structures extended from endoplasmic reticulum. A proteomic analysis of HCV RNP complexes revealed the association of oxysterol binding protein (OSBP) as one of the components of these complexes. OSBP interacted with the N-terminal domain I of the HCV NS5A protein and colocalized to the Golgi compartment with NS5A. An OSBP-specific short hairpin RNA that partially downregulated OSBP expression resulted in a decrease of the HCV particle release in culture supernatant with little effect on viral RNA replication. The pleckstrin homology (PH) domain located in the N-terminal region of OSBP targeted this protein to the Golgi apparatus. OSBP deletion mutation in the PH (ΔPH) domain failed to localize to the Golgi apparatus and inhibited the HCV particle release. These studies suggest a possible functional role of OSBP in the HCV maturation process.
Hepatitis C virus (HCV) infection is one of the leading causes of chronic hepatitis. HCV infection is associated with cirrhosis, steatosis, and hepatocellular carcinoma (33). The HCV RNA genome of ∼9.6 kb is translated via an internal ribosome entry site element on the rough endoplasmic reticulum (ER) as a polyprotein precursor of about 3,010 amino acids that is co- and posttranslationally processed by cellular and viral proteases into mature structural and nonstructural (NS) proteins (33). HCV replicates within ribonucleoprotein (RNP) complexes associated with modified ER membranous structures (15). Recent work implicated lipid droplets that emanate from the ER as sites of RNA replication (28, 44). Almost all of the HCV NS proteins along with a variety of cellular factors are associated with the RNP complexes engaged in viral RNA replication (37). It is likely that these NS proteins not only participate in replication process but also are involved in the various steps of virion morphogenesis and assembly. Membrane-associated RNP complexes are generally composed of viral proteins, replicating RNA, host proteins, and altered cellular membranes (1). In this respect, a growing body of evidence implicates the functional role of NS5A in early steps of virion assembly and morphogenesis (3, 27, 45). NS5A is a phosphoprotein that migrates in sodium dodecyl sulfate gels as 56-kDa (basally phosphorylated) and 58-kDa (hyperphosphorylated) forms of proteins. The C-terminal domain III region of NS5A and the phosphorylated residue (Ser457) are important for virion maturation (3, 27, 45). NS5A domain III contains the binding site for viral core protein, indicating the possible involvement of NS5A protein in virus assembly (27). NS5A anchors to the ER membrane by an N-terminal hydrophobic α-helix, and this attachment is needed for its key role(s) in viral replication (10). Studies suggest that phosphorylation of NS5A plays a functional role in viral replication (12). The hyperphosphorylated NS5A reduces its interaction with the human vesicle-associated membrane protein-associated protein A (VAP-A) (12). VAP-A binds both NS5A and NS5B (13, 17). These associations are important for RNA replication (13, 17).
HCV alters lipid homeostasis to benefit its infectious processes. Host lipids and their synthesis affect viral infectious process (21, 40, 51, 57). HCV RNA replication can be induced by saturated and monounsaturated fatty acids and inhibited by polyunsaturated fatty acids (18, 21). HCV gene expression induces lipogenesis by stimulating the activation of the sterol regulatory element binding proteins, the master regulators of lipid/fatty acid biosynthetic pathways (51). Reagents that interfere with host lipid biosynthetic pathways abrogate viral replication (21, 57). It has been suggested that HCV utilizes the very-low-density lipoprotein (VLDL) secretion pathway for its viral particle release (14, 19). These studies collectively suggest that host lipid metabolism plays a key role in the viral life cycle including replication, virion assembly, and secretion (56).
In the present study, we focus on the functional role of oxysterol binding protein (OSBP) that was identified by proteomic analysis as one of the host factors associated with the HCV RNP complexes. OSBP belongs to a family of the OSBP-related proteins. Originally discovered as a major cytosolic receptor for oxidized cholesterols, it undergoes translocation from the cytosolic/vesicular compartment to the Golgi apparatus upon ligand (hydroxycholesterol) binding (38). OSBP also binds to VAP-A via its FFAT motif (53). Golgi apparatus translocation of OSBP is regulated by the pleckstrin homology (PH) domain. This domain also harbors binding sites for phosphatidylinositol 4-phosphate (PI4P) and phosphatidylinositol 4,5-biphosphate (PI4,5P2) (25). OSBP and OSBP-related proteins are implicated in cholesterol homeostasis, phospholipid metabolism, vesicular transport, and cell signaling (55). OSBP functions as sterol sensor that regulates the transport of ceramide from the ER to the Golgi apparatus for de novo synthesis of sphingomyelin by coordinated action with ceramide transport protein (CERT) (36). OSBP also functions as a scaffolding protein for two phosphatases (phosphatase 2A/HePTP) (49). This complex regulates the activity of extracellular signal-regulate kinase. This cytosolic 440-kDa complex disassembles by the addition of 25-hydroxycholesterol (25-HC) or depletion of cholesterol, both of which cause OSBP translocation to the Golgi compartment (49). Thus, in addition to its role in intracellular trafficking, OSBP appears to regulate cell signaling. We investigated the functional significance of OSBP association with HCV RNP complexes. RNA interference studies support a functional role of OSBP in virion morphogenesis and release process. The OSBP PH domain deletion mutant (ΔPH) failed to localize to the Golgi apparatus and caused an inhibition of the HCV particle release. Our work described herein also demonstrates that the association of OSBP with NS5A may also contribute to the overall HCV maturation process.
MATERIALS AND METHODS
Plasmids.
The plasmids pJFH1, pSGR-JFH1, and pSGR-Luc-JFH1 were the generous gift of T. Wakita (22, 48). pSGR-JFH1-5A1ST, in which NS5A gene contains the One-STrEP tag (IBA, Göttingen, Germany), was generated by PCR-mediated mutagenesis using oligonucleotides described in Table S1 in the supplemental material. pFL-Luc-Jc1, an analogue to Luc-Jc1 (23), was constructed as described in Table S1 in the supplemental material. For constructing the human OSBP expression vector pFLAG-CMV-OSBP (where CMV is cytomegalovirus), OSBP1 cDNA was amplified from total RNA extracted from Huh7 cells by reverse transcription-PCR (RT-PCR), using oligonucleotides described in Table S1 in the supplemental material, and amplified product was digested with both HindIII and BamHI and cloned into the corresponding sites in the pFLAG-CMV2 vector (Sigma-Aldrich, St. Louis, MO). Expression vectors for mutant forms of OSBPs were constructed as described in Table S1 in the supplemental material. The VAP-A expression vector, pEF-FLAG-VAP-A, was kindly provided by Y. Matsuura (17). Generation of NS5A deletion mutants has been described previously (20). The NS5A derived from HCV JFH1 (genotype 2a) was cloned in the pEF1/Myc-His vector (Invitrogen, Carlsbad, CA) at the KpnI and XbaI sites by using a PCR-amplified fragment using primers described in Table S1 in the supplemental material. Lentiviral vectors, L-CMV-GFP-NheI (where GFP is green fluorescent protein), and packaging plasmids pMDL, pVSV-G (where VSV-G is vesicular stomatitis virus glycoprotein), and pREV were kindly provided by I. Verma (Salk Institute, La Jolla, CA) (46) and used for cloning short hairpin RNAs (shRNAs). The oligonucleotides used for constructing a scrambled shRNA (unrelated) and the OSBP-specific shRNA-1 and shRNA-2 are described in Table S1 in the supplemental material.
Cell culture.
Human hepatoma cell lines Huh7 and Huh7.5.1 were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin, and 1 mM minimal essential medium with nonessential amino acids (Invitrogen, Carlsbad, CA). The Huh7.5.1 cell line was a kind gift of F. Chisari (Scripps Institute, La Jolla, CA).
In vitro RNA transcription and RNA transfection.
The plasmid encoding HCV (JFH1) was linearized by XbaI digestion, followed by mung bean nuclease treatment to blunt the XbaI-digested termini, and served as a template for RNA synthesis using a RiboMAX Large Scale RNA production system-T7 (Promega, Madison, WI). Synthesized RNAs were extracted by the acid guanidium thiocyanate phenol-chloroform (AGPC) method prior to transfection (7). Electroporation was used for RNA transfection. Subconfluent Huh7 or Huh7.5.1 cells were trypsinized and washed with ice-cold phosphate-buffered saline (PBS) and resuspended at 1 × 107 cells/ml in Cytomix buffer (47). Synthesized RNAs were mixed in 0.4 ml of cell suspension in a 4-mm gap electroporation cuvette (Genesee Scientific, San Diego, CA), pulsed at 260 V for 25 ms in a square wave mode of Gene Pulser Xcell electroporation system (Bio-Rad, Hercules, CA). For developing subgenomic replicon cell lines, Huh7 cells were transfected with in vitro synthesized replicon RNAs, incubated for 48 h in complete medium, and maintained in the presence of G418 (300 μg/ml). Stably expressing subgenomic replicon colonies were isolated after 3 weeks of growth in the presence of G418.
Proteomics analysis.
Subgenomic replicon cell lines were established by transfecting SGR-JFH1 or SGR-JFH1-5A1ST RNA into Huh7 cells as described above, and isolated cell lines were designated SGR-JFH1 and SGR-JFH1-5A1ST, respectively. The NS5A protein complexes were purified by affinity chromatography from whole-cell lysates of the SGR-JFH1-5A1ST clone. Briefly, semiconfluent cell monolayers on 100-mm dishes were lysed with 4 ml of lysis buffer (0.2% deoxycholic acid, 0.25 M sucrose, 0.1 mM EDTA, and 3 mM Tris-HCl, pH 7.4) and incubated on ice for 20 min. The protein lysates were centrifuged at 25,000 × g for 30 min at 4°C. The cleared lysates were dialyzed against binding buffer (100 mM Tris-HCl, 150 mM NaCl, and 0.1% octylglucoside) and centrifuged again at 25,000 × g for 30 min at 4°C. The cleared lysates were loaded onto a StrepTactin-Sepharose column (1-ml bed volume; IBA) by filtering through 0.45-μm-pore-size polyethersulfone filter (Corning) and fractionated according to the manufacturer's instructions. The peak NS5A fraction from 30 preparations (12.5 μg) was collected and concentrated by acetone-methanol precipitation and subjected to multidimensional protein identification technology (MudPIT) analysis (5, 8, 9, 11, 26, 34, 39, 41). For the details of MudPIT analysis, see Materials and Methods in the supplemental material.
HCV infection and focus-forming unit assay.
HCV JFH1 strain (48) was used for the production of HCV infectious viral particles. Huh7.5.1 cells were transfected with the in vitro synthesized RNA by electroporation. Ten days posttransfection cultured supernatants were collected. The infectious virion titers of collected supernatants were determined by a focus-forming-unit assay as described previously (58). HCV infection was performed at a multiplicity of infection (MOI) of 1.
Real-time RT-PCR.
Total cellular RNAs were purified by the AGPC method (7). Viral RNAs were extracted from 100 μl of supernatant by the AGPC method. Five micrograms of Saccharomyces cerevisiae tRNA was added as a carrier (Sigma-Aldrich). HCV RNA was quantified on an ABI Prism 7000 sequence detection system (Applied Biosystems, Foster City, CA) as described previously (43). For the quantification of OSBP mRNA, 100 ng of total cellular RNA was subjected to cDNA synthesis using Improm II reverse transcriptase (Promega) oligo(dT) as a primer. Molecular copy number of OSBP and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA was quantified by real-time PCR using SYBR premix Ex Taq (Takara Mirus Bio, Madison, WI) in an absolute quantification manner. The following set of primers was used for quantitative PCR: OSBP sense, 5′-AGAATACCCTTCGGACCCTCTC-3′; OSBP antisense, 5′-TCTTTTCATTGCTCTCAGCAGG-3′; GAPDH sense, 5′-GCCATCAATGACCCCTTCATT-3′; and GAPDH antisense, 5′-TTGACGGTGCCATGGAATTT-3′.
Western blotting analysis and immunoprecipitation.
Huh7 cells or HCV-infected Huh7 cells were transfected with the indicated expression vectors by lipofection using TransIT-LT1 reagent (Mirus Bio, Madison, WI). Cells grown in a 35-mm dish were transiently transfected and incubated for 24 h, washed once with ice-cold PBS, and lysed on ice with 500 μl of lysis buffer (0.2% [wt/vol] deoxycholic acid, 20 mM Tris-HCl, 150 mM NaCl, 0.1 mM EDTA, 250 mM sucrose), supplemented with 1× Halt protease inhibitor single-use cocktail (Thermo Scientific, Rockford, IL). The cellular lysates were subjected to brief sonication, followed by centrifugation at 13,400 × g for 10 min. The clarified lysates were incubated with 5 μg of anti-FLAG M2 monoclonal antibody (Sigma-Aldrich) for 1 h. Five microliters of protein G-Sepharose 4 Fast Flow was added (GE Healthcare, Piscataway, NJ) for 2 h. Sepharose beads were collected by centrifugation and washed four times with 1 ml of lysis buffer. The immunoprecipitates were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Immunoblotting analysis was performed as described previously (52). The following antibodies were used for Western blotting analysis: mouse monoclonal anticore (Affinity Bioreagents, Golden, CO), goat polyclonal anti-OSBP (Novus biological, Littleton, CO), goat polyclonal anti-ApoB (Chemicon International, Temecula, CA), rabbit polyclonal anti-human albumin (MP Biomedicals, Solon, OH), rabbit polyclonal anti-adipose differentiation-related protein (Novus Biological), mouse monoclonal anti-fatty acid synthase (BD Bioscience), mouse monoclonal anti-NS3 (Virogen), and monoclonal antibody 9E10/A3 for NS5A (a generous gift from C. Rice).
Immunofluorescence microscopy.
Infected and/or transfected cells were grown on glass coverslips, washed twice with PBS, and fixed in 3% paraformaldehyde in PBS supplemented with 2 mM MgCl2 and 1.25 mM EGTA for 20 m at room temperature. Fixed cells were permeabilized and blocked in antibody-binding buffer (PBS, 0.2% [wt/vol] saponin, 0.2% [wt/vol] nonfat dry milk, 1% [wt/vol] bovine serum albumin, and 0.02% sodium azide) for 2 h at 4°C. Fixed cells were incubated in the presence of primary antibodies overnight. The primary antibodies used for immunofluorescence were mouse monoclonal 9E10/A3 for NS5A, goat polyclonal anti-OSBP (Novus), rabbit polyclonal anti-TGN46 (trans-Golgi network protein 46 kDa; Sigma-Aldrich), and rabbit polyclonal anti-FLAG (Anaspec, San Jose, CA) to detect FLAG-tagged proteins. Coverslips were washed three times with PBS and once with antibody-binding buffer, followed by the incubation with corresponding secondary antibodies for the detection of primary antibodies. These were donkey antibodies against mouse (DyLight 488), rabbit (DyLight 549), and goat (DyLight 649; Rockland Immunochemicals, Gilbertsville, PA). The slides were analyzed by confocal laser scanning microscopy (Zeiss LSM 510), using 488-nm, 543-nm, and 633-nm laser lines. The nuclei were counterstained by DAPI (4′,6′-diamidino-2-phenylindole) (Invitrogen, Carlsbad, CA).
RESULTS
Identification of host factors associated with RNP complexes.
To identify host factors involved in HCV RNA replication, we employed an affinity-based purification procedure to isolate RNP complexes under nondenaturing conditions. We inserted an affinity tag peptide called One-STrEP-tag (28 amino acid residues) in the C-terminal region of NS5A of subgenomic replicon constructs (see Fig. S1A in the supplemental material) and established stable replicon cell lines. This insertion did not interfere with the replication of HCV replicon RNA (see Fig. S1B in the supplemental material) and/or developing G418-resistant clones (see Fig. S1C in the supplemental material). We developed, a one-step, nondenaturing affinity column chromatography procedure for the isolation of NS5A-binding proteins or protein complexes. Purified protein complexes were analyzed by Western blot assay (see Fig. S2A and B in the supplemental material). Viral protein complexes purified by this method were subjected to proteomic MudPIT analysis, which revealed a large repertoire of cellular factors (see Tables S2 to S7 in the supplemental material) and included all the viral NS proteins. Western blot analysis of the isolated proteins demonstrated the association of host factors that were identified by proteomic analysis (see Fig. S2C, right panels, in the supplemental material). In this analysis, we chose proteins whose peptide spectrum counts were significantly high. Figure S2C in the supplemental material shows the association of ApoB, OSBP, adipose differentiation-related protein, and fatty acid synthase along with the representative HCV NS proteins, NS5A and NS3. In contrast, these proteins, including NS5A, were not detected in the eluted fractions prepared from wild-type replicon (without the One-STrEP-tag) lysates (see Fig. S2C, left panels, in the supplemental material). In this study, we focused on the association of OSBP and investigated its functional relevance in the cycles of HCV infection.
Downregulation of OSBP protein affects HCV replication and viral particle release.
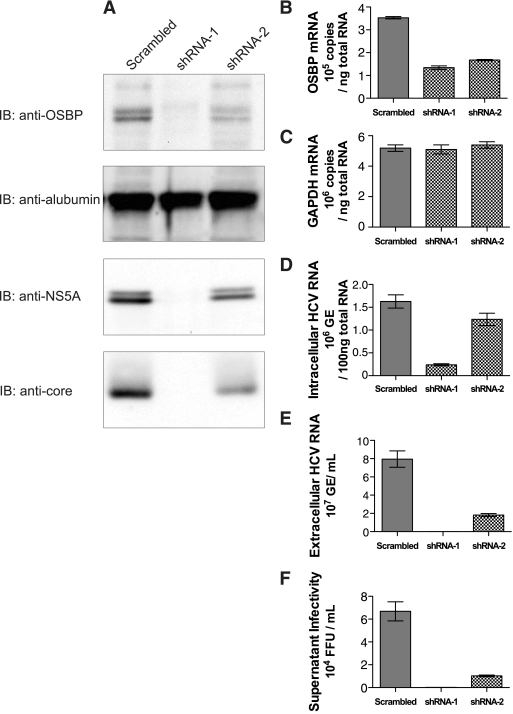
To demonstrate the functional role of this interaction, we employed RNA interference strategy. We developed lentiviral vectors encoding two OSBP-specific shRNAs (shRNA-1 and -2). These shRNAs have been previously reported to suppress OSBP mRNA synthesis effectively (31, 36). Huh7.5.1 cells were first infected with either of the lentiviral particle-encoding shRNAs and subsequently challenged with tissue culture-grown HCV particles at an MOI of 1. A lentivirus encoding a scrambled shRNA was used as a negative control. OSBP expression was monitored by Western blot assays (Fig. 1A) and by RT-PCR analysis (Fig. 1B). Cells expressing OSBP shRNA-1 and -2 displayed differential levels of suppression of OSBP protein (Fig. 1A, top). OSBP protein was barely detectable in shRNA-1-expressing cells. On the other hand, the reduction of OSBP was modest in shRNA-2-expressing cells. OSBP protein appears as a doublet representing differentially phosphorylated forms. Albumin levels were uniformly expressed in these lysates (Fig. 1A, second panel). We examined the expression of viral core and NS5A proteins, which reflected the profile of OSBP protein expression. Both viral proteins were barely detectable in shRNA-1-expressing cells, where OSBP proteins were severely depleted, indicating a functional role of OSBP in viral translation/replication. shRNA-2, on the other hand, modestly reduced their expression (Fig. 1, lower panels). We next performed quantitative RT-PCR analyses of OSBP mRNA and viral RNAs isolated from cellular extracts (intracellular) and culture supernatants (extracellular). Intracellular RNA levels indicate viral replication, whereas extracellular viral RNA is a measure of released viral particles in the culture supernatant. The results show that both shRNAs were effective in suppressing OSBP mRNA synthesis (63% and 53%, respectively, for shRNA-1 and -2) without affecting GAPDH mRNA levels (Fig. 1B and C). The unrelated scrambled shRNA (control) did not have any effect on OSBP mRNA synthesis (Fig. 1C). shRNA-1, which was more potent in silencing the OSBP mRNA (63% reduction) (Fig. 1B), significantly suppressed intracellular levels of HCV RNA (85% reduction) (Fig. 1D). shRNA-2, on the other hand, which caused a modest reduction of OSBP mRNA (53%) compared to the shRNA-1 (Fig. 1B), reduced the intracellular HCV RNA replication levels only modestly (14%) (Fig. 1D). When levels of extracellular virion RNA were analyzed, both shRNA-1 and -2 effectively blocked the release of viral particles in the culture medium (99.8% and 87.2% reduction, respectively) (Fig. 1E). More importantly, the shRNA-2, which has little effect on intracellular HCV RNA replication levels, dramatically reduced HCV release (Fig. 1E). We further confirmed these results by determining the number of focus-forming units of the cultured supernatant as described previously (58). The culture supernatant from HCV-infected cells, which was previously infected with lentiviral vector expressing scrambled (negative control) shRNA, yielded 6 × 104 focus-forming units per ml of supernatant, whereas supernatants from shRNA-1- and shRNA-2-expressing cells displayed significantly reduced infectious virion titers, indicating that OSBP depletion effectively attenuated the rate of infectious virion release (99.8% and 84.7%, respectively) (Fig. 1F). This result confirms the previous observation that shRNA-2, which does not affect HCV replication levels (intracellular levels), dramatically affected the accumulation of extracellular viral particles (Fig. 1, compare D with E and F). Taken together, these results indicate that OSBP expression is required for both viral replication and release of viral particles.
FIG. 1.
Effect of silencing OSBP on HCV replication and viral particle release. Huh7.5.1 cells were infected with lentiviral vectors encoding an shRNA expression cassette of scrambled shRNA, shRNA-1, and shRNA-2. Four days after lentiviral infection, cells were infected with HCV (JFH1) at an MOI of 1. (A) Western blot analysis of HCV-infected cells with indicated shRNAs at day 6. IB, immunoblotting carried out using indicated antibodies. (B) Quantitative RT-PCR analysis of OSBP mRNA. (C) Quantitative RT-PCR analysis of GAPDH mRNA. (D) Intracellular HCV RNA levels measured by quantitative RT-PCR analysis. (E) Accumulation of extracellular HCV viral RNA in the culture medium as measured by quantitative RT-PCR analysis. (F) Supernatant infectivity assay. The infectivity of cultured supernatant at day 6 was determined as described in Materials and Methods and previously (58). Means and standard errors of at least triplicate measurements are shown. GE, genomic copies.
OSBP affects virus secretion.
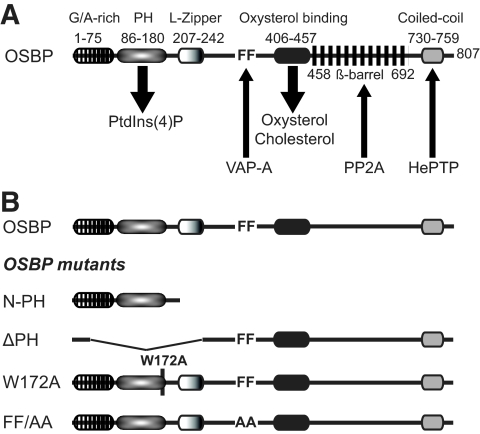
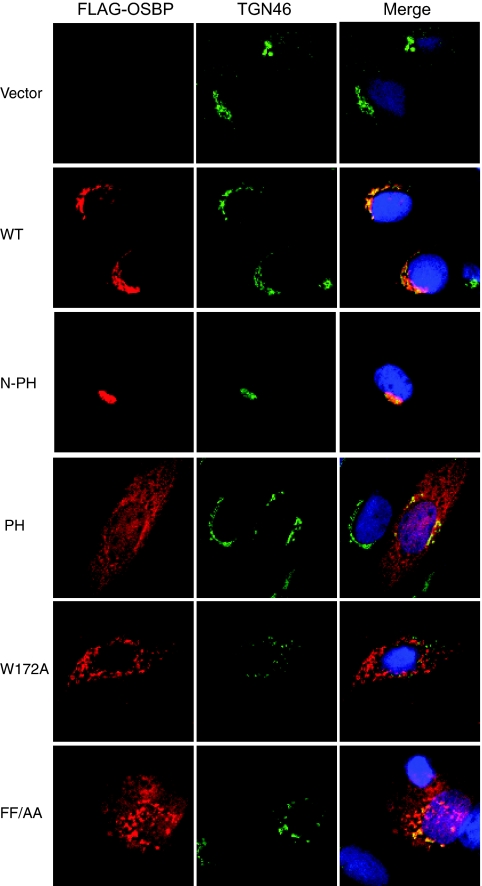
One of the unique characteristics of the OSBP protein is its ability to translocate to the Golgi apparatus upon ligand binding. The translocalization of OSBP is regulated by its PH domain, which binds to Golgi lipids such as PI4P (Fig. 2A). The RNA interference studies described above suggest that partial depletion of OSBP can cause inhibition of viral particle secretion. To elucidate the functional role of OSBP in virion maturation processes further, we generated several mutants of OSBP (Fig. 2B). These include an N-terminal region of OSBP containing only the PH domain fragment (N-PH), a deletion in the PH domain (ΔPH), a base substitution mutation in the PH domain (W172A) (53), and mutation of FF to AA in the FFAT motif (FF/AA) (54) (Fig. 2B). Wild-type OSBP and the mutants were transiently transfected into Huh7 cells and analyzed for their subcellular distribution by indirect fluorescence microscopy using anti-FLAG antibody (OSBP and all OSBP mutants contain a FLAG tag at their N termini). Cells were counterstained with TGN46 antibody and DAPI. Both wild-type OSBP and the PH domain-containing mutant (N-PH) predominantly localized to the Golgi compartment (Fig. 3). The N-PH mutant displayed Golgi compartment localizations as a consequence of its exposed PH domain. Other mutants including ΔPH and the base substitution mutants (W172A and FF/AA) in general displayed a diffuse cytosolic pattern of OSBP distribution. Both W172A and FF/AA mutants showed a punctate pattern of OSBP distribution in the cytosol and appeared to induce a distortion of TGN (Fig. 3). The OSBP mutant FF/AA displayed partial Golgi compartment localization. The substitutions of alanines for two phenylalanine residues in the FFAT motif abolish its ability to bind VAP-A (53, 54). VAP-A has been previously shown to bind NS5A (13, 17).
FIG. 2.
Schematic representation of various OSBP domains. (A) The domain organizations of OSBP and amino acid regions are indicated on the figure. G/A-rich, glycine- and alanine-rich region; PH, binds to PI4P [PtdIns(4)P]; L-Zipper, leucine zipper domain; FF, FFAT; oxysterol-binding domain, binds to oxysterols and cholesterols. The numbering of amino acids is based on human OSBP (NM_002556). (B) OSBP mutants. The coding for the N-PH mutant extends from aa 1 to 208. The ΔPH mutant lacks aa 35 to 273. Mutant W172A carries a point mutation of a conserved tryptophan residue at 172 within the PH domain. Mutant FF/AA contains two phenylalanine residues in the FFAT motif replaced with alanine residues, required for VAP-A association. PP2A, phosphatase 2A; HePTP, tyrosine phosphatase.
FIG. 3.
Subcellular localization of OSBP and OSBP mutants. Huh7 cells were transfected with vectors encoding the FLAG-tagged wild-type (WT) and indicated mutant OSBP genes. Cells were analyzed by confocal immunofluorescence microscopy with anti-FLAG and anti-TGN46 antibodies (see Materials and Methods). Panels in the left column show FLAG-tagged OSBP (red). Panels in the center column show subcellular localization of TGN46 as a marker of the TGN. Panels in the right column show superimposed images of OSBP and TGN46.
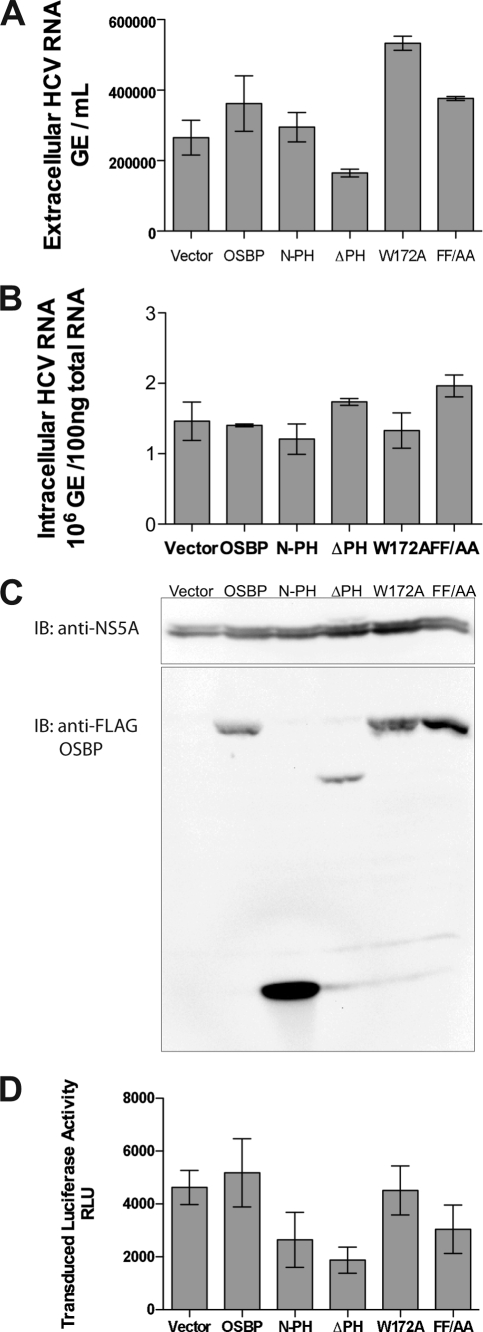
These OSBP mutants were also introduced into HCV-infected cells and examined for their effect on replication and viral particle release. Figure 4C shows a Western blot analysis of HCV-infected cells expressing ectopically expressed OSBP and its mutants. None of the OSBP mutants affected intracellular HCV RNA replication to any significant degree (Fig. 4B). However, the extracellular accumulation of viral RNA was affected by the ΔPH OSBP mutant (Fig. 4A). The PH domain is essential for the OSBP translocalization to the Golgi apparatus (24). Other mutants did not have any significant effect on the viral particle release except the W172A mutant. Surprisingly, the W172A mutant showed a modest increase in viral particle release. At present, we have no explanation for this observed result, but further characterization of this mutant will be needed. To further confirm the effect of OSBP mutants on HCV release, a chimeric HCV vector containing a luciferase reporter gene, analogous to Luc-Jc1 (23), was constructed and tested for virus production in the presence of OSBP mutants (Fig. 4D). Again, the ΔPH mutant consistently showed an inhibitory effect on viral particle release as assayed by luciferase activity of the released reporter virus. Based on these observations, the failure of the ΔPH OSBP mutant to maintain wild-type levels of viral particle release indicates that OSBP is likely involved in viral particle release via the Golgi compartments.
FIG. 4.
Effect of OSBP mutants on HCV replication and secretion. Huh7 cells were infected with HCV at an MOI of 0.5, maintained for 8 days, transfected with OSBP expression vector by electroporation, and analyzed after 48 h by RT-PCR (A and B) and Western blot assays (C). (A) Accumulation of HCV RNA in the culture supernatant. (B) The level of intracellular HCV RNA. (C) Western blot analysis of the HCV-infected cells using anti-NS5A (upper panel) and anti-FLAG for the detection of OSBP and mutants (lower panel). (D) Production of chimeric reporter HCV. Huh7.5.1 cells were transfected with both Luc-Jc1 RNA (luciferase reporter virus) and wild-type and mutant OSBP expression vectors, as indicated. Culture supernatants were collected at 72 h after transfection and used to infect naïve Huh7.5.1 cells. Cellular lysates from these secondary infections were assayed at 42 h for luciferase activity to determine the level of infectious reporter virion titer. Means and standard errors of at least triplicate measurements are shown. Expression vectors used for all the experiments are indicated. GE, genomic copies.
NS5A binds OSBP.
OSBP was identified in this study as an NS5A-associated protein (see Fig. S2C in the supplemental material). NS5A has been previously shown to interact with VAP-A and VAP-B (17). VAP-A also has been reported to bind OSBP (53). We have confirmed these interactions by immunoprecipitation studies. NS5A was shown to interact with OSBP and VAP-A (see Fig. S3A in the supplemental material). We further determined that OSBP interacts with NS5A irrespective of its genotypic origin. NS5A derived from genotype 1b (con1) or 2a (JFH1) binds to OSBP (see Fig. S3B in the supplemental material).
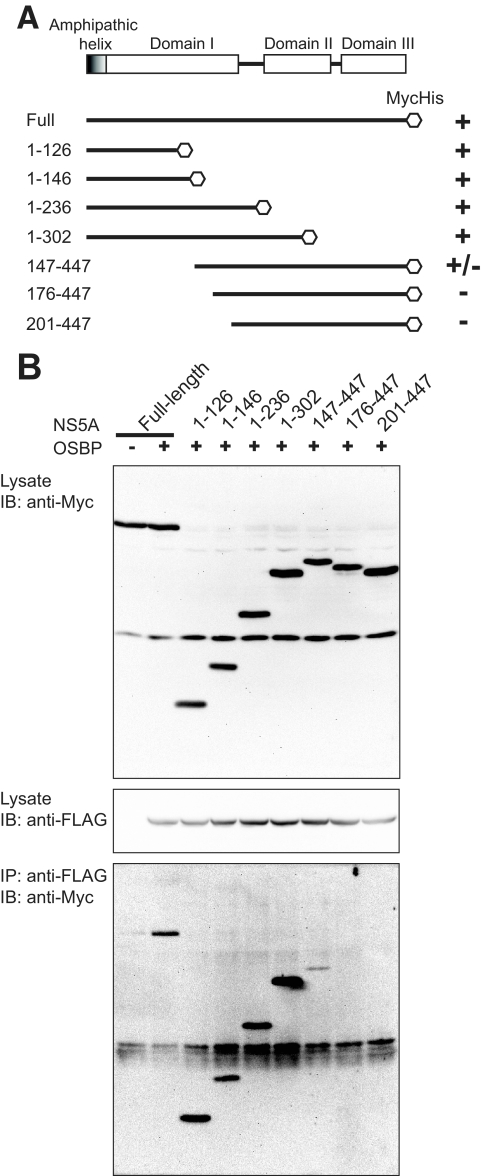
We next mapped the binding site(s) of OSBP within the NS5A protein. Huh7 cells were cotransfected with FLAG-tagged OSBP vector and various NS5A deletion mutants. All NS5A expression vectors contain a Myc/His tag (Fig. 5A). Cellular lysates were immunoprecipitated with anti-FLAG antibody to capture OSBP, followed by immunoblotting with anti-Myc antibody to probe for NS5A. Cellular lysates were examined for NS5A and OSBP expression by Western blot assays (Fig. 5B, upper and middle panels, respectively). The results show that N-terminal amino acid residues of NS5A in the region of amino acids (aa) 126 to 302 remained associated with OSBP (Fig. 5B, lower panel). These preliminary mapping studies identify domain I of NS5A as the approximate binding site(s) for OSBP. A more clearly defined motif of the NS5A region harboring OSBP binding needs to be identified. The interaction between NS5A and OSBP was also supported by the merged images of these proteins by confocal immunofluorescence microscopy, which displays a Golgi network-like pattern (see Fig. S4 in the supplemental material). The slightly diffuse pattern of OSBP seen here may reflect the state of the HCV-infected cell which may have contributed to the altered ER-Golgi apparatus pattern.
FIG. 5.
Mapping of OSBP binding site(s) within the NS5A protein. (A) Schematic representation of wild-type and deletion mutants of NS5A encoded by pEF-NS5A vectors. Various domains of NS5A are shown. Huh7 cells were cotransfected with OSBP (pFLAG-CMV-OSBP), wild-type NS5A (pEF1-NS5A), or NS5A deletion mutants. Wild-type and mutant NS5A expression vectors contain a Myc/His tag. (B) Expression of wild-type NS5A, NS5A deletion mutant proteins (upper panel), and OSBP (middle panel) was analyzed by Western blot assays. For OSBP-NS5A protein-protein interaction studies, cellular lysates were immunoprecipitated (IP) using anti-FLAG antibody (OSBP), followed by immunoblotting (IB) using anti-Myc antibody (lower panel).
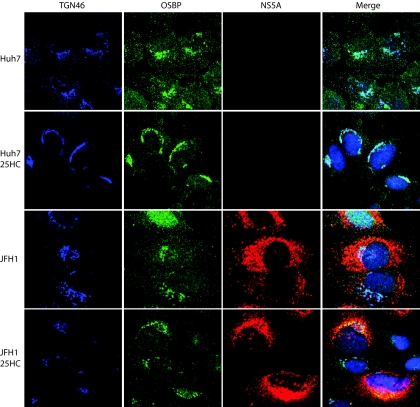
Oxysterol stimulates Golgi translocalization of OSBP and NS5A.
25-HC represents an oxidized sterol species and the most potent ligand for OSBP. The ligand binding to the OSBP triggers its Golgi translocalization, especially in the TGN. In Huh7 cells, the subcellular distribution of endogenous OSBP protein was cytosolic/vesicular as well as associated with the Golgi apparatus. OSBP association with the Golgi apparatus is seen as superimposed images in light blue (Fig. 6, top row). When Huh7 cells were stimulated by 25-HC, OSBP displayed a discrete and predominant Golgi compartment localization (Fig. 6, second row). When we analyzed the subcellular distribution of NS5A upon 25-HC stimulation in the HCV (JFH1)-infected cells, NS5A, in addition to its typical punctate ER localization, also displayed a discrete localization to the TGN. The merged image of OSBP and NS5A localization in the Golgi network can be seen in yellow (Fig. 6, bottom row).
FIG. 6.
25-HC stimulates Golgi compartment localization of OSBP and NS5A. Uninfected and HCV (JFH1)-infected Huh7 cells were treated with or without 10 μM 25-HC and incubated for 12 h prior to immunostaining. The immunofluorescence assay was performed as described in Materials and Methods.
We also examined this phenomenon in cells expressing the NS5A gene via an ectopic expression vector encoding NS5A. As can be seen, the addition of 25-HC again triggered its Golgi compartment distribution, suggesting that NS5A via its interaction with OSBP is targeted to the Golgi compartment (see Fig. S4B in the supplemental material). This result suggests that NS5A can localize to the Golgi compartment in the absence of other viral proteins or without the context of HCV infection. These associations clearly implicate NS5A's involvement in the HCV maturation/release processes.
DISCUSSION
HCV RNA replicates within a RNP complex in the modified membranous structures originated from the ER. Lipid droplets and rafts have been implicated in supporting HCV replication (13, 28). Host lipid synthesis has been shown to play an essential role in the viral reproduction process (40). Several inhibitors of lipid/fatty acid biosynthesis affect viral RNA synthesis and most likely the secretion of HCV. Our proteomic analysis of HCV viral protein complexes revealed the association of OSBP along with other factors involved in lipid/fatty acid biosynthetic pathways. Here, we investigated the functional role of OSBP in HCV replication and postreplicative processes. OSBP, a cytosolic lipid binding protein that translocates to the Golgi compartment upon ligand binding, plays a functional role in the ER to Golgi lipid trafficking.
VAP-A, which has been previously shown to bind viral proteins NS5A and NS5B, has emerged as a key player in this process. VAP-A, which is mostly localized in the ER, is known to interact with both OSBP and CERT thorough the FFAT motif, a motif shared by these lipid-binding proteins. It is interesting that the NS5A staining pattern with VAP-A and OSBP mirrors the distribution of these cellular proteins in the merged images (see Fig. S4A in the supplemental material). Both CERT and OSBP proteins contain the PH domain and are targeted to the Golgi apparatus and interact with each other at the ER-Golgi membrane contact sites (35, 36). PI transported to the Golgi compartment by Nir-2 is phosphorylated in the Golgi compartment by PI4 kinase, producing PI4P, which recruits OSBP and CERT by direct binding to their PH domains (35). Through these actions, CERT then transfers ceramide from the ER to the Golgi apparatus, which enables the production of sphingomyelin (SM) and diacylglycerol in the TGN. These studies establish the functional role of OSBP in regulation of lipid transport from the ER to the Golgi apparatus and stimulation of SM synthesis. In this context, a recent study showed that HPA-12, an inhibitor SM synthesis, blocked HCV secretion, thus lending support to the model that HCV maturation/secretion may be occurring through the Golgi compartment (2). In this respect, two recent reports have demonstrated an indispensable role of PI4 kinases in HCV replication (4, 42).
Using RNA interference studies, it is shown that partial depletion of OSBP did not affect HCV replication but dramatically inhibited the accumulation of HCV particles (Fig. 1B to E). OSBP-specific shRNA-1, which severely depleted OSBP protein, affected both viral RNA replication and virion secretion. Such a depletion of OSBP by shRNA-1 also affected viral gene expression (Fig. 1A), suggesting that OSBP may be involved in the regulation of viral gene expression (translation/replication). The exact mechanism(s) by which this may occur remains to be characterized. Complete depletion of OSBP has been shown to cause Golgi apparatus fragmentation and inhibition of transport of vesicular stomatitis virus glycoprotein from the ER (30). Partial depletion of OSBP by shRNA-2 gave different results. The viral gene expression was proportionally affected by this shRNA (Fig. 1A). Intracellular levels of viral RNA were not affected, but the accumulation of extracellular RNA was severely reduced. This result suggested the involvement of OSBP in virion secretion.
Mutations within the functional domains of OSBP that affect its various functions have been described (50). We developed a few of these OSBP mutations and ectopically expressed them in HCV-infected cells (Fig. 2B). Most of these mutations displayed diffuse patterns of OSBP with reduced Golgi compartment localizations (W172A and FF/AA). The PH domain deletion mutant, ΔPH, failed to localize to the Golgi compartment, impairing the virion release (Fig. 1D and E). Overall, results of this study point more emphatically to a major role of OSBP in the regulation of virion secretion.
Several studies support the notion that HCV may utilize the VLDL secretory pathway (6, 14, 19, 29). The most compelling evidence comes from the use of microsomal transfer protein inhibitor and ApoB silencing studies, both of which affect the VLDL particle assembly (14). Microsomal transfer protein inhibitor and ApoB small interfering RNA affect the accumulation of extracellular viral RNA while not affecting intracellular viral RNA replication (14). Use of the antioxidant flavonoid naringenin, which inhibits VLDL assembly, similarly inhibited virion release (29). Huang and colleagues showed that secretion of HCV infectious particles is dependent on active secretion of VLDL, showing that the VLDL assembly/secretion pathway is being utilized by HCV (19). Pre-VLDL particles are assembled in the ER, and mature VLDL particle formation occurs in the Golgi apparatus (16). While it is not known whether OSBP localization and/or trafficking to the Golgi apparatus is needed for VLDL secretion, we show here that NS5A along with OSBP is localized to the Golgi apparatus as well as to its characteristic localization in the ER. Our confocal microscopy studies show that a notable fraction of NS5A is localized to the Golgi compartment (Fig. 6). These data implicate a pivotal role of NS5A in a virion maturation process, which likely occurs in the Golgi compartments. NS5A interacts with OSBP via its N-terminal residues, and this interaction may be needed for NS5A's localization to the Golgi compartment as well. The preliminary data presented here map the binding site(s) of OSBP within domain I of NS5A (Fig. 4). Exact motifs involved in this interaction remain to be characterized. NS5A has three functional domains (I, II, and III). Domains I and II are relevant to the regulation of HCV RNA replication. Several studies point to the role of domain III in HCV particle secretion (3, 27, 45). The serine 457 residue in this domain, in particular, which is also the site of casein kinase II phosphorylation, has been shown to be essential for virion production (45). It will be of interest to determine the Golgi localization of an NS5A Ser457 mutant. The importance of Golgi trafficking for the HCV life cycle is further supported by the observations that HCV envelope proteins traffic thorough the cisternae of the Golgi compartment for glycosylation (32). Little is known about the HCV assembly and secretion processes. The studies described herein represent attempts to probe into these processes. Clearly, the delineation of these pathways is needed for the elucidation of the HCV life cycle and its relevance to infectious processes.
In summary, we have demonstrated that OSBP function(s) is relevant to the HCV maturation process. Our studies also suggest that NS5A localizes to the Golgi compartment and that OSBP and NS5A together aid in the viral assembly and or secretion processes. Further studies are needed to characterize, in depth, the exact role(s) of OSBP in the various steps of the HCV life cycle.
Supplementary Material
Acknowledgments
We thank T. Wakita for the generous gift of the infectious JFH1 molecular clone and the subgenomic construct, F. Chisari (Scripps Research Institute, La Jolla, CA) for the kind gift of Huh7.5.1 cells, C. Rice and C. Cameron for anti-NS5A antibodies, I. Verma (Salk Institute, La Jolla, CA) for providing lentiviral plasmids, and Y. Matsuura for providing VAP-A expression vector.
This study was supported by NIH grants DK077704 and U19066313 to A.S. and P41 RR011823 to J.Y.
Footnotes
Published ahead of print on 1 July 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Ahlquist, P., A. O. Noueiry, W. M. Lee, D. B. Kushner, and B. T. Dye. 2003. Host factors in positive-strand RNA virus genome replication. J. Virol. 77:8181-8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aizaki, H., K. Morikawa, M. Fukasawa, H. Hara, Y. Inoue, H. Tani, K. Saito, M. Nishijima, K. Hanada, Y. Matsuura, M. M. Lai, T. Miyamura, T. Wakita, and T. Suzuki. 2008. Critical role of virion-associated cholesterol and sphingolipid in hepatitis C virus infection. J. Virol. 82:5715-5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Appel, N., M. Zayas, S. Miller, J. Krijnse-Locker, T. Schaller, P. Friebe, S. Kallis, U. Engel, and R. Bartenschlager. 2008. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog. 4:e1000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger, K. L., J. D. Cooper, N. S. Heaton, R. Yoon, T. E. Oakland, T. X. Jordan, G. Mateu, A. Grakoui, and G. Randall. 2009. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc. Natl. Acad. Sci. USA 106:7577-7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bern, M., D. Goldberg, W. H. McDonald, and J. R. Yates, 3rd. 2004. Automatic quality assessment of peptide tandem mass spectra. Bioinformatics 20(Suppl. 1):i49-i54. [DOI] [PubMed] [Google Scholar]
- 6.Chang, K. S., J. Jiang, Z. Cai, and G. Luo. 2007. Human apolipoprotein e is required for infectivity and production of hepatitis C virus in cell culture. J. Virol. 81:13783-13793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chomczynski, P., and N. Sacchi. 2006. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: twenty-something years on. Nat. Protoc. 1:581-585. [DOI] [PubMed] [Google Scholar]
- 8.Cociorva, D., L. T. D., and J. R. Yates. 2007. Validation of tandem mass spectrometry database search results using DTASelect. Curr. Protoc. Bioinformatics Chapter 13:Unit 13.4. [DOI] [PubMed] [Google Scholar]
- 9.Diop, S. B., K. Bertaux, D. Vasanthi, A. Sarkeshik, B. Goirand, D. Aragnol, N. S. Tolwinski, M. D. Cole, J. Pradel, J. R. Yates III, R. K. Mishra, Y. Graba, and A. J. Saurin. 2008. Reptin and pontin function antagonistically with PcG and TrxG complexes to mediate Hox gene control. EMBO Rep. 9:260-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elazar, M., K. H. Cheong, P. Liu, H. B. Greenberg, C. M. Rice, and J. S. Glenn. 2003. Amphipathic helix-dependent localization of NS5A mediates hepatitis C virus RNA replication. J. Virol. 77:6055-6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eng, J., A. McCormack, and J. R. Yates, 3rd. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5:976-989. [DOI] [PubMed] [Google Scholar]
- 12.Evans, M. J., C. M. Rice, and S. P. Goff. 2004. Phosphorylation of hepatitis C virus nonstructural protein 5A modulates its protein interactions and viral RNA replication. Proc. Natl. Acad. Sci. USA 101:13038-13043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao, L., H. Aizaki, J. W. He, and M. M. Lai. 2004. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J. Virol. 78:3480-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gastaminza, P., G. Cheng, S. Wieland, J. Zhong, W. Liao, and F. V. Chisari. 2008. Cellular determinants of hepatitis C virus assembly, maturation, degradation, and secretion. J. Virol. 82:2120-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gosert, R., D. Egger, V. Lohmann, R. Bartenschlager, H. E. Blum, K. Bienz, and D. Moradpour. 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 77:5487-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gusarova, V., J. Seo, M. L. Sullivan, S. C. Watkins, J. L. Brodsky, and E. A. Fisher. 2007. Golgi-associated maturation of very low density lipoproteins involves conformational changes in apolipoprotein B, but is not dependent on apolipoprotein E. J. Biol. Chem. 282:19453-19462. [DOI] [PubMed] [Google Scholar]
- 17.Hamamoto, I., Y. Nishimura, T. Okamoto, H. Aizaki, M. Liu, Y. Mori, T. Abe, T. Suzuki, M. M. Lai, T. Miyamura, K. Moriishi, and Y. Matsuura. 2005. Human VAP-B is involved in hepatitis C virus replication through interaction with NS5A and NS5B. J. Virol. 79:13473-13482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang, H., Y. Chen, and J. Ye. 2007. Inhibition of hepatitis C virus replication by peroxidation of arachidonate and restoration by vitamin E. Proc. Natl. Acad. Sci. USA 104:18666-18670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang, H., F. Sun, D. M. Owen, W. Li, Y. Chen, M. Gale, Jr., and J. Ye. 2007. Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc. Natl. Acad. Sci. USA 104:5848-5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inubushi, S., M. Nagano-Fujii, K. Kitayama, M. Tanaka, C. An, H. Yokozaki, H. Yamamura, H. Nuriya, M. Kohara, K. Sada, and H. Hotta. 2008. Hepatitis C virus NS5A protein interacts with and negatively regulates the non-receptor protein tyrosine kinase Syk. J. Gen. Virol. 89:1231-1242. [DOI] [PubMed] [Google Scholar]
- 21.Kapadia, S. B., and F. V. Chisari. 2005. Hepatitis C virus RNA replication is regulated by host geranylgeranylation and fatty acids. Proc. Natl. Acad. Sci. USA 102:2561-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kato, T., T. Date, M. Miyamoto, M. Sugiyama, Y. Tanaka, E. Orito, T. Ohno, K. Sugihara, I. Hasegawa, K. Fujiwara, K. Ito, A. Ozasa, M. Mizokami, and T. Wakita. 2005. Detection of anti-hepatitis C virus effects of interferon and ribavirin by a sensitive replicon system. J. Clin. Microbiol. 43:5679-5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koutsoudakis, G., A. Kaul, E. Steinmann, S. Kallis, V. Lohmann, T. Pietschmann, and R. Bartenschlager. 2006. Characterization of the early steps of hepatitis C virus infection by using luciferase reporter viruses. J. Virol. 80:5308-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lagace, T. A., D. M. Byers, H. W. Cook, and N. D. Ridgway. 1997. Altered regulation of cholesterol and cholesteryl ester synthesis in Chinese-hamster ovary cells overexpressing the oxysterol-binding protein is dependent on the pleckstrin homology domain. Biochem. J. 326:205-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levine, T. P., and S. Munro. 2002. Targeting of Golgi-specific pleckstrin homology domains involves both PtdIns 4-kinase-dependent and -independent components. Curr. Biol. 12:695-704. [DOI] [PubMed] [Google Scholar]
- 26.MacCoss, M. J., C. C. Wu, and J. R. Yates, 3rd. 2002. Probability-based validation of protein identifications using a modified SEQUEST algorithm. Anal. Chem. 74:5593-5599. [DOI] [PubMed] [Google Scholar]
- 27.Masaki, T., R. Suzuki, K. Murakami, H. Aizaki, K. Ishii, A. Murayama, T. Date, Y. Matsuura, T. Miyamura, T. Wakita, and T. Suzuki. 2008. Interaction of hepatitis C virus nonstructural protein 5A with core protein is critical for the production of infectious virus particles. J. Virol. 82:7964-7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyanari, Y., K. Atsuzawa, N. Usuda, K. Watashi, T. Hishiki, M. Zayas, R. Bartenschlager, T. Wakita, M. Hijikata, and K. Shimotohno. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 9:1089-1097. [DOI] [PubMed] [Google Scholar]
- 29.Nahmias, Y., J. Goldwasser, M. Casali, D. van Poll, T. Wakita, R. T. Chung, and M. L. Yarmush. 2008. Apolipoprotein B-dependent hepatitis C virus secretion is inhibited by the grapefruit flavonoid naringenin. Hepatology 47:1437-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ngo, M., and N. D. Ridgway. 2009. Oxysterol binding protein (OSBP)-related protein 9 (ORP9) is a cholesterol transfer protein that regulates Golgi structure and function. Mol. Biol. Cell 20:1388-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishimura, T., T. Inoue, N. Shibata, A. Sekine, W. Takabe, N. Noguchi, and H. Arai. 2005. Inhibition of cholesterol biosynthesis by 25-hydroxycholesterol is independent of OSBP. Genes Cells 10:793-801. [DOI] [PubMed] [Google Scholar]
- 32.Op De Beeck, A., C. Voisset, B. Bartosch, Y. Ciczora, L. Cocquerel, Z. Keck, S. Foung, F. L. Cosset, and J. Dubuisson. 2004. Characterization of functional hepatitis C virus envelope glycoproteins. J. Virol. 78:2994-3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pawlotsky, J. M. 2004. Pathophysiology of hepatitis C virus infection and related liver disease. Trends Microbiol. 12:96-102. [DOI] [PubMed] [Google Scholar]
- 34.Peng, J., J. E. Elias, C. C. Thoreen, L. J. Licklider, and S. P. Gygi. 2003. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J. Proteome Res. 2:43-50. [DOI] [PubMed] [Google Scholar]
- 35.Peretti, D., N. Dahan, E. Shimoni, K. Hirschberg, and S. Lev. 2008. Coordinated lipid transfer between the endoplasmic reticulum and the Golgi complex requires the VAP proteins and is essential for Golgi-mediated transport. Mol. Biol. Cell 19:3871-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perry, R. J., and N. D. Ridgway. 2006. Oxysterol-binding protein and vesicle-associated membrane protein-associated protein are required for sterol-dependent activation of the ceramide transport protein. Mol. Biol. Cell 17:2604-2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Randall, G., M. Panis, J. D. Cooper, T. L. Tellinghuisen, K. E. Sukhodolets, S. Pfeffer, M. Landthaler, P. Landgraf, S. Kan, B. D. Lindenbach, M. Chien, D. B. Weir, J. J. Russo, J. Ju, M. J. Brownstein, R. Sheridan, C. Sander, M. Zavolan, T. Tuschl, and C. M. Rice. 2007. Cellular cofactors affecting hepatitis C virus infection and replication. Proc. Natl. Acad. Sci. USA 104:12884-12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ridgway, N. D., P. A. Dawson, Y. K. Ho, M. S. Brown, and J. L. Goldstein. 1992. Translocation of oxysterol binding protein to Golgi apparatus triggered by ligand binding. J. Cell Biol. 116:307-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sadygov, R. G., J. Eng, E. Durr, A. Saraf, H. McDonald, M. J. MacCoss, and J. R. Yates III. 2002. Code developments to improve the efficiency of automated MS/MS spectra interpretation. J. Proteome Res. 1:211-215. [DOI] [PubMed] [Google Scholar]
- 40.Su, A. I., J. P. Pezacki, L. Wodicka, A. D. Brideau, L. Supekova, R. Thimme, S. Wieland, J. Bukh, R. H. Purcell, P. G. Schultz, and F. V. Chisari. 2002. Genomic analysis of the host response to hepatitis C virus infection. Proc. Natl. Acad. Sci. USA 99:15669-15674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tabb, D. L., W. H. McDonald, and J. R. Yates III. 2002. DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1:21-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tai, A. W., Y. Benita, L. F. Peng, S. S. Kim, N. Sakamoto, R. J. Xavier, and R. T. Chung. 2009. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe 5:298-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takeuchi, T., A. Katsume, T. Tanaka, A. Abe, K. Inoue, K. Tsukiyama-Kohara, R. Kawaguchi, S. Tanaka, and M. Kohara. 1999. Real-time detection system for quantification of hepatitis C virus genome. Gastroenterology 116:636-642. [DOI] [PubMed] [Google Scholar]
- 44.Targett-Adams, P., S. Boulant, and J. McLauchlan. 2008. Visualization of double-stranded RNA in cells supporting hepatitis C virus RNA replication. J. Virol. 82:2182-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tellinghuisen, T. L., K. L. Foss, and J. Treadaway. 2008. Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog 4:e1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tiscornia, G., O. Singer, and I. M. Verma. 2006. Design and cloning of lentiviral vectors expressing small interfering RNAs. Nat. Protoc. 1:234-240. [DOI] [PubMed] [Google Scholar]
- 47.van den Hoff, M. J., A. F. Moorman, and W. H. Lamers. 1992. Electroporation in “intracellular” buffer increases cell survival. Nucleic Acids Res. 20:2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, P. Y., J. Weng, and R. G. Anderson. 2005. OSBP is a cholesterol-regulated scaffolding protein in control of ERK 1/2 activation. Science 307:1472-1476. [DOI] [PubMed] [Google Scholar]
- 50.Wang, P. Y., J. Weng, S. Lee, and R. G. Anderson. 2008. The N terminus controls sterol binding while the C terminus regulates the scaffolding function of OSBP. J. Biol. Chem. 283:8034-8045. [DOI] [PubMed] [Google Scholar]
- 51.Waris, G., D. J. Felmlee, F. Negro, and A. Siddiqui. 2007. Hepatitis C virus induces proteolytic cleavage of sterol regulatory element binding proteins and stimulates their phosphorylation via oxidative stress. J. Virol. 81:8122-8130. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 52.Waris, G., J. Turkson, T. Hassanein, and A. Siddiqui. 2005. Hepatitis C virus (HCV) constitutively activates STAT-3 via oxidative stress: role of STAT-3 in HCV replication. J. Virol. 79:1569-1580. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53.Wyles, J. P., C. R. McMaster, and N. D. Ridgway. 2002. Vesicle-associated membrane protein-associated protein-A (VAP-A) interacts with the oxysterol-binding protein to modify export from the endoplasmic reticulum. J. Biol. Chem. 277:29908-29918. [DOI] [PubMed] [Google Scholar]
- 54.Wyles, J. P., and N. D. Ridgway. 2004. VAMP-associated protein-A regulates partitioning of oxysterol-binding protein-related protein-9 between the endoplasmic reticulum and Golgi apparatus. Exp. Cell Res. 297:533-547. [DOI] [PubMed] [Google Scholar]
- 55.Yan, D., and V. M. Olkkonen. 2008. Characteristics of oxysterol binding proteins. Int. Rev. Cytol. 265:253-285. [DOI] [PubMed] [Google Scholar]
- 56.Ye, J. 2007. Reliance of host cholesterol metabolic pathways for the life cycle of hepatitis C virus. PLoS Pathog. 3:e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ye, J., C. Wang, R. Sumpter, Jr., M. S. Brown, J. L. Goldstein, and M. Gale, Jr. 2003. Disruption of hepatitis C virus RNA replication through inhibition of host protein geranylgeranylation. Proc. Natl. Acad. Sci. USA 100:15865-15870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 102:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.