Abstract
Vibrio parahaemolyticus O3:K6 pandemic strains recovered in Chile frequently possess a 42-kb plasmid which is the prophage of a myovirus. We studied the prototype phage VP58.5 and show that it does not integrate into the host cell chromosome but replicates as a linear plasmid (Vp58.5) with covalently closed ends (telomeres). The Vp58.5 replicon coexists with other plasmid prophages (N15, PY54, and ΦKO2) in the same cell and thus belongs to a new incompatibility group of telomere phages. We determined the complete nucleotide sequence (42,612 nucleotides) of the VP58.5 phage DNA and compared it with that of the plasmid prophage. The two molecules share the same nucleotide sequence but are 35% circularly permuted to each other. In contrast to the hairpin ends of the plasmid, VP58.5 phage DNA contains 5′-protruding ends. The VP58.5 sequence is 92% identical to the sequence of phage VHML, which was reported to integrate into the host chromosome. However, the gene order and termini of the phage DNAs are different. The VHML genome exhibits the same gene order as does the Vp58.5 plasmid. VHML phage DNA has been reported to contain terminal inverted repeats. This repetitive sequence is similar to the telomere resolution site (telRL) of VP58.5 which, after processing by the phage protelomerase, forms the hairpin ends of the Vp58.5 prophage. It is discussed why these closely related phages may be so different in terms of their genome ends and their lifestyle.
Most temperate bacteriophages integrate into the host chromosome during lysogeny. However, there are some phages (telomere phages) whose prophages are linear plasmids with covalently closed ends. Members of this group of phages are the siphoviruses N15, PY54, and ΦKO2 isolated from Escherichia coli, Yersinia enterocolitica, and Klebsiella oxytoca, respectively, and the recently described myoviruses ΦHAP-1 of Halomonas aquamarina and VP882 of Vibrio parahaemolyticus (6, 20, 23, 26, 37). Despite their different origins (enterobacteria versus marine bacterium) and morphologies, all known telomere phages share similar genome organizations and some protein similarities. The linear DNA of each phage is a circular permutation of the respective linear plasmid prophage. For the generation of the terminal hairpins of the linear plasmid, the protelomerase (Tel) is essential (8). This enzyme has cleaving/joining activity; its target is a large palindromic DNA sequence called the telomere resolution site (telRL) located upstream of tel on the phage genome. After cleaving telRL by staggered cuts, the resulting self-complementary single-stranded DNA overhangs fold back and are rejoined by the protelomerase (9). Besides tel, all telomere phages possess the gene repA, encoding a multifunctional replication protein. repA of N15 and PY54 was shown to harbor the prophage replication origin and to function as a circular minimal replicon (35, 42). Compatibility studies demonstrated that the N15 and ΦKO2 plasmids belong to the same incompatibility group, whereas the PY54 plasmid is able to coexist with these two prophages in doubly lysogenic E. coli and Y. enterocolitica hosts (19).
There are some reports on the presence of tel and repA in prophages (VP882, VHML, and Vp58.5) of marine Vibrio strains (28, 41). V. parahaemolyticus phage VP882 is a close relative of the Halomonas phage ΦHAP-1 (26). VHML was isolated from a toxin-producing Vibrio harveyi strain, pathogenic for some crustaceans and fish (30). Similarly to ΦHAP-1 and VP882, VHML has a myovirus-like morphology. The phage contains genes for products similar to Tel and RepA, suggesting that its prophage is a linear plasmid with terminal hairpins. However, it was surmised that VHML integrates into the Vibrio chromosome (28, 29). Phage VP58.5 was isolated from a V. parahaemolyticus strain belonging to the serovar O3:K6 pandemic clonal complex (41). During the last several years, this clone has been associated with many seafood-borne diarrhea outbreaks in Southeast Asia and South America, particularly Chile (5, 12, 13, 15). Up to 33% of the Chilean isolates harbored a 42-kb plasmid which was shown to be the prophage of a myovirus inducible by mitomycin C. VP58.5 is the prototype of these phages.
In this work we demonstrate that VP58.5 is closely related to the V. harveyi phage VHML but that its prophage is a linear plasmid with covalently closed ends. The Vp58.5 prophage belongs to a new incompatibility group of telomere phages.
MATERIALS AND METHODS
Bacteriophages, bacterial strains, media, and growth conditions.
Phages and strains utilized in this study are listed in Table S1 in the supplemental material. The E. coli strain Genehogs (Invitrogen) was employed for standard cloning procedures. E. coli and V. parahaemolyticus strains were cultivated in Luria-Bertani (LB) broth (38) at 37°C under shaking conditions (200 to 225 rpm). For the cultivation of V. parahaemolyticus, LB broth was supplemented with sodium chloride to a final concentration of 3% (wt/vol) (41). Solid media contained 1.8% (wt/vol) agar. When required, kanamycin and ampicillin were used as supplements at 100 μg ml−1 and chloramphenicol and tetracycline were used at 12.5 μg ml−1.
Isolation and purification of VP58.5 phages.
Phages were recovered by mitomycin C (Sigma-Aldrich) induction of V. parahaemolyticus strain PMC58.5. At an adsorption (A600) of 0.4, mitomycin C (30 ng ml−1) was added and shaking was continued for 4 h (41). Bacterium-free phage lysates were obtained by centrifugation at 12,000 × g for 30 min and filtration of the supernatant through 0.45-μm- and 0.2-μm-pore-size sterile filters (Schleicher und Schüll). Between the filtration steps, 10 mM MgCl2, 1 μg ml−1 DNase I, and RNase A (Roche) were applied to the lysate, which was incubated at 37°C for 2 h. Phage particles were concentrated by ultracentrifugation at 230,000 × g for 2 h. Phage pellets were suspended in SM buffer and purified through discontinuous cesium chloride (CsCl) gradients (1.3 to 1.7 g ml−1), as described by Sambrook and Russell (38).
Transmission electron microscopy.
Phage particles were adsorbed onto carbon-coated grids for 5 min and negatively stained with 1% (wt/vol) uranyl acetate (pH 4.5) for 1 min according to the method of Steven et al (39). Micrographs were taken with a JEM-1010 transmission electron microscope (JEOL, Japan) at 80 kV.
Extraction and characterization of phage VP58.5 DNA.
Phage DNA was extracted from VP58.5 virus particles by proteinase K-sodium dodecyl sulfate (SDS) treatment followed by phenol-chloroform extraction, as described by Sambrook and Russell (38). To determine the presence of cohesive ends, phage DNA was digested with Van91I (Fermentas). Restriction patterns of unheated and heated phage DNAs were compared. Additionally, phage DNA treated with T4 ligase was analyzed by the same procedure. Protruding nucleotides in the phage DNA were determined by runoff sequencing using the primers cos-F (5′-CAGTAAAGTCCCAATCGCTC) and cos-R (5′-GTCCATTCCGTCAGTTAGTTTC).
VP58.5 genome sequencing.
Based on the previous determination of partial DNA sequences of phage VP58.5, approximately 100 oligonucleotide primers deduced from the annotated sequence of VHML were employed for genome sequencing (28, 41). One hundred to two hundred nanograms of DNA from CsCl-purified phages was used as template. Gaps between contigs were closed by primer walking. The direct sequencing strategy was also performed for the determination of the cohesive ends. Similarly, the circularly permutated prophage DNA was partially sequenced with the same set of oligonucleotide primers.
Sequence analysis of the VP58.5 genome.
Sequence analyses and alignments were carried out using the MacVector 8.0 software of the Oxford Molecular Group (Campbell, CA). Putative open reading frames (ORFs) were defined using the algorithms of the software packages MacVector, Accelrys Gene v2.5 (Accelrys Inc.), and ORF Finder (NCBI). BLAST searches were performed at the NCBI homepage (GenBank database) using BLASTp (http://www.ncbi.nlm.nih.gov/BLAST/) for calculating similarity and identity values (1).
Construction of linear and circular miniplasmid derivatives of Vp58.5.
To generate the linear miniplasmid pJH1210 (12.4 kb), phage DNA was digested with the restriction endonuclease BglII (Fermentas). After the ends of the restriction fragments were filled in with T4 polymerase, the fragments were ligated to the tetracycline resistance gene of transposon Tn5 that had been amplified by PCR using the vector pBR329 as template (7). The recombinant DNA preparation was introduced into the E. coli strain Genehogs. Similarly the circular miniplasmid pJH1201 (6.1 kb) was isolated by digestion of phage VP58.5 DNA with EcoRV (New England Biolabs). The composition of the miniplasmids pJH1201 and pJH1210 was verified by restriction analysis and DNA sequencing.
Compatibility studies.
Compatibility of the telomere phages was studied with circular miniplasmid derivatives of the prophages retaining replicative competence (see Table S1 in the supplemental material). The Vp58.5 miniplasmid pJH1201 (Tcr) was introduced into E. coli C-1a and Y. enterocolitica 83/88/2. Thereafter, transformants were lysogenized with the N15 mutant C02 (Cmr) and PY54 mutant A (Kmr), respectively (19). Selection of lysogens containing the respective prophage and the miniplasmid pJH1201 was achieved by plating infected bacteria on agar containing tetracycline and kanamycin (pJH1201 and PY54-A) or tetracycline and chloramphenicol (pJH1201 and N15-C02). The presence of the linear prophages and the Vp58.5 miniplasmid in the lysogens was examined by restriction analysis. Similarly, the compatibility of PY54, N15, and ΦKO2 miniplasmids with phage VP58.5 was studied in V. parahaemolyticus. For the introduction of the miniplasmids pSH120 (PY54, Kmr), pJH320 (N15, Cmr), and pJH520 (ΦKO2, Tcr) into the V. parahaemolyticus strain PMC58.5, the transformation protocol of Hamashima et al. was used (18).
SDS-PAGE and protein analysis by mass spectrometry.
SDS-polyacrylamide gel electrophoresis (PAGE) was performed according to the method of Laemmli (22). Structural proteins of VP58.5 were separated at 20 mA on a one-dimensional 15% (wt/vol) SDS gel at 15°C. After staining by Coomassie brilliant blue R-250 (Bio-Rad), protein bands were excised from the gel, washed twice with 50% (vol/vol) acetonitrile in 5 mM ammonium bicarbonate for 1 h, digested with porcine trypsin (Merck), and purified as described previously (21). For the analysis of the proteins, high-pressure liquid chromatography coupled to mass spectrometry was used, and automated tandem mass spectrometry fragmentation was performed during the high-pressure liquid chromatography run. To determine peptide sequences, tandem mass spectrometry spectra were obtained using a QStar XL hybrid mass spectrometer (Applied Biosystems) with a nanoelectrospray source. The obtained data were submitted to the Mascot web server database (http://www.matrixscience.com) (34).
Nucleotide sequence accession numbers.
The sequence of phage VP58.5 has been deposited in the GenBank database under accession number FN297812. The GenBank accession numbers of the phage genomes used in this study are as follows: VHML, NC_004456; VP882, NC_009016; ΦHAP-1, EU399241; N15, NC_001901; ΦKO2, NC_005857; and PY54, NC_005069 (6, 21, 23, 26, 28, 36).
RESULTS
VP58.5 is a close relative of phage VHML.
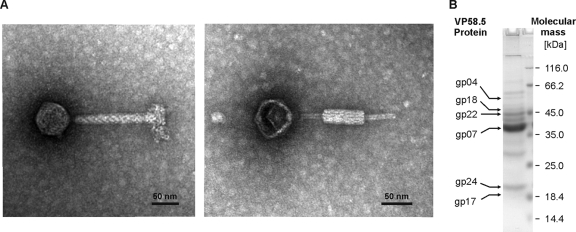
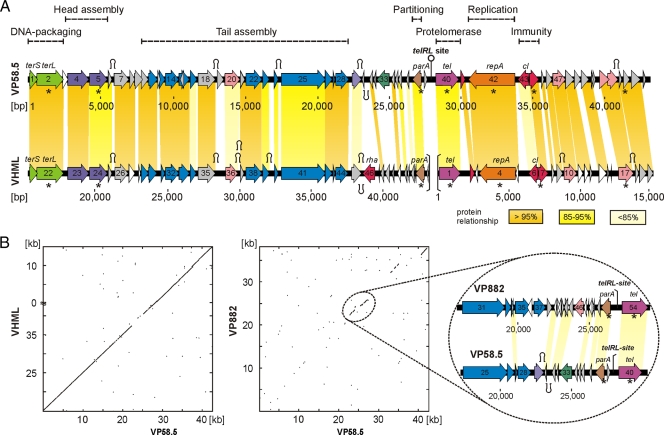
In a previous study, small DNA fragments of myophage VP58.5 (Fig. 1A) obtained by random molecular cloning and PCR were shown to be highly homologous to sequences of phage VHML (41). Based on this finding, we determined the whole VP58.5 genomic sequence (42,612 nucleotides, 50.9% G+C content) by a direct approach using phage DNA as template and primers deduced from the VHML sequence (see Materials and Methods). Except for an approximately 3-kb-long stretch (positions 22200 to 25200) on the VP58.5 genome that exhibits significant homology to the V. parahaemolyticus phage VP882, the remaining VP58.5 DNA is very similar to that of VHML (Fig. 2). On average, the VP58.5 and VHML DNAs are 92% identical. Out of 58 gene products that have been predicted for VP58.5, 44 products revealed values of identity to VHML proteins of more than 91%. In addition, nine other VP58.5 products are similar to VHML proteins, with identity values between 38% and 86%. Like VHML, the capsid proteins and tail proteins of VP58.5 exhibit some relatedness to proteins of lambda-like phages and P2-like phages, respectively. Other nonstructural VP58.5 products (e.g., protelomerase, RepA, and prophage repressor) are similar to telomere phage proteins. Hence, both VHML and VP58.5 are chimeric phages belonging to several phage clusters according to the classification by Lima-Mendez et al (24). Nonetheless, the VP58.5 and VHML genomes differ with respect to their gene order. While the VHML genome has been reported to start with the protelomerase gene which is located close to the center of the VP58.5 genome, the first two ORFs of VP58.5 probably code for the subunits of the terminase (Fig. 2A). The overall genome organization of VP58.5 is strongly reminiscent of those of telomere phages. The genome can be divided into two arms separated by a long (88-bp) palindromic sequence comparable to the telomere resolution sites (telRL) of N15, PY54, and ΦKO2. Similar to these phages and also to ΦHAP-1 and VP882, the left arm of the VP58.5 genome mainly consists of genes for structural proteins and phage assembly. The VP58.5 virion protein profile is composed of at least 12 proteins (Fig. 1B). Each of the protein bands was excised from the gel and analyzed by peptide mass fingerprinting. Six of the proteins could be allocated to VP58.5 gene products predicted by in silico analysis (Fig. 1B). We identified the products of ORF 4 (putative portal protein); ORFs 7, 17, and 18 (functions unknown); ORF 22 (putative tail tube protein); and ORF 24 (putative tail protein).
FIG. 1.
Morphology and protein composition of phage VP58.5. (A) Transmission electron micrographs of VP58.5 particles. Phages were isolated from V. parahaemolyticus pandemic strain PMC58.5 (serovar O3:K6) by induction with mitomycin C. (B) Mass spectrometric analysis of VP58.5 structural proteins. CsCl-purified phages were analyzed by peptide mass fingerprinting. Proteins were separated by SDS-PAGE and assigned to VP58.5 gene products predicted by in silico analysis.
FIG. 2.
Relationship of VP58.5 to VHML and VP882. (A) Genome organization of the phages VP58.5 and VHML. To facilitate the comparison of the phage genomes, the VMHL DNA is depicted in the same gene order as VP58.5, starting with the terminase genes. The proposed ends of the VHML genome are indicated by brackets. Numbering of VHML ORFs and that of the nucleotide positions on the VHML genome were adopted from the work of Oakey et al (28). The colors indicate predicted functions of gene products as follows: green, DNA packaging; violet, head assembly; blue, tail assembly; brown, plasmid stability/partitioning; mauve, telomere resolution; orange, replication; red, repressor; pink, product with other function; gray, putative protein; white, unknown. Related genes of the phages are connected by colored shading. Selected ORFs are denoted as follows: terS/terL, small and large terminase subunit; parA, partitioning; tel, protelomerase; repA, replication; cI, prophage repressor. Genes whose products show similarities to N15-like phages (N15, PY54, and ΦKO2) are marked by asterisks (see Table S2 in the supplemental material). The positions of putative Rho-independent transcription terminators are indicated by the character Ω (see Table S3 in the supplemental material) (4). (B) Dot plot matrices of bacteriophage VP58.5 with the Vibrio phages VHML and VP882. The axes of abscissas and ordinates show the coordinates of the respective phage genomes. Nucleotide alignments were performed with MacVector (version 8.0; Oxford Molecular Group). On the right, the DNA region similar in VP58.5 and VP882 is shown in detail.
Similarly to ΦHAP-1 and VP882, the VP58.5 genome contains a single ORF (ORF 38) at the right end of the left arm whose product (ParA) might be part of a partitioning system. In N15, PY54, and ΦKO2, partitioning systems are composed of two proteins (ParA and ParB) encoded by an operon, plus centromere-like sites (parC) to which the partitioning proteins bind.
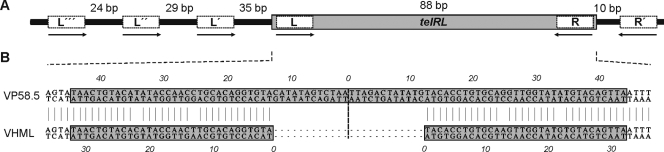
Telomere phages possess a palindromic telRL site situated between the two arms of the genome, contiguous to the partitioning genes on the left and to the protelomerase gene on the right. The telomere resolution site is the target for the protelomerase, which generates the hairpin ends of the prophage by processing telRL. On the VP58.5 genome, the perfect 88-bp palindrome is located 106 bp upstream of the putative tel gene (ORF 40, Fig. 2). Moreover, similarly to telRL of phage PY54, the VP58.5 palindrome is flanked by additional inverted repeats (Fig. 3A). Both the palindromes and the predicted tel gene products of VP58.5 and VHML are highly homologous (Fig. 3B; see also Fig. S1 in the supplemental material). The protelomerases of the two phages contain identical clusters (motifs A to C) and a pentad (“RKHRH”) which are found in several protelomerases and integrases as well (11, 16). In addition, a tyrosine residue (“Y”) pivotal for the enzyme's catalytic activity is present at the same position (9). Comparison of the palindromes disclosed that the central 24 bp of the VP58.5 telRL site are missing in the otherwise nearly identical VHML sequence (Fig. 3B).
FIG. 3.
Alignment of the VP58.5 and VHML inverted repeats situated upstream from the tel genes. (A) telRL region of phage VP58.5. The 88-bp palindrome (telRL) is flanked by additional inverted repeats (L and R). (B) Comparison of the VP58.5 palindrome with the terminal inverted repeat of the VHML genome (28). The central 24 bp of the VP58.5 telRL site are missing in the VHML sequence.
The upstream region of tel contains some ORFs whose proposed functions and order are typical for telomere phages. A large ORF (ORF 42) is located to the right of the predicted protelomerase gene, in an orientation opposite that of tel. Its possible product has 96% identity to the putative replication protein RepA of VHML and also some similarity to RepA of telomere phages. Moreover, some motifs conserved in bacterial helicases and primases have also been detected in the VP58.5 and VHML replication proteins (data not shown) (17). Next to repA, there are two ORFs (ORFs 43 and 44) on the VP58.5 genome that apparently belong to an immunity region. While the product of ORF 43 is similar to the putative prophage repressors of VHML and VP882, ORF 44 might encode the Cro repressor, 100% identical to the corresponding VHML protein. We additionally searched for possible operator sites which are most often situated between the repressor genes. However, within the 21-bp intergenic region we could not detect any operator-like sequence, while a number of possible promoters have been identified there (data not shown).
Among the remaining ORFs in the right arm of the VP58.5 genome, there are only a few candidates for which a functional assignment could be made. However, many gene products in VP58.5 are probably analogous to proteins in the closely related phage VHML (28). The search for possible ρ-independent transcription terminators on the VP58.5 genome revealed a number of inverted repeats located immediately downstream from stop codons (Fig. 2A). Most of the repeats have the potential (confidence value = 100%) to form a G+C-rich stem with a loop in the mRNA (10), followed by a run of several thymidine bases, and have also been identified in VHML (see Table S3 in the supplemental material). Hence, it is likely that the two phages possess similar sets of transcripts.
Phage VP58.5 genomic DNA has 5′-protruding ends.
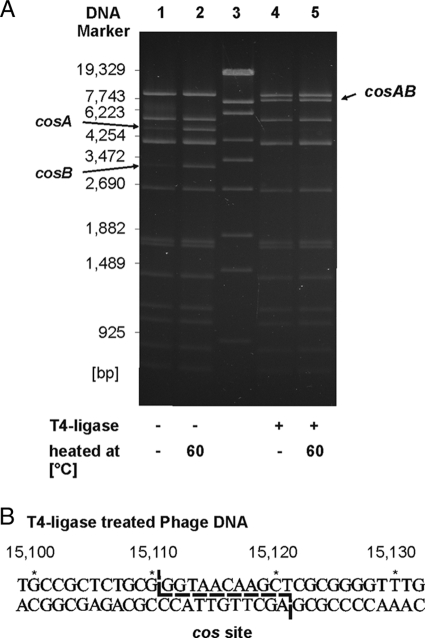
Cleavage of the VP58.5 genome by the restriction endonuclease Van91I revealed two DNA fragments (4.8 kb and 3.2 kb) which appeared as weak bands on an agarose gel (Fig. 4A, lane 1). After heating of the digested phage DNA at 60°C for 5 min and immediate chilling on ice, the two particular restriction fragments had the same intensities as those of other Van91I fragments, suggesting that they contain cohesive ends (Fig. 4A, lane 2). This was confirmed by treatment of the DNA with T4 ligase prior to digestion. A new restriction fragment of 8 kb appeared, while the two smaller fragments were not detectable, even after heating (Fig. 4A, lanes 4 and 5). To define cos on the phage DNA, we sequenced the ends of the genome using primers (cos-F and cos-R) deduced from adjacent regions. Alignment of the sequences revealed an overlap of 11 nucleotides, which means that the phage genome possesses 11 nucleotides protruding at the 5′ ends.
FIG. 4.
Analysis of the VP58.5 cos region. (A) Van91I restriction patterns of VP58.5 phage DNA. Lane 1, restriction pattern of untreated phage DNA; lane 2, same DNA as in lane 1 but heated for 5 min at 60°C before being loaded onto the gel; lane 3, DNA size marker (λ DNA Eco130I); lanes 4 and 5, unheated and heated phage DNA, respectively, that had been treated with T4 ligase before digestion. (B) Sequence of the cos region after treatment of the phage DNA with T4 ligase. The overhanging nucleotides determined in untreated phage DNA are marked.
The Vp58.5 prophage is a linear plasmid with covalently closed ends.
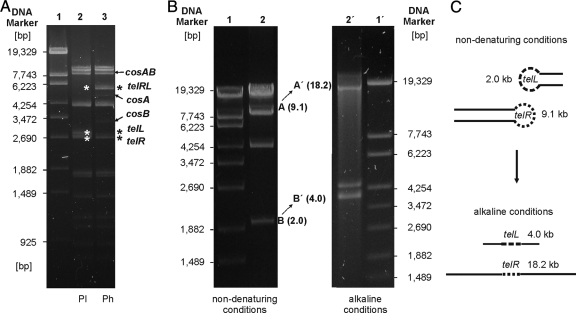
It has been reported that the Vp58.5 prophage exists as an extrachromosomal DNA which can be recovered by alkaline extraction of cells (41). The plasmid showed restriction patterns similar to those of the phage genome, indicating that both molecules are circularly permuted. In this study, we investigated the circular permutation of the VP58.5 phage genome and plasmid as well as the structure of the plasmid ends in more detail. Figure 5A shows the Van91I restriction patterns of VP58.5 phage and plasmid DNA. As described above, the phage DNA displayed two weak cos fragments (cosA and cosB) lacking in the plasmid. On the other hand, two restriction fragments (telL, 2.8 kb, and telR, 2.7 kb) were present in the prophage but missing in the phage DNA. Instead, a 5.5-kb restriction fragment (telRL) was visible in the digest of the phage genome. Based on the different restriction patterns and on the complete sequence of the VP58.5 genome, physical maps of the linear phage DNA and prophage have been devised (data not shown). The molecules are 35% circularly permuted.
FIG. 5.
Determination of the covalently closed ends of the Vp58.5 plasmid. (A) Comparison of Van91I restriction patterns of the Vp58.5 plasmid (Pl) and phage DNA (Ph). Lane 1, DNA size marker (λ Eco130I); lane 2, Van91I-digested plasmid; lane 3, Van91I-digested phage DNA. Asterisks indicate the terminal restriction fragments of the plasmid and the corresponding fragment of the phage DNA. cos fragments of the phage DNA are also marked. (B) BglII-digested prophage DNA was separated on 0.8% agarose gels under nondenaturing (left panel) and alkaline (right panel) conditions. Restriction fragments containing the left and right telomeres of the prophage plasmid exhibited twice the length under denaturing conditions. (C) Schematic presentation of the structure of the terminal prophage fragments under native and alkaline conditions.
To elucidate whether the Vp58.5 prophage contains terminal hairpins, restriction analyses under native and denaturing conditions have been performed (14, 25). For this study, the restriction endonuclease BglII was used because this enzyme cleaves the linear plasmid into only four fragments. Figure 5B shows that the two terminal BglII fragments (9.1 kb and 2.0 kb) exhibited twice the length on an alkaline agarose gel while the remaining restriction fragments showed the same length as that under nondenaturing conditions. The same result was obtained with prophage DNA that had been treated with proteinase K prior to analysis. From these experiments, it can be inferred that the ends of the plasmid prophage are not bound to protein but contain an additional covalent linkage that must be part of a hairpin-like structure.
VP58.5 belongs to a new incompatibility group of telomere phages.
To learn more about the replicative functions of VP58.5 genes, we designed linear and circular miniplasmid derivatives of the prophage retaining replicative competence. Plasmid pJH1210 is composed of the 11.1-kb BglII phage DNA fragment (nucleotide positions 25511 to 36631) and tet. It contains 11 VP58.5 ORFs, including parA, tel, repA, and the putative repressor genes cI and cro. Restriction of the plasmid by BglII resulted in three fragments, namely, tet and the two terminal 9.1-kb and 2.0-kb fragments of the prophage which harbor the telomeres (data not shown). Hence, pJH1210 possesses all genetic elements essential for linear plasmid replication.
The circular miniplasmid pJH1201 comprises the 4.8-kb EcoRV fragment of the prophage (nucleotide positions 30285 to 35109) with the genes repA and cI plus a number of ORFs of unknown function lying in between these genes (see Table S2 in the supplemental material). Transformation experiments revealed that the plasmid is able to replicate in V. parahaemolyticus and also in E. coli and Y. enterocolitica. To study the compatibility between pJH1201 and other telomere phage replicons, E. coli and Yersinia transformants harboring pJH1201 were lysogenized with the phage mutants N15-C02 (Cmr) and PY54-A (Kmr), respectively, described in a previous study (19). Doubly resistant lysogens which appeared on selective agar were analyzed with respect to their plasmid content. All of them contained plasmid pJH1201 and the respective prophage. We also investigated the compatibility of PY54, N15, and ΦKO2 miniplasmids with phage VP58.5 in V. parahaemolyticus. This was accomplished by introduction of the miniplasmids pSH120 (PY54, Kmr), pJH320 (N15, Cmr), and pJH520 (ΦKO2, Tcr) into the lysogenic V. parahaemolyticus strain PMC58.5. Similarly to the situation in E. coli and Yersinia, the Vp58.5 prophage coexisted with the other plasmids in Vibrio. Therefore, VP58.5 belongs to a new incompatibility group of telomere phages.
DISCUSSION
V. parahaemolyticus O3:K6 pandemic strains isolated in Chile frequently possess a 42-kb plasmid which was shown to be the prophage of a myovirus (41). The determination of the complete DNA sequence of the prototype phage VP58.5 revealed very strong similarity to phage VHML. Besides V. harveyi strains, VHML has been reported to lyse the strains Vibrio alginolyticus ACMM102 and Vibrio cholerae ATCC 14035, but there are no published data on the infectiousness for V. parahaemolyticus (33). On the other hand, we could not find a V. harveyi strain susceptible to VP58.5. The question arises why VP58.5 and VHML infect a different spectrum of bacteria. The close relationship suggests that the phages have a common ancestor. They share similar morphologies, and most of their gene products are 92 to 100% identical. However, the predicted tail fiber proteins of VP58.5 and VHML are only 77% identical and might therefore account for the divergent host range of the phages by binding to different receptors on the cell surface. Adjacent to the tail assembly module, the VP58.5 genome contains some genes (ORFs 30, 31, 32, and 35, Fig. 2B) whose products are similar to proteins of the V. parahaemolyticus phage VP882 isolated from an O3:K6 strain in Taiwan. The function of these genes is unknown, and it cannot be excluded that they play a role in host specificity. Yet, VP882 has a broader host range than does VP58.5 and infects a high proportion of V. parahaemolyticus, Vibrio vulnificus, and V. cholerae strains (23).
On the basis of the strong sequence similarity between VP58.5 and VHML, it is tempting to speculate that the two phages have the same lifestyle. Thus, it is notable that the Vp58.5 prophage is a linear plasmid with covalently closed ends whereas VHML was suggested to integrate into the host cell's chromosome (28, 29). Linear plasmid replication of telomere phages requires four components: (i) a multifunctional replication protein (RepA), (ii) an origin of replication (ori) located within repA, (iii) a protelomerase (Tel), and (iv) a telomere resolution site (telRL), which is the target of Tel (37). All of these elements exist in VP58.5 and VHML. The predicted RepA proteins of the phages are 96% identical. The construction of the replicative circular miniplasmid pJH1201 comprising a 4.8-kb restriction fragment of the Vp58.5 prophage indicated that the origin of this phage is located close to or even within repA, similarly to the replication genes of N15, PY54, and ΦKO2 (35, 42). Plasmid pJH1201 coexists with the aforementioned prophages in the same cell. Thus, there are currently three incompatibility groups of telomere phages: (i) N15/ΦKO2, (ii) PY54, and (iii) VP58.5. Due to the close relationship of VHML and VP58.5, it is likely that VHML and VP58.5 belong to the same group, while the more distantly related phages ΦHAP-1 and VP882 may constitute a fourth incompatibility group.
For the conversion of the linear phage genome into the linear plasmid and for linear plasmid replication, a protelomerase, which processes the telomere resolution site telRL, is essential (8). Both elements can be identified in VP58.5 and VHML. The putative protelomerases are 92% identical and contain all motifs conserved in other protelomerases and integrases (see Fig. S1 in the supplemental material) (11). Upstream of the respective tel gene, both phages harbor a large inverted repeat of 88 bp (VP58.5) or 66 bp (VHML) (28). The VP58.5 repeat is a perfect palindrome and apparently the target for the protelomerase of this phage (Fig. 3). Although its overall similarity to telRL sites of other telomere phages is low, an alternating 11-bp Pur/Pyr sequence is located close to the palindrome center that is likely to form a region of Z-DNA structure (37). The VHML repeat is very similar to the VP58.5 sequence. However, 24 nucleotides in the center of the VP58.5 palindrome are absent in VHML. The core region of the telomere resolution site is crucial for processing (9). Therefore, it is conceivable that the VHML inverted repeat may not function as a substrate for the VHML protelomerase to generate a linear plasmid. Interestingly, the two arms of the inverted repeat were reported to be the termini of the VHML phage genome (28). In contrast, VP58.5 phage DNA contains cohesive ends. The cos site of VP58.5 also exists in VHML, and its flanking DNA sequences, which harbor some repeats, are almost identically present in the V. harveyi phage. Only 4 out of 500 nucleotides are divergent within this DNA region. In view of the fact that the predicted terminase subunits of the phages are 98% and 99% identical, it is puzzling why the VHML phage DNA does not possess cohesive ends but a terminal inverted repeat which corresponds to the ends of the Vp58.5 plasmid prophage. Oakey et al. speculated that, upon infection of a susceptible host, the phage VHML genome becomes integrated into the bacterial chromosome by a transposition mechanism, similar to that of phage Mu (28). In this model, the terminal inverted repeat of the phage DNA was suggested to function as the core sequence recognized by a recombinase, the possible VHML ORF 10 product. A transposase and the protelomerase of VHML were also reported to be implicated in the integration of the phage genome (29). It is noteworthy that VP58.5 contains two ORFs (ORF 20 and ORF 47) whose possible products might function as a recombinase and a transposase, 99% and 80% identical to the corresponding VHML proteins, respectively. Nevertheless, our data clearly demonstrate that the Vp58.5 prophage is a linear plasmid with terminal hairpins, whereas integration into the V. parahaemolyticus chromosome was not observed (data not shown). Therefore, it remains open why the two closely related phages exhibit such different properties. The intriguing question arises whether the central part of the large VP58.5 palindrome which is lacking in the terminal VHML repeat may be responsible for the different lifestyles of the phages. Lima-Mendez et al. supposed that the VHML prophage may be a linear plasmid as well because it encodes a protelomerase and a plasmid-partitioning protein (24). We would have liked to verify this assumption by suitable experiments, but unfortunately VHML was not made available for this study. It has been reported elsewhere that the phage was lost (26). Thus, to elucidate the importance of the central nucleotides of the VP58.5 palindrome for linear plasmid replication, a deletion could be introduced into the tel site of this phage by homologous recombination. Alternatively, the VP58.5 protelomerase could be purified and used for in vitro assays with the tel sites of VP58.5 and VHML.
Phage VHML has been associated with the virulence of V. harveyi strains for some marine animals (2, 27, 30, 31). It has been reported elsewhere that after infection with VHML, the phenotypic profile of the bacteria changed (40). Production of hemolysin was upregulated, and some toxin-related proteins were secreted by the lysogens (27). Oakey et al. speculated that the VHML ORF 17 product, a putative adenine methyltransferase (Dam), might be involved in virulence by acting as a transcriptional regulator. In addition, this protein contains a motif similarly detected in some ADP-ribosylating toxins (28, 29). A Dam methylase was also reported to be associated with the virulence of V. parahaemolyticus strains harboring phage VP882 (23). Phage VP58.5 possesses two overlapping ORFs whose possible products are 98% and 100% identical, respectively, to the proposed VHML adenine methyltransferase. However, a possible relation of VP58.5 or its methyltransferases with the virulence of the pandemic V. parahaemolyticus strain has not been studied yet. Nonetheless, oceanic phages are generally critical components of the global ecosystem, where they play a major role in bacterial mortality, gene expression, and evolution (32). Temperate phages can increase the host's sensitivity to environmental factors, as demonstrated for the enhancement of UV light sensitivity mediated by VP58.5 (41). In addition, phages participate in the evolution of marine bacteria as an efficient horizontal gene transfer factor (3).
In conclusion there are seven temperate phages known to possess a protelomerase gene. According to their morphology, these phages can be divided into two subgroups, the siphoviruses N15, PY54, and ΦKO2 isolated from the Enterobacteriaceae and the marine myoviruses ΦHAP-1, VP882, VP58.5, and VHML (23, 26, 29, 41). In spite of the fact that siphoviruses and myoviruses belong to different phage families, telomere phages described so far are very similar in terms of their gene content and genome organization (24). Additional sequence data on new telomere phages will make it easier to answer the question of whether all members of this group share a common ancestor.
Supplementary Material
Acknowledgments
We thank Jochen Reetz and Maria Vargas from the Bundesinstitut für Risikobewertung (BfR) for electron microscopy of phage VP58.5 and Erich Lanka for critical reading of the manuscript. We are also indebted to Sebastian Beck from the Humboldt-Universität Berlin for his support in mass spectrometry.
Beatriz Zabala acknowledges a scholarship from Deutscher Akademischer Austausch Dienst (DAAD). Her work was supported in part by a grant from FONDECYT, 1070658. The work of J. A. Hammerl was supported by a grant from the Deutsche Forschungsgemeinschaft (DFG).
Footnotes
Published ahead of print on 8 July 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Austin, B., A. C. Pride, and G. A. Rhodie. 2003. Association of a bacteriophage with virulence in Vibrio harveyi. J. Fish Dis. 26:55-58. [DOI] [PubMed] [Google Scholar]
- 3.Breitbart, M., and F. Rohwer. 2005. Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 13:278-284. [DOI] [PubMed] [Google Scholar]
- 4.Brown, C. M., M. E. Dalphin, P. A. Stockwell, and W. P. Tate. 1993. The translational termination signal database. Nucleic Acids Res. 21:3119-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cabello, F. C., R. T. Espejo, M. C. Hernandez, M. L. Rioseco, J. Ulloa, and J. A. Vergara. 2007. Vibrio parahaemolyticus O3:K6 epidemic diarrhea, Chile, 2005. Emerg. Infect. Dis. 13:655-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casjens, S. R., E. B. Gilcrease, W. M. Huang, K. L. Bunny, M. L. Pedulla, M. E. Ford, J. M. Houtz, G. F. Hatfull, and R. W. Hendrix. 2004. The ΦKO2 linear plasmid prophage of Klebsiella oxytoca. J. Bacteriol. 186:1818-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Covarrubias, L., and F. Bolivar. 1982. Construction and characterization of new cloning vehicles. VI. Plasmid pBR329, a new derivative of pBR328 lacking the 482-base-pair inverted duplication. Gene 17:79-89. [DOI] [PubMed] [Google Scholar]
- 8.Deneke, J., G. Ziegelin, R. Lurz, and E. Lanka. 2000. The protelomerase of temperate Escherichia coli phage N15 has cleaving-joining activity. Proc. Natl. Acad. Sci. USA 97:7721-7726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deneke, J., G. Ziegelin, R. Lurz, and E. Lanka. 2002. Phage N15 telomere resolution. Target requirements for recognition and processing by the protelomerase. J. Biol. Chem. 277:10410-10419. [DOI] [PubMed] [Google Scholar]
- 10.Ermolaeva, M. D., H. G. Khalak, O. White, H. O. Smith, and S. L. Salzberg. 2000. Prediction of transcription terminators in bacterial genomes. J. Mol. Biol. 301:27-33. [DOI] [PubMed] [Google Scholar]
- 11.Esposito, D., and J. J. Scocca. 1997. The integrase family of tyrosine recombinases: evolution of a conserved active site domain. Nucleic Acids Res. 25:3605-3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuenzalida, L., L. Armijo, B. Zabala, C. Hernandez, M. L. Rioseco, C. Riquelme, and R. T. Espejo. 2007. Vibrio parahaemolyticus strains isolated during investigation of the summer 2006 seafood related diarrhea outbreaks in two regions of Chile. Int. J. Food Microbiol. 117:270-275. [DOI] [PubMed] [Google Scholar]
- 13.Fuenzalida, L., C. Hernandez, J. Toro, M. L. Rioseco, J. Romero, and R. T. Espejo. 2006. Vibrio parahaemolyticus in shellfish and clinical samples during two large epidemics of diarrhoea in southern Chile. Environ. Microbiol. 8:675-683. [DOI] [PubMed] [Google Scholar]
- 14.Fuerste, J. P., W. Pansegrau, G. Ziegelin, M. Kroeger, and E. Lanka. 1989. Conjugative transfer of promiscuous IncP plasmids: interaction of plasmid-encoded products with the transfer origin. Proc. Natl. Acad. Sci. USA 86:1771-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez-Escalona, N., V. Cachicas, C. Acevedo, M. L. Rioseco, J. A. Vergara, F. Cabello, J. Romero, and R. T. Espejo. 2005. Vibrio parahaemolyticus diarrhea, Chile, 1998 and 2004. Emerg. Infect. Dis. 11:129-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Groth, A. C., and M. P. Calos. 2004. Phage integrases: biology and applications. J. Mol. Biol. 335:667-678. [DOI] [PubMed] [Google Scholar]
- 17.Hall, M. C., and S. W. Matson. 1999. Helicase motifs: the engine that powers DNA unwinding. Mol. Microbiol. 34:867-877. [DOI] [PubMed] [Google Scholar]
- 18.Hamashima, H., M. Iwasaki, and T. Arai. 1995. A simple and rapid method for transformation of Vibrio species by electroporation. Methods Mol. Biol. 47:155-160. [DOI] [PubMed] [Google Scholar]
- 19.Hammerl, J. A., I. Klein, B. Appel, and S. Hertwig. 2007. Interplay between the temperate phages PY54 and N15, linear plasmid prophages with covalently closed ends. J. Bacteriol. 189:8366-8370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hertwig, S., I. Klein, R. Lurz, E. Lanka, and B. Appel. 2003. PY54, a linear plasmid prophage of Yersinia enterocolitica with covalently closed ends. Mol. Microbiol. 48:989-1003. [DOI] [PubMed] [Google Scholar]
- 21.Hertwig, S., I. Klein, V. Schmidt, S. Beck, J. A. Hammerl, and B. Appel. 2003. Sequence analysis of the genome of the temperate Yersinia enterocolitica phage PY54. J. Mol. Biol. 331:605-622. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 23.Lan, S. F., C. H. Huang, C. H. Chang, W. C. Liao, I. H. Lin, W. N. Jian, Y. G. Wu, S. Y. Chen, and H. C. Wong. 2009. Characterization of a new plasmid-like prophage in a pandemic Vibrio parahaemolyticus O3:K6 strain. Appl. Environ. Microbiol. 75:2659-2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lima-Mendez, G., J. van Helden, A. Toussaint, and R. Leplae. 2008. Reticulate representation of evolutionary and functional relationships between phage genomes. Mol. Biol. Evol. 25:762-777. [DOI] [PubMed] [Google Scholar]
- 25.McDonell, M. W., M. N. Simon, and F. W. Studier. 1977. Analysis of restriction fragments of T7 DNA and determination of molecular weights by electrophoresis in neutral and alkaline gels. J. Mol. Biol. 110:119-146. [DOI] [PubMed] [Google Scholar]
- 26.Mobberley, J. M., R. N. Authement, A. M. Segall, and J. H. Paul. 2008. The temperate marine phage ΦHAP-1 of Halomonas aquamarina possesses a linear plasmid-like prophage genome. J. Virol. 82:6618-6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Munro, J., J. Oakey, E. Bromage, and L. Owens. 2003. Experimental bacteriophage-mediated virulence in strains of Vibrio harveyi. Dis. Aquat. Organ. 54:187-194. [DOI] [PubMed] [Google Scholar]
- 28.Oakey, H. J., B. R. Cullen, and L. Owens. 2002. The complete nucleotide sequence of the Vibrio harveyi bacteriophage VHML. J. Appl. Microbiol. 93:1089-1098. [DOI] [PubMed] [Google Scholar]
- 29.Oakey, H. J., B. R. Cullen, and L. Owens. 2005. A hypothetical model for VHML phage conversion of Vibrio harveyi. Dis. Asian Aquac. V:457-464. [Google Scholar]
- 30.Oakey, H. J., and L. Owens. 2000. A new bacteriophage, VHML, isolated from a toxin-producing strain of Vibrio harveyi in tropical Australia. J. Appl. Microbiol. 89:702-709. [DOI] [PubMed] [Google Scholar]
- 31.Owens, L., and N. Busico-Salcedo. 2006. Vibrio harveyi: pretty problems in paradise, p. 266-280. In F. L. Thompson, B. Austin, and J. Swings (ed.), The biology of the vibrios. ASM Press, Washington, DC.
- 32.Paul, J. H. 2008. Prophages in marine bacteria: dangerous molecular time bombs or the key to survival in the seas? ISME J. 2:579-589. [DOI] [PubMed] [Google Scholar]
- 33.Payne, M., J. Oakey, and L. Owens. 2004. The ability of two different Vibrio spp. bacteriophages to infect Vibrio harveyi, Vibrio cholerae and Vibrio mimicus. J. Appl. Microbiol. 97:663-672. [DOI] [PubMed] [Google Scholar]
- 34.Perkins, D. N., D. J. Pappin, D. M. Creasy, and J. S. Cottrell. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551-3567. [DOI] [PubMed] [Google Scholar]
- 35.Ravin, N. V., V. V. Kuprianov, E. B. Gilcrease, and S. R. Casjens. 2003. Bidirectional replication from an internal ori site of the linear N15 plasmid prophage. Nucleic Acids Res. 31:6552-6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ravin, V., N. Ravin, S. Casjens, M. E. Ford, G. F. Hatfull, and R. W. Hendrix. 2000. Genomic sequence and analysis of the atypical temperate bacteriophage N15. J. Mol. Biol. 299:53-73. [DOI] [PubMed] [Google Scholar]
- 37.Rybchin, V. N., and A. N. Svarchevsky. 1999. The plasmid prophage N15: a linear DNA with covalently closed ends. Mol. Microbiol. 33:895-903. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 39.Steven, A. C., B. L. Trus, J. V. Maizel, M. Unser, D. A. Parry, J. S. Wall, J. F. Hainfeld, and F. W. Studier. 1988. Molecular substructure of a viral receptor-recognition protein. The gp17 tail-fiber of bacteriophage T7. J. Mol. Biol. 200:351-365. [DOI] [PubMed] [Google Scholar]
- 40.Vidgen, M., J. Carson, M. Higgins, and L. Owens. 2006. Changes to the phenotypic profile of Vibrio harveyi when infected with the Vibrio harveyi myovirus-like (VHML) bacteriophage. J. Appl. Microbiol. 100:481-487. [DOI] [PubMed] [Google Scholar]
- 41.Zabala, B., K. Garcia, and R. T. Espejo. 2009. Enhancement of UV light sensitivity of a Vibrio parahaemolyticus O3:K6 pandemic strain due to natural lysogenization by a telomeric phage. Appl. Environ. Microbiol. 75:1697-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ziegelin, G., N. Tegtmeyer, R. Lurz, S. Hertwig, J. Hammerl, B. Appel, and E. Lanka. 2005. The repA gene of the linear Yersinia enterocolitica prophage PY54 functions as a circular minimal replicon in Escherichia coli. J. Bacteriol. 187:3445-3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.