Abstract
Naturally occurring hepatitis C virus (HCV) subgenomic RNAs have been found in several HCV patients. These subgenomic deletion mutants, mostly lacking the genes encoding envelope glycoproteins, were found in both liver and serum, where their relatively high abundance suggests that they are capable of autonomous replication and can be packaged and secreted in viral particles, presumably harboring the envelope proteins from wild type virus coinfecting the same cell. We recapitulated some of these natural subgenomic deletions in the context of the isolate JFH-1 and confirmed these hypotheses in vitro. In Huh-7.5 cells, these deletion-containing genomes show robust replication and can be efficiently trans-packaged and infect naïve Huh-7.5 cells when cotransfected with the full-length wild-type J6/JFH genome. The genome structure of these natural subgenomic deletion mutants was dissected, and the maintenance of both core and NS2 regions was proven to be significant for efficient replication and trans-packaging. To further explore the requirements needed to achieve trans-complementation, we provided different combinations of structural proteins in trans. Optimal trans-complementation was obtained when fragments of the polyprotein encompassing core to p7 or E1 to NS2 were expressed. Finally, we generated a stable helper cell line, constitutively expressing the structural proteins from core to p7, which efficiently supports trans-complementation of a subgenomic deletion encompassing amino acids 284 to 732. This cell line can produce and be infected by defective particles, thus representing a powerful tool to investigate the life cycle and relevance of natural HCV subgenomic deletion mutants in vivo.
Hepatitis C virus (HCV) is an enveloped virus belonging to the family Flaviviridae. The virus genome is a positive-stranded RNA of about 9,600 nucleotides, which contains a single open reading frame (ORF) encoding both structural (core, E1, E2, and p7) and nonstructural (NS2, NS3, NS4A, NS4B, NS5A, and NS5B) proteins (47). Two highly conserved untranslated regions (UTRs) are found at the 5′ and 3′ ends, which play critical roles in both viral translation and replication (13, 14, 23).
HCV is estimated to infect 170 million people worldwide (1, 43) and in a high percentage of individuals causes a chronic liver infection that frequently evolves into an array of diseases, including cirrhosis (12, 15, 38) and hepatocellular carcinoma (4, 7, 17, 30, 38, 49).
The HCV RNA-dependent RNA polymerase (NS5B) has a high frequency of incorrect nucleotide insertions, in the range of 10−4 to 10−5 base substitutions per site, which can result in the rapid generation of HCV quasispecies (37, 45). Because of this huge genetic diversity, HCV is currently classified into six major genotypes and more than 80 subtypes (44). Recombination may be another mechanism exploited by HCV to increase genetic diversity: naturally occurring intergenotypic recombinant viruses that often have their recombination points in the trans-membrane domains of NS2 were recently identified (20, 21, 26, 27, 33, 35).
Recent publications have reported the presence of natural HCV subgenomic RNAs in serum and liver of infected patients, mostly containing large in-frame deletions from E1 up to NS2, always found together with the full-length wild-type (wt) RNAs (5, 16, 36, 54). These mutant viral genomes persist for a long time (at least 2 years), and sequence analysis suggests that subgenomic (the predominant species during this period) and full-length HCV evolve independently (54). The relative abundance and persistence of such subgenomic RNAs in vivo suggests that (i) they are capable of autonomous replication and (ii) they can be packaged and secreted in infectious viral particles, presumably harboring the envelope proteins from wt virus coinfecting the same cell.
Analysis of the genetic structure of 18 independent subgenomic deletion-containing RNAs (5, 16, 36, 54) strongly suggests that the possibility of recombination and/or deletion is restricted to specific regions.
As expected, the 5′ UTR, the 3′ UTR, and the region coding from NS3 to NS5B are always conserved, in line with the notion that these regions are the minimal requirements for RNA replication. In addition to the regions required for RNA replication, however, naturally occurring subgenomic HCV RNA invariably contains an intact core region and the protease domain of NS2. In fact, the 5′ deletion boundary falls between the codons for amino acid 189 of core and amino acid 4 of E1 in 33% of cases and between the codons for amino acids 21 and 29 of E1 in 39% of cases, such that in 72% of cases overall, the 5′ boundary is between the codons for amino acid 189 of core and amino acid 29 of E1 (see Fig. S1 in the supplemental material). Likewise, the 3′ boundary shows a distinct localization, occurring between the codons for amino acids 51 and 79 of NS2 in 56% of cases.
In the present study, we modified the infectious isolate JFH-1 in order to recapitulate in vitro the genetic structure of two of the most representative in-frame natural subgenomic deletion-containing RNAs found circulating in patients (28, 51, 57). Using this system, we analyzed natural subgenomic variants for their ability to replicate autonomously and demonstrated that the natural subgenomic deletion mutants are replication competent and are trans-packaged into infectious virions when coexpressed together with wt virus. Furthermore, our data suggest that the presence of the NS2 protease domain is required in order to generate the correct NS3 N terminus, required for RNA replication. Unexpectedly, the presence of NS2 generates, in turn, a strict cis requirement for the core region in order to allow efficient trans-packaging of the subgenomic RNA, revealing a complex interplay between the NS2 and the core viral genes. Finally, we performed trans-complementation studies of the subgenomic deletions with different HCV structural proteins to gain insight into the minimal requirements for the assembly and release of HCV virions.
MATERIALS AND METHODS
Cell lines and culture conditions.
The human hepatoma-derived cell line Huh-7.5 (6) was grown in high-glucose Dulbecco's modified Eagle medium (DMEM; Life Technologies) supplemented with 2 mM l-glutamine, 100 U/ml of penicillin, 100 μg/ml of streptomycin, and 10% fetal bovine serum. A stable cell line expressing the region core-p7 was generated by transfection with 2.5 μg of pEF6/V5-core-p7 plasmid in Huh-7.5 cells that were subsequently cultured in the presence of 3 μg/ml of blasticidin S HCl (Invitrogen). Blasticidin S-resistant clones were isolated and expanded. Clone 9 was selected following immunostaining with anticore antibody and a functional assay. Cells were subcultivated twice per week, with a 1:4 split ratio.
Manipulation of nucleic acids and construction of recombinant plasmids.
Nucleic acids were manipulated according to standard protocols, and plasmid DNA was prepared from overnight cultures in Luria-Bertani broth using the CsCl method (41). Deletions mutants were constructed in the context of the J6/JFH chimera using a PCR-based strategy and employing the following oligonucleotides: for J6/JFH-Δ284-736, E1-p7rev (5′-GATGAGCATCCATAAGCAGGCGCACATCACCCCACCGCAGAGGT-3′) and E1-p7for (5′-A CCTCTGCGGTGGGGTGATGTGCGCCTGCTTATGGATGCTCATC -3′); for J6/JFH-Δ212-890, 212-886rev (5′-GCCAAGAGCCACTTGGTTATGTCAAAACCAATGCTGTCATTGGTGCAGTCGTTAGT-3′) and 212-886for (5′-ACTAACGACTGCACCAATGACAGCATTGGTTTTGACATAACCAAGTGGCTCTTGGC-3′); for J6/JFH-ΔE1E2, 217-567for (5′-GACT GCACCAATGACAGCATTACCTGGCAGCTCGCACCACCCTGCCGTA CTAGAGCTGACT-3′) and 217-567rev (5′-AGTCAGCTCTGATACGGCAGGGTGGTGCGAGCTGCCAGGTAATGCTGTCATTGGTGCAGT-3′).
Expression plasmids were constructed by using a PCR-based strategy and employing the appropriate oligonucleotides for cloning in the pEF6/V5His TOPO expression vector (Invitrogen). For core-NS2, Core2afor (5′-ACCATGAGCACAAATCCTAAACCT-3′) and NS22arev (5′-CTAGAGAAGACTCCACCCCTTGGA-3′); for core-p7, Core2afor and p72arev (5′-CTAAGCATAAGCCTGTTGGGGCAA-3′); for core-E2, Core2afor and E2Stop (5′-CTATGCTTCGGCCTGGCCCAACAAGATGAGCATCCATAAGCA-3′); for spE1-NS2, spCoreATG (5′-ACCATGGCAACAGGGAACTTACCCGGTTGCTCCTTTTCTATCT-3′) and NS2-2arev; for spE1-p7, spCoreATG and p72arev; for spE1-E2, spCoreATG and E2Stop.
Subgenomic deletions containing luciferase reporter gene followed by the 2A protease of foot-and-mouth disease virus (FMDV) or ubiquitin monomer (UBI) were constructed by using a PCR-based strategy and employing the following oligonucleotides: LUC-UBI-DIR (5′-GCCAAGAAGGGCGGAAAGATCGCCGTGATGCAGATCTTCGTGAAGACCCTGACG-3′), LUC-UBI-REV (5′-CGTCA GGGTCTTCACGAAGATCTGCATCACGGCGATCTTTCCGCCCTTCTTGG C-3′), UBI-NS3-DIR (5′-GCACCTGGTGCTGCGTCTCCGCGGTGGTGCTCCCATCACTGCTTATGCCCAGCAA-3′), UBI-NS3-REV (5′-TTGCTGGGCATA AGCAGTGATGGGAGCACCACCGCGGAGACGCAGCACCAGGTG C-3′), UBI-NS2-Dir (5′-GCACCTGGTGCTGCGTCTCCGCGGTGGTTATGACGCATCTGTGCATGGCCAGATA-3′), UBI-NS2-REV (5′-TATCTGGCCATGC ACAGATGCGTCATAACCACCGCGGAGACGCAGCACCAGGTG C-3′), LUC-2A-NS2Dir (5′-AGCTGGCCGGCGACGTGGAAAGCAACCCAGGCCCATATGACGCATCTGTGCATGGCCAGAT-3′), LUC-2A-NS2REV (5′- TTCCACGTCGCCGGCCAGCTTCAGCAGGTCGAAGTTCAGCAGCTGCA CGGCGATCTTTCCGCCCTTCTTGGC-3′), core-luc-dir (5′-GAAAAACCAAA AGAAACACCAACCGTCGCCCAATGGAAGACGCCAAAAACATAAAGG AAG-3′), core-luc-REV (5′-CTTCCTTTATGTTTTTGGCGTCTTCCATTGGGCGACGGTTGGTGTTTCTTTTGGTTTTTC-3′), 212E1-luc-dir (5′-TAACGACT GCACCAATGACAGCATTATGGAAGACGCCAAAAACATAAAGGAAGG C-3′), and 212E1-luc-REV (5′-GCCTTCCTTTATGTTTTTGGCGTCTTCCATAATGCTGTCATTGGTGCAGTCGTTA-3′). The firefly luciferase gene was originally cloned from pGL2 vector (Promega).
FMDV2A (60 nucleotides, corresponding to the 19-amino-acid sequence QLLNFDLLKLAGDVESNPGP) and a UBI monomer DNA fragment (228 nucleotides, corresponding to the 76-amino-acid sequence MQIFVKTLTGKTITLEVEPSDTI ENVKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLSDYNIQKESTLHLVLRL RGG) were assembled in vitro with small oligonucleotides. All the PCR-amplified fragments and final constructs were verified by sequencing.
In vitro transcription and electroporation.
Plasmid DNAs encoding full-length HCV and HCV with subgenomic deletions were linearized with XbaI and transcribed in vitro by T7 RNA polymerase using an Ambion Megascript kit under nuclease-free conditions following the manufacturer's instructions. Transcription mixtures were treated with DNase I (except for cotransfections with DNA plasmid) and purified on Microspin G25 columns (GE Healthcare) according to the manufacturer's instructions. RNA eluted in nuclease-free water was quantified by measurement of the optical density at 260 nm, immediately frozen in dry ice, and stored at −80°C. Denaturing gel electrophoresis was used to check RNA integrity.
For electroporation of HCV RNA, confluent cells from 15-cm-diameter plates were split 1:2, and after 20 h, cells were recovered in 5 ml medium, washed twice with DMEM (Life Technologies), filtered with Cell Strainer filters (Falcon), and diluted in diethyl pyrocarbonate-treated phosphate-buffered saline (PBS) at a concentration of 107 cells/ml. The appropriate amount of RNA was mixed with 200 μl of cell suspension and then electroporated using a Bio-Rad Genepulser II in a cuvette with a gap width of 0.2 cm (Bio-Rad), with two pulses at 25 μF and 0.4 V. For cotransfections of wt J6/JFH and virus with subgenomic deletions, 3 μg of each RNA was used. For RNA and DNA cotransfections, 3 μg deletion mutants or replicon RNA plus 1.5 μg structural protein DNA were used. Immediately after electric pulses, cells were incubated for 10 min at 37°C, diluted in complete DMEM, and plated.
Replication and infection assays.
For replication assays, transfected cells were plated on 96-well-plates at 105, 6.6 × 104, 5 × 104, and 3.3 × 104 cells/well, for 24, 48, 72, and 120 h, respectively, accordingly to the time frame of the assay. Cellular RNA was purified with a 96-well total RNA isolation kit (Applied Biosystems) on a Starlet workstation (Hamilton) following the manufacturer's instructions. RNA eluted in nuclease-free water was quantified by measurement of the optical density at 260 nm, and denaturing gel electrophoresis was used to check RNA integrity.
For the infection assay, 50 μl of the relevant supernatant was transferred onto 6 × 103 naïve Huh-7.5 or clone 9 cells plated in a 96-well-plate in 50 μl of complete DMEM. After 2 days, total RNA was extracted, or cells were fixed for the immunostaining assay. Polyclonal antibody α-CD81 (clone 1.3.3.22; Santa Cruz Biotechnology) at 1 μg/ml was used to inhibit infection, while NS3 protease inhibitor (PI) at 1 μM was used to inhibit replication (29).
Quantitative detection of HCV RNA and luciferase assay.
The number of HCV genomes was determined by quantitative PCR (qPCR) TaqMan assay. Briefly, a 384-well-plate-based assay was performed with 10 to 50 ng of total RNA per well. To quantify HCV in RNA samples, the following oligonucleotides and probe sets were used: HCV-IRES (general set that recognize all constructs), sense (5′-GCGAAAGGCCTTGTGGTACT-3′), antisense (5′-CACGGTCTACGAGACCTCCC-3′), and probe (5′-CCTGATAGGGTGCTTGCGAGTGCC-3′, 5′ 6-carboxyfluorescein [FAM]/3′ 6-carboxytetramethylrhodamine [TAMRA]); J6/JFH-Δ212-890 specific, sense (5′-TGCCGAAGTGAAGAACATCAGT-3′), antisense (5′-AGCCACTTGGTTATGTCAAAACC-3′), and probe (5′-CGGCTACATGGTGACTAACGACTGCACC-3′, 5′ FAM/3′ TAMRA); J6/JFH-Δ284-736 specific, sense (5′-CGGTGGGGTGATGTGCG-3′), antisense (5′-GCGTGCAAGATGACCAGCTT-3′), and probe (5′-TCATCTTGTTGGGCCAGGCCGA-3′, 5′FAM/3′TAMRA); J6/JFH-ΔE1E2 specific, sense (5′-TGCCGAAGTGAAGAACATCAGT-3′), antisense (5′-GGTGCGAGCTGCCAGGTA-3′), and probe (5′-TGACTAACGACTGCACCAATGACAGC-3′, 5′FAM/3′TAMRA); full-length J6/JFH specific, sense (5′-CACAGCTTCAACTCGTCAGGAT-3′), antisense (5′-ACCCGGAAGGCCTCGATAC-3′), and probe (5′-TCCCGAACGCATGTCCGCC-3′, 5′ FAM/3′ TAMRA); JFH-neo-replicon specific (neomycin), sense (5′-GATGGATTGCACGCAGGTT-3′), antisense (5′-GCCCAGTCATAGCCGAATAGC-3′), and probe (5′-TCCGGCCGCTTGGGTGGAG, 5′ FAM/3′ TAMRA). The primers and the probe were used at 0.9 pmol and 0.25 pmol, respectively, per 20-μl reaction mixture. All reactions were run in triplicate with ABI PRISM 7900 (Applied Biosystems) under the following conditions: 30 min at 50°C (reverse transcriptase step), 10 min at 95°C, 40 cycles of 15 s at 95°C and 1 min at 60°C. Quantitative calculations were obtained either using the comparative cycle threshold method (described in user bulletin 2 of the ABI PRISM 7700 sequence detection system; Applied Biosystems, December 1997) with the level of rRNA (rRNA 18S mix; Applied Biosystems) or by quantification with an appropriate transcript-specific RNA titration curve. Quantification of rRNA was used for normalization of HCV genome copies that were expressed per 50 ng of total RNA. The luciferase assay was performed with a Bright-Glo luciferase assay system (Promega) following the manufacturer's instructions.
Transfections in wt J6/JFH-infected Huh-7.5 cells.
Huh-7.5 cells transfected with the wt J6/JFH transcript (10 μg RNA) were cultured for 6 days, mixed 1:1 with naïve Huh-7.5 cells, cultured for an additional 6 days, and finally transfected again with 5 μg of subgenomic deletion mutants.
Immunostaining.
Cells were fixed for 20 min with ice-cold isopropanol, washed three times with PBS, blocked with 5% nonfat dry milk in PBSTS (PBS, 0.1% Triton X-100, and 0.02% sodium dodecyl sulfate), and incubated overnight at 4°C with a monoclonal anticore (mouse anti-HCV core antigen immunoglobulin G [IgG] fraction, clone 4H2-2; Austral Biologicals) or anti-NS5A (mouse anti-HCV NS5 antigen IgG fraction, clone 2F6/G11; Austral Biologicals) diluted in milk-PBSTS. After five washes with PBSTS, cells were incubated for 3 h at room temperature with anti-human IgG conjugated to alkaline phosphatase (Sigma) in milk-PBSTS. After five more washes with PBSTS and one with PBS, bound antibodies were revealed with the alkaline phosphatase substrates p-nitroblue tetrazolium chloride and 5-bromo-4-chloro-3-indolylphosphate. Foci were counted, and the results are the averages from four wells.
RESULTS
Generation of J6/JFH deletion mutants.
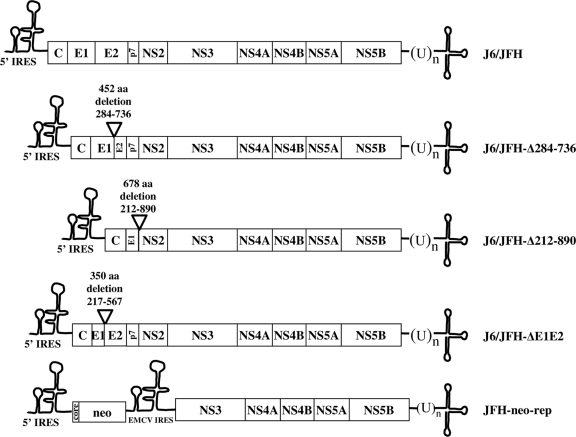
For our study, we selected two representative natural subgenomic deletions (36): Δ284-732 (corresponding to Δ284-736 in JFH-1, due to a 4-amino-acid insertion in E2 in this isolate), resulting in an in-frame 448-amino-acid deletion spanning E1 and E2 (from amino acid 284 in E1 to amino acid 732 in E2), and Δ212-886 (corresponding to Δ212-890 in JFH-1), resulting in an in-frame 674-amino-acid deletion from position 212 in E1 to amino acid 886 in NS2. While the former deletion spans the coding regions of E1 and E2, the latter completely removes E2 and p7. We generated these deletions in the J6/JFH background (J6/JFH-Δ284-736 and J6/JFH-Δ212-890 [Fig. 1]), since it efficiently supports both replication and virus production in Huh-7.5 cells (28). Of note, the NS2 boundary (amino acid 890) in J6/JFH-Δ212-890 is located at the junction between the membrane-spanning and protease domains of NS2 (55), more exactly inside the third trans-membrane domain and downstream of the C4 junction (amino acid 876), reported to be important for efficient generation of intra- and intergenotypic HCV chimeras (40). According to the hypothesized topology of NS2 (55), this deletion has a defective NS2 that cannot be membrane associated and is impaired for virus production, as already shown by Jirasko and coworkers (18). We also generated the deletion mutant J6/JFH-ΔE1E2 (deletion from the codon for amino acid 217 in E1 to that for amino acid 567 in E2), previously used as a nonpackaging control RNA transcript (3, 40, 42, 51).
FIG. 1.
Schematic representation of subgenomic HCV RNAs used in this study. The 5′ NTR, 3′ NTR, and EMCV IRES (solid line) structures are shown, and ORFs are depicted as open boxes with the polyprotein cleavage products indicated. Parental (wt) J6/JFH genome (encoding amino acids 1 to 1030 [J6] and 1031 to 3034 [JFH1]) is shown on top, and the JFH-1 replicon (JFH-neo-rep) is at the bottom.
J6/JFH subgenomes replicate efficiently in human hepatoma Huh-7.5 cells.
RNA transcripts harboring the subgenomic deletions J6/JFH-Δ284-736 and J6/JFH-Δ212-890 were transfected into naïve Huh-7.5 cells to verify whether they were replication competent. J6/JFH-ΔE1E2 and wt J6/JFH were used as controls. Total cellular RNA was extracted at 24, 48, 72, and 120 h posttransfection (p.t.), and qPCR was performed to measure the number of RNA copies. The results demonstrate that subgenomic deletion mutants are replication competent but with slightly different profiles (Fig. 2).
FIG. 2.
Replication efficiency of subgenomic deletion mutants in Huh-7.5 cells. RNA transcripts were transfected into Huh-7.5 cells, and total cellular RNA was extracted at different times from 24 to 120 h p.t. RNA was quantified by qPCR utilizing rRNA for normalization. Values are medians from at least three independent experiments. Error bars indicate standard deviations.
J6/JFH-Δ212-890 and J6/JFH-Δ284-736 replicate more efficiently than the wt construct at 24 h p.t., with a peak at 48 h followed by a rapid decline, to the extent that RNA cannot be detected at 120 h p.t. This might reflect a cytotoxic effect due to the accumulation of core protein and/or replication complexes in the cells (42). A similar replication profile is observed with J6/JFH-ΔE1E2, although with lower efficiency. J6/JFH, as expected, showed a steady progression, starting moderately low at 24 h and increasing considerably at 5 days due to efficient virus production and spreading, as demonstrated by assaying supernatant for infection (data not shown). As expected, all the subgenomic deletion mutants behaved as bona fide replicon units but lacked the ability to produce infectious particles (see Fig. 10; also data not shown).
FIG. 10.
Efficiency of infection and replication in naïve Huh-7.5 or clone 9 cells of subgenomic deletion mutants packaged in naïve Huh-7.5 or clone 9 cells. Supernatants of transfections at 48 h p.t. from the experiment described in the legend to Fig. 9 were used to infect either naïve Huh-7.5 (A) or clone 9 (B) cells. Total cellular RNA was extracted at 24, 48, and 72 h postinfection. RNA quantification was performed by qPCR. Quantification of rRNA was used for normalization. Values are numbers of HCV genome copies per 50 ng of total RNA and are averages from at least three independent experiments. Error bars indicate standard deviations.
Subgenomic deletion mutants are trans-packaged in Huh-7.5 cells infected with wt J6/JFH virus.
Having demonstrated that the naturally occurring HCV subgenomes are capable of autonomous replication in Huh-7.5 cells, we investigated whether they could be packaged into infectious virions in the presence of the wt virus.
With this aim, we transfected Huh-7.5 cells with J6/JFH RNA and passaged cells for 9 days, a time sufficient to obtain approximately 100% infected cells (57). These cells were subsequently transfected with subgenomic deletion mutants and replicon (JFH-neo-rep) RNA transcripts (22). Supernatants from each transfection were collected at 24, 48, and 72 h p.t. and transferred onto naïve Huh-7.5 cells for an infection assay. After 2 days, total cellular RNA was extracted and analyzed by real-time qPCR. In order to identify the different constructs, we used specific sets of qPCR probes that enabled us to quantify precisely each different RNA genome.
Figure 3A shows the specific J6/JFH wt RNA quantification in cells infected with the different supernatants. As expected, we observed that similar amounts of wt J6/JFH virus were present in all the supernatants from the different transfection experiments. The observed decline of J6/JFH in the supernatant collected 72 h p.t. probably resulted from the competition of replication and packaging factors between the two transfected genomes. Quantification of each individual deletion construct in the corresponding transfection (Fig. 3B) showed that both the subgenomic deletion mutants and JFH-neo-rep were trans-packaged and, upon release in the supernatants, were able to infect naïve Huh-7.5. It is worth noting that J6/JFH-Δ212-890, which is defective in NS2, could be trans-complemented not only by wt E1, E2, and p7 but also by wt NS2 expressed by wt J6/JFH. In all cases, replication and bona fide infection were demonstrated by the efficacy of a replication inhibitor (an NS3 PI) (Fig. 3) and by treatment with an anti-CD81 monoclonal antibody (data not shown), respectively.
FIG. 3.
trans-packaging efficiency of subgenomic deletion mutants in Huh-7.5 infected with wt J6/JFH. Subgenomic deletion mutants and JFH-1 replicon (JFH-neo-rep) RNA were transfected into J6/JFH-infected Huh-7.5 cells. Supernatants of each transfection experiment were collected at 24, 48, and 72 h p.t. and used to infect naïve Huh-7.5 cells. Total cellular RNA was extracted at 48 h postinfection, and RNA quantification was performed by quantitative qPCR with J6/JFH (A) or individual subgenome-specific (B) oligonucleotide-probe sets. Quantification of rRNA was used for normalization, and values are numbers of HCV genome copies per 50 ng of total RNA. Values are medians from at least three independent experiments. Error bars indicate standard deviations.
The ratio between packaged subgenomic deletion mutants and wt J6/JFH RNA was very low, about 1:40 (on average, 4 × 106 copies of J6/JFH and 1 × 105 copies of subgenomic deletion mutants per 50 ng of total RNA). As expected, with the exception of wt J6/JFH, the second passage of supernatants onto naïve Huh-7.5 cells resulted in no productive infection, presumably due to the small chance of coinfection of a single naïve cell with wt and deletion-containing genome virus. In evaluating the results, we should consider the possibility that they might have been negatively affected by many factors, such as efficiency of transfection in the infected cells, interference with or competition of the transfected RNA by the resident wt genome, and replication efficiency of each subgenomic deletion. Altogether, these results demonstrate that subgenome deletion mutants can be trans-packaged in cell culture by a wt virus.
Subgenomic deletion mutants are efficiently trans-packaged when cotransfected with wt J6/JFH RNA into Huh-7.5 cells.
Next, we decided to explore whether cotransfection of the deletion-containing genomes with wt J6/JFH RNA could result in their trans-packaging. This experimental setting would allow us to better quantify the relative replication and packaging efficiencies of the different RNA genomes. Thus, naïve Huh-7.5 cells were cotransfected with equal amounts of wt J6/JFH and individual subgenomic RNA (3 μg), and supernatants were collected at 24, 48, and 72 h p.t. and transferred onto naïve Huh-7.5 cells for a 2-day infection assay. Cellular RNA was then extracted and analyzed by qPCR to monitor genome replication as a consequence of infection.
As shown in Fig. 4A, in all cases we could measure the infection of J6/JFH-wt virus. As expected, the number of HCV copies increased over time due to the virus spreading. Considerable differences appeared when the infection of packaged subgenomic deletion mutants was measured. The deletion mutant most efficient in trans-packaging was J6/JFH-Δ212-890, whose amount was 3- and 1.6-fold higher than that of the wt J6/JFH at 24 and 48 h, respectively, followed by J6/JFH-Δ284-736 (1.3- and 0.4-fold) and J6/JFH-ΔE1E2 (0.6- and 0.2-fold) (Fig. 4B). Though measurable, the signal of the trans-packaged JFH-neo-rep was very poor compared to signals of the other subgenomic deletion mutants, being only 0.1- and 0.08-fold that of the wt genome at 24 and 48 h, respectively. Replication and infection were confirmed by treating the cells with a PI and an anti-CD81 monoclonal antibody, respectively.
FIG. 4.
Efficiency of packaging of subgenomic deletion mutants and JFH-1 replicon when cotransfected with wt J6/JFH RNA. Subgenomic deletion mutants or the JFH-1 replicon (JFH-neo-rep) was cotransfected (3 μg RNA each per 2 × 106 cells) into Huh-7.5 cells with an equal amount of wt J6/JFH RNA. Supernatants of each transfection were collected at 24, 48, and 72 h p.t. and used to infect naïve Huh-7.5 cells. Total cellular RNA was extracted at 48 h, and RNA of different genomes was quantified by quantitative qPCR with construct-specific oligonucleotide-probe sets. Quantification of rRNA was used for normalization, and HCV RNA amounts are expressed as number of genome copies per 50 ng of total RNA. (A) Results of the infection with supernatants collected at the indicated times p.t. Values are medians from at least three independent experiments. Error bars indicate standard deviations. (B) Ratio of the copy number of subgenomic deletion mutants to that of J6/JFH RNA.
It is worth noting that in the cotransfection experiments, the RNAs corresponding to the natural subgenomic deletion mutants were more efficiently trans-packaged than the in-house-generated J6/JFH-ΔE1E2 RNA. This might suggest that in vivo, selection of HCV subgenomic RNA follows the evolutionary rule of survival of the fittest. It is conceivable that natural subgenomic deletion mutants sustain the competition of wt virus for both replication and packaging, thereby achieving efficient spreading and stability in patients.
Core and NS2 contribute to replication and trans-packaging efficiency.
In order to determine the contributions of core and NS2 to both replication and trans-packaging efficiency of subgenomic deletion mutants, we generated the constructs depicted in Fig. 5 in which the core-E1 and/or NS2 coding sequences was selectively removed. To facilitate the detection of replication and infection, a firefly luciferase reporter gene was inserted upstream of NS2 or NS3, separated by the FMDV2A autoprotease (9) or a UBI monomer (6, 39) in order to guarantee the correct processing of their N termini. We were unable to generate replication-competent constructs with FMDV2A autoprotease located upstream of NS3, likely due to the incorrect generation of the NS3 N terminus. These constructs were individually tested for replication efficiency in naïve Huh-7.5 cells. After RNA transfection, luciferase activity was measured at 5, 24, 48, and 72 h (Fig. 6). Although the same amount of RNA was transfected for all constructs, luciferase readings at 5 h (measuring transient translation of the transfected RNA) were substantially different. Constructs containing the core coding region (namely, Luc-coreΔ212-NS2 and Luc-coreΔ212-NS3) showed a lower luciferase signal than those without the core region. The same results were obtained in many independent experiments, and it is therefore conceivable that the differences observed do reflect different translation efficiencies of the various constructs rather than simply different transfection efficiencies. This finding is not surprising, given that the core region has been suggested to negatively influence internal ribosome entry site (IRES)-dependent translation (8, 52, 56).
FIG. 5.
Schematic representation of subgenomic HCV mutants expressing a luciferase reporter gene. The 5′ NTR, 3′ NTR, and EMCV IRES (solid line) structures are shown, and ORFs are depicted as open boxes with the polyprotein cleavage products indicated. The luciferase-2A reporter gene was inserted after the codons for amino acid 212 of E1 and amino acid 19 of core in Luc-coreΔ212-NS2 and Luc-nocore-NS2, respectively, and before the codon for amino acid 814 of NS2 in both. The luciferase-UBI reporter gene was inserted after the codons for amino acid 212 of E1 and amino acid 19 of core in Luc-coreΔ212-NS3 and Luc-nocore-NS3, respectively, and before the codon for amino acid 1031 of NS3 in both.
FIG. 6.
Replication of subgenomic deletion mutants expressing a luciferase reporter gene. RNA transcripts were transfected into Huh-7.5 cells, and luciferase activity was measured at different times from 5 to 72 h p.t. Values are medians from at least three independent experiments. Error bars indicate standard deviations. (A) Efficiency of replication of each RNA, measured in luciferase units. (B) Results from panel A, reported as percentages of the value at 5 h for each RNA.
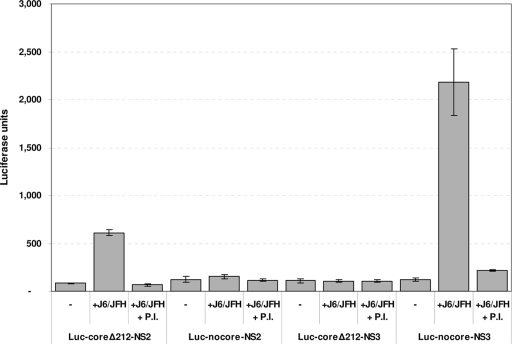
After a sharp decline at 24 h p.t., the luciferase signal associated with cells transduced with Luc-coreΔ212-NS2, Luc-nocore-NS2, and Luc-nocore-NS3 increased to levels higher than those observed at 5 h p.t., indicating amplification of the transfected RNA. Replication of the RNA constructs at 72 h was confirmed by the inhibitory effect of an NS3 PI (Fig. 6A). Unexpectedly, Luc-coreΔ212-NS3 failed to replicate. According to the relative luciferase signals, at 72 h p.t., the Luc-coreΔ212-NS2 RNA was amplified 8.3-fold compared to the 5-h reading. Conversely, for the two constructs lacking the core region, namely, Luc-nocore-NS2 and Luc-nocore-NS3, the luciferase signals at 72 h p.t. were 0.7- and 3.3-fold higher than the 5-h measurement. These data suggest that the presence of the core sequence could positively affect the replication capability of a subgenomic HCV RNA. To assess the potential of the same constructs to be trans-packaged by the wt helper virus, we cotransfected them with equal amounts of wt J6/JFH RNA into Huh-7.5 cells. After 48 h we collected the supernatants and transferred these onto naïve Huh-7.5 cells for a 2-day infection assay. As expected, no infectious particles were produced without cotransfection with J6/JFH (Fig. 7). Conversely, when cotransfected with J6/JFH, only Luc-coreΔ212-NS2 and Luc-nocore-NS3 were able to be trans-packaged by J6/JFH. Interestingly, these constructs either have both core and NS2 or have neither. In contrast, construct Luc-nocore-NS2, which showed a good replication capability, was not able to be trans-packaged efficiently. Finally, Luc-coreΔ212-NS3 was unable to replicate (even in the presence of wt J6/JFH) and consequently to be trans-packaged. In conclusion, these data reveal a complex interplay between the core region and the NS2 regions in the regulation of trans-packaging and possibly RNA replication.
FIG. 7.
trans-packaging of subgenomic deletion mutants expressing a luciferase reporter gene. Subgenomic deletion mutants containing a luciferase reporter were cotransfected (3 μg RNA each per 2 × 106 cells) in Huh-7.5 cells alone or with equal amounts of wt J6/JFH RNA. Transfection supernatants were collected at 48 h p.t. and used to infect naive Huh-7.5 cells. Luciferase activities of each infection were measured at 48 h postinfection. Values are medians from at least three independent experiments. Error bars indicate standard deviations.
Cloning of structural proteins into an expression vector and trans-complementation of subgenomes.
In order to elucidate whether trans-packaging of subgenomes could also be achieved by providing structural proteins outside the context of a replicating wt virus, we cloned them into a plasmid DNA expression vector (pEF6/V5; Invitrogen). This approach also gave us the opportunity to analyze in detail the minimal requirements for efficient trans-packaging. With this aim, we cloned the coding region from core to NS2, in various arrangements (Fig. 8). In the constructs lacking core, correct expression and localization of E1-E2 structural proteins was achieved by including the terminal region of core protein (27 amino acids) as a signal peptide (10, 11).
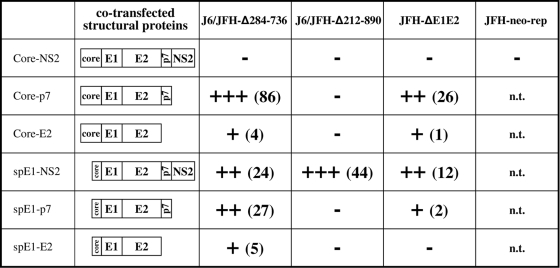
FIG. 8.
Summary of naïve Huh-7.5 infections with supernatants generated by DNA-RNA cotransfections of subgenomic deletion mutants with structural protein expression vectors at 48 h p.t. Subgenomic deletion mutants used in the cotransfection experiments are at the tops of the columns, and expression vectors are on the left. The qualitative evaluation of packaging efficiency is proportionally expressed with plus signs. Numbers in parentheses are the numbers of foci (two to four cells each) detected by immunostaining. The results are the averages from four independent experiments. n.t., not tested.
We tested the ability of these constructs to trans-package different subgenomic deletion mutants by DNA-RNA cotransfections in hepatoma cells. After cotransfection of the expression plasmids and subgenomic RNA transcripts in Huh-7.5 cells, supernatants were collected at different times and applied to naïve Huh-7.5 cells for a 2-day infection followed by immunostaining using an anticore antibody (or anti NS5A in the case of JFH-neo-rep) in order to estimate the amount of infected cells. Bona fide infection versus passage of naked (or complexed) RNA was guaranteed by the lack of positive cells when infection was performed in the presence of anti-CD81. Representative results obtained with supernatants collected at 48 h p.t. are summarized in Fig. 8.
Among all the cotransfections performed, some were not expected to produce viral particles. This concerned all the cases in which parts of the HCV coding regions were missing both in the genome RNAs and in the vectors expressing the structural proteins. On this basis, JFH-neo-rep could in theory be trans-packaged only with core-NS2 (the only construct we tested) and J6/JFH-Δ212-890, which lacks p7 and the trans-membrane domain of NS2, only with core-NS2 and spE1-NS2. The results obtained, besides confirming some of these predictions, revealed some additional interesting findings. In fact, core-NS2, which spans all structural proteins from core to the whole NS2, was not able to trans-package any of the subgenomic deletion mutants or JFH-neo-rep.
Interestingly, exclusion of NS2, as in the case of core-p7, efficiently confers complementation capacity to the expressed proteins with both subgenomic deletion mutants J6/JFH-Δ284-736 and J6/JFH-ΔE1E2, the former being more efficiently trans-packaged. Furthermore, when the core-coding region was excluded, as in spE1-NS2, J6/JFH-ΔE1E2, J6/JFH-Δ284-736, and J6/JFH-Δ212-890 were all trans-packaged. Again, it is worth noting that infectivity of the NS2-defective mutant J6/JFH-Δ212-890 was rescued by the expression of wt NS2 (namely, spE1-NS2).
Together with these results, the observation that core-NS2 is also not able to trans-complement any subgenomic deletion mutants (J6/JFH-ΔE1E2, J6/JFH-Δ284-736, or J6/JFH-Δ212-890) suggests that the presence of both core and NS2 in the expression vector does not lead to productive trans-packaging. It is sufficient to exclude one of the two, however, to restore this capability. These results suggest that the mechanism underlying trans-complementation of subgenomic deletions with wt virus (J6/JFH) is different from that achieved by simply providing structural proteins with an expression vector which is thus outside the viral context.
A stable cell line expressing core-p7 structural proteins supports both trans-packaging of deletion-containing RNA genomes and spreading of the defective virus.
The possibility of complementing subgenomic deletions with structural proteins expressed in trans prompted us to explore whether a stable cell line supporting multiple rounds of infection by defective particles containing subgenomic deletions could be generated.
Taking advantage of the presence of the blasticidin resistance gene in the expression vector described above, we selected a cell line, clone 9, stably expressing core-p7. We then tested this clone for the capacity to trans-complement the packaging of J6/JFH-Δ284-736 and J6/JFH-ΔE1E2.
If clone 9 were to prove capable of trans-packaging subgenomic RNA and of being infected, a full cycle of virus production and infection could be established, allowing defective virus to be spread. In addition, the release of virus particles might also reduce the toxicity derived from accumulation of viral proteins, such as core (see above). Taking all these hypotheses together, by transfecting subgenomes in clone 9, we expected to obtain replication profiles similar to those obtained by transfecting J6/JFH in naïve Huh-7.5.
RNA transcripts of the subgenomic deletion mutants J6/JFH-Δ284-736, J6/JFH-ΔE1E2, and wt J6/JFH were transfected into both Huh-7.5 and clone 9 cells. A large amount (10 μg) of RNA was used to achieve maximal replication and packaging. RNA replication was followed by real-time qPCR at 24, 48, and 72 h p.t.
As predicted (and shown in Fig. 2), in Huh-7.5 cells, the subgenomic deletion mutants J6/JFH-Δ284-736 and J6/JFH-ΔE1E2 replicated efficiently starting as early as at 24 h, peaking at 48 h, and declining rapidly thereafter, while wt J6/JFH increased constantly (Fig. 9). However, the transfection of clone 9 showed different kinetics. In this case, J6/JFH-Δ284-736 and J6/JFH-ΔE1E2 RNA levels also steadily increased with time, clearly demonstrating that the signal was more than the result of simple replication and likely involved production and reinfection of defective particles.
FIG. 9.
Efficiency of replication of J6/JFH-Δ284-736, J6/JFH-ΔE1E2, and J6/JFH in naïve Huh-7.5 and clone 9 cells. RNA transcripts were transfected (10 μg RNA per 2 × 106 cells) in naïve Huh-7.5 and clone 9 cells, and total cellular RNA was extracted at 24, 48, and 72 h p.t. RNA was quantified by qPCR, and quantification of rRNA was used for normalization. Values are numbers of HCV genome copies per 50 ng of total RNA and are averages from at least three independent experiments. Error bars indicate standard deviations.
In addition to monitoring RNA replication, we collected supernatants of each transfection at 2 days p.t., at which time a sufficient amount of virus particle production had been observed in previous experiments. Each supernatant was transferred onto naïve Huh-7.5 (Fig. 10A) and clone 9 (Fig. 10B) cells, and HCV replication and infection at 1, 2, and 3 days postinfection were monitored by real-time PCR. As shown in Fig. 10, with the exception of J6/JFH-Δ284-736 and J6/JFH-ΔE1E2 transfection into naïve Huh-7.5 cells, we observed infection by all supernatants, demonstrating that clone 9 produces defective particles and therefore behaves exactly as predicted. In all cases, anti-CD81 and PI completely abolished any signal, demonstrating that the signals measured derived from infection.
Similarly to transfection of naked RNA, the time-dependent increasing signal in clone 9 transfected with deletion mutants strongly suggests that clone 9 was able to support production, as well as spreading, of the defective virus particles. This was confirmed by the observation that by immunostaining with anticore antibody, single infected cells were seen in Huh-7.5 while groups of infected cells were visible in clone 9 (data not shown).
Altogether, these experiments demonstrate that clone 9 supports virus production from a transfected subgenomic HCV RNA, and this defective virus is capable of spreading in this cell line. Thus, clone 9 can be considered a helper cell line. It is noteworthy that this defective virus is still capable of infecting naïve Huh-7.5 cells, and the RNA replicates inside the infected cells but is not able to produce viruses.
DISCUSSION
In this study, we attempted to elucidate the poorly understood phenomenon of the presence of HCV deletion mutants circulating in chronically infected patients (36), liver transplant recipients (16), and patients with immunosilent infections (5). To date, the clinical relevance of these naturally occurring subgenomic deletion mutants has not been clarified. A constant feature of such deletion mutants is that they are present in large amounts both in serum and in liver, where the ratio between deletion-containing genomes and wt could be as high as 500:1 (54). This probably depends on subgenomes having to compete with the wt virus for replication space inside the hepatic cell, which, combined with the dependence on wt for trans-packaging, might have restricted the size and boundary of the deletions.
It is noteworthy that in almost all subgenomic deletion mutants reported (5, 16, 36, 54), the integrity of the core protein is maintained, as are some amino acid sequences of E1 that might be necessary for the correct core processing. We cannot exclude the possibility that this could merely increase the amount of core protein in the host cell, thus increasing the chance of subgenome trans-packaging, but our results suggest that the conservation of this region might have additional significance.
The idea that the core region contains some important RNA structures has already been explored. In attempt to elucidate the role of the core alternative reading frame, McMullan and coworkers found that when an H77 RNA transcript containing mutations that destroyed two RNA structures (stem-loops V and VI) in the core region was inoculated into a chimpanzee, this gave rise to an atypical attenuated infection (31). After a few weeks, mutations in stem-loop VI reverted to wt, suggesting that this structure plays an important role in the HCV life cycle (31). Very recently, another study reached analogous conclusions with the JFH-1 isolate, also determining the importance of other RNA structures in the same region that play important roles in translating the genome and are required for robust viral production both in vitro and in vivo (50). Also in this case, reversal of these mutations was observed upon inoculation in the uPA-SCID mouse model (50).Therefore, RNA sequences contained in the core region are cis-acting elements important but not essential for RNA replication. In line with this notion, both the Luc-nocore-NS3 and the Luc-nocore-NS2 RNA constructs replicated efficiently in cell culture. Despite efficient RNA replication, however, the former construct was also efficiently trans-packaged, while the latter was not trans-packaged or was trans-packaged at very low efficiency. The only difference between these two constructs is the presence of NS2 in Luc-nocore-NS2. Thus, in this particular context, the presence of NS2 appears to hinder the ability of a subgenomic HCV RNA to be efficiently trans-packaged by the wt helper virus. Interestingly, however, the impairment due to the presence of NS2 was rescued by the presence of core, pointing to an important cross talk between these two viral genes.
A similar conclusion could be drawn from a comparison of the monocistronic constructs containing core region: Luc-coreΔ212-NS2 and Luc-coreΔ212-NS3. The former was replication and trans-packaging competent, while the latter was totally impaired, confirming that the presence or absence of NS2 makes the difference.
Consistent with our finding, the genomic region in addition to core that is almost always maintained in natural subgenomic deletion mutants is NS2 and, in particular, its catalytic domain. In fact, NS2 plays a critical function in the processing of the NS2/NS3 junction (24, 53), and the correct NS3 N terminus is absolutely necessary for HCV replication (19). Thus, it is conceivable that it may be extremely difficult to obtain a precise boundary at the NS3 N terminus by whatever natural mechanism leads to the generation of internal deletions in vivo (e.g., recombination). Therefore, the presence of a catalytically active NS2 protease domain guarantees a precise cleavage of the NS2/NS3 junction and offers some flexibility at the 3′ recombination point, locating it almost anywhere between E1-E2 and the end of the trans-membrane NS2 domain. It is notable that in both the NS3-NS5B replicon and in the Luc-nocore-NS3 construct reported here (both without core and NS2 regions and, nevertheless, efficiently trans-packaged), the correct NS3 N terminus is obtained via the encephalomyocarditis virus (EMCV) IRES and via the presence of a UBI monomer, respectively. In addition, although NS2 has been demonstrated to be dispensable for replication, it is essential for virus production even independently of its protease activity (18, 19, 40), likely playing some important structural role. Altogether, these experiments further confirm the strict positive relationship of core and NS2 in both replication and trans-packaging efficiency. The precise nature of this interplay has yet to be elucidated, but important core/NS2 direct or indirect interactions have also been implied by the observation that an alanine-scanning mutant in core (amino acids 69 to 72) could be efficiently rescued for virus production by an NS2-A880P compensating mutation (34).
In summary, we speculate that the specific genome regions core and NS2 must be maintained for the efficient replication and trans-packaging of natural subgenomes. NS2 at least is necessary for the correct determination of the NS3 N terminus and its presence, for an efficient trans-packaging, requires the presence of the core region.
trans-complementation of HCV proteins has been demonstrated in vitro in a few cases for both structural and nonstructural proteins. The latter have been extensively investigated with the HCV replicon system, and the only protein that can be provided in trans is NS5A (2, 48). trans-complementation of core has been demonstrated with a 17- to 163-amino-acid core deletion in JFH-1 which is complemented and trans-packaged by a core-expressing DNA plasmid (32). trans-complementation of larger structural protein deletions has been demonstrated with JFH-1 by Jones et al. and, more recently, by Steinmann and coworkers (19, 46). In the work of Jones et al., two JFH-1 luciferase constructs, one with E1-E2 deleted (identical deletion to our J6/JFH-ΔE1E2) and the other in p7, both replication competent, were cotransfected into Huh-7.5 cells with the rescue of virus production. A more extensive study was performed by Steinmann and coworkers in which both helper viruses and a stable cell line were utilized for complementation in trans (46). Although in overall agreement with their results, the study presented here reveals some discrepancies.
When wt JFH-1 was cotransfected together with either Luc-ΔE1E2 (an IRES-luc-JFH-1ΔE1E2 construct) or Luc-NS3-5B (an IRES-luc-NS3/5B replicon construct), the latter was packaged much more efficiently than the former (46). In contrast, we found that in similar cotransfection experiments, with wt J6/JFH as the helper, J6/JFH-ΔE1E2 is trans-packaged with an efficiency similar to or better than that of the JFH-neo-rep construct (Fig. 4). This might be explained by the different structure of the packaged genomes. It is known that Luc-ΔE1/E2 replicates with substantially delayed kinetics compared to JFH-1 (25). More likely, as discussed above with regard to the boundaries of natural subgenomic deletions, the deletion of important core RNA sequences might be detrimental to trans-packaging efficiency.
Steinmann et al. generated a stable cell line expressing core-E1 and E2-p7-NS2 from two distinct genetic units (46). This packaging cell line was able to trans-complement both a Δp7 deletion of JFH-1 (Jc1Δp7) and a Luc replicon (Luc-SG), although the former was much more efficient than the latter (about 10-fold).
Using our core-NS2 expression plasmid, we were unable to achieve efficient trans-packaging of any of the subgenomic deletion mutants we investigated, including the JFH-1 replicon. This discrepancy may be due to the different genetic organization of packaging cell lines: two genetic units (separately encoding core-E1 and E2-p7-NS2) as opposed to a single RNA messenger encoding core-NS2 proteins, in our case processed from a common polyprotein precursor. The fact that core-NS2 polyprotein could be correctly processed and topologically oriented in cells is demonstrated by the finding that inserting the EMCV IRES between NS2 and NS3 did not interfere with RNA replication and reduced infectivity titers only very moderately (19).
It is tempting to speculate that our core-NS2 expression vector might generate structural proteins that might not easily be available for trans-packaging. In DNA-RNA cotransfections with subgenomic J6/JFH-Δ284-736 (which contains a wt NS2), the removal of either core or NS2 from the core-NS2 expression vector restores the capacity for trans-complementation (Fig. 8). In the first case, E1-E2-p7-NS2 could not be targeted independently to lipid droplets due to the absence of core (32). In the second case, that of core-E1-E2-p7, the lack of NS2 might play an important role.
Upon analysis of these trans-complementation experiments, one might speculate that NS2 participates in two independent complexes: one that includes structural proteins (NS2 plus core-E1-E2-p7) and one that includes NS3-NS5B/RNA (NS2 plus replication complex). These complexes probably have different localizations: around lipid droplets for the former and diffusely on the ER membranes for the latter (32). Once these two complexes are separately formed, they cannot interact to assemble a virus. When NS2 is removed from any of these complexes (or the removal of core affects assembly complex formation or lipid droplet localization), the reciprocal interaction is restored. NS2 might behave as an exclusive alternative bridge between these two complexes.
It is conceivable that replication of such subgenomes in the hepatocyte in the absence of coinfection by wt virus could lead to an abnormal accumulation of core proteins in lipid droplets inside the host cells, as demonstrated in vitro with genomes that are poorly packaged (42). It would be interesting to study in detail whether core protein accumulation as a consequence of the efficient replication of these subgenomes and the lack of egress of virus could influence and contribute to liver pathogenesis during the progression of HCV-associated chronic hepatitis.
In conclusion, we have studied the in vitro behavior of two representative in-frame subgenomic deletion mutants found circulating in patients. The study of natural subgenomic deletion mutants, as opposed to artificial constructs generated in laboratories, gives us the unique opportunity to study the HCV life cycle with a wider range of physiological reagents and provides insight into the requirements of HCV for survival in vivo. This is a significant step forward in comparison with other studies of trans-packaging performed so far, confirming the hypotheses that naturally occurring deletion-containing HCV genomes can be competent for replication and packaged into infectious virions by coexistence with wt virus. In addition, we have found that the boundaries of the natural subgenomic deletions correspond to precise requirements for efficient replication and trans-packaging. We have established trans-packaging protein requirements as well as a trans-packaging and infectious cell line capable of mimicking the effect of wt virus coinfection. In addition, the variety of structural proteins provided for trans-packaging has highlighted important features that might help dissect the interaction of the replication complex with structural proteins, and this will form the basis of future studies.
Supplementary Material
Acknowledgments
We are grateful to Takaji Wakita for the JFH-1 replicon and to Charles Rice for Huh-7.5 cells. We also thank Janet Clench for editorial assistance. We thank Nicola La Monica, Licia Tomei, Matthew Bottomley, and Maria Teresa Catanese for valuable advice, critical discussion, and input on the manuscript.
Footnotes
Published ahead of print on 8 July 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Alter, M. J. 2007. Epidemiology of hepatitis C virus infection. World J. Gastroenterol. 13:2436-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Appel, N., U. Herian, and R. Bartenschlager. 2005. Efficient rescue of hepatitis C virus RNA replication by trans-complementation with nonstructural protein 5A. J. Virol. 79:896-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Appel, N., M. Zayas, S. Miller, J. Krijnse-Locker, T. Schaller, P. Friebe, S. Kallis, U. Engel, and R. Bartenschlager. 2008. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog. 4:e1000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barazani, Y., J. R. Hiatt, M. J. Tong, and R. W. Busuttil. 2007. Chronic viral hepatitis and hepatocellular carcinoma. World J. Surg. 31:1243-1248. [DOI] [PubMed] [Google Scholar]
- 5.Bernardin, F., S. L. Stramer, B. Rehermann, K. Page-Shafer, S. Cooper, D. R. Bangsberg, J. Hahn, L. Tobler, M. Busch, and E. Delwart. 2007. High levels of subgenomic HCV plasma RNA in immunosilent infections. Virology 365:446-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blight, K. J., J. A. McKeating, and C. M. Rice. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blonski, W., and K. R. Reddy. 2008. Hepatitis C virus infection and hepatocellular carcinoma. Clin. Liver Dis. 12:661-674. [DOI] [PubMed] [Google Scholar]
- 8.Boni, S., J. P. Lavergne, S. Boulant, and A. Cahour. 2005. Hepatitis C virus core protein acts as a trans-modulating factor on internal translation initiation of the viral RNA. J. Biol. Chem. 280:17737-17748. [DOI] [PubMed] [Google Scholar]
- 9.de Felipe, P., G. A. Luke, L. E. Hughes, D. Gani, C. Halpin, and M. D. Ryan. 2006. E unum pluribus: multiple proteins from a self-processing polyprotein. Trends Biotechnol. 24:68-75. [DOI] [PubMed] [Google Scholar]
- 10.Drummer, H. E., A. Maerz, and P. Poumbourios. 2003. Cell surface expression of functional hepatitis C virus E1 and E2 glycoproteins. FEBS Lett. 546:385-390. [DOI] [PubMed] [Google Scholar]
- 11.Duvet, S., A. Op De Beeck, L. Cocquerel, C. Wychowski, R. Cacan, and J. Dubuisson. 2002. Glycosylation of the hepatitis C virus envelope protein E1 occurs posttranslationally in a mannosylphosphoryldolichol-deficient CHO mutant cell line. Glycobiology 12:95-101. [DOI] [PubMed] [Google Scholar]
- 12.Everson, G. T. 2005. Management of cirrhosis due to chronic hepatitis C. J. Hepatol 42(Suppl.):S65-S74. [DOI] [PubMed] [Google Scholar]
- 13.Friebe, P., and R. Bartenschlager. 2002. Genetic analysis of sequences in the 3′ nontranslated region of hepatitis C virus that are important for RNA replication. J. Virol. 76:5326-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friebe, P., V. Lohmann, N. Krieger, and R. Bartenschlager. 2001. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 75:12047-12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hollinger, F. B. 1999. Factors contributing to the evolution and outcome of cirrhosis in hepatitis C. Clin. Liver Dis. 3:741-755. [DOI] [PubMed] [Google Scholar]
- 16.Iwai, A., H. Marusawa, Y. Takada, H. Egawa, K. Ikeda, M. Nabeshima, S. Uemoto, and T. Chiba. 2006. Identification of novel defective HCV clones in liver transplant recipients with recurrent HCV infection. J. Viral Hepat. 13:523-531. [DOI] [PubMed] [Google Scholar]
- 17.Jin, D. Y. 2007. Molecular pathogenesis of hepatitis C virus-associated hepatocellular carcinoma. Front. Biosci. 12:222-233. [DOI] [PubMed] [Google Scholar]
- 18.Jirasko, V., R. Montserret, N. Appel, A. Janvier, L. Eustachi, C. Brohm, E. Steinmann, T. Pietschmann, F. Penin, and R. Bartenschlager. 2008. Structural and functional characterization of nonstructural protein 2 for its role in hepatitis C virus assembly. J. Biol. Chem. 283:28546-28562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones, C. T., C. L. Murray, D. K. Eastman, J. Tassello, and C. M. Rice. 2007. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J. Virol. 81:8374-8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kageyama, S., D. M. Agdamag, E. T. Alesna, P. S. Leano, A. M. Heredia, I. P. Abellanosa-Tac-An, L. D. Jereza, T. Tanimoto, J. Yamamura, and H. Ichimura. 2006. A natural inter-genotypic (2b/1b) recombinant of hepatitis C virus in the Philippines. J. Med. Virol. 78:1423-1428. [DOI] [PubMed] [Google Scholar]
- 21.Kalinina, O., H. Norder, S. Mukomolov, and L. O. Magnius. 2002. A natural intergenotypic recombinant of hepatitis C virus identified in St. Petersburg. J. Virol. 76:4034-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kato, T., T. Date, M. Miyamoto, A. Furusaka, K. Tokushige, M. Mizokami, and T. Wakita. 2003. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 125:1808-1817. [DOI] [PubMed] [Google Scholar]
- 23.Kolykhalov, A. A., S. M. Feinstone, and C. M. Rice. 1996. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 70:3363-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolykhalov, A. A., K. Mihalik, S. M. Feinstone, and C. M. Rice. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 74:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koutsoudakis, G., A. Kaul, E. Steinmann, S. Kallis, V. Lohmann, T. Pietschmann, and R. Bartenschlager. 2006. Characterization of the early steps of hepatitis C virus infection by using luciferase reporter viruses. J. Virol. 80:5308-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurbanov, F., Y. Tanaka, D. Avazova, A. Khan, F. Sugauchi, N. Kan, D. Kurbanova-Khudayberganova, A. Khikmatullaeva, E. Musabaev, and M. Mizokami. 2007. Detection of hepatitis C virus natural recombinant RF1_2k/1b strain among intravenous drug users in Uzbekistan. Hepatol. Res. 38:457-464. [DOI] [PubMed] [Google Scholar]
- 27.Legrand-Abravanel, F., J. Claudinon, F. Nicot, M. Dubois, S. Chapuy-Regaud, K. Sandres-Saune, C. Pasquier, and J. Izopet. 2007. New natural intergenotypic (2/5) recombinant of hepatitis C virus. J. Virol. 81:4357-4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 29.Ludmerer, S. W., D. J. Graham, M. Patel, K. Gilbert, M. Stahlhut, and D. B. Olsen. 2008. A transient cell-based phenotype assay for hepatitis C NS3/4A protease: application to potency determinations of a novel macrocyclic inhibitor against diverse protease sequences isolated from plasma infected with HCV. J. Virol. Methods 151:301-307. [DOI] [PubMed] [Google Scholar]
- 30.Marrero, C. R., and J. A. Marrero. 2007. Viral hepatitis and hepatocellular carcinoma. Arch. Med. Res. 38:612-620. [DOI] [PubMed] [Google Scholar]
- 31.McMullan, L. K., A. Grakoui, M. J. Evans, K. Mihalik, M. Puig, A. D. Branch, S. M. Feinstone, and C. M. Rice. 2007. Evidence for a functional RNA element in the hepatitis C virus core gene. Proc. Natl. Acad. Sci. USA 104:2879-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyanari, Y., K. Atsuzawa, N. Usuda, K. Watashi, T. Hishiki, M. Zayas, R. Bartenschlager, T. Wakita, M. Hijikata, and K. Shimotohno. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 9:1089-1097. [DOI] [PubMed] [Google Scholar]
- 33.Moreau, I., S. Hegarty, J. Levis, P. Sheehy, O. Crosbie, E. Kenny-Walsh, and L. J. Fanning. 2006. Serendipitous identification of natural intergenotypic recombinants of hepatitis C in Ireland. Virol. J. 3:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murray, C. L., C. T. Jones, J. Tassello, and C. M. Rice. 2007. Alanine scanning of the hepatitis C virus core protein reveals numerous residues essential for production of infectious virus. J. Virol. 81:10220-10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noppornpanth, S., T. X. Lien, Y. Poovorawan, S. L. Smits, A. D. Osterhaus, and B. L. Haagmans. 2006. Identification of a naturally occurring recombinant genotype 2/6 hepatitis C virus. J. Virol. 80:7569-7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noppornpanth, S., S. L. Smits, T. X. Lien, Y. Poovorawan, A. D. Osterhaus, and B. L. Haagmans. 2007. Characterization of hepatitis C virus deletion mutants circulating in chronically infected patients. J. Virol. 81:12496-12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okamoto, H., M. Kojima, S. Okada, H. Yoshizawa, H. Iizuka, T. Tanaka, E. E. Muchmore, D. A. Peterson, Y. Ito, and S. Mishiro. 1992. Genetic drift of hepatitis C virus during an 8.2-year infection in a chimpanzee: variability and stability. Virology 190:894-899. [DOI] [PubMed] [Google Scholar]
- 38.Pekow, J. R., A. K. Bhan, H. Zheng, and R. T. Chung. 2007. Hepatic steatosis is associated with increased frequency of hepatocellular carcinoma in patients with hepatitis C-related cirrhosis. Cancer 109:2490-2496. [DOI] [PubMed] [Google Scholar]
- 39.Pickart, C. M., and I. A. Rose. 1985. Ubiquitin carboxyl-terminal hydrolase acts on ubiquitin carboxyl-terminal amides. J. Biol. Chem. 260:7903-7910. [PubMed] [Google Scholar]
- 40.Pietschmann, T., A. Kaul, G. Koutsoudakis, A. Shavinskaya, S. Kallis, E. Steinmann, K. Abid, F. Negro, M. Dreux, F. L. Cosset, and R. Bartenschlager. 2006. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc. Natl. Acad. Sci. USA 103:7408-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 42.Shavinskaya, A., S. Boulant, F. Penin, J. McLauchlan, and R. Bartenschlager. 2007. The lipid droplet binding domain of hepatitis C virus core protein is a major determinant for efficient virus assembly. J. Biol. Chem. 282:37158-37169. [DOI] [PubMed] [Google Scholar]
- 43.Shepard, C. W., L. Finelli, and M. J. Alter. 2005. Global epidemiology of hepatitis C virus infection. Lancet Infect. Dis. 5:558-567. [DOI] [PubMed] [Google Scholar]
- 44.Simmonds, P., J. Bukh, C. Combet, G. Deleage, N. Enomoto, S. Feinstone, P. Halfon, G. Inchauspe, C. Kuiken, G. Maertens, M. Mizokami, D. G. Murphy, H. Okamoto, J. M. Pawlotsky, F. Penin, E. Sablon, I. T. Shin, L. J. Stuyver, H. J. Thiel, S. Viazov, A. J. Weiner, and A. Widell. 2005. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42:962-973. [DOI] [PubMed] [Google Scholar]
- 45.Smith, D. B., S. Pathirana, F. Davidson, E. Lawlor, J. Power, P. L. Yap, and P. Simmonds. 1997. The origin of hepatitis C virus genotypes. J. Gen. Virol. 78(Pt. 2):321-328. [DOI] [PubMed] [Google Scholar]
- 46.Steinmann, E., C. Brohm, S. Kallis, R. Bartenschlager, and T. Pietschmann. 2008. Efficient trans-encapsidation of hepatitis C virus RNAs into infectious virus-like particles. J. Virol. 82:7034-7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suzuki, T., H. Aizaki, K. Murakami, I. Shoji, and T. Wakita. 2007. Molecular biology of hepatitis C virus. J. Gastroenterol. 42:411-423. [DOI] [PubMed] [Google Scholar]
- 48.Tong, X., and B. A. Malcolm. 2006. Trans-complementation of HCV replication by non-structural protein 5A. Virus Res. 115:122-130. [DOI] [PubMed] [Google Scholar]
- 49.Trinchet, J. C., N. Ganne-Carrie, P. Nahon, G. N′Kontchou, and M. Beaugrand. 2007. Hepatocellular carcinoma in patients with hepatitis C virus-related chronic liver disease. World J. Gastroenterol. 13:2455-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vassilaki, N., P. Friebe, P. Meuleman, S. Kallis, A. Kaul, G. Paranhos-Baccala, G. Leroux-Roels, P. Mavromara, and R. Bartenschlager. 2008. Role of the hepatitis C virus core+1 open reading frame and core cis-acting RNA elements in viral RNA translation and replication. J. Virol. 82:11503-11515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, T. H., R. C. Rijnbrand, and S. M. Lemon. 2000. Core protein-coding sequence, but not core protein, modulates the efficiency of cap-independent translation directed by the internal ribosome entry site of hepatitis C virus. J. Virol. 74:11347-11358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Welbourn, S., R. Green, I. Gamache, S. Dandache, V. Lohmann, R. Bartenschlager, K. Meerovitch, and A. Pause. 2005. Hepatitis C virus NS2/3 processing is required for NS3 stability and viral RNA replication. J. Biol. Chem. 280:29604-29611. [DOI] [PubMed] [Google Scholar]
- 54.Yagi, S., K. Mori, E. Tanaka, A. Matsumoto, F. Sunaga, K. Kiyosawa, and K. Yamaguchi. 2005. Identification of novel HCV subgenome replicating persistently in chronic active hepatitis C patients. J. Med. Virol. 77:399-413. [DOI] [PubMed] [Google Scholar]
- 55.Yamaga, A. K., and J. H. Ou. 2002. Membrane topology of the hepatitis C virus NS2 protein. J. Biol. Chem. 277:33228-33234. [DOI] [PubMed] [Google Scholar]
- 56.Zhang, J., O. Yamada, H. Yoshida, T. Iwai, and H. Araki. 2002. Autogenous translational inhibition of core protein: implication for switch from translation to RNA replication in hepatitis C virus. Virology 293:141-150. [DOI] [PubMed] [Google Scholar]
- 57.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 102:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.