Abstract
Begomoviruses (family Geminiviridae) cause major losses to crops throughout the tropical regions of the world. Begomoviruses originating from the New World (NW) and the Old World (OW) are genetically distinct. Whereas the majority of OW begomoviruses have monopartite genomes and whereas most of these associate with a class of symptom-modulating satellites (known as betasatellites), the genomes of NW begomoviruses are exclusively bipartite and do not associate with satellites. Here, we show for the first time that a betasatellite (cotton leaf curl Multan betasatellite [CLCuMuB]) associated with a serious disease of cotton across southern Asia is capable of interacting with a NW begomovirus. In the presence of CLCuMuB, the symptoms of the NW cabbage leaf curl virus (CbLCuV) are enhanced in Nicotiana benthamiana. However, CbLCuV was unable to interact with a second betasatellite, chili leaf curl betasatellite. Although CbLCuV can transreplicate CLCuMuB, satellite accumulation levels in plants were low. However, progeny CLCuMuB isolated after just one round of infection with CbLCuV contained numerous mutations. Reinoculation of one such progeny CLCuMuB with CbLCuV to N. benthamiana yielded infections with significantly higher satellite DNA levels. This suggests that betasatellites can rapidly adapt for efficient transreplication by a new helper begomovirus, including begomoviruses originating from the NW. Although the precise mechanism of transreplication of betasatellites by begomoviruses remains unknown, an analysis of betasatellite mutants suggests that the sequence(s) required for maintenance of CLCuMuB by one of its cognate begomoviruses (cotton leaf curl Rajasthan virus) differs from the sequences required for maintenance by CbLCuV. The significance of these findings and, particularly, the threat that betasatellites pose to agriculture in the NW, are discussed.
Viruses of the family Geminiviridae are distinct in having genomes of circular, single-stranded DNA (ssDNA) contained within twinned quasi-isometric (“geminate”) virions from which they derive their name. Geminiviruses are divided into four genera based on the organization of their genomes, biological properties, type of insect vector (either whitefly, leafhopper, or treehopper), and host range (either mono- or dicotyledonous hosts) (37). The genus Begomovirus contains the vast majority of the identified geminivirus species, and these are transmitted exclusively by the whitefly Bemisia tabaci (Gennadius) to dicotyledonous plants. All begomoviruses native to the New World (NW) and a small number originating from the Old World (OW) have bipartite genomes (with components known as DNA-A and DNA-B). The majority of the OW begomoviruses have genomes consisting of a single component homologous to the DNA-A component of the bipartite viruses. Begomoviruses from the NW and OW are genetically distinct. They segregate separately in phylogenetic analyses, and the OW viruses show a greater genetic diversity and have an additional, absolutely conserved gene (known as V2 for the monopartite and AV2 for the bipartite viruses) that is absent in the NW begomoviruses.
The global trade in agricultural products is leading to the spread of many viruses. The prime example here is tomato yellow leaf curl virus. This monopartite begomovirus has its origins in the Middle East/Mediterranean region but has been inadvertently introduced to the NW, with serious consequences for tomato production across the southern United States (24, 26). Similarly, the NW begomovirus squash leaf curl virus from the southwestern United States has been introduced into the Middle East (2, 20).
The majority of OW monopartite begomoviruses are associated with additional ssDNA molecules. The first evidence for this came with the report by Dry et al. (14) of an ssDNA satellite associated with tomato leaf curl virus (ToLCV) occurring in Australia. This molecule was later shown to be a defective (truncated) version of a much larger group of subviral components associated with begomoviruses that are now known collectively as betasatellites (6). Betasatellites are approximately half the size of a begomovirus component (∼1,360 nucleotides [nt]) and are required by the helper begomovirus for efficient infection of some hosts (9, 30, 31). Betasatellites have been shown to be associated with an increasing number of diseases caused by begomoviruses, including many of the most significant, economically damaging diseases occurring in the OW. The most noteworthy of these diseases is cotton leaf curl disease (CLCuD). CLCuD was epidemic during the 1990s across Pakistan and continues to be so in northern India. The disease is caused by a complex consisting of representatives of at least seven distinct begomovirus species and a specific betasatellite (23).
Betasatellites have a highly conserved structure although their sequences are highly diverse, with distinct species showing as little as 50% sequence identity (6, 11, 42). They contain a single coding sequence (known as βC1), a region of sequence rich in adenine, and a ∼150-nt region, known as the satellite conserved region (SCR), that is highly conserved between all betasatellites. The SCR contains a predicted hairpin structure that contains within the loop a nonanucleotide sequence (TAATATTAC) that for geminiviruses marks the origin of virion-strand DNA replication. The βC1 gene is a pathogenicity determinant (27, 28, 33) and encodes all satellite functions identified so far, including suppression of RNA-mediated host defense (13) and possibly a role in virus movement (29). For many of the monopartite begomoviruses, the betasatellite is essential for inducing typical disease symptoms in the hosts from which they were isolated (6, 9, 30). However, recently some viruses with less dependence on interaction with their betasatellites have been identified (6).
When betasatellites were first identified, their ability to interact with NW begomoviruses was investigated, but no evidence for interaction was found (R.W. Briddon, unpublished results). Here, we report a positive interaction between a betasatellite and the NW cabbage leaf curl virus (CbLCuV). We show that the interaction between the betasatellite and this NW begomovirus leads to rapid sequence changes in the satellite, which enhances its interaction with the virus.
MATERIALS AND METHODS
Virus and betasatellite clones.
A full-length clone of cotton leaf curl Rajasthan virus (CLCuRaV; clone Dav isolated from Multan, Pakistan, in 2006 [PK:Mul:Dav:06], from Gossypium davidsonii; accession number EU365616) and full-length clones of cotton leaf curl Multan betasatellite (CLCuMuB) (accession numbers EU384587 [PK:Mul:Pun7s:06], FJ607041 [PK:Mul:08]; and EU384595 [PK:Mul:Dav85:06]) were isolated from Gossypium punctatum, Gossypium hirsutum and G. davidsonii plants, respectively, and amplified by rolling-circle amplification (RCA) from field-infected plants originating from Multan, Pakistan in 2006 (Nawaz-ul-Rehman, unpublished data). CLCuMuB-(PK:Mul:Pun7s:06) (EU384587) is a typical betasatellite that was associated with the CLCuD epidemic in Pakistan during the 1990s (reviewed in reference 8). We shall henceforth refer to this as the Multan strain of CLCuMuB (CLCuMuBMul). CLCuMuB-(PK:Mul:08) (FJ607041) is a recombinant CLCuMuB that contains a small fragment of the SCR originating from a tomato leaf curl betasatellite and is typical of the betasatellite associated with the ongoing epidemic of a resistance-breaking strain of CLCuMuB (1). We shall henceforth refer to this as the Burewala strain of CLCuMuB (CLCuMuBBur). CLCuMuB-(PK:Mul:Dav85:06) (EU384595) is a recombinant CLCuMuB in which the SCR has been replaced by the intergenic region of CLCuRaV; this contains the origin of replication of the virus. By definition this is not a betasatellite since satellites have no significant levels of sequence identity to their helper viruses (10). We shall henceforth refer to this as the CLCuMuB/CLCuRaV recombinant (CLCuMuBrMul/Raj).
A chili leaf curl betasatellite (ChLCB; [PK:Mul:07]; accession number FJ515274) was PCR amplified from field-infected tomato samples collected during 2006 from Multan (Pakistan) using universal betasatellite primers (7). Clones of the DNA-A (U65529) and DNA-B (U65530) components of CbLCuV (CbLCuVA and CbLCuVB, respectively) were kindly provided by Dominique Robertson (39).
Agrobacterium-mediated inoculation.
All inoculations of plants with CLCuRaV were conducted by Agrobacterium-mediated inoculation. A partial direct repeat construct of CLCuRaV was produced by digesting the clone with XbaI and KpnI to release an approximately 2,216-bp fragment. This was cloned into XbaI-KpnI-restricted pUC19. The resultant partial clone was linearized with XbaI, and the full-length XbaI insert of the CLCuRaV clone was ligated into this. Finally, the partial repeat construct was transferred from pUC19 into the binary vector pCAMBIA2300 (Cambia) as a HindIII-SacI fragment.
Complete direct repeat constructs of CLCuMuBMul, CLCuMuBBur, and CLCuMuBrMul/Raj were prepared by gel, and their clone inserts were isolated following SalI digestion. These were then circularized by ligation and amplified by RCA using φ29 polymerase (illustra TempliPhi 100; GE Healthcare). Concatameric RCA products were electrophoresed on ethidium bromide-stained 0.8% (wt/vol) agarose gels after partial digestion with SalI. Fragments of approximately 2.8 kb (equivalent to two copies of the satellite molecule) were gel isolated and cloned into pUC19. The dimeric inserts of these clones were then excised as HindIII-SacI fragments and ligated into pCAMBIA2300. A dimeric clone of ChLCB in pCAMBIA2300 was produced in the same way as the CLCuMuBs but using a unique KpnI restriction site instead of SalI.
Binary vector plasmids harboring dimeric or partial direct repeat constructs were transformed into Agrobacterium tumefaciens strain GV3101. Agrobacterium cultures for inoculation were produced and infiltrated into Nicotiana benthamiana as described previously (41).
Production of a CLCuMuB deletion mutant.
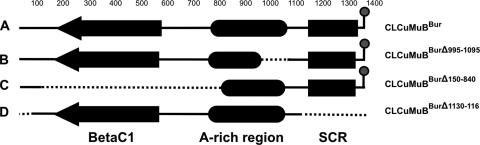
A deletion mutant of CLCuMuBBur was produced by PCR amplification using the circularized clone insert as the template. A mutant of CLCuMuBBur with a deletion from position 1130 to 116 (nucleotide numbering is according to CLCuMuB FJ607041) (CLCuMuBBurΔ1130-116) was produced using primers Δ1130-116-F (5′-AAGCTTATGGCCTTTTATGGGTTATGG-3′) and Δ1130-116-R (5′-CAAGCTTAGGTAATTTATGAGTCCCCAATTG-3′) which contain HindIII restriction sites (underlined).
HindIII-restricted PCR product was self-ligated and subjected to φ29 amplification to produce a concatameric product. This was partially restricted with HindIII, and ∼1-kb fragments were purified from an agarose gel and ligated into the binary vector pGreen0029 (19) linearized with HindIII.
Dimeric constructs of the additional mutants CLCuMuBBurΔ150-840 and CLCuMuBBurΔ995-1095 were produced as described in the previous section (based on a KpnI restriction site) in the binary vector pGreen0029.
Production of constructs for the inoculation of CbLCuV.
The CbLCuV components were introduced into plants by biolistic inoculation. Plasmids containing partial direct repeat constructs of CbLCuVA and CbLCuVB (39) were amplified by φ29 polymerase amplification. For coinoculation with betasatellites, the CbLCuV component φ29 polymerase products were mixed with concatameric RCA products obtained from the satellites produced as described earlier. Approximately equal amounts of each component to be inoculated were mixed to a final concentration of 100 ng/μl of DNA. This was coated on 1-μm gold particles and inoculated to N. benthamiana plants using a Helios gene gun (Bio-Rad) as described previously (5).
Analysis of virus accumulation in plants.
The accumulation of virus and betasatellite DNA in plants was assessed by Southern blot hybridization. DNA was extracted from leaf tissue using a Qiagen plant DNA isolation kit. Approximately 500 ng of each DNA sample was electrophoresed on 1.5% agarose gels and transferred to nylon membrane Hybond (Hybond N+; Amersham). Digoxigenin-labeled PCR products were produced and used as a probe for each component as recommended by the manufacturer (PCR DIG Probe Synthesis Kit; Roche).
Sequencing and sequence analysis.
The sequences of cloned betasatellites were determined by dideoxynucleotide chain termination sequencing on an automated DNA sequencer (Applied Biosystems models 3130 and 3730 DNA sequencers). Sequences were assembled and analyzed by the Lasergene sequence analysis package (version 8; DNAStar Inc., Madison, WI). Sequence alignments were produced using the MegAlign program of the Lasergene package.
RESULTS
An NW bipartite begomovirus supports the replication and systemic movement of a betasatellite.
To investigate the potential for NW begomoviruses to productively interact with betasatellites, two distinct betasatellites, CLCuMuB and ChLCB, were inoculated with the components of CbLCuV. Inoculation of CbLCuVA in the absence of its cognate DNA-B did not yield infection of N. benthamiana. In contrast, coinoculation of the DNA-A and DNA-B components (CbLCuVA+B) yielded efficient infection (Table 1) with typical symptoms, consisting of yellowing, leaf deformation, and stunting, approximately 12 days postinoculation (Fig. 1G). This confirms the obligate interaction of the CbLCuVA and CbLCuVB for infection.
TABLE 1.
Infectivity for genomic components of CLCuRaV and CbLCuV in the presence of betasatellites to N. benthamiana
| Inoculum | No. of plants infected/No. of plants inoculated |
Symptom severityb | Betasatellite replication (no. of plants showing betasatellite replication) | ||
|---|---|---|---|---|---|
| Expt 1 | Expt 2 | Expt 3 | |||
| CLCuRaV | 0/9 | 0/9 | 0/9 | 0 | |
| CLCuRaV + CLCuMuBMul | 9/9 | 9/9 | 9/9 | 5 | Yes (27) |
| CLCuRaV + CLCuMuBrMul/Raj | 9/9 | 9/9 | 9/9 | 4 | Yes (27) |
| CLCuRaV + CLCuMuBBur | 9/9 | 9/9 | 9/9 | 5 | Yes (27) |
| CLCuRaV + ChLCB | 9/9 | 9/9 | 9/9 | 4 | Yes (27) |
| CLCuRaV + CLCuMuBBurΔ995-1095 | 9/9 | 6/9 | 9/12 | 4 | Yes (24) |
| CLCuRaV + CLCuMuBBurΔ150-840 | 0/9 | 0/9 | 0/12 | 0 | Yes (30)a |
| CLCuRaV + CLCuMuBBurΔ1130-116 | 0/9 | 0/9 | 0/12 | 0 | No |
| CbLCuVA | 0/9 | 0/9 | 0/9 | 0 | |
| CbLCuVA+B | 7/9 | 9/9 | 9/9 | 3 | |
| CbLCuVA + CLCuMuBMul | 0/9 | 0/9 | 0/9 | 0 | No |
| CbLCuVA + CLCuMuBrMul/Raj | 0/9 | 0/9 | 0/9 | 0 | No |
| CbLCuVA+B + CLCuMuBMul | 6(2)/9c | 7(2)/9c | 7(1)/9c | 4(5)c | Yes (25) |
| CbLCuVA+B + ChLCB | 7/9 | 9/9 | 9/9 | 3 | No |
| CbLCuVA+B + CLCuMuBrMul/Raj | 8/9 | 9/9 | 9/9 | 3 | No |
| CbLCuVA+B + CLCuMuBBur | 7/9 | 8/9 | 9/9 | 4 | Yes (24) |
| CbLCuVA+B +CLCuMuBBurΔ995-1095 | 7/9 | 9/9 | 8/9 | 4 | Yes (24) |
| CbLCuVA+B +CLCuMuBBurΔ150-840 | 7/9 | 8/9 | 9/9 | 3 | Yes (24) |
| CbLCuVA+B +CLCuMuBBurΔ1130-116 | 7/9 | 9/9 | 9/9 | 3 | No |
| CbLCuVA+B + CLCuMuBMul/Mut | 9/9 | 8/9 | 9/9 | 5 | Yes (26) |
Replication of CLCuMuBBurΔ150-840 was assessed only in the infiltrated leaves.
Symptom severity is indicated on a scale of 0 to 5, with 5 indicating severe symptoms associated with infection of N. benthamiana with CLCuRaV and CLCuMuBMul (see Materials and Methods).
For the CbLCuVA+B with CLCuMuBMul inoculations, plants exhibiting two distinct symptom severity levels were noted. The figures in parentheses are the numbers of plants showing the more severe symptoms (severity level of 5).
FIG. 1.
Symptoms exhibited by N. benthamiana plants inoculated to examine the ability of viruses to interact with betasatellites: mock inoculated (A), CLCuRaV and CLCuMuBMul (B), CLCuRaV and CLCuMuBBur (C), CLCuRaV and CLCuMuBrMul/Raj (D), CLCuRaV and CLCuMuBBurΔ995-1095 (E), CLCuRaV and ChLCB (F), CbLCuVA+B (G), CbLCuVA+B and CLCuMuBMul (H), CbLCuVA+B and CLCuMuBBur (I), and CbLCuVA+B and CLCuMuBMul/Mut (J).
Inoculation of CbLCuVA with CLCuMuBMul also did not lead to infection, indicating that the betasatellite is unable to complement the missing movement functions encoded on DNA-B. This contrasts with plants inoculated with CLCuRaV, one of the viruses which associates with CLCuMuB in the field (36), and either CLCuMuBMul or ChLCB. Both of these combinations were highly infectious to N. benthamiana and produced severe symptoms. Symptoms in the presence of CLCuMuBMul infection were vein yellowing, leaf crumpling, and a downward curling of the leaves (Fig. 1B) and are indistinguishable from the symptoms reported previously for this betasatellite infecting N. benthamiana with CLCuMuV (22). In conjunction with ChLCB infection the symptoms were milder, with a reduction in leaf size and mild leaf curling (Fig. 1F). Surprisingly, CLCuRaV was not infectious to N. benthamiana in the absence of a betasatellite (Table 1). This is unusual since all of the other CLCuD-associated monopartite begomoviruses that have previously been investigated are infectious to N. benthamiana, producing relatively mild symptoms in the absence of the betasatellite (22).
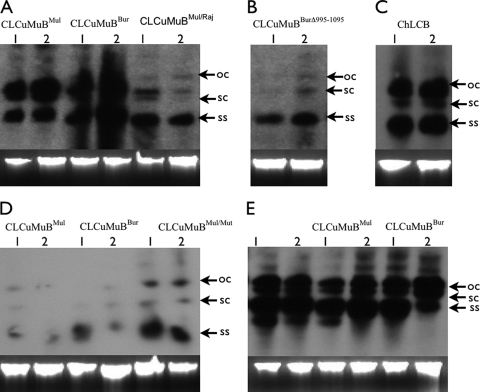
Although inoculation of ChLCB with CbLCuVA+B yielded infection, both Southern blot analysis and PCR diagnostics indicated that the betasatellite was not maintained (results not shown). The efficiency of infection, as well as the symptoms induced in this case, was equivalent to inoculation with only CbLCuVA+B (Table 1). In contrast, inoculation of N. benthamiana with CbLCuVA+B and CLCuMuBMul yielded plants with severe symptoms within 12 days postinoculation. The symptoms in this case were more severe (severity rating of 4; symptom severity is indicated on a scale of 0 to 5, with 0 indicating no symptoms and 5 indicating severe symptoms) than in plants inoculated with only CbLCuVA+B (severity rating of 3) and consisted of a significant reduction in leaf size, more prominent chlorotic patches, and pronounced downward curling of the edges. For a small number of plants inoculated with CbLCuVA+B and CLCuMuBMul, the symptoms were more severe, with a severity rating of 5 (Table 1). One such plant is shown in Fig. 1H. Southern blot analysis of representative plants showed that only very low levels of CLCuMuBMul were maintained (Fig. 2D), and these were considerably lower than levels maintained in plants infected with CLCuRaV and CLCuMuBMul (Fig. 2A). Probing of Southern blots of DNA extracts from plants infected with CbLCuVA+B and CLCuMuBMul for the presence of CbLCuVA showed the viral DNA levels to be approximately equivalent to those in plants infected with only the components of CbLCuV (Fig. 2E).
FIG. 2.
Southern hybridization for the detection of betasatellites and CbLCuV in N. benthamiana plants. Blots were probed with labeled DNA fragments containing the βC1 gene of CLCuMuBMul (A, B, and D), the βC1 gene of ChLCB (C) or the AC2 gene of CbLCuV (E). Plants were inoculated with either CLCuRaV (A to C) or CbLCuV (D and E) and either CLCuMuBMul, CLCuMuBBur, CLCuMuBrMul/Raj, CLCuMuBBurΔ995-1095, CLCuMuBMul/Mut or ChLCB, as indicated above the blots. In each case, DNA (approximately 0.5 μg) extracted from two plants (1 and 2) was loaded. In each case a photograph of the ethidium bromide-stained gel is shown below the blot as a loading control. Viral DNA forms are indicated as single-stranded (ss), supercoiled (sc), and open circular (oc).
Two further betasatellite clones were assessed for their ability to interact with CbLCuV. CLCuMuBBur is a recombinant consisting for the most part of sequence derived from CLCuMuBMul but with some (nt 1090 to 1260) of the SCR originating from a tomato leaf curl betasatellite. This recombinant has previously been described and is now the prevalent betasatellite affecting cotton in the field in Pakistan (1). CLCuMuBrMul/Raj is a naturally occurring recombinant isolated from cotton plants. It also consists for the most part of sequences derived from CLCuMuB, but in this case the SCR (the presumed origin of betasatellite replication) is replaced with the origin of replication (intergenic region sequences) of CLCuRaV (Fig. 3 and 4). Similar recombinant molecules have previously been described from a number of betasatellites (38), including CLCuMuB containing the origin of replication of CLCuMuV (9, 31). Inoculation of CbLCuVA with CLCuMuBBur did not lead to infection, whereas both CLCuMuBBur and CLCuMuBrMul/Raj were efficiently maintained by CLCuRaV infection, inducing symptoms similar to those induced by this virus in association with CLCuMuBMul (Fig. 1B, C, and D). The symptoms for CLCuRaV infection in the presence of CLCuMuBrMul/Raj were slightly milder than those associated with CLCuMuBBur. Southern blots probed for the presence of CLCuMuB sequences showed that in both cases (CLCuRaV infection with either CLCuMuBBur or CLCuMuBMul) approximately equal amounts of the satellite were present, whereas plants infected with CLCuRaV and CLCuMuBMul/Raj contained less satellite DNA (Fig. 2A). Inoculation of CbLCuVA+B with CLCuMuBrMul/Raj led to a normal CbLCuV infection with no betasatellite sequences detectable in plants (results not shown). This suggests that the sequences required for maintenance of the satellite are not present in CLCuMuBrMul/Raj. In contrast, CLCuMuBBur was efficiently maintained by a CbLCuVA+B infection (Table 1). As in the case of an infection with CbLCuVA+B and CLCuMuBMul, the symptoms were more severe than with a conventional CbLCuVA+B infection (Fig. 1H and I), and the DNA levels of the satellite in plants were low (Fig. 2D).
FIG. 3.
Graphical representation of the genetic organization of CLCuMuB showing the sequence deletions identified in the study: CLCuMuB (A), CLCuMuBBurΔ995-1095 (B), CLCuMuBBurΔ150-840 (C), and CLCuMuBBurΔ1130-116 (D). Deleted sequences are indicated with a dashed line. The diagram shows the position of the βC1 coding sequence, the SCR, the A-rich region, and the conserved hairpin structure.
FIG. 4.
Alignment of the sequences between the A-rich region and the 3′ region of the hairpin structure of betasatellites and mutants used in the analysis. The approximate positions of the 3′ end of the A-rich region and the SCR are highlighted in green and gray, respectively. The SCR contains the predicted hairpin structure containing the nonanucleotide (TAATATTAC) sequence. The sequence of CLCuMuBrMul/Raj derived from CLCuRaV is shown in red letters. This contains the presumed TATA box of the CLCuRaV Rep gene promoter (highlighted in yellow) surrounded by the predicted Rep iteron sequences of CLCuRaV (highlighted in purple). The sequence of AYVB identified by Saunders et al. (32) as important for maintenance by AYVV is underlined. Also indicated are the AYVV iteron-like sequences identified by the authors (highlighted in bright green). The corresponding iteron-like sequences of the betasatellites (and mutants) used in the present study are highlighted in bright green. Sequences with similarity to the predicted iterons of CbLCuV are highlighted as white text on a black background. Nucleotide changes between CLCuMuBMul/Mut (FJ861370), the mutant betasatellite derived from an infection with CbLCuV with severe symptoms, and CLCuMuB EU384587, the parental molecule, are highlighted in bright blue. The nucleotide numbering of the ruler corresponds to the numbering of the full sequence alignment.
To ensure that transreplication and maintenance of the betasatellite by CbLCuVA+B were not due to recombination between the satellite and the virus, transferring the viral origin of replication, the betasatellites from representative infections involving CbLCuVA+B were PCR amplified (7), cloned, and sequenced (results not shown). In none of the plants analyzed was there evidence for the exchange of the viral origin of replication.
Transreplication of CLCuMuB by CbLCuV results in rapid accumulation of mutations.
Using universal primers, betasatellites were PCR amplified from an N. benthamiana plant infected with CbLCuVA+B in the presence of CLCuMuBBur and cloned. Sequence analysis of two clones indicated that they contained distinct deletions. The deletion in clone CLCuMuBBurΔ150-840 (FJ861369) was 665 nt in length and spanned the entire βC1 gene up to the 5′ end of the A-rich region (Fig. 3). In N. benthamiana this clone was unable to support the systemic infection of CLCuRaV, although transreplication of the satellite by the begomovirus was evident in inoculated leaves (results not shown). These findings are consistent with a requirement of βC1 for systemic movement of this CLCuRaV clone. When inoculated with CbLCuVA+B, CLCuMuBBurΔ150-840 was efficiently maintained but at low levels (results not shown). The plants in this case exhibited the milder phenotype typical of a CbLCuVA+B infection, confirming the importance of the βC1 gene in mediating enhancement of symptoms.
The second clone, CLCuMuBBurΔ995-1095 (FJ861368), contained a 100-nt deletion extending from the center of the A-rich region into the sequence between the A-rich region and the SCR (Fig. 3 and 4). This mutant was able to support the infection of N. benthamiana by CLCuRaV and induced a phenotype indistinguishable from that induced by the intact satellite in association with this virus (Table 1). Inoculation of CLCuMuBBurΔ995-1095 with CbLCuVA+B led to maintenance of the satellite and the enhanced symptom phenotype associated with the intact CLCuMuBBur. However, the betasatellite was below the detection threshold of Southern blotting and was detected only by PCR. Thus, even at low levels, this mutant betasatellite is able to enhance symptoms. The results with CLCuMuBBurΔ995-1095 infection suggest that the sequences required for efficient transreplication by CbLCuVA+B have been affected by this mutation. The difference seen between CLCuRaV and CbLCuV in the maintenance of CLCuMuBBurΔ995-1095 also suggests that the sequence(s) required for maintenance of the satellite by these two viruses differs.
N. benthamiana plants that were inoculated with CbLCuVA+B and CLCuMuBMul showed two distinct phenotypes (results not shown). From an N. benthamiana plant showing the more severe symptom phenotype (Fig. 2I), PCR with universal betasatellite primers was used to obtain clones of the satellite to investigate the possible basis for this enhancement in symptom severity. Three clones containing potentially full-length molecules were selected and sequenced in their entirety (these sequences are available in the databases under accession numbers FJ861370, FJ861371, and FJ861372). A summary of the changes detected in sequence comparisons with the sequence of the betasatellite with which the plant was inoculated (CLCuMuB EU384587) is shown in Table 2. This shows that the majority of the sequence changes occurred in the sequence between the A-rich region and the SCR (25 changes over a stretch of approximately 267 nt; 1 in 11 nt mutated) as well as in the SCR (9 changes over 124 nt; 1 change per 14 nt). Only three sequence changes were detected in the βC1 gene (for clone CLCuMuB FJ861371) which resulted in two amino acid changes (three changes in 356 nt; one change per 110 nt). For the remaining betasatellite sequences, approximately equal numbers of sequence changes were detected: one change per 24 nt for the sequence between the SCR and the 3′-end of the βC1 gene, one change in 15 nt for the βC1 promoter region (sequence between the 5′-end of the βC1 gene and the A-rich region), and one change per 18 nt in the A-rich region. Although based on only three clones from a single plant, these results suggest that, at least when maintained by CbLCuV, CLCuMuB accumulates more mutations in the sequences between the A-rich region and the SCR than in any other part of the molecule. As might be expected from a coding sequence, significantly fewer mutations occurred in the βC1 gene.
TABLE 2.
List of point mutations detected in the satellite after coinfection of CbLCuVA+B with the CLCuMuB parent strain
| Residue in the parent CLCuMuB (EU384587)a | Mutation in the indicated CLCuMuB variant (accession no.)a,b |
Nucleotide coordinate(s)c | ||
|---|---|---|---|---|
| FJ861370 | FJ861372 | FJ861371 | ||
| T | — | — | C | 23 |
| T | C | — | — | 99 |
| T | — | — | C | 121 |
| T | del | del | del | 125 |
| A | G | — | — | 155 |
| C | — | — | A | 253 |
| T | — | — | C | 361 |
| T | — | — | G | 536 |
| G | — | — | A | 568 |
| T | — | — | A | 599 |
| C | — | — | T | 689 |
| C | — | — | A | 714 |
| A | G | G | — | 726 |
| T | — | — | C | 742 |
| T | — | — | C | 751 |
| A | — | — | G | 753 |
| A | G | G | — | 812 |
| C | — | — | T | 828 |
| A | — | — | C | 876 |
| A | G | G | — | 894 |
| A | G | G | — | 904 |
| A | G | G | — | 914 |
| ins | — | — | A | 918 |
| A | — | — | del | 960 |
| TGT | — | — | AAC | 1028-1030 |
| G | — | — | A | 1043 |
| G | — | — | A | 1045 |
| G | — | — | T | 1049 |
| A | C | — | — | 1047 |
| G | — | — | A | 1052 |
| T | — | — | G | 1055 |
| C | T | — | — | 1073 |
| TGT | — | — | GAG | 1087-1089 |
| T | — | — | C | 1102 |
| AGAA | — | — | TACT | 1105-1108 |
| G | — | — | A | 1116 |
| TC | — | — | AG | 1120-1121 |
| A | — | — | T | 1126 |
| ins | — | — | T | 1130 |
| C | T | — | — | 1137 |
| A | — | C | — | 1143 |
| AGA ins | TTTT | — | — | 1162-1165 |
| ins | A | — | — | 1168 |
| C | A | — | — | 1171 |
| A | — | — | G | 1233 |
| AA | GT | GT | GT | 1289-1290d |
| 4 × ins | GCTAC | GCTAC | GCTAC | 1321-1325d |
| C | T | — | — | 1345 |
ins, insertion; del, deletion; 4×, nucleotide insertion due to primers.
—, residue unchanged.
The nucleotide coordinates given are of the parent molecule (CLCuMuB EU384587).
The sequence change is derived from the primers used to amplify the component and is not included in the overall analysis.
One clone (CLCuMuB FJ861370; henceforth referred to as CLCuMuBMul/Mut) isolated from a plant inoculated with CbLCuVA+B and CLCuMuBMul that exhibited severe symptoms, was back-inoculated to N. benthamiana with CbLCuVA+B. This betasatellite molecule was maintained by CbLCuVA+B, and the infection induced symptoms that were more severe than those induced by the parental betasatellite molecule in the presence of CbLCuVA+B (Fig. 1J; Table 1). These severe symptoms were mainly characterized by more severely stunted plant growth and also an increased level of foliar chlorosis. Southern blot analysis showed the levels of the betasatellite in this case to be higher than those of the parental molecule in the presence of CbLCuVA+B (Fig. 2D).
Analysis of transreplication specificity.
A deletion mutant of CLCuMuBBur was produced by PCR amplification with specific primers. Clone CLCuMuBBurΔ1130-116 contained a deletion from position 1130 to 116 (nucleotide numbering is according to CLCuMuB FJ607041, the molecule from which the mutant was derived). This region extends from approximately 140 nt from the 3′ end of the A-rich region to 80 nt 5′ of the stop codon of the βC1 gene and includes the entire SCR and hairpin structure (Fig. 3 and 4). CbLCuVA+B was not able to maintain this molecule when inoculated in plants; a normal CbLCuVA+B infection ensued, inducing the symptoms typical of this virus (Table 1). Similarly, CLCuMuBBurΔ1130-116 inoculated with CLCuRaV to N. benthamiana was unable to induce a systemic infection. This indicates that sequences between nucleotide positions 1130 and 116 are important for the transreplication of the satellite by these viruses. The deletion includes the conserved hairpin structure that contains the nonanucleotide sequence. Although it has yet to be shown experimentally, it is likely that the hairpin structure forms part of the origin of virion-strand DNA replication for betasatellites. The hairpin structure is similar to the origin of virion-strand DNA replication of geminiviruses. For geminiviruses the nonanucleotide sequence, which forms part of the loop of the predicted hairpin structure, is nicked by the cognate Rep to initiate rolling-circle replication (reviewed in reference 18). It is thus likely that CLCuMuBBurΔ1130-116 is impaired for virion-sense DNA replication because it lacks the site of Rep nicking.
The recombinant betasatellite CLCuMuBrMul/Raj is a naturally occurring molecule that has all satellite sequences between the A-rich region and approximately 30 nt downstream of the hairpin structure replaced with the intergenic region of CLCuRaV (Fig. 4). When the virus was inoculated to N. benthamiana in the presence of CLCuRaV, symptoms appeared within 12 days of inoculation and were severe and indistinguishable from those induced by CLCuRaV infection in the presence of CLCuMuBMul, with the exception that the infection with CLCuMuBrMul/Raj did not induce the leaf-like enations typical of both CLCuMuBMul and CLCuMuBBur infections. The levels of the satellite were approximately equal to those of CLCuMuBMul. However, CLCuMuBrMul/Raj was not maintained by CbLCuVA+B, and transreplication of this molecule by CbLCuVA+B was not detectable in inoculated tissues (results not shown). This indicates that the sequences of the betasatellite required for maintenance by CbLCuVA+B are not present in CLCuMuBrMul/Raj. In contrast, maintenance by CLCuRaV is possible due to the transfer of the origin of replication into the satellite molecule, essentially creating a DNA-B-like molecule.
DISCUSSION
Begomoviruses cause serious losses of many dicot crops throughout the warmer parts of the world, and intensive agriculture, as well as the global trade in agricultural products, has led to an ever-increasing significance of these viruses (35, 40). There are distinct differences between the begomoviruses of the NW and OW, the most significant of which is the association of symptom-modulating satellites with the majority of these viruses in the OW but not the NW (6, 10). Here, we have investigated the capacity of a typical NW begomovirus to interact with betasatellites.
The results of the transreplication studies show that, of the two betasatellite species used, one has the capacity to be transreplicated and maintained by the NW CbLCuV. The precise mechanism by which the Rep encoded by the helper begomovirus recognizes the putative origin of replication of the betasatellites remains unknown. Replication of DNA-A and DNA-B components relies on an interaction between the DNA-A-encoded Rep and reiterated motifs (termed iterons) located within the origin of replication (3, 16, 17), which usually limits Rep transreplication to its own DNA-A and DNA-B components. In contrast, the ability of betasatellites to be transreplicated by distinct begomoviruses (14, 32, 34) indicates that productive origin recognition is more relaxed for the betasatellite. The betasatellites, for the most part, do not contain the iteron sequences of their helper viruses, and when they do, these do not have the typical iteron arrangement that has been shown to be a requirement for initiation of geminivirus virion-sense DNA replication (3). We can propose two, not necessarily mutually exclusive, hypotheses to explain this situation. The first suggests that the Rep proteins encoded by betasatellite-associated begomoviruses might have more relaxed origin recognition properties; this is the “universal Rep” hypothesis. The second suggests that betasatellites might contain a sequence or sequences that allow them to be recognized by a greater range of Reps; this is the “universal iteron” hypothesis. Recently, Saunders et al. (32) used deletion mutagenesis to identify sequences on Ageratum yellow vein betasatellite (AYVB) required for transreplication by Ageratum yellow vein virus (AYVV). They identified a sequence between the A-rich region and the SCR (coordinates 1047 to 1146 of AYVB), which contains sequences with similarity (but not identity) to the predicted iterons of AYVV, to be important for replication of the satellite. Interestingly this region aligns well with a sequence of the ToLCV satellite identified by Dry et al. (14) that contains a ToLCV iteron-like sequence that is a high-affinity Rep binding site but is not essential for replication (21). Our analysis of naturally occurring mutants of CLCuMuB are consistent with the findings of Saunders et al. (32) and earlier studies (8, 42). Sequences spanning the βC1 gene and its promoter (sequences between the βC1 gene and the A-rich region) are not essential for transreplication by begomoviruses. However, deletion of these sequences abolishes the ability of the betasatellite to upregulate virus levels in plants, and the symptoms of CbLCuV infection were not enhanced. This is consistent with the idea that βC1 is an important symptom determinant (13, 27, 28, 33). The CLCuRaV clone used here is unusual in not being infectious to N. benthamiana in the absence of the betasatellite. All other CLCuD-associated begomoviruses that have been examined are infectious to N. benthamiana in the absence of a betasatellite (22), including other clones of CLCuRaV (M. S. Shahid, personal communication). The clone used here may thus be a mutant that requires the betasatellite for spread in this host. Recently, the βC1 of CLCuMuB has been shown to have possible virus movement function (29). It is therefore likely that the CLCuRaV clone used may be dependent on the movement functions provided by the βC1 protein for systemic movement in N. benthamiana.
Deletion of satellite sequences from position 1130 to 116 abolished the satellite's ability to be transreplicated and maintained by both CLCuRaV and CbLCuV. The deleted sequences span the conserved (between all betasatellites) hairpin structure that contains the nonanucleotide sequence. For geminiviruses a similar structure is an essential part of the virion sense origin of replication that is recognized and nicked (within the nonanucleotide sequence) by Rep to initiate rolling-circle replication of the virion strand. It is thus likely that the betasatellite mutant lacking the hairpin structure is compromised for Rep-mediated initiation of virion strand replication.
The CLCuMuB with a deletion of the sequence between coordinates 995 and 1095 gave somewhat conflicting results. Maintenance and levels of transreplication by CLCuRaV remained unaffected, and normal, severe symptoms were induced. However, this mutant was not replicated at high levels by CbLCuV although symptoms were enhanced, indicating that even at low betasatellite levels the βC1 can affect symptom development. This finding suggests that betasatellite sequences required for maintenance by CLCuRaV and for maintenance by CbLCuV differ, implying that their Reps recognize distinct sequences on the satellite. The deletion covers the left arm of the sequence identified by Saunders et al. (32) as important for transreplication of AYVB by AYVV. However, for CLCuMuB the sequences that align with this have no recognizable similarity to the predicted iterons of CLCuRaV (GGTGAT). Rather, 3′ of this sequence, and thus outside the deletion in CLCuMuBBurΔ995-1095, are several CLCuRaV iteron-like sequences (GGGGT and GGGGA) that align well with a similar sequence identified by Saunders et al. (32). This suggests that, at least for transreplication of CLCuMuB by CLCuRaV, we can narrow the area identified for AYVB transreplication by AYVV. However, for transreplication by CbLCuV the situation seems a little more complex. The deletion of sequence at position 995 to 1095 severely reduces the transreplication efficiency, suggesting that these sequences do not play an essential part but are nevertheless important for the interaction. CLCuMuB contains two instances of the iteron sequences which we would predict for CbLCuV (GGTGT; based upon the original theoretical work of Arguello-Astorga and Ruiz-Medrano [3, 4] and experimental definition of iterons for other viruses [12]) in the sequence between the A-rich region and the hairpin. One of these occurs within the SCR and is thus unlikely to be the determining factor in transreplication since it also occurs in ChLCB (position 1306 of the alignment in Fig. 4). The second (Fig. 4; position 1227) is also unlikely to be the determining factor in transreplication since this does not occur in either CLCuMuBMul/Mut (FJ861370) or CLCuMuB EU3845870, both of which can be transreplicated by CbLCuV. However, the sequence deleted in CLCuMuBBurΔ995-1095 does contain sequences resembling the predicted CbLCuV iterons [GGTTT and GGTG(A/C/T)] that could possibly interact with CbLCuV Rep (Fig. 4). This would be consistent with the reduction in, but not total loss of, transreplication for CLCuMuBBurΔ995-1095 by CbLCuV. Also, these motifs do not occur in the sequence of ChLCB (Fig. 4), which may explain why this is not transreplicated and maintained by CbLCuV. However, what the essential sequences are for transreplication of these betasatellites by CbLCuV remains unclear.
The point mutations that accumulated in CLCuMuB following just one passage through N. benthamiana are suggestive of adaptation of the betasatellite to the Rep encoded by CbLCuV. The mutant molecule, isolated following infection with CbLCuVA+B, is capable of replication to higher levels and induces more severe symptoms when inoculated with the CbLCuV components. This finding supports the universal iteron hypothesis for begomovirus-betasatellite interaction. However, although sequence changes in CLCuMuBMul/Mut occurred adjacent to the region of sequence shown by Saunders et al. (32) to be important for transreplication of AYVB (Table 2 and Fig. 4), none of these changes creates clear CbLCuV-like iterons, and the effect may thus be indirect by improving the presumed iteron-like sequence(s) and/or their relationship to the nonanucleotide-containing hairpin structure so as to enhance Rep recognition and, hence, transreplication. An analysis of a greater number of such mutants, isolated over a greater number of passages through plants, could potentially allow the identification of sequences important for the interaction of betasatellites with begomoviruses.
The region of betasatellite sequence between the A-rich region and the SCR, which has been identified as likely to contain the Rep recognition sequences (32), has been noted previously as being hypervariable (1). It was suggested that the A-rich region might play a role in maintaining this variability by promoting misincorporation during virion strand DNA synthesis. This would be a useful adaptation for a satellite molecule, allowing it to rapidly adapt to a new helper virus. The results presented here, that interaction with CbLCuV leads to sequence changes in the satellite and selection for mutants which interact more effectively with the virus, are wholly consistent with this hypothesis.
The findings presented in this study have serious implications for biosecurity. We shown not only that, at least for some combinations, OW betasatellites can interact with NW begomoviruses but also that when they do, the betasatellite may rapidly adapt to yield a molecule which is transreplicated more efficiently and enhances symptom severity. The global trade in agricultural products has led to the introduction of the OW monopartite begomovirus tomato yellow leaf curl into the NW (25), probably on two separate occasions (15). The virus rapidly spread to affect much of southern North America and central and northern South America, with serious losses to tomato cultivation. Across the warmer parts of the OW, begomovirus-betasatellite complexes are serious pathogens of many of the crops that have their origins in the NW, indicating that there is little, if any, natural resistance to them in the germplasm. These crops include tomato, peppers, and potato. In light of our findings, it is clear that frontier plant protection controls should be extended to include all betasatellites. It may also be desirable to investigate the availability of natural resistance to these pathogens and introgress this into local elite crop varieties for deployment as a first line of defense in the event of an introduction. Similarly, a study of the potential of NW weeds to act as reservoirs, and thus likely to be important in the epidemiology of an introduced betasatellite, could provide information useful in efforts to control spread while resistant varieties are deployed.
Acknowledgments
We thank D. Robertson for providing the CbLCuV clones and the greenhouse staff at Donald Danforth Plant Science Center for maintaining the plants and making them available for this study.
This project was supported by the Pak-US Linkage Program under contract PGA-7251-05-007. R.W.B. is supported by the Higher Education Commission, Government of Pakistan, under the Foreign Faculty Hiring Program.
Footnotes
Published ahead of print on 1 July 2009.
REFERENCES
- 1.Amin, I., S. Mansoor, L. Amrao, M. Hussain, S. Irum, Y. Zafar, S. E. Bull, and R. W. Briddon. 2006. Mobilisation into cotton and spread of a recombinant cotton leaf curl disease satellite. Arch. Virol. 151:2055-2065. [DOI] [PubMed] [Google Scholar]
- 2.Antignus, Y. L., O. Pearlman, M. Omer, S. Yunis, H. Messika, Y. Uko, and O. Koren, A. 2003. Squash leaf curl geminivirus-a new illegal immigrant from the western hemisphere and a threat to cucurbit crops in Israel. Phytoparasitica 31:415. [Google Scholar]
- 3.Argüello-Astorga, G., L. Herrera-Estrella, and R. Rivera-Bustamante. 1994. Experimental and theoretical definition of geminivirus origin of replication. Plant Mol. Biol. 26:553-556. [DOI] [PubMed] [Google Scholar]
- 4.Arguello-Astorga, G. R., and R. Ruiz-Medrano. 2001. An iteron-related domain is associated to Motif 1 in the replication proteins of geminiviruses: identification of potential interacting amino acid-base pairs by a comparative approach. Arch. Virol. 146:1465-1485. [DOI] [PubMed] [Google Scholar]
- 5.Ariyo, O. A., G. I. Atiri, A. G. Dixon, and S. Winter. 2006. The use of biolistic inoculation of cassava mosaic begomoviruses in screening cassava for resistance to cassava mosaic disease. J. Virol. Methods 137:43-50. [DOI] [PubMed] [Google Scholar]
- 6.Briddon, R., J. K. Brown, E. Moriones, J. Stanley, M. Zerbini, X. Zhou, and C. Fauquet. 2008. Recommendations for the classification and nomenclature of the DNA-β satellites of begomoviruses. Arch. Virol. 153:763-781. [DOI] [PubMed] [Google Scholar]
- 7.Briddon, R., S. Bull, S. Mansoor, I. Amin, and P. Markham. 2002. Universal primers for the PCR-mediated amplification of DNA β. Mol. Biotechnol. 20:315-318. [DOI] [PubMed] [Google Scholar]
- 8.Briddon, R. W., S. E. Bull, I. Amin, A. M. Idris, S. Mansoor, I. D. Bedford, P. Dhawan, N. Rishi, S. S. Siwatch, A. M. Abdel-Salam, J. K. Brown, Y. Zafar, and P. G. Markham. 2003. Diversity of DNA β, a satellite molecule associated with some monopartite begomoviruses. Virology 312:106-121. [DOI] [PubMed] [Google Scholar]
- 9.Briddon, R. W., S. Mansoor, I. D. Bedford, M. S. Pinner, K. Saunders, J. Stanley, Y. Zafar, K. A. Malik, and P. G. Markham. 2001. Identification of DNA components required for induction of cotton leaf curl disease. Virology 285:234-243. [DOI] [PubMed] [Google Scholar]
- 10.Briddon, R. W., and J. Stanley. 2006. Subviral agents associated with plant single-stranded DNA viruses. Virology 344:198-210. [DOI] [PubMed] [Google Scholar]
- 11.Bull, S. E., W. S. Tsai, R. W. Briddon, P. G. Markham, J. Stanley, and S. K. Green. 2004. Diversity of begomovirus DNA ß satellites of non-malvaceous plants in East and Southeast Asia. Arch. Virol. 149:1193-1200. [DOI] [PubMed] [Google Scholar]
- 12.Chatterji, A., M. Padidam, R. N. Beachy, and C. M. Fauquet. 1999. Identification of replication specificity determinants in two strains of tomato leaf curl virus from New Delhi. J. Virol. 73:5481-5489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui, X., G. Li, D. Wang, D. Hu, and X. Zhou. 2005. A Begomovirus DNAβ-encoded protein binds DNA, functions as a suppressor of RNA silencing, and targets the cell nucleus. J. Virol. 79:10764-10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dry, I. B., L. R. Krake, J. E. Rigden, and M. A. Rezaian. 1997. A novel subviral agent associated with a geminivirus: the first report of a DNA satellite. Proc. Natl. Acad. Sci. USA 94:7088-7093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duffy, S., and E. C. Holmes. 2007. Multiple introductions of the Old World begomovirus tomato yellow leaf curl virus into the New World. Appl. Environ. Microbiol. 73:7114-7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fontes, E. P., H. J. Gladfelter, R. L. Schaffer, I. T. Petty, and L. Hanley-Bowdoin. 1994. Geminivirus replication origins have a modular organization. Plant Cell 6:405-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gladfelter, H. J., P. A. Eagle, E. P. B. Fontes, L. A. Batts, and L. Hanley-Bowdoin. 1997. Two domains of the AL1 protein mediate geminivirus origin recognition. Virology 239:186-197. [DOI] [PubMed] [Google Scholar]
- 18.Gutierrez, C. 1999. Geminivirus DNA replication. Cell. Mol. Life Sci. 56:313-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hellens, R. P., E. A. Edwards, N. R. Leyland, S. Bean, and P. M. Mullineaux. 2000. pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42:819-832. [DOI] [PubMed] [Google Scholar]
- 20.Idris, A. M., A. Abdel-Salam, and J. K. Brown. 2006. Introduction of the New World squash leaf curl virus to squash (Cucurbita pepo) in Egypt: a potential threat to important food crops. Plant Dis. 90:1262. [DOI] [PubMed] [Google Scholar]
- 21.Lin, B., S. A. Akbar Behjatnia, I. B. Dry, J. W. Randles, and M. A. Rezaian. 2003. High-affinity Rep-binding is not required for the replication of a geminivirus DNA and its satellite. Virology 305:353-363. [DOI] [PubMed] [Google Scholar]
- 22.Mansoor, S., R. W. Briddon, S. E. Bull, I. D. Bedford, A. Bashir, M. Hussain, M. Saeed, Y. Zafar, K. A. Malik, C. Fauquet, and P. G. Markham. 2003. Cotton leaf curl disease is associated with multiple monopartite begomoviruses supported by single DNA β. Arch. Virol. 148:1969-1986. [DOI] [PubMed] [Google Scholar]
- 23.Mansoor, S., Y. Zafar, and R. W. Briddon. 2006. Geminivirus disease complexes: the threat is spreading. Trends Plant Sci. 11:209-212. [DOI] [PubMed] [Google Scholar]
- 24.McGlashan, D., J. E. Polston, and D. Bois. 1994. Tomato yellow leaf curl virus in Jamaica. Plant Dis. 78:1219. [Google Scholar]
- 25.Morales, F. J. 2006. History and current distribution of begomoviruses in Latin America. Adv. Virus. Res. 67:127-162. [DOI] [PubMed] [Google Scholar]
- 26.Polston, J. E., D. Bois, C. A. Serra, and S. Concepcion. 1994. First report of a tomato yellow leaf curl-like geminivirus in the western hemisphere. Plant Dis. 78:831. [Google Scholar]
- 27.Qazi, J., I. Amin, S. Mansoor, M. J. Iqbal, and R. W. Briddon. 2007. Contribution of the satellite encoded gene βC1 to cotton leaf curl disease symptoms. Virus Res. 128:135-139. [DOI] [PubMed] [Google Scholar]
- 28.Saeed, M., S. A. A. Behjatnia, S. Mansoor, Y. Zafar, S. Hasnain, and M. A. Razaian. 2005. A single complementary-sense transcript of a geminiviral DNA β satellite is determinant of pathogenicity. Mol. Plant Microbe Interact. 18:7-14. [DOI] [PubMed] [Google Scholar]
- 29.Saeed, M., Y. Zafar, J. W. Randles, and M. A. Rezaian. 2007. A monopartite begomovirus-associated DNA β satellite substitutes for the DNA B of a bipartite begomovirus to permit systemic infection. J. Gen. Virol. 88:2881-2889. [DOI] [PubMed] [Google Scholar]
- 30.Saunders, K., I. D. Bedford, R. W. Briddon, P. G. Markham, S. M. Wong, and J. Stanley. 2000. A unique virus complex causes Ageratum yellow vein disease. Proc. Natl. Acad. Sci. USA 97:6890-6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saunders, K., I. D. Bedford, and J. Stanley. 2001. Pathogenicity of a natural recombinant associated with Ageratum yellow vein disease: implications for geminivirus evolution and disease aetiology. Virology 282:38-47. [DOI] [PubMed] [Google Scholar]
- 32.Saunders, K., R. W. Briddon, and J. Stanley. 2008. Replication promiscuity of DNA-β satellites associated with monopartite begomoviruses; deletion mutagenesis of the Ageratum yellow vein virus DNA-β satellite localizes sequences involved in replication. J. Gen. Virol. 89:3165-3172. [DOI] [PubMed] [Google Scholar]
- 33.Saunders, K., A. Norman, S. Gucciardo, and J. Stanley. 2004. The DNA β satellite component associated with Ageratum yellow vein disease encodes an essential pathogenicity protein (βC1). Virology 324:37-47. [DOI] [PubMed] [Google Scholar]
- 34.Saunders, K., N. Salim, V. R. Mali, V. G. Malathi, R. Briddon, P. G. Markham, and J. Stanley. 2002. Characterisation of Sri Lankan cassava mosaic virus and Indian cassava mosaic virus: evidence for acquisition of a DNA B component by a monopartite begomovirus. Virology 293:63-74. [DOI] [PubMed] [Google Scholar]
- 35.Seal, S. E., F. vandenBosch, and M. J. Jeger. 2006. Factors influencing begomovirus evolution and their increasing global significance: implications for sustainable control. Crit. Rev. Plant Sci. 25:23-46. [Google Scholar]
- 36.Shahid, M. S., S. Mansoor, and R. W. Briddon. 2007. Complete nucleotide sequences of cotton leaf curl Rajasthan virus and its associated DNA β molecule infecting tomato. Arch. Virol. 152:2131-2134. [DOI] [PubMed] [Google Scholar]
- 37.Stanley, J., D. M. Bisaro., R. W. Briddon., J. K. Brown., C. M. Fauquet., B. D. Harrison., E. P. Rybicki, and D. C. Stenger. 2005. Geminiviridae, p. 301-326. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy: classification and nomenclature of viruses. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press, London, United Kingdom.
- 38.Tao, X., and X. Zhou. 2008. Pathogenicity of a naturally occurring recombinant DNA satellite associated with tomato yellow leaf curl China virus. J. Gen. Virol. 89:306-311. [DOI] [PubMed] [Google Scholar]
- 39.Turnage, M. A., N. Muangsan, C. G. Peele, and D. Robertson. 2002. Geminivirus-based vectors for gene silencing in Arabidopsis. Plant J. 30:107-114. [DOI] [PubMed] [Google Scholar]
- 40.Van Den Bosch, F., F. Akudibilah, S. Seal, and M. Jeger. 2006. Host resistance and the evolutionary response of plant viruses. J. Appl. Ecol. 43:506-516. [Google Scholar]
- 41.Yang, Y., R. Li, and M. Qi. 2000. In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J. 22:543-551. [DOI] [PubMed] [Google Scholar]
- 42.Zhou, X., Y. Xie, X. Tao, Z. Zhang, Z. Li, and C. M. Fauquet. 2003. Characterization of DNA-beta associated with begomoviruses in China and evidence for coevolution with their cognate viral DNA-A. J. Gen. Virol. 84:237-247. [DOI] [PubMed] [Google Scholar]