Abstract
Subacute sclerosing panencephalitis (SSPE) is a demyelinating central nervous system disease caused by a persistent measles virus (MV) infection of neurons and glial cells. There is still no specific therapy available, and in spite of an intact innate and adaptive immune response, SSPE leads inevitably to death. In order to select effective antiviral short interfering RNAs (siRNAs), we established a plasmid-based test system expressing the mRNA of DsRed2 fused with mRNA sequences of single viral genes, to which certain siRNAs were directed. siRNA sequences were expressed as short hairpin RNA (shRNA) from a lentiviral vector additionally expressing enhanced green fluorescent protein (EGFP) as an indicator. Evaluation by flow cytometry of the dual-color system (DsRed and EGFP) allowed us to find optimal shRNA sequences. Using the most active shRNA constructs, we transduced persistently infected human NT2 cells expressing virus-encoded HcRed (piNT2-HcRed) as an indicator of infection. shRNA against N, P, and L mRNAs of MV led to a reduction of the infection below detectable levels in a high percentage of transduced piNT2-HcRed cells within 1 week. The fraction of virus-negative cells in these cultures was constant over at least 3 weeks posttransduction in the presence of a fusion-inhibiting peptide (Z-Phe-Phe-Gly), preventing the cell fusion of potentially cured cells with persistently infected cells. Transduced piNT2 cells that lost HcRed did not fuse with underlying Vero/hSLAM cells, indicating that these cells do not express viral proteins any more and are “cured.” This demonstrates in tissue culture that NT2 cells persistently infected with MV can be cured by the transduction of lentiviral vectors mediating the long-lasting expression of anti-MV shRNA.
The neurodegenerative human disease subacute sclerosing panencephalitis (SSPE) occurs with an incidence rate of approximately 1:10,000 after infection with wild-type measles virus (MV) (4, 38). The course of the illness is quite variable, usually lasting from 1 to 3 years. Much more rapid forms that lead to death within a few months as well as prolonged courses with a duration of more than 20 years have been described (40). Neuropathological findings include diffuse encephalitis, affecting both the gray and white matters, characterized by perivascular cuffing and diffuse lymphocytic infiltrations. Neurons, oligodendrocytes, fibrous astrocytes, and some brain microvascular endothelial cells contain large aggregates of intranuclear inclusion bodies consisting of MV nucleocapsid structures (1, 16). In these persistently infected cells, viral ribonucleoprotein particles (RNPs) replicate intracellularly, whereas the budding of complete viruses and cell-cell fusion are not observed. A characteristic feature of this central nervous system disease is that the expression of viral envelope proteins (matrix [M], fusion [F], and hemagglutinin [H] proteins) is restricted by various means. In particular, the M protein and the cytoplasmic part of the F protein harbor single or hypermutations or deletions, which prevent their proper expression (2, 3, 9, 10). The lack of M reduces budding, supports cell fusion, and enhances the intracellular replication of RNPs (7, 8, 32, 37). As far as is known, the cell-to-cell spread of infectivity in the human brain occurs in the presence of normal cellular and strong humoral antiviral immune responses with very high anti-MV antibody titers in the cerebrospinal fluid. This, however, cannot prevent virus spread.
A variety of approaches to the treatment of SSPE have been attempted, but an evaluation of their efficiency has been extremely difficult, since clinical trials are based on small numbers of patients, the course of SSPE is highly variable, and spontaneous remissions may also occur. Intrathecal or intraventricular administration of alpha interferon, inosiplex, and/or ribavirin is a common regimen, but despite many efforts, the establishment of an effective therapy has not been possible. Since the immune systems of the patients appear normal, and given the fact that virus spreads in the form of intracellular RNPs, a promising specific therapy must target this intracellular replication of MV.
RNA interference (RNAi) may provide such a means and has already been used successfully to inhibit the expression of a number of viral infections, including the Ebola, influenza A, hepatitis B and C, human immunodeficiency, respiratory syncytial, and West Nile viruses, and several RNAi-based therapeutics are already in preclinical test phases (for reviews, see references 6 and 24). Small interfering RNAs (siRNAs) have also been described to be active against MV (20, 29, 32), including an MV isolate from an SSPE patient (SSPE-Kobe-1) (28). In the latter approach, the authors generated recombinant adenoviruses (rAdV) expressing siRNA against MV L mRNA and assessed them in freshly infected Vero/SLAM cells. In contrast to this work, we constructed lentiviral vectors expressing short hairpin RNAs (shRNAs) and transduced persistently infected human NT2 cells with these vectors. This lentiviral approach provided the proof of principle that a preexisting persistent MV infection can be cured by shRNA.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
Human embryonal kidney 293T, HeLa, and African green monkey Vero cells were cultivated in Dulbecco's modified Eagle's medium containing 5% fetal calf serum. Human NT2 cells are a pluripotent human testicular embryonic cell line, which can be differentiated in cells with neuronal appearance (30). NT2 cells persistently infected with rMVHcRed-CAMH (piNT2-HcRed) were grown in Dulbecco's modified Eagle's medium containing 10% fetal calf serum, 100 U/ml penicillin-streptomycin, and nonessential amino acids. The recombinant virus rMVHcRed-CAMH was generated by the insertion of an additional transcription unit containing the open reading frame for HcRed (Clontech) upstream of the N gene in p(+)MV and the substitution of the H open reading frame for that of the CAM-RB strain (15, 22). Recombinant MV rMVHcRed-CAMH and rMV-EGFP-CAMH (where EGFP is enhanced green fluorescent protein) (36a) were propagated using Vero cells. The cells were infected at a multiplicity of infection (MOI) of 0.01, and virus was harvested when maximal giant cell formation was observed as previously described (36). Monoclonal antibody (MAb) against the MV N protein (clone F227) was generated (39), produced, and purified using protein G columns (Pharmacia) in our laboratory.
Cloning of shRNA-expressing lentiviral vectors and preparation of pseudotyped lentiviral particles.
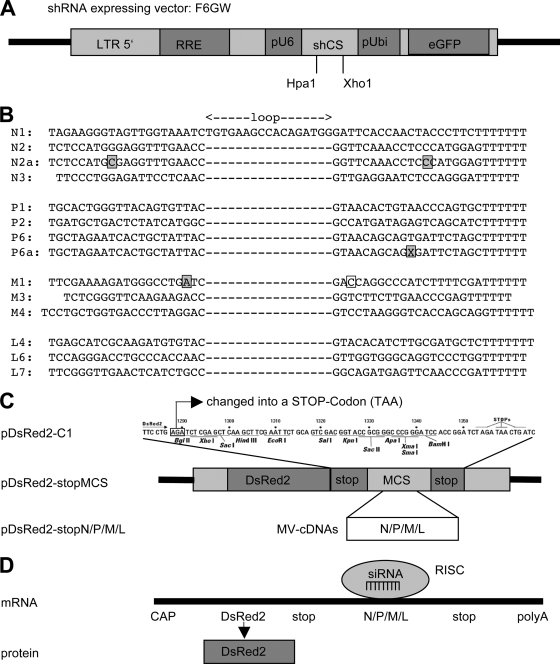
The shRNA-expressing vector F6GW (Flap, U6, GFP, woodchuck posttranscriptional enhancer element) is based on F6GW (14, 23, 35), in which the U6 promoter was introduced in PacI sites 5′ of the ubiquitin promoter, which directs the expression of EGFP (Fig. 1A). Primers for the siRNA sequences (AA-[N]18-TT, AA-[N]19-TT, or AA-[N]20-TT) for shRNA expression were selected using the program iRNA 2.0 (http://mekentosj.com/irnai/). The selected gene-specific sequences (Fig. 1B) correspond to nucleotides (starting with 1 at the respective ATGs) as follows: N1 (607 to 625), N2 (1035 to 1053), N3 (116 to 134), M1 (722 to 739), M3 (820 to 837), M4 (567 to 586), P1 (311 to 329), P2 (373 to 391), P6 (1005 to 1023), L4 (2544 to 2562), L6 (4864 to 4882), and L7 (5280 to 5298). To improve the selection of the target-recognizing guide strand and the degradation of the passenger strand, nucleotide 19 of M1 (Fig. 1B) was altered to destabilize the duplex and to create thermodynamic asymmetry. In order to enhance gene silencing, we used the loop of the cellular human miR30 pre-micro-RNA backbone in our shRNA constructs (TGTGAAGCCACAGATGG) (5). As an inactive control shRNA, we used the random sequence 5′ TCTCAACGGTCAACATATTAC (TGTGAAGCCACAGATGG) GTAATATGTTGACCGTTGAGTTTTTT 3′. The sequences of all constructs were verified by sequencing. Two shRNA constructs with mutations (N2a with a G→C exchange at position 9 and P6a with the deletion of nucleotide 41) were used as controls.
FIG. 1.
(A) The shRNA expressing vector F6GW contains the U6 promoter and HpaI and XhoI sites for cloning of the shRNA-encoding oligonucleotides and a ubiquitin promoter directing the expression of EGFP. (B) Oligonucleotide sequences for MV-Edmonson-specific shRNAs that were cloned in the vector F6GW. Boxed residues indicate a mismatch in the short hairpin sequence N2a, a deletion in P6a, and a mismatch in M1. (C) A stop codon was introduced in the plasmid pDsRed2-C1, and the cDNAs for the viral N, P, M, and L mRNAs were cloned into the multiple cloning site (MCS). (D) The mRNAs generated from the pDsRed2-stopN, -P, -M, and -L constructs contain an open reading frame for DsRed, followed by an MV-specific sequence. These MV-specific sequences act as target sequences for the RNA-inducing silencing complex-containing virus-specific siRNAs. When the mRNA is degraded due to interaction with an MV-specific siRNA, levels of the DsRed protein are reduced.
Vesicular stomatitis virus envelope protein G (VSV-G)-pseudotyped lentiviral particles were produced by the transfection of 293T cells with plasmids F6GW-shRNA (N1 to L7), pMDLg/pRRE (gag, pol), pRSV-Rev (rev for nuclear export of RNAs), and pMD.G (VSV-G) using polyethylenimine (25 K; Polysciences, Inc.). After 2 days, the supernatant was harvested and filter sterilized using 0.4-μm filters. The particle preparations were titrated using HeLa cells. To transduce the NT2 target cells, 1 × 104 cells were seeded in 48-well dishes. After 24 h, the cells were transduced using an MOI of 1 and, 24 h later, split into larger dishes as required. We tested virus-specific siRNAs for off-target effects earlier and did not find effects on cell growth or the induction of alpha/beta interferon (32). No identities between the siRNA sequences and the cellular genes were detected. After the transduction of the cells with shRNA-expressing lentiviral vectors, no effects on cell morphology and proliferation were observed.
Cloning of DsRed indicator plasmids.
We first introduced a stop codon at nucleotide positions 1288 to 1290 (AGA→TAA) in plasmid pDsRed2-C1 (Invitrogen) using a site-directed mutagenesis kit (QuikChange II; Stratagene), resulting in pDsRed-stopMCS. Then, cDNA fragments of the N, P, M, and L genes of the Edmonston strain of MV were cut out of plasmids pSC-N (MfeI, 1,763 bp), pSC-P (MfeI, 1,742 bp), pCG-M (MfeI, 1,644 bp), and pEMC-La (EcoRI-SalI, 6,449 bp [31]) and cloned into the EcoRI site of the multiple cloning site of pDsRed-stopMCS. Clones with inserts inserted in the right orientation were selected by restriction enzyme analysis, and sequences were verified by sequencing. The functional expression of DsRed was tested by the transfection of the plasmids in 293T cells and analysis by flow cytometry.
Flow cytometry.
To stain cells with MAb against the viral N protein, 1 × 105 cells were fixed and permeabilized with 3.7% paraformaldehyde and 0.25% Triton X-100 in fluorescence-activated cell sorter (FACS) buffer (phosphate-buffered saline containing 0.4% bovine serum albumin and 0.02% sodium azide) and then incubated for 1 h on ice with 1 μg of MAb F227 in 100 μl FACS buffer. Cells were washed twice in FACS buffer and incubated with 200 μl of a 1:100 dilution of Alexa488-conjugated goat anti-mouse immunoglobulin on ice for a further 1 h. After three washes with FACS buffer, flow cytometric analysis was performed on a FACScan (Becton Dickinson). The data were evaluated using the program FlowJo (Cytek Development).
RESULTS
Selection of siRNA sequences, cloning of lentiviral vectors, and evaluation of shRNA constructs.
We decided to use a lentiviral shRNA expression system because pseudotyped lentiviral particles can infect and integrate their genome into postmitotic neurons and thus can potentially be used to treat infected brains in vivo. The vector F6GW (Fig. 1A) contains a U6 promoter for the expression of shRNA, followed by an EGFP expression cassette. Using the open-source siRNA design program iRNAi 2.0, we selected oligonucleotides with high probabilities to express functional siRNAs against MV mRNAs (Fig. 1B). The oligonucleotides encoding shRNAs were cloned into the HpaI/XhoI sites of F6GW, and the insertions were verified by sequencing.
In order to test the efficacy of the selected shRNAs, we constructed an indicator system for FACS analysis based on a two-color system consisting of one plasmid (indicator) expressing the target mRNA and DsRed and a second plasmid (F6GW-shRNA) expressing shRNA and EGFP. The cDNAs of the viral target mRNAs were cloned into the DsRed indicator plasmids so that the DsRed protein was translated from an mRNA fused with the viral target RNA (Fig. 1C). The interaction of this target RNA with siRNAs will induce the degradation of the complete mRNA and thus reduce the expression of DsRed (Fig. 1D).
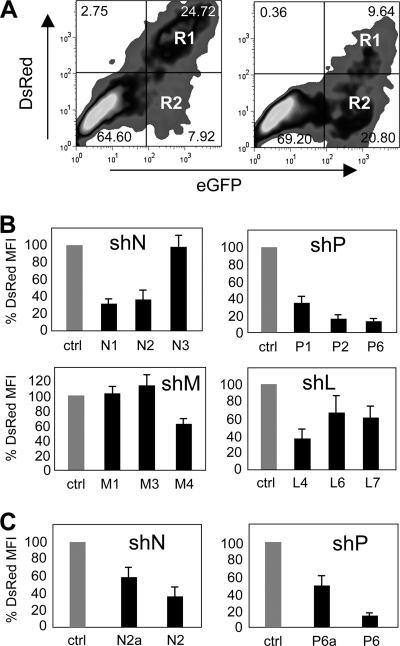
The reduction of DsRed by shRNAs was quantified by flow cytometry in transfected 293T cells (Fig. 2). To analyze the DsRed reduction, cells were cotransfected with the indicator plasmid and the shRNA-expressing plasmid for 2 days, before the red mean fluorescence intensity (MFI) measured in GFP-positive cells (regions R1 and R2) was evaluated. The example shown in Fig. 2A shows that after the transfection of shRNA (P6), the percentage of double-positive cells in R1 dropped from 24.72 to 9.64%. The MFIs in R1 and R2 were used to quantify the effects of the shRNAs. When the MFI of the control transfection is set to 100%, the example given in Fig. 2A for shRNA P6 (shP6) equals a reduction of 14%. The selected shRNAs led to maximal reductions of 67% by shN1 against N mRNA, 32% by shM4 against M mRNA, 86% by shP6 against P mRNA, and 60% by shL4 against L mRNA containing indicator constructs (Fig. 2B). Interestingly, shRNAs with single point mutations (N2a and P6a) led to reductions of only approximately 50% of the corresponding perfectly complementary shRNAs (Fig. 2C).
FIG. 2.
Flow cytometry analysis of 293T cells cotransfected with the pDsRed2-stopN, -P, -M, and -L indicator and corresponding F6GW-based MV-specific shRNA-expressing plasmids. (A) 293T cells transfected with pDsRed2-stopN and F6GW expressing control shRNA (left panel) and 293T cells transfected with pDsRed2-stopP and F6GW expressing shP6 (F6GW-P6-EGFP) (right panel). The percentages of cells in each region are shown. (B) 293T cells were cotransfected with the pDsRed2-stop indicator plasmids containing the cDNAs for N, P, M, and L and the F6GW vector expressing control shRNA or the corresponding MV-specific shRNAs (n = 5). Results are presented as percentages of the MFI of control shRNA. (C) The shRNAs with point mutations (N2a and P6a) in comparison to the 100% homologous shRNAs (N2 and P6) show a reduced inhibitory effect.
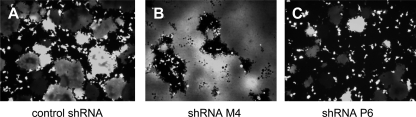
To exclude the possibility that the shRNAs per se were cytotoxic in target cells, we performed a cell viability assay. After the transfection of cells with shRNA-expressing plasmids, the fraction of healthy cells was between 78 and 85% for most constructs including the control shRNA plasmid (not shown). One exception was shL7 with a lower fraction of healthy cells (61%); this was excluded from further experiments. As another control for determining the function of the shRNAs, we also transfected 293T cells with shRNA-expressing vectors and simultaneously infected them with MV expressing GFP (MV-EGFP-CAMH) at an MOI of 0.1 to evaluate the shRNA effect on syncytium formation in the microscope. With respect to the control plasmid, we observed considerably larger syncytia in the case of shM4 and considerably smaller syncytia in the case of shP6 (Fig. 3), shN1, and shL4 (not shown). These results confirmed the effectiveness of the shRNA sequences selected for N1, M4, P6, and L4 in freshly MV-infected 293T cells.
FIG. 3.
Effect of shRNAs on syncytium formation and EGFP expression. 293T cells were infected at an MOI of 0.1 with MV-Edtag-CAMH-EGFP and 1 h later transfected with the F6GW control vector (A) or vectors expressing shRNAs against M (M4) (B) and P (P6) (C) and incubated for 24 h.
Generation of piNT2-HcRed cells.
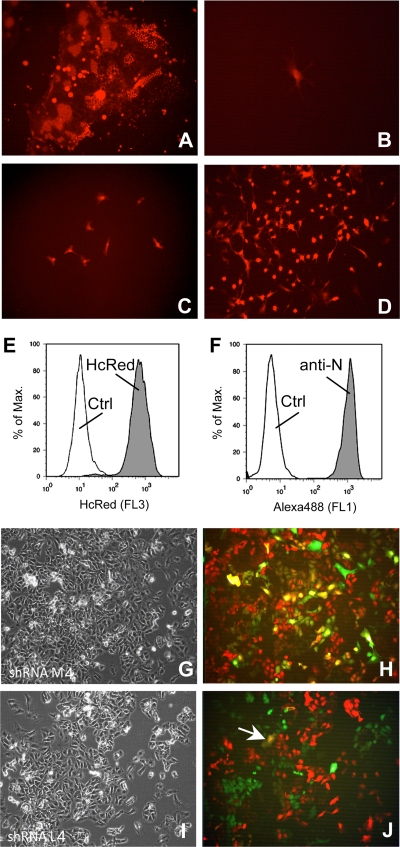
The interesting question remained of whether the expression of shRNA after transduction is able to reduce or to eliminate completely a persistent MV infection from target cells. In order to achieve this goal, we generated persistently infected NT2 cells using rMVHcRed-CAMH. NT2 cells were chosen, as they are a well-characterized pluripotent human testicular embryonic cell line which can be differentiated with retinoic acid in cells with neuronal appearance and which had been persistently infected with MV earlier (25, 30). The cells were infected at an MOI of 0.1 and cultured until most of the cells died. After 1 to 2 weeks, a few remaining cells continued to grow (Fig. 4A to D). These cells were analyzed by flow cytometry for their expression of HcRed and the viral N protein (Fig. 4E and F, respectively). Practically all surviving cells were HcRed and N protein positive and can be cultured as piNT2-HcRed cells.
FIG. 4.
Generation of persistently infected NT2 cells. NT2 cells were infected with rMVHcRed-CAMH at an MOI of 0.1. (A) Syncytium formation was apparent after 24 h. (B) After further cultivation, most of the cells died (4 days postinfection [d.p.i.]). (C) A small percentage of the cells survive and are persistently infected (7 d.p.i.). (D) These persistently infected cells can be grown and passaged but do not form syncytia (21 d.p.i.). The expression of HcRed (E) by piNT2-HcRed cells was compared with the expression of the viral N protein (F) after staining with MAb F227 and secondary antibodies by flow cytometry. Persistently infected NT2 cells were transduced with shM (G, H)- and shL (I, J)-producing lentiviral particles and incubated for 12 days. The phase-contrast pictures and fluorescence overlays with red (infection) and green (lentiviral vectors) are shown, respectively. The arrow in J points to a yellow cell (transduced and still infected).
“Therapy” of piNT2-HcRed cells.
piNT2-HcRed cells were transduced with VSV-G-pseudotyped particles carrying lentiviral genomes that integrate in the cellular genome and mediate the expression of shRNAs. The MOI of 1 used for transduction was chosen so that transduced and nontransduced cells can be compared in one culture. Examples of transduced cultures analyzed after 12 days by fluorescence microscopy are shown in Fig. 4G and H. When cells were transduced with vectors expressing shRNA against M, which does not affect the persistent infection, nontransduced cells appear red and most transduced cells yellow, due to the simultaneous expression of HcRed and GFP (Fig. 4H). In contrast, when the cells were transduced with vectors expressing shRNA against L, there are either red or green cells, due to the fact that the infection is downregulated in transduced cells (Fig. 4J). Only a very small number of transduced cells remained infected at this time and appear yellow (Fig. 4J, arrow).
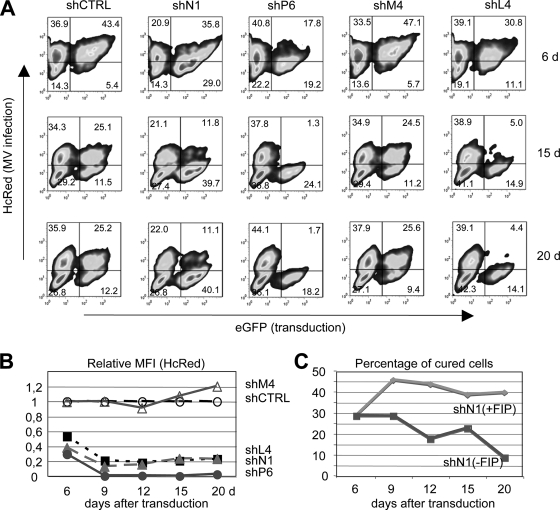
To quantify the effect of transduction, aliquots were analyzed after 6 to 20 days by flow cytometry (Fig. 5A). Since persistently infected NT2 cells do not fuse to each other but maintain their capacity to fuse with uninfected cells expressing appropriate receptors (25), transduced cells were cultivated either in the absence or presence of a fusion-inhibiting peptide (FIP; Z-Phe-Phe-Gly) (33) to prevent the potential fusion of infected cells with “cured” cells. In cells expressing random control and M4 shRNA, the percentage of HcRed/EGFP double-positive cells (top right quadrant) stays nearly constant, whereas in cells expressing shN1, shP6, and shL4, the double-positive cells disappeared and EGFP single-positive cells (bottom right quadrant) appeared. These cells in the bottom right quadrant, which do not express HcRed anymore, are potentially cured cells. Interestingly, in the case of shN1, a significant proportion of the cells stayed HcRed/EGFP double positive (11.09%), whereas no such population remained in shP6- and shL4-treated cells, in which all transduced cells became HcRed negative at 20 days posttransduction. Similar results were obtained in the presence (Fig. 5A) and absence (not shown) of FIP. In Fig. 5B, the MFIs of HcRed in the transduced cultures in relation to that in the control shRNA-transduced culture are presented. The curves illustrate that after 9 days posttransduction, HcRed is significantly degraded in shN1-, shP6-, and shL4-transduced cells and that the effect is maintained over a longer period of time.
FIG. 5.
Transduction of piNT2-HcRed cells with pseudotyped lentiviral particles expressing anti-MV shRNAs. (A) Persistently infected NT2 cells were transduced with pseudotyped particles mediating the expression of control (shCTRL), N1, P6, M4, or L4 shRNAs at an MOI of 2.5 and cultivated for 6, 15, and 20 days in the presence of FIP (Z-Phe-Phe-Gly). Cell aliquots were analyzed by flow cytometry. HcRed-positive cells are infected (top left and right quadrants), EGFP-positive cells are transduced (top and bottom right quadrants), and double-positive cells are infected and transduced (top right quadrant). The relative MFIs of HcRed in transduced EGFP-positive cells (top and bottom right quadrants) are shown in relation to the control shRNA in panel B. The HcRed signal stays high after treatment of the cells with control shRNA (shCTRL, unfilled circle [set to 1]) and shRNA against M (shM, unfilled triangle), whereas shRNAs against L, N, and P (shL4, black square; shN1, gray triangle; shP6, gray circle) led to a decrease of HcRed. (C) The percentage of EGFP-positive/HcRed-negative cured cells (bottom right quadrants in panel A) is shown over time for shN-treated cells in the presence (+FIP) and absence (−FIP) of FIP.
The percentages of potentially cured cells over time (bottom right quadrant) are shown in Fig. 5C. At 6 days posttransduction, there is still a considerable number of double-positive cells, indicating that the siRNA-mediated degradation of the viral RNA and the subsequent disappearance of the proteins were not completed. At day 9 posttransduction, the percentage of cured cells in the cultures was maximal. These percentages also reflect the initial transduction efficiency of the individual experiments. At later times (15 and 20 days posttransduction), the percentage of cured cells stayed nearly constant in FIP-treated cultures, whereas it decreased in cultures without FIP in which cell fusion was allowed. This suggests that cells in which the viral replication is inhibited reexpress normally distributed viral receptors (CD46) and can fuse with infected neighboring cells.
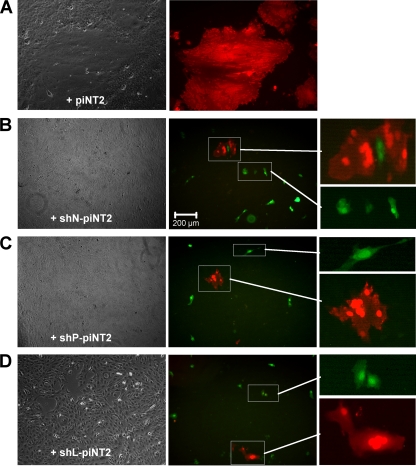
To prove that HcRed-negative/EGFP-positive cells obtained after the transductions do not contain infectious virus anymore, we used an overlay assay as described for persistently infected NT2 cells. When these cells were overlaid (in a 1:1,000 dilution) on a monolayer of uninfected Vero cells, cell-to-cell fusion was readily observed (25). We therefore assessed the presence of infection at 15 days in the shN1-, shP6-, and shL4-transduced piNT2-HcRed cultures containing infected (red) and potentially cured (green) cells. The cells were trypsinized, diluted, and overlaid on a monolayer of Vero cells, and the formation of syncytia was observed by microscope after 24 to 72 h (Fig. 6). Whereas the red cells readily fused and formed large infected syncytia with underlying Vero cells, the green cells did not fuse and remained single green cells on top of the Vero cell monolayer. This indicated that MV was indeed eliminated from the green transduced NT2 cells and that these cells can be designated as cured.
FIG. 6.
Detection of infectivity in transduced cells by a syncytium formation assay with Vero cells. Ninety percent confluent Vero cells were overlaid with piNT2 cells transduced for 12 days and nontransduced and incubated at 37°C to allow cell fusion. At 72 h, large syncytia were formed with nontransduced piNT2-HcRed cells (A), whereas smaller syncytia were formed with HcRed-positive cells and no syncytia were formed with EGFP-positive cells in overlays with shN1 (B)-, shP6 (C)-, and shL4 (D)-transduced cells. Enlargements as indicated show red syncytia and green single cells present in the cultures.
DISCUSSION
We demonstrated here that a persistent MV infection can successfully be cured by siRNAs against N, P, and L mRNAs. In contrast, in the case of siRNA against M, no reduction of infection and an enhancement of cell fusion with uninfected cells were detected. This supports earlier findings that M siRNA stimulated cell-cell fusion and intracellular RNP replication (32). The persistent infection was successfully eliminated by lentivirus-expressed shRNAs against N, P, and L within 2 weeks after transduction. Interestingly, shRNAs against all three RNP components were effective in eliminating the infection from piNT2 cells. In cultures treated with shRNA against P and L, transduced cells became completely HcRed negative, indicating the complete elimination of viral replication. In cells transduced with shRNA against N, a remaining population of HcRed-positive cells was still present after 20 days. The reason for that may be that the siRNA sequence chosen was not effective enough or that the amount of siRNA present in these cells is not sufficient to induce the degradation of this viral mRNA, which is, according to the expression gradient of negative-stranded RNA viruses, present in the highest concentration beyond viral mRNAs. Nevertheless, our data indicate that the transduction of persistently infected cells with lentiviral vectors expressing shRNAs against the viral RNP mRNAs can successfully induce the elimination of infectivity.
We are aware of the fact that the culture system does not reflect the situation in the brain and that at present, an optimal way of applying siRNA in vivo in the human brain is not yet available (21, 24). The lentiviral system may not be optimal for human use because of the random integration in the genome and potential activation of oncogenic properties but can be used to introduce a long-lasting RNAi-based gene knockdown into nondividing postmitotic cells (11, 19, 23, 27). It may be used in the mouse model to further investigate the therapeutic potential of shRNAs. Our data deliver the proof of principle and support the view that RNAi may be a promising therapy of persistent infections, which cannot be eliminated by the immune system, and against which no other specific antiviral treatment is available. This is the case for SSPE patients, who develop an extremely strong humoral immune response against MV but fail to eliminate the virus (13, 17, 26).
An rAdV vector system was used in an attempt to inhibit MV expression in Vero/SLAM cells freshly infected with virus isolates from a clinical MV case (K52) and an SSPE patient (Kobe-1) (28). In this study, three sequences in the MV L mRNA were targeted, with success only when cells were infected with rAdV at a high MOI (30, 300, and 3,000) before the infection with MV but not when the cells were infected with rAdV 6 h or later after MV infection (28). This seems to be a problem of fresh infections, since we also made the observation that freshly MV-infected cells cannot be successfully treated when the siRNA is applied after the infection (unpublished data). In contrast, the persistently infected NT2 cells, as described in the present publication, could successfully be cured with the lentiviral vector, providing a long-lasting expression of shRNA.
Human NT2 cells persistently infected with similar recombinant MV as we used in this study have been generated and characterized before (25). These cell cultures are characterized by infection of most, if not all, of the cells. They divide normally and a constant level of virus replication and gene expression is observed, whereas infectious particles in the supernatant were not detected. Persistence and the absence of cell fusion are maintained in the presence of fully functional viral fusion complexes. Cell surface reorganization of the MV receptor CD46 plays an important role in this process. When such persistently infected cells were overlaid on uninfected cells, they readily fused (25). shRNA-treated cured cells probably redistribute CD46 on their surfaces, reachieve the normal even distribution of receptors, and, as observed (Fig. 5C), may fuse with persistently infected adjacent cells. Therefore, we cultivated transduced cells in the presence of FIP to inhibit the potential fusion of infected cells with cured cells and the subsequent disappearance of cured cells. The different percentages of cured cells in the absence or presence of FIP demonstrated that cell fusion does play a role in these cultures. When cell fusion was allowed, the number of cured cells was reduced over time. The situation in infected brains is different. Similar effects can most likely be excluded in infected brains in vivo, since cell fusion is usually not observed in the central nervous system and the expression of viral envelope proteins is reduced by other mechanisms.
A complication for any in vivo application is certainly that the sequence of MV in an individual SSPE brain is derived from a certain wild-type virus and may be altered by mutations (34). Due to the required sequence specificity of siRNA, the sequence must be known exactly before a potential treatment with shRNA-expressing vectors can be performed. Therefore, a highly conserved region of the viral genome must be targeted. In addition, due to the high mutation rates conferred by RNA polymerases, RNA viruses may escape from single siRNAs. This was found, e.g., for HIV infections using siRNA against the Nef protein (12) and for poliovirus (18). However, using a pool of siRNAs to simultaneously target multiple sites in the viral genome prevented the emergence of resistant viruses (18). This has important implications for therapeutic approaches using siRNA. The application of multiple siRNAs in the brain may have an advantage over single siRNAs by preventing the emergence of resistant viruses. On the other hand, also in the presence of few mutations, a certain siRNA may impair the translation of viral proteins according to the mechanism of microRNAs, which do not induce the degradation of a target mRNA but induce a block in translation. Nevertheless, the therapeutic usage of siRNA for persistently infected brain cells may require the use of multiple or at least several siRNAs with a variety of sequences to prevent the establishment of escape mutants. However, a still-unresolved problem is how to apply multiple siRNAs in vivo in the brain for therapeutic approaches. Applications in the brain using adenoviral or lentiviral vectors, which can direct shRNA expression in postmitotic neurons, should be further investigated in animal models.
Acknowledgments
We thank the Deutsche Forschungsgemeinschaft for financial support.
Footnotes
Published ahead of print on 8 July 2009.
REFERENCES
- 1.Allen, I. V., S. McQuaid, J. McMahon, J. Kirk, and R. McConnell. 1996. The significance of measles virus antigen and genome distribution in the CNS in SSPE for mechanisms of viral spread and demyelination. J. Neuropathol. Exp. Neurol. 55:471-480. [DOI] [PubMed] [Google Scholar]
- 2.Baczko, K., M. Carter, M. A. Billeter, and V. ter Meulen. 1984. Measles virus gene expression in subacute sclerosing panencephalitis. Virus Res. 1:585-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baczko, K., U. G. Liebert, M. A. Billeter, R. Cattaneo, H. Budka, and V. ter Meulen. 1986. Expression of defective measles virus genes in brain tissues of patients with subacute sclerosing panencephalitis. J. Virol. 59:472-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellini, W. J., J. S. Rota, L. E. Lowe, R. S. Katz, P. R. Dyken, S. R. Zaki, W.-J. Shieh, and P. A. Rota. 2005. Subacute sclerosing panencephalitis: more cases of this fatal disease are prevented by measles immunization than previously recognized. J. Infect. Dis. 192:1686-1693. [DOI] [PubMed] [Google Scholar]
- 5.Boden, D., O. Pusch, R. Silbermann, F. Lee, L. Tucker, and B. Ramratnam. 2004. Enhanced gene silencing of HIV-1 specific siRNA using microRNA designed hairpins. Nucleic Acids Res. 32:1154-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caplen, N. J. 2003. RNAi as a gene therapy approach. Expert Opin. Biol. Ther. 3:575-586. [DOI] [PubMed] [Google Scholar]
- 7.Cathomen, T., B. Mrkic, D. Spehner, R. Drillien, R. Naef, J. Pavlovic, A. Aguzzi, M. A. Billeter, and R. Cattaneo. 1998. A matrix-less measles virus is infectious and elicits extensive cell fusion: consequences for propagation in the brain. EMBO J. 17:3899-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cathomen, T., H. Y. Naim, and R. Cattaneo. 1998. Measles viruses with altered envelope protein cytoplasmic tails gain cell fusion competence. J. Virol. 72:1224-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cattaneo, R., G. Rebmann, A. Schmid, K. Baczko, V. ter Meulen, and M. A. Billeter. 1987. Altered transcription of a defective measles virus genome derived from a diseased human brain. EMBO J. 6:681-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cattaneo, R., A. Schmid, D. Eschle, K. Baczko, V. ter Meulen, and M. A. Billeter. 1988. Biased hypermutation and other genetic changes in defective measles viruses in human brain infections. Cell 55:255-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Consiglio, A., A. Gritti, D. Dolcetta, A. Follenzi, C. Bordignon, F. H. Gage, A. L. Vescovi, and L. Naldini. 2004. Robust in vivo gene transfer into adult mammalian neural stem cells by lentiviral vectors. Proc. Natl. Acad. Sci. USA 101:14835-14840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Das, A. T., T. R. Brummelkamp, E. M. Westerhout, M. Vink, M. Madiredjo, and R. Bernards. 2004. Human immunodeficiency virus type 1 escapes from RNA interference-mediated inhibition. J. Virol. 78:2601-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dörries, R., and V. ter Meulen. 1984. Detection and identification of virus specific oligoclonal IgG in unconcentrated cerebrospinal fluid by immunoblot technique. J. Neuroimmunol. 7:77-89. [DOI] [PubMed] [Google Scholar]
- 14.Dull, T., R. Zufferey, M. Kelly, R. J. Mandel, M. Nguyen, D. Trono, and L. Naldini. 1998. A third generation lentivirus vector with a conditional packaging system. J. Virol. 72:8463-8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duprex, W. P., I. Duffy, S. McQuaid, L. Hamill, J. Schneider-Schaulies, L. Cosby, M. Billeter, V. ter Meulen, and B. Rima. 1999. The H gene of rodent brain-adapted measles virus confers neurovirulence to the Edmonston vaccine strain. J. Virol. 73:6916-6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Esiri, M. M., D. R. Oppenheimer, B. Brownell, and M. Haire. 1981. Distribution of measles antigen and immunoglobulin-containing cells in the CNS in subacute sclerosing panencephalitis (SSPE) and atypical measles encephalitis. J. Neurol. Sci. 53:29-43. [DOI] [PubMed] [Google Scholar]
- 17.Fujinami, R. S., and M. B. A. Oldstone. 1979. Antiviral antibody reacting on the plasma membrane alters measles virus expression inside the cell. Nature 279:529-530. [DOI] [PubMed] [Google Scholar]
- 18.Gitlin, L., J. K. Stone, and R. Andino. 2005. Poliovirus escape from RNA interference: short interfering RNA-target recognition and implications for therapeutic approaches. J. Virol. 79:1027-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hughes, S. M., F. Moussavi-Harami, S. L. Sauter, and B. L. Davidson. 2002. Viral-mediated gene transfer to mouse primary neural progenitor cells. Mol. Ther. 5:16-24. [DOI] [PubMed] [Google Scholar]
- 20.Keita, D., R. Servan de Almeida, G. Libeau, and E. Albina. 2008. Identification and mapping of a region on the mRNA of Morbillivirus nucleoprotein susceptible to RNA interference. Antivir. Res. 80:158-167. [DOI] [PubMed] [Google Scholar]
- 21.Kumar, P., H. Wu, J. L. McBride, K. E. Jung, M. H. Kim, B. L. Davidson, S. K. Lee, P. Shankar, and N. Manjunath. 2007. Transvascular delivery of small interfering RNA to the central nervous system. Nature 448:39-43. [DOI] [PubMed] [Google Scholar]
- 22.Liebert, U. G., and D. Finke. 1995. Measles infections in rodents. Curr. Top. Microbiol. Immunol. 191:149-166. [DOI] [PubMed] [Google Scholar]
- 23.Lois, C., E. J. Hong, S. Pease, E. J. Brown, and D. Baltimore. 2002. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science 295:868-872. [DOI] [PubMed] [Google Scholar]
- 24.Lopez-Fraga, M., N. Wright, and A. Jimenez. 2008. RNA interference-based therapeutics: new strategies to fight infectious disease. Infect. Disord. Drug Targets 8:262-273. [DOI] [PubMed] [Google Scholar]
- 25.Ludlow, M., S. McQuaid, S. L. Cosby, R. Cattaneo, B. K. Rima, and W. P. Duprex. 2005. Measles virus superinfection immunity and receptor redistribution in persistently infected NT2 cells. J. Gen. Virol. 86:2291-2303. [DOI] [PubMed] [Google Scholar]
- 26.Mehta, P. D., J. Kulczycki, S. P. Mehta, W. Sobczyk, P. K. Coyle, E. A. Sersen, and H. M. Wisniewski. 1992. Increased levels of beta 2-microglobulin, soluble interleukin-2 receptor, and soluble CD8 in patients with subacute sclerosing panencephalitis. Clin. Immunol. Immunopathol. 65:53-59. [DOI] [PubMed] [Google Scholar]
- 27.Naldini, L., U. Blomer, P. Gallay, D. Ory, R. Mulligan, F. H. Gage, I. M. Verma, and D. Trono. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263-267. [DOI] [PubMed] [Google Scholar]
- 28.Otaki, M., D. P. Jiang, M. Sasayama, M. Nagano-Fujii, and H. Hotta. 2007. Generation of recombinant adenovirus expressing siRNA against the L mRNA of measles virus and subacute sclerosing panencephalitis virus. Microbiol. Immunol. 51:985-991. [DOI] [PubMed] [Google Scholar]
- 29.Otaki, M., K. Sada, H. Kadoya, M. Nagano-Fujii, and H. Hotta. 2006. Inhibition of measles virus and subacute sclerosing panencephalitis virus by RNA interference. Antivir. Res. 70:105-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pleasure, S. J., and V. M. Lee. 1993. NTera 2 cells: a human cell line which displays characteristics expected of a human committed neuronal progenitor cell. J. Neurosci. Res. 35:585-602. [DOI] [PubMed] [Google Scholar]
- 31.Radecke, F., P. Spielhofer, H. Schneider, K. Kaelin, M. Huber, C. Dötsch, G. Christiansen, and M. A. Billeter. 1995. Rescue of measles virus from cloned DNA. EMBO J. 14:5773-5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reuter, T., B. Weissbrich, S. Schneider-Schaulies, and J. Schneider-Schaulies. 2006. RNA interference with measles virus N, P, and L mRNAs efficiently prevents and with matrix protein mRNA enhances viral transcription. J. Virol. 80:5951-5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richardson, C. D., and P. W. Choppin. 1983. Oligopeptides that specifically inhibit membrane fusion by paramyxoviruses: studies on the site of action. Virology 131:518-532. [DOI] [PubMed] [Google Scholar]
- 34.Rima, B. K., J. A. P. Earle, K. Baczko, V. ter Meulen, J. Carabana, M. Caballero, M. L. Celma, and R. Fernandez-Munoz. 1997. Sequence divergence of measles virus haemagglutinin during natural evolution and adaptation to cell culture. J. Gen. Virol. 78:97-106. [DOI] [PubMed] [Google Scholar]
- 35.Rubinson, D. G., C. P. Dillon, A. V. Kwiatkowski, C. Sievers, L. Yang, J. Kopinja, M. Zhang, M. T. McManus, F. B. Gertler, M. L. Scott, and L. van Parijs. 2003. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat. Genet. 33:401-406. [DOI] [PubMed] [Google Scholar]
- 36.Schneider-Schaulies, J., S. Schneider-Schaulies, and V. Ter Meulen. 1993. Differential induction of cytokines by primary and persistent measles virus infections in human glial cells. Virology 195:219-228. [DOI] [PubMed] [Google Scholar]
- 36a.Schubert, S., K. Möller-Ehrlich, K. Singethan, S. Wiese, W. P. Duprex, B. K. Rima, S. Niewiesk, and J. Schneider-Schaulies. 2006. A mouse model of persistent brain infection with recombinant measles virus. J. Gen. Virol. 87:2011-2019. [DOI] [PubMed] [Google Scholar]
- 37.Suryanarayana, K., K. Baczko, V. ter Meulen, and R. R. Wagner. 1994. Transcription inhibition and other properties of matrix proteins expressed by M genes cloned from measles viruses and diseased human brain tissue. J. Virol. 68:1532-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takasu, T., J. M. Mgone, C. S. Mgone, K. Miki, K. Komase, H. Namae, Y. Saito, Y. Kokubun, T. Nishimura, R. Kawanishi, T. Mizutani, T. J. Markus, J. Kono, P. G. Asuo, and M. P. Alpers. 2003. A continuing high incidence of subacute sclerosing panencephalitis (SSPE) in the eastern highlands of Papua New Guinea. Epidemiol. Infect. 131:887-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.ter Meulen, V., S. Loffler, M. J. Carter, and J. R. Stephenson. 1981. Antigenic characterization of measles and SSPE virus haemagglutinin by monoclonal antibodies. J. Gen. Virol. 57:357-364. [DOI] [PubMed] [Google Scholar]
- 40.Weissbrich, B., J. Schneider-Schaulies, and V. ter Meulen. 2003. Measles and its neurological complications. Marcel Dekker, New York, NY.