Abstract
TRF2, a component of the shelterin complex, functions to protect telomeres. TRF2 contains an N-terminal basic domain rich in glycines and arginines, similar to the GAR motif that is methylated by protein arginine methyltransferases. However, whether arginine methylation regulates TRF2 function has not been determined. Here we report that amino acid substitutions of arginines with lysines in the basic domain of TRF2 induce telomere dysfunction-induced focus formation, leading to induction of cellular senescence. We have demonstrated that cells overexpressing TRF2 lysine mutants accumulate telomere doublets, indicative of telomere instability. We uncovered that TRF2 interacts with PRMT1, and its arginines in the basic domain undergo PRMT1-mediated methylation both in vitro and in vivo. We have shown that loss of PRMT1 induces growth arrest in normal human cells but has no effect on cell proliferation in cancer cells, suggesting that PRMT1 may control cell proliferation in a cell type-specific manner. We found that depletion of PRMT1 in normal human cells results in accumulation of telomere doublets, indistinguishable from overexpression of TRF2 lysine mutants. PRMT1 knockdown in cancer cells upregulates TRF2 association with telomeres, promoting telomere shortening. Taken together, these results suggest that PRMT1 may control telomere length and stability in part through TRF2 methylation.
The integrity of telomeres is vital to cell survival and proliferation. Mammalian telomeric DNA is coated with a telomere-specific complex, referred to as shelterin (18, 41). Shelterin, consisting of TRF1, TRF2, TIN2, RAP1, TPP1, and POT1, functions to control telomere length and stability (18, 41). Disruption or depletion of the shelterin complex and its interacting proteins has been shown to induce a variety of telomere abnormalities, such as telomere loss, telomere end-to-end fusions, telomere-containing double-minute chromosomes, and telomere doublets (more than one telomeric signal at a single chromatid end) (14, 45, 55, 56, 58, 66). Telomeres containing these abnormalities have been shown to be associated with DNA damage response factors, such as 53BP1, forming nuclear structures that are referred to as telomere dysfunction-induced foci (TIFs) (15, 30, 50, 55, 58).
TRF2, a component of the shelterin complex, binds to telomeric DNA as a dimer and has been shown to play a crucial role in telomere length maintenance and telomere protection. TRF2 contains an N-terminal basic domain rich in glycines and arginines, a central TRFH dimerization domain, and a C-terminal Myb DNA-binding domain (56). Overexpression of TRF2 has been shown to induce telomere shortening (2, 26, 37, 48). Loss of TRF2 from telomeres either through overexpression of a TRF2 dominant-negative allele (TRF2ΔBΔM) or depletion of TRF2 leads to an accumulation of telomere end-to-end fusions, resulting in ATM- and p53-dependent growth arrest or apoptosis depending upon the cell type (10, 25, 50, 56). Inhibition of TRF2 function at telomeres through overexpression of TRF2 lacking the basic domain has been shown to promote DNA recombination at telomeres, leading to the induction of telomere loss (58). TRF2 has been shown to interact with many proteins involved in DNA repair, recombination, and replication, including Apollo (20, 30, 55), WRN (39), FEN1 (36), and ORC (3). While loss of WRN or inhibition of FEN1 results in telomere loss (14, 45), depletion of Apollo induces telomere doublets (55). Although DNA recombination is thought to play an important role in the formation of telomere loss, the mechanism underlying the formation of telomere doublets remains elusive. Furthermore, it has not been determined whether TRF2 inhibition may induce telomere doublets.
Protein arginine methyltransferases (PRMTs) represent a family of enzymes that utilize S-adenosyl methionine as a methyl donor and catalyze the direct transfer of the methyl group to one or two of the guanidino nitrogen atoms of arginine (5, 34). In mammalian cells, 11 PRMTs have been identified, and the majority of them are able to catalyze not only the formation of a monomethylated arginine intermediate but also the production of a dimethylated arginine (4, 40). Based on their substrate specificity, mammalian PRMTs can be classified into type I or type II enzymes. Type I enzymes, including PRMT1 (32), PRMT3 (53), CARM1 (11), PRMT6 (19), and PRMT8 (29), catalyze arginine dimethylation asymmetrically. On the other hand, type II enzymes, including PRMT5 (44), PRMT7 (21, 35), and PRMT9 (12), catalyze arginine dimethylation symmetrically.
PRMT1, the predominant mammalian type I enzyme (52, 54), accounts for more than 85% of all arginine methylation reactions in human cells (54). PRMT1 methylates a diverse range of proteins involved in transcription (1, 57), RNA processing (13, 24, 47), and DNA damage repair (8, 9). Recently PRMT1 has also been shown to be associated with human telomeres (17). Most of its substrates contain a characteristic motif rich in glycines and arginines, referred to as the GAR motif (6, 7, 38). TRF2 contains an N-terminal basic domain rich in glycines and arginines, similar to the GAR motif. However whether TRF2 is a substrate of PRMT1 has not been determined.
In this report, we show that the basic domain of TRF2 undergoes arginine methylation both in vitro and in vivo. We show that PRMT1 interacts with TRF2 and is the main enzyme responsible for methylating the basic domain of TRF2 both in vivo and in vitro. Overexpression of TRF2 carrying amino acid changes of arginines to lysines in the basic domain results in the formation of telomere doublets in hTERT-BJ cells, suggesting that arginines in the basic domain of TRF2 are essential for maintaining telomere stability. Knockdown of PRMT1 induces growth arrest in normal human cells but has no effect on cell proliferation in cancer cells, suggesting that PRMT1 may control cell growth in a cell type-dependent manner. We find that depletion of PRMT1 affects both telomere length and stability. While a loss of PRMT1 in cancer cells promotes telomere shortening, PRMT1 knockdown in normal human cells leads to an accumulation of telomere doublets, resembling the phenotype observed with overexpression of TRF2 mutants carrying amino acid changes of arginines to lysines. In addition, we show that depletion of PRMT1 mitigates TRF2 methylation but upregulates TRF2 association with telomeric DNA, suggesting that TRF2 methylation by PRMT1 may control its association with telomeric DNA. We propose that PRMT1 regulates telomere length and stability in part through TRF2 methylation.
MATERIALS AND METHODS
DNA constructs.
The constructs expressing wild-type TRF2 (pLPC-TRF2) and TRF2 lacking the basic domain (pLPC-TRF2ΔB) were generously provided by Titia de Lange, Rockefeller University. All TRF2 mutants except for TRF2-RK5-8 were first made in the pKS vector (Stratagene), followed by subcloning into the retroviral pLPC vector. Annealed oligonucleotides encoding the first 18 amino acids of TRF2 (TRF21-18), which either is wild type or contains amino acid substitutions of arginines to lysines at positions 13, 17, and 18, were ligated to BamHI- and HindIII-linearized pKS vector, generating an intermediate construct of pKS-TRF21-18 or pKS-TRF21-18-RK1-3, respectively. TRF219-500-RK4, which lacks the first 18 amino acids and contains an amino acid change of arginine to lysine at position 21, was obtained by PCR using a forward primer containing the mutation. Ligation of TRF219-500-RK4 into HindIII- and XhoI-linearized pKS-TRF21-18-RK1-3 gave rise to pKS-TRF2-RK1-4, which contains substitutions of arginines with lysines at positions 13, 17, 18, and 21. TRF219-500-RK4-8 was made by PCR using a forward primer encoding TRF2 carrying amino acid changes of arginines to lysines at positions 21, 25, 27, 28, and 30. Subsequent ligation of TRF219-500-RK4-8 to HindIII- and XhoI-linearized pKS-TRF21-18-RK1-3 gave rise to pKS-TRF2-RK, which contains substitutions of arginines with lysines at positions 13, 17, 18, 21, 25, 27, 28, and 30. pKS-TRF2-RA was cloned in a similar manner except that oligonucleotides encoding amino acid changes of arginines to alanines were used. pLPC-TRF2-RK5-8 was made by PCR from the pLPC-TRF2 expression construct using a forward primer encoding TRF2 carrying amino acid changes of arginines to lysines at positions 25, 27, 28, and 30. The presence of all TRF2 mutations was verified by DNA sequencing.
Wild-type TRF2 and various TRF2 mutants were also subcloned into the bacterial expression vector pGST-Parallel-2 (46) (a gift from Murray Junop, McMaster University).
The oligonucleotides encoding small interfering RNAs directed against TRF2, PRMT1, PRMT5, and PRMT6 have been described previously (49, 51, 62, 63). The annealed oligonucleotides were ligated into the pRetroSuper vector (kindly provided by Titia de Lange, Rockefeller University), giving rise to pRetroSuper-shPRMT1, pRetroSuper-shPRMT5, and pRetroSuper-shPRMT6.
Cell culture and retroviral infection.
Cells were grown in Dulbecco's modified Eagle medium with 10% fetal bovine serum for HT1080, Phoenix, 293T, and SV40-transformed skin fibroblast GM637 (Coriell) cells supplemented with nonessential amino acids, l-glutamine, 100 U/ml penicillin, and 0.1 mg/ml streptomycin. Supplemented Dulbecco's modified Eagle medium plus 15% fetal bovine serum was used to culture normal primary fibroblasts (IMR90, MRC5, and GM08399) (Coriell), Nbs1-deficient primary fibroblasts (GM07166) (Coriell), and hTERT-immortalized BJ (hTERT-BJ) cells (a kind gift from Titia de Lange, Rockefeller University). Retroviral gene delivery was carried out essentially as described previously (26). Phoenix amphotropic retroviral packaging cells were transfected with the desired DNA constructs. At 36, 48, 60, 72, and 84 h posttransfection, the virus-containing medium was collected and used to infect cells in the presence of polybrene (4 μg/ml). Twelve hours after the last infection, puromycin (2 μg/ml) was added to the medium, and the cells were maintained in the selection medium for the entirety of the experiments.
Production of PRMT1 and TRF2 proteins.
PRMT1, wild-type TRF2, and TRF2 mutants were expressed as glutathione S-transferase (GST) fusion proteins in Escherichia coli strain BL21(DE3)pLysS. The expression construct for the GST-PRMT1 fusion protein was generously provided by Stéphane Richard, McGill University (Montreal, Canada). Expression of GST-PRMT1 was induced with 0.1 mM isopropyl-1-thio-β-d-galactopyranoside for 3 h at 37°C, whereas expression of GST-TRF2 fusion proteins was induced with 0.3 mM isopropyl-1-thio-β-d-galactopyranoside overnight at room temperature. Following a wash with phosphate-buffered saline (PBS), the cell pellet for GST-PRMT1 was resuspended in PBS containing 0.5% Triton X-100 and 1 mM phenylmethylsulfonyl fluoride and lysed by sonication. For GST-TRF2 proteins, the cell pellet was resuspended in buffer containing 50 mM potassium phosphate buffer (pH 7.0), 5 mM EDTA, 100 mM NaCl, and 1% Triton X-100 and then lysed by sonication. The lysate was subjected to centrifugation, and the supernatant was incubated with a 50% slurry of glutathione-Sepharose 4B (GE Life Sciences) for 4 h at 4°C. For GST-PRMT1, bound proteins were eluted with 40 mM reduced glutathione in buffer containing 50 mM Tris-HCl (pH 8.0), 200 mM NaCl, and 10 mM dithiothreitol and stored in aliquots at −80°C. For GST-TRF2 proteins, TEV protease was added to the mixture to release TRF2 proteins from GST. While GST remained bound to the beads, free TRF2 proteins were recovered in the supernatant.
In vitro methylation assays.
Five to seven micrograms of the recombinant TRF2 protein was incubated with 5 μg purified GST-PRMT1 in a 30-μl reaction mixture containing 1 μCi of S-adenosyl-l-[methyl-3H]methionine (Perkin Elmer) and 25 mM Tris-HCl (pH 7.5) for 2 h at 37°C. Following sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis, the gel was stained with Coomassie blue, destained with 20% (vol/vol) methanol and 8% (vol/vol) acetic acid, and treated with EN3HANCE solution (Perkin Elmer) according to the manufacturer. The gel was then dried and exposed to Kodak BioMax X-ray film at −80°C.
Identification of methylation sites in the basic domain of TRF2.
Endogenous TRF2 was immunoprecipitated from whole-cell extracts of approximately 1.7 × 109 HeLaI.2.11 cells as described previously (65). The beads were washed five times in cold buffer D (20 mM HEPES-KOH, pH 7.9, 20% glycerol, 100 mM KCl, 0.2 mM EDTA, 0.2 mM EGTA, 1 mM dithiothreitol, and 0.5 mM phenylmethylsulfonyl fluoride). Mass spectrometry analysis of TRF2 was done through service provided by WEMB Biochem. Inc., Toronto, Canada, as described previously (64). Briefly, TRF2 bound to beads was digested with both trypsin and chymotrypsin overnight at room temperature. The sample was then acidified to pH 2, and the solution was treated with C18 ZipTip pipette tips prior to liquid chromatography-tandem mass spectrometry analysis with an LCQ Deca XP spectrometer (Thermo Finnegan). The data were analyzed using the Xcalibur software program, and peptide sequence data were rechecked manually.
Mass spectrometry analysis of in vitro-methylated TRF2 was done through the MALDI Mass Spectrometry Facility, University of Western Ontario, London, Ontario, Canada. Following in vitro methylation assays, recombinant wild-type TRF2 was separated by SDS-polyacrylamide gel electrophoresis, visualized with Coomassie blue, and excised. In-gel digestion was performed with trypsin using a MassPREP automated digester station (Perkin Elmer). Mass spectrometry data were obtained using a 4700 proteomics analyzer matrix-assisted laser desorption ionization time-of-flight TOF/TOF system (Applied Biosystems). Data acquisition and data processing, respectively, were done using the 4000 Series Explorer (Applied Biosystems) and Data Explorer (Applied Biosystems) software packages.
Immunoprecipitation and immunoblotting.
Immunoprecipitation was performed with 750 μg of HeLa nuclear extract and nonimmune serum, anti-TRF2 antibody, or anti-PRMT1 antibody as described previously (65). Immunoblotting was carried out with whole-cell extracts as described previously (65). Extracts were fractionated by 8% SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose that was then immunoblotted. Rabbit anti-2meR17 antibody (no. 5510) was developed by Biosynthesis Inc. against a TRF2 peptide containing asymmetrically dimethylated arginine 17 (AAG-RMe2-RASRSSGRARRGRH). Antibodies to TRF1, TRF2, and TIN2 were kind gifts from Titia de Lange, Rockefeller University. Antibodies to PRMT1 and Sam68 were generously provided by Stéphane Richard, McGill University. Antibodies to PRMT1 and PRMT5 were from Upstate. Anti-PRMT6 was from Bethyl Laboratory, and anti-γ-tubulin (GTU88) was from Sigma.
ChIP.
Chromatin immunoprecipitation (ChIP) assays were carried out essentially as described previously (33, 59, 60). Cells were fixed with 1% (wt/vol) formaldehyde in PBS, followed by sonication (10 cycles of 20 s each, 50% duty, and output of 5). Cell lysate of 200 μl (equivalent to 2 × 106 cells) was used in each ChIP. Fifty microliters of cell lysate (equivalent to one-quarter of the amount used for immunoprecipitation) was used for quantifying the total telomeric DNA and processed along with the immunoprecipitation samples at the step of reversing the cross-links. One-fifth of the immunoprecipitated DNA was loaded on the dot blots, whereas 5% of total DNA was loaded in the input lane. The signals on the dot blots were quantified by ImageQuant analysis (GE Healthcare). The ratio of the signal from each ChIP to the signal from the input lane was multiplied by 5% (representing 5% of total DNA) and a factor of 5 (since only one-fifth of the precipitated DNA was loaded for each ChIP) to give the percentage of total telomeric DNA recovered from each ChIP.
Immunofluorescence.
Immunofluorescence was performed essentially as described previously (50, 65). Cells were fixed at room temperature (RT) for 10 min in 3% paraformaldehyde and 2% sucrose, followed by permeablization at RT for 10 min in Triton X-100 buffer (0.5% Triton X-100, 20 mM HEPES-KOH, pH 7.9, 50 mM NaCl, 3 mM MgCl2, 300 mM sucrose). Fixed cells were blocked with 0.5% bovine serum albumin (Sigma) and 0.2% gelatin (Sigma) in PBS and then incubated at RT for 2 h with both rabbit anti-TRF1 (a kind gift from Titia de Lange) and mouse anti-53BP1 (BD Biosciences). Following a wash in PBS, cells were incubated with both fluorescein isothiocyanate (FITC)-conjugated donkey anti-rabbit and tetramethylrhodamine isocyanate-conjugated donkey anti-mouse antibodies (1:100 dilution; Jackson Laboratories) at RT for 1 h. DNA was stained with 4,6-diamidino-2-phenylindole (DAPI) (0.2 μg/ml). Cell images were then recorded on a Zeiss Axioplan 2 microscope with a Hammamatsu C4742-95 camera and processed using the Openlab software package.
Metaphase chromosome spreads and fluorescence in situ hybridization (FISH).
Metaphase chromosome spreads were essentially prepared as described previously (56, 66). hTERT-BJ cells (∼80%) were arrested in colcemid (0.1 μg/ml) for 120 to 180 min. Following arrest, cells were harvested by trypsinization, incubated for 7 min at 37°C in 75 mM KCl, and fixed in freshly made methanol-glacial acidic acid (3:1). Cells were stored overnight at 4°C, dropped onto wet slides, and air dried overnight in a chemical hood.
FISH was carried out essentially as described previously (28, 66). Slides with chromosome spreads were incubated with 0.5 μg/ml FITC-conjugated-(CCCTAA)3 PNA probe (Biosynthesis Inc.) for 2 h at room temperature. Following incubation, slides were washed, counterstained with 0.2 μg/ml DAPI, and embedded in 90% glycerol-10% PBS containing 1 mg/ml p-phenylene diamine (Sigma).
In-gel G-overhang assay and telomere blots.
In-gel G-overhang assay was performed essentially as described previously (23, 66). Genomic DNA isolated from cells was digested with RsaI and HinfI and loaded onto a 0.7% agarose gel in 0.5× Tris-borate-EDTA (TBE). Following electrophoresis, gels were dried down at 50°C and prehybridized at 50°C for 1 h in Church mix (0.5 M Na2PO4 [pH 7.2], 1 mM EDTA, 7% SDS, and 1% bovine serum albumin). Hybridization was done overnight at 50°C with an end-labeled (CCCTTA)4 oligonucleotide. After hybridization, gels were washed and exposed overnight to PhosphorImager screens. For telomere blots, gels were alkali denatured (0.5 M NaOH and 1.5 M NaCl), neutralized (3 M NaCl and 0.5 M Tris-HCl [pH 7.0]), rinsed with dH2O, and hybridized with the end-labeled (CCCTTA)4 oligonucleotide as described previously (23, 66). The average length of telomeric restriction fragments was determined by PhosphorImager analysis using the ImageQuant and Microsoft Excel software programs as described previously (31, 61).
For pulsed-field gel electrophoresis, genomic DNA isolated from hTERT-BJ cells expressing various TRF2 proteins was digested with RsaI and HinfI. Plugs containing the digested DNA were equilibrated with 0.5× TBE and loaded on a 1% agarose gel in 0.5× TBE. Gels were run for 20 h at 5.4 V/cm at a constant pulse time of 5 s using a CHEF DR-II pulsed-field apparatus (Bio-Rad). Following electrophoresis, the gel was processed as described above.
Growth curve assay and other assays.
The growth curve assay and other assays were conducted essentially as described previously (59). For HT1080 cells expressing the vector, wild-type TRF2, or TRF2 mutants, 2.5 × 105 cells per 10-cm plate were seeded in duplicate on the third day of selection in puromycin. Cells were counted every 2 days using a Coulter counter (Beckman). Cells expressing wild-type TRF2 or the vector alone were split 1:8 and 1:16 on the fifth day and the seventh day after seeding, respectively, whereas cells expressing TRF2 mutants were split 1:4 once on the fifth day after seeding. For hTERT-BJ cells expressing the vector, wild-type TRF2, or TRF2 mutants, 2 × 104 cells per well were seeded in duplicate on 12-well plates on day three of selection. Cells were counted every 2 days, and medium was changed every 4 days. For hTERT-BJ and MRC5 cells expressing short-hairpin RNA constructs shPRMT1, shPRMT5, or shPRMT6, 4.8 × 104 cells per well were seeded in triplicate on 12-well plates on the day before infection. Cells were first counted 2 days after selection in puromycin, followed by counting every 2 days.
For the senescence-associated β-galactosidase (β-Gal) assays, hTERT-BJ cells expressing TRF2 mutants were fixed on the 13th day of selection whereas IMR90 cells expressing knockdown constructs were fixed on the 7th day. The senescence-associated β-Gal assays were performed on the fixed cells using the senescence-associated β-Gal senescence kit (Cell Signaling).
The activity of telomerase in cells was determined using a Trapeze telomerase detection kit (Chemicon) according to the manufacturer's protocol. PCR amplification was performed for 31 cycles. The products were separated on a 12.5% nondenaturing polyacrylamide gel in 0.5× TBE buffer and visualized using SYBR green (Invitrogen).
RESULTS
Amino acid substitutions of arginines with lysines or alanines in the basic domain of TRF2 induce TIF formation and cellular senescence.
Overexpression of TRF2 lacking the basic domain (TRF2ΔB) has been shown to promote recombination-dependent telomere loss (58), leading to induction of cellular senescence (56, 58). It has been suggested that TRF2 may inhibit DNA recombination at telomeres through its basic domain (58). The basic domain of TRF2 contains nine arginines that are conserved between mouse and human (Fig. 1A) (56); however, whether these arginines are important for TRF2 function has not been determined. To address this question, we generated four TRF2 mutants (TRF2-RK, TRF2-RA, TRF2-RK1-4, and TRF2-RK5-8). In the TRF2-RK or TRF2-RA mutant, the first eight arginines were simultaneously changed to either lysines (TRF2-RK) or alanines (TRF2-RA). In the TRF2-RK1-4 mutant, arginines at positions 13, 17, 18, and 21 were replaced with lysines, whereas arginines at positions 25, 27, 28, and 30 were replaced with lysines in the TRF2-RK5-8 mutant. Expression of various TRF2 mutants was comparable to that of wild-type TRF2 (Fig. 1B). We found that overexpression of TRF2-RK, TRF2-RA, TRF2-RK1-4, or TRF2-RK5-8 led to induction of slow growth and cellular senescence in both human fibrosarcoma HT1080 cells and hTERT-immortalized normal human fibroblast hTERT-BJ cells (Fig. 1C to F and data not shown), similar to that of TRF2ΔB (Fig. 1C to F). These results suggest that maintaining the charge alone in the basic domain is insufficient for TRF2 function. These results further imply that more than one arginine is required for TRF2 function.
FIG. 1.
Arginines in the basic domain of TRF2 are crucial for its function. (A) Schematic diagram of human TRF2. Arginines in the basic domain are highlighted in red, and they are conserved between human and mouse. (B) Western blot analysis of expression of the wild type and various TRF2 mutants. Whole-cell extracts made from 200,000 cells were used, and immunoblotting was performed with anti-TRF2 antibody. The γ-tubulin blot was used as a loading control. (C) Growth curve of HT1080 cells expressing TRF2 carrying amino acid changes of arginines to lysines or alanines. Cells were infected with retrovirus expressing either wild-type TRF2, various TRF2 mutants, or the vector alone. Following the last infection, cells were selected with puromycin (2 μg/ml) and maintained in the selection medium for 13 days. (D) Growth curve of hTERT-BJ cells expressing TRF2 carrying amino acid changes of arginines to lysines. Cells were infected with the indicated retrovirus. Following the last infection, cells were selected with puromycin (2 μg/ml) and maintained in the selection medium for 15 days. Standard deviations derived from three independent experiments are indicated. (E) Overexpression of TRF2 carrying amino acid changes of arginines to lysines induces senescence. hTERT-BJ cells infected with the indicated viruses were stained for senescence-associated β-Gal on day 13. (F) Quantification of percentage of cells staining positive for senescence-associated β-Gal. A total of more than 1,500 cells from three independent experiments were scored. Standard deviations derived from three independent experiments are indicated.
Disruption of TRF2 function is known to result in induction of TIFs (50, 58). TIFs are sites where telomeres colocalize with DNA damage response factors, such as 53BP1 (50). To investigate whether arginines in the basic domain of TRF2 are essential for maintaining telomere integrity, we performed coimmunostaining in TRF2-RK-expressing cells using anti-53BP1 and anti-TRF1 antibodies. Compared to the vector alone, we observed a sharp increase in TIF formation in TRF2-RK-expressing cells (Fig. 2A and B). About 20% of TRF2-RK-expressing cells displayed TIFs, whereas less than 1% of the vector-expressing cells showed TIF formation (Fig. 2B). In addition, we found that overexpression of TRF2-RK led to a reduction in telomeric association of TIN2 (Fig. 2C and D), a component of shelterin (27). Overexpression of TRF2-RK had no impact on the level of TIN2 expression (Fig. 2E). Together, these results reveal that arginines in the basic domain of TRF2 are important for telomere integrity as well as shelterin association with telomeres.
FIG. 2.
(A) Overexpresssion of TRF2-RK induces TIF formation. Indirect immunofluorescence using anti-TRF1 in conjunction with anti-53BP1 was performed with fixed hTERT-BJ cells expressing either TRF2-RK or the vector alone. Arrowheads indicate sites of colocalization of 53BP1 and TRF1. (B) Quantification of percentage of cells with more than five TIFs. For each cell line, a total of more than 800 cells from three independent experiments were scored. Standard deviations derived from three independent experiments are indicated. (C) Dot blots of ChIPs. ChIPs were performed with either anti-TIN2 or anti-IgG antibody in cell extracts from HT1080 cells overexpressing TRF2-RK or the vector alone. Precipitated DNA was analyzed for the presence of TTAGGG repeats and Alu repeats by Southern blotting. (D) Quantification of anti-TIN2 ChIPs. The signals were quantified by ImageQuant analysis. The percentage of precipitated DNA was calculated relative to the input signals. Standard deviations derived from three independent experiments are indicated. (E) Western blot analysis of TRF2 protein expression. Whole-cell extracts made from 200,000 cells were used, and immunoblotting was performed with anti-TRF2 and anti-TIN2 antibodies. The γ-tubulin blot was used as a loading control. (F) Genomic blot of telomeric restriction fragments of hTERT-BJ cells expressing various proteins as indicated above the blot. About 3 μg of RsaI/HinfI-digested genomic DNA from each sample was used for gel electrophoresis. The DNA molecular size markers are shown on the left of the blot. The bottom panel, taken from an ethidium bromide-stained agarose gel, is used as a loading control. (G) Genomic blot of telomeric restriction fragments of hTERT-BJ cells expressing various proteins as indicated above the blot. About 3 μg of RsaI/HinfI-digested genomic DNA from each sample was separated on a CHEF gel. The picture of an ethidium bromide (EtBr)-staining gel is shown at the right of the blot, whereas the DNA molecular size markers are shown at the left of the blot.
Overexpression of TRF2 carrying amino acid changes of arginines to lysines promotes the formation of telomere doublets.
We have shown that overexpression of TRF2-RK led to induction of TIFs (Fig. 2A and B). To gain further insight into the role of arginines in the basic domain of TRF2, we decided to examine whether overexpression of TRF2-RK, TRF2-RK1-4, or TRF2-RK5-8 may affect telomere length or stability. While no substantial change in the average telomere length was detected in hTERT-BJ or HT1080 cells overexpressing TRF2-RK, TRF2-RK1-4, or TRF2-RK5-8 (Fig. 2F and G and data not shown), we found that overexpression of TRF2-RK, TRF2-RK1-4, or TRF2-RK5-8 promotes the formation of telomere doublets (Fig. 3A and B). FISH analysis revealed a significant increase in telomere doublets in cells overexpressing TRF2-RK (about fourfold; P = 0.005), TRF2-RK1-4 (more than threefold; P = 0.003), and TRF2-RK-5-8 (more than threefold; P = 0.002) compared to results with the vector alone (Fig. 3B). Accumulation of telomere doublets was not observed in cells overexpressing TRF2ΔB, whereas there was only a slight increase in telomere doublets in cells overexpressing TRF2 compared to results with the vector alone (Fig. 3B). These results suggest that TRF2-RK, TRF2-RK1-4, and TRF2-RK5-8 may affect telomere stability in a manner that is distinctive from that of TRF2ΔB.
FIG. 3.
Overexpression of TRF2 carrying amino acid changes of arginines to lysines promotes the formation of telomere doublets in both hTERT-BJ and Nbs1-deficient GM07166 cells. (A) Metaphase spreads from hTERT-BJ cells expressing TRF2-RK, TRF2ΔB, or the vector alone. Chromosomes were stained with DAPI and false colored in red. Telomeric DNA was detected by FISH using a FITC-conjugated (CCCTAA)3-containing PNA probe (green). Arrows indicate telomere doublets. Enlarged images of chromosomes with telomere doublets are shown. (B) Quantification of telomere doublets in hTERT-BJ cells stably infected with retrovirus as indicated. For each cell line, a total of 1,600 to 1,900 chromosomes from 42 to 45 metaphase cells were scored. Standard deviations derived from at least three independent experiments are indicated. (C) Quantification of telomere loss in hTERT-BJ cells expressing TRF2 alleles as indicated. For each cell line, a total of 1,600 to 1,900 chromosomes from 42 to 45 metaphase cells were scored. Standard deviations derived from at least three independent experiments are indicated. (D) Quantification of telomere doublets in Nbs1-deficient GM07166 cells stably infected with retrovirus as indicated. For each cell line, a total of 1,372 to 1,650 chromosomes from 38 to 40 metaphase cells were scored. Standard deviations derived from three independent experiments are indicated.
Overexpression of TRF2ΔB led to a drastic increase in telomere loss (more than 16-fold; P ≪ 0.0001) compared to results with the vector alone or wild-type TRF2 (Fig. 3A and C), consistent with previous findings (58). However, only a slight increase in telomere loss was observed in cells overexpressing TRF2-RK, TRF2-RK1-4, and TRF2-RK5-8 compared to results with the vector alone (Fig. 3C). This increase in telomere loss was indistinguishable from that in cells overexpressing wild-type TRF2 (Fig. 3C). Taken together, these results suggest that arginines in the basic domain of TRF2 are essential for blocking the formation of telomere doublets.
TRF2ΔB-induced telomere loss has been shown to result from homologous recombination at telomeres (58). Loss of Nbs1, a component of the conserved Mre11/Rad50/Nbs1 complex essential for DNA recombination (22), has been shown to abrogate TRF2ΔB-induced telomere loss (58). To address whether TRF2-RK-induced telomere doublets require Nbs1-dependent DNA recombination, we performed FISH analysis with Nbs1-deficient GM07166 cells expressing either TRF2-RK or the vector alone. We found that overexpression of TRF2-RK resulted in about a threefold increase in telomere doublets in Nbs1-deficient GM07166 cells (Fig. 3D), similar to that seen in Nbs1-proficient hTERT-BJ cells (Fig. 3B). These results suggest that Nbs1-dependent DNA recombination may not be required for the formation of telomere doublets. These results further imply that the basic domain of TRF2 may modulate telomere stability through multiple mechanisms.
PRMT1 interacts with TRF2 both in vivo and in vitro.
Our findings that amino acid substitutions of arginines with lysines in the N terminus of TRF2 abrogate its function suggest that posttranslational modification, such as methylation on arginines, may be important for TRF2 function. To investigate whether TRF2 is methylated in vivo, we immunoprecipitated endogenous TRF2 from HeLa cells and subjected it to mass spectrometric analysis. Mass spectrometry analysis revealed that arginine at position 17 and arginine at position 18 of TRF2 were dimethylated and monomethylated, respectively (Fig. 4), demonstrating that the basic domain of TRF2 is methylated in vivo.
FIG. 4.
Arginines of TRF2 at positions 17 and 18 are methylated in vivo. Liquid chromatography-tandem mass spectrometry analysis was performed with endogenous TRF2 immunoprecipitated from HeLa cells. The spectra of two peptides identified to contain methylated arginines in the basic domain are shown, with the relative abundance plotted against the monoisotopic mass (m/z). The m/z peaks from both b-type and y-type ions are indicated. The peptide presented in panel A contained dimethylated arginine at position 17 and monomethylated arginine at position 18, whereas the peptide presented in panel B contained dimethylated arginine at position 17.
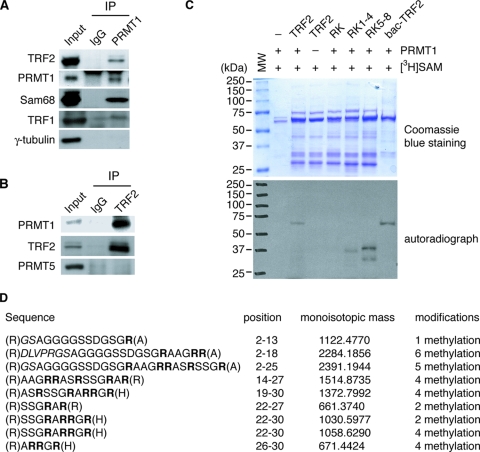
PRMT1, the major protein arginine methyltransferase in mammalian cells (54), has been shown to be associated with telomeres (17). We decided to examine whether PRMT1 is responsible for methylating arginines in the N terminus of TRF2 in vivo. Coimmunoprecipitation using anti-PRMT1 or anti-immunoglobulin G (IgG) antibody showed that TRF2 was associated with PRMT1 but not IgG (Fig. 5A). Anti-PRMT1 immunoprecipitation brought down Sam68, a known substrate of PRMT1 (13) (Fig. 5A). A very weak interaction between PRMT1 and TRF1 was also observed (Fig. 5A). PRMT1 interaction with TRF2 appears to be specific, since anti-PRMT1 immunoprecipitation did not bring down γ-tubulin (Fig. 5A), an abundant protein in the cells. The interaction of PRMT1 with TRF2 was also detected in reverse immunoprecipitation using anti-TRF2 antibody (Fig. 5B). Addition of ethidium bromide to protein extracts prior to coimmunoprecipitation failed to disrupt TRF2 interaction with PRMT1 (data not shown), indicating that its interaction with PRMT1 is unlikely to be mediated through DNA. Furthermore, PRMT5, a type II PRMT, was not found to be associated with TRF2 (Fig. 5B). These results suggest that PRMT1 specifically interacts with TRF2 in vivo.
FIG. 5.
Interaction of PRMT1 with TRF2. (A) Association of PRMT1 with TRF2 in vivo. Coimmunoprecipitations with HeLa nuclear extracts were conducted with either anti-IgG or anti-PRMT1 antibody. Western analysis was done with anti-PRMT1, anti-TRF2, anti-TRF1, anti-Sam68, and anti-γ-tubulin antibodies. (B) Association of TRF2 with PRMT1 in vivo. Coimmunoprecipitations with HeLa nuclear extracts were conducted with either anti-IgG or anti-TRF2 antibody. Western analysis was done with anti-PRMT1, anti-TRF2, and anti-PRMT5 antibodies. (C) PRMT1 methylates arginines in the basic domain of TRF2. In vitro methylation assays were conducted using [3H]SAM, GST-PRMT1, and recombinant TRF2 as indicated. Both the Coomassie blue-staining gel (top panel) and autoradiograph (bottom panel) are shown. The protein molecular mass markers are shown on the left of the gel and blot. All proteins except for bac-TRF2 were expressed in bacteria. Bac-TRF2 (a gift from Titia de Lange) refers to wild-type TRF2 derived from baculovirus. (D) PRMT1 methylates multiple arginines of the basic domain in vitro. Bacterium-expressed wild-type TRF2 was methylated in vitro and then subjected to matrix-assisted laser desorption ionization mass spectrometry analysis. Peptides containing methylated arginines are shown. Arginines in the basic domain are highlighted in bold.
To investigate whether PRMT1 methylates arginines in the basic domain of TRF2, we performed in vitro methylation assays using [3H]S-adenosylmethionine, recombinant GST-tagged PRMT1, and various TRF2 proteins. As shown in Fig. 5C, we found that recombinant wild-type TRF2 derived from either bacteria or baculovirus was methylated by PRMT1. However, PRMT1 failed to methylate full-length recombinant TRF2-RK, TRF2-RK1-4, and TRF2-RK5-8 (Fig. 5C), suggesting that lysine substitutions of arginines in the basic domain abrogate methylation by PRMT1. Some methylation was observed on proteins migrating at or below a 37-kDa marker in the lanes containing TRF2-RK1-4 or TRF2-RK5-8 (Fig. 5C), which is likely nonspecific since anti-TRF2 antibody failed to recognize these proteins (data not shown). In addition, mass spectrometry analysis of in vitro-methylated wild-type TRF2 showed that PRMT1 was able to methylate essentially every arginine in the basic domain of TRF2 (Fig. 5D). Taken together, these results demonstrate that PRMT1 methylates multiple arginines in the basic domain of TRF2 in vitro.
To gain further evidence that PRMT1 is responsible for methylating arginines in the basic domain of TRF2, we raised an antibody (5510) against a TRF2 peptide containing asymmetrically dimethylated arginine 17. Anti-2meR17 antibody 5510 showed a much higher affinity for the methylated TRF2 peptide than for the unmethylated peptide (Fig. 6A). To address whether 5510 specifically recognizes the methylated TRF2 peptide, we performed peptide competition assays. Although preincubation with unmethylated peptide completely abrogated binding of 5510 to unmethylated peptide (Fig. 6B), such preincubation failed to abolish binding of 5510 to methylated peptide (Fig. 6B and C). In contrast, preincubation with methylated peptide completely diminished 5510 binding to methylated peptide (Fig. 6C). These results together demonstrate that 5510 specifically recognizes the methylated TRF2 peptide.
FIG. 6.
PRMT1 is the main enzyme responsible for methylating TRF2 in vivo. (A) Affinity-purified 5510 antibody specifically recognizes TRF2 peptide containing asymmetrically dimethylated arginine 17 (R17). Increasing amounts of peptide carrying either unmodified R17 or asymmetrically dimethylated R17 were spotted onto a nitrocellulose membrane, followed by immunoblotting with affinity-purified 5510 or crude serum (5510CS). The amounts of peptide spotted, from left to right, are 0.5 μg, 1.3 μg, and 2.7 μg. (B) Peptide competition assays. Affinity-purified 5510 antibody was incubated with 5.5 μg of unmodified peptide prior to immunoblotting. Crude serum 5510 was used to show the presence of the TRF2 peptide on the nitrocellulose membrane. Amounts of peptide spotted, from left to right, for both modified and unmodified, are 0.5 μg, 1.3 μg, and 2.7 μg. (C) Peptide competition assays. Prior to immunoblotting TRF2 peptide carrying asymmetrically dimethylated R17, affinity-purified 5510 antibody was incubated with 5.5 μg of either unmodified or modified peptide as indicated. Crude serum 5510 was used to show the presence of the methylated TRF2 peptide. (D) Western analysis of methylated TRF2. Twenty micrograms of the whole-cell extract from hTERT-BJ cells was immunoblotted with affinity-purified 5510 antibody. (E) Depletion of endogenous TRF2 leads to loss of methylated TRF2 bound by 5510 antibody. hTERT-BJ cells were infected with retrovirus expressing either shTRF2 or the vector pRS alone. Western analysis was performed with 5510 and anti-TRF2. The γ-tubulin blot was used as a loading control. (F) Western analysis of methylated TRF2. Affinity-purified 5510 antibody was incubated with 5.5 μg of unmodified peptide prior to immunoblotting. Twenty micrograms of the whole-cell extract from several cell lines, as indicated, was used for immunoblotting. (G) 5510 recognizes immunoprecipitated TRF2. Endogenous TRF2 was immunoprecipitated from HeLa cells using anti-TRF2 antibody. Immunoblotting was performed with anti-TRF2 antibody or affinity-purified 5510 antibody that had been preincubated with 5.5 μg of unmodified peptide. (H) Depletion of endogenous PRMT1 decreases asymmetrical dimethylation of R17. hTERT-BJ, HT1080, and 293T cells were infected with retrovirus expressing either shPRMT1 or the vector pRS alone. For detection of methylated TRF2, Western analysis was done with 5510 that had been preincubated with 5.5 μg of unmodified peptide. Western analysis was also done with anti-TRF2 and anti-PRMT1. The γ-tubulin blot was used as a loading control. (I) Depletion of endogenous PRMT6 has no effect on dimethylation of R17. hTERT-BJ cells were infected with retrovirus expressing either shPRMT6 or the vector pRS alone. Western analysis was done with 5510, anti-TRF2, and anti-PRMT6. The γ-tubulin blot was used as a loading control.
When incubated with whole-cell extracts from cells, 5510 predominantly recognized a single protein band with an apparent molecular weight indistinguishable from that of endogenous TRF2 (Fig. 6D). Depletion of TRF2 led to a loss of TRF2 recognized by 5510 antibody, indicating that 5510 recognizes TRF2 in vivo (Fig. 6E). To investigate whether 5510 recognizes methylated TRF2 in vivo, we incubated 5510 with unmethylated peptide prior to Western analysis. As shown in Fig. 6F, preincubation with unmethylated peptide did not abrogate the ability of 5510 to recognize endogenous TRF2 from a number of human cell lines that are either primary, immortalized, or transformed, suggesting 5510 recognizes methylated TRF2 in vivo. The 5510 antibody also recognized immunoprecipitated TRF2 in the presence of unmethylated peptide (Fig. 6G). We estimate that dimethylation of arginine 17 is present in about 1 to 5% of endogenous TRF2 in HeLa cells (Fig. 6G).
We have shown that PRMT1 methylates TRF2 in vitro (Fig. 5C). To examine whether PRMT1 methylates TRF2 in vivo, we carried out Western analysis on PRMT1-depleted cells using 5510 antibody that had been preincubated with unmethylated peptide. We found that depletion of PRMT1 mitigated R17 methylation in hTERT-BJ, HT1080, 293T, and HeLa cells (Fig. 6H and data not shown). Knockdown of PRMT1 had no effect on the level of TRF2 expression (Fig. 6H). Methylation of R17 was not affected by depletion of PRMT6 (19), also a type I enzyme (Fig. 6I). These results suggest that PRMT1 is the main enzyme responsible for methylating TRF2 R17 in vivo.
Depletion of PRMT1 promotes the formation of telomere doublets, leading to induction of growth arrest in normal human cells.
We have shown that PRMT1 interacts with TRF2 and is responsible for methylating TRF2 R17 in vivo, suggesting that PRMT1 may play a role in telomere maintenance. To address this question, we stably knocked down PRMT1 in a number of normal human fibroblast cell lines (hTERT-BJ, IMR90, MRC5, and GM08399) (Fig. 7A). As a control, we also generated hTERT-BJ cell lines stably expressing shPRMT5, shPRMT6, or the vector alone (Fig. 7B and C). Compared to results with cells expressing the vector alone, knockdown of PRMT5 or PRMT6 had no impact on cell proliferation (Fig. 7D). In contrast, we found that depletion of PRMT1 resulted in growth arrest within 3 to 5 days after infection in hTERT-BJ, IMR90, MRC5, and GM08399 cells (Fig. 7D to F). These results suggest that PRMT1 is essential for cell proliferation in normal human cells.
FIG. 7.
Depletion of PRMT1 results in induction of growth arrest in normal human fibroblast cells. (A) Western analysis of PRMT1 expression. Whole-cell extracts made from 200,000 cells were used, and immunoblotting was performed with anti-PRMT1 antibody. The γ-tubulin blot was used as a loading control. (B) Western analysis of PRMT5 expression. Immunoblotting was conducted with anti-PRMT5 antibody, and the γ-tubulin blot was used as a loading control. (C) Western analysis of PRMT6 expression. Immunoblotting was conducted with anti-PRMT6 and anti-γ-tubulin antibodies, the latter serving as a loading control. (D) Growth curve of hTERT-BJ cells infected with indicated virus. hTERT-BJ cells expressing shPRMT1 undergo growth arrest within 3 days of infection, and seeding after infection was associated with massive cell loss. To overcome this problem, 4.8 × 104 cells were seeded in triplicate on day −4. Cells were infected five times with indicated virus over 12-h intervals between day −3 and day −1. Selection with puromycin (2 μg/ml) started on day 0, and cells were maintained in the selection medium for 9 days. Standard deviations derived from three independent experiments are indicated. (E) Growth curve of normal primary fibroblast MRC5 cells infected with indicated virus. MRC5 cells expressing shPRMT1 undergo growth arrest. Seeding, retroviral infection, and selection of MRC5 were done as described for panel D. Standard deviations derived from three independent experiments are indicated. (F) Loss of PRMT1 results in growth arrest in normal human cells. Live cell images show hTERT-BJ, IMR90, or GM08399 cells infected with retrovirus expressing shPRMT1 or the vector pRS alone. Images were taken 5 days after infection. (G) Genomic blots of telomeric restriction fragments from hTERT-BJ, IMR90, and MRC5 cells expressing either shPRMT1 or pRS. About 3 μg of RsaI/HinfI-digested genomic DNA from each sample was used for gel electrophoresis. The DNA molecular size markers are shown to the left of the blots.
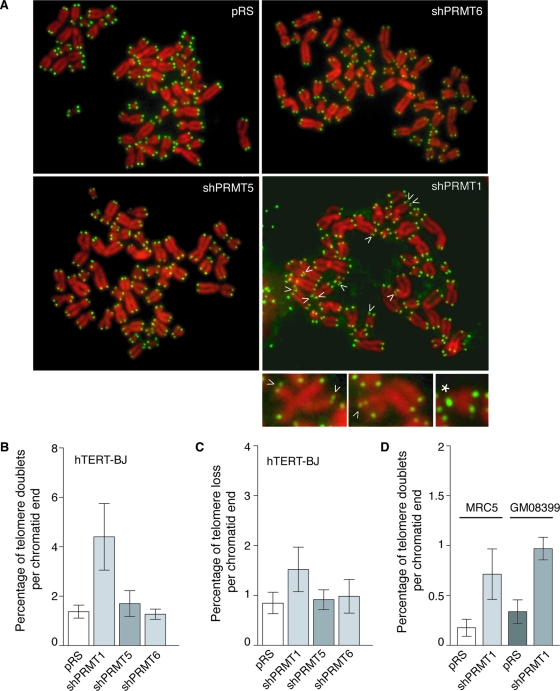
No substantial change in telomere length was detected in hTERT-BJ, IMR90, and MRC5 cells expressing shPRMT1 (Fig. 7G). However, FISH analysis revealed that loss of PRMT1 induced the formation of telomere doublets (Fig. 8). Compared to hTERT-BJ cells expressing the vector alone, we observed about a fourfold increase (P = 0.004) in telomere doublets in hTERT-BJ cells expressing shPRMT1 (Fig. 8A and B). This increase was not observed in hTERT-BJ cells expressing shPRMT5 or shPRMT6 (Fig. 8A and B). Induction of telomere doublets resulting from depletion of PRMT1 was also detected in MRC5 and GM08399 cells (Fig. 8D). Compared to MRC5 cells expressing the vector alone, MRC5 cells expressing shPRMT1 exhibited a fourfold increase (P = 0.023) in telomere doublets (Fig. 8D). Similarly, GM08399 cells expressing shPRMT1 showed a more than threefold increase (P = 0.002) in telomere doublets compared to results for the vector-expressing cells (Fig. 8D). A 1.8-fold increase (P = 0.034) in telomere loss was also observed in hTERT-BJ cells expressing shPRMT1 compared to results with the vector alone (Fig. 8C). Taken together, these results suggest that PRMT1 is required for maintaining telomere stability in normal human cells.
FIG. 8.
Depletion of PRMT1 in human normal cells results in telomere instability, promoting the formation of telomere doublets. (A) Analysis of metaphase chromosomes from hTERT-BJ cells infected with indicated virus. Chromosomes were stained with DAPI and false colored in red. Telomeric DNA was detected by FISH using a FITC-conjugated (CCCTAA)3-containing PNA probe (green). Arrows indicate telomere doublets, and the asterisk represents telomere loss. Enlarged images of chromosomes with telomere doublets are shown at the bottom. (B and C) Quantification of telomere doublets and telomere loss in hTERT-BJ cells stably infected with the indicated virus. For each cell line, a total of more than 2,700 chromosomes from at least 60 metaphase cells were scored. Standard deviations derived from at least three independent experiments are indicated. (D) Quantification of telomere doublets in MRC5 and GM08399 cells stably infected with the indicated virus. For each cell line, a total of more than 1,600 chromosomes from 40 to 44 metaphase cells were scored. Standard deviations derived from at least three independent experiments are indicated.
Depletion of PRMT1 alters telomeric association of TRF2, promoting telomere shortening in cancer cells.
To examine the effect of depletion of PRMT1 in telomere maintenance in transformed cells, several transformed cell lines (HT1080, 293T, and GM637) were infected with retrovirus expressing either shPRMT1 or the vector alone, generating six stable cell lines (HT1080-pRS, HT1080-shPRMT1, 293T-pRS, 293T-shPRMT1, GM637-pRS, and GM637-shPRMT1) (Fig. 9A). Knockdown of PRMT1 had no effect on cell proliferation in the transformed cells (Fig. 9B to D), suggesting that PRMT1 may not be essential for cell proliferation in cancer cells.
FIG. 9.
Depletion of PRMT1 does not affect cell proliferation in human cancer cells. (A) Western analysis of PRMT1 expression in HT1080, 293T, and GM637 cells expressing shPRMT1 or pRS. The γ-tubulin blot was used as a loading control. (B) Growth curve of 293T cells expressing shPRMT1 or pRS. The number of PDs was plotted against days in culture. (C) Growth curve of HT1080 cells expressing shPRMT1 or pRS. The number of PDs was plotted against days in culture. (D) Growth curve of GM637 cells expressing shPRMT1 or pRS. The number of PDs was plotted against days in culture.
To assess telomere length dynamics, cells stably expressing either shPRMT1 or the vector alone were subjected to long-term culturing. While HT1080 cells expressing the vector alone experienced little change in their telomere length for 84 population doublings (PDs) (Fig. 10A and B), we detected a decline in telomere length at a rate of about 18 bp/PD in HT1080 cells expressing shPRMT1 (Fig. 10A and B). This decrease in telomere length resulting from PRMT1 knockdown appears to be specific. since we did not observe telomere shortening in HT1080 cells expressing shPRMT6 (data not shown). Furthermore, PRMT1 knockdown in HT1080 cells had no impact on telomerase activity (Fig. 10C) and 3′ G-strand overhang (data not shown). The negative impact of PRMT1 depletion on telomere length maintenance was also observed in GM637 and 293T cells (Fig. 10D and E and data not shown). While the vector-expressing GM637 and 293T cells experienced growth in their telomere length during the long-term culturing (Fig. 10D and E), PRMT1 knockdown suppressed their telomere lengthening (Fig. 10D and E). The rebound in telomere length in shPRMT1-expressing GM637 cells after 30 PDs was likely due to the loss of PRMT1 knockdown observed in these cells at later passages (Fig. 9A). Taken together, these results suggest that PRMT1 is a positive regulator of telomere length maintenance.
FIG. 10.
PRMT1 regulates telomere length maintenance in human cancer cells. (A) Genomic blot of telomere restriction fragments from HT1080 cells expressing shPRMT1 or the vector pRS alone after indicated numbers of PDs. About 5 μg of RsaI/HinfI-digested genomic DNA from each sample was used for gel electrophoresis. The DNA molecular size markers are shown to the left of the blot. The bottom panel, taken from an ethidium bromide-stained agarose gel, is used as a loading control. (B) Average telomere length of HT1080 expressing shPRMT1 or pRS was plotted against PDs. (C) Depletion of endogenous PRMT1 has no impact on telomerase activity. Ten thousand HT1080 cells expressing either shPRMT1 or pRS were used to measure telomerase activity. TSR8 was used as a positive control, whereas 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate buffer was used as a negative control. (D) Average telomere length of GM637 cells expressing shPRMT1 or pRS was plotted against PDs. (E) Average telomere length of 293T cells expressing shPRMT1 or pRS was plotted against PDs.
Overexpression of TRF2 has been shown to promote telomere shortening (2, 26, 37, 48). We decided to investigate whether PRMT1 may modulate TRF2 association with telomeric DNA. ChIP using anti-TRF2 antibody was performed with HT1080 cells expressing shPRMT1 or the vector alone. ChIP analysis showed that cells expressing shPRMT1 exhibited a 65% increase (P = 0.0125) in TRF2 association with telomeric DNA compared to cells expressing the vector alone (Fig. 11A and B). Depletion of PRMT1 had no impact on telomeric association of TRF1 and TIN2, two other shelterin proteins (Fig. 11C to E). Furthermore, PRMT1 knockdown had no effect on TRF2, TRF1, and TIN2 expression in cells (Fig. 6H and data not shown). Altogether, these results suggest that PRMT1 negatively regulates TRF2 binding to telomeric DNA.
FIG. 11.
(A) Dot blots of ChIPs. ChIPs were performed with either anti-TRF2 or anti-IgG antibody in cell extracts from HT1080 cells expressing shPRMT1 or the vector pRS alone. Precipitated DNA was analyzed for the presence of TTAGGG repeats and Alu repeats by Southern blotting. (B) Quantification of anti-TRF2 ChIPs. The signals were quantified by ImageQuant analysis. The percentage of precipitated DNA was calculated relative to the input signals. Standard deviations derived from three independent experiments are indicated. (C) Dot blots of anti-TRF1 and anti-TIN2 ChIPs from HT1080 cells expressing shPRMT1 or the vector pRS alone. Precipitated DNA was analyzed for the presence of TTAGGG repeats and Alu repeats by Southern blotting. (D) Quantification of anti-TRF1 ChIPs from dot blots in panel C. Standard deviations derived from three independent experiments are indicated. (E) Quantification of anti-TIN2 ChIPs from dot blots in panel C. Standard deviations derived from three independent experiments are indicated.
DISCUSSION
In this report, we have shown that arginines in the basic domain of TRF2 undergo methylation both in vitro and in vivo. Amino acid changes of arginines to lysines in the basic domain of TRF2 alter telomeric association of the shelterin protein TIN2, promoting the formation of TIFs and telomere doublets (more than one telomeric signal at a single chromatid end), indicative of dysfunctional telomeres. We have further revealed that PRMT1 interacts with TRF2 and is the main enzyme responsible for methylating TRF2. We have shown that depletion of PRMT1 in transformed cells leads to telomere shortening whereas removal of PRMT1 in normal human cells induces the formation of telomere doublets. Our results suggest that arginine methylation by PRMT1 plays a crucial role in regulating telomere length and stability, perhaps in part through TRF2 methylation.
We have shown that depletion of PRMT1 or overexpression of TRF2 carrying amino acid changes of arginines to lysines in the basic domain (TRF2-RK, TRF2-RK1-4, or TRF2-RK5-8) promotes the formation of telomere doublets. Telomere doublets have been seen at a very low frequency in human cells (43). An increase (∼3-fold) in the formation of telomere doublets has been shown to be associated with induction of growth arrest and cellular senescence (55). Consistent with this previous finding, we also observed an induction of growth arrest and cellular senescence in normal human cells depleted for PRMT1 or overexpressing TRF2-RK, TRF2-RK1-4, or TRF2-RK5-8, and these cells display about a three- to fourfold increase in telomere doublets.
Telomere doublets have been suggested to result from a DNA replication defect (55) (T. de Lange, personal communication). Consistent with this view, we find that Nbs1, a protein known to be involved in DNA recombination and repair (16, 22), is not required for the formation of TRF2-RK-induced telomere doublets. Overexpression of TRF2 lacking the basic domain (TRF2ΔB) has been shown to induce drastic telomere loss (58), which is dependent upon Nbs1-mediated DNA recombination at telomeres (58). Taken together, our results suggest that arginine methylation may play an important role in telomere replication and that the basic domain of TRF2 may be involved in multiple mechanisms to ensure telomere integrity.
PRMT1 is known to methylate multiple arginines in the GAR motif (8, 9, 13). Through mass spectrometry analysis, we have shown that arginines 17 and 18 are methylated in vivo. We have shown that lysine substitutions of four arginines at positions 13 to 21 or at positions 25 to 30 abrogate TRF2 function in vivo. Furthermore, we have shown that lysine substitutions of either eight or four arginines abolish methylation of full-length TRF2 by PRMT1 in vitro. Our data suggest that PRMT1 may methylate multiple arginines in the basic domain of TRF2, including arginines at positions 25 to 30 in vivo. Using specific anti-2meR17 antibody, we have estimated that about 1 to 5% of endogenous TRF2 in HeLa cells contains dimethylated R17, suggesting a low abundance of methylated TRF2 in cells. Although whether all arginines in the basic domain are methylated equally in vivo requires further investigation, the low abundance of methylated TRF2 may in part account for our failure to detect arginine methylation at positions 25 to 30 in vivo.
PRMT1 is involved in a wide range of cellular processes (4, 5). Null mutations in PRMT1 leads to early embryonic lethality in mice shortly after implantation, whereas mouse embryonic stem cells lacking PRMT1 are viable (42). We have shown that knockdown of PRMT1 does not affect cell proliferation in multiple cancer cell lines (HT1080, 293T, GM637, and HeLa). However, loss of PRMT1 results in growth arrest and cellular senescence in four normal human fibroblasts tested (hTERT-BJ, IMR90, MRC5, and GM08399). These results suggest that PRMT1 may be dispensable for cell proliferation in transformed cells but essential in normal human cells. However, we cannot rule out the possibility that PRMT1 depletion is less severe in transformed cells than that in normal cells. The residual level of PRMT1 may also account for the observed discrepancy in growth phenotype between TRF2 lysine mutants and PRMT1 knockdown in transformed cells. Alternatively, arginines in the basic domain of TRF2 might also have a function that is independent of PRMT1-mediated methylation. Our finding that depletion of PRMT1 results in growth arrest in telomerase-immortalized BJ cells implies that exogenous expression of telomerase in normal human cells is insufficient to bypass the requirement of PRMT1 for cell proliferation.
We have shown that loss of PRMT1 affects both telomere length and stability. Depletion of PRMT1 in normal human cells promotes telomere doublets in a manner indistinguishable from overexpression of TRF2 carrying lysine substitutions of arginines in the basic domain, suggesting that PRMT1 may control telomere stability in part through TRF2 methylation. In human fibrosarcoma HT1080 cells, we find that knockdown of PRMT1 leads to telomere shortening. Depletion of PRMT1 also suppresses telomere lengthening associated with long-term culturing of 293T and GM637 cells. Although loss of PRMT1 does not appear to affect telomerase activity in HT1080 cells, we find that telomeric association of TRF2 is upregulated in PRMT1-depleted cells. Excess TRF2 has been shown to lead to telomere shortening (2, 26, 37, 48). Our results suggest that PRMT1-mediated methylation may control TRF2 association with telomeres, which in turn modulates telomere length maintenance.
We have shown that about 1 to 5% of endogenous TRF2 is methylated. How such a small fraction of methylated TRF2 contributes to regulation of telomere function is unknown. TRF2 has been shown to interact with many nonshelterin proteins important for maintaining telomere length and stability, including Mre11/Rad50/Nbs1 (65), XPF/ERCC1 (66), Apollo (20, 30, 55), WRN (39), and FEN1 (36). A small fraction (about 1 to 5%) of TRF2 has been shown to interact with Mre11/Rad50/Nbs1 (65) and XPF/ERCC1 (66), raising the possibility that TRF2 methylation by PRMT1 might be important for its interaction with nonshelterin proteins.
PRMT1 has been shown to be associated with human telomeres (17). We have identified that TRF2, a shelterin protein, is a substrate of PRMT1. Our finding that PRMT1 is required for telomere length maintenance and stability is consistent with the fact that TRF2 plays a multiple role in regulating telomere length maintenance and telomere protection (18). Our results suggest that PRMT1 may control telomere length and stability, perhaps in part through TRF2 methylation. Identification of additional PRMT1 substrates at telomeres may further our understanding of its role in telomere maintenance.
Acknowledgments
We are grateful to Titia de Lange for various reagents. We thank Stéphane Richard for communicating unpublished data and providing the expression construct GST-PRMT1, as well as anti-PRMT1 and -Sam68 antibodies. We thank Rulin Zhang from WEMB Biochem Inc. and Kristina Jurcic from the University of Western Ontario for performing the mass spectrometry analysis. We thank John R. Walker and members of the Zhu laboratory for their critical comments.
X.D.Z. is a Canadian Institutes of Health Research New Investigator. This work was supported by the Ontario Early Researcher Award program and funding from Canadian Institutes of Health Research to X.-D.Z.
Footnotes
Published ahead of print on 13 July 2009.
REFERENCES
- 1.An, W., J. Kim, and R. G. Roeder. 2004. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117735-748. [DOI] [PubMed] [Google Scholar]
- 2.Ancelin, K., M. Brunori, S. Bauwens, C. E. Koering, C. Brun, M. Ricoul, J. P. Pommier, L. Sabatier, and E. Gilson. 2002. Targeting assay to study the cis functions of human telomeric proteins: evidence for inhibition of telomerase by TRF1 and for activation of telomere degradation by TRF2. Mol. Cell. Biol. 223474-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atanasiu, C., Z. Deng, A. Wiedmer, J. Norseen, and P. M. Lieberman. 2006. ORC binding to TRF2 stimulates OriP replication. EMBO Rep. 7716-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bedford, M. T. 2007. Arginine methylation at a glance. J. Cell Sci. 1204243-4246. [DOI] [PubMed] [Google Scholar]
- 5.Bedford, M. T., and S. Richard. 2005. Arginine methylation an emerging regulator of protein function. Mol. Cell 18263-272. [DOI] [PubMed] [Google Scholar]
- 6.Boisvert, F. M., C. A. Chenard, and S. Richard. 2005. Protein interfaces in signaling regulated by arginine methylation. Sci. STKE 2005re2. [DOI] [PubMed] [Google Scholar]
- 7.Boisvert, F. M., J. Cote, M. C. Boulanger, and S. Richard. 2003. A proteomic analysis of arginine-methylated protein complexes. Mol. Cell Proteomics 21319-1330. [DOI] [PubMed] [Google Scholar]
- 8.Boisvert, F. M., U. Dery, J. Y. Masson, and S. Richard. 2005. Arginine methylation of MRE11 by PRMT1 is required for DNA damage checkpoint control. Genes Dev. 19671-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boisvert, F. M., A. Rhie, S. Richard, and A. J. Doherty. 2005. The GAR motif of 53BP1 is arginine methylated by PRMT1 and is necessary for 53BP1 DNA binding activity. Cell Cycle 41834-1841. [DOI] [PubMed] [Google Scholar]
- 10.Celli, G. B., and T. de Lange. 2005. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat. Cell Biol. 7712-718. [DOI] [PubMed] [Google Scholar]
- 11.Chen, D., H. Ma, H. Hong, S. S. Koh, S. M. Huang, B. T. Schurter, D. W. Aswad, and M. R. Stallcup. 1999. Regulation of transcription by a protein methyltransferase. Science 2842174-2177. [DOI] [PubMed] [Google Scholar]
- 12.Cook, J. R., J. H. Lee, Z. H. Yang, C. D. Krause, N. Herth, R. Hoffmann, and S. Pestka. 2006. FBXO11/PRMT9, a new protein arginine methyltransferase, symmetrically dimethylates arginine residues. Biochem. Biophys. Res. Commun. 342472-481. [DOI] [PubMed] [Google Scholar]
- 13.Cote, J., F. M. Boisvert, M. C. Boulanger, M. T. Bedford, and S. Richard. 2003. Sam68 RNA binding protein is an in vivo substrate for protein arginine N-methyltransferase 1. Mol. Biol. Cell 14274-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crabbe, L., R. E. Verdun, C. I. Haggblom, and J. Karlseder. 2004. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science 3061951-1953. [DOI] [PubMed] [Google Scholar]
- 15.d'Adda di Fagagna, F., P. M. Reaper, L. Clay-Farrace, H. Fiegler, P. Carr, T. Von Zglinicki, G. Saretzki, N. P. Carter, and S. P. Jackson. 2003. A DNA damage checkpoint response in telomere-initiated senescence. Nature 426194-198. [DOI] [PubMed] [Google Scholar]
- 16.D'Amours, D., and S. P. Jackson. 2002. The Mre11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat. Rev. Mol. Cell Biol. 3317-327. [DOI] [PubMed] [Google Scholar]
- 17.Dejardin, J., and R. E. Kingston. 2009. Purification of proteins associated with specific genomic loci. Cell 136175-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Lange, T. 2005. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 192100-2110. [DOI] [PubMed] [Google Scholar]
- 19.Frankel, A., N. Yadav, J. Lee, T. L. Branscombe, S. Clarke, and M. T. Bedford. 2002. The novel human protein arginine N-methyltransferase PRMT6 is a nuclear enzyme displaying unique substrate specificity. J. Biol. Chem. 2773537-3543. [DOI] [PubMed] [Google Scholar]
- 20.Freibaum, B. D., and C. M. Counter. 2006. hSnm1B is a novel telomere-associated protein. J. Biol. Chem. 28115033-15036. [DOI] [PubMed] [Google Scholar]
- 21.Gros, L., C. Delaporte, S. Frey, J. Decesse, B. R. de Saint-Vincent, L. Cavarec, A. Dubart, A. V. Gudkov, and A. Jacquemin-Sablon. 2003. Identification of new drug sensitivity genes using genetic suppressor elements: protein arginine N-methyltransferase mediates cell sensitivity to DNA-damaging agents. Cancer Res. 63164-171. [PubMed] [Google Scholar]
- 22.Haber, J. E. 1998. The many interfaces of Mre11. Cell 95583-586. [DOI] [PubMed] [Google Scholar]
- 23.Hemann, M. T., and C. W. Greider. 1999. G-strand overhangs on telomeres in telomerase-deficient mouse cells. Nucleic Acids Res. 273964-3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herrmann, F., M. Bossert, A. Schwander, E. Akgun, and F. O. Fackelmayer. 2004. Arginine methylation of scaffold attachment factor A by heterogeneous nuclear ribonucleoprotein particle-associated PRMT1. J. Biol. Chem. 27948774-48779. [DOI] [PubMed] [Google Scholar]
- 25.Karlseder, J., D. Broccoli, Y. Dai, S. Hardy, and T. de Lange. 1999. p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science 2831321-1325. [DOI] [PubMed] [Google Scholar]
- 26.Karlseder, J., A. Smogorzewska, and T. de Lange. 2002. Senescence induced by altered telomere state, not telomere loss. Science 2952446-2449. [DOI] [PubMed] [Google Scholar]
- 27.Kim, S. H., P. Kaminker, and J. Campisi. 1999. TIN2, a new regulator of telomere length in human cells. Nat. Genet. 23405-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lansdorp, P. M., N. P. Verwoerd, F. M. van de Rijke, V. Dragowska, M. T. Little, R. W. Dirks, A. K. Raap, and H. J. Tanke. 1996. Heterogeneity in telomere length of human chromosomes. Hum. Mol. Genet. 5685-691. [DOI] [PubMed] [Google Scholar]
- 29.Lee, J., J. Sayegh, J. Daniel, S. Clarke, and M. T. Bedford. 2005. PRMT8, a new membrane-bound tissue-specific member of the protein arginine methyltransferase family. J. Biol. Chem. 28032890-32896. [DOI] [PubMed] [Google Scholar]
- 30.Lenain, C., S. Bauwens, S. Amiard, M. Brunori, M. J. Giraud-Panis, and E. Gilson. 2006. The Apollo 5′ exonuclease functions together with TRF2 to protect telomeres from DNA repair. Curr. Biol. 161303-1310. [DOI] [PubMed] [Google Scholar]
- 31.Li, B., and T. de Lange. 2003. Rap1 affects the length and heterogeneity of human telomeres. Mol. Biol. Cell 145060-5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin, W. J., J. D. Gary, M. C. Yang, S. Clarke, and H. R. Herschman. 1996. The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J. Biol. Chem. 27115034-15044. [DOI] [PubMed] [Google Scholar]
- 33.Loayza, D., and T. De Lange. 2003. POT1 as a terminal transducer of TRF1 telomere length control. Nature 4231013-1018. [DOI] [PubMed] [Google Scholar]
- 34.McBride, A. E., and P. A. Silver. 2001. State of the arg: protein methylation at arginine comes of age. Cell 1065-8. [DOI] [PubMed] [Google Scholar]
- 35.Miranda, T. B., M. Miranda, A. Frankel, and S. Clarke. 2004. PRMT7 is a member of the protein arginine methyltransferase family with a distinct substrate specificity. J. Biol. Chem. 27922902-22907. [DOI] [PubMed] [Google Scholar]
- 36.Muftuoglu, M., H. K. Wong, S. Z. Imam, D. M. Wilson III, V. A. Bohr, and P. L. Opresko. 2006. Telomere repeat binding factor 2 interacts with base excision repair proteins and stimulates DNA synthesis by DNA polymerase beta. Cancer Res. 66113-124. [DOI] [PubMed] [Google Scholar]
- 37.Munoz, P., R. Blanco, J. M. Flores, and M. A. Blasco. 2005. XPF nuclease-dependent telomere loss and increased DNA damage in mice overexpressing TRF2 result in premature aging and cancer. Nat. Genet. 371063-1071. [DOI] [PubMed] [Google Scholar]
- 38.Najbauer, J., B. A. Johnson, A. L. Young, and D. W. Aswad. 1993. Peptides with sequences similar to glycine, arginine-rich motifs in proteins interacting with RNA are efficiently recognized by methyltransferase(s) modifying arginine in numerous proteins. J. Biol. Chem. 26810501-10509. [PubMed] [Google Scholar]
- 39.Opresko, P. L., C. Von Kobbe, J. P. Laine, J. Harrigan, I. D. Hickson, and V. A. Bohr. 2002. Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J. Biol. Chem. 27741110-41119. [DOI] [PubMed] [Google Scholar]
- 40.Pal, S., and S. Sif. 2007. Interplay between chromatin remodelers and protein arginine methyltransferases. J. Cell Physiol. 213306-315. [DOI] [PubMed] [Google Scholar]
- 41.Palm, W., and T. de Lange. 2008. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 42301-334. [DOI] [PubMed] [Google Scholar]
- 42.Pawlak, M. R., C. A. Scherer, J. Chen, M. J. Roshon, and H. E. Ruley. 2000. Arginine N-methyltransferase 1 is required for early postimplantation mouse development, but cells deficient in the enzyme are viable. Mol. Cell. Biol. 204859-4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Philippe, C., P. Coullin, and A. Bernheim. 1999. Double telomeric signals on single chromatids revealed by FISH and PRINS. Ann. Genet. 42202-209. [PubMed] [Google Scholar]
- 44.Pollack, B. P., S. V. Kotenko, W. He, L. S. Izotova, B. L. Barnoski, and S. Pestka. 1999. The human homologue of the yeast proteins Skb1 and Hsl7p interacts with Jak kinases and contains protein methyltransferase activity. J. Biol. Chem. 27431531-31542. [DOI] [PubMed] [Google Scholar]
- 45.Saharia, A., L. Guittat, S. Crocker, A. Lim, M. Steffen, S. Kulkarni, and S. A. Stewart. 2008. Flap endonuclease 1 contributes to telomere stability. Curr. Biol. 18496-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sheffield, P., S. Garrard, and Z. Derewenda. 1999. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr. Purif. 1534-39. [DOI] [PubMed] [Google Scholar]
- 47.Smith, W. A., B. T. Schurter, F. Wong-Staal, and M. David. 2004. Arginine methylation of RNA helicase a determines its subcellular localization. J. Biol. Chem. 27922795-22798. [DOI] [PubMed] [Google Scholar]
- 48.Smogorzewska, A., B. van Steensel, A. Bianchi, S. Oelmann, M. R. Schaefer, G. Schnapp, and T. de Lange. 2000. Control of human telomere length by TRF1 and TRF2. Mol. Cell. Biol. 201659-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stagno D'Alcontres, M., A. Mendez-Bermudez, J. L. Foxon, N. J. Royle, and P. Salomoni. 2007. Lack of TRF2 in ALT cells causes PML-dependent p53 activation and loss of telomeric DNA. J. Cell Biol. 179855-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takai, H., A. Smogorzewska, and T. de Lange. 2003. DNA damage foci at dysfunctional telomeres. Curr. Biol. 131549-1556. [DOI] [PubMed] [Google Scholar]
- 51.Tan, C. P., and S. Nakielny. 2006. Control of the DNA methylation system component MBD2 by protein arginine methylation. Mol. Cell. Biol. 267224-7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang, J., A. Frankel, R. J. Cook, S. Kim, W. K. Paik, K. R. Williams, S. Clarke, and H. R. Herschman. 2000. PRMT1 is the predominant type I protein arginine methyltransferase in mammalian cells. J. Biol. Chem. 2757723-7730. [DOI] [PubMed] [Google Scholar]
- 53.Tang, J., J. D. Gary, S. Clarke, and H. R. Herschman. 1998. PRMT 3, a type I protein arginine N-methyltransferase that differs from PRMT1 in its oligomerization, subcellular localization, substrate specificity, and regulation. J. Biol. Chem. 27316935-16945. [DOI] [PubMed] [Google Scholar]
- 54.Tang, J., P. N. Kao, and H. R. Herschman. 2000. Protein-arginine methyltransferase I, the predominant protein-arginine methyltransferase in cells, interacts with and is regulated by interleukin enhancer-binding factor 3. J. Biol. Chem. 27519866-19876. [DOI] [PubMed] [Google Scholar]
- 55.van Overbeek, M., and T. de Lange. 2006. Apollo, an artemis-related nuclease, interacts with TRF2 and protects human telomeres in S phase. Curr. Biol. 161295-1302. [DOI] [PubMed] [Google Scholar]
- 56.van Steensel, B., A. Smogorzewska, and T. de Lange. 1998. TRF2 protects human telomeres from end-to-end fusions. Cell 92401-413. [DOI] [PubMed] [Google Scholar]
- 57.Wang, H., Z. Q. Huang, L. Xia, Q. Feng, H. Erdjument-Bromage, B. D. Strahl, S. D. Briggs, C. D. Allis, J. Wong, P. Tempst, and Y. Zhang. 2001. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science 293853-857. [DOI] [PubMed] [Google Scholar]
- 58.Wang, R. C., A. Smogorzewska, and T. de Lange. 2004. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell 119355-368. [DOI] [PubMed] [Google Scholar]
- 59.Wu, Y., T. R. Mitchell, and X. D. Zhu. 2008. Human XPF controls TRF2 and telomere length maintenance through distinctive mechanisms. Mech. Ageing Dev. 129602-610. [DOI] [PubMed] [Google Scholar]
- 60.Wu, Y., S. Xiao, and X. D. Zhu. 2007. MRE11-RAD50-NBS1 and ATM function as co-mediators of TRF1 in telomere length control. Nat. Struct. Mol. Biol. 14832-840. [DOI] [PubMed] [Google Scholar]
- 61.Wu, Y., N. J. Zacal, A. J. Rainbow, and X. D. Zhu. 2007. XPF with mutations in its conserved nuclease domain is defective in DNA repair but functions in TRF2-mediated telomere shortening. DNA Repair (Amsterdam) 6157-166. [DOI] [PubMed] [Google Scholar]
- 62.Xie, B., C. F. Invernizzi, S. Richard, and M. A. Wainberg. 2007. Arginine methylation of the human immunodeficiency virus type 1 Tat protein by PRMT6 negatively affects Tat interactions with both cyclin T1 and the Tat transactivation region. J. Virol. 814226-4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu, L., and E. H. Blackburn. 2004. Human Rif1 protein binds aberrant telomeres and aligns along anaphase midzone microtubules. J. Cell Biol. 167819-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang, R., L. Barker, D. Pinchev, J. Marshall, M. Rasamoelisolo, C. Smith, P. Kupchak, I. Kireeva, L. Ingratta, and G. Jackowski. 2004. Mining biomarkers in human sera using proteomic tools. Proteomics 4244-256. [DOI] [PubMed] [Google Scholar]
- 65.Zhu, X. D., B. Kuster, M. Mann, J. H. Petrini, and T. Lange. 2000. Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat. Genet. 25347-352. [DOI] [PubMed] [Google Scholar]
- 66.Zhu, X. D., L. Niedernhofer, B. Kuster, M. Mann, J. H. Hoeijmakers, and T. de Lange. 2003. ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol. Cell 121489-1498. [DOI] [PubMed] [Google Scholar]