Abstract
Histone methylation plays key roles in regulating chromatin structure and function. The recent identification of enzymes that antagonize or remove histone methylation offers new opportunities to appreciate histone methylation plasticity in the regulation of epigenetic pathways. Peptidylarginine deiminase 4 (PADI4; also known as PAD4) was the first enzyme shown to antagonize histone methylation. PADI4 functions as a histone deiminase converting a methylarginine residue to citrulline at specific sites on the tails of histones H3 and H4. This activity is linked to repression of the estrogen-regulated pS2 promoter. Very little is known as to how PADI4 silences gene expression. We show here that PADI4 associates with the histone deacetylase 1 (HDAC1). Kinetic chromatin immunoprecipitation assays revealed that PADI4 and HDAC1, and the corresponding activities, associate cyclically and coordinately with the pS2 promoter during repression phases. Knockdown of HDAC1 led to decreased H3 citrullination, concomitantly with increased histone arginine methylation. In cells with a reduced HDAC1 and a slightly decreased PADI4 level, these effects were more pronounced. Our data thus suggest that PADI4 and HDAC1 collaborate to generate a repressive chromatin environment on the pS2 promoter. These findings further substantiate the “transcriptional clock” concept, highlighting the dynamic connection between deimination and deacetylation of histones.
Until recently, it was unclear whether enzymes capable of antagonizing histone methylation existed. However, recent studies have revealed a growing number of lysine demethylases that can reverse histone lysine methylation, such as LSD1/KDM1 and the JmjC-domain-containing proteins (16, 21, 31, 37, 43, 45, 47).
In addition to the methylation of lysine, histones can also be methylated on arginine (6, 34). Recent work has identified the JMJD6 protein as an H3R2 and H4R3 demethylase (5). An alternative pathway for the reversal of arginine methylation has been identified in mammals. In this pathway, a methyl group is removed from a methylarginine residue by conversion of this residue to citrulline. The reaction is termed deimination because the methyl group is removed along with the imine group of arginine (7, 42). The enzyme that catalyzes this reaction is peptidylarginine deiminase 4 (PADI4; also known as PAD4) (7, 42). PADI4 has a relatively broad substrate specificity, since the enzyme can deiminate multiple arginine sites on histones H3 (R2, R8, R17, and R26) and H4 (R3) (42). PADI4 is a Ca2+-dependent enzyme (1). Functionally, the induction of PADI4-mediated deimination has been best studied as part of the estrogen signaling pathway, particularly in the context of the estrogen-regulated pS2 promoter.
Chromatin immunoprecipitation (ChIP)-based kinetic analyses performed on human breast cancer cell lines have shown that in the presence of estrogen (E2), pS2 expression is controlled by the estrogen receptor alpha (ERα), which cycles on its promoter. Not only does the receptor cycle, it also follows a sequence of cofactor recruitment (20, 30). For example, during transcriptional activation by the estrogen receptor, arginine methylation of histone H3 appears transient and cyclic (20). After the initial hormone-induced transcriptional activation phase, PADI4 is recruited to the promoter region of pS2, where its presence correlates with loss of arginine methylation, acquisition of citrullinated histones, and disengagement of RNA polymerase II (Pol II) from the gene (7, 42). Thus, methylation of arginine residues followed by deimination by PADI4 seems to participate in the normal cyclic on-and-off regulation of pS2 transcription. Although these studies have begun to shed light on how PADI4-mediated histone deimination silences gene expression, much remains to be discovered in order to understand the mechanistic basis of this process.
In the present study we have sought to better understand how PADI4 represses transcription. Our starting point was an effort to identify proteins that associate with PADI4. We report that PADI4 interacts with histone deacetylase 1 (HDAC1) both in vitro and in vivo. We provide evidence that PADI4 and HDAC1, and their corresponding enzymatic activities, associate cyclically with the pS2 promoter and are present simultaneously on this promoter in the presence of estradiol. Finally, RNA interference (RNAi) experiments suggest a coordinated action of the PADI4 and HDAC1 enzymes on the pS2 promoter.
MATERIALS AND METHODS
Expression plasmids.
We cloned full-length PADI4 into pcDNA3.1-HA and PADI4 fragments into the vector pGex (Pharmacia) using appropriate sets of primers. We also cloned various domains of HDAC1 into the pGex vector by PCR using appropriate primers. The following plasmids were used: pGex PADI4 full-length (23), pcDNA3 Flag RbBP5 (Addgene), pcDNA3 HDAC1-Flag (3), and pING14A-HDAC1 (10).
GST fusion, in vitro translation, and pulldown assays.
We expressed glutathione S-transferase (GST) and GST fusion proteins in Escherichia coli Top10 or BL21 using the pGex (Pharmacia) vector system. Bacteria were disrupted by using an ultrasonicator (Bioruptor; Diagenode), and proteins from crude bacterial lysates were purified using the glutathione-Sepharose 4B (Pharmacia) used according to the manufacturer's instructions. We used the TNT system (Promega) to carry out in vitro transcription and translation. The PADI4, HDAC1, and control RbBP5 genes were in vitro transcribed and/or translated from pcDNA3.1-HA PADI4, pcDNA3 HDAC1-Flag, and pcDNA3 Flag RbBP5, respectively. The GST pulldown experiments were performed as described previously (40).
Cell culture and transient transfections.
293T cells were maintained in Dulbecco modified Eagle medium (DMEM; Sigma) supplemented with 5% fetal calf serum (FCS; BioWest) and antibiotics (Roche) at 37°C under 5% CO2. MDA-MB231 cells stably expressing ER (MDA::ERα) (19) were grown in DMEM complemented with 5% FCS. 17β-estradiol (E2) was purchased from Sigma. Transfections were performed with polyethylene imine (Euromedex) as described previously (9).
Immunoprecipitations and Western blot analyses.
293T cells were transiently transfected in culture dishes (10-cm diameter) with a total of 6 μg of plasmids as described previously (9). Standard procedures were used for coimmunoprecipitations and Western blotting (9). Antihemagglutinin (anti-HA; ab18181; Abcam), anti-Flag (M2; Sigma), and anti-HDAC1 (pAb-053-050; Diagenode) antibodies were used for immunoprecipitations from preparations of transfected 293T cells. For immunoprecipitations of endogenous proteins from preparations of MDA::ERα cells, antibodies were incubated at 4°C with MDA::ERα whole nuclear extracts in IPH buffer (4). Antibodies to HDAC1 were purchased from Diagenode (pAb-053-050). Immunoglobulin G (IgG; ab3745; Abcam), IgG (sc-2027; Santa Cruz), HA (ab9110; Abcam), and RNA Pol II (sc-899; Santa Cruz) were used as control antibodies. PADI4 antibody has been described previously (24).
Histone deimination assay.
The assay was done essentially as described previously (7). 293T cells were transiently transfected with mammalian expression plasmids encoding HA-PADI4 or HDAC1 or with both of these plasmids. Whole-cell extracts were prepared in IPH lysis buffer (4) and incubated overnight at 4°C with the relevant antibodies. Antibody complexes were collected on protein A/G-Sepharose beads, washed, and then tested for histone deiminase activity. The reaction mixture containing 100 mM Tris-HCl (pH 7.6), 5 mM dithiothreitol (DTT), 10 mM CaCl2, and 10 μg of histones (Roche) was incubated at 37°C for 1 h. Reaction products were then resolved by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and Western blotted. Anti-histone H3 (citrulline 17+2+8) antibody (ab5103; Abcam) was used to reveal the presence of deiminated histones. Levels of PADI4 and HDAC1 in the immunoprecipitates were checked by Western blot analysis with anti-HA (ab18184; Abcam) and anti-HDAC1 (pAb-053-050; Diagenode) antibodies, respectively.
ChIPs and Re-ChIPs.
ChIP assays were performed essentially as described previously (20). A total of 5 × 107 MDA::ERα cells were synchronized by 3 days of culture in DMEM-0.5% dextran-charcoal-treated FCS (BioWest) and then treated with 2.5 μM α-amanitin (Sigma) for 2 h, followed by exposure to 10−8 M E2 (Sigma) or ethanol. After cross-linking for 10 min with 1.5% formaldehyde (Sigma) at room temperature, the cells were collected in 5 ml of collection buffer (100 mM Tris-HCl [pH 9.4] and 100 mM DTT) and incubated first on ice for 10 min and then at 30°C for 10 min. The cells were then lysed sequentially, with pipetting and a 5-min centrifugation (at 2,000 × g and 4°C) between steps. The lysis steps were as follows: (i) 5 ml of phosphate-buffered saline; (ii) 5 ml of buffer A (10 mM EDTA, 0.5 mM EGTA, 10 mM HEPES [pH 6.5], and 0.25% Triton X-100); (iii) 5 ml of buffer B (1 mM EDTA, 0.5 mM EGTA, 10 mM HEPES [pH 6.5], and 200 mM NaCl); (iv) lysis for 15 min at room temperature in 2.5 ml of lysis buffer (10 mM EDTA, 50 mM Tris-HCl [pH 8.0], 1% SDS, 0.5% Empigen BB [Sigma]). Then, 500-μl aliquots were sonicated three times by 15 pulses at 50% duty and power 3 (Sonifier cell disruptor; Branson). After centrifugation, 50 μl of the combined supernatants was used as inputs, and the remainder was diluted 2.5-fold in immunoprecipitation buffer (2 mM EDTA, 100 mM NaCl, 20 mM Tris-HCl [pH 8.1], and 0.5%Triton X-100). This fraction was first precleared by incubation for 3 h at 4°C with 15 μg of sheared salmon sperm DNA, and 1 ml of 50% protein A-Sepharose beads (Amersham Pharmacia Biosciences) slurry. A portion (1/20) of the precleared chromatin was then subjected to immunoprecipitation overnight at 4°C under rocking with appropriate antibodies (purchased from Santa Cruz, Upstate Biotechnology, or Abcam). Complexes were recovered after 2 h of incubation at 4°C with 2 μg of sheared salmon sperm DNA and 50 μl of protein A-Sepharose. Precipitates were then serially washed with 300 μl of washing buffers WB I (2 mM EDTA, 20 mM Tris-HCl [pH 8.1], 0.1% SDS, 1% Triton X-100, 150 mM NaCl), WB II (2 mM EDTA, 20 mM Tris-HCl [pH 8.1], and specific combinations of detergent and salt, as indicated in Table 1), and WB III (1 mM EDTA, 10 mM Tris-HCl [pH 8.1], 1% NP-40, 1% deoxycholate, 0.25 M LiCl) and then twice with 1 mM EDTA-10 mM Tris-HCl (pH 8.1). Precipitated complexes were removed from the beads by three sequential 10-min incubations with 50 μl of 1% SDS-0.1 M NaHCO3.
TABLE 1.
Antibodies used and ChIP conditions
| Protein | Provider (catalog no.) or reference | Amt (μg) | Washing conditionsa |
|---|---|---|---|
| ERα | Santa Cruz (sc-543) | 1.0 | WB I, WB II (Det. I, 500 mM NaCl [×1]), WB III |
| Pol II | Santa Cruz (sc-899) | 1.0 | WB I, WB II (Det. II, 500 mM NaCl [×2]) |
| HA | Santa Cruz (sc-805) | 1.0 | WB I, WB II (Det. I, 500 mM NaCl [×1]), WB III |
| H3K14ac | Upstate Biotech (06-911) | 0.5 | WB I, WB II (Det. I, 500 mM NaCl [×1]), WB III |
| H3R17me2 | Upstate Biotech (07-214) | 0.5 | WB I, WB II (Det. II, 500 mM NaCl [×2]) |
| H3R(17+2+8)cit | Abcam (ab5103) | 0.5 | WB I, WB II (Det. I, 500 mM NaCl [×1]), WB III |
| H4R3me2 | Upstate Biotech (07-213) | 0.5 | WB I, WB II (Det. I, 250 mM NaCl [×2]) |
| H4R3cit | Upstate Biotech (07-596) | 0.5 | WB I, WB II (Det. I, 250 mM NaCl [×2]) |
| PADI4 | Nakashima et al. (24) | 0.2 | WB I, WB II (Det. I, 250 mM NaCl [×3]) |
| HDAC1 | Abcam (ab7028) | 1.5 | WB I, WB II (Det. II, 500 mM NaCl [×2]) |
Washings used three washing buffers (WB), with WB II containing different salt concentrations and detergent (Det.) mixes as indicated. Det. I = 0.1% SDS, 1% Triton X-100. Det. II = 1% Triton X-100. The numbers in brackets indicate the number of washes done with WB II.
In Re-ChIP assays, DNA-protein complexes were extracted twice from the beads by adding 25 μl of 10 mM DTT, followed by incubation for 20 min at 37°C with vortexing every 5 min. Supernatants were then diluted 10 times in immunoprecipitation buffer, and the immunoprecipitation procedure was performed again. Cross-linking was reversed by an overnight incubation at 65°C. DNA was purified on Qiaquick columns (Qiagen). In subsequent semiquantitative PCR analysis (Taq; Qiagen), performed on a total volume of 25 μl including 15 mM MgCl2 (final concentration), 2 μl of input material or 3 μl of ChIP samples was used, with the following primers (Proligo): left, 5′-GTTGTCAGGCCAAGCCTTTT-3′; and right, 5′-GAGCGTTAGATAACATTTGCC-3′ (54°C; 15 mM MgCl2 [final concentration]).
In ChIP assays, quantitative PCRs were performed on 1 μl of input or ChIP sample DNA, with a Bio-Rad MyiQ apparatus and Bio-Rad iQ SYBR green supermix, in a total volume of 20 μl containing 10 mM MgCl2, with the following primers (Proligo): left pS2 promoter, 5′-TTCCGGCCATCTCTCACTAT-3′; right pS2 promoter, 5′-CGGGGATCCTCTGAGACA-3′; forward unrelated control region (cf. Fig. 4B), 5′-AAGCTGCGTGTTGTCAGATG-3′; and reverse unrelated control region, 5′-AAGGAAGGGCCCTCTATTCA-3′.
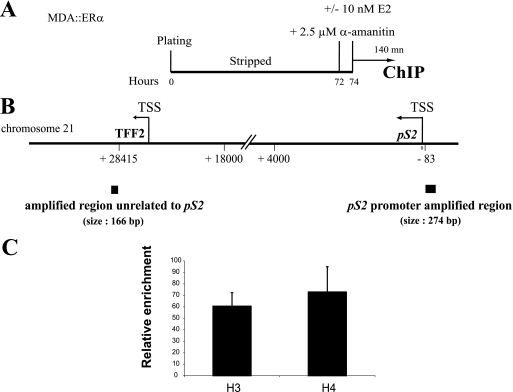
FIG. 4.
Cyclic binding of HDAC1 and PADI4 to the pS2 promoter. (A) Schematic description of kinetic ChIP assays performed on MDA::ERα cells. The cells were synchronized by 3 days of culture and then treated with 2.5 μM α-amanitin for 2 h, followed by exposure to 10 nM E2. Chromatin was then prepared from cells sampled at 5-min intervals over a 140-min period. (B) Locations of the control and pS2 promoter regions on which ChIP experiments focused. Transcription start sites (TSS) of the pS2 gene and upstream TFF2 gene are shown, as is the unrelated amplified control region. (C) ChIP assays showing enrichment in histones H3 and H4 in the unrelated control region were performed. Chromatin samples from MDA::ERα cells were immunoprecipitated with antibodies raised against histone H3 or histone H4. The amount of immunoprecipitated unrelated control region was quantified by real-time PCR, and determined on the basis of the threshold cycle (i.e., the CT value) for each PCR. Calculation details are described in Materials and Methods. The final relative enrichment values shown are means of three separate experiments. (D) A chromatin sample as depicted in panel A was subjected to immunoprecipitation with antibodies raised against ERα and RNA Pol II. The amount of immunoprecipitated pS2 promoter was quantified by real-time PCR and determined on the basis of the threshold cycle (CT) for each PCR. Calculation details are described in Materials and Methods. The final relative enrichment values shown are means of three separate experiments. (E) Chromatin samples were subjected, as depicted in panel A, to immunoprecipitation with anti-AcH3, anti-dimeH3R17, or anti-dimeH4R3. The amount of immunoprecipitated pS2 promoter was quantified by real-time PCR. The relative enrichment values shown are means of three separate experiments. (F) Kinetic ChIPs indicate that PADI4 and HDAC1 are recruited to the pS2 promoter within the same time-frame. Chromatin samples were subjected, as depicted in panel A, to immunoprecipitation with antibodies raised against HDAC1, PADI4, or CitH3 [H3R(17+2+8) citrullinated]. The amount of immunoprecipitated pS2 promoter was evaluated on the basis of the threshold cycle (CT) for each PCR (see reference 20 and Materials and Methods for details). The relative enrichment values shown are means of three separate experiments.
The relative levels of the fragments of interest in the immunoprecipitated DNA were determined from the threshold cycle (CT) for each PCR. To ensure the reliability of our ChIP data, two control samples specific for the ChIP experiment have been included, as we have previsously reported (20): the input sample (indicative of the presence and amount of chromatin used in the ChIP reaction) and the control antibody (HA antibody) sample (indicative of the amount of background signal generated by the chromatin preparations and ChIP procedure). The calculations of the relative enrichment values were as described below. (i) We normalized the quantitative PCR signals obtained from the immunoprecipitated ChIP sample to the input sample, i.e., CT input - CT ChIP. The PCR efficiency, corresponding to the different sets of primers used in our quantitative PCR, was then raised to the power of this CT difference, i.e., (primer PCR efficiency)(CT input − CT ChIP). (ii) The enrichment (n-fold) of the immunoprecipitated sequence of interest was obtained by normalizing the values to the ChIP background (relative to IP HA antibody). (iii) To ensure that the observed binding of the tested proteins or histone marks reflect specific binding to the pS2/TFF1 promoter, we also amplified an unrelated control region (cf. Fig. 4B) in a quantitative PCR. The relative enrichment values were calculated by dividing the enrichment (n-fold) derived from the sequence of interest (pS2/TFF1 locus) by the signal derived from this control locus (unrelated control region).
RNAi and retroviral infection.
RNAi PADI4 and RNAi HDAC1 were generated as previously described (2, 40). Briefly, the target sequence used to silence HDAC1 or PADI4 was inserted as a short hairpin into the pRetroSuper (pRS) retroviral vector according to the manufacturer's (OligoEngine) recommendations to form RNAi PADI4 or RNAi HDAC1, respectively. Retrovirus production by 293T GP cells and infection of target MDA::ERα were performed as described. Infected cells were selected with 1 μg of puromycin (Sigma)/ml. The primer sequences are available from the authors upon request.
RNA purification and RT-PCR analyses.
Extraction of total RNA was carried out with TriPure reagent (Roche) according to the manufacturer's instructions. DNase treatment was performed with a DNA-free DNase kit (Ambion) according to the manufacturer's protocol. cDNA was reverse transcribed from 1 μg of RNA using random hexamers (Amersham/Pharmacia Biotech) and Superscript II reverse transcriptase (Life Technologies, Inc.). The reverse transcription (RT) reaction mixture was diluted with diethyl pyrocarbonate water (Fluka Biochemika) before addition to the PCR. To quantitatively evaluate expression levels under different conditions, real-time PCRs were performed in a LightCycler 480 system (Roche). The primers sequences for PADI4 and HDAC1 amplifications were as follows: left PADI4, 5′-GACAAAGTGAGGGTGTTTCA-3′; right PADI4, 5′-AGAAGTCCATGTTGTGCTTT-3′; left HDAC1, 5′-GGATCGGTTAGGTTGCTTCA-3′; and right HDAC1, 5′-AGCATCAGCATAGGCAGGTT-3′. SDHA was amplified as an internal control to measure the amounts of the cDNAs (39).
Formaldehyde-assisted isolation of regulatory elements (FAIRE).
After cross-linking for 10 min with 1.5% formaldehyde (Sigma) at room temperature, cells were collected in 5 ml of collection buffer (100 mM Tris-HCl [pH 9.4], 100 mM DTT) and incubated first for 10 min and then at 30°C for 10 min. The cells were then washed in 1 ml of PBS, lysed for 15 min at room temperature in 300 μl of lysis buffer (10 mM EDTA, 50 mM Tris-HCl [pH 8.0], 1% SDS, 0.5% Empigen BB [Sigma]), and sonicated for 14 min in a BioRuptor apparatus (Diagenode), with 30-s on/off cycles. After centrifugation for 10 min at 10,000 × g, 30 μl of the supernatant was used as input, and the remainder was subjected to three consecutive phenol-chloroform-isoamyl ethanol (25:24:1) extractions. Inputs and extracted samples were then incubated overnight at 65°C to reverse formaldehyde cross-linking linking with 5 μg of proteinase K (Sigma). After a subsequent incubation of the samples with 2 μg of RNase (Sigma) for 1 h at 37°C, the DNA was then purified on NucleoSpin columns (Macherey-Nagel) in NTB buffer and quantified with a Nanodrop apparatus (Thermo Scientific). Then, 20 ng of input or purified DNA was used for quantitative PCRs. Quantitative PCR conditions and primers were as described above. We typically calculated the relative enrichment values using the comparative CT method. First, a ratio was calculated using the signal from the FAIRE sample relative to the signal from the input sample. Second, consistent with other reports using the FAIRE technique (11, 12), all ratios are then normalized to the unrelated control region (cf. Fig. 4B).
Statistical analysis.
In ChIP and FAIRE experiments, statistical analysis of the results was performed by using one-way analysis of variance, followed by Dunnett (bilateral) post hoc comparisons and by using the Kruskal-Wallis test, followed by Mann-Whitney comparisons when the conditions of the parametric tests were not satisfied. The statistical software used was SPSS Statistics 17.0.
RESULTS
PADI4 interacts with HDAC1 in vitro and in vivo.
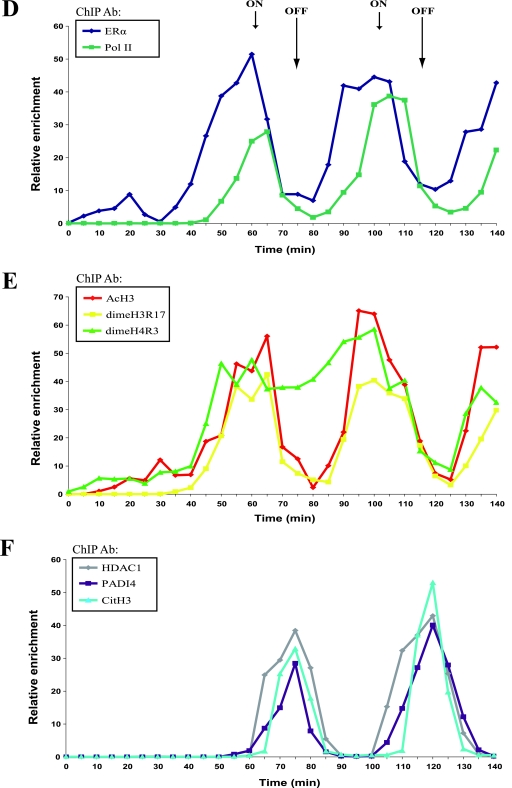
To identify proteins that associate with PADI4, we used an in vitro GST pull-down assay. We found that PADI4 associates with HDAC1. As shown in Fig. 1A, full-length PADI4 (residues 1 to 663) fused to GST was able to bind in vitro-translated (IVT) radiolabeled full-length HDAC1 (lane 3), whereas GST did not. As a further control for the specificity of the interaction, we show that in vitro translation of the transcriptional regulator RbBP5 (IVT RbBP5) was unable to bind to GST PADI4 (Fig. 1A). Mapping experiments revealed that the binding of PADI4 to HDAC1 is mediated primarily by its C-terminal part, spanning residues 460 to 663 (Fig. 1B). As shown in Fig. 1B (lower part), the gel was Coomassie blue stained to check that the amounts of GST fusion proteins used were equal.
FIG. 1.
The histone deiminase PADI4 interacts with the histone deacetylase HDAC1 in vitro. (A) The full-length PADI4 peptidylarginine deiminase protein was fused to GST and tested in GST pull-down experiments using IVT full-length HDAC1 (lanes 1 to 3) or IVT RbBP5 (as a negative control; lanes 4 to 6). Lanes 1 and 4, inputs (5%) of radiolabeled IVT-HDAC1 and IVT-RbBP5, respectively. A Coomassie blue-stained gel shows the input of GST fusion proteins used. (B) HDAC1 binds specific regions of PADI4. A schematic representation of the PADI4, with its known domains highlighted, is shown in the upper part of the panel. The depicted PADI4 protein fragments were fused to GST and tested in GST pull-down experiments using IVT full-length HDAC1. Lane 1: radiolabeled IVT-HDAC1 Input (5%). The results are summarized on the right (from “++” [strong interaction] to “-” [no interaction]). The lower part shows a Coomassie blue-stained gel showing that equivalent GST fusion proteins were used. (C) Representation of the HDAC1 regions fused to GST (upper panel). These proteins were tested for interaction with IVT-PADI4 in GST pull-down assays. The Coomassie blue-stained gel shows the input of GST fusion proteins used.
We next sought to determine which part of HDAC1 mediates the observed association with PADI4. To this end, various GST fragments spanning the HDAC1 protein were incubated with IVT full-length PADI4. Figure 1C shows that the association with IVT PADI4 is mediated by the N-terminal portion of HDAC1, encompassing residues 1 to 90. The Coomassie blue-stained gel shows the input of GST fusion proteins used (Fig. 1C, lower part).
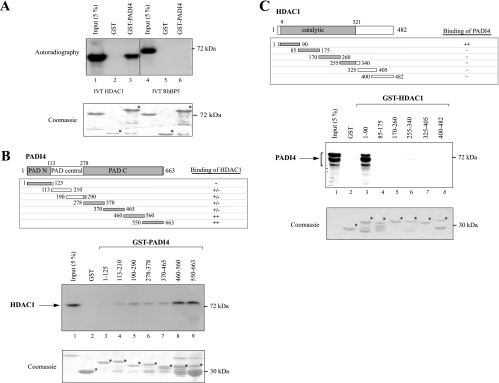
To further substantiate the interaction between PADI4 and HDAC1, we used coimmunoprecipitation. We cotransfected 293T mammalian cells with a vector expressing HA-tagged full-length PADI4, a vector expressing Flag-tagged full-length HDAC1, or with both vectors. We first analyzed the cell lysates by immunoprecipitation with anti-HA antibody (for PADI4), followed by Western blotting with anti-Flag antibody (for HDAC1). Figure 2A shows that PADI4 interacts with HDAC1 (lane 3). No precipitate was detected after transfection of cells with either the HA-PADI4-encoding plasmid or the HDAC1-Flag-encoding plasmid alone (lanes 1 and 2, respectively). The input levels were checked by Western blotting with anti-Flag or anti-HA antibodies (input controls). The converse experiment, i.e., immunoprecipitation of HDAC1-Flag followed by Western blotting with HA-PADI4 detection, also revealed specific association of the proteins (Fig. 2B).
FIG. 2.
PADI4 binds to HDAC1 in vivo. (A) 293T cells were transiently transfected with the expression vector(s) for HA-tagged full-length PADI4 and/or Flag-tagged full-length HDAC1. Cell extracts were precipitated with anti-HA antibody, and HDAC1-Flag was detected in the immunoprecipitates by Western blot analysis with anti-Flag antibody. The respective amounts of the different proteins present in the inputs were checked by Western blotting with the appropriate antibodies (Input controls). (B) Anti-Flag antibody was used to immunoprecipitate HDAC1-Flag, and anti-HA was used to probe immunoblots for HA-PADI4. The levels of the different proteins in the inputs were checked by Western blotting with anti-HA or anti-Flag antibody (Input controls). (C) PADI4 coimmunoprecipitates with HDAC1 from untransfected cells. MDA::ERα cells treated with E2 for 4 h were lysed in IPH buffer. Whole-cell extracts were then immunoprecipitated with anti-HDAC1 or with the following control antibodies: anti-RNA Pol II, anti-HA, IgG, or the beads only. Precipitates were then probed with anti-PADI4 antibody.
The interaction between HDAC1 and PADI4 was also demonstrated in untransfected cells. In this experiment, we used MDA-MB231 cells stably expressing ERα (MDA::ERα), as previously described (19). We stimulated the cells with estrogen, since this is known to increase their PADI4 level (7). We first immunoprecipitated endogenous HDAC1 from estrogen-stimulated MDA::ERα cells and detected PADI4 in the immunoprecipitates by Western blotting with anti-PADI4 antibody. Figure 2C shows that endogenous PADI4 associates with native HDAC1 (lane 1). This association is specific, as demonstrated by the following controls: no precipitation of PADI4 was observed with an antibody against another transcriptional regulator (RNA Pol II, lane 2), the unrelated anti-HA antibody (lane 3), IgG (lanes 4 and 5), or the beads only (lane 6). Taken together, our data show that the histone deiminase PADI4 interacts with HDAC1 both in vitro and in vivo.
HDAC1 associates with PADI4-mediated histone deiminase activity.
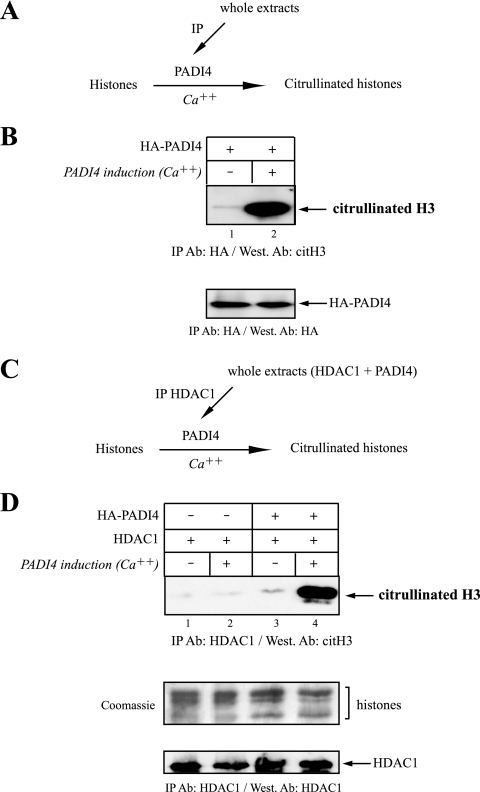
Given the ability of PADI4 to deiminate histones when presented with core histones (7, 42) and, in the light of the above data showing an interaction between PADI4 and HDAC1 (Fig. 1 and 2), we decided to test whether HDAC1 associates with PADI4-mediated histone deiminase activity (removal of an imino group from peptidyl arginine to produce peptidyl citrulline). The first step was to measure this activity in 293T cells overexpressing HA-PADI4. We used these cells rather than MDA::ERα cells because they can be transfected at high efficiency. Whole-cell extracts were then precipitated with anti-HA antibody, and the immune complexes were added to purified core histones. Histone deiminase activity was then monitored in the presence or absence of calcium (Fig. 3A), since binding of calcium ions is essential to PADI4 activation (41). The presence of deiminated histones H3 was revealed by Western blotting with an antibody recognizing citrullinated H3. This anti-CitH3 antibody specifically recognizes the tail of histone H3 when arginines R2, R8, and R17 are replaced by citrulline (citrulline 17+2+8) (7). In our experiment, as shown in Fig. 3B, this antibody detected histone H3 when overexpressed PADI4 (lane 2) was added to the histones. The observed histone deiminase activity was further shown to require calcium (Fig. 3B, lane 1). The levels of PADI4 in the citrullination reaction are shown (Fig. 3B, bottom panel). Thus, in keeping with earlier reports (7, 42), PADI4 is able to specifically deiminate histone H3 when presented with core histones as substrates.
FIG. 3.
HDAC1 associates with PADI4-mediated histone deiminase activity. (A) Outline of the experimental procedure used to monitor histone deiminase activity by PADI4. 293T cells were transiently transfected with HA-PADI4. Whole-cell extracts were then immunoprecipitated with anti-HA antibody (IP PADI4), and the complexes were incubated with histones in the presence or absence of calcium (Ca2+). This ion is required for the PADI4-catalyzed reaction involving removal of an imine group from a peptidyl arginine of histone to produce peptidyl citrulline. (B) Using the approach described in panel A, the histone deiminase activity was assayed by Western blotting with an antibody recognizing citrullinated residues (citrulline 17+2+8) in the tail of histone H3 (7). This anticitrulline antibody detects histone H3 only when overexpressed PADI4 is added to the histones. Levels of PADI4 in the deiminase reaction were checked by Western blotting with anti-HA antibody. (C) Outline of the experiment revealing association of HDAC1 with PADI4-mediated histone deiminase activity. 293T cells were transiently transfected with HDAC1 alone or with HA-PADI4. Whole-cell extracts were then immunoprecipitated with anti-HDAC1 antibody, and the complexes were incubated with histones in the presence or absence of calcium (Ca2+). (D) Histone deiminase activity was assayed by Western blotting with anti-histone H3 (citrulline 17+2+8). Immunoprecipitation of HDAC1 purified histone deiminase activity only when the cells were cotransfected with PADI4. The levels of HDAC1 in the reaction were checked by Western blotting with anti-HDAC1. The Coomassie blue-stained gel shows that equal amounts of histones were used in all reactions.
The next step was to test the association of PAD14 deiminase activity with HDAC1. We first transfected 293T cells with HDAC1, alone or together with HA-PADI4. Whole-cell extracts were then precipitated with anti-HDAC1 antibody, and the immune complexes were tested for histone deiminase activity in the presence or absence of calcium (Fig. 3C). Deiminated histone H3 was revealed by Western blotting with anti-CitH3 antibody. Figure 3C shows that immunoprecipitation of HDAC1 purified histone deiminase activity only when the cells were cotransfected with PADI4 (lane 4). No HDAC1-associated histone deiminase activity was observed after transfection with HDAC1 alone (lane 2). As a control, we checked levels of HDAC1 in the reaction by Western blotting with anti-HDAC1 antibody (Fig. 3D, lower part). Equal amounts of histone H3 were used in all reactions (Fig. 3D, middle part), and we performed Western blotting with anti-HA to detect HA-PADI4 (data not shown). Together, these results show that HDAC1 associates in vivo with PADI4 histone deiminase activity.
Cycling of HDAC1 and PADI4 on the estrogen-responsive promoter pS2.
Next, we investigated the functional consequences of the PADI4-HDAC1 association. More precisely, we tested whether PADI4 and HDAC1 can be recruited concomitantly to the same target gene promoter. It is known that in the presence of estrogen, pS2 gene expression is controlled by ERα, which cycles on its promoter. Not only does the receptor cycle, it also follows a sequence of cofactor recruitment ultimately resulting in binding of RNA Pol II (26). Moreover, recent work has linked recruitment of PADI4 to the pS2 promoter with downregulation of the gene (7, 42). These observations prompted us to probe whether PADI4, together with HDCA1, might associate with the pS2 promoter. To this end, we performed ChIP kinetics at 5-min resolution with chromatin prepared from 3-day-synchronized MDA::ERα cells treated with 10−8 M estradiol (Fig. 4A). Synchronization, done with α-amanitin, was needed to obtain a synchronized cell population whose pS2 promoters were devoid of trans-acting factors and whose local histones were not acetylated (26). In keeping with our previous observations (26), we found the endogenous pS2 promoter to become cyclically permissive to the binding of E2-liganded ERα at a frequency of 40 to 45 min, after an initial 20-min cycle (Fig. 4D). Although Pol II is not recruited during the first cycle, afterward it lags 10 min behind ERα association, defining transcriptionally productive cycles (Fig. 4D). We also observed that histone H3 becomes modified by acetylation (ac H3) and histone H4 becomes modified by dimethylation on arginine 3 (H4R3me2) during the initial nonproductive cycle, so as to induce transcriptional competence within the pS2 promoter (Fig. 4E). Furthermore, methylation of arginine 17 of H3 (di-meH3R17) shows kinetics similar to our previous observations (20). After this first transcriptionally silent cycle, the level of pS2 promoter bound to acetylated H3, along with dimethylation on H3R17, increases dramatically at each cycle after ERα binding (Fig. 4E). Of note, we found in our previous work that the presence on the pS2 promoter of not only H4R3me2 but also of certain factors, such as TATA box binding protein, persists over two cycles. In addition, rearrangement of nucleosome phasing changes at completion of every double cycle (20). We believe that these events reflect a sequential difference in the clearance phase of alternating transcriptionally productive cycles, in which complete resetting of chromatin organization correlates, at least in part, with removal of TATA box binding protein.
It is worth mentioning that the observed kinetics of recruitment to the pS2 promoter does not appear to reflect nonspecific binding of the tested proteins or histone marks to all DNA sequences, as indicated by an important ChIP control that extends our initial findings (19, 20): we used an unrelated DNA sequence, located several kb upstream from the pS2 gene (Fig. 4B). Kinetic ChIPs for ERα, RNA Pol II, AcH3, H3R17me2, and H4R3me2 showed only background binding to this control region (these values were used to normalize our ChIP data on pS2; see Materials and Methods).
We next examined PADI4 and HDAC1 binding to the pS2 promoter. ChIP assays revealed that both PADI4 and HDAC1 associate cyclically with this promoter when it becomes refractory to ERα (Fig. 4F). The presence of PADI4 enzymatic activity was determined with antibodies to citrullinated H3 (citH3). These ChIP assays demonstrated that histone H3 becomes modified by citrullination when PADI4 and HDAC1 are associated with the pS2 promoter (Fig. 4F).
These bindings to the pS2 promoter are specific, since they were not detected on an unrelated control region (values used to normalize the ChIP data on pS2; see Materials and Methods). Taken together, these results demonstrate that PADI4 and HDAC1, and their corresponding enzymatic activities, associate specifically with the estrogen-responsive pS2 promoter (during the transcriptionally unproductive phase) and cycle on it.
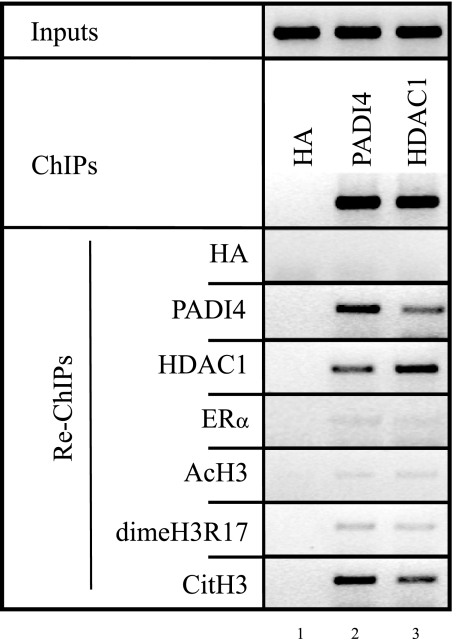
PADI4 and HDAC1 bind together to the pS2 promoter.
To investigate whether PADI4 and HDAC1 bind independently or coordinately to the pS2 promoter in vivo, we performed sequential ChIP (Re-ChIP). In these experiments, MDA::ERα cells were grown in the absence of estrogen for 2 days and then treated with estrogen. A first round of immunoprecipitation was carried out with anti-PADI4 antibody (Fig. 5, lane 2). Then, immunoprecipitated cross-linked DNA-protein complexes were isolated and reimmunoprecipitated with antibodies to HDAC1. Figure 5, lane 2, shows that the pS2 promoter region was successfully amplified from both the fraction reimmunoprecipitated with anti-HDAC1 and the fraction reimmunoprecipitated with anti-citH3, whereas only weak signal was detected when antibodies against ER, acH3, or dimethylated H3R17 were used in the second round of immunoprecipitation. Control reactions performed with nonspecific anti-HA antibodies gave, as expected, no signal (Fig. 5, lane 1). Similar results were obtained when anti-HDAC1 was used in the first round of immunoprecipitation (Fig. 5, lane 3): a signal was observed with the fractions reimmunoprecipitated with either anti-PADI4 or anti-CitH3. Weak signal was detected with anti-ER or anti-acH3 antibodies used in the second round of immunoprecipitation (Fig. 5, lane 3).
FIG. 5.
Simultaneous association of PADI4 and HDAC1 with the pS2 promoter in the presence of estradiol. Chromatin prepared from MDA::ERα cells treated with 10−8 M E2 was subjected to the ChIP procedure with anti-HA (used as a negative control), anti-PADI4, or anti-HDAC1 and then immunoprecipitated again (Re-ChIP) with the antibody shown on the left. The semiquantitative PCR results shown are representative of experiments performed at least three times.
Together, these data strongly suggest that the histone deiminase PADI4 and the histone deacetylase HDAC1 are simultaneously and cyclically present on the pS2 promoter, with maximum occupation as the pS2 promoter is becoming refractory to ERα, when the gene is disengaged from RNA polymerase II and both histone acetylation and arginine dimethylation of H3 are at a minimum.
Collaboration between PADI4 and HDAC1 in generating a repressive chromatin environment on pS2.
To further examine the relationship between PADI4 and HDAC1, we used a short hairpin RNA (shRNA) knockdown strategy in which MDA::ERα cells were infected with retroviruses expressing shRNA directed against PADI4 or HDAC1 (RNAi PADI4 and RNAi HDAC1, respectively). Under conditions similar to those described in Fig. 5 for the Re-ChIP experiments, the cells were grown without estrogen for 2 days before the addition of estrogen.
Focusing first on RNAi for HDAC1, RNAi infected cells showed a significantly (although not completely) reduced level of HDAC1 expression (Fig. 6A, left panel). Expression of PADI4 was unaffected, as shown by real-time RT-PCR (Fig. 6A, right panel). ChIP assays (Fig. 6B, lane 3) showed that RNAi HDAC1 caused decreased binding of HDAC1 to the pS2 promoter. The levels of acetylated histone H3 on pS2 were unaffected by HDAC1 depletion (Fig. 6C, lane 3). This may be due to functional redundancy among HDAC enzymes, since several HDACs can bind to pS2 (20). Interestingly, HDAC1 knockdown resulted in decreased PADI4 binding and decreased H3 citrullination at the pS2 promoter (Fig. 6B, lanes 7 and 11, respectively). In contrast, dimethylation of R17 of H3 and R3 of H4 was slightly increased after HDAC1-infected cells (Fig. 6C, lanes 7 and 11). ChIP assays performed on an unrelated control region demonstrated the specificity of these observations (these values are used to normalize the ChIP data on pS2; cf. Materials and Methods). These results show that HDAC1 favors the presence of PADI4 and histone citrullination on pS2.
FIG. 6.
Collaboration between PADI4 and HDAC1 in generating a repressive pS2 chromatin environment. (A) Specific-RNAi-encoding vectors decrease HDAC1 expression substantially and PADI4 expression moderately. MDA::ERα cells were stably infected with the pRS/control vector (RNAi), the pRS/PADI4 vector (RNAi PADI4), the pRS/HDAC1 vector (RNAi HDAC1), or both pRS/PADI4 and pRS/HDAC1 (RNAi PADI4 + RNAi HDAC1). After selection the cells were harvested, and quantitative RT-PCR was performed on their mRNA, with primers corresponding to HDAC1 (left panel) or PADI4 (right panel). All transcript levels were normalized with respect to SDHA and then divided by the normalized level recorded for control cells. Values are means of three independent experiments. (B) Chromatin was prepared from MDA::ERα cells expressing a control RNAi, PADI4 RNAi, HDAC1 RNAi, or both PADI4 RNAi and HDAC1 RNAi (RNAi PADI4 + RNAi HDAC1) and treated with 10−8 M E2. Cross-linked chromatin samples were immunoprecipitated with antibodies to HDAC1 (lanes 1 to 4), PADI4 (lanes 5 to 8), or CitH3 [H3R(17+2+8) citrullinated] (lanes 9 to 12). The amount of immunoprecipitated pS2 promoter was quantified by real-time PCR on the basis of the threshold cycle value (CT). Calculation details were done as previously described (20) (see also Materials and Methods). The data are presented as the means ± the standard errors of four independent experiments, and asterisks indicate statistical significance as determined by Dunnett and Mann-Whitney comparisons (*, P < 0.05; **, P < 0.01; ***, P < 0.001). (C) Chromatin samples were subjected, as described in panel B, to immunoprecipitation with antibodies raised against AcH3 (lanes 1 to 4), dimeH3R17 (lanes 5 to 8), or dimeH4R3 (lanes 9 to 12). The immunoprecipitated pS2 promoter was quantified by real-time PCR on the basis of the CT for each PCR (see reference 20 and Materials and Methods for calculation details). The relative enrichment values shown are means of four independent experiments. Asterisks indicate the statistical significance as determined by Dunnett and Mann-Whitney comparisons (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Regarding PADI4 knockdown in MDA::ERα cells, and despite tremendous efforts, we were able to reduce PADI4 expression only slightly (Fig. 6A, right panel). The PADI4 RNAi construct we used is specific for PADI4, since we could use it to strongly and specifically decrease PADI4 levels in the U2OS cell line (data not shown). The reason for this difference between cell types is currently unclear. Consistent with the slight effect of our construct on the PADI4 level in MDA::ERα cells, this knockdown approach barely affected the presence of PADI4, HDAC1 or their corresponding enzymatic activities on the pS2 promoter (Fig. 6B, lanes 2, 6, and 10 and 6C).
We subsequently investigated whether PADI4 and HDAC1 might functionally collaborate on the pS2 promoter. We reasoned that the limited reduction of PADI4 expression observed after infection of MDA::ERα cells with the shRNA for PADI4 (Fig. 6A, right panel) could provide a useful tool for testing whether PADI4 collaborates with HDAC1. We generated PADI4 and HDAC1 doubly infected MDA::ERα cells displaying decreased PADI4 and HDAC1 levels comparable to those observed in the corresponding singly infected cells (Fig. 6A). Simultaneous expression of PADI4 and HDAC1 RNAi, compared to expression of HDAC1 RNAi alone, led to (i) a further decrease in H3 citrullination (Fig. 6B, lane 12), (ii) a substantial increase in acetylated H3 (Fig. 6C, lane 4), and (iii) an enhanced level of dimethylated R17 of histone H3 and R3 of histone H4 (Fig. 6C, lanes 8 and 12). Control ChIP assays performed on an unrelated region showed no such effects (these values were used to normalize ChIP assays on pS2; cf. Materials and Methods). Together, these observations suggest that PADI4 and HDAC1 may favor H3 citrullination and deacetylation of the pS2 promoter.
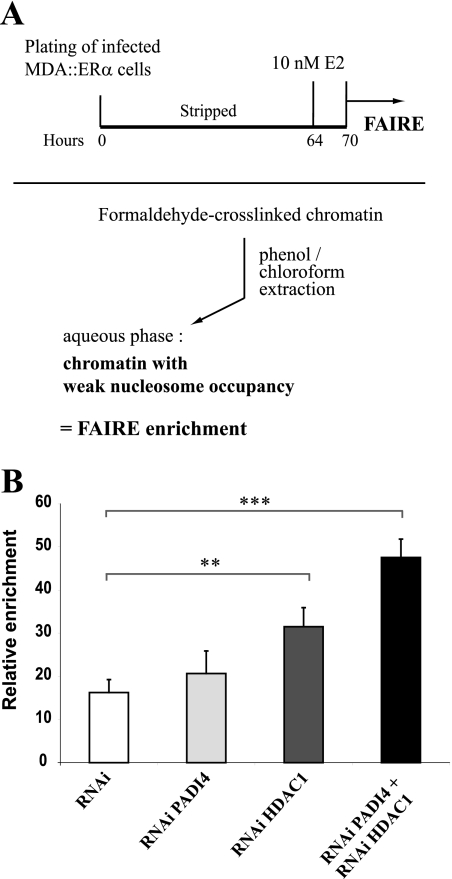
To complement the data described above, we used the technique called FAIRE. This technique starts like a standard ChIP: chromatin is cross-linked with formaldehyde, sheared by sonication, and phenol-chloroform extracted. After cross-link reversion, the DNA is purified, and a real-time PCR is carried out on the sequence of interest. DNA segments that actively regulate transcription are typically characterized by eviction of nucleosomes from chromatin, and this can be facilitated in part by increased acetylation of the nucleosome before activation of transcription (27). Since DNA covalently linked to proteins (consisting mainly of histones) is sequestered in the organic phase, leaving only protein-free DNA fragments in the aqueous phase, the FAIRE technique provides an estimate of nucleosome occupancy or of its relative loose association with DNA at a given region, correlating directly with its transcriptional status (Fig. 7A) (14, 22). We performed FAIRE on the pS2 promoter, which contains two phased nucleosomes (29, 33), in PADI4 and HDAC1 single- and dual-interference MDA::ERα cells treated with E2 as before (Fig. 5). On the basis of the results presented in Fig. 6, we hypothesized that a double infection of cells with our shRNA would lead to enhanced FAIRE enrichment. This hypothesis proved correct. After FAIRE, as shown in Fig. 7B, preparations from PADI4 and HDAC1 dual-interference cells proved to be more strongly enriched in pS2 promoter than preparations derived from single-interference cells. This was not observed with an unrelated control region (these values were used to normalize the FAIRE data on pS2; cf. Materials and Methods). These results indicate that the association of nucleosomes at the pS2 promoter is looser in PADI4 and HDAC1 dual-interference cells compared to controls, supporting the view that there is functional cross talk between these two enzymes at the pS2 promoter.
FIG. 7.
Effects of PADI4 and HDAC1 on pS2 nucleosome occupancy. (A) Schematic description of the FAIRE technique. Infected MDA::ERα cells were placed for 2 days in an E2-free medium and then exposed to 10 nM E2 for 6 h (upper part). Chromatin was then prepared and phenol-chloroform extracted without previous reverse cross-linking (bottom part). (B) MDA::ERα chromatin samples were prepared from cells expressing control RNAi, PADI4 RNAi, HDAC1 RNAi, or PADI4 RNAi + HDAC1 RNAi (RNAi PADI4 + RNAi HDAC1). FAIRE enrichment in pS2 promoter was quantified by real-time PCR on the basis of the threshold cycle value (CT). Calculation details are described in Materials and Methods. The data are presented as means ± the standard errors of four independent experiments. Asterisks indicate the statistical significance as determined by Dunnett post hoc comparisons (**, P < 0.01; ***, P < 0.001).
Taken together, these data suggest a functional interconnection between the PADI4 and HDAC1 enzymes in the regulation of the pS2 gene.
DISCUSSION
The discovery of the histone deiminase PADI4, converting methylarginine to citrulline, has revealed that histone arginine methylation is not a static modification, as previously thought (7, 42). However, understanding how this newly discovered enzyme functions in chromatin biology remains an important challenge. We here report a close connection between the PADI4 histone deiminase and HDAC1. We show that PADI4 binds to HDAC1 and that HDAC1 associates with PADI4-mediated histone deiminase activity. The results of our kinetic ChIP assays show that PADI4 and HDAC1 appear transiently and in a cyclic manner on the estrogen-responsive promoter pS2, in the presence of estradiol. Their presence correlates with a loss of arginine methylation, acquisition of citrulline, histone deacetylation, and disengagement of RNA polymerase II from the pS2 promoter.
We have previously proposed the concept of a “transcriptional clock” that synchronizes sequential waves of promoter accessibility for given cofactors and thereby dictates an ordered and dynamic sequence of events that initiate and sustain transcription (20). Periodic limitation of transcription is caused by events that clear the pS2 promoter of transcription factors. The precise mechanisms underlying this promoter clearance and the termination of each cycle are still unclear. Our current work offers important insights into the molecular basis of the clearance phase of the pS2 promoter. We found that the joint processes of PADI4-mediated histone deimination and HDAC1-mediated histone deacetylation contribute, perhaps partially but importantly, to this phase. These concerted repressive histone modifications appear to participate in marking the end of each cycle, allowing the pS2 promoter to be reset and making it possible for subsequent cycles to proceed. Histone deimination and deacetylation may thus be particularly important in enabling the cell to adjust continuously the transcription rate of the pS2 gene according to the level of estrogen. We do not know, at this stage, which factors recruit PADI4 and HDAC1 when the pS2 promoter is becoming refractory to ERα. The Sp proteins might be involved, since these transcription factors are reported to interact with HDACs and since, in the absence of the ERα, their presence on the pS2 promoter coincides with binding of HDACs to the promoter (13, 35).
Our findings may illustrate nicely the concept of cross talk between histone modifications leading to modulation of gene expression. For example, deacetylation of lysine 9 of H3 is reported to be required to allow methylation of this residue (25). This is due, at least in some instances, to the concerted and coordinated action of the SU(VAR)3-9 methyltransferase and HDAC1 (8). Recent studies have shown an interplay between histone deacetylation and the LSD1/KDM1 H3K4 histone demethylase: lysine 9 deacetylation by HDAC1 renders histone H3 more susceptible to LSD1/KDM1-mediated H3K4 demethylation, and LSD1/KDM1 cofactors are required for optimal HDAC1 activity (18, 32). Here, we show that HDAC1 plays a role in PADI4-mediated histone citrullination at the pS2 promoter. Furthermore, our data suggest a functional link between the PADI4 and HDAC1 enzymes in the regulation of the pS2 promoter. The challenge for the future will be to unravel the precise sequence of events of the coordinated action of PADI4 and HDAC1 at the pS2 promoter. Although our data suggest that this coordinated action may be related to the physical association we have identified between PADI4 and HDAC1, further work will be needed to formally prove a direct functional link between these two enzymes. Furthermore, the biological or regulatory roles of the PADI4-HDAC1 association remain unclear and defining these roles will deserve future studies.
The deiminating activity of PADI4 has been implicated in the pathophysiology of rheumatoid arthritis (RA) (36), one of the most common human systemic autoimmune diseases. In addition, assays of antibodies against citrullinated peptides can be used as valuable diagnostic tools (28, 38). Histone deacetylase inhibitors are emerging as effective and valuable tools for both chemotherapy and chemoprevention of cancer (46). In addition to the efficacy of such inhibitors in cancer therapy, recent work points to the HDAC inhibitor trichostatin A as a potential therapeutic agent in the treatment of RA (15). It seems that trichostatin A sensitizes RA-related synovial fibroblasts to TRAIL-induced apoptosis (15). We reveal here coordinated histone deimination and deacetylation, through the combined action of the PADI4 and HDAC1 enzymes. Whether inhibition of histone deimination is an important component of the anti-RA effect is not known, but our present findings raise this attractive possibility. Future studies aimed at developing PADI4-inhibiting drugs should make it possible to test this possibility and may contribute to progress in the medical care of RA.
Acknowledgments
H.D. and R.D. were funded by the FNRS. F.F. is a Chercheur Qualifié du FNRS of the Belgian Fonds National de la Recherche Scientifique. Grants to R.M. were from the CNRS, University of Rennes I and from the Association pour la Recherche contre le Cancer. This study was also funded by grants to F.F. from the Fondation contre le Cancer and the FNRS, grants from the Action de Recherche Concertée de la Communauté Française de Belgique and from the Interuniversity Attraction Poles (IAP P6/28), by EU grant CANCERDIP FP7-200620, and by the European Molecular Biology Organization Young Investigator Programme.
We thank A. Cook and T. Kouzarides for HDAC1 constructs.
Footnotes
Published ahead of print on 6 July 2009.
REFERENCES
- 1.Arita, K., H. Hashimoto, T. Shimizu, K. Nakashima, M. Yamada, and M. Sato. 2004. Structural basis for Ca2+-induced activation of human PAD4. Nat. Struct. Mol. Biol. 11777-783. [DOI] [PubMed] [Google Scholar]
- 2.Bernard, D., J. Gil, P. Dumont, S. Rizzo, D. Monte, B. Quatannens, D. Hudson, T. Visakorpi, F. Fuks, and Y. de Launoit. 2006. The methyl-CpG-binding protein MECP2 is required for prostate cancer cell growth. Oncogene 251358-1366. [DOI] [PubMed] [Google Scholar]
- 3.Brehm, A., E. A. Miska, D. J. McCance, J. L. Reid, A. J. Bannister, and T. Kouzarides. 1998. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature 391597-601. [DOI] [PubMed] [Google Scholar]
- 4.Brenner, C., R. Deplus, C. Didelot, A. Loriot, E. Vire, C. De Smet, A. Gutierrez, D. Danovi, D. Bernard, T. Boon, P. G. Pelicci, B. Amati, T. Kouzarides, Y. de Launoit, L. Di Croce, and F. Fuks. 2005. Myc represses transcription through recruitment of DNA methyltransferase corepressor. EMBO J. 24336-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, B., Y. Chen, Y. Zhao, and R. K. Bruick. 2007. JMJD6 is a histone arginine demethylase. Science 318444-447. [DOI] [PubMed] [Google Scholar]
- 6.Chen, D., H. Ma, H. Hong, S. S. Koh, S. M. Huang, B. T. Schurter, D. W. Aswad, and M. R. Stallcup. 1999. Regulation of transcription by a protein methyltransferase. Science 2842174-2177. [DOI] [PubMed] [Google Scholar]
- 7.Cuthbert, G. L., S. Daujat, A. W. Snowden, H. Erdjument-Bromage, T. Hagiwara, M. Yamada, R. Schneider, P. D. Gregory, P. Tempst, A. J. Bannister, and T. Kouzarides. 2004. Histone deimination antagonizes arginine methylation. Cell 118545-553. [DOI] [PubMed] [Google Scholar]
- 8.Czermin, B., G. Schotta, B. B. Hulsmann, A. Brehm, P. B. Becker, G. Reuter, and A. Imhof. 2001. Physical and functional association of SU(VAR)3-9 and HDAC1 in Drosophila. EMBO Rep. 2915-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deplus, R., C. Brenner, W. A. Burgers, P. Putmans, T. Kouzarides, Y. de Launoit, and F. Fuks. 2002. Dnmt3L is a transcriptional repressor that recruits histone deacetylase. Nucleic Acids Res. 30:3831-3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuks, F., W. A. Burgers, A. Brehm, L. Hughes-Davies, and T. Kouzarides. 2000. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat. Genet. 2488-91. [DOI] [PubMed] [Google Scholar]
- 11.Giresi, P. G., J. Kim, R. M. McDaniell, V. R. Iyer, and J. D. Lieb. 2007. FAIRE (formaldehyde-assisted isolation of regulatory elements) isolates active regulatory elements from human chromatin. Genome Res. 17877-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giresi, P. G., and J. D. Lieb. 2009. Isolation of active regulatory elements from eukaryotic chromatin using FAIRE (formaldehyde assisted isolation of regulatory elements). Methods 48233-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He, S., J. M. Sun, L. Li, and J. R. Davie. 2005. Differential intranuclear organization of transcription factors Sp1 and Sp3. Mol. Biol. Cell 164073-4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hogan, G. J., C. K. Lee, and J. D. Lieb. 2006. Cell cycle-specified fluctuation of nucleosome occupancy at gene promoters. PLoS Genet. 2e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jungel, A., V. Baresova, C. Ospelt, B. R. Simmen, B. A. Michel, R. E. Gay, S. Gay, C. A. Seemayer, and M. Neidhart. 2006. Trichostatin A sensitizes rheumatoid arthritis synovial fibroblasts for TRAIL-induced apoptosis. Ann. Rheum. Dis. 65910-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klose, R. J., K. Yamane, Y. Bae, D. Zhang, H. Erdjument-Bromage, P. Tempst, J. Wong, and Y. Zhang. 2006. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature 442312-316. [DOI] [PubMed] [Google Scholar]
- 17.Reference deleted.
- 18.Lee, M. G., C. Wynder, D. A. Bochar, M. A. Hakimi, N. Cooch, and R. Shiekhattar. 2006. Functional interplay between histone demethylase and deacetylase enzymes. Mol. Cell. Biol. 266395-6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Metivier, R., G. Penot, R. P. Carmouche, M. R. Hubner, G. Reid, S. Denger, D. Manu, H. Brand, M. Kos, V. Benes, and F. Gannon. 2004. Transcriptional complexes engaged by apo-estrogen receptor-alpha isoforms have divergent outcomes. EMBO J. 233653-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Metivier, R., G. Penot, M. R. Hubner, G. Reid, H. Brand, M. Kos, and F. Gannon. 2003. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115751-763. [DOI] [PubMed] [Google Scholar]
- 21.Metzger, E., M. Wissmann, N. Yin, J. M. Muller, R. Schneider, A. H. Peters, T. Gunther, R. Buettner, and R. Schule. 2005. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437436-439. [DOI] [PubMed] [Google Scholar]
- 22.Nagy, P. L., M. L. Cleary, P. O. Brown, and J. D. Lieb. 2003. Genomewide demarcation of RNA polymerase II transcription units revealed by physical fractionation of chromatin. Proc. Natl. Acad. Sci. USA 1006364-6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakashima, K., T. Hagiwara, A. Ishigami, S. Nagata, H. Asaga, M. Kuramoto, T. Senshu, and M. Yamada. 1999. Molecular characterization of peptidylarginine deiminase in HL-60 cells induced by retinoic acid and 1α,25-dihydroxyvitamin D3. J. Biol. Chem. 27427786-27792. [DOI] [PubMed] [Google Scholar]
- 24.Nakashima, K., T. Hagiwara, and M. Yamada. 2002. Nuclear localization of peptidylarginine deiminase V and histone deimination in granulocytes. J. Biol. Chem. 27749562-49568. [DOI] [PubMed] [Google Scholar]
- 25.Rea, S., F. Eisenhaber, D. O'Carroll, B. D. Strahl, Z. W. Sun, M. Schmid, S. Opravil, K. Mechtler, C. P. Ponting, C. D. Allis, and T. Jenuwein. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406593-599. [DOI] [PubMed] [Google Scholar]
- 26.Reid, G., M. R. Hubner, R. Metivier, H. Brand, S. Denger, D. Manu, J. Beaudouin, J. Ellenberg, and F. Gannon. 2003. Cyclic, proteasome-mediated turnover of unliganded and liganded ERα on responsive promoters is an integral feature of estrogen signaling. Mol. Cell 11695-707. [DOI] [PubMed] [Google Scholar]
- 27.Reinke, H., and W. Horz. 2003. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol. Cell 111599-1607. [DOI] [PubMed] [Google Scholar]
- 28.Schellekens, G. A., H. Visser, B. A. de Jong, F. H. van den Hoogen, J. M. Hazes, F. C. Breedveld, and W. J. van Venrooij. 2000. The diagnostic properties of rheumatoid arthritis antibodies recognizing a cyclic citrullinated peptide. Arthritis Rheum. 43155-163. [DOI] [PubMed] [Google Scholar]
- 29.Sewack, G. F., and U. Hansen. 1997. Nucleosome positioning and transcription-associated chromatin alterations on the human estrogen-responsive pS2 promoter. J. Biol. Chem. 27231118-31129. [DOI] [PubMed] [Google Scholar]
- 30.Shang, Y., X. Hu, J. DiRenzo, M. A. Lazar, and M. Brown. 2000. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103843-852. [DOI] [PubMed] [Google Scholar]
- 31.Shi, Y., F. Lan, C. Matson, P. Mulligan, J. R. Whetstine, P. A. Cole, R. A. Casero, and Y. Shi. 2004. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119941-953. [DOI] [PubMed] [Google Scholar]
- 32.Shi, Y. J., C. Matson, F. Lan, S. Iwase, T. Baba, and Y. Shi. 2005. Regulation of LSD1 histone demethylase activity by its associated factors. Mol. Cell 19857-864. [DOI] [PubMed] [Google Scholar]
- 33.Sparmann, A., and M. van Lohuizen. 2006. Polycomb silencers control cell fate, development and cancer. Nat. Rev. Cancer 6846-856. [DOI] [PubMed] [Google Scholar]
- 34.Strahl, B. D., S. D. Briggs, C. J. Brame, J. A. Caldwell, S. S. Koh, H. Ma, R. G. Cook, J. Shabanowitz, D. F. Hunt, M. R. Stallcup, and C. D. Allis. 2001. Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Curr. Biol. 11996-1000. [DOI] [PubMed] [Google Scholar]
- 35.Sun, J. M., V. A. Spencer, L. Li, H. Yu Chen, J. Yu, and J. R. Davie. 2005. Estrogen regulation of trefoil factor 1 expression by estrogen receptor alpha and Sp proteins. Exp. Cell Res. 30296-107. [DOI] [PubMed] [Google Scholar]
- 36.Suzuki, A., R. Yamada, X. Chang, S. Tokuhiro, T. Sawada, M. Suzuki, M. Nagasaki, M. Nakayama-Hamada, R. Kawaida, M. Ono, M. Ohtsuki, H. Furukawa, S. Yoshino, M. Yukioka, S. Tohma, T. Matsubara, S. Wakitani, R. Teshima, Y. Nishioka, A. Sekine, A. Iida, A. Takahashi, T. Tsunoda, Y. Nakamura, and K. Yamamoto. 2003. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat. Genet. 34395-402. [DOI] [PubMed] [Google Scholar]
- 37.Tsukada, Y., J. Fang, H. Erdjument-Bromage, M. E. Warren, C. H. Borchers, P. Tempst, and Y. Zhang. 2006. Histone demethylation by a family of JmjC domain-containing proteins. Nature 439811-816. [DOI] [PubMed] [Google Scholar]
- 38.van Boekel, M. A., E. R. Vossenaar, F. H. van den Hoogen, and W. J. van Venrooij. 2002. Autoantibody systems in rheumatoid arthritis: specificity, sensitivity and diagnostic value. Arthritis Res. 487-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vandesompele, J., K. De Preter, F. Pattyn, B. Poppe, N. Van Roy, A. De Paepe, and F. Speleman. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vire, E., C. Brenner, R. Deplus, L. Blanchon, M. Fraga, C. Didelot, L. Morey, A. Van Eynde, D. Bernard, J. M. Vanderwinden, M. Bollen, M. Esteller, L. Di Croce, Y. de Launoit, and F. Fuks. 2006. The Polycomb group protein EZH2 directly controls DNA methylation. Nature 439871-874. [DOI] [PubMed] [Google Scholar]
- 41.Vossenaar, E. R., A. J. Zendman, W. J. van Venrooij, and G. J. Pruijn. 2003. PAD, a growing family of citrullinating enzymes: genes, features and involvement in disease. Bioessays 251106-1118. [DOI] [PubMed] [Google Scholar]
- 42.Wang, Y., J. Wysocka, J. Sayegh, Y. H. Lee, J. R. Perlin, L. Leonelli, L. S. Sonbuchner, C. H. McDonald, R. G. Cook, Y. Dou, R. G. Roeder, S. Clarke, M. R. Stallcup, C. D. Allis, and S. A. Coonrod. 2004. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science 306279-283. [DOI] [PubMed] [Google Scholar]
- 43.Whetstine, J. R., A. Nottke, F. Lan, M. Huarte, S. Smolikov, Z. Chen, E. Spooner, E. Li, G. Zhang, M. Colaiacovo, and Y. Shi. 2006. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125467-481. [DOI] [PubMed] [Google Scholar]
- 44.Reference deleted.
- 45.Yamane, K., C. Toumazou, Y. Tsukada, H. Erdjument-Bromage, P. Tempst, J. Wong, and Y. Zhang. 2006. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 125483-495. [DOI] [PubMed] [Google Scholar]
- 46.Yoo, C. B., and P. A. Jones. 2006. Epigenetic therapy of cancer: past, present and future. Nat. Rev. Drug Discov. 537-50. [DOI] [PubMed] [Google Scholar]
- 47.Zhang, Y. 2004. Molecular biology: no exception to reversibility. Nature 431637-639. [DOI] [PubMed] [Google Scholar]