Abstract
We recently showed that a leukemia inhibitory factor (LIF)-engaged signaling pathway consisting of JAK1, STAT1, and STAT3 plays dual roles in myogenic differentiation: while it participates in myoblast proliferation, it also actively represses differentiation. Downregulation of this pathway is required at the onset of differentiation. However, it remained unclear how this is achieved mechanistically. We now show that SOCS1, SOCS3, and PIAS1 promote myogenic differentiation by specifically inhibiting the LIF-induced JAK1/STAT1/STAT3 pathway via distinct targets; whereas SOCS1 and SOCS3 selectively bind and inhibit JAK1 and gp130, respectively, PIAS1 targets mainly the activated STAT1 and prevents its binding to DNA. We further demonstrated that the SUMO E3-ligase activity of PIAS1 is dispensable for its role in myogenic differentiation. Collectively, our current study revealed a molecular mechanism that explains how the LIF-induced JAK1/STAT1/STAT3 pathway is downregulated upon myogenic differentiation.
Myogenic differentiation is a fundamental cellular process governing the formation of skeletal muscles in both vertebrates and invertebrates (2, 4, 31, 33). Due to intensive studies by many laboratories over the past 2 decades, myogenic differentiation has become one of the best-characterized differentiation systems. In mammals, two key families of transcription factors are known to play important roles in myogenic differentiation: one is made up of myogenic regulatory factors (MRFs), which consist of MyoD, Myf5, MRF4, and myogenin (37, 40), and the other includes myocyte enhancer binding factor 2s (MEF2), which consist of MEF2A, MEF2B, MEF2C, and MEF2D (3, 30). MRFs serve as muscle-specific master regulatory factors. When ectopically expressed in many non-muscle cells, MRFs can induce a cascade of genes, ultimately resulting in the conversion of these non-muscle cells into the skeletal muscle lineage (40). Unlike MRFs, MEF2A and MEF2D are more ubiquitously expressed, while MEF2C is preferentially expressed in skeletal muscles and in the heart, spleen, and brain (3). MEF2 can physically bind to and cooperate with MRFs to synergistically induce many muscle-specific genes, as the binding sites for MRFs and MEF2s are frequently found in the promoter/or enhancer regions of these genes (26). In addition, MEF2 and MRFs can also mutually regulate the expression of each other (26).
In proliferating myoblasts grown in cell cultures, MyoD or Myf5 is already present at low levels, while myogenin and MRF4 are absent (40). At the early onset of differentiation, cell cycle inhibitors, including p21Cip1, p27Kip1, and p57Kip2, are known to be upregulated first, which helps to arrest the cell cycle and to facilitate the cell cycle exit (10, 12-14, 29, 44). Subsequently, cells start to express myogenin, which is often used as one of the earliest known differentiation markers. The appearance of myogenin signals that cells have irreversibly withdrawn from the cell cycle and entered the differentiation program. The mononucleated differentiating cells are also called myocytes. At the late stage of differentiation, myocytes start to align and fuse with each other to form multinucleated myotubes. Many muscle structural proteins, including myosin heavy chains (MHC), are abundantly expressed at this late stage.
In the past decade, several intracellular signaling pathways, including the p38 mitogen-activated protein kinase-mediated pathway and the insulin-like growth factor/phosphatidylinositol 3-kinase/Akt-mediated pathway have been found to critically control the onset of myogenic differentiation (11, 20, 24, 43). These pathways function via diverse mechanisms, ultimately resulting in changes in the expression of an array of target genes, including the myogenin gene. Recently, we also found that two Janus kinase (JAK)/signal transducer and activator of transcription (STAT)-mediated pathways are involved in mammalian myogenic differentiation in opposite manners: whereas one pathway consisting of JAK1/STAT1/STAT3 inhibits differentiation, the other pathway consisting of JAK2/STAT2/STAT3 is required for myogenic differentiation (36, 39). Interestingly, the JAK/STAT pathway has also been implicated in muscle development in both Drosophila melanogaster and zebrafish (23), which raises the possibility that some key JAK/STAT-dependent regulatory circuitries are conserved in different species. Leukemia inhibitory factor (LIF) is known to stimulate myoblast proliferation and inhibit its differentiation (6). As a member of the interleukin-6 family of cytokines, LIF exerts its biological function by binding to a receptor complex consisting of LIF receptor (LIFR) and gp130 (15, 25). Downstream of the LIFR/gp130, both the JAK/STAT-mediated pathway and the extracellular signal-regulated kinase-mediated pathway have been shown to be capable of mediating the effects of LIF (15). In myoblasts, we have shown that LIF engages JAK1, STAT1, and STAT3 to promote cell proliferation and to repress myogenic differentiation (36). Therefore, it is apparent that the LIF-mediated JAK1/STAT1/STAT3 pathway has to be downregulated at the onset of differentiation. Indeed, we have shown that the JAK1 kinase activity decreases upon differentiation (36). However, it remained unclear how exactly JAK1 is inhibited and how the whole pathway is turned off in myoblasts at the onset of differentiation. To address this question, we turn our attention to known negative regulators of the JAK/STAT pathways. At present, three families of proteins are known to negatively regulate the JAK/STAT pathways: one is the SH2-containing phosphatase (SHP) family, the second is the suppressor of the cytokine signaling (SOCS) family of proteins, and the third is the protein inhibitor of the activated STAT (PIAS) family of proteins (41). In both mouse and human genomes, there are two SHPs (i.e., SHP-1 and SHP-2), eight SOCS (SOCS1 to SOCS7 and CIS), and four PIAS (PIAS1, -3, -x, and -y) (8, 34, 41). Of the two mammalian SHPs, SHP-2 has been shown to be involved in myogenic differentiation (21). However, our preliminary small interfering RNA (siRNA)-based analysis suggested that SHP-2 is not involved in downregulating the JAK1/STAT1/STAT3 pathway in myoblasts (our unpublished data). Therefore, we decided to focus on SOCS and PIAS proteins. Here, we provide evidence that SOCS1, SOCS3, and PIAS1 are three key regulators in myoblasts that specifically inhibit the LIF-induced JAK1/STAT1/STAT3 pathway via distinct targets; while SOCS1 and SOCS3 selectively target JAK1 and gp130, respectively, PIAS1 targets mainly the activated STAT1 and prevents it from binding to DNA. Thus, our current study has provided a clear molecular mechanism to explain how the LIF-induced JAK1/STAT1/STAT3 pathway is downregulated upon myogenic differentiation.
MATERIALS AND METHODS
Cell culture, DNA constructs, and reagents.
C2C12 cells (ATCC, Manassas, VA) were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 20% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin (growth medium [GM]) in a 37°C incubator with 5% CO2. To induce differentiation, near-confluent cells were grown in DMEM with 2% horse serum (differentiation medium [DM]). Constructs encoding Flag-SOCS1, Flag-SOCS2, and Flag-SOCS3 (mouse) were kindly provided by G. Zhu (Hong Kong University of Science and Technology, Hong Kong, China). Myc-PIAS1 and Flag-PIAS1(C350S) were gifts from S. C. Lin (Xiamen University, China) and Cory Abate-Shen (Columbia University, NY), respectively. G133-Luc, MCK-Luc, 4RE-Luc, and 3MEF2-Luc were described previously (36). LIF was purchased from Millipore (Billerica, MA).
RNA interference.
For siRNA transfection, 40 to 60% confluent C2C12 cells were transfected with 100 nM siRNA by Lipofectamine 2000 following the manufacturer's instructions (Invitrogen Carlsbad, CA). All siRNAs were designed at Dharmacon, Inc. (Thermo Scientific) and synthesized at RiboBio Co. Ltd. (Guangzhou, China). Their sequences were listed as follows (only the top-strand sequences were shown, from 5′ to 3′): SOCS1 (siRNA set 1, CTACCTAGTTCCTTCCCCTT; siRNA set 2, GAGACCTTCGACTGCCTTT), SOCS3 (1, GACCCAGTCTGGGACCAAG; 2, GGAGCAAAAGGGUCAGAGG), PIAS1 (1, AATCCG GATCATTCTAGAGCT; 2, CTTCAGAGGTTACGAGCAA), and enhanced green fluorescent protein (GFP) (GCTGACCCTGAAGTTCATC).
Transfection and cell lysis.
Transient transfection of plasmids was performed by using Lipofectamine Plus reagents from Invitrogen as instructed by the manufacturer. Cells were lysed in the lysis buffer (50 mM HEPES, pH 7.6, 10% glycerol, 1% Triton X-100, 150 mM NaCl, 1 mM EGTA, 1.5 mM MgCl2, 100 mM NaF, 20 mM p-nitrophenyl phosphate, 20 mM β-glycerol phosphate, 2 mM dithiothreitol, 50 μM sodium vanadate, 0.5 mM phenylmethylsulfonyl fluoride, 2 μg/ml aprotinin, 0.5 μg/ml leupeptin, 0.7 μg/ml pepstatin) for 10 min on ice. Soluble whole-cell extracts (WCE) were prepared by centrifugation, followed by removal of cell debris.
Antibodies and Western blot analysis.
Antibodies against myogenin, MEF2, MyoD, p21Cip1, p27Kip1, gp130, LIFR, JAK1, STAT1, STAT3, SOCS3, PIAS1, anti-Myc, and β-actin were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA); anti-phospho-JAK1 (Tyr1022/Tyr1023), anti-phopho-STAT1 (Tyr705), and anti-phospho-STAT3 (Tyr705) were from Cell Signaling (Danvers, MA); anti-SOCS1 was from Immuno-Biological Laboratories Co. Ltd. (Gumma, Japan); anti-Flag (M2) was from Sigma-Aldrich (St. Louis, MO); anti-phospho-tyrosine (4G10) was from Millipore; anti-MHC (MF20) was from the Developmental Studies Hybridoma Bank (Iowa City, IA); and polyclonal STAT3 antibody was a gift from Z. Wen (Hong Kong University of Science and Technology). Western blotting was conducted as previously described (36).
Reporter assays.
For reporter assays, 50% to 70% confluent C2C12 cells in 12-well dishes were cotransfected with 0.5 μg of a reporter plasmid and 100 nM of an siRNA by Lipofectamine 2000. All the samples were prepared in triplicate. The luciferase activity was determined with a LB9507 luminometer (Berthold Technologies, Bad Wilbad, Germany) by adding 10 μl of WCE to 150 μl of freshly made luciferase buffer (0.4 μM luciferin, 13.3 mM ATP, 0.1 M Tris-HCl, pH 7.8, 1 mM EDTA, pH 8.0, 10 mM MgOAc2). Luciferase units were normalized against the total protein amount present in each sample as determined by using protein assay reagent from Bio-Rad (Hercules, CA).
Immunoprecipitation and coimmunoprecipitation assays.
For immunoprecipitation, 1 to 2 μg of an antibody was incubated with 0.3 to 1 mg of C2C12 WCE together with 30 μl (bed volume) of protein A-Sepharose beads (GE Healthcare) at 4°C overnight with rotation. For coimmunoprecipitation, C2C12 cells were first cross-linked with 200 μg/ml of dithiobis(succinimidyl propionate) (Thermo Scientific) for 5 min and then harvested and lysed in radioimmunoprecipitation assay (RIPA) buffer (25 mM HEPES, pH 7.4, 1% Nonidet P-40, 0.1% sodium dodecyl sulfate, 0.5% sodium deoxycholate, 0.5 mM phenylmethylsulfonyl fluoride, 2 μg/ml aprotinin, 0.5 μg/ml leupeptin, 0.7 μg/ml pepstatin). A total of 30 μl (bed volume) of protein G-Sepharose 4B beads (Invitrogen) was incubated with 500 μg of WCE and 1 μg of an appropriate antibody in RIPA buffer overnight at 4°C with rotation. After extensive washing with the lysis buffer (for immunoprecipitation) or RIPA buffer (for coimmunoprecipitation), the bound proteins were eluted by boiling in 1× sodium dodecyl sulfate sample buffer and subjected to Western blot analysis.
Immunostaining and microscopic imaging.
Cells were fixed in 4% paraformaldehyde for 15 min, permeabilized by 0.2% Triton X-100 in phosphate-buffered saline (PBS) for another 15 min, and incubated with a specific primary antibody overnight at 4°C. After removal of the primary antibody followed by repeated washing, a fluorescein isothiocyanate- or rhodamine-conjugated second antibody was incubated with cells for 1 h at room temperature. 4′,6′-Diamidino-2-phenylindole (DAPI) was used to counterstain the nuclei. The images were captured by a fluorescent microscope (IX70; Olympus) coupled with a charge-coupled device camera. Images were subsequently processed by SPOT software (Diagnostic Instruments, Sterling Heights, MI) and Photoshop 6.0 (Adobe).
RNA preparation and RT-PCR.
C2C12 cells were lysed by TRIzol (Invitrogen), and total RNA was subsequently isolated following the manufacturer's protocol. Reverse transcription-PCR (RT-PCR) was performed by using a two-step method. First, single-stranded cDNA was generated from 0.5 μg of total RNA by the ImProm-II RT system (Promega, Madison, WI) with oligo(dT) as a primer according to the manufacturer's instructions. Amplification of DNA was performed in a thermocycler with 40 ng of cDNA in a total volume of 25 μl. PCR products were analyzed by using 1% agarose gel electrophoresis. The sequences of primers for each gene are listed as follows: SOCS1 forward (5′-ATGGTAGCACACAACCAGGTG-3′), SOCS1 reverse (5′-TCAAATCTGGAAGGGGAAGGA-3′); SOCS3 forward (5′-CTCAAGACCTTCAGCTCCAA-3′), SOCS3 reverse (5′ TTCTCATAGGAGTCCAGGTG-3′); PIAS1 forward (5′-GCAGGTCCAATTAAGGTTTTG-3′), PIAS1 reverse (5′-GTAACCTGGAAGGCTGCAAG-3′); myogenin forward (5′-CAGGAGATCATTTGCTCG-3′), myogenin reverse (5′-GGGCATGGTTTCGTCTGG-3′); and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) forward (5′-TGATGCTGGTGCTGAGTATGTCGTG-3′), GAPDH reverse (5′-TCCTTGGAGGCCATGTAGGCCAT-3′).
EMSA.
An electrophoretic mobility shift assay (EMSA) was performed as described previously (5). The top strand sequence of the sis-inducible element probe is 5′-GTCGACATTTCCCGTAAATC-3′.
Preparation of primary myoblasts.
Primary myoblasts were isolated as described previously with minor modifications (1, 32). Briefly, skeletal muscles from 10- to 15-day-old C57BL/6J mice were dissected, minced, and incubated at 37°C in a shaking water bath with 1 mg/ml Pronase (Calbiochem) in DMEM for 1.5 h. After two to three cycles of centrifugation and resuspension, myoblast-containing solutions were passed through a 40-μm cell sieve (BD Biosciences) to remove debris. Myoblasts were further enriched by differential sedimentation, in which fibroblasts were allowed to attach to uncoated tissue culture dishes for 1 h while myoblasts in supernatant were removed and transferred to another dish precoated with 4 mg/ml Matrigel (BD Biosciences). Myoblasts were maintained in GM (Ham's F10 medium with 20% FBS, 2.5 ng/ml recombinant human basic fibroblast growth factor, and 2% chicken embryo extracts) and induced to differentiate in DM (DMEM with 5% horse serum).
Cell infection by adenovirus.
The BLOCK-iT adenoviral expression system (Invitrogen) was used to generate adenoviruses following the manufacturer's instructions. To facilitate tracking of the infected cells, we modified the pENTR/U6 entry vector by inserting a fragment (obtained from pEGFP-N1) encoding enhanced GFP together with an immediate early promoter of cytomegalovirus (CMV) and SV40 polyadenylation signals in a two-step PCR with the following primers: CMV-GFP forward (5′-GGAACCAATTCAGTCGACTGGAATTAATAGTAATCAATTACGG-3′), GFP-SV40 poly(A) reverse (5′-ACGCGTTAAGATACATTGATGAG-3′), pU6-attL1 forward (5′-CTAGACCCAGCTTTCTTGTAC-3′), and pU6-attL2 reverse (5′-CCGTAATTGATTACTATTAATTCCAGTCGACTGAATTGGTTCC-3′). Then SOCS1, SOCS3, and PIAS1 cDNAs were subcloned into the modified viral entry vector. Primary myoblasts were infected with viruses at a multiplicity of infection of 50 for 1.5 h. After the removal of viruses, cells were allowed to differentiate in DM for another 24 h before being fixed, permeabilized, and subjected to immunostaining.
RESULTS
The expression profiles of SOCS and PIAS during myogenic differentiation.
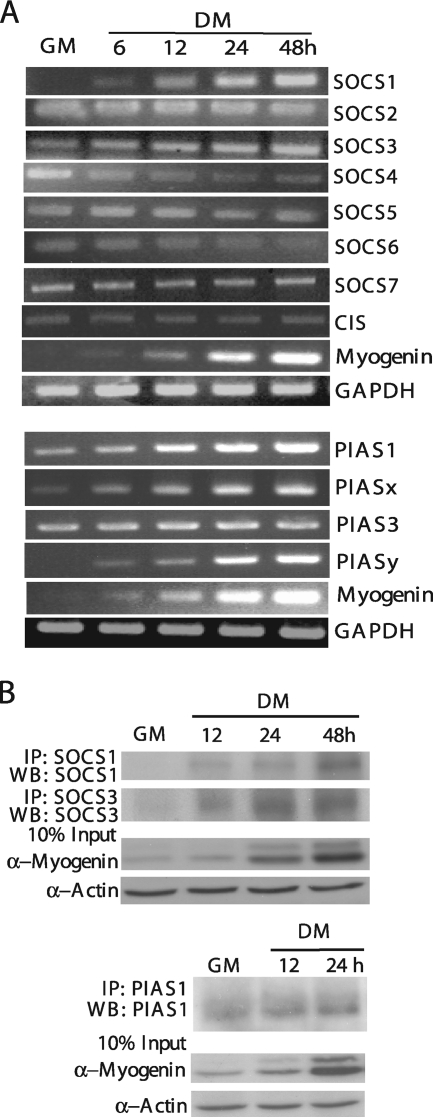
In both human and mouse genomes, there are eight different SOCS genes and four different PIAS genes (8, 34). To explore the potential involvement of different SOCS and PIAS proteins in myogenic differentiation, we first examined the mRNA expression pattern of these genes in C2C12 cells before and after differentiation by using RT-PCR. As shown in Fig. 1A, among eight different SOCS genes, the mRNA levels of SOCS1 and SOCS3 gradually increased during differentiation. The mRNA levels of SOCS2, SOCS5, SOCS7, and CIS remained relatively constant, while those of SOCS4 and SOCS6 decreased during differentiation. As for the members of the PIAS family, the mRNA levels of PIAS1, PIASx, and PIASy gradually increased during differentiation, whereas that of PIAS3 remained constant. Because the aim of the current study was to identify the negative regulators of the LIF-induced JAK1/STAT1/STAT3 pathway, we decided to focus on those SOCS and PIAS members (i.e., SOCS1, SOCS3, PIAS1, PIASx, and PIASy) whose expression levels increased during differentiation. As the knockdown of PIASx and PIASy had no obvious effect on myogenic differentiation (our unpublished data), we focused mainly on PIAS1 in the subsequent studies. By using Western blot analysis, we showed that the protein levels of SOCS1, SOCS3, and PIAS1 also increased during C2C12 differentiation (Fig. 1B), which was consistent with the expression pattern of their mRNA.
FIG. 1.
The expression profiles of SOCS and PIAS during myogenic differentiation. (A) Total RNA was isolated from C2C12 cells harvested at different time points as indicated and then subjected to semiquantitative RT-PCR analysis to detect the mRNA levels of eight SOCS family members and four PIAS family members. (B) WCE was prepared from C2C12 cells harvested at different time points as indicated. SOCS1, SOCS3, and PIAS1 proteins were separately immunoprecipitated (IP) from 500 μg of WCE and then subjected to Western blotting (WB) with the same set of antibodies for detection. For both (A) and (B), myogenin was used as an early differentiation marker. Several independent experiments were performed with similar results, and a representative is shown.
Individual knockdown of SOCS1, SOCS3, or PIAS1 inhibits differentiation in both primary myoblasts and immortalized myogenic cells.
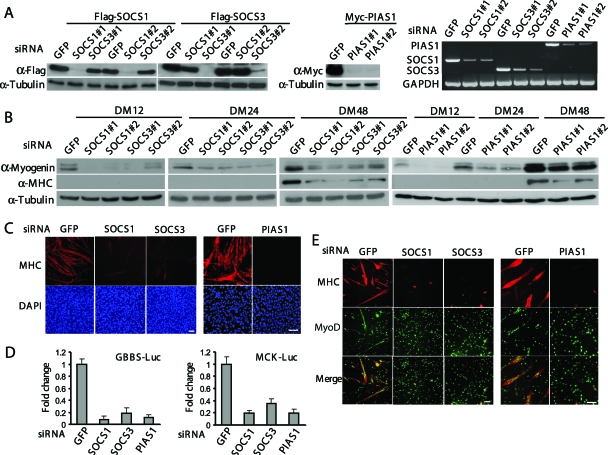
To further explore the function of SOCS1, SOCS3, and PIAS1 in myogenic cells, we decided to use the RNA interference technique to individually knock down these genes. For each gene, we designed two siRNAs to target different regions of the gene. All siRNAs used were found to be effective and specific in knocking down their intended target genes, as judged by both Western blot analysis and RT-PCR (Fig. 2A). Most importantly, when these siRNAs were separately transfected into C2C12 cells with an siRNA against the green fluorescent protein gene (GFP) as a control, they all inhibited the expression of myogenin and MHC that are two representative differentiation-specific proteins often used as an early and late differentiation marker, respectively (Fig. 2B). Since two different sets of siRNAs for each gene showed very similar effects, to avoid unnecessary duplication we would present only data obtained with one set of siRNAs in the following sections. We then examined the morphology of the siRNA-transfected C2C12 cells undergoing differentiation by immunostaining using an antibody (MF20) specifically recognizing the sarcomeric MHC. We found that the cells transfected with the control siRNA (i.e., GFP-siRNA) underwent normal differentiation to form multiple MHC-positive multinucleated myotubes (Fig. 2C). In contrast, the cells transfected with the siRNAs targeting SOCS1, SOCS3, or PIAS1 had a drastically reduced number of MHC-positive myotubes (Fig. 2C). Consistently, we found that knockdown of SOCS1, SOCS3, or PIAS1 considerably repressed the activity of two luciferase reporter genes controlled by the mouse myogenin gene promoter (i.e., GBBS-Luc) and the muscle creatine kinase gene (MCK) promoter (i.e., MCK-Luc), respectively (Fig. 2D). Next, we asked whether these siRNAs had similar effects in primary myoblasts. We isolated mouse primary myoblasts and transfected them with different siRNAs. By immunostaining for MHC, we found that cells transfected with an siRNA against SOCS1, SOCS3, or PIAS1 were indeed defective in myogenic differentiation, as indicated by a drastically reduced number of MHC-positive multinucleated myotubes (Fig. 2E). Thus, our siRNA-based data suggest that SOCS1, SOCS3, and PIAS1 are all required for myogenic differentiation.
FIG. 2.
Individual knockdown of SOCS1, SOCS3, or PIAS1 genes inhibits differentiation both in primary myoblasts and C2C12 cells. (A) C2C12 cells were transfected with either siRNA alone or siRNA plus cDNA expression vectors as indicated. Cells were harvested 24 h after transfection. Either WCE or total RNA was prepared and subjected to Western blot analysis or RT-PCR. #1 and #2, different sets of siRNAs. (B and C) C2C12 cells were transfected with various siRNAs as indicated. (B) WCE was collected at different time points. Fifty micrograms of WCE was subjected to Western blot analysis with various antibodies. (C) Twenty-four hours after siRNA transfection, C2C12 cells were cultured in DM for another 48 h before being fixed and subjected to immunostaining with an anti-MHC antibody (MF20). The nuclei were counterstained with DAPI. Bars, 100 μm. (D) C2C12 cells were cotransfected with siRNA and luciferase reporter constructs in different combinations as indicated. Twenty-four hours after transfection, cells were allowed to grow in DM for another 24 h (for GBBS-Luc) or 48 h (for MCK-Luc) before being harvested. WCE was collected and subjected to luciferase assays. All the samples were prepared in triplicate, and the results are presented as the mean ± the standard deviation (error bars). The change was calculated as the ratio of the luciferase activity in SOCS (or PIAS)-siRNA-transfected cells to that in GFP-siRNA-transfected cells. (E) Freshly isolated primary myoblasts were separately transfected with SOCS1-, SOCS3-, or PIAS1-siRNA. Cells were fixed after 24 h in DM and subjected to double immunostaining with anti-MyoD and anti-MHC antibodies. Bars, 100 μm.
Overexpression of SOCS1, SOCS3, or PIAS1 promotes myogenic differentiation.
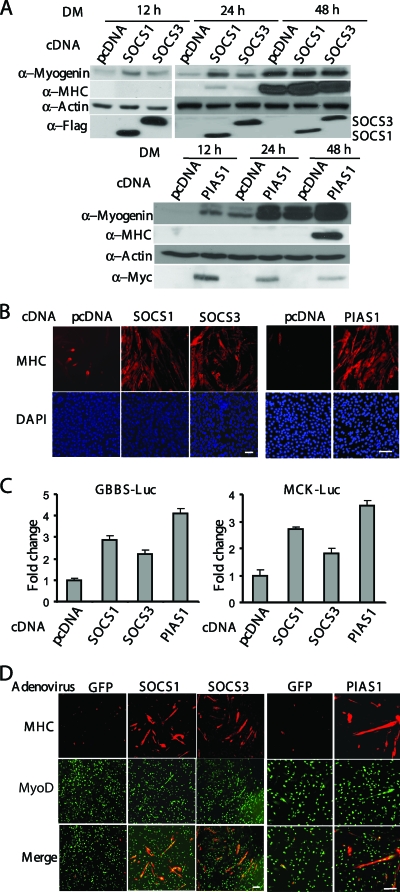
As a complementary approach to our siRNA-based experiments, we first individually overexpressed SOCS1, SOCS3, or PIAS1 in C2C12 cells. By using Western blot analysis, we found that overexpression of PIAS1 or SOCS1 promoted myogenic differentiation, as evidenced by increased expression of myogenin and MHC in cells transfected with PIAS1 or SOCS1 (Fig. 3A). Although SOCS3 had less of a stimulatory effect than SOCS1 compared to the control (i.e., cells transfected with an empty vector), its stimulatory effect on myogenin expression could still be observed (Fig. 3A). These observations were confirmed by both MHC-based immunostaining (Fig. 3B) and GBBS-Luc-based or MCK-Luc-based reporter assays (Fig. 3C). Furthermore, we also separately infected the mouse primary myoblasts with recombinant adenoviruses expressing GFP (negative control), SOCS1, SOCS3, or PIAS1. Consistently, we found that overexpression of SOCS1, SOCS3, or PIAS1 accelerated myogenic differentiation in primary myoblasts (Fig. 3D).
FIG. 3.
Overexpression of SOCS1, SOCS3, or PIAS1 promotes myogenic differentiation. (A and B) C2C12 cells were transiently transfected with pcDNA3.1, SOCS1, SOCS3 or PIAS1 cDNA construct. (A) Fifty micrograms of WCE harvested at different time points was subjected to Western blot analysis. (B) After 48 h in DM, C2C12 cells were fixed and subjected to immunostaining for MHC. (C) C2C12 cells were cotransfected with various cDNA and luciferase reporter constructs as indicated. Twenty-four hours after transfection, cells were switched to DM for another 24 h (for GBBS-Luc) or 48 h (for MCK-Luc) before harvest. WCE was prepared and subjected to luciferase assays. All the samples were prepared in triplicate, and the results are presented as the mean ± the standard deviation (error bars). The activation was calculated as the ratio of the luciferase activity in SOCS/PIAS- transfected cells to that in pcDNA3.1-transfected cells. (D) Primary myoblasts were separately infected with adenovirus expressing GFP, SOCS1, SOCS3, or PIAS1. Two days after infection, myoblasts were switched to DM for another 24 h before being fixed and subjected to immunostaining for MHC and MyoD. Bars, 100 μm.
SOCS1, SOCS3, and PIAS1 promote myogenic differentiation by inhibiting the LIF-mediated JAK1/STAT1/STAT3 pathway.
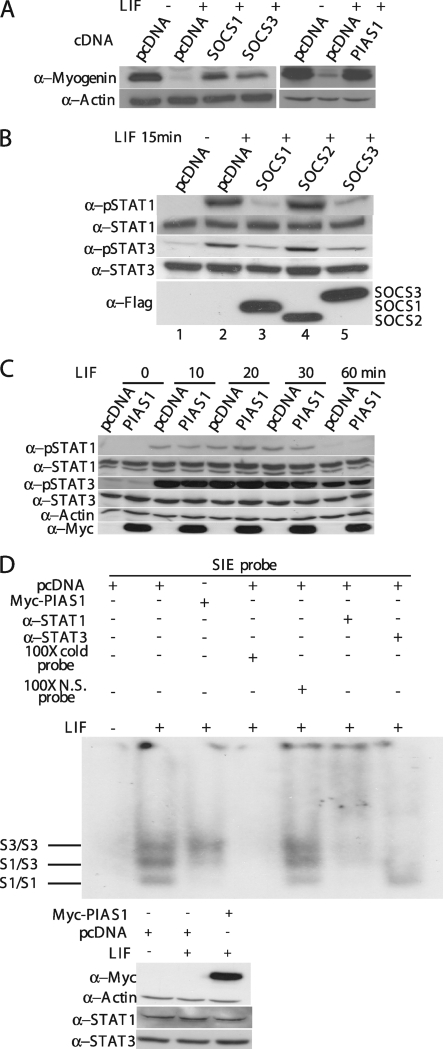
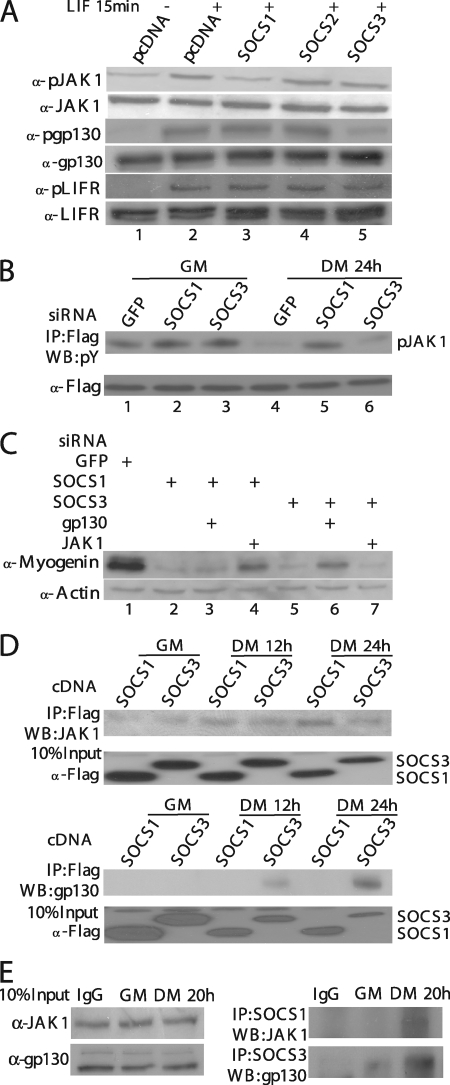
We previously showed that the LIF-induced JAK1/STAT1/STAT3 pathway potently represses myogenic differentiation and needs to be downregulated at the onset of differentiation (36). The promyogenic effect of SOCS1, SOCS3, and PIAS1 described above suggests that these molecules could function by specifically targeting the JAK1/STAT1/STAT3 pathway. Indeed, we found that LIF-mediated repression of myogenin expression could be partially rescued by overexpression of SOCS1, SOCS3, or PIAS1 (Fig. 4A). To examine whether the LIF-induced activation of STAT1 and STAT3 was affected by SOCS1, SOCS3, and PIAS1, we first transfected C2C12 cells with an empty vector or with individual expression vectors encoding SOCS1, SOCS2, or SOCS3, followed by LIF treatment for 15 min. As expected, LIF treatment activated STAT1 and STAT3 and led to the formation of their tyrosine-phosphorylated forms (Fig. 4B, lane 2) (36). Overexpression of SOCS1 and SOCS3, but not SOCS2, abolished LIF-induced tyrosine phosphorylation of STAT1 and STAT3 (Fig. 4B, lanes 3 and 5). Unlike SOCS1 and SOCS3, the overexpressed PIAS1 had no obvious effect on LIF-induced phosphorylation of STAT1 and STAT3 (Fig. 4C). As PIAS was capable of inhibiting activated STATs by preventing them from binding to DNA (34), we then examined whether LIF-induced STAT1/3 binding to DNA was affected by overexpression of PIAS1. As shown in Fig. 4D, LIF treatment induced the formation of three distinct complexes: the STAT1 homodimer, the STAT3 homodimer, and the STAT1/3 heterodimer (7). Overexpression of PIAS1 selectively abolished DNA binding by the STAT1-containing complexes (i.e., the STAT1 homodimer and STAT1/3 heterodimer) without affecting DNA binding by the STAT3 homodimer. As a control, we also showed that overexpression of PIAS1 did not affect the expression level of either STAT1 or STAT3 (Fig. 4D, bottom).
FIG. 4.
SOCS1, SOCS3, and PIAS1 promote myogenic differentiation by inhibiting the LIF-mediated JAK1/STAT1/STAT3 pathway. (A to D) C2C12 cells were separately transfected with various cDNAs as indicated and cultured in GM for 24 h. (A) Cells were switched to DM for 24 h with or without 10 ng/ml of LIF. (B) Cells were treated with LIF (10 ng/ml) for 15 min before being harvested. (C) Cells were treated with LIF (10 ng/ml) for various times as indicated. WCE (60 μg) as shown in panels A, B, and C was subjected to Western blot analysis with various antibodies as indicated. (D) Cells were treated with buffer (i.e., PBS) or LIF (10 ng/ml) for 15 min, and WCE was prepared. WCE (20 μg) was subjected to EMSA (top) using a double-stranded oligonucleotide containing the sis-inducible element (SIE) as a probe. The expression levels of the endogenous STAT1 and STAT3 and the exogenous Myc-PIAS1 in various samples used in the EMSA were revealed by using Western blot analysis (bottom). p-STAT1/STAT3, phospho-STAT1/STAT3; N.S., nonspecific; S1/S1 and S3/S3, STAT1 and STAT3 homodimers; S1/S3, STAT1/STAT3 heterodimer.
SOCS1 and SOCS3 target JAK1 and gp130, respectively.
To understand mechanistically how SOCS1 and SOCS3 inhibit the LIF-induced activation of STAT1 and STAT3, we took the lysates from the experiment shown in Fig. 4B and analyzed the activation status of JAK1, LIFR, and gp130, as these are essential upstream molecules indispensable for the LIF-induced STAT1/3 activation (15, 25, 36). As shown in Fig. 5A, treatment of C2C12 cells with LIF induced phosphorylation of JAK1, gp130, and LIFR (Fig. 5A, lane 2). Interestingly, the LIF-induced phosphorylation of JAK1 was selectively inhibited by overexpressed SOCS1 (Fig. 5A, lane 3), while that of gp130 was preferentially abolished by overexpressed SOCS3 (Fig. 5A, lane 5). None of the three overexpressed SOCSs had any effect on the LIF-induced phosphorylation of LIFR. Moreover, three additional pieces of evidence support our notion that SOCS1 and SOCS3 selectively target JAK1 and gp130, respectively. First, to examine the kinase activity of JAK1 before and after differentiation, we transfected C2C12 cells with a vector encoding Flag-JAK1 together with an siRNA against GFP, SOCS1, or SOCS3 and then measured the JAK1 kinase activity before and after differentiation. In control cells transfected with GFP-siRNA, the kinase activity of Flag-JAK1 significantly decreased upon differentiation (Fig. 5B, compare lanes 1 and 4), which was consistent with our previous findings (36). Knockdown of SOCS1 effectively prevented the reduction of JAK1 kinase activity upon differentiation (Fig. 5B, compare lanes 2 and 5), while knockdown of SOCS3 had no such effect (Fig. 5B, compare lanes 3 and 6). Second, to further understand the connection of different SOCS proteins with JAK1 and gp130, we transfected C2C12 cells with siRNAs against GFP, SOCS1, SOCS3, JAK1, and gp130 in different combinations. Consistent with that shown in Fig. 2, knockdown of either SOCS1 or SOCS3 alone drastically reduced myogenin expression levels compared to cells transfected with GFP-siRNA (Fig. 5C, lanes 2 and 5). Simultaneous knockdown of SOCS1 and gp130, or SOCS3 and JAK1, could not reelevate myogenin expression (Fig. 5C, lanes 3 and 7). In contrast, simultaneous knockdown of SOCS1 and JAK1, or SOCS3 and gp130, partially rescued myogenin expression (Fig. 5C, lanes 4 and 6). Third, to study the physical interaction of different SOCS proteins with JAK1 and gp130, we transfected C2C12 cells separately with Flag-tagged SOCS1 and SOCS3. At different time points before or after differentiation, we harvested cells and subjected equal amounts of the whole-cell lysates to immunoprecipitation with the anti-Flag antibody (M2), followed by immunoblotting for the endogenous JAK1 and gp130. As shown in Fig. 5D, we found that the endogenous JAK1 preferentially coprecipitated with SOCS1 (Fig. 5D, top), while the endogenous gp130 specifically coprecipitated with SOCS3 (Fig. 5D, bottom). Interestingly, both pairs of interaction were detected mainly in the lysates prepared from differentiating cells (i.e., Fig. 5D, DM 12 h and 24 h) but not from proliferating cells (i.e., Fig. 5D, GM). Similar results were also obtained by directly immunoprecipitating the endogenous SOCS1 and SOCS3 from cell lysates prepared from nontransfected C2C12 cells before or after differentiation (Fig. 5E). Our data described above strongly suggested that SOCS1 selectively targets JAK1, whereas SOCS3 preferentially targets gp130 upon myogenic differentiation.
FIG. 5.
SOCS1 and SOCS3 target JAK1 and gp130, respectively. (A) C2C12 cells transiently transfected with various plasmids were treated with LIF (10 ng/ml) for 15 min before harvest. WCE (50 μg) was subjected to Western blot analysis (WB) for various molecules as indicated. (B) WCE from C2C12 cells cotransfected with various siRNAs and Flag-JAK1 were prepared at different time points as indicated. Flag-JAK1 was immunoprecipitated (IP) from 500 μg of WCE and subjected to Western blot analysis with an anti-P-tyrosine antibody (4G10). (C) C2C12 cells were transfected with siRNAs in different combinations as indicated. After 24 h in GM, cells were switched to DM for 24 h before being harvested. WCE was prepared, and the expression level of myogenin was revealed by Western blot analysis. (D) C2C12 cells were first transfected with constructs encoding Flag-SOCS1 or Flag-SOCS3 and cultured in GM for 24 h. Cells were then switched to DM for 12 or 24 h. Flag-SOCS1 or SOCS3 was immunoprecipitated from 500 μg of WCE. (E) An equal amount of WCE (400 μg), which was prepared from C2C12 cells grown in GM or DM for 20 h, was subjected to immunoprecipitation using specific antibodies against either SOCS1 or SOCS3. The rabbit preimmune immunoglobulin G (IgG) was used as a negative control. After extensive washing, the immunoprecipitates shown in panels D and E were subjected to Western blot analysis with antibodies against JAK1 and gp130.
SOCS1, SOCS3, and PIAS1 regulate the expression of MyoD, MEF2, p21Cip1, and p27Kip1.
We previously showed that the JAK1/STAT1/STAT3 pathway regulates myoblast proliferation and differentiation by selectively regulating the expression of the genes p21Cip1, p27Kip1, MyoD, and MEF2 (36). If SOCS1, SOCS3, and PIAS1 indeed target this pathway in myoblasts, one would expect that the same set of target genes will be affected when we deliberately alter the expression levels of SOCS1, SOCS3, and PIAS1. To test whether this is indeed the case, we first separately transfected C2C12 cells with siRNAs against GFP, SOCS1, SOCS3, or PIAS1 and then let cells differentiate for various times. As shown in Fig. 6A, individual knockdown of SOCS1, SOCS3, or PIAS1 led to reduced expression of the genes MEF2, MyoD, p21Cip1, and p27Kip1. Consistently, when we individually overexpressed SOCS1, SOCS3, or PIAS1 in C2C12 myoblasts, we detected increased expression of genes MEF2 and MyoD (Fig. 6B). Alteration of the expression levels of SOCS1, SOCS3, and PIAS1 by either siRNA-mediated gene knockdown or overexpression also had similar effects on the activity of the MRF- and MEF2-dependent reporter genes (i.e., 4RE-Luc and 3xMEF2-Luc) (Fig. 6C).
FIG. 6.
SOCS1, SOCS3, and PIAS1 regulate the expression of MyoD, MEF2, p21Cip1, and p27Kip1. (A) WCE was collected at the indicated time points from C2C12 cells transfected with various siRNAs. Thirty micrograms of WCE was subjected to Western blot analysis with various antibodies. (B) C2C12 cells were transiently transfected with pcDNA3.1, SOCS1, SOCS3, or PIAS1 constructs. Fifty micrograms of WCE prepared at different time points was subjected to Western blot analysis for detection of MEF2 and MyoD. (C) C2C12 cells in triplicate were cotransfected with either siRNAs or plasmids together with luciferase reporter constructs (3xMEF2-Luc or 4RE-Luc). Twenty-four hours after transfection, cells were cultured in DM for another 24 h before being harvested. The WCE was subjected to luciferase assays. Results are presented as the mean ± the standard deviation (error bars). The change was calculated the same way as described in legends for Fig. 2D and 3C.
The SUMO E3 ligase activity of PIAS1 is not required for its myogenic effect.
PIAS1 can serve as a SUMO E3 ligase, with both cysteine 350 (i.e., C350) and tryptophan 372 (i.e., W372) in the RING domain being essential for its SUMO E3 ligase activity (22). To test whether the SUMO E3 ligase activity of PIAS1 is required for its myogenic effect, we transfected C2C12 cells with vectors encoding either an empty vector or the wild-type PIAS1, PIAS1(C350S), or PIAS1(W372A), the latter two being the SUMO E3 ligase-defective mutants of PIAS1 (22), and let the cells differentiate for 12 or 24 h. As shown in Fig. 7A, at both time points, both PIAS1(C350S) and PIAS1(W372A) were as effective as the wild-type PIAS1 in enhancing the expression of myogenin. As PIAS1 had been implicated in sumoylating STAT1 (38), it prompted us to test whether PIAS1 could promote STAT1 sumoylation in C2C12 cells. As a positive control, we first showed that PIAS1 could promote p53 sumoylation in C2C12 cells cotransfected with p53, PIAS1, SUMO1, and Ubc9 (i.e., the SUMO E2) (Fig. 7B, left) (18). Two SUMO E3 ligase-defective PIAS1 mutants (i.e., C350S and W372A) failed to promote p53 sumoylation. Importantly, under the same conditions, no STAT1 sumoylation could be detected in C2C12 cells with or without LIF stimulation (Fig. 7B, right). Our data suggested that the SUMO E3 ligase activity of PIAS1 is not essential for its myogenic effect.
FIG. 7.
The SUMO E3 ligase activity of PIAS1 is not required for its myogenic effect. (A) C2C12 cells transiently transfected with pcDNA3.1, wild-type (wt) PIAS1, PIAS1(C350S), or PIAS1(W372A) plasmids were induced to differentiate for 12 or 24 h before being harvested. (B) C2C12 cells were cotransfected with various plasmids as indicated and harvested 24 h after transfection. WCE as shown in panels A and B was prepared and subjected to Western blot analysis with various antibodies. CS, C350S; WA, W372A. (C) A schematic drawing showing the differential activation status of the LIF-induced JAK1/STAT1/STAT3 pathway in proliferating myoblasts (left) and differentiating myocytes/myotubes (right) and the distinct targets of SOCS1, SOCS3, and PIAS1.
DISCUSSION
Distinct targets of SOCS1, SOCS3, and PIAS1 in myogenic differentiation.
We show in this report that SOCS1, SOCS3, and PIAS1 promote myogenic differentiation by targeting distinct molecules of the LIF-induced JAK1/STAT1/STAT3 pathway (Fig. 7C). In addition, we find that both SOCS1 and SOCS3 genes, but not PIAS1, are induced by LIF in a STAT1- and STAT3-dependent manner (our unpublished data). Our findings are consistent with the current knowledge about these molecules (8, 34). For example, although both SOCS1 and SOCS3 contain a similar kinase inhibitory region N-terminal to their SH2 domains, only SOCS1 is known to directly inhibit JAK (9, 28). SOCS3, on the other hand, preferentially associates with gp130 and other cytokine receptors (8, 27). Moreover, both SOCS1 and SOCS3 are indeed known to be rapidly induced by cytokines in a STAT-dependent manner by a classical feedback inhibition mechanism. Questions may arise as to why three different negative regulators are needed to shut down the same pathway. An obvious answer is that the coordinated action of these three molecules could achieve maximal inhibition efficiency; while SOCS1 and SOCS3 associate with and inhibit JAK1 and gp130, respectively, near the plasma membrane to prevent cytoplasmic STATs from being activated, PIAS1 specifically targets the existing activated STATs in the nucleus to ensure that the pathway can be effectively turned off. Another possible answer could be that each of the three negative regulators just exerts a partial inhibitory effect in cells without being overexpressed. Only with the combined action of all three negative regulators can a cytokine (e.g., LIF)-induced JAK/STAT pathway be completely inhibited.
Involvement of SOCS1, SOCS3, and PIAS1 in myogenic differentiation.
Our current study identifies SOCS1, SOCS3, and PIAS1 as key molecules that promote myogenic differentiation by inhibiting the LIF-induced JAK1/STAT1/STAT3 pathway. For SOCS3, its promyogenic effect seen by us is consistent with that in a previous report (35). However, in that report, SOCS3 was studied mainly in the context of IGF1 stimulation: the authors showed that the SOCS3 promoter was induced by IGF1, but they did not show how SOCS3 promoted myogenic differentiation mechanistically. In addition, most of the data in the report were obtained based on the overexpression of SOCS3.
For PIAS1, there are a few conflicting reports in the literature. In smooth muscle cells, PIAS1 was found to promote differentiation by directly interacting with and activating E12, E2-2, and the serum response factor (19). However, the molecular basis for such an activating effect of PIAS1 remains unclear. It is possible that PIAS1 may have multiple functions: in addition to its inhibitory effect on the JAK/STAT pathway, it may also function, independently of the JAK/STAT pathway, by interacting with other cellular proteins to modulate their activities. Indeed, in skeletal muscle cells, PIAS1 was shown to interact with either SnoN or Msx1 that in turn may regulate myogenic differentiation (16, 22, 42). In the case of SnoN, PIAS1 was found to interact with and directly sumoylate SnoN, which enhances the ability of SnoN to repress myogenin gene expression and myogenic differentiation (16, 42). The SUMO E3 ligase activity of PIAS1 was shown to be required for SnoN to function in myogenic differentiation. In the case of Msx1, PIAS1 was also shown to interact with and sumoylate Msx1, which confers DNA-binding specificity on Msx1 to allow it to bind to a defined region in the MyoD promoter and to repress MyoD gene expression (22). A point that differs from the SnoN case is that the SUMO E3 ligase activity of PIAS1 was found to be dispensable here for Msx1 to function in myogenic differentiation. Another confusing point is about the effect of small hairpin RNAs of PIAS1 (PIAS1-shRNA) on myogenin gene expression: whereas one report found that a PIAS1-shRNA slightly enhanced myogenin promoter-driven luciferase reporter activity (16), the other showed that it had no obvious effect on myogenin expression when used alone (22). In our experience, the transfection efficiency of a plasmid-based shRNA is always lower than that of an oligonucleotide-based siRNA. This may partially account for the discrepancy between these results and ours.
As for SOCS1, it was found to suppress myogenic differentiation based on enhanced differentiation seen in both SOCS1−/− myoblasts and C2C12 cells infected with the SOCS1-expressing adenovirus (17). However, we noticed that there were many more MyoD-positive myoblasts in the SOCS1−/− group than in the control wild-type group in Fig. 3A of the paper by Inaba et al. (17). As the confluence of myoblasts is known to correlate with the extent of differentiation in cell cultures (i.e., the more confluent the myoblasts are, the faster and better they differentiate), the difference in the extent of differentiation observed between SOCS1−/− and wild-type myoblasts could be simply due to a cell confluence issue rather than the absence or presence of SOCS1. In addition, the experimental schemes used by Inaba et al. are quite different from what we used here, which may also contribute to the discrepancy between their results and ours (17). If SOCS1 indeed inhibits differentiation, it is counterintuitive to see that its mRNA and protein levels increase upon differentiation (Fig. 1). Moreover, in our experience, SOCS1 consistently displays more potent promyogenic effects than SOCS3 does in multiple assays (Fig. 3A, 4A, and 6B).
In our study, we have combined both the overexpression approach with the siRNA-mediated knockdown approach, and both sets of results are consistent. Multiple siRNAs against distinct regions of the target genes are employed to minimize the chance of misinterpreting the siRNA-based data due to the potential “off-target” effect. Multiple complementary assays (e.g., Western blotting, immunostaining, RT-PCR, and reporter assays) are also employed to assess the role of SCOS1, SCOS3, and PIAS1 in myogenic differentiation. In addition, both an immortalized myogenic cell line (i.e., C2C12) and primary myoblasts are used to make sure the observed effects are reproducible and not an artifact from the established cell line. Furthermore, the current study complements and extends a previous study from our group (36). Thus, our current work not only helps to fill in a missing gap in our understanding of the regulation of the LIF-induced JAK1/STAT1/STAT3 pathway in myoblasts but also demonstrates that modulation of the activation status of this pathway could significantly influence the outcome of myogenic differentiation.
Acknowledgments
We thank Guang Zhu, Shengcai Lin, and Cory Abate-Shen for SOCS and PIAS1 constructs and Zilong Wen for the STAT3 antibody.
This project was supported by research grants from the Hong Kong Research Grant Council (663007, HKUST6496/06 M, and HKUST1/06C to Z.W.) and an Area of Excellence Scheme (AoE/B-15/01) of the University Grants Council.
Footnotes
Published ahead of print on 20 July 2009.
REFERENCES
- 1.Allen, R. E., C. J. Temm-Grove, S. M. Sheehan, and G. Rice. 1997. Skeletal muscle satellite cell cultures. Methods Cell Biol. 52155-176. [DOI] [PubMed] [Google Scholar]
- 2.Baylies, M. K., and A. M. Michelson. 2001. Invertebrate myogenesis: looking back to the future of muscle development. Curr. Opin. Genet. Dev. 11431-439. [DOI] [PubMed] [Google Scholar]
- 3.Black, B. L., and E. N. Olson. 1998. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell Dev. Biol. 14167-196. [DOI] [PubMed] [Google Scholar]
- 4.Buckingham, M. 2001. Skeletal muscle formation in vertebrates. Curr. Opin. Genet. Dev. 11440-448. [DOI] [PubMed] [Google Scholar]
- 5.Chan, J. K., L. Sun, X. J. Yang, G. Zhu, and Z. Wu. 2003. Functional characterization of an amino-terminal region of HDAC4 that possesses MEF2 binding and transcriptional repressive activity. J. Biol. Chem. 27823515-23521. [DOI] [PubMed] [Google Scholar]
- 6.Chargé, S. B., and M. A. Rudnicki. 2004. Cellular and molecular regulation of muscle regeneration. Physiol. Rev. 84209-238. [DOI] [PubMed] [Google Scholar]
- 7.Chung, C. D., J. Liao, B. Liu, X. Rao, P. Jay, P. Berta, and K. Shuai. 1997. Specific inhibition of Stat3 signal transduction by PIAS3. Science 2781803-1805. [DOI] [PubMed] [Google Scholar]
- 8.Croker, B. A., H. Kiu, and S. E. Nicholson. 2008. SOCS regulation of the JAK/STAT signalling pathway. Semin. Cell Dev. Biol. 19414-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Endo, T. A., M. Masuhara, M. Yokouchi, R. Suzuki, H. Sakamoto, K. Mitsui, A. Matsumoto, S. Tanimura, M. Ohtsubo, H. Misawa, T. Miyazaki, N. Leonor, T. Taniguchi, T. Fujita, Y. Kanakura, S. Komiya, and A. Yoshimura. 1997. A new protein containing an SH2 domain that inhibits JAK kinases. Nature 387921-924. [DOI] [PubMed] [Google Scholar]
- 10.Figliola, R., and R. Maione. 2004. MyoD induces the expression of p57Kip2 in cells lacking p21Cip1/Waf1: overlapping and distinct functions of the two cdk inhibitors. J. Cell Physiol. 200468-475. [DOI] [PubMed] [Google Scholar]
- 11.Florini, J. R., D. Z. Ewton, and S. A. Coolican. 1996. Growth hormone and the insulin-like growth factor system in myogenesis. Endocr. Rev. 17481-517. [DOI] [PubMed] [Google Scholar]
- 12.Franklin, D. S., V. L. Godfrey, H. Lee, G. I. Kovalev, R. Schoonhoven, S. Chen-Kiang, L. Su, and Y. Xiong. 1998. CDK inhibitors p18(INK4c) and p27(Kip1) mediate two separate pathways to collaboratively suppress pituitary tumorigenesis. Genes Dev. 122899-2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo, K., J. Wang, V. Andres, R. C. Smith, and K. Walsh. 1995. MyoD-induced expression of p21 inhibits cyclin-dependent kinase activity upon myocyte terminal differentiation. Mol. Cell. Biol. 153823-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halevy, O., B. G. Novitch, D. B. Spicer, S. X. Skapek, J. Rhee, G. J. Hannon, D. Beach, and A. B. Lassar. 1995. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science 2671018-1021. [DOI] [PubMed] [Google Scholar]
- 15.Heinrich, P. C., I. Behrmann, S. Haan, H. M. Hermanns, G. Muller-Newen, and F. Schaper. 2003. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem. J. 3741-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu, Y. H., K. P. Sarker, I. Pot, A. Chan, S. J. Netherton, and S. Bonni. 2006. Sumoylated SnoN represses transcription in a promoter-specific manner. J. Biol. Chem. 28133008-33018. [DOI] [PubMed] [Google Scholar]
- 17.Inaba, M., H. Saito, M. Fujimoto, S. Sumitani, T. Ohkawara, T. Tanaka, H. Kouhara, S. Kasayama, I. Kawase, T. Kishimoto, and T. Naka. 2005. Suppressor of cytokine signaling 1 suppresses muscle differentiation through modulation of IGF-I receptor signal transduction. Biochem. Biophys. Res. Commun. 328953-961. [DOI] [PubMed] [Google Scholar]
- 18.Kahyo, T., T. Nishida, and H. Yasuda. 2001. Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol. Cell 8713-718. [DOI] [PubMed] [Google Scholar]
- 19.Kawai-Kowase, K., M. S. Kumar, M. H. Hoofnagle, T. Yoshida, and G. K. Owens. 2005. PIAS1 activates the expression of smooth muscle cell differentiation marker genes by interacting with serum response factor and class I basic helix-loop-helix proteins. Mol. Cell. Biol. 258009-8023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keren, A., Y. Tamir, and E. Bengal. 2006. The p38 MAPK signaling pathway: a major regulator of skeletal muscle development. Mol. Cell Endocrinol. 252224-230. [DOI] [PubMed] [Google Scholar]
- 21.Kontaridis, M. I., S. Eminaga, M. Fornaro, C. I. Zito, R. Sordella, J. Settleman, and A. M. Bennett. 2004. SHP-2 positively regulates myogenesis by coupling to the Rho GTPase signaling pathway. Mol. Cell. Biol. 245340-5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee, H., J. C. Quinn, K. V. Prasanth, V. A. Swiss, K. D. Economides, M. M. Camacho, D. L. Spector, and C. Abate-Shen. 2006. PIAS1 confers DNA-binding specificity on the Msx1 homeoprotein. Genes Dev. 20784-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, Y. H., J. S. Jakobsen, G. Valentin, I. Amarantos, D. T. Gilmour, and E. E. Furlong. 2009. A systematic analysis of Tinman function reveals Eya and JAK-STAT signaling as essential regulators of muscle development. Dev. Cell 16280-291. [DOI] [PubMed] [Google Scholar]
- 24.Lluís, F., E. Perdiguero, A. R. Nebreda, and P. Munoz-Canoves. 2006. Regulation of skeletal muscle gene expression by p38 MAP kinases. Trends Cell Biol. 1636-44. [DOI] [PubMed] [Google Scholar]
- 25.Metcalf, D. 2003. The unsolved enigmas of leukemia inhibitory factor. Stem Cells 215-14. [DOI] [PubMed] [Google Scholar]
- 26.Molkentin, J. D., and E. N. Olson. 1996. Defining the regulatory networks for muscle development. Curr. Opin. Genet. Dev. 6445-453. [DOI] [PubMed] [Google Scholar]
- 27.Nicholson, S. E., D. De Souza, L. J. Fabri, J. Corbin, T. A. Willson, J. G. Zhang, A. Silva, M. Asimakis, A. Farley, A. D. Nash, D. Metcalf, D. J. Hilton, N. A. Nicola, and M. Baca. 2000. Suppressor of cytokine signaling-3 preferentially binds to the SHP-2-binding site on the shared cytokine receptor subunit gp130. Proc. Natl. Acad. Sci. USA 976493-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicholson, S. E., T. A. Willson, A. Farley, R. Starr, J. G. Zhang, M. Baca, W. S. Alexander, D. Metcalf, D. J. Hilton, and N. A. Nicola. 1999. Mutational analyses of the SOCS proteins suggest a dual domain requirement but distinct mechanisms for inhibition of LIF and IL-6 signal transduction. EMBO J. 18375-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parker, S. B., G. Eichele, P. Zhang, A. Rawls, A. T. Sands, A. Bradley, E. N. Olson, J. W. Harper, and S. J. Elledge. 1995. p53-independent expression of p21Cip1 in muscle and other terminally differentiating cells. Science 2671024-1027. [DOI] [PubMed] [Google Scholar]
- 30.Potthoff, M. J., and E. N. Olson. 2007. MEF2: a central regulator of diverse developmental programs. Development 1344131-4140. [DOI] [PubMed] [Google Scholar]
- 31.Puri, P. L., and V. Sartorelli. 2000. Regulation of muscle regulatory factors by DNA-binding, interacting proteins, and post-transcriptional modifications. J. Cell Physiol. 185155-173. [DOI] [PubMed] [Google Scholar]
- 32.Rando, T. A., and H. M. Blau. 1994. Primary mouse myoblast purification, characterization, and transplantation for cell-mediated gene therapy. J. Cell Biol. 1251275-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sabourin, L. A., and M. A. Rudnicki. 2000. The molecular regulation of myogenesis. Clin. Genet. 5716-25. [DOI] [PubMed] [Google Scholar]
- 34.Shuai, K., and B. Liu. 2005. Regulation of gene-activation pathways by PIAS proteins in the immune system. Nat. Rev. Immunol. 5593-605. [DOI] [PubMed] [Google Scholar]
- 35.Spangenburg, E. E. 2005. SOCS-3 induces myoblast differentiation. J. Biol. Chem. 28010749-10758. [DOI] [PubMed] [Google Scholar]
- 36.Sun, L., K. Ma, H. Wang, F. Xiao, Y. Gao, W. Zhang, K. Wang, X. Gao, N. Ip, and Z. Wu. 2007. JAK1-STAT1-STAT3, a key pathway promoting proliferation and preventing premature differentiation of myoblasts. J. Cell Biol. 179129-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tapscott, S. J. 2005. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development 1322685-2695. [DOI] [PubMed] [Google Scholar]
- 38.Ungureanu, D., S. Vanhatupa, N. Kotaja, J. Yang, S. Aittomaki, O. A. Janne, J. J. Palvimo, and O. Silvennoinen. 2003. PIAS proteins promote SUMO-1 conjugation to STAT1. Blood 1023311-3313. [DOI] [PubMed] [Google Scholar]
- 39.Wang, K., C. Wang, F. Xiao, H. Wang, and Z. Wu. 2008. JAK2/STAT2/STAT3 are required for myogenic differentiation. J. Biol. Chem. 28334029-34036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weintraub, H. 1993. The MyoD family and myogenesis: redundancy, networks, and thresholds. Cell 751241-1244. [DOI] [PubMed] [Google Scholar]
- 41.Wormald, S., and D. J. Hilton. 2004. Inhibitors of cytokine signal transduction. J. Biol. Chem. 279821-824. [DOI] [PubMed] [Google Scholar]
- 42.Wrighton, K. H., M. Liang, B. Bryan, K. Luo, M. Liu, X. H. Feng, and X. Lin. 2007. Transforming growth factor-beta-independent regulation of myogenesis by SnoN sumoylation. J. Biol. Chem. 2826517-6524. [DOI] [PubMed] [Google Scholar]
- 43.Xu, Q., and Z. Wu. 2000. The insulin-like growth factor-phosphatidylinositol 3-kinase-Akt signaling pathway regulates myogenin expression in normal myogenic cells but not in rhabdomyosarcoma-derived RD cells. J. Biol. Chem. 27536750-36757. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, P., C. Wong, D. Liu, M. Finegold, J. W. Harper, and S. J. Elledge. 1999. p21(CIP1) and p57(KIP2) control muscle differentiation at the myogenin step. Genes Dev. 13213-224. [DOI] [PMC free article] [PubMed] [Google Scholar]