Abstract
Menin, the product of the MEN1 (multiple endocrine neoplasia type 1) tumor suppressor gene, is involved in activation of gene transcription as part of an MLL1 (mixed-lineage leukemia 1)/MLL2 (KMT2A/B)-containing protein complex which harbors methyltransferase activity for lysine 4 of histone H3 (H3K4). As MEN1 patients frequently develop lipomas and peroxisome proliferator-activated receptor γ (PPARγ) is expressed in several MEN1-related tumor types, we investigated regulation of PPARγ activity by menin. We found that menin is required for adipocyte differentiation of murine 3T3-L1 cells and PPARγ-expressing mouse embryonic fibroblasts. Menin augments PPARγ target gene expression through recruitment of H3K4 methyltransferase activity. Menin interacts directly with the activation function 2 transcription activation domain of PPARγ in a ligand-independent fashion. Ligand-dependent coactivation, however, is dependent on the LXXLL motif of menin and the intact helix 12 of PPARγ. We propose that menin is an important factor in PPARγ-mediated adipogenesis and that loss of PPARγ function may contribute to lipoma development in MEN1 patients.
Inactivating germ line mutations of the MEN1 gene are causal to multiple endocrine neoplasia type 1 (MEN1), an inherited tumor syndrome with high penetrance and a variable expression. MEN1 is characterized by the combined occurrence of tumors in the parathyroid glands, pancreas and duodenum, pituitary glands, and adrenal glands as well as neuroendocrine tumors. Other manifestations include angiofibromas, collagenomas, and lipomas (5). The mechanism of lipoma formation in MEN1 patients is currently unknown.
Peroxisome proliferator-activated receptor γ (PPARγ) is a member of the PPAR nuclear receptor (NR) family (28). Polyunsaturated fatty acids, eicosanoids, components of oxidized low-density lipoproteins, and nitro-oleic acid are natural low-affinity ligands for PPARγ. Synthetic thiazolidinediones, such as pioglitazone and rosiglitazone, have high affinities for PPARγ. Two isoforms of PPARγ exist, with the PPARγ1 isoform being expressed in several tissues, including the pituitary gland and pancreatic islets (10, 36), while expression of the PPARγ2 isoform is largely restricted to adipose tissue. Both PPARγ isoforms heterodimerize with the retinoid X receptors (RXRs) to bind to PPAR-responsive elements (PPREs) in promoters and regulate transcription of target genes (46). Ectopic expression of PPARγ in mouse fibroblasts can result in full adipogenesis (47). Mutations in the PPARG gene are causative of an inherited form of partial lipodystrophy (41). Furthermore, PPARγ has been shown to have antiproliferative effects in adipocytes, by inhibiting DNA binding of the cell cycle transcription factor E2F/DP (3). In conclusion, PPARγ is a key regulator in adipocyte differentiation, which acts at the level of gene transcription.
Menin is the product of the MEN1 tumor suppressor gene. Menin is expressed ubiquitously as a nuclear protein and can take part in many cellular processes, including transcription regulation, DNA replication, and DNA repair (29). As a transcriptional regulator, menin is involved in both repression and activation of transcription. Through association with a histone-deacetylating complex, menin acts as a corepressor for JunD-mediated transcription (21). Menin was demonstrated to attenuate transcription of the hTERT gene, possibly by this mechanism (30). Activation of transcription by menin also involves histone modifications. Menin is an integral component of MLL1(KMT2A) or MLL2(KMT2B) protein complexes which have histone methyltransferase (HMT) activity specific for lysine 4 of histone H3 (H3K4) (17, 55). Trimethylation of this residue (H3K4me3) is associated with active transcription (39). Several downstream effector proteins reading the H3K4me3 mark via plant homeodomain fingers such as BPTF/NURF and TFIID are involved in transcription activation (50, 53). The menin-HMT complex is an activator of several Hox genes and the p18Ink4c and p27Kip1 cyclin-dependent kinase inhibitor genes (17, 32, 55). Analysis of genome-wide DNA occupancy revealed that menin is located at 5′ regions of many active genes and also at 3′ regions and intergenic loci (1, 40).
Promoter recruitment of menin depends on interactions with DNA sequence-specific transcription factors: for example, via JunD in the repressing complex or via Smad transcription factors as an activator (43). Furthermore, promoter association may be stabilized by the low-affinity sequence-independent interaction of menin with DNA (24). The menin-HMT complex was recently demonstrated to be recruited to the c-Myc gene enhancer by interacting with the C-terminal activation domain of β-catenin (42). A complex containing MLL1(KMT2A) and possibly menin was found to couple E2F function to H3K4me3 (49). Previously, we have demonstrated that menin is able to coactivate estrogen receptor α (ERα)- and vitamin D receptor (VDR)-mediated transcription. In this case, menin recruits H3K4 HMT activity to the estrogen-responsive TFF1/pS2 gene (9).
Approximately 30% of MEN1 patients develop lipomatous tumors (8). In addition, it has been shown that PPARγ1 is expressed in MEN1-associated tumor types, such as pituitary adenomas and neuroendocrine tumors (15, 31). We hypothesized that deficient differentiation dependent on PPARγ plays a role in lipoma tumorigenesis and that menin is involved in this differentiation. Therefore, to further explore the role of menin as a coactivator for NRs and to determine the biological relevance of these interactions, we examined PPARγ as a potential menin-interacting protein. In this report, we show that menin is indeed a transcriptional coactivator for PPARγ and that menin is important for PPARγ-dependent transcription and adipocyte differentiation. Menin can recruit histone H3K4 trimethylation to PPARγ target genes, through a direct interaction with the activation function 2 (AF2) domain of PPARγ. These results extend the role of menin as a coactivator for NR-mediated transcription to PPARγ and provide new insight into the tissue-specific manifestations of MEN1.
MATERIALS AND METHODS
Patient material.
Normal adipose tissue (from paracolic, pararenal, breast, and peritoneal origin) and lipoma samples (five intramuscular lipomas, one subcutaneous lipoma, one osteochondrolipoma, and one soft tissue lipoma) were obtained from the tissue bank of the Department of Pathology/UMCU Biobank and used in accordance with the hospital scientific committee regulations and the code “Proper Secondary Use of Human Tissue,” as installed by the Federation of Biomedical Scientific Societies (www.federa.org).
Plasmids and siRNA oligonucleotides.
Construction of vectors B42-PPARγ, B42-RXRα, Gal4-RXRα, Gal4-PPARγ(AF2) (containing AF2), Gal4-PPARγ(AF1), glutathione S-transferase (GST)-PPARγ(AF2), GST-PPARγ(AF2-L468A/E471A) (containing AF2 with L468A and E471A mutations), pBabeHygroMenin, pBabeHygroMeninL22R, pEG202NLS-menin, pMSCV-PPARγ2, pSG5-HA-CARM1, and pSG5-SRC1e has been described previously (7, 9, 12, 17-19, 37, 52). The menin expression vector pCDNA3.1 M+ was a gift from G. Weber. The menin L22R, LLWAA (mutated residues in italic), and Gal4-PPARγ(AF2-L468A/E471A) mutations were introduced by site-directed mutagenesis. Control (D-001810-10), mMEN1#1 (D-042675-02), mMEN1#2 (D-042675-04), and mPPARG (J-040712-05) small interfering RNA (siRNA) oligonucleotides were purchased from Dharmacon.
Antibodies and immunoblotting.
The following antibodies were used: anti-α-tubulin (CP06; Calbiochem), anti-Gal4 (SC-510; Santa Cruz), anti-GST (SC-138; Santa Cruz), anti-H3K4me3 (ab-8580; Abcam), antimenin (A300-105A; Bethyl), anti-PPARγ (SC-7196 [Santa Cruz] or C26H12,#2435 [Cell Signaling Technology]), anti-FABP4 (SC-18661; Santa Cruz), and antihemagglutinin (anti-HA) (3F10; Roche). Immunoblotting was carried out as described before (9).
Cell lines and stable and transient protein expression.
Cos7 (African green monkey kidney) cells and murine 3T3-L1 cells were routinely cultured in Dulbecco's modified Eagle's medium, supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. Generation of wild-type and MEN1−/− (MEN1 T/T) mouse embryonic fibroblasts (MEFs) has been described previously (4). MEFs were kindly provided by C. Zhang. MEFs were maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, 2 mM l-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and 100 μM β-mercaptoethanol.
For polyclonal PPARγ-expressing MEF cell lines, wild-type and MEN1−/− MEFs were infected with retroviruses collected from 293T Phoenix cells that had been transiently transfected with pMSCV-PPARγ2 or pMSCV and were maintained under selection of 2 μg/ml puromycin.
For menin reexpression studies, MEN1−/− MEFs expressing PPARγ were infected with viruses obtained from Phoenix cells transiently transfected with pBabeHygro, pBabeHygroMenin, and pBabeHygroMeninL22R. Cells were grown in medium containing 2 μg/ml puromycin and 500 μg/ml hygromycin, and monoclonal cell lines were isolated.
Luciferase reporter experiments were carried out as described previously (9). DNA (750 ng per well, in a 12-well format) was transfected using FuGene 6 reagent (Roche). Transfection mixtures consisted of 200 ng luciferase reporter, 25 ng pCMV-Renilla, 5 ng Gal4-PPARγ(AF2), 25 ng Gal4-PPARγ(AF1), or 10 ng Gal4-PPARγ(AF2-L468A/E471A) vector and supplemented with a maximum of 520 ng pCDNA3.1 M+ and/or empty pCDNA3 plasmid. Twenty-four hours after transfection, the medium was changed to medium containing the appropriate ligand—1 μM rosiglitazone (in dimethyl sulfoxide [DMSO]), 1 μM 9-cis-retinoic acid (in ethanol)—or vehicle. Luciferase and Renilla activities were measured 24 h afterwards. Total cell lysates were used for immunoblotting.
Cell differentiation and siRNA transfection assays.
Differentiation of 3T3-L1 cells was performed as described previously (18). For siRNA experiments, cells were grown to 80% confluence and transfected with the indicated siRNA oligonucleotides using Dharmafect 3 reagent. siRNA transfections were repeated every 3 days during the differentiation assay. Polyclonal MEFs expressing PPARγ were seeded at 5 × 103 cells/ml in a 24-well plate. Cells were grown in medium containing 1 μM rosiglitazone. The medium was changed every 2 days. After 10 days, cells were subjected to oil red O staining. Monoclonal MEN1−/− MEF cell lines were grown to confluence. After 2 days, 1 μM rosiglitazone was added to the medium, which was refreshed every 2 days for 14 days before cells were subjected to oil red O staining.
Protein expression and purification.
The expression and purification of baculovirus-expressed menin have been described previously (9). Plasmids encoding GST, GST-PPARγ(AF2), and GST-PPARγ(AF2-L468A/E471A) were transformed into the Escherichia coli BL21-Codon Plus strain. Cells were grown at 30°C with shaking. Expression and lysis procedures have been described previously (2).
Yeast two-hybrid analysis.
EGY48 cells were transformed with the B42-PPARγ(AF2) constructs and the indicated LexA-menin constructs. Cells were grown overnight at 30°C in 2% galactose-1% sucrose-containing SC medium lacking the appropriate amino acids and in the presence of vehicle, 1 μM rosiglitazone, and 1 μM 9-cis-retinoic acid. Lysates were prepared, and LacZ activity was determined by a liquid β-galactosidase assay, as described previously (2).
Protein binding experiments.
Soluble lysates containing equivalent amounts of GST, GST-PPARγ(AF2), and GST-PPARγ(AF2-L468A/E471A) were bound to glutathione agarose beads (Sigma) for 3 h at 4°C in binding buffer (50 mM Tris-HCl [pH 8.0], 10% glycerol, 140 mM NaCl, 10 mM MgCl2, 1 mM NaF, 1 mM Na3Vo4, 0.5 mM dithiothreitol, and protease inhibitors). After washing with binding buffer, purified menin protein (6 μg/ml) was added. Binding experiments with GST and GST-PPARγ(AF2) were carried out in the presence of 100 nM rosiglitazone or DMSO for 3 h at 4°C. Glutathione beads were washed three times with excess binding buffer, and eluted proteins were analyzed by immunoblotting.
Analysis of PPARγ target gene expression.
RNA from differentiated and undifferentiated 3T3-L1 cells was isolated using the RNeasy kit (Qiagen). Two hundred fifty nanograms of total RNA was used for cDNA synthesis using random primers. mRNA levels were analyzed by quantitative PCR on a Chromo4-equipped PCR cycler (Bio-Rad) and normalized against a standard reference cDNA.
A 1 μM concentration of rosiglitazone or DMSO was added to the MEF medium 6 h prior to cell harvesting. Total RNA was extracted using the RNeasy kit (Qiagen) and subsequently treated with DNase. After homogenization, RNA from human samples was isolated using the RNeasy lipid tissue kit (Qiagen). Two hundred nanograms of total RNA was used for cDNA synthesis using oligo(dT). The following PCR primers were used: hB-actin-F (AGAAAATCTGGCACCACACC), hB-actin-R (AGAGGCGTACAGGGATAGCA), hMEN1-F (GCTCTGGCTGCTCTATGACC), hMEN1-R (TAGAGGGTGAGTGGGTCTGG), hPPARG-F (GCTGGCCTCCTTGATGAATA), hPPARG-R (TTGGGCTCCATAAAGTCACC), hFABP4-F (AACCTTAGATGGGGGTGTCC), hFABP4-R (GTGGAAGTGACGCCTTTCAT), hAQP7-F (GGAAGGTCAGCAGGAGCAC), hAQP7-R (GGTGCTCAGCCAATGAGG), mB-actin-F (GATCTGGCACCACACCTTCT), mB-actin-R (GGGGTGTTGAAGGTCTCAAA), mFabp4-F (GAAAACGAGATGGTGACAAGC), mFabp4-R (TTGTGGAAGTCACGCCTTT), mAqp7-F (GGCTTCTCCCTTCCTCTAGTTT), mAqp-R (AAGGCCACTGAGGAAGTCATT), mP18-F (CTGGAGTTCCAGGCTGATGT), mP18-R (GCAGGCTGTGTGCTTCATAA), mP27-F (AACTAACCCGGGACTTGGAG), mP27-R (CCAGGGGCTTATGATTCTGA), mMen1-F (CGATCTTCACACTGACTCTTTGG), mMen1-R (AGGTCTGCCAAGTTCCCTAGC), mPparg-F (GATGGAAGACCACTCGCATT), and mPparg-R (AACCATTGGGTCAGCTCTTG).
ChIP experiments.
The chromatin immunoprecipitation (ChIP) procedure followed has essentially been described previously (9), with the following adjustments. Subconfluent cultures of MEF cells were treated with 1 μM rosiglitazone or DMSO for 6 h. Cells were cross-linked by addition of 1% formaldehyde in phosphate-buffered saline for 10 min at 37°C. Glycine was added to a final concentration of 125 mM. After lysis, the cell lysate was sonicated three times for 30 s in a Bioruptor (Diagenode), yielding DNA fragments of about 200 bp. After reverse cross-linking and proteinase K treatment, DNA was purified by phenol-chloroform-isoamyl alcohol treatment (twice), followed by a chloroform-isoamyl alcohol cleaning and ethanol precipitation. Binding of DNA was assessed by quantitative PCR and normalized against input samples from the same experiment. The following primers were used: Fabp4(PPRE)-F (GACAAAGGCAGAAATGCACA), Fabp4(PPRE)-R (AATGTCAGGCATCTGGGAAC), Fabp4(ORF)-F (CAGCTTAAGGGACCGAGATG), Fabp4(ORF)-R (TCGCATTCTCAACAATCAGG), Aqp7(PPRE)-F (CTTGGCTTAAGTGGCATGGG), Aqp7(PPRE)-R (CCAGGGTGGCTGTGAGTTCT), Myo-F (CACCCTGAGACCCTGGATAA), and Myo-R (GCAGCATGTTGTCCCTTCTT).
RESULTS
Expression of the PPARγ target gene FABP4 is reduced in sporadic lipomas.
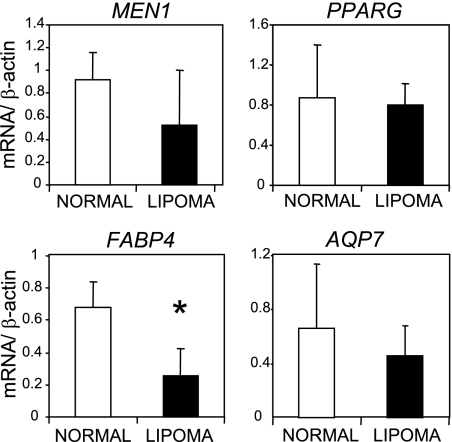
To investigate our hypothesis that PPARγ-dependent transcription is affected in lipomas, we carried out a reverse transcription-PCR (RT-PCR) analysis on a series of four normal adipose tissue samples and eight sporadic lipomas. We measured mRNA levels of total PPARG, MEN1, and β-actin genes as well as two known PPARγ target genes: those coding for fatty acid binding protein 4 (FABP4/Ap2) and aquaporin 7 (AQP7) (22, 46). We found that whereas PPARG, MEN1, and AQP7 mRNA levels did not differ significantly between these groups (using a two-tailed Student's t test), relative FABP4 mRNA levels were reduced significantly (P = 0.004) in lipomas (Fig. 1). From these observations, we suggest that loss of PPARγ-dependent gene expression may be a feature of sporadic lipomas. In addition, we show that MEN1 is expressed in adipose tissue.
FIG. 1.
Expression of the PPARγ target gene FABP4 is reduced in sporadic lipomas. mRNA levels of MEN1, PPARG, FABP4, and AQP7 were determined by quantitative RT-PCR in four normal adipose tissue samples (white columns) and eight sporadic lipomas (black columns). The values shown are averages and standard deviations of mRNA levels normalized against β-actin mRNA. The asterisk indicates statistical significance (P = 0.004).
Menin promotes PPARγ-dependent adipocyte differentiation.
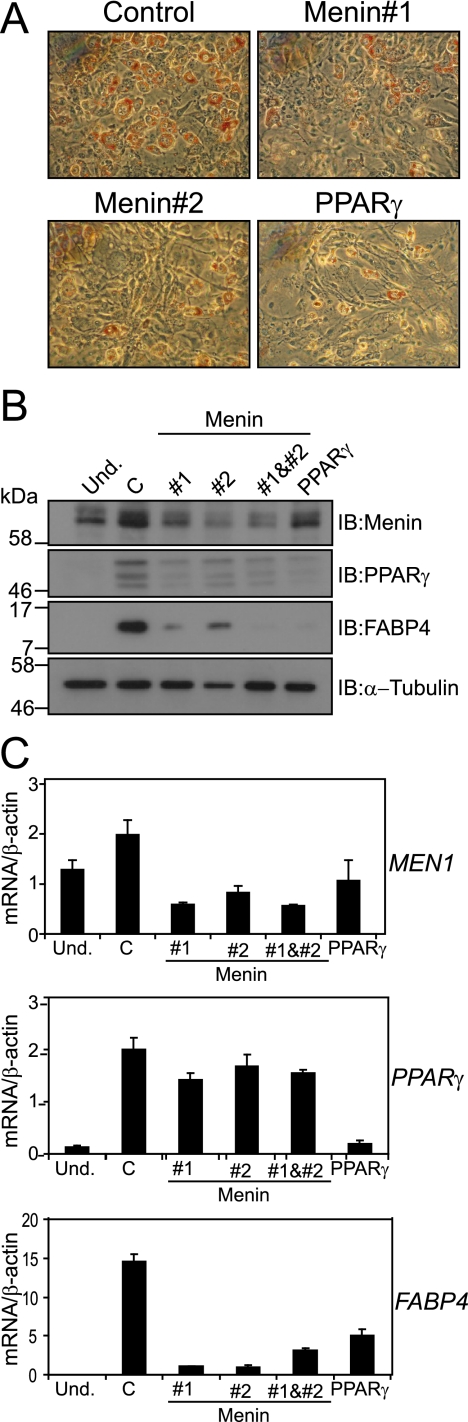
Based on the results shown above, we decided to further explore a potential role for menin in adipocyte differentiation. Unfortunately, no collection of MEN1 patient-derived lipomas is available to us, because of the rarity of the disease and the infrequent need to perform surgery on lipomas. Therefore, we chose to study the relevance of menin for the biological function of PPARγ using two different controlled mouse cell systems. First, we studied adipocyte differentiation of 3T3-L1 murine fibroblasts. We used siRNA oligonucleotides directed against MEN1 and as a control against Pparg to determine the importance of menin for adipogenesis. Knockdown of menin levels showed a clear reduction in adipogenic potential of the 3T3-L1 cells compared to cells treated with the control siRNA, as illustrated by oil red O staining of triglycerides at day 6 of differentiation (Fig. 2A and B). Both knockdown of menin and PPARγ resulted in reduced protein levels of the adipocyte marker FABP4 (Fig. 2B). To investigate whether the effect of menin knockdown on adipocyte differentiation results from an altered transcription, mRNA expression was examined in the knockdown cells by quantitative RT-PCR. This showed that in cells treated with siRNAs against Men1, Fabp4 mRNA levels were lower, irrespective of Pparg mRNA levels (Fig. 2C). These findings suggest that menin is involved in transcription regulation of the Fabp4 gene.
FIG. 2.
Knockdown of menin interferes with differentiation of murine 3T3-L1 cells. (A) 3T3-L1 cells were transfected with the indicated siRNA oligonucleotides. After 6 days of differentiation, oil red O staining was performed to assess lipid accumulation. (B) Immunoblot (IB) analysis of global menin, PPARγ (the Cell Signaling Technology antibody recognizes multiple forms of PPARγ), and FABP4 levels in undifferentiated (Und.) cells and siRNA-treated cells was performed. α-Tubulin was used as a loading control. The positions of molecular mass markers, with their respective molecular masses in kilodaltons, are indicated next to the blots. (C) RT-PCR analysis of total mRNA levels of Men1, Pparg, and Fabp4 in siRNA-treated 3T3-L1 cells. The values shown represent averages and standard deviations. mRNA levels were normalized against β-actin mRNA.
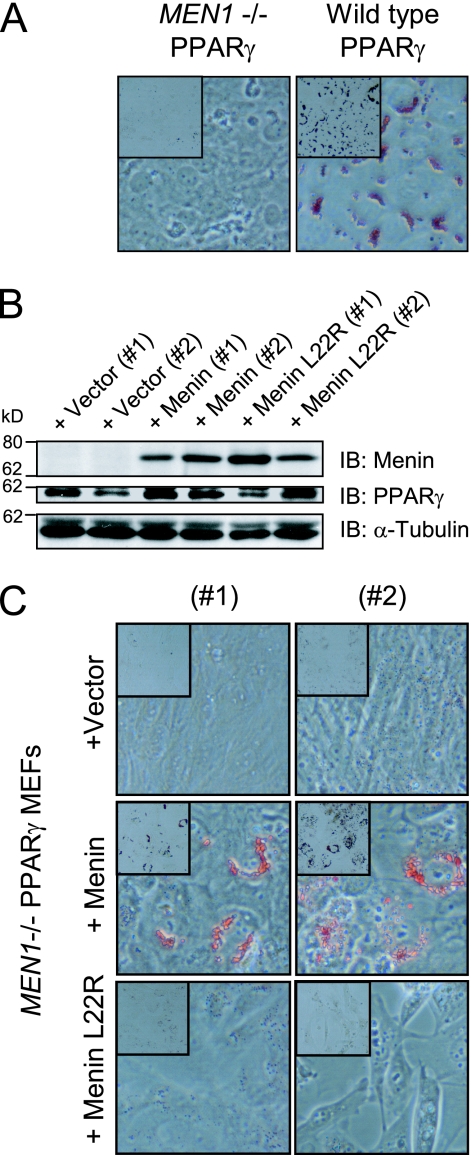
The second cell system used were the MEFs as they have the capacity to differentiate into adipocytes upon ectopic expression of PPARγ (47). Wild-type and MEN1−/− MEFs were first retrovirally transduced with wild-type mouse PPARγ2 (mPPARγ2). Both polyclonal cell lines were treated with rosiglitazone. We noticed that MEN1−/− MEFs failed to undergo adipogenesis compared to wild-type MEFs as visualized by oil red O staining (Fig. 3A). To exclude that this was due to differences between the cell lines unrelated to the MEN1 gene status, the wild-type MEN1 gene was reexpressed in the MEN1−/− PPARγ-expressing cells and monoclonal cell lines were generated (Fig. 3B). Additionally, cell lines expressing a MEN1 gene missense mutation derived from a MEN1 patient with a lipoma were established (Fig. 3B) (51). This mutation leads to a change from leucine to arginine at position 22 (hereafter, menin L22R). Menin protein levels were found to be similar to those of the wild-type MEF cell line (data not shown). After 14 days of treatment with rosiglitazone, marked lipid accumulation was detected in menin-expressing cells as opposed to cells expressing menin L22R or the empty vector (vector)-transfected cells (Fig. 3C). Furthermore, we noted that introduction of wild-type menin reduces the proliferation rate of MEN1−/− MEFs (data not shown). These results show that the presence of wild-type menin is important for proper PPARγ-dependent adipogenesis in this MEF system.
FIG. 3.
Menin is required for PPARγ-dependent adipocyte differentiation. (A) Polyclonal MEN1−/− and wild-type MEF cells that ectopically express PPARγ were treated with 1 μM rosiglitazone for 10 days. Lipid vesicle formation was assessed by oil red O staining (40× magnification, inset represents 10× magnification). (B) Menin and menin L22R were reexpressed in MEN1−/− MEFs expressing PPARγ. The presence of menin and PPARγ (SC antibody recognizes PPARγ2 specifically) was visualized by immunoblotting (IB) using α-tubulin as a loading control in total cell lysates of two monoclonal cell lines carrying the empty vector, two cell lines reexpressing wild-type menin, and two cell lines expressing menin L22R that had been treated with 1 μM rosiglitazone for 14 days. The positions of molecular mass markers, with their respective molecular masses in kilodaltons, are indicated next to the blots. (C) Oil red O staining of the cell lines described in panel B.
PPARγ target gene expression is regulated by menin.
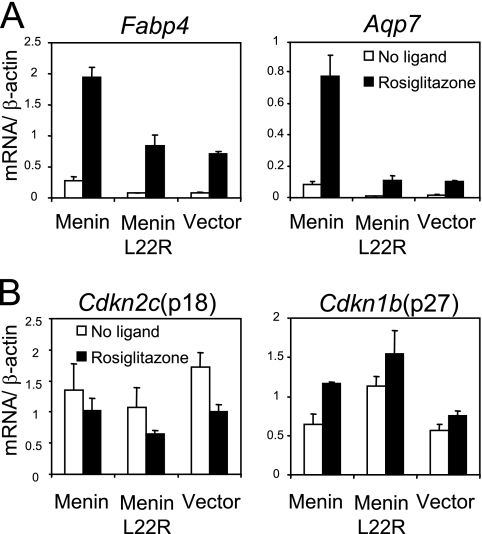
To investigate further how menin affects PPARγ function in the MEF system, we studied the expression of the PPARγ target genes, Fabp4 and Aqp7. Monoclonal cell lines expressing similar levels of menin and PPARγ were selected for this analysis. Cells were treated for 6 h with rosiglitazone or DMSO and subsequently subjected to quantitative RT-PCR analysis. Compared to cells expressing wild-type menin, ligand-induced mRNA levels of Fabp4 and Aqp7 were found to be lower in cells expressing menin L22R or the cells carrying only the vector (Fig. 4A).
FIG. 4.
Menin regulates transcription of PPARγ target genes. (A) The menin (no. 1), menin L22R (no. 2), and vector (no. 1) cell lines were treated with DMSO (white columns) or 1 μM rosiglitazone (black columns) for 6 h. mRNA levels of Fabp4 and Aqp7 were measured by quantitative RT-PCR. Averages and standard deviations relative to β-actin mRNA are shown from a representative experiment performed in triplicate. (B) Relative levels of p18/Cdkn2c and p27/Cdkn1b mRNA were determined in the same experiment.
The genes for p18Ink4c(Cdkn2c) and p27Kip1(Cdkn1b) are well-established menin target genes, which can restrict cell proliferation (32). Moreover, p18Ink4c was found to be upregulated in mouse fibroblasts that ectopically expressed PPARγ (34). We hypothesized that mRNA levels of Cdkn2c and Cdkn1b would be higher in menin-expressing cells. However, expression of both genes did not differ between these cell lines, and treatment with rosiglitazone did not clearly affect transcription of these genes (Fig. 4B). RT-PCR analysis of expression of the above-mentioned genes in polyclonal MEN1−/− and wild-type MEF cell lines ectopically expressing PPARγ yielded similar results (data not shown). Altogether, these experiments show that in the absence of menin the expression of PPARγ target genes is downregulated and that in our system p18Ink4c and p27Kip1 are probably not involved in adipocyte differentiation.
Histone H3K4 trimethylation on PPARγ target genes is largely dependent on menin.
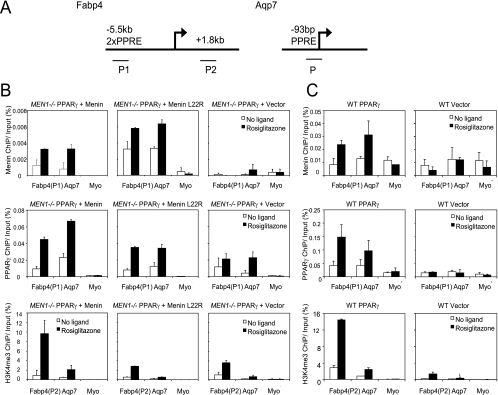
Upon ligand stimulation, PPARγ target genes show increased H3K4 trimethylation levels (44, 45). To determine the H3K4me3 status of PPARγ target genes in the presence or absence of menin, we performed ChIP experiments in the monoclonal MEN1−/− MEF cell lines ectopically expressing PPARγ. We used antibodies directed at menin, PPARγ, or the trimethylated form of histone H3K4 to study their presence on the Fabp4 and Aqp7 genes (Fig. 5A). The heterochromatic myoglobin (Myo) gene was used as an internal negative control. In both wild-type menin- and menin L22R-reexpressing cells, menin could be detected on the PPREs of the Fabp4 and Aqp7 genes after cells had been treated with rosiglitazone for 6 h (Fig. 5B, upper panels). No menin recruitment could be detected at the transcription start site of Fabp4 (data not shown). In Fig. 5B (middle panels), we show that when ligand is present approximately equal quantities of PPARγ are recruited. However, both menin L22R-expressing and vector control cells showed a clear reduction of ligand-induced H3K4me3 levels in the open reading frame of Fabp4 and at the transcription start site of Aqp7 (Fig. 5B, lower panels). From these data, we conclude that both wild-type menin and mutant menin colocalize with PPARγ at PPREs and that menin is needed for recruitment of adequate H3K4 trimethyltransferase activity to two different PPARγ target genes.
FIG. 5.
Menin recruits H3K4 trimethylation activity to PPARγ target genes. (A) Schematic representation of the Fabp4 and Aqp7 genes. Fabp4 contains two PPREs in an upstream enhancer element. For ChIP analysis, two primer pairs were used: P1 for amplifying the region of the PPREs for studying menin and PPARγ occupancy and P2 in the open reading frame to measure H3K4me3. The PPRE of Aqp7 is located close to the transcription start site; one primer pair was used for ChIP analysis. (B and C) Cell lines used for mRNA expression analysis (B) and polyclonal wild-type (WT) MEFs expressing either PPARγ or vector (C) were used. Cells were stimulated for 6 h with 1 μM rosiglitazone or DMSO. Averages of immunoprecipitated DNA, as a percentage of input, and standard errors of the means of two independent experiments are shown. (B and C, upper and middle panels) Menin and PPARγ recruitment was assessed on the PPREs of Fabp4 and Aqp7. (B and C, lower panels) H3K4me3 levels were measured in the open reading frame of Fabp4 and near the transcription start site of Aqp7. The myoglobin (Myo) gene was used as an internal negative control. (B, upper right panel) The MEN1−/− vector cell line that does not express menin serves as a negative control for the menin antibody. (C, right panels) The wild-type MEF cells that do not express exogenous PPARγ serve as a control for PPARγ in menin recruitment.
To ascertain that menin recruitment on PPARγ-responsive promoters is dependent on PPARγ, we performed additional ChIP experiments. Wild-type MEFs expressing endogenous menin were used that had been infected with PPARγ2 or vector control viruses. As shown in Fig. 5C (upper panels), menin is recruited exclusively to the Fabp4 and Aqp7 promoters when liganded PPARγ is present (Fig. 5C, middle panels). In the absence of PPARγ, H3K4 trimethylation levels are severely reduced (Fig. 5C, lower panels). These experiments show that PPARγ is required for bringing the menin HMT complex to PPARγ target genes.
Menin can interact directly with the AF2 domain of PPARγ in a ligand-independent fashion.
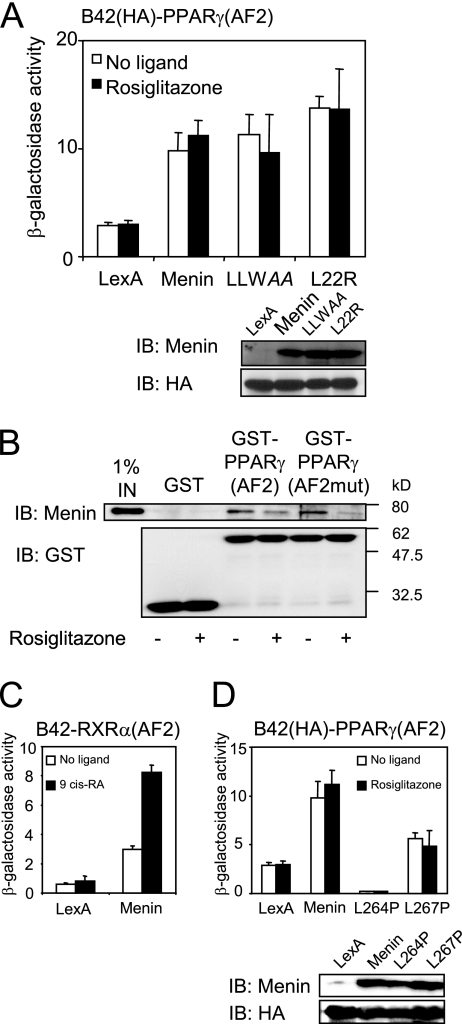
We previously found that menin can bind to ERα in a ligand-dependent manner (9). To investigate whether menin and PPARγ can interact directly, we carried out yeast two-hybrid and GST pulldown assays. In yeast two-hybrid assays, we found a ligand-independent interaction between wild-type menin fused to LexA and B42-coupled PPARγ(AF2). Menin contains one potential LXXLL NR interaction motif (amino acids 263 to 267; LLWLL). Mutation of this motif disrupts the canonical NR coactivator interaction (16). Interestingly, both the LLWAA and L22R menin mutants retained PPARγ binding (Fig. 6A). In GST pulldown experiments, purified wild-type menin was incubated with equal amounts of a GST-PPARγ(AF2) fusion protein. Consistent with the yeast two-hybrid result, menin could bind to GST-PPARγ(AF2), albeit with low affinity (Fig. 6B). In fact, in this assay we reproducibly found a small decrease of the interaction in the presence of ligand. Also in this assay, the menin-PPARγ interaction did not depend on the presence of ligand. The L468A E471A double mutant in helix 12 in the AF2 domain of PPARγ does not bind to the coactivators CBP and SRC1, but is still able to bind ligand (14). This L468A E471A mutation—hereafter PPARγ(AF2mut)— was introduced into the GST-PPARγ(AF2) expression plasmid. As shown in Fig. 6B, menin was able to bind to a GST-PPARγ(AF2mut) protein, indicating that the integrity of helix 12 of PPARγ is not required for menin binding. PPARγ binds to PPREs in a heterodimeric form with RXR isoforms. To determine if menin can bind to RXR, we performed yeast two-hybrid assays. Coexpression in yeast of LexA-menin and B42-fused RXRα revealed an interaction between menin and RXRα, which is increased in the presence of its ligand (Fig. 6C). These findings suggest that besides by binding to PPARγ, menin can potentially also be recruited to PPREs via liganded RXRα.
FIG. 6.
Menin and PPARγ can interact directly. (A) Mutations resulting in LLWAA and L22R menin mutants were introduced in the pEG202-LexAMenin yeast expression vector. EGY48 cells transformed with the indicated LexA constructs and B42-HA-PPARγ(AF2) were treated with 1 μM rosiglitazone or DMSO for 12 h. Cells were lysed, and β-galactosidase activity was determined and normalized for the protein content of the lysates. Averages and standard deviations of a representative experiment performed in triplicate are shown. Expression levels of the menin proteins and the levels of B42-HA-PPARγ(AF2) were verified by immunoblotting (IB). (B) Glutathione beads were coated with purified GST, GST-PPARγ(AF2), or GST-PPARγ(AF2mut) proteins and incubated with purified recombinant menin in the absence or presence of 100 nM rosiglitazone. Retention of menin protein was determined by immunoblotting. (C) Together with a LexA-menin expression construct, a B42-fused ligand binding domain of RXRα was transformed into EGY48 cells. Cells were incubated with 1 μM 9-cis-retinoic acid (9 cis-RA) or vehicle for 12 h, and β-galactosidase activities were determined. (D) Two patient-derived mutations, L264P and L267P, were introduced into the LexA-menin fusion plasmid and transformed for a yeast two-hybrid experiment as described.
Two patient-derived missense mutations leading to leucine-to-proline substitutions at positions 264 or 267 in menin severely disrupt the capacity of menin to bind to ERα (9). These mutations are both in the predicted NR interaction motif, leading to LPWLL and LLWLP, respectively (mutated residue in italic), and have more severe structural consequences than the LLWAA mutation. In a yeast two-hybrid setup, both the L264P and L267P mutants showed reduced menin binding (Fig. 6D). From these observations, we conclude that the interaction between menin and PPARγ is not a classical LXXLL-helix 12 interaction. Rather, a larger domain of menin which includes LLWLL may be required for menin-PPARγ binding.
Menin enhances PPARγ-dependent transcription.
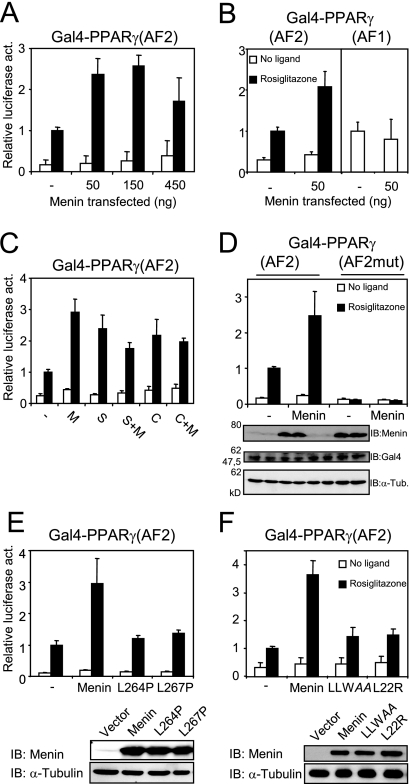
To study in more detail the mechanism of coactivation of PPARγ by menin, we performed luciferase assays. To this end, Cos7 cells were transiently transfected with increasing amounts of MEN1 expression vector together with the expression vector for a fusion protein containing the AF2 domain of PPARγ fused to the DNA binding domain of the yeast Gal4 activator. A thymidine kinase promoter-luciferase construct with five Gal4 binding sites was used as a reporter gene. Overexpression of menin increased ligand-induced transcription up to approximately threefold, depending on the amount of menin that was cotransfected (Fig. 7A). This is similar to the effect of overexpression of menin on ligand-dependent transcription driven by a Gal4-ERα fusion protein (9). We performed similar experiments using a Gal4-RXRα fusion protein expression vector and found that menin can also coactivate 9-cis-retinoic acid-induced transcription (data not shown). To assess the specificity of menin for liganded PPARγ, the MEN1 expression vector was cotransfected with a Gal4-PPARγ(AF1) expression plasmid. Gal4-PPARγ(AF1)-dependent transcription was not affected by menin (Fig. 7B). The coactivation of AF2 observed by menin was also similar to the effect of two known PPARγ coactivators, SRC1 and CARM1 (Fig. 7C) (35, 54). In this setup, we did not detect synergy between menin and these known coactivators (Fig. 7C).
FIG. 7.
Menin coactivates PPARγ-mediated transcription in a ligand-dependent fashion. (A) Cos7 cells were transiently transfected with a 5× GalTK-luciferase construct and an expression plasmid for Gal4-PPARγ(AF2) and treated with 1 μM rosiglitazone or DMSO for 24 h. Increasing amounts of menin were cotransfected. Firefly luciferase readings were normalized against Renilla luciferase, which served as an internal control. Columns represent averages and standard deviations of two independent experiments performed in triplicate. All luciferase assays shown were performed in this fashion. (B) Menin was overexpressed in cells cotransfected with Gal4-PPARγ(AF2) and in cells coexpressing Gal4-PPARγ(AF1). (C) Along with 50 ng of the menin (M) expression plasmid, cells were transfected with 450 ng of pSG5-SRC1e (S), 450 ng pSG5-HA-CARM1 (C), or a combination of these factors and menin. (D) A Gal4-PPARγ(AF2) construct bearing two mutations in helix 12 (L468A E471A) was transiently transfected into Cos7 cells. Protein levels of menin, Gal4-PPARγ(AF2), and α-tubulin (α-Tub.) were determined by immunoblotting (IB). Lanes on the gel correspond to the columns shown in the panel above. (E) Expression plasmids encoding the L264P and L267P menin mutants were cotransfected with Gal4-PPARγ(AF2). Menin protein levels were determined by immunoblotting with α-tubulin as a loading control. (F) The LLWAA and L22R mutations were also introduced in the MEN1 expression plasmid and transfected into Cos7 cells for luciferase assays. Protein levels were determined as in panel E.
We introduced the L468A E471A double mutation into the Gal4-PPARγ(AF2) vector. As shown in Fig. 7D, Gal4-PPARγ(AF2mut) does not activate transcription upon addition of ligand. Overexpression of menin did not affect luciferase activity driven by this PPARγ mutant (Fig. 7D). Cotransfection of the menin L264P and L267P mutants did not result in any coactivation (Fig. 7E). The menin LLWAA and L22R mutations were also introduced into the MEN1 expression vector. Strikingly, menin LLWAA could also not stimulate rosiglitazone-induced transcription (Fig. 7F). Finally, as expected, menin L22R, the mutant that failed to recruit HMT activity to the Fabp4 and Aqp7 gene promoters (Fig. 5D) or restore PPARγ-driven adipogenesis (Fig. 3C), was also not able to coactivate PPARγ(AF2) in the luciferase setup (Fig. 7F).
We conclude from these results that menin acts as a coactivator for the AF2 function of PPARγ to an extent that is comparable to those of other well-established coactivators. The integrity of helix 12 of PPARγ as well as the LLWLL motif in menin is required for coactivation. The MEN1 lipoma-derived menin L22R mutant failed to coactivate PPARγ-dependent transcription.
DISCUSSION
The hallmark of the MEN1 syndrome is the development of multiple tumors in endocrine organs. A large proportion of MEN1 patients also develop lipomas (8). Our results link the function of menin to the nuclear receptor PPARγ. This receptor has been reported to be expressed in liposarcomas (malignant lipomatous tumors), and treatment of cultured liposarcoma cells with the synthetic PPARγ ligand pioglitazone led to increased differentiation (48). To our knowledge, no study has addressed yet the role of PPARγ or menin in lipoma development. In a series of sporadic lipomas, we found that expression of the PPARγ target gene FABP4 was lower than that of normal adipose tissue (Fig. 1). As PPARγ is involved in adipocyte differentiation and proliferation, it is tempting to speculate that loss of PPARγ signaling plays a role in lipoma development. Inactivation of menin, which functions as a PPARγ coactivator, could in this way play a direct role in loss of PPARγ-dependent differentiation, resulting in MEN1 lipomas. Possibly, MEN1 control of cell proliferation also contributes to this. In sporadic lipomas, inactivation of this pathway may occur through alternative mechanisms, since we found no significant differences in MEN1 and PPARG levels between normal and lipomatous tissues (Fig. 1).
Besides its expression in lipomas, several studies have shown that PPARγ is expressed in MEN1-related tumors, like pituitary tumors and neuroendocrine tumors (15, 31) and also neuroendocrine tumors of the thymus in MEN1 patients (K. M. A. Dreijerink and H. T. M. Timmers, unpublished results). In addition, pioglitazone has been shown to inhibit proliferation of a lung neuroendocrine tumor cell line (13), while rosiglitazone treatment of mice that had been subcutaneously injected with pituitary adenoma cells led to attenuation of tumor growth (15). Furthermore, the recent finding that menin plays a role in suppression of adaptive pancreatic β-cell proliferation in obese mice, in which PPARγ is also involved, raises the interesting possibility that the interaction of menin and PPARγ plays a role in this process as well (20, 38). Taken together, the relevance of the menin-PPARγ interaction could move beyond that of a role in lipoma development.
Menin and adipocyte differentiation.
Our findings provide more insight into the most intriguing question pending for MEN1: how do germ line mutations in a ubiquitously expressed gene cause an endocrine tumor phenotype? The Cdkn2c gene (encoding p18Ink4c) and the Cdkn1b gene (encoding p27Kip1) are targets of the menin-HMT complex (32). These cyclin-dependent kinase-inhibiting genes are inhibitors of cell proliferation. Mice lacking either p18Ink4c, p21CIP1/Waf1, or p27Kip1 develop tumor phenotypes that show significant overlap with the manifestations of the MEN1 syndrome (11). However, why these knockout mice display endocrine tissue-specific phenotypes is not clear. Expression levels of Cdkn2c and Cdkn1b were not clearly affected in our MEF system, rendering it unlikely that deregulation of these genes contributes to the effect of menin on adipocyte differentiation. We found that murine 3T3-L1 cells and embryonic fibroblasts lacking functional menin are defective in PPARγ-dependent adipocyte differentiation. Reexpression of menin in MEN1−/− MEFs restored the adipogenic potential (Fig. 3). In this study, we used a system in which adipocyte differentiation is maximally dependent on PPARγ activity. Previously, Sowa et al. showed that transient knockdown of menin in multipotential mouse mesenchymal stem cells did not affect bone morphogenic protein 2 (BMP-2)-induced adipocyte differentiation (43). This indicates that menin is involved in selective differentiation pathways.
Recruitment of H3K4-HMT complexes by menin.
Menin is an integral component of HMT complexes that contain MLL1(KMT2A) or MLL2(KMT2B) (17, 55). Our results extend the evidence that menin can be tethered to DNA through interaction with NRs, which results in increased H3K4me3 levels on target genes.
In MEN1−/− cells, we found that ligand-induced H3K4 trimethylation on the Fabp4 and Aqp7 genes is reduced but not completely absent (Fig. 5B). The identification of several other protein complexes that can recruit HMT activity directed at H3K4 to NRs raises the possibility of redundancy of these complexes. Mo et al. reported that an MLL4(KMT2D)-HMT complex, which lacks menin, can coactivate ERα-mediated transcription (33). Furthermore, the ASCOM complex, which contains the NR coactivator activating signal cointegrator 2 (ASC-2), MLL3(KMT2C), and MLL4(KMT2D), which also lacks menin, can recruit HMT activity to the retinoic acid receptor (RAR) and the liver X receptor (LXR) (26, 27). Similar to menin-HMT, the ASCOM complex was recently demonstrated to regulate adipogenesis and H3K4me3 on a PPARγ target gene (25). MLL3 knockout cells showed partial loss of H3K4me3 on the FABP4 gene. Therefore, other H3K4me3 complexes could very well be responsible for the remaining H3K4me3 observed on the PPARγ target genes in the MEN1−/− MEFs (Fig. 5B). Loss of menin in tumors of MEN1 patients does not lead to global loss of H3K4 trimethylation in these tumors, indicating that menin plays a gene-specific function (R. A. Varier, J. A. Kummer, and H. T. M. Timmers, unpublished results).
We found that a MEN1 lipoma-derived mutant, menin L22R, retained the ability to bind to PPARγ in a yeast two-hybrid assay (Fig. 6A) but could not coactivate PPARγ in a luciferase assay (Fig. 7F). Consistent with these findings, we could detect colocalization of menin L22R with PPARγ on the PPREs of the Fabp4 and Aqp7 genes (Fig. 5B). Rosiglitazone-induced H3K4me3 levels at these genes, however, were found to be reduced (Fig. 5B). Hughes et al. showed that menin L22R can be incorporated into an enzymatically active MLL complex (17). Our results demonstrate that menin L22R bound to PPARγ on DNA does not recruit HMT activity. Possibly, this is due to a conformational change in menin L22R, which prevents menin from recruiting HMT activity in the chromatin context.
Menin interacts with PPARγ.
We studied the interaction between menin and the AF2 domain of PPARγ. Using yeast two-hybrid and GST pulldown experiments, we found that this interaction is not dependent on the presence of ligand (Fig. 6A and B). In the liganded situation, the affinity of the interaction seemed slightly weaker than without ligand (Fig. 6B). This is in contrast to the interactions of menin with ERα and RXR (Fig. 6C), which are ligand dependent and represent classical NR coactivator interactions (9). Unlike a mutation of ERα(AF2) helix 12 (9), a mutation of helix 12 of PPARγ(AF2) did not affect the interaction with menin (Fig. 6B). Based on structural differences within the ligand binding domains, NRs can be divided into two classes (6). These structural differences between the AF2 domains of PPARγ (which is a class II receptor) and those of ERα and RXR (which are class I receptors) could be related to differential binding to menin. In support of this, yeast two-hybrid studies with menin and the ligand binding domain of VDR, which also belongs to the class II receptors, showed a similar ligand-independent binding (K. M. A. Dreijerink and H. T. M. Timmers, unpublished results).
The role of the LLWLL (amino acids 263 to 267) motif of menin for PPARγ function is not entirely clear. Whereas the LLWAA mutant affects the coactivator function of menin, binding to AF2 of PPARγ is not disturbed (Fig. 6A and 7F). In contrast, the MEN1 patient-derived mutants LPWLL (L264P) and LLWLP (L267P) are defective both for PPARγ binding and the coactivator function (Fig. 6D and 7E). Interestingly, the PPARγ interaction pattern of menin resembles that of another known PPARγ coactivator, CBP. CBP also does not interact with PPARγ in a classical manner as it is also able to bind to PPARγ irrespective of the presence of ligand. Nuclear magnetic resonance studies revealed that the segment within CBP required for the interaction with PPARγ is larger than the LXXLL motif only (23). Possibly, the PPARγ interaction function of menin also extends beyond the LLWLL motif, as indicated by the patient-derived mutations. It would be interesting to obtain a structural model for the domain within menin that interacts with PPARγ.
In conclusion, we demonstrate that menin is a direct coactivator for PPARγ-mediated transcription by recruiting HMT activity to PPARγ target genes. The lipoma-derived menin L22R mutant, which failed to recruit HMT activity, is defective in supporting the capacity of PPARγ to induce adipogenesis. Our results couple PPARγ-dependent adipocyte differentiation to MEN1-related lipoma development and possibly other tissue-specific manifestations of MEN1. The functional linkage of PPARγ and menin could form the basis for future therapeutic interventions for MEN1 patients.
Acknowledgments
We thank the members of the Timmers laboratory, especially P. de Graaf, F. van Werven, and P. Pijnappel, for advice and P. Pijnappel for critical reading of the manuscript. We thank A. Gijsbers-Bruggink for assistance with collecting human samples and A. Koppen, N. Hamers, K. Mulder, and K. van Veghel for technical assistance. The MEF cell lines are a kind gift of C. Zhang. G. Folkers, K. Ge, G. Weber, and M. Stallcup provided plasmids.
This work was supported by The Netherlands Organization for Health Research and Development (ZonMw; AGIKO 920-03-231 to C.J.M.L., H.T.M.T., and K.M.A.D.), The Netherlands Proteomics Center (to H.T.M.T. and K.M.A.D.), and The Netherlands Metabolomics Center (to O.V.B., E.H.J., J.W.M.H., and E.K.).
Footnotes
Published ahead of print on 13 July 2009.
REFERENCES
- 1.Agarwal, S. K., S. Impey, S. McWeeney, P. C. Scacheri, F. S. Collins, R. H. Goodman, A. M. Spiegel, and S. J. Marx. 2007. Distribution of menin-occupied regions in chromatin specifies a broad role of menin in transcriptional regulation. Neoplasia 9101-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albert, T. K., H. Hanzawa, Y. I. Legtenberg, M. J. de Ruwe, F. A. van den Heuvel, M. A. Collart, R. Boelens, and H. T. Timmers. 2002. Identification of a ubiquitin-protein ligase subunit within the CCR4-NOT transcription repressor complex. EMBO J. 21355-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altiok, S., M. Xu, and B. M. Spiegelman. 1997. PPARgamma induces cell cycle withdrawal: inhibition of E2F/DP DNA-binding activity via down-regulation of PP2A. Genes Dev. 111987-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertolino, P., I. Radovanovic, H. Casse, A. Aguzzi, Z. Q. Wang, and C. X. Zhang. 2003. Genetic ablation of the tumor suppressor menin causes lethality at mid-gestation with defects in multiple organs. Mech. Dev. 120549-560. [DOI] [PubMed] [Google Scholar]
- 5.Brandi, M. L., R. F. Gagel, A. Angeli, J. P. Bilezikian, P. Beck-Peccoz, C. Bordi, B. Conte-Devolx, A. Falchetti, R. G. Gheri, A. Libroia, C. J. Lips, G. Lombardi, M. Mannelli, F. Pacini, B. A. Ponder, F. Raue, B. Skogseid, G. Tamburrano, R. V. Thakker, N. W. Thompson, P. Tomassetti, F. Tonelli, S. A. Wells, Jr., and S. J. Marx. 2001. Guidelines for diagnosis and therapy of MEN type 1 and type 2. J. Clin. Endocrinol. Metab. 865658-5671. [DOI] [PubMed] [Google Scholar]
- 6.Brelivet, Y., S. Kammerer, N. Rochel, O. Poch, and D. Moras. 2004. Signature of the oligomeric behaviour of nuclear receptors at the sequence and structural level. EMBO Rep. 5423-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, D., H. Ma, H. Hong, S. S. Koh, S. M. Huang, B. T. Schurter, D. W. Aswad, and M. R. Stallcup. 1999. Regulation of transcription by a protein methyltransferase. Science 2842174-2177. [DOI] [PubMed] [Google Scholar]
- 8.Darling, T. N., M. C. Skarulis, S. M. Steinberg, S. J. Marx, A. M. Spiegel, and M. Turner. 1997. Multiple facial angiofibromas and collagenomas in patients with multiple endocrine neoplasia type 1. Arch. Dermatol. 133853-857. [PubMed] [Google Scholar]
- 9.Dreijerink, K. M., K. W. Mulder, G. S. Winkler, J. W. Hoppener, C. J. Lips, and H. T. Timmers. 2006. Menin links estrogen receptor activation to histone H3K4 trimethylation. Cancer Res. 664929-4935. [DOI] [PubMed] [Google Scholar]
- 10.Dubois, M., F. Pattou, J. Kerr-Conte, V. Gmyr, B. Vandewalle, P. Desreumaux, J. Auwerx, K. Schoonjans, and J. Lefebvre. 2000. Expression of peroxisome proliferator-activated receptor gamma (PPARgamma) in normal human pancreatic islet cells. Diabetologia 431165-1169. [DOI] [PubMed] [Google Scholar]
- 11.Franklin, D. S., V. L. Godfrey, D. A. O'Brien, C. Deng, and Y. Xiong. 2000. Functional collaboration between different cyclin-dependent kinase inhibitors suppresses tumor growth with distinct tissue specificity. Mol. Cell. Biol. 206147-6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ge, K., M. Guermah, C. X. Yuan, M. Ito, A. E. Wallberg, B. M. Spiegelman, and R. G. Roeder. 2002. Transcription coactivator TRAP220 is required for PPAR gamma 2-stimulated adipogenesis. Nature 417563-567. [DOI] [PubMed] [Google Scholar]
- 13.Goke, R., A. Goke, B. Goke, W. S. El-Deiry, and Y. Chen. 2001. Pioglitazone inhibits growth of carcinoid cells and promotes TRAIL-induced apoptosis by induction of p21waf1/cip1. Digestion 6475-80. [DOI] [PubMed] [Google Scholar]
- 14.Gurnell, M., J. M. Wentworth, M. Agostini, M. Adams, T. N. Collingwood, C. Provenzano, P. O. Browne, O. Rajanayagam, T. P. Burris, J. W. Schwabe, M. A. Lazar, and V. K. Chatterjee. 2000. A dominant-negative peroxisome proliferator-activated receptor gamma (PPARgamma) mutant is a constitutive repressor and inhibits PPARgamma-mediated adipogenesis. J. Biol. Chem. 2755754-5759. [DOI] [PubMed] [Google Scholar]
- 15.Heaney, A. P., M. Fernando, and S. Melmed. 2003. PPAR-gamma receptor ligands: novel therapy for pituitary adenomas. J. Clin. Investig. 1111381-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heery, D. M., E. Kalkhoven, S. Hoare, and M. G. Parker. 1997. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387733-736. [DOI] [PubMed] [Google Scholar]
- 17.Hughes, C. M., O. Rozenblatt-Rosen, T. A. Milne, T. D. Copeland, S. S. Levine, J. C. Lee, D. N. Hayes, K. S. Shanmugam, A. Bhattacharjee, C. A. Biondi, G. F. Kay, N. K. Hayward, J. L. Hess, and M. Meyerson. 2004. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol. Cell 13587-597. [DOI] [PubMed] [Google Scholar]
- 18.Jeninga, E. H., O. van Beekum, A. D. van Dijk, N. Hamers, B. I. Hendriks-Stegeman, A. M. Bonvin, R. Berger, and E. Kalkhoven. 2007. Impaired peroxisome proliferator-activated receptor gamma function through mutation of a conserved salt bridge (R425C) in familial partial lipodystrophy. Mol. Endocrinol. 211049-1065. [DOI] [PubMed] [Google Scholar]
- 19.Kalkhoven, E., J. E. Valentine, D. M. Heery, and M. G. Parker. 1998. Isoforms of steroid receptor co-activator 1 differ in their ability to potentiate transcription by the oestrogen receptor. EMBO J. 17232-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karnik, S. K., H. Chen, G. W. McLean, J. J. Heit, X. Gu, A. Y. Zhang, M. Fontaine, M. H. Yen, and S. K. Kim. 2007. Menin controls growth of pancreatic beta-cells in pregnant mice and promotes gestational diabetes mellitus. Science 318806-809. [DOI] [PubMed] [Google Scholar]
- 21.Kim, H., J. E. Lee, E. J. Cho, J. O. Liu, and H. D. Youn. 2003. Menin, a tumor suppressor, represses JunD-mediated transcriptional activity by association with an mSin3A-histone deacetylase complex. Cancer Res. 636135-6139. [PubMed] [Google Scholar]
- 22.Kishida, K., I. Shimomura, H. Nishizawa, N. Maeda, H. Kuriyama, H. Kondo, M. Matsuda, H. Nagaretani, N. Ouchi, K. Hotta, S. Kihara, T. Kadowaki, T. Funahashi, and Y. Matsuzawa. 2001. Enhancement of the aquaporin adipose gene expression by a peroxisome proliferator-activated receptor gamma. J. Biol. Chem. 27648572-48579. [DOI] [PubMed] [Google Scholar]
- 23.Klein, F. A., R. A. Atkinson, N. Potier, D. Moras, and J. Cavarelli. 2005. Biochemical and NMR mapping of the interface between CREB-binding protein and ligand binding domains of nuclear receptor: beyond the LXXLL motif. J. Biol. Chem. 2805682-5692. [DOI] [PubMed] [Google Scholar]
- 24.La, P., A. C. Silva, Z. Hou, H. Wang, R. W. Schnepp, N. Yan, Y. Shi, and X. Hua. 2004. Direct binding of DNA by tumor suppressor menin. J. Biol. Chem. 27949045-49054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, J., P. K. Saha, Q. H. Yang, S. Lee, J. Y. Park, Y. Suh, S. K. Lee, L. Chan, R. G. Roeder, and J. W. Lee. 2008. Targeted inactivation of MLL3 histone H3-Lys-4 methyltransferase activity in the mouse reveals vital roles for MLL3 in adipogenesis. Proc. Natl. Acad. Sci. USA 10519229-19234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, S., D. K. Lee, Y. Dou, J. Lee, B. Lee, E. Kwak, Y. Y. Kong, S. K. Lee, R. G. Roeder, and J. W. Lee. 2006. Coactivator as a target gene specificity determinant for histone H3 lysine 4 methyltransferases. Proc. Natl. Acad. Sci. USA 10315392-15397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, S., J. Lee, S. K. Lee, and J. W. Lee. 2008. Activating signal cointegrator-2 is an essential adaptor to recruit histone H3 lysine 4 methyltransferases MLL3 and MLL4 to the liver X receptors. Mol. Endocrinol. 221312-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lehrke, M., and M. A. Lazar. 2005. The many faces of PPARgamma. Cell 123993-999. [DOI] [PubMed] [Google Scholar]
- 29.Lemos, M. C., and R. V. Thakker. 2008. Multiple endocrine neoplasia type 1 (MEN1): analysis of 1336 mutations reported in the first decade following identification of the gene. Hum. Mutat. 2922-32. [DOI] [PubMed] [Google Scholar]
- 30.Lin, S. Y., and S. J. Elledge. 2003. Multiple tumor suppressor pathways negatively regulate telomerase. Cell 113881-889. [DOI] [PubMed] [Google Scholar]
- 31.Messager, M., C. Carriere, X. Bertagna, and Y. de Keyzer. 2006. RT-PCR analysis of corticotroph-associated genes expression in carcinoid tumours in the ectopic-ACTH syndrome. Eur. J. Endocrinol. 154159-166. [DOI] [PubMed] [Google Scholar]
- 32.Milne, T. A., C. M. Hughes, R. Lloyd, Z. Yang, O. Rozenblatt-Rosen, Y. Dou, R. W. Schnepp, C. Krankel, V. A. Livolsi, D. Gibbs, X. Hua, R. G. Roeder, M. Meyerson, and J. L. Hess. 2005. Menin and MLL cooperatively regulate expression of cyclin-dependent kinase inhibitors. Proc. Natl. Acad. Sci. USA 102749-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mo, R., S. M. Rao, and Y. J. Zhu. 2006. Identification of the MLL2 complex as a coactivator for estrogen receptor alpha. J. Biol. Chem. 28115714-15720. [DOI] [PubMed] [Google Scholar]
- 34.Morrison, R. F., and S. R. Farmer. 1999. Role of PPARgamma in regulating a cascade expression of cyclin-dependent kinase inhibitors, p18(INK4c) and p21(Waf1/Cip1), during adipogenesis. J. Biol. Chem. 27417088-17097. [DOI] [PubMed] [Google Scholar]
- 35.Nolte, R. T., G. B. Wisely, S. Westin, J. E. Cobb, M. H. Lambert, R. Kurokawa, M. G. Rosenfeld, T. M. Willson, C. K. Glass, and M. V. Milburn. 1998. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature 395137-143. [DOI] [PubMed] [Google Scholar]
- 36.Occhi, G., N. Albiger, S. Berlucchi, M. Gardiman, M. Scanarini, R. Scienza, A. Fassina, F. Mantero, and C. Scaroni. 2007. Peroxisome proliferator-activated receptor gamma in the human pituitary gland: expression and splicing pattern in adenomas versus normal pituitary. J. Neuroendocrinol. 19552-559. [DOI] [PubMed] [Google Scholar]
- 37.Pijnappel, W. W., H. F. Hendriks, G. E. Folkers, C. E. van den Brink, E. J. Dekker, C. Edelenbosch, P. T. van der Saag, and A. J. Durston. 1993. The retinoid ligand 4-oxo-retinoic acid is a highly active modulator of positional specification. Nature 366340-344. [DOI] [PubMed] [Google Scholar]
- 38.Rosen, E. D., R. N. Kulkarni, P. Sarraf, U. Ozcan, T. Okada, C.-H. Hsu, D. Eisenman, M. A. Magnuson, F. J. Gonzalez, C. R. Kahn, and B. M. Spiegelman. 2003. Targeted elimination of peroxisome proliferator-activated receptor γ in β cells leads to abnormalities in islet mass without compromising glucose homeostasis. Mol. Cell. Biol. 237222-7229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruthenburg, A. J., C. D. Allis, and J. Wysocka. 2007. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol. Cell 2515-30. [DOI] [PubMed] [Google Scholar]
- 40.Scacheri, P. C., S. Davis, D. T. Odom, G. E. Crawford, S. Perkins, M. J. Halawi, S. K. Agarwal, S. J. Marx, A. M. Spiegel, P. S. Meltzer, and F. S. Collins. 2006. Genome-wide analysis of menin binding provides insights into MEN1 tumorigenesis. PLoS Genet. 2e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Semple, R. K., V. K. Chatterjee, and S. O'Rahilly. 2006. PPAR gamma and human metabolic disease. J. Clin. Investig. 116581-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sierra, J., T. Yoshida, C. A. Joazeiro, and K. A. Jones. 2006. The APC tumor suppressor counteracts beta-catenin activation and H3K4 methylation at Wnt target genes. Genes Dev. 20586-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sowa, H., H. Kaji, L. Canaff, G. N. Hendy, T. Tsukamoto, T. Yamaguchi, K. Miyazono, T. Sugimoto, and K. Chihara. 2003. Inactivation of menin, the product of the multiple endocrine neoplasia type 1 gene, inhibits the commitment of multipotential mesenchymal stem cells into the osteoblast lineage. J. Biol. Chem. 27821058-21069. [DOI] [PubMed] [Google Scholar]
- 44.Steger, D. J., M. I. Lefterova, L. Ying, A. J. Stonestrom, M. Schupp, D. Zhuo, A. L. Vakoc, J.-E. Kim, J. Chen, M. A. Lazar, G. A. Blobel, and C. R. Vakoc. 2008. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol. Cell. Biol. 282825-2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takada, I., M. Mihara, M. Suzawa, F. Ohtake, S. Kobayashi, M. Igarashi, M. Y. Youn, K. Takeyama, T. Nakamura, Y. Mezaki, S. Takezawa, Y. Yogiashi, H. Kitagawa, G. Yamada, S. Takada, Y. Minami, H. Shibuya, K. Matsumoto, and S. Kato. 2007. A histone lysine methyltransferase activated by non-canonical Wnt signalling suppresses PPAR-gamma transactivation. Nat. Cell Biol. 91273-1285. [DOI] [PubMed] [Google Scholar]
- 46.Tontonoz, P., E. Hu, R. A. Graves, A. I. Budavari, and B. M. Spiegelman. 1994. mPPAR gamma 2: tissue-specific regulator of an adipocyte enhancer. Genes Dev. 81224-1234. [DOI] [PubMed] [Google Scholar]
- 47.Tontonoz, P., E. Hu, and B. M. Spiegelman. 1994. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell 791147-1156. [DOI] [PubMed] [Google Scholar]
- 48.Tontonoz, P., S. Singer, B. M. Forman, P. Sarraf, J. A. Fletcher, C. D. Fletcher, R. P. Brun, E. Mueller, S. Altiok, H. Oppenheim, R. M. Evans, and B. M. Spiegelman. 1997. Terminal differentiation of human liposarcoma cells induced by ligands for peroxisome proliferator-activated receptor gamma and the retinoid X receptor. Proc. Natl. Acad. Sci. USA 94237-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tyagi, S., A. L. Chabes, J. Wysocka, and W. Herr. 2007. E2F activation of S phase promoters via association with HCF-1 and the MLL family of histone H3K4 methyltransferases. Mol. Cell 27107-119. [DOI] [PubMed] [Google Scholar]
- 50.Vermeulen, M., K. W. Mulder, S. Denissov, W. W. Pijnappel, F. M. van Schaik, R. A. Varier, M. P. Baltissen, H. G. Stunnenberg, M. Mann, and H. T. Timmers. 2007. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 13158-69. [DOI] [PubMed] [Google Scholar]
- 51.Vortmeyer, A. O., R. Boni, E. Pak, S. Pack, and Z. Zhuang. 1998. Multiple endocrine neoplasia 1 gene alterations in MEN1-associated and sporadic lipomas. J. Natl. Cancer Inst. 90398-399. [DOI] [PubMed] [Google Scholar]
- 52.Winkler, G. S., K. W. Mulder, V. J. Bardwell, E. Kalkhoven, and H. T. Timmers. 2006. Human Ccr4-Not complex is a ligand-dependent repressor of nuclear receptor-mediated transcription. EMBO J. 253089-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wysocka, J., T. Swigut, H. Xiao, T. A. Milne, S. Y. Kwon, J. Landry, M. Kauer, A. J. Tackett, B. T. Chait, P. Badenhorst, C. Wu, and C. D. Allis. 2006. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 44286-90. [DOI] [PubMed] [Google Scholar]
- 54.Yadav, N., D. Cheng, S. Richard, M. Morel, V. R. Iyer, C. M. Aldaz, and M. T. Bedford. 2008. CARM1 promotes adipocyte differentiation by coactivating PPARgamma. EMBO Rep. 9193-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yokoyama, A., Z. Wang, J. Wysocka, M. Sanyal, D. J. Aufiero, I. Kitabayashi, W. Herr, and M. L. Cleary. 2004. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol. Cell. Biol. 245639-5649. [DOI] [PMC free article] [PubMed] [Google Scholar]