Abstract
Nucleophosmin (NPM1) is a multifunctional protein involved in the regulation of centrosome duplication, ribosome biogenesis, genomic stability, histone chaperone function, and transcription. Overexpression of NPM1 is associated with cancers of diverse histological origins. Here, we have found that p300-mediated acetylation of NPM1 modulates its subcellular localization and augments its oncogenic potential. Acetylated NPM1 is predominantly localized in the nucleoplasm, where it associates with transcriptionally active RNA polymerase II. Deacetylation of NPM1 is brought about by human SIRT1 and reduces its transcriptional activation potential. Remarkably, increased levels of acetylated NPM1 were found in grade II and III oral squamous cell carcinoma (OSCC) patient samples. Small interfering RNA (siRNA)-mediated knockdown of NPM1 in an OSCC cell line, followed by microarray analysis and chromatin immunoprecipitation experiments, revealed that some of the genes involved in oral cancer malignancy are regulated by NPM1 and have acetylated NPM1 localized at their promoters. Either suppression of p300 by siRNA or mutation of acetylatable lysine residues of NPM1 resulted in reduced occupancy of acetylated NPM1 on the target gene promoter concomitant with its decreased transcript levels. These observations suggest that acetylated NPM1 transcriptionally regulates genes involved in cell survival and proliferation during carcinogenesis.
NPM1 (nucleophosmin or B23) is a multifaceted nucleolar protein which is involved in several cellular processes, including ribosome biogenesis (15), nucleocytoplasmic transport, centrosome duplication (12, 19), embryonic development (4), histone chaperone function, and transcriptional regulation (14, 18). NPM1 undergoes a variety of posttranslational modifications, such as phosphorylation, acetylation, sumoylation, ubiquitination, and poly(ADP-ribosyl)ation, which direct its various cellular functions (13). Phosphorylation of NPM1 by CDK2/cyclin E regulates the initiation of centrosome duplication (12), whereas p300-mediated acetylation of NPM1 enhances its histone chaperone activity and transcriptional activation potential. Acetylated NPM1 has an enhanced ability to interact with acetylated core histones and activate transcription from the chromatin template (18). NPM1 has a role in cell proliferation and transformation and is overexpressed in human cancers of diverse histological origins (3). Aberrant gene function and altered patterns of gene expression induced by epigenetic events are key features of cancer (6). NPM1 inactivation leads to unrestricted centrosome duplication and genomic instability, implying that NPM1 is essential for embryonic development and the maintenance of genomic stability (4). Thus, NPM1 may serve as a tumor suppressor because abrogation of its function leads to tumorigenic phenotypes (10). By contrast, NPM1 also regulates cell proliferation and may have oncogenic potential when overexpressed. A study using clinical cancer samples has shown a correlation between the level of NPM1 and cancerous growth (21), suggesting that NPM1 functions as a positive regulator of cell proliferation during carcinogenesis (10). However, the posttranslational modification status of NPM1 during such malignancies is not known. In this study, we have found that acetylated NPM1 associates with transcriptionally active foci in cells. SIRT1 deacetylates NPM1, which reduces its potency as an activator of gene expression. In our studies with grade II oral cancer patient samples, we have observed a significant enhancement of the levels of acetylated NPM1 in malignant oral tumor tissues with respect to adjacent normal tissues. Additionally, we have also found higher expression of p300 in tumors. Small interfering RNA (siRNA)-mediated knockdown of NPM1 resulted in the differential regulation of gene expression globally and alteration of several genes associated with cancers. By using chromatin immunoprecipitation (ChIP) assays, we demonstrate that acetylated NPM1 occupies the promoter and directly regulates the transcriptional activity of some of the genes associated with oral cancer manifestation. A fine balance between SIRT1 and p300 activity maintains the NPM1 acetylation status in cells. Depletion of either SIRT1 or p300 results in altered levels of acetylated NPM1 at target gene promoters and consequently influences the expression of those genes.
MATERIALS AND METHODS
Identification of the in vivo acetylation sites of NPM1.
HEK293T whole-cell extracts were incubated with a highly specific anti-NPM1 monoclonal antibody (Sigma). The immune complex was precipitated with protein G-Sepharose and processed for matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) analysis as described previously (18).
Generation of polyclonal antibodies specific for acetylated NPM1.
Based on the identified acetylation sites, two different peptides having acetylated lysine residues (AcK) with N-terminal keyhole limpet hemocyanin (KLH) conjugation were designed, i.e., AcNPM1-01 [(KLH)-C-NG(AcK)DS(AcK)PSSTPRSKGQESF(AcK)(AcK)Q (residues 210 to 231)] and AcNPM1-02 [(KLH)-C-MQASIE(AcK)GGSLPKVEA(AcK)FI (residues 251 to 269)]. Polyclonal antibodies against these two peptides were raised in rabbits. The specificity of the antibodies was confirmed by acetylated-NPM1 peptide blocking studies. Western blotting and immunofluorescence studies were also done with rabbit preimmune serum (PIS) as a negative control for anti-AcNPM1-01 and anti-AcNPM1-02 antibodies.
Site-directed mutagenesis and plasmid constructs.
Lysine-to-arginine point mutations of His6-NPM1 were made by the site-directed mutagenesis technique (QuikChange II XL site-directed mutagenesis kit; Stratagene). The mutations were confirmed by sequencing. The functional nature of the 7K-7R mutant NPM1 protein was confirmed by histone pulldown assays at 300 mM salt (data not shown). The wild-type (WT) and 7K-7R mutant NPM1 proteins were subcloned into a pCMV-Flag mammalian expression vector (Sigma), where the Flag epitope was used to tag the N terminus. The WT and 7K-7R mutant NPM1 proteins were also subcloned into the enhanced green fluorescent protein N1 vector (see Fig. S4 in the supplemental material). Transient transfections were carried out with 60 to 80% confluent cells for 36 to 48 h by using Effectene transfection reagent in accordance with the manufacturer's instructions (Qiagen).
Protein purification.
The His6-tagged WT and 7K-7R mutant NPM1 proteins, the His6-tagged Tip60-HAT domain, and full-length His6-tagged GCN5 were purified from Escherichia coli cells with an Ni-nitrilotriacetic acid affinity column (18). Full-length p300 was purified from recombinant baculovirus-infected Sf21 cells as a His6-tagged protein, and Flag epitope-tagged CREB binding protein (CBP), p 300/CBP-associated factor, and histone deacetylase 1 (HDAC1) were purified from recombinant baculovirus-infected Sf21 insect cells by immunoaffinity purification with Flag-M2-agarose resin (Sigma) as described previously (18). Glutathione S-transferase (GST)-tagged proteins yeast Sir2 (ySir2) and human SIRT1 (hSIRT1) were purified with GST bind resin (Novagen) and eluted with GST elution buffer (10 mM reduced glutathione, 50 mM Tris-HCl, pH 8.0).
Histone acetyltransferase (HAT) and HDAC assays.
In vitro acetylation and deacetylation assays were performed as described previously (1), with 1 μg of bacterially expressed His6-NPM1. The activities of different HATs and HDACs were normalized with 2 μg of core histones (purified from HeLa cells) as the substrate.
Cell culture and immunofluorescence studies.
HEK293T and KB cells were grown on polylysine-coated coverslips for 24 to 48 h in the presence of Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum. Cells were fixed with 4% paraformaldehyde and permeabilized with 1% Triton X-100. Anti-AcNPM1-01, anti-NPM1 (Zymed and Sigma), anti-RNA polymerase II (anti-RNAP II; Santa Cruz and Abcam), and anti-Flag (Sigma) antibodies were used as primary antibodies in this study. Fluorescently labeled anti-mouse Alexa 568- and anti-rabbit Alexa 488 (Invitrogen)-conjugated antibodies were used as secondary antibodies. Alexa and Hoechst fluorescence was visualized with a Carl Zeiss confocal laser scanning microscope (Axioskop 2 Plus). Images were captured with an AxioCam MRc camera. AxioVision 3.1 software was used to process the images and calculate the weighted colocalization coefficients. The images for comparative studies were captured at identical microscope settings.
Luciferase assay.
The mammalian expression constructs of Flag-tagged WT NPM1, Flag-tagged 7K-7R mutant NPM1, Gal4-VP16, and p53 used in this study were placed under the control of a cytomegalovirus promoter. The Gal4-VP16-responsive reporter construct pG10luc contains 10 Gal4-binding sites in tandem, followed by the luciferase gene. The p53-responsive reporter construct pG13luc contains 13 tandemly arranged p53-binding sites, followed by a polyomavirus promoter, which drives the luciferase gene. The pCMV-β-galactosidase construct was used as an internal control. HEK293T cells were grown to 80% confluence in a 30-mm dish and transfected with different plasmid constructs by using Effectene reagent (Qiagen). The total amount of DNA was kept constant for each transfection. Luciferase and β-galactosidase activities were measured 48 h after transfection according to the manufacturer's protocol (Promega, Madison, WI). Values are representative of at least three independent biological replicates (transfection). The same volume of cell lysates used for taking luciferase counts was checked for the expression of Flag-tagged proteins through Western blot analysis. The glyceraldehyde-3-phosphate dehydrogenase level was used as a loading control.
Immunohistochemistry (IHC) analysis.
Oral cancer patient samples were collected at the Bangalore Institute of Oncology. The Bangalore Institute of Oncology ethical clearance committee agreed to this investigation, and informed consent was obtained from all patients. Tumor and adjacent normal tissues from 18 patient samples (see Table S2 in the supplemental material) were dehydrated, paraffin embedded, and sectioned with a microtome (Leica). Briefly, after blocking, the sections were incubated with different primary antibodies, i.e., anti-NPM1 (Zymed), anti-AcNPM1-01, anti-SIRT1 (Santa Cruz), and anti-p300-C-20 (Santa Cruz) antibodies. IHC analysis was performed with a Strept-Avidin Biotin kit (Dako). Immunoreactivity (brown precipitate) was developed in the chromogen diaminobenzidine tetrahydrochloride (Sigma), and counterstaining was done with hematoxylin. The staining intensity of the images was evaluated by two independent pathologists.
Statistical analysis.
Five independent fields (around 100 cells) from normal and malignant IHC sections of tissue samples were scored for the percentage of cells positive for acetylated NPM1 and similarly scored for the percentage of cells positive for nucleoplasmic NPM1. Paired t tests were performed with GraphPad Prism software to analyze differences between means. Results were considered significant when the P value was <0.05 in a paired t test.
Tissue lysate preparation and RNA isolation.
Whole-cell proteins were extracted from patient tissue samples in lysis buffer (50 mM Tris HCl [pH 8.0], 125 mM NaCl, 1 mM EDTA, 0.5% NP-40, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride). TRIzol reagent (Invitrogen) was used for RNA isolation from tissues stored in RNAlater solution (Ambion).
siRNA knockdown of NPM1, p300, and SIRT1.
The two siRNAs (Qiagen) used for NPM1 knockdown are NPM1 siRNAs 1 (sense, 5′-GAAUUGCUUCCGGAUGACU-dTdT; antisense, 5′-AGUCAUCCGGAAGCAAUUC-dTdT) and 2 (sense, 5′-AGGUGGUUCUCUUCCCAAA-dTdT; antisense, 5′-UUUGGGAAGAGAACCACCU-dTdT). The siRNAs (Sigma) used to knock down p300 and SIRT1 are p300 siRNA (sense, 5′-CUAGAGACACCUUGUAGUA-dTdT; antisense, 5′-UACUACAAGGUGUCUCUAG-dTdT) and SIRT1 siRNA (sense, 5′-GUGUC AUGGUUCCUUUGCA-dTdT; antisense, 5′-UGCAAAGGAACCAUGACAC- dTdT).
Alexa 488-conjugated all-star negative control siRNA (Human Starter Kit; Qiagen) was used as a control. siRNA transfections of KB and HEK293T cells were done for 48 h with Hiperfect transfection reagent (Qiagen) according to the manufacturer's instructions. The p300 and SIRT1 siRNA transfections resulted in almost 1.5-fold reductions in their transcript levels (see Fig. S3 in the supplemental material). The efficiency of knockdown by NPM1 siRNA in a real-time reverse transcription (RT)-PCR was around three- to fourfold, while at the protein level it was more than twofold.
The primers used in this study are TNF-α Forward (5′-ATGAGCACTGAAAGCATGATCCGGG-3′), TNF-α Reverse (5′-ACCTGGGAGTAGATGAGGTACAGGCCC-3′), IL6ST Forward (5′-ATGTTGACGTTGCAGACTTGGCTAGTG-3′), IL6ST Reverse (5′-TCAAGCTGTCCGAATGTAAGAATGTTGC-3′), CTNB1 Forward (5′-ATGGCTACTCAAGCTGATTTGATGGAG-3′), CTNB1 Reverse (5′-GCATGCCCTCATCTAATGTCTCAGGG-3′), NPM1 Forward (5′-ATGGAAGATTCGATGGACATGG-3′), NPM1 Reverse (5′-CGAGAAGAGACTTCCTCCACTGC-3′), p300 Forward (5′-ATGGCCGAGAATGTGGTG-3′), p300 Reverse (5′-ATTATCCCTTGTCCATTGCC-3′), SIRT1 Forward (5′-AAGATGGCGGACGAGGCGGCCCTC-3′), SIRT1 Reverse (5′-GAAGGTTATCTCGGTACCCAATCGC-3′), Actin Forward (5′-GTGGGGCGCCCCAGGCACCA-3′), and Actin Reverse (5′-CTCCTTAATGTCACGCACGATTTC-3′).
ChIP assays.
HEK293T and KB cells were maintained in Dulbecco modified Eagle medium and 10% fetal bovine serum to 80% confluence. The pulldowns for ChIP assays were performed with anti-NPM1 antibody (Zymed and Sigma) and purified anti-AcNPM1-01, anti-RNAP II (Abcam and Santa Cruz), anti-pS5-RNAP II (Abcam), anti-acetylated histone H3 (H3AcK9AcK14; Calbiochem), anti-SIRT1 (Santa Cruz), anti-p300-C-20 (Santa Cruz), and anti-Flag M2-agarose (Sigma) antibodies. ChIP assays were done as described earlier (7). For sequential ChIP (reChIP) assays, the specific DNA-protein complexes from the first immunoprecipitation were extracted twice by adding 25 μl of 10 mM dithiothreitol, followed by incubation for 20 min at 37°C with vortexing every 5 min. The supernatants were pooled and diluted 10 times with reChIP buffer (20 mM Tris-HCl [pH 8.0], 2 mM EDTA, 150 mM NaCl, 0.1% Triton X-100), following which a second immunoprecipitation was performed. Real-time PCR analysis was done with primers specific for the tumor necrosis factor alpha (TNF-α) upstream promoter (+1 to −170). Paired t tests were performed with GraphPad Prism software to analyze the differences between means. The sequences of the primers used in this study are as follows: TNF-α promoter Forward, 5′-CTCCCTCAGCAAGGACAGCAGAGGAC-3′; TNF-α promoter Reverse, 5′-CATGGTGTCCTTTCCAGGGGAGAG-3′.
RESULTS
Acetylated NPM1 localizes to the nucleoplasm and associates with transcriptionally active RNAP II.
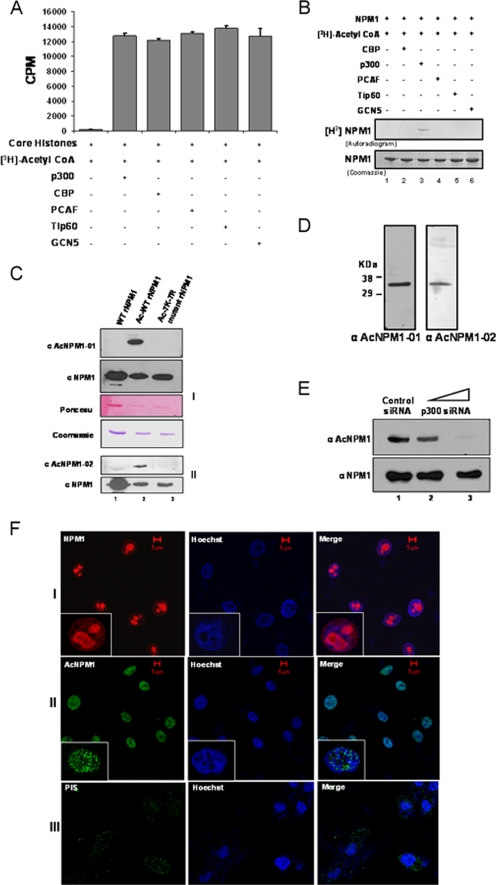
NPM1 is acetylated by p300 in vitro at multiple lysine residues (18). In order to find out the enzyme specificity of NPM1 acetylation, the activities of different classes of HATs, like p300, CBP, p 300/CBP-associated factor, GCN5, and Tip60, were normalized with core histones (purified from HeLa cells) as the substrate (Fig. 1A; see Fig. S1, left panel, in the supplemental material) and the HATs were subsequently tested for the ability to acetylate NPM1 in vitro. Apart from p300, none of the other HATs were able to acetylate NPM1 (Fig. 1B, lane 3 versus lanes 2 and 4 to 6; see Fig. S1, right panel, in the supplemental material), suggesting that NPM1 is specifically acetylated by p300. In order to identify the lysine residues which are acetylated in vivo, the endogenous NPM1 protein was immunoprecipitated from HEK293T cells with a highly specific monoclonal antibody against NPM1 and subjected to MALDI-TOF analysis (see Table S1 in the supplemental material). Using this strategy, we found that K212, K215, K229, K230, K257, K267, and K292 are the acetylation sites of NPM1. However, acetylation of K292 was detected only in vitro (18). Based on this analysis, acetylated peptides spanning two different regions of NPM1 were designed (see Materials and Methods) and used to raise highly specific polyclonal antibodies named anti-AcNPM1-01 and anti-AcNPM1-02. The anti-AcNPM1-01 antibody was mainly used for further experiments. The specificity of both of the antibodies was characterized by Western blot analysis. Both of the antibodies recognized acetylated NPM1 but not unmodified NPM1 (Fig. 1C, compare lane 1 with lane 2). To further confirm the sites, all of the acetylatable lysine residues of NPM1 (as identified by MALDI-TOF analysis) were replaced with arginine by site-directed mutagenesis. The acetylation-defective 7K-7R mutant NPM1 protein (after being subjected to an in vitro acetylation reaction) was not recognized by anti-acetylated NPM1 antibodies (Fig. 1C, lane 3). Both of the antibodies recognized a single band from HEK293T whole-cell lysates (Fig. 1D), and ablation of NPM1 by RNA interference also resulted in reduced acetylated NPM1 levels in the cells (see Fig. S2 in the supplemental material [compare lane 1 with lane 4]). Additionally, we also performed siRNA-mediated silencing of p300 for 48 h (see Materials and Methods; see Fig. S3 in the supplemental material), which resulted in a drastic reduction in NPM1 acetylation status (Fig. 1E, upper panel, lane 3). However, there was no significant drop in the NPM1 protein level. These data suggest that NPM1 is specifically acetylated by p300 even in the cellular system.
FIG. 1.
p300 acetylates NPM1 in the nucleoplasm. (A) Results of a filter binding assay performed to normalize the activities of different HATs with core histones (purified from HeLa cells) as substrates are represented as bar graphs. CoA, coenzyme A. (B) An in vitro HAT assay was performed with 1 μg recombinant NPM1 (rNPM1) as a substrate for different acetyltransferases. (C) Characterization of polyclonal antibodies raised against two acetylated NPM1 peptides, AcNPM1-01 (panel 1) and AcNPM1-02 (panel 2), by Western blotting along with the corresponding NPM1 control. The loading control of the proteins is also shown by Ponceau staining of the membrane, and the same amount of each protein was resolved separately and visualized by Coomassie staining. WT recombinant NPM1 (lane 1), in vitro acetylated WT recombinant NPM1 (lane 2), and 7K-7R mutant recombinant NPM1 were subjected to an in vitro acetylation reaction (lane 3). (D) HEK293T whole-cell extract was subjected to Western blot analysis with anti-AcNPM1-01 and anti-AcNPM1-02 antibodies. (E) Western blot analysis was done with anti-AcNPM1 antibody at 48 h posttransfection of p300 siRNA (upper panel). A corresponding loading control was done with anti-NPM1 antibody (lower panel). (F) Immunofluorescence staining of NPM1 (red) in HEK293T cells with anti-NPM1 monoclonal antibody (panel I) and acetylated NPM1 (green) with an anti-AcNPM1-01 polyclonal antibody (panel II). Rabbit PIS was used as a negative control for anti-Ac-NPM1-01 staining (panel III). DNA staining (blue) was done with Hoechst. Scale bar, 5 μm.
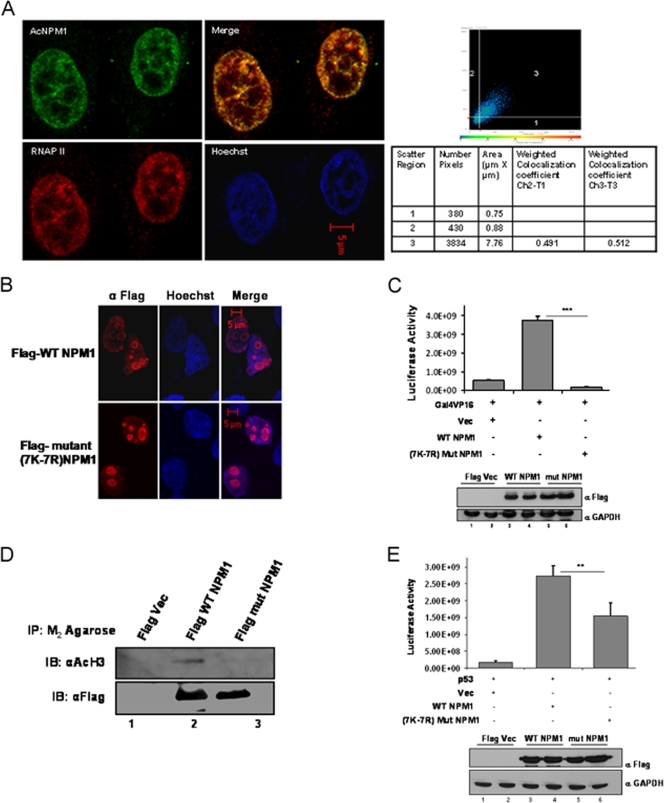
NPM1 is a nuclear protein; however, it shuttles between the nucleolus and the nucleoplasm (9). An immunofluorescence image showed that NPM1 is predominantly localized in the nucleolus and a minor fraction is in the nucleoplasm of interphase cells (Fig. 1F, panel I, inset). Interestingly, we observed that acetylated NPM1 was present in the nucleoplasm in a speckled manner (Fig. 1F, panel II, inset). Rabbit PIS (used for immunization) was used as a negative control to show the specificity of anti-Ac-NPM1 antibody staining (Fig. 1F, panel III). Taken together, these data show that NPM1 is specifically acetylated by p300 and that acetylated NPM1 is predominantly distributed in the nucleoplasm. Acetylation of NPM1 is necessary for its transcriptional activation potential (18), possibly by localizing at the active transcription foci. We carried out coimmunofluorescence analysis to study the distribution of AcNPM1 and RNAP II in cells. Quadrant 3 of the scatter plot shows the colocalized pixels of AcNPM1 and RNAP II (Fig. 2A). The calculated weighted colocalization coefficient determined with Carl Zeiss software for RNAP II is 0.491 (Fig. 2A, tabular data for Ch2-T1), and that for AcNPM1 is 0.512 (Fig. 2A, tabular data for Ch3-T3), both of which are greater than 0.3 and are thus considered significant (according to Pearson's correlation coefficient). The coimmunofluorescence studies illustrated significant colocalization of acetylated NPM1 with RNAP II (Fig. 2A), probably at the actively transcribed regions of the chromatin. Hence, the acetylatable lysine residues of NPM1 would be very essential for its function as a transcriptional enhancer. Flag-tagged mammalian expression constructs of 7K-7R mutant and WT NPM1 were made to verify this possibility. Both the Flag-tagged WT and Flag-tagged 7K-7R mutant NPM1 proteins predominantly localized to the nucleolus in interphase cells (Fig. 2B; see Fig. S4 in the supplemental material). In order to assay for their activity, we used the luciferase reporter assay system employing a Gal4-VP16 (activator)-responsive promoter construct. The transient transfection of Flag-tagged WT NPM1 resulted in around sevenfold transcriptional activation (Fig. 2C, compare lane 1 with lane 2). However, an equivalent amount of Flag-tagged 7K-7R mutant NPM1 transfection (Fig. 2C, bottom panel) failed to induce the activator-dependent transcription any further (Fig. 2C, lane 3). These observations indicate that acetylation of NPM1 is essential to induce transcription in a cellular system. We have previously shown that acetylated NPM1 interacts preferentially with acetylated histones in an in vitro system (18). We probed the in vivo interaction of WT NPM1 and 7K-7R mutant NPM1 with histones to elucidate the mechanism by which WT NPM1 activated Gal4-VP16-driven transcription severalfold, whereas the mutant could not enhance activator-dependent transcription at all. M2-agarose pulldown assays were performed with Flag-tagged WT and 7K-7R mutant NPM1-transfected cells. The immunopulldown complex was probed with anti-acetylated H3 antibody. It was found that WT NPM1 (which is amenable to acetylation) has a higher affinity for interaction with acetylated histone H3 than does 7K-7R mutant NPM1 (Fig. 2D, compare lanes 2 and 3). Acetylated NPM1 could probably facilitate disassembly of acetylated nucleosomes at active promoters and augment the transcriptional output.
FIG. 2.
Acetylated NPM1 colocalizes with RNAP II. (A) Interphase cell nuclei stained with anti-AcNPM1 (green) and anti-RNAP II (red) antibodies. A merged coimmunofluorescence image is shown. DNA staining with Hoechst is blue. The scatter plot denotes weighted colocalization coefficient measurements of the coimmunofluorescence image of AcNPM1 with RNAP II. (B) Immunofluorescence image illustrating the predominant nucleolar localization of both the Flag-tagged WT and 7K-7R mutant NPM1 proteins stained with anti-Flag antibody at 48 h posttransfection in HEK293T cells. DNA was stained with Hoechst (blue). Scale bar, 5 μm. (C) A Gal4-VP16-responsive luciferase assay (pG10luc) was performed with 1 μg of the Flag vector (Vec) alone (lane 1), Flag-tagged WT NPM1 (lane 2), or Flag-tagged 7K-7R mutant NPM1 (lane 3) in HEK293T cells transiently transfected with pG10luc (100 ng), Gal4-VP16 (10 ng), and pCMV-gal (100 ng). Values are means ± standard deviations of triplicates. The data were analyzed by paired t test (***, P < 0.001). Equal expression levels of both the WT and 7K-7R mutant NPM1 proteins of a replicate experiment are depicted by Western blotting with anti-Flag antibody (Sigma). GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (D) An anti-Flag (M2-agarose) pulldown assay was performed with HEK293T cells transfected with either Flag-tagged WT NPM1 or Flag-tagged 7K-7R mutant NPM1. Flag vector-transfected cells were used as a negative control. Following immunoprecipitation, immunoblotting (IB) was done with anti-acetylated histone H3 antibody (AcH3; Calbiochem). Western blotting with anti-Flag antibody was done to show the presence of almost equal amounts of Flag-tagged WT and 7K-7R mutant NPM1 proteins in the pulldown complex. (E) A p53-responsive luciferase assay (pG13luc) was performed with 1 μg of the Flag vector alone (lane 1), Flag-tagged WT NPM1 (lane 2), or Flag-tagged 7K-7R mutant NPM1 (lane 3) in HEK293T cells transiently transfected with pG13luc (100 ng), p53 (10 ng), and pCMV-gal (100 ng). Values are means ± standard deviations of triplicates. The data were analyzed by paired t test (**, P < 0.01). Similar expression levels of both the WT and 7K-7R mutant NPM1 proteins in a replicate experiment are depicted by Western blotting with an anti-Flag antibody.
We have also performed similar experiments with a p53-responsive reporter construct, where we found that WT NPM1 activated p53-dependent reporter gene expression severalfold, whereas 7K-7R mutant NPM1 activated it to a lesser extent than the WT did (Fig. 2E). There could be a differential requirement for acetylated NPM1 to activate diverse genes, which is indicated by the capacity of 7K-7R mutant NPM1 to enhance either Gal4-VP16-dependent or p53-dependent gene expression. The direct interaction of p53 and NPM1 may also contribute to transcriptional activation, which may be independent of its acetylation status.
The class III HDAC SIRT1 deacetylates NPM1.
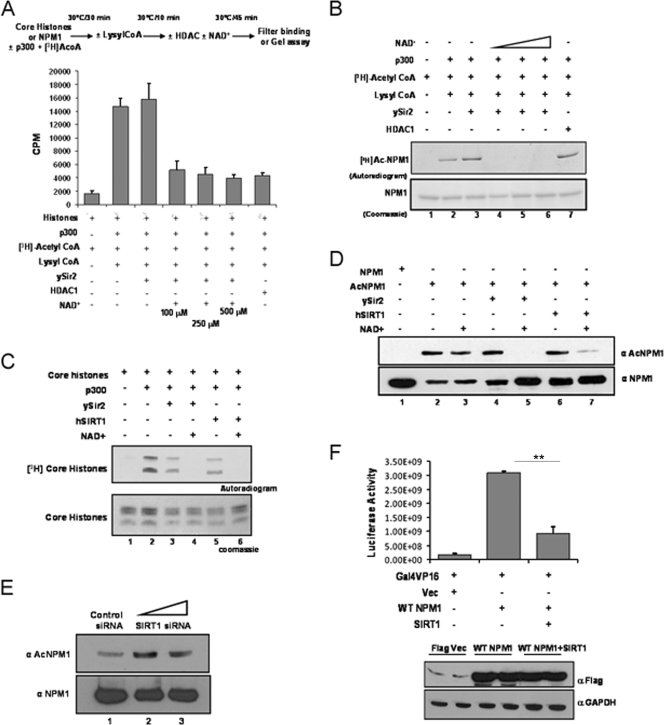
It is well known that the phenomenon of acetylation is a highly dynamic and reversible process. There are several HDACs which catalyze the removal of acetyl groups from histone, as well as nonhistone, protein substrates. Deacetylation of NPM1 by a candidate cellular deacetylase would result in attenuation of its acetylation-dependent function and bring about tighter regulation. In vitro deacetylation assays were performed with different classes (I and III) of HDACs. The HDAC activities of different deacetylases were normalized with core histones (purified from HeLa cells) as the substrate (Fig. 3A). It was found that ySir2, belonging to HDAC class III, could reverse the p300-mediated acetylation of NPM1 in an NAD+-dependent manner (Fig. 3B, lanes 4 to 6), whereas the class I deacetylase HDAC1 failed to do so (Fig. 3B, lane 7). The closest mammalian homologue of ySir2 is SIRT1. The activities of bacterially purified, GST-tagged ySir2 and hSIRT1 (see Fig. S5 in the supplemental material) were normalized with core histones (Fig. 3C). The deacetylation assay was subsequently performed with acetylated NPM1, and the reaction product was probed with anti-AcNPM1-01 antibody. SIRT1 efficiently deacetylated NPM1 comparably to ySir2 (Fig. 3D, compare lane 5 with lane 7). We also performed SIRT1 siRNA transfection and checked the status of NPM1 acetylation in the cells 48 h later (see Materials and Methods; see Fig. S3 in the supplemental material). Western blot analysis revealed that after 48 h of transfection, acetylation of NPM1 was enhanced in SIRT1 knockdown cells compared to that in control siRNA-transfected cells (Fig. 3E), which clearly suggests that SIRT1 is a bona fide NPM1 deacetylase. In order to test the effect of SIRT1-mediated deacetylation on the transcriptional activation ability of NPM1, a luciferase reporter gene expression assay was performed. Transiently transfected Flag-WT NPM1 enhanced transcription from the Gal4-VP16-responsive promoter more than sixfold (Fig. 3F, compare lane 1 with lane 2). However, upon cotransfection of SIRT1, Flag-WT NPM1 enhanced activator-driven transcription minimally (Fig. 3F, compare lane 1 with lane 3). These results indicate that the transcriptional activation potential of NPM1 is determined by its acetylation status, whereas deacetylation of NPM1 drastically reduces its capacity to promote transcription of the target gene. Presumably, acetyltransferase p300 and class III deacetylase SIRT1 dynamically modulate the acetylation status of NPM1 and, as a consequence, profoundly influence an array of biological and pathological processes carried out by NPM1.
FIG. 3.
hSIRT1 deacetylates NPM1. (A) Results of a filter binding assay done to normalize the HDAC activities of ySir2 and HDAC1 with core histones (purified from HeLa cells) as the substrate are represented as bar graphs. The details of the reaction are given as a schematic (upper panel). The p300 inhibitor lysyl coenzyme A (CoA) was added at the end of the acetylation step to prevent reacetylation of proteins in the subsequent deacetylation reaction. (B) Acetylated (by p300) NPM1 was subjected to deacetylation with ySir2 with or without NAD+ and HDAC1. Unlabeled NPM1 (lane 1), 3H-labeled acetylated NPM1 (lane 2), acetylated NPM1 incubated with ySir2 (lane 3), acetylated NPM1 incubated with ySir2 in the presence of increasing concentrations of NAD+ (lanes 4 to 6), and acetylated NPM1 incubated with HDAC1 (lane 7) are shown. (C) Gel picture depicting the normalization of HDAC activity of ySir2 and hSIRT1 with core histones (purified from HeLa cells) as the substrate. (D) Acetylated NPM1 was incubated with the indicated deacetylases with or without NAD+, followed by Western blotting with anti-AcNPM1-01. A corresponding loading control is shown after reprobing of the same blot with anti-NPM1 antibody. (E) A Western blot analysis of SIRT1 siRNA with anti-AcNPM1 antibody was done at 48 h posttransfection (upper panel). A Western blot analysis with anti-NPM1 antibody was done to show the corresponding loading control (lower panel). (F) A luciferase assay was done at 48 h posttransfection with 1 μg of either the Flag vector (Vec) alone (lane 1) or Flag-tagged WT NPM1 (lane 2) and also upon the cotransfection of SIRT1 with Flag-tagged WT NPM1 (lane 3) in HEK293T cells along with pG10luc (100 ng), Gal4-VP16 (10 ng), and pCMV-gal (100 ng). Values are means ± standard deviations of triplicates. The data were analyzed by paired t test (**, P < 0.01). GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Levels of acetylated NPM1 increase in oral cancer.
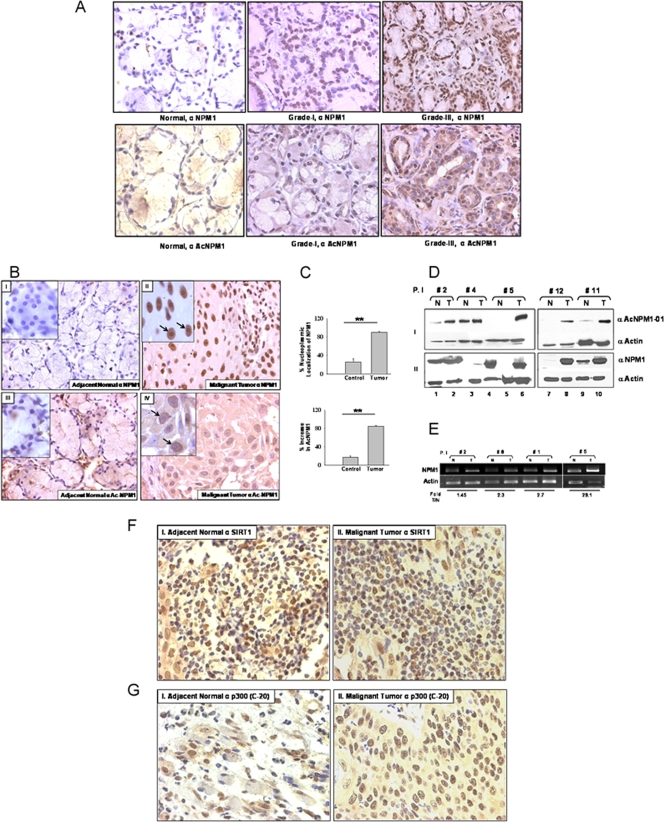
Manifestation of cancer is often found to be associated with alteration of epigenetic marks of chromatin, which include local DNA hypermethylation, global hypomethylation, and alteration of histone acetylation/deacetylation balance (8). NPM1 is reported to be overexpressed in highly proliferative cells compared to normal resting cells and also in cancers of diverse histological origins (3). However, almost no reports are available to give insight into the posttranslational modification status of NPM1 during such malignancies. We investigated the expression levels and acetylation status of NPM1 in precancerous oral lesions (grade I, n = 3) and highly malignant (grade III, n = 3) oral cancer patient samples with respect to normal oral tissue by an IHC technique. We found a drastic difference in both NPM1 levels (Fig. 4A, upper panel) and the acetylation status of NPM1 (Fig. 4A, lower panel) from early to advanced grades of oral tumors. The enhancement of the levels of acetylated NPM1 protein correlated with oral cancer malignancy. We further investigated the expression and acetylation status of NPM1 in 12 patient samples belonging to grade II oral squamous cell carcinoma (OSCC; as tumor samples belonging to this grade were readily available). IHC analysis showed higher nuclear positivity for NPM1 in malignant tissues compared to adjacent normal tissue (Fig. 4B, panel I versus panel II). The enhanced expression of NPM1 was observed in the nucleolus, as well as in the nucleoplasm (Fig. 4B, panel II, inset), and was found to be statistically significant (Fig. 4C, upper graph; **, P < 0.01; n = 5). A drastic increase in the acetylation status of NPM1 was also seen in the nucleoplasm of malignant tissue (Fig. 4B, panel III versus panel IV), with almost no nucleolar localization (Fig. 4B, panel IV, inset). The predominant increase in acetylated NPM1 levels in the oral tumors was highly significant (Fig. 4C, lower graph; **, P < 0.01; n = 5). Western analysis of patient samples showed a marked increase in NPM1 acetylation in malignant tumor tissue compared to that in corresponding normal tissue (Fig. 4D, panel I). Overexpression of NPM1 was also detected in malignant oral tumors (Fig. 4D, panel II). Real-time RT-PCR analysis of RNA isolated from patient samples showed higher NPM1 transcript levels in malignant tissues than in adjacent normal tissue (Fig. 4E). In order to elucidate the factors contributing to the enhanced acetylation status of NPM1, an IHC analysis was performed with anti-SIRT1 antibody and four selected pairs of oral patient samples. The intensity of SIRT1 staining observed in a few normal tissue sections was relatively low but was not significantly different from that in malignant tumors (Fig. 4F). Additionally, no difference from early to advanced-grade tumors was observed (see Fig. S6, upper panel, in the supplemental material). We also carried out IHC analysis with the same patient samples using an antibody against p300 (anti-p300-C-20). The oral tumor sections had relatively higher expression of p300 than did the corresponding normal tissues (Fig. 4G; *, P < 0.05; n = 4). However, levels of p300 were not significantly different in lower (grade I)- versus higher (grade III)-grade oral tumors (see Fig. S6, lower panel, in the supplemental material). Taken together, these observations suggest that hyperacetylation of NPM1, which is at least partly caused by the overexpression of p300, is associated with oral cancer manifestation, where the acetylation status of NPM1 could be contributing to its oncogenic potential during tumorigenesis.
FIG. 4.
NPM1 is overexpressed and is predominantly acetylated in grade II OSCC. (A) Representative images of an IHC analysis performed with normal oral tissue and grade I and III oral tumor samples with an anti-NPM1 antibody (upper panel) and an anti-AcNPM1 antibody (lower panel) at ×40 magnification. (B) Representative images of IHC analysis of malignant oral tumor and adjacent normal tissues done with anti-NPM1 antibody at ×40 magnification (panels II and I, respectively). Similarly, IHC analysis was done with an anti-AcNPM1-01 antibody (panels IV and III, respectively). A higher magnification (×100) of the same region is included (inset). Nucleoli are indicated by arrows. (C) A paired t test was plotted for mean difference in the intensity of nucleoplasmic NPM1 observed in normal versus tumor tissue samples from 12 oral cancer patients (upper graph) and similarly for the increase in acetylated NPM1 levels (lower graph) (**, P < 0.01; n = 5). (D) Acetylated NPM1 (panel I) and NPM1 (panel II) were compared in paired patient samples by Western blot analysis. Expression of β-actin was used as a loading control. P.I, patient identification number; N, adjacent normal tissue; T, malignant oral tumor tissue. (E) Real-time RT-PCR analysis of NPM1 transcripts in paired samples from oral cancer patients with a corresponding β-actin control. Fold T/N denotes the n-fold NPM1 expression increase in tumor versus normal tissue. (F) IHC analysis of malignant oral tumor and adjacent normal tissues was done with anti-hSIRT1 antibody and imaged at ×40 magnification (panels I and II, respectively). (G) IHC analysis of malignant oral tumor and adjacent normal tissues was done with anti-p300-C-20 antibody (panels I and II, respectively) and imaged at ×40 magnification. The data were analyzed by paired t test (*, P < 0.05).
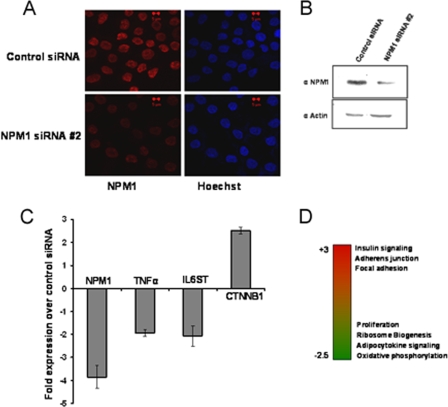
Knockdown of NPM1 alters global gene expression in an OSCC cell line.
The enhanced levels of acetylated NPM1 observed in grade II OSCC patients could be a hallmark of malignant transformation. In order to look in greater detail into NPM1-regulated global gene expression levels in OSCC, siRNA-mediated knockdown of NPM1 was carried out with KB (OSCC) cells. The knockdown efficiency was around 75% at the RNA level (Fig. 5C) and 60% at the protein level (Fig. 5A and B). Microarray analysis upon knockdown of NPM1 showed that depletion of NPM1 resulted in the differential regulation of several genes which have wide-ranging functions in the cell. The major pathways which were downregulated belonged to that of ribosome biogenesis, proliferation, and oxidative phosphorylation, whereas genes involved in focal adhesion, adherens junctions, insulin signaling, and histidine and butanoate metabolism were significantly upregulated. Additionally, expression of some of the genes involved in apoptosis and cell death was increased (Fig. 5D). Apart from the global effect on gene expression, several genes implicated in cancer manifestation were also found to be downregulated upon knockdown of NPM1. Significantly, we found that the expression of genes like those for TNF-α, the TNF receptor, the interleukin-6 (IL-6) receptor, and IL-6ST (the IL-6 signal transducer) were reduced as a consequence of NPM1 silencing (selected genes represented in Fig. 5C).
FIG. 5.
Knockdown of NPM1 results in altered expression of genes involved in carcinogenesis. (A) KB cells stained with anti-NPM1 antibody after 48 h of NPM1 siRNA no. 2 transfection. Scale bar, 5 μm. (B) Western blot analysis of NPM1 after 48 h of NPM1 siRNA no. 2 transfection. Expression of β-actin was used as a loading control. (C) Real-time quantitative RT-PCR analysis of gene expression upon siRNA-mediated knockdown of NPM1 in KB cells. The changes in the transcript levels of selected genes upon NPM1 siRNA transfection over those in the control are plotted as graphs. Internal normalization was done with β-actin levels. Values are means ± standard deviations of three independent NPM1 siRNA transfections. (D) Heat map depicting differential regulation of cellular pathways upon NPM1 knockdown. Red indicates upregulated genes, and green denotes downregulated genes.
Interestingly, it has been reported earlier that TNF-α, IL-6, and IL-6 receptor levels increase substantially in OSCC (11, 20). Significant upregulation was also observed in genes like CTNNA1 (alpha-1 catenin [cadherin-associated protein]) and CTNNB1 (beta-1 catenin), which encode components of the cell-cell adhesion complex (5), suggesting a possible role for NPM1 in facilitating the metastasis process during carcinogenesis. Our results show that NPM1 influences the expression of genes involved in cancer. Presumably, transcriptional activation of these genes could be directly regulated by acetylated NPM1. This was further verified by studying the occupancy and acetylation status of NPM1 at human gene promoters globally.
Acetylated NPM1 activates genes during oral carcinogenesis.
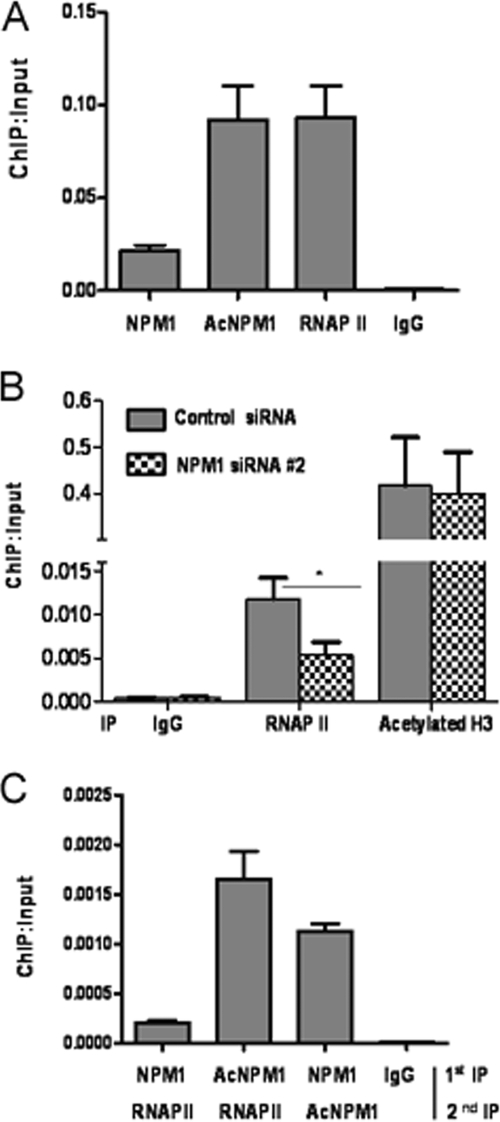
Microarray analysis revealed that a significant proportion of genes were differentially affected upon NPM1 knockdown where some of the genes could be a direct target of NPM1. In order to address this aspect, a ChIP-on-chip microarray was done to identify the global occupancy of NPM1 across human promoters in HEK293T cells. Interestingly, NPM1 was found to be enriched on a number of human gene promoters, such as those for IL-encoding genes, the TNF-α-encoding gene, and Homeobox-encoding genes (data not shown), indicating that NPM1 is directly involved in the transcriptional regulation of specific genes, as no global enrichment of NPM1 on all gene promoters could be detected. Higher levels of acetylated NPM1 in cancer would possibly result in its increased occupancy of such gene promoters. We identified genes which were downregulated upon NPM1 knockdown and were directly regulated by NPM1 in OSCC cells. ChIP assays revealed that acetylated NPM1 occupies the promoter of the gene for TNF-α in KB cells. RNAP II was used as a positive control (Fig. 6A). A ChIP assay done after siRNA-mediated knockdown of NPM1 resulted in reduced recruitment of RNAP II on the TNF-α promoter compared to that in control siRNA-transfected cells. However, knockdown of NPM1 did not alter the histone H3 acetylation level (Fig. 6B). reChIP assays in which the first chromatin pulldown was done with either an anti-NPM1 or an anti-AcNPM1-01 antibody, followed by an anti-RNAP II antibody, showed that acetylated NPM1 and RNAP II colocalized at the TNF-α promoter. A reChIP assay was also done with an anti-NPM1 antibody, followed by an anti- AcNPM1 antibody, to estimate the acetylation status of NPM1 (Fig. 6C). We have done further experiments with antibodies which recognize the phosphorylated form of RNA Pol II (phospho-S5-RNAP II). reChIP assays were carried out in which the first IP was performed with an anti-acetylated NPM1 antibody, followed by a second IP with pS5-RNAP II to look into their association at the TNF-α promoter. reChIP assays showed that pS5-RNAP II colocalized with acetylated NPM1 (see Fig. S7 in the supplemental material). Subsequently, we repeated the earlier experiment in which we silenced NPM1 by siRNA transfection for 48 h, followed by a ChIP assay with a pS5-RNAP II antibody, which showed a reduction in the active RNAP II recruitment at the TNF-α promoter (see Fig. S8 in the supplemental material). These experiments showed that acetylated NPM1 associates with active RNAP II at the promoter of TNF-α, which results in its enhanced expression.
FIG. 6.
Acetylated NPM1 occupies the TNF-α gene promoter. (A) A ChIP assay was done with anti-NPM1, anti-AcNPM1-01, and anti-RNAP II antibodies along with an immunoglobulin G (IgG) control in KB cells at the TNF-α promoter. Values are relative to the immunoprecipitated input DNA. (B) A ChIP assay was performed following 48 h of either control siRNA or NPM1 siRNA no. 2 transfection of KB cells with anti-RNAP II and anti-acetylated histone H3 antibodies. Results and error bars are averages and standard deviations of PCRs from three independent ChIP experiments. The data were analyzed by paired t test (*, P < 0.05). (C) A reChIP assay was done by carrying out first immunoprecipitation with either anti-NPM1 or anti-AcNPM1-01 antibody and then anti-RNAP II antibody at the TNF-α promoter. A reChIP assay was also done with anti-NPM1 antibody, followed by anti-AcNPM1 antibodies. Crude chromatin was used as the input for reChIP assays. Results and error bars are averages and standard deviations of at least three independent PCRs from two independent reChIP experiments.
p300 and SIRT1 regulate the expression of NPM1 target genes.
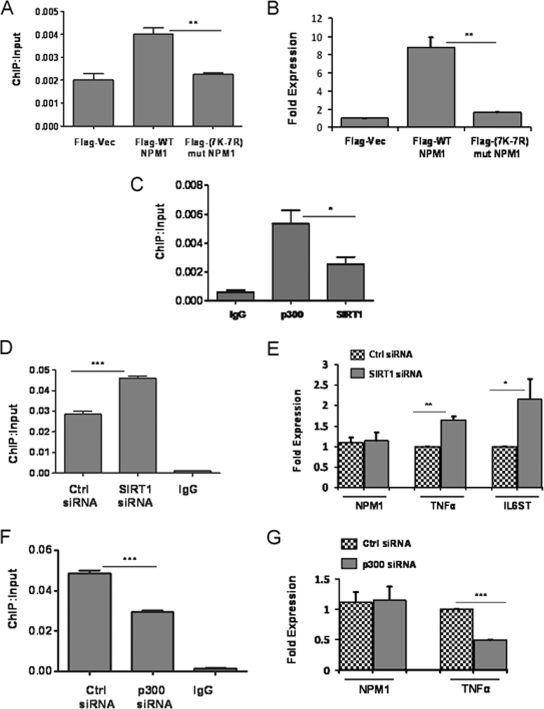
In order to further establish that the acetylation status of NPM1 is essential for transcriptional activation of its target genes, we focused on the TNF-α gene, which is regulated by NPM1. Flag-tagged WT and 7K-7R mutant NPM1 constructs were transiently transfected into cells, and their recruitment on the TNF-α promoter was monitored through ChIP assays with an anti-Flag M2-agarose antibody. The results indicate that WT NPM1 has almost twofold higher recruitment than 7K-7R mutant NPM1 (Fig. 7A). Additionally, overexpression of WT NPM1 resulted in a TNF-α expression level fivefold higher than that produced by 7K-7R mutant NPM1 transfection (Fig. 7B). We also performed ChIP assays to address the recruitment of p300 and SIRT1 at the TNF-α promoter, which showed that p300 is present in a greater amount than SIRT1 (Fig. 7C). Overall, these results imply that p300-mediated acetylation of NPM1 is indeed essential for transcriptional activation of the TNF-α gene. Subsequently, we performed siRNA-mediated knockdown of SIRT1 and monitored the expression of NPM1 target genes. Our earlier experiments had shown that SIRT1 suppression resulted in a significant increase in the overall acetylation status of NPM1 without altering NPM1 expression (Fig. 3E). Additionally, we also carried out ChIP assays and showed that SIRT1 knockdown resulted in greater occupancy of acetylated NPM1 at the TNF-α promoter (Fig. 7D). The expression of genes such as those for TNF-α and IL-6ST was enhanced as a consequence of SIRT1 suppression (Fig. 7E). Similar experiments were performed under p300 knockdown conditions, which resulted in decreased occupancy of acetylated NPM1 at the TNF-α promoter (Fig. 7F) and also correlated with reduced expression of the TNF-α gene (Fig. 7G).
FIG. 7.
p300 and SIRT1 regulate expression of NPM1 target genes. (A) A ChIP assay was done with anti-Flag antibody following transient transfection of Flag-tagged WT NPM1 or 7K-7R mutant (mut) NPM1 at the TNF-α promoter. Flag vector (Vec) transfection was used as a negative control. Values are relative to the immunoprecipitated input DNA. Results and error bars are averages and standard deviations of at least three independent PCRs from two independent ChIP experiments. The data were analyzed by paired t test (**, P < 0.01). (B) Real-time RT-PCR analysis of TNF-α gene expression upon transient transfection of the Flag vector, WT NPM1, or 7K-7R mutant NPM1 for 48 h. The data were analyzed by paired t test (**, P < 0.01). (C) A ChIP assay was done with equal amount of anti-SIRT1 and anti-p300 antibodies at the TNF-α promoter. Values are relative to the immunoprecipitated input DNA. Results and error bars are averages and standard deviations of at least three independent PCRs from two independent ChIP experiments (*, P < 0.05). IgG, immunoglobulin G. (D) A ChIP assay was done with an anti-AcNPM1 antibody at the TNF-α promoter following SIRT1 siRNA transfection for 48 h. The data were analyzed by paired t test (***, P < 0.001). Ctrl, control. (E) Real-time RT-PCR analysis of NPM1, TNF-α (**, P < 0.01), and IL-6ST (*, P < 0.05) gene expression upon siRNA-mediated knockdown of SIRT1. Internal normalization was done with β-actin levels. Values are means ± standard deviations of three independent siRNA transfections. (F) A ChIP assay was done with an anti-AcNPM1 antibody at the TNF-α promoter following p300 siRNA transfection for 48 h. The data were analyzed by paired t test (***, P < 0.001). (G) Real-time RT-PCR analysis of NPM1 and TNF-α gene expression upon siRNA-mediated suppression of p300 (***, P < 0.001). Values are means ± standard deviations of three independent siRNA transfections.
These results show that acetylation of NPM1 is a prerequisite for its transcriptional activation property (as neither p300 nor SIRT1 siRNA transfection resulted in downregulation of NPM1 expression). These enzymatic machineries could be dynamically regulating the acetylation-dependent functions of NPM1 in cells. Any deregulation of this fine balance would contribute to its aberrant acetylation status and, in turn, contribute to the oncogenic functions of NPM1 during malignancies.
DISCUSSION
NPM1 is a multifunctional histone chaperone protein that is involved in several cellular processes like ribosome biogenesis, development, centrosome duplication, and genomic stability and is also linked with cell proliferation and transformation associated with solid tumors. NPM1 is frequently overexpressed in several human cancers. It is also aberrantly translocated to the cytoplasm in acute myeloid leukemia (3). Recently, it was found to be essential for androgen-dependent transcriptional activation in prostate cancer cells (9). Apart from NPM1, there is increasing evidence which links other members of the histone chaperone family with cancer manifestation. The highly abundant nucleolar protein nucleolin was directly linked to human papillomavirus type 18-induced cervical carcinogenesis (2). The histone chaperone CAF-1 was found to be overexpressed in a breast cancer cell line compared to that in a cell line derived from normal cells (16). NPM1 is strongly linked with carcinogenesis and has been ascribed both tumor-suppressive and oncogenic functions. Alterations of NPM1 function by loss of function, mutation, or translocation contribute to oncogenesis by losing the potential of NPM1 to regulate the array of biological processes. Additionally, NPM1 is also reported to have oncogenic potential when it is overexpressed in cancers, where it may function to translate or amplify multiple oncogenic signaling mechanisms during carcinogenesis (10). There are several reports of enzymes that modify histones showing altered activity in cancer. For instance, missense mutations of p300 HATs and loss of heterozygosity at the p300 locus are associated with colorectal and breast cancers and with glioblastomas. Additionally, p300 is involved in androgen receptor transactivation, with a potentially important function in the progression of prostate cancer (17). Apart from targeting histones, p300 can also acetylate several nonhistone cellular proteins, which has wide-ranging functional consequences. In this report, we have demonstrated that p300-mediated acetylation regulates the oncogenic potential of NPM1. We have also identified SIRT1 as the candidate deacetylase for NPM1 which can reverse its acetylation status and, as a result, regulate its transcriptional activation properties. Elevated levels of NPM1 were observed in OSCC, which was predominantly acetylated in the nuclei of the malignant cells. ChIP assays have revealed that acetylated NPM1 occupied the promoter of the TNF-α gene (Fig. 6), probably in a complex with active RNAP II and p300, and enhances gene expression during carcinogenesis. We show here for the first time that the posttranslational modification status (in this case, acetylation) of the histone chaperone NPM1 could be involved in oral cancer manifestation through the activation of critical genes required for cell proliferation and survival. An aberrant increase in acetylated NPM1 could be a consequence of both a higher substrate (NPM1) concentration and a substantial increase in p300 expression. As has been reported earlier (18), acetylated NPM1 has a higher affinity for acetylated histones, which may facilitate histone exchange at the active gene promoters. Possibly, p300-mediated enhanced acetylation of histones and NPM1 would, together, increase the proliferative capacity of dedifferentiated tumors, which is associated with increased transcription. In this study, we have found a positive correlation between NPM1 acetylation and oncogenesis. The acetylation status of NPM1 may serve as a diagnostic marker, as well as a possible therapeutic target against malignant transformation. It would be interesting to find out the status of NPM1 modification (acetylation) in other cancers where it is reported to be overexpressed.
Supplementary Material
Acknowledgments
We thank M. Horikoshi for providing GST-ySir2 and His6-Tip60 (HAT domain) constructs and B. M. T. Burgering for mammalian and GST-hSIRT1 clones. We thank B. S. Suma, Confocal Facility, JNCASR, and G. Nagashanker for technical help.
This work was supported by JNCASR and Department of Biotechnology, India. J.S. is a Senior Research Fellow of CSIR, Government of India.
Footnotes
Published ahead of print on 6 July 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Balasubramanyam, K., R. A. Varier, M. Altaf, V. Swaminathan, N. B. Siddappa, U. Ranga, and T. K. Kundu. 2004. Curcumin, a novel p300/CREB-binding protein-specific inhibitor of acetyltransferase, represses the acetylation of histone/nonhistone proteins and histone acetyltransferase-dependent chromatin transcription. J. Biol. Chem. 27951163-51171. [DOI] [PubMed] [Google Scholar]
- 2.Grinstein, E., P. Wernet, P. J. Snijders, F. Rösl, I. Weinert, W. Jia, R. Kraft, C. Schewe, M. Schwabe, S. Hauptmann, M. Dietel, C. J. Meijer, and H. D. Royer. 2002. Nucleolin as activator of human papillomavirus type 18 oncogene transcription in cervical cancer. J. Exp. Med. 1961067-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grisendi, S., C. Mecucci, B. Falini, and P. P. Pandolfi. 2006. Nucleophosmin and cancer. Nat. Rev. Cancer 6493-505. [DOI] [PubMed] [Google Scholar]
- 4.Grisendi, S., R. Bernardi, M. Rossi, K. Cheng, L. Khandker, K. Manova, and P. P. Pandolfi. 2005. Role of nucleophosmin in embryonic development and tumorigenesis. Nature 437147-153. [DOI] [PubMed] [Google Scholar]
- 5.Inge, L. J., S. A. Rajasekaran, D. Wolle, S. P. Barwe, S. Ryazantsev, C. M. Ewing, W. B. Isaacs, and A. K. Rajasekaran. 2008. α-Catenin overrides Src-dependent activation of β-catenin oncogenic signaling. Mol. Cancer Ther. 71386-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones, P. A., and S. B. Baylin. 2007. The epigenomics of cancer. Cell 128683-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kishore, A. H., K. Batta, C. Das, S. Agarwal, and T. K. Kundu. 2007. p53 regulates its own activator: transcriptional co-activator PC4, a new p53-responsive gene. Biochem. J. 406437-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lafon-Hughes, L., M. V. DiTomaso, L. Méndez-Acuña, and W. Martínez-López. 2008. Chromatin-remodelling mechanisms in cancer. Mutat. Res. 658191-214. [DOI] [PubMed] [Google Scholar]
- 9.Léotoing, L., L. Meunier, M. Manin, C. Mauduit, M. Decaussin, G. Verrijdt, F. Claessens, M. Benahmed, G. Veyssière, L. Morel, and C. Beaudoin. 2008. Influence of nucleophosmin/B23 on DNA binding and transcriptional activity of the androgen receptor in prostate cancer cell. Oncogene 272858-2867. [DOI] [PubMed] [Google Scholar]
- 10.Lim, M. J., and X. W. Wang. 2006. Nucleophosmin and human cancer. Cancer Detect. Prev. 30481-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakano, Y., W. Kobayashi, S. Sugai, H. Kimura, and S. Yagihashi. 1999. Expression of tumor necrosis factor-alpha and interleukin-6 in oral squamous cell carcinoma. Jpn. J. Cancer Res. 90858-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okuda, M., H. F. Horn, P. Tarapore, Y. Tokuyama, A. G. Smulian, P. K. Chan, E. S. Knudsen, I. A. Hofmann, J. D. Snyder, K. E. Bove, and K. Fukasawa. 2000. Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication. Cell 103127-140. [DOI] [PubMed] [Google Scholar]
- 13.Okuwaki, M. 2008. The structure and functions of NPM1/nucleophsmin/B23, a multifunctional nucleolar acidic protein. J. Biochem. 143441-448. [DOI] [PubMed] [Google Scholar]
- 14.Okuwaki, M., K. Matsumoto, M. Tsujimoto, and K. Nagata. 2001. Function of nucleophosmin/B23, a nuclear acidic protein, as a histone chaperone. FEBS Lett. 506272-276. [DOI] [PubMed] [Google Scholar]
- 15.Okuwaki, M., M. Tsujimoto, and K. Nagata. 2002. The RNA binding activity of a ribosome biogenesis factor, nucleophosmin/B23, is modulated by phosphorylation with a cell cycle-dependent kinase and by association with its subtype. Mol. Biol. Cell 32016-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polo, S. E., and G. Almouzni. 2005. Histone metabolic pathways and chromatin assembly factors as proliferation markers. Cancer Lett. 2201-9. [DOI] [PubMed] [Google Scholar]
- 17.Seligson, D. B., S. Horvath, T. Shi, H. Yu, S. Tze, M. Grunstein, and S. K. Kurdistani. 2005. Global histone modification patterns predict risk of prostate cancer recurrence. Nature 4351262-1266. [DOI] [PubMed] [Google Scholar]
- 18.Swaminathan, V., A. H. Kishore, K. K. Febitha, and T. K. Kundu. 2005. Human histone chaperone nucleophosmin enhances acetylation-dependent chromatin transcription. Mol. Cell. Biol. 257534-7545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tarapore, P., M. Okuda, and K. Fukasawa. 2002. A mammalian in vitro centriole duplication system: evidence for involvement of CDK2/cyclin E and nucleophosmin/B23 in centrosome duplication. Cell Cycle 175-81. [PubMed] [Google Scholar]
- 20.Vairaktaris, E., C. Yapijakis, Z. Serefoglou, D. Avgoustidis, E. Critselis, S. Spyridonidou, A. Vylliotis, S. Derka, S. Vassiliou, E. Nkenke, and E. Patsouris. 2008. Gene expression polymorphisms of interleukins-1β, -4, -6, -8, -10, and tumor necrosis factors-α, -β: regression analysis of their effect upon oral squamous cell carcinoma. J. Cancer Res. Clin. Oncol. 134821-832. [DOI] [PubMed] [Google Scholar]
- 21.Zhang, Y. 2004. The ARF-B23 connection: implications for growth control and cancer treatment. Cell Cycle 3259-262. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.