Abstract
Transcription of the MluI cell cycle box (MCB) motif-containing genes at G1 phase is regulated by the MCB-binding factors (MBF) (also called DSC1) in Schizosaccharomyces pombe. Upon S-phase arrest, the MBF transcriptional activity is induced through the accumulation of the MBF activator Rep2. In this study, we show that the turnover of Rep2 is attributable to ubiquitin-mediated proteolysis. Levels of Rep2 oscillate during the cell cycle, with a peak at G1 phase, coincident with the MBF activity. Furthermore, we show that Rep2 ubiquitination requires the function of the E3 ligase anaphase-promoting complex/cyclosome (APC/C). Ste9 can be phosphorylated by the checkpoint kinase Cds1 in vitro, and its inhibition/phosphorylation at S-phase arrest is dependent on the function of Cds1. Our data indicate that the Cds1-dependent stabilization of Rep2 is achieved through the inhibition/phosphorylation of APC/C-Ste9 at the onset of S-phase arrest. Stabilization of Rep2 is important for stimulating transcription of the MBF-dependent genes to ensure a sufficient supply of proteins essential for cell recovery from S-phase arrest. We propose that oscillation of Rep2 plays a role in regulation of periodic transcription of the MBF-dependent genes during cell cycle progression.
The cell cycle is the series of events that occur in eukaryotic cells, such as synthesis of genomic DNA (S phase) and subsequent segregation of chromosomes (M phase), leading to cell proliferation. Successful proliferation requires temporal coordination of a number of critical events. Cyclin-dependent protein kinases (CDKs) play a central role in cell cycle control. Timely destruction of CDK subunits and their upstream regulators permits proper progression of the cell cycle (43). This is achieved by the evolutionarily conserved pathway known as ubiquitin-mediated proteolysis, which includes at least two major steps (29). The first is the covalent linkage between ubiquitins and the target protein catalyzed by ubiquitin-protein ligases (E3 ligases). The second is the destruction of the polyubiquitinated proteins by the 26S proteasome (60). Two distinct multiple-subunit E3 ligases, the anaphase-promoting complex/cyclosome (APC/C) and the Skp1-cullin-F-box protein-related (SCF) complex, are known to play pivotal roles in regulation of the cell cycle (55, 56). The APC/C is believed to be required for destruction of B-type cyclins and triggers the degradation of chromosome cohesion at the metaphase-anaphase transition (28, 30). On the other hand, the SCF complex is thought to be involved in destruction of CDK inhibitors such as p27 in mammalian systems (13), Rum1 in Schizosaccharomyces pombe (32) or Sic1 in Saccharomyces cerevisiae (3) at passage through the restriction point G0 (or START in yeast) late in G1 phase. It has been noted that APC/C also plays a role in suppression of mitotic cyclins to prevent premature entry into S phase during G1 phase (1, 11, 62).
The cell cycle appears to be also regulated and/or accompanied by oscillating transcriptional activities that induce several hundred genes in the genome at various stages across the entire duration of the cell cycle in mammalian cell line systems (15, 59) and yeasts (14, 44, 45, 51, 53). One of these activities is the well-characterized MluI-binding factor transcriptional complex (MBF) in S. pombe, which is similar to the MBF (Swi6 and Mbp1) and SBF (Swi6 and Swi4) in S. cerevisiae. MBF and SBF in S. cerevisiae play a redundant role in induction of MluI cell cycle box (MCB) and Swi6/Swi4 cell cycle box motif-containing genes at G1 phase (14, 53). In S. pombe, the MBF, consisting of Cdc10 (Swi6 homolog), Res1 (Mbp1 homolog), Res2 (Mbp1 homolog), Rep2, and Nrm1 (Nrm1 homolog), is essential for induction of ∼15 MBF-dependent genes at G1 phase through the binding at the MCB-like promoter motif(s) (44, 45, 51, 19). The majority of the MBF-dependent genes encode proteins involved in initiation of DNA replication, nucleotide biogenesis, and regulation of S phase.
Checkpoints involved in the response to DNA damage and DNA replication blockage are conserved in most eukaryotes (3, 12, 36). When DNA lesions are detected, the ATM-like kinase Rad3 in fission yeast activates the CHEK1-like kinase Chk1 to arrest cells at the G2/M boundary (57, 58). Conversely, when DNA replication forks stall, Rad3 activates the CHEK2-like kinase Cds1 to block cells at S phase (34, 38). Both Chk1 and Cds1 regulate CDK activity via inactivation of the Cdc25 phosphatase or activation of the Wee1 or Mik1 protein kinase, leading to cell cycle arrest at the G2/M boundary and S phase, respectively (24, 9, 50). The Rad3-Cds1-mediated S-phase checkpoint is also found to play a role in transcriptional induction of the MBF-dependent genes at the onset of S-phase arrest induced by treatment with hydroxyurea (HU), a compound that induces DNA replication blockage (16, 21, 18). Presumably the checkpoint activates the MBF activity to ensure a sufficient supply of proteins, such as the licensing factor Cdc18, whose activity is necessary for recovery from the S-phase block (27, 39).
Cdc10 activation/phosphorylation by Cds1 has been found to be required for the transcriptional induction of the MBF-dependent genes (21). On the other hand, inhibition/phosphorylation of Nrm1, a negative regulator of MBF, by Cds1 is also important for this transcriptional induction (18). In a previous study, we have shown that this transcriptional modulation is mediated through the accumulation of Rep2, an activator of MBF that induces the MBF-dependent genes at G1 phase (16). Most likely, the S-phase checkpoint acts through multiple pathways to ensure that MBF is active at S-phase arrest.
In this study, we show that Rep2 is degraded via ubiquitin-mediated proteolysis. Accumulation of Rep2 at the onset of S-phase arrest is a result of the Cds1-dependent protection from ubiquitination by APC/C-Ste9. Moreover, we show that Rep2 stabilization stimulates transcription of the MBF-dependent genes, which allows accumulation of their encoded proteins such as Cdc18 at S-phase arrest. Together, our data indicate that the S-phase checkpoint modulates the MBF transcriptional activity through the inhibition of the APC/C-Ste9 E3 ligase for accumulation of proteins encoded by the MBF-dependent genes that are likely to be required for recovery from S-phase arrest. We further propose that Rep2 oscillation plays a role in regulation of periodic transcription of the MBF-dependent genes in cell cycle progression.
MATERIALS AND METHODS
Yeast strains, media, and culture manipulation.
Strains used in this study are listed in Table 1. Deletion or epitope-tagging alleles were constructed using the PCR-mediated gene-disruption or epitope-tagging protocol (2). Tetrad dissection was applied for desired allele combinations by crossing and selection of nonparental ditype spores. The standard rich medium yeast extract plus supplements (YES) and the minimal medium Edinburgh minimal medium in liquid or solid (supplemented with 2% Bacto agar) forms were used for cell growth (37). To test cellular responses to HU treatment, fresh log-phase cells were subjected to treatment with HU at 8 mM unless indicated otherwise. To test protein stability, cells were subjected to treatment with 100 μg/ml cycloheximide (CHX). Synchronous cultures were generated by releasing cdc25-22 or cdc10-V50 cells to the permissive temperature after growth block at the restrictive temperature for 4 h.
TABLE 1.
Strains used in this study
| Strain | Relevant genotype | Source |
|---|---|---|
| KGY574 | h−leu1-32 mts3-1 | K. Gould's lab |
| KGY1150 | h−ura4-D18 leu1-32 ade6-M21X lid1-6 | K. Gould's lab |
| AL1-A4 | h−ura4-D18 leu1-32 skp1-A4 | T. Toda's lab |
| LJY0100 | h+ura4-D18 leu1-32 ade6-M216 | Lab stock |
| LJY0162 | h−ura4-D18 leu1-32 his3-D1 ade6-M210 cdc25-22 | Lab stock |
| LJY0188 | h−ura4-D18 leu1-32 | Lab stock |
| LJY1687 | h−ura4-D18 leu1-32 cds1Δ::ura4+ | Lab stock |
| LJY1902 | h−ura4-D18 leu1-32 rep2+-3HA-6His::ura4+ | Lab stock |
| LJY2031 | h−ura4-D18 leu1-32 cdc10+-3HA-6His::ura4+ | Lab stock |
| LJY2112 | h−ura4-D18 leu1-32 res2+-3HA-6His::ura4+ | Lab stock |
| LJY2168 | h−ura4-D18 leu1-32 res1+-3HA-6His::ura4+ | Lab stock |
| LJY2378 | h−ura4-D18 leu1-32 cds1+-3HA-6His::ura4+ | Lab stock |
| LJY2742 | h−ura4-D18 leu1-32 ade6-M216 cds1Δ::ura4+rep2+-3HA-6His::ura4+ | Lab stock |
| LJY2743 | h−ura4-D18 leu1-32 rad3Δ::LEU2 rep2+-3HA-6His::ura4+ | Lab stock |
| LJY2753 | h−ura4-D18 leu1-32 his3-D1 ade6-M210 cdc25-22 rep2+-3HA-6His::ura4+ | This study |
| LJY2785 | h+ura4-D18 leu1-32 ade6-M216 mts3-1 | This study |
| LJY2794 | h+ura4-D18 leu1-32 mts3-1 rep2+-3HA-6His::ura4+ | This study |
| LJY2797 | h−ura4-D18 leu1-32 mts3-1 rep2+-3HA-6His::ura4+ | This study |
| LJY2944 | h+ura4-D18 leu1-32 rep2+-3HA-6His::ura4+ | This study |
| LJY2946 | h−ura4-D18 leu1-32 lid1-6 mts3-1 rep2+-3HA-6His::ura4+ | This study |
| LJY2947 | h−ura4-D18 leu1-32 skp1-A4 mts3-1 rep2+-3HA-6His::ura4+ | This study |
| LJY2986 | h−ura4-D18 leu1-32 lid1-6 rep2+-3HA-6His::ura4+ | This study |
| LJY2988 | h−ura4-D18 leu1-32 skp1-A4 rep2+-3HA-6His::ura4+ | This study |
| LJY2990 | h−ura4-D18 leu1-32 rep2-CΔ53-3HA-6his::ura4+ | This study |
| LJY3043 | h−ura4-D18 leu1-32 rep2+-3HA-6His::ura4+lid1+-13myc::LEU2+ | This study |
| LJY3045 | h−ura4-D18 leu1-32 lid1+-13myc::LEU2+ | This study |
| LJY3106 | h−ura4-D18 leu1-32 mts3-1 rep2-CΔ53-3HA-6His::ura4+ | This study |
| LJY3752 | h−ura4-D18 leu1-32 ste9Δ::kanr rep2+-3HA-6His::ura4+ | This study |
| LJY3755 | h−ura4-D18 leu1-32 ste9Δ::kanr rad3Δ::LEU2 rep2+-3HA-6His::ura4+ | This study |
| LJY3758 | h−ura4-D18 leu1-32 ste9Δ::kanr cds1Δ::ura4+rep2+-3HA-6His::ura4+ | This study |
| LJY3850 | h−ura4-D18 leu1-32 rep2-CΔ21-3HA-6His::ura4+ | This study |
| LJY3853 | h−ura4-D18 leu1-32 rep2-CΔ94-3HA-6His::ura4+ | This study |
| JLY3859 | h−ura4-D18 leu1-32 cds1Δ::ura4+ste9+-3HA-6His::LEU2 | This study |
| LJY3863 | h−ura4-D18 leu1-32 ste9+-3HA-6His::LEU2 | This study |
| LJY3988 | h−ura4-D18 leu1-32 cds1Δ::ura4+lid1+-13myc::LEU2 | This study |
| LJY4035 | h−ura4-D18 leu1-32 cdc25-22 ste9Δ::kanr rep2+-3HA-6His::ura4+ | This study |
| LJY4050 | h−ura4-D18 leu1-32 cdc10-V50 rep2+-3HA-6His::ura4+ | This study |
| LJY4055 | h−ura4-D18 leu1-32 cdc10-V50 cds1Δ::kanr rep2+-3HA-6His::ura4+ | This study |
| LJY4072 | h−ura4-D18 leu1-32 ste9Δ::kanr mts3-1 rep2+-3HA-6His::ura4+ | This study |
| LJY2858 | h−ura4-D18 leu1-32 res2Δ::LEU2 rep2+-3HA-6His::ura4+ | This study |
To test the function of the Rep2 C-terminal sequences, various truncations at the C terminus were constructed as a sole chromosomal copy under the control of the Rep2 native promoter. In brief, the C-terminal epitope-tagging approach was applied. Instead of tagging at the last amino acid residue as in C-terminal epitope tagging, rep2-CΔ21 was generated by tagging at the residue that is 21 amino acids upstream of the last amino acid. Similarly, rep2-CΔ53 and rep2-CΔ94 were generated by tagging at the residue 53 and 94 amino acids, respectively, upstream of the last amino acid using the primers 5′-CCCCTAACTTAATTTGCTCAAAATGCAATACCACGTTCAACCATTCAACGGCTCTTATGATGCATGAAGCAACCTGCTTG TGGATCCCCGGGTTAATTAA-3′ (for CΔ21), 5′-AGAATCACTCTTCTGATGCTCGTGATGCTATTAAATCTGCCCTTCGAAGGAGAATGCGCCGTCGTGAAGGCCGTGTTCAATGGATCCCCGGGTTAATTAA-3′ (for CΔ53), and 5′-GAAAACTTGGAATTGCTCAACTCATTCGTCAACAAACTGATCCTCCTCAAATTATCCATAGAAAGCAAGACAAGGGCTTA TGGATCCCCGGGTTAATTAA-3′ (for CΔ94) paired with 5′-TCATTAATAAGCGATAATTATTACATATCCGTAATGCTACTAGATACGAAAGCTGCTTCTTTGTATGAACGGAAATATTACTCACTATAGGGCGAATTGG-3′.
Plating and recovery assays.
To determine hypersensitivities to HU treatment, plating assays were applied. In brief, overnight cultures at 30°C were inoculated into fresh YES medium to generate log-phase cells after growing for ∼2 generations or ∼5 h. Tenfold serially diluted log-phase cells were spotted on YES plates containing 2.5 to 8 mM HU (Sigma-Aldrich, St. Louis, MO). To determine viability after transient treatment with HU, log-phase cultures were subjected to treatment with 2.5 to 8 mM HU for 3 h. Treated cells were washed twice and subsequently plated on YES medium after various dilutions. CFU in the recovery assay were calculated based on the average from at least three independent experiments.
Western blot analysis.
Standard Western blot analysis was performed as described elsewhere (16). The primary antibodies against hemagglutinin (HA) (monoclonal; Santa Cruz Biotechnology Inc., Santa Cruz, CA), myc (Santa Cruz Biotechnology Inc.), ubiquitin protein (Biomol International Inc.), Cig2 (Abcam, Cambridge, United Kingdom), PSTAIR (Upstate Biotechnology Inc., Waltham, MA), Cdc18 (a gift of P. Nurse) (42), and Ste9 (a gift of S. Moreno) (8) were used according to the manufacturer's instructions. Anti-mouse and anti-rabbit immunoglobulin G conjugated with horseradish peroxidase (Amersham, Buckinghamshire, United Kingdom) were used as secondary antibodies. The chemiluminescence was developed and captured using the ECL horseradish peroxidase system (Amersham).
Coimmunoprecipitation assays.
To test the interaction between two different proteins, coimmunoprecipitation assays were applied. In brief, cell extracts (∼2.5 mg proteins) in 50 mM HEPES-KOH (pH 7.5)-140 mM NaCl-1 mM EDTA (pH 7.5)-1% (vol/vol) Triton X-100-0.1% (wt/vol) sodium deoxycholate were mixed with agarose-coupled mouse monoclonal anti-myc antibody or anti-HA antibody and rotated overnight at 4°C. To remove nonspecifically bound proteins, the resins were washed once with 50 mM HEPES-KOH (pH 7.5)-140 mM NaCl-1 mM EDTA (pH 7.5)-1% (vol/vol) Triton X-100-0.1% (wt/vol) sodium deoxycholate, twice with 50 mM HEPES-KOH (pH 7.5)-500 mM NaCl-1 mM EDTA (pH 7.5)-1% (vol/vol) Triton X-100-0.1% (wt/vol) sodium deoxycholate, and twice with 10 mM Tris-Cl (pH 8.0)-250 mM LiCl-1 mM EDTA (pH 7.5)000.5% (vol/vol) Nonidet P-40-0.5% (wt/vol) sodium deoxycholate. Each washing process was performed in the cold for 10 min. The extensively washed beads were boiled in the standard sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer for Western blot analysis.
In vivo ubiquitin assays.
To determine if a protein is polyubiquitinated in vivo, in vivo ubiquitin assays were performed as described previously (5). In brief, the relevant strains were transformed with the pREP1-6His-ubi plasmid (a gift of K. Gould). Following growth in the absence of thiamine for ∼22 h, cells were shifted to the nonpermissive temperature for 4 h and lysed in buffer containing 8 M urea, 100 mM sodium phosphate (pH 8.0), and 5 mM imidazole. Cell extracts were clarified by centrifugation for 5 min, and the protein concentration in the supernatants was determined using the Bradford method (10). Six milligrams of extract proteins was mixed with Ni-nitrilotriacetic acid (NTA) beads (Qiagen, Hilden, Germany) and incubated for 4 h at room temperature. The resin was washed three times with urea buffer containing 8 M urea and 0.1 M sodium phosphate (pH 8.0); subsequently washed with 1:1 (vol/vol) and 1:3 (vol/vol) mixtures of urea buffer and glycerol-NP-40 buffer containing 50 mM sodium phosphate (pH 8.0), 100 mM KCl, 20% glycerol, and 0.2% NP-40; and finally washed with glycerol-NP-40 buffer supplemented with 10 mM imidazole. Each washing step was carried out in the cold for ∼10 min. Subsequently, the extensively washed resin was resuspended in glycerol-NP-40 buffer and boiled for 5 min, and ubiquitinated forms of total proteins and rep2+-HA protein were detected by Western blotting using mouse monoclonal antibody to ubiquitin proteins (BIOMOL International, Plymouth Meeting, PA) and anti-HA antibodies (Santa Cruz Biotechnology Inc.), respectively.
Alkaline phosphatase assay.
The alkaline phosphatase assay was done as described previously (52) with some modification. In brief, 3 × 108 cells were lysed in urea buffer containing 8 M urea, 0.1 M sodium phosphate, and 50 mM Tris-HCl (pH 8.0). The resulting lysate was clarified by centrifugation for 10 min at room temperature and subsequently mixed with Ni-NTA beads (Qiagen) and incubated on a STR1 roller mixer (Barloworld Scientific, Staffordshire, United Kingdom) for 60 min at room temperature. The Ni-NTA beads were washed with urea buffer and then wash with a series of urea buffers containing decreasing concentrations of urea (6, 4, 2, and 0.5 M). After being washed twice with alkaline phosphatase buffer containing 5 mM Tris-HCl (pH 8.0)-0.5 mM MgCl2, the beads were incubated with 50 U of alkaline phosphatase (Roche Pharmaceuticals, Stockholm, Sweden) for 30 min at 37°C in the presence or absence of phosphatase inhibitors at a final concentration of 1% of the stock solution (Sigma-Aldrich). The reactions were stopped by addition of SDS-PAGE loading buffer and boiling for 3 min. Samples were displayed on a 4% to 20% SDS-polyacrylamide gradient gel and subjected to Western blot analysis.
In vitro kinase assays with recombinant proteins.
Substrate glutathione S-transferase (GST)-Ste9 fusion protein or GST proteins serving as a negative control were expressed in Escherichia coli strain BL21 and enriched with glutathione Sepharose 4B resin (Amersham) according to the manufacturer's instructions.
Cells bearing a cds1+-HA allele (∼1 liter) were treated with 8 mM HU for ∼3 h for activation of Cds1 kinase activity and subsequently harvested by centrifugation. The cell pellet was resuspended in lysis buffer containing 50 mM Tris-HCl (pH 7.6), 150 mM NaCl, 2 mM EDTA, 1× phosphorylation inhibitor cocktail (Sigma-Aldrich), 0.1% NP-40, and 1× proteinase inhibitor cocktail (Roche Pharmaceuticals) and subjected to the glass bead beater FastPrep system (Bio 101, Vista, CA). The resulting lysate was clarified by centrifugation at 43,000 × g and 4°C for 20 min and mixed with 100 μl anti-HA antibody-coupled agarose beads (Santa Cruz Biotechnology Inc.). The mixture was incubated at 4°C overnight and subsequently washed five times with washing buffer containing 25 mM Tris-HCl (pH 7.6), 250 mM NaCl, and 0.1% Tween 20 and resuspended in kinase assay buffer containing 20 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 1 mM dithiothreitol, 75 mM KCl, and 50 μM [γ-35S]ATP (Perkin-Elmer, Waltham, MA). The substrate GST-Ste9 or GST was added to the mixture and incubated for 30 min as described previously (61). The reaction was stopped by adding the standard SDS-PAGE loading buffer and boiled for 5 min. The supernatant was displayed on SDS-polyacrylamide gels for autoradiography.
Expression microarray analysis.
Expression profiling was performed using open reading frame-specific oligonucleotide microarrays as previously described (16). Time course transcriptional levels were normalized based on the 0-min samples (cells prior to HU treatment) to show the relative transcriptional level change after HU addition. Average linkage hierarchical gene cluster analysis with uncentered correlation metrics was performed using Gene Cluster v3 (22). The TreeView software (22) was used for visualization of the cluster results.
Microarray data accession number.
The original microarray data sets were submitted to the GEO database with the accession number GSE10901.
RESULTS
Rep2 is an unstable protein whose levels oscillate during the cell cycle with a peak at G1 phase.
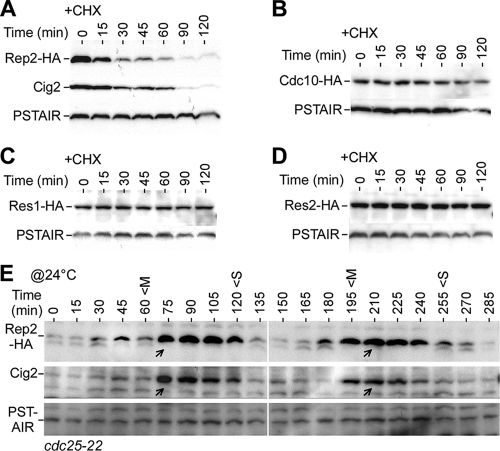
We have previously shown that Rep2, but not the other MBF components Cdc10, Res1, and Res2, accumulates in cells upon HU treatment (16). To test whether Rep2 accumulation was due to the increased stability at the onset of S-phase arrest, we examined the stabilities of Rep2 and other MBF components. To this end, strains bearing a chromosomal copy of HA-tagged rep2+, cdc10+, res1+, or res2+ were utilized. Due to the low level of Rep2 in log-phase (asynchronous) culture, the cells were pretreated with 8 mM HU for 4 h to allow Rep2 accumulation in cells. The HU-treated cells were subjected to treatment with 100 μg/ml CHX to block protein synthesis and sampled at various time points for a period of 120 min. Western blot analysis of the CHX-treated samples indicated that the Rep2 level was reduced by ∼50% in ∼15 min after addition of CHX, slightly faster than the turnover of the S-phase cyclin Cig2 (63) (Fig. 1A). In contrast, the levels of Cdc10, Res1, and Res2 in HU-arrested cells were hardly decreased after CHX treatment (Fig. 1B-D). The stabilities of the Cdc10, Res1, and Res2 proteins were not altered in cells without HU pretreatment (data not shown). These results indicate that Rep2, but not Cdc10, Res1, or Res2, is an unstable protein.
FIG. 1.
Rep2 is an unstable protein whose levels oscillate along with cell cycle. (A) Rep2 is an unstable protein. Western blot analysis using anti-HA antibodies shows decreased levels of Rep2-HA in cells at various time points after treatment with CHX. The S-phase cyclin Cig2 is a known unstable protein that served as a positive control. PSTAIR (Cdc2 conserved sequences, regardless of phosphorylation status at the residue 15 tyrosine) was used as loading control. (B to D) Cdc10, Res1, and Res2 are stable proteins. (E) Levels of Rep2 oscillate with the cell cycle. The level of Rep2 in synchronous cdc25-22 culture was monitored at various time points after release to the permissive temperature (@24°C). Peaks of mitotic indexes (M) and septation indexes (S) are indicated. The arrows indicate the peaks of the oscillated Rep2 or Cig2 levels.
Given that the Rep2 level was correlated with the transcription level of the MBF-dependent genes at the onset of S-phase arrest (16), we wanted to know whether Rep2 protein levels would oscillate with the transcription levels of the MBF-dependent genes during cell cycle progression (44, 45, 51). For this reason, the rep2+-HA cdc25-22 strain was constructed. Cdc25 is a phosphatase that activates Cdc2 by dephosphorylation at its tyrosine-15 residue (7). Inactivation of cdc25-22 at the restrictive temperature would block cells at the G2/M boundary (20). Upon release to the permissive temperature, the cells would undergo synchronous waves of mitotic cell division cycle. We adopted this block-and-release protocol to monitor the level of Rep2 in synchronous cultures. Clearly, levels of Rep2 oscillated coincident with those of Cig2; the peak was at G1 phase as judged by the fact that it was after the peak of mitosis (indicated by the peak of mitotic indexes) and before the peak of S phase (indicated by the peak of septation indexes) (Fig. 1E). This result implies that the level of Rep2, the activator of MBF, is correlated with the level of the MBF transcriptional activity during the cell cycle. Interestingly, the cell cycle oscillation pattern of rep2+ transcription was apparent in elutriation-synchronized wild-type cells but less apparent in block-and-release-synchronized cdc25-22 cells (44, 45, 51). It was noted that the profile of rep2+ mRNA was different from that of the MBF-dependent genes.
Rep2 turnover is dependent on the function of the 26S proteasome.
To test if Rep2 turnover was attributable to the ubiquitin-mediated protein degradation pathway, we examined the level of Rep2 in cells bearing a temperature-sensitive allele of mts3, a gene encoding an essential component of the 26S proteasome (25). Western blot analysis indicated that the level of Rep2 increased in cells carrying the mts3-1 allele after a temperature shift to 36°C (the restrictive temperature for the mts3-1 allele) but not in those carrying the wild-type allele (Fig. 2A and data not shown), indicating that the Mts3-associated 26S proteasome plays a role in degradation of Rep2. Next, we wanted to determine if Rep2 turnover would be diminished in cells after inactivation of the 26S proteasome. For this reason, mts3-1 cells were grown at the restrictive temperature for 4 h prior to addition of 100 μg/ml CHX. Rep2 levels were determined in cells after CHX treatment using Western blot analysis. It was clear that Rep2 turnover was largely diminished in mts3-1 cells but not in wild-type cells (Fig. 2B). These results indicate that Rep2 degradation is attributable to the Mts3-associated 26S proteasome, an essential protease complex responsible for degradation of ubiquitin-conjugated proteins.
FIG. 2.
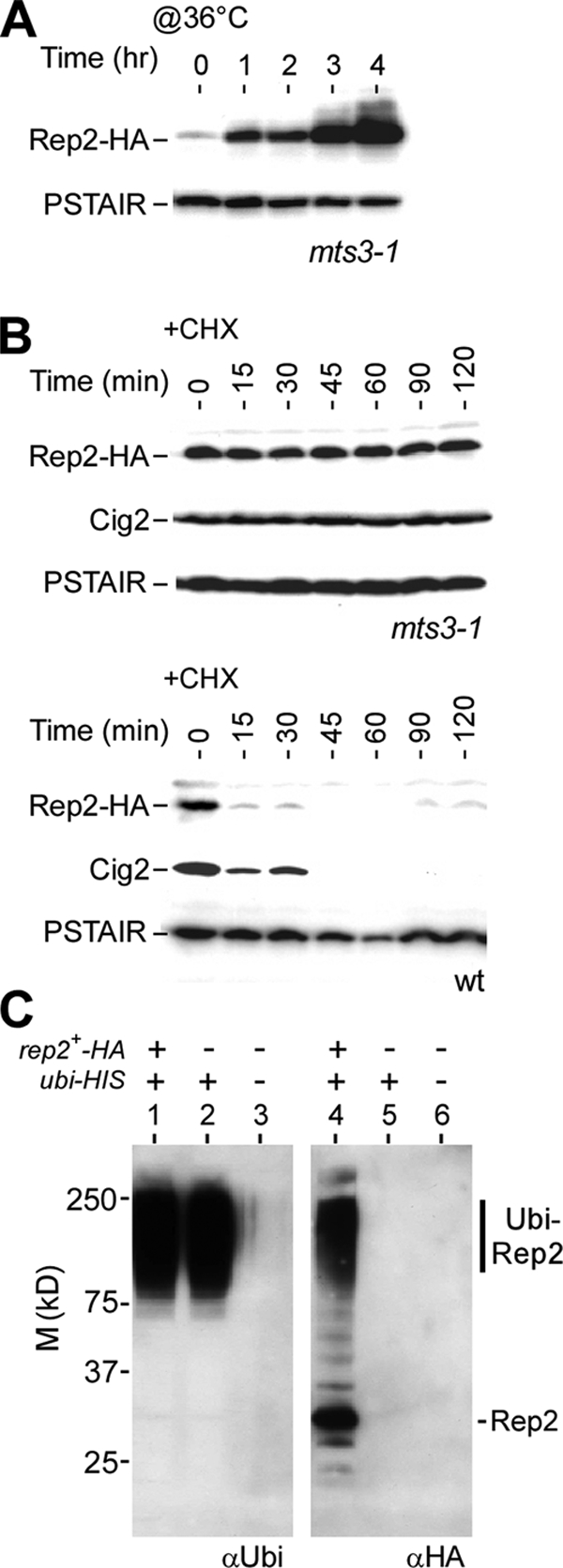
Degradation of Rep2 depends on the ubiquitin-mediated proteolytic pathways. (A) Rep2 accumulation in cells defective in the Mts3-mediated proteolytic pathway. Western blot analysis of Rep2 levels in mts3-1 cells at various time points after a shift to the restrictive temperature (@36°C) is shown. PSTAIR served as loading control. (B) Rep2 degradation requires the function of the Mts3-mediated proteolytic pathway. Western blot analysis of Rep2 levels in HU-treated mts3-1 or wild-type (wt) cells at various time points after addition of CHX is shown. (C) Rep2 is ubiquitinated in cells. An in vivo ubiquitination assay shows the Ni-TNA bead-enriched HIS-tagged polyubiquitin-conjugated proteins using antiubiquitin antibodies (left panel). The blot was stripped and rehybridized with anti-HA antibodies (right panel). Polyubiquitin-conjugated Rep2-HA proteins (Ubi-Rep2) that migrate slower than unmodified Rep2-HA are indicated. M, molecular mass.
It was noted previously that not all target proteins of the 26S proteasome were polyubiquitinated (47). To test if Rep2 was polyubiquitinated in cells, we performed in vivo ubiquitination assays (5), for which we constructed a strain bearing the rep2+-HA mts3-1 alleles and a pREP1 plasmid containing a HIS-tagged ubiquitin gene (see Materials and Methods). After induction of HIS-tagged ubiquitin for ∼22 h, cells were shifted to 36°C for 4 h to inactivate the Mts3-mediated 26S proteasome. This would lead to the accumulation of polyubiquitinated proteins in cells. Poly-HIS-ubiquitinated proteins from the cell lysate were enriched through the Ni-NTA affinity column and subsequently displayed on SDS-polyacrylamide gels for Western blot analysis. The location of polyubiquitinated proteins was determined by Western blot analysis using antiubiquitin antibodies (Fig. 2C). Slowly migrating Rep2 at the location of the polyubiquitinated proteins was detected when anti-HA antibodies were used, suggesting that Rep2 is polyubiquitinated in vivo. The unmodified Rep2 was also detected.
Rep2 degradation is mediated through the APC/C but not the SCF complex.
Two major E3 ligases, APC/C and the SCF complex, were known to conjugate ubiquitins to distinct sets of target proteins. We wanted to know if APC/C or the SCF complex was responsible for the Rep2 degradation. For this reason, we constructed rep2+-HA lid1-6 and rep2+-HA skp1-A4 strains. Lid1 is an essential component of the APC/C (6), whereas Skp1 is a nondispensable subunit of the SCF complex (31). Rep2 turnover in these strains was determined after growth at the restrictive temperature to inactivate Lid1-6 or Skp1-A4. Western blot analysis showed that the Rep2 turnover was clearly diminished in lid1-6 cells (Fig. 3A), indicating that the Lid1-associated APC/C plays a role in the instability of Rep2. On the other hand, the Rep2 turnover was still noticeable in skp1-A4 cells (Fig. 3B), although the half-life was a bit longer than that in wild-type cells (see Fig. 1A). This result implies that the Skp1-associated SCF complex was not a major player in regulation of Rep2 turnover.
FIG. 3.
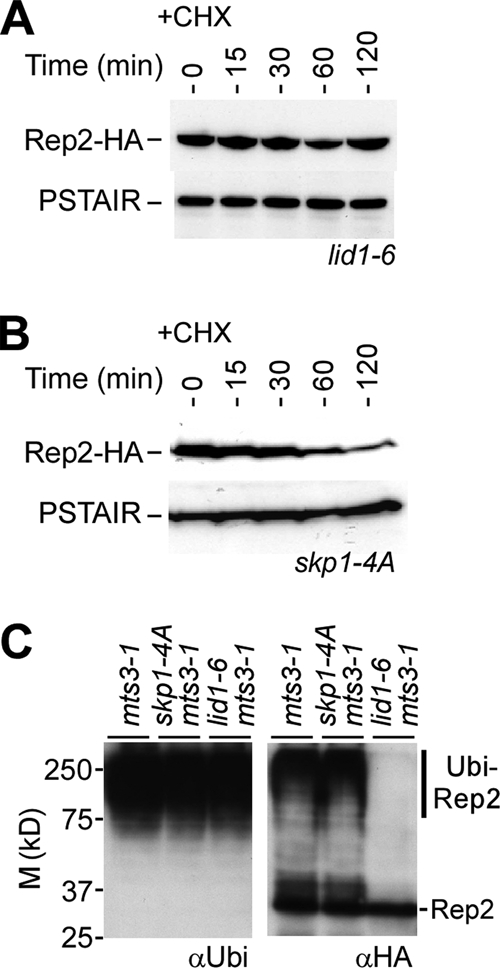
Degradation of Rep2 is mediated through APC/C but not the SCF complex. (A) Rep2 is stabilized in lid1-6 cells. Western blot analysis of Rep2-HA levels in lid1-6 cells at various time points after addition of CHX is shown. Prior to CHX addition, cells were grown at 36°C for 4 h to inactivate lid1-6. (B) Rep2 is unstable in skp1-4A cells. Western blot analysis of Rep2-HA levels in skp1-4A cells after addition of CHX is shown. Prior to CHX addition, cells were grown at 36°C for 4 h to inactivate skp1-4A. (C) Inactivation of Lid1 but not Skp1 diminishes the polyubiquitination of Rep2. In vivo ubiquitin assays show the Ni-NTA bead-enriched polyubiquitin-conjugated proteins using antiubiquitin antibodies (left panel) and after rehybridization with anti-HA antibodies (right panel). The position of slowly migrating polyubiquitinated Rep2 (Ubi-Rep2) is indicated. M, molecular mass.
We next wanted to know whether the diminished turnover of Rep2 was associated with the lack of Rep2 polyubiquitination in lid1-6 cells. Therefore, we performed in vivo ubiquitin assays. As expected, polyubiquitinated proteins were still detected in cells defective in either lid1-6 or skp1-4A (Fig. 3C). The in vivo ubiquitin assay showed that the slowly migrating polyubiquitinated Rep2 was clearly present in skp1-4A mts3-1 or mts3-1 cells, suggesting that the ubiquitination of Rep2 does not require the function of Skp1 or Mts3. On the other hand, polyubiquitinated Rep2 protein was hardly detected in lid1-6 cells, indicating that the Lid1-associated APC/C plays a major role in Rep2 polyubiquitination.
Interaction with Res2 is important for Rep2 degradation.
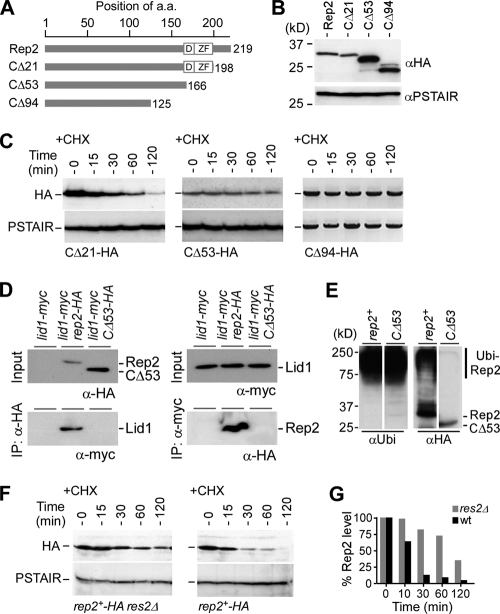
It was proposed that the C terminus of Rep2 harbors sequences that are responsible for physical interaction with Res2, leading to transcriptional induction of the MBF-dependent genes (40). To test if the Rep2 C terminus was essential for Rep2 ubiquitination, we constructed various C-terminally truncated alleles of Rep2. We found that the turnover of Rep2 was less apparent in cells whose rep2 transcription was controlled by the inducible nmt promoters in plasmids such as pREP41 (data not shown). To ensure that the expression of all rep2 truncated alleles was under the control of the native promoter in the chromosome, various lengths of the Rep2 C-terminal sequences were deleted directly from the wild-type chromosomal copy of rep2+ using the PCR-mediated gene disruption protocol (2) (Fig. 4A). All variants of truncated Rep2 were tagged with HA epitopes at the C terminus, and the expected changes in sizes of the various alleles were apparent in Western blot analysis (Fig. 4B).
FIG. 4.
The C terminus of Rep2 is essential for physical interaction with Lid1 but not Skp1. (A) Physical maps of various C-terminally truncated Rep2 proteins. The number of amino acid (a.a.) residues for each Rep2 allele is indicated. D and ZF, noncanonical D-box and zinc finger domains, respectively. (B) Various alleles of C-terminally truncated Rep2 protein. Western blot analysis reveals different molecular masses of various truncated Rep2 molecules. The loading control was PSTAIR. (C) Stabilities of various alleles of Rep2. Western blot analysis shows the levels of various truncated Rep2 proteins at various time points after addition of CHX. (D) The C-terminal truncation protein Rep2-Δ53 fails to interact with Lid1. The results of coimmunoprecipitation (IP) assays using anti-HA (left panel) or anti-myc (right panel) antibodies are shown. (E) In vivo ubiquitin assays showing the Ni-NTA bead-enriched polyubiquitin-conjugated proteins using antiubiquitin antibodies (left panel) or anti-HA antibodies (right panel). The position of polyubiquitinated Rep2 (Ubi-Rep2) is indicated. (F) Rep2 degradation is impeded in the res2Δ strain. Western blot analysis shows the level of Rep2 after addition of CHX in wild-type and res2Δ cells. (G) Quantification of the relative levels of Rep2 in wild-type and res2Δ cells (D). The relative Rep2 level (i.e., Rep2 level/PSTAIR level) at the starting point (0 min) is set to 100%.
The turnover of various truncated Rep2 proteins was determined using Western blot analysis after addition of CHX. Rep2-CΔ21 turnover was apparent, with a half-life close to that of full-length Rep2 in wild-type cells (Fig. 4C), suggesting that the C-terminal residues 198 to 219 are not required for Rep2 ubiquitination. On the other hand, the turnover of the C-terminally truncated Rep2-CΔ53 protein was largely diminished, suggesting that the 32-amino-acid sequence at the C-terminal residues 167 to 198 are important for Rep2 ubiquitination.
We wanted to test if the reduced turnover of Rep2-CΔ53 was a result of failure in the physical interaction with APC/C component Lid1. For this reason, we performed coimmunoprecipitation assays to determine the physical interaction between Rep2-CΔ53 and Lid1. Lid1 was hardly detected in proteins that were associated with Rep2-CΔ53 (Fig. 4D). On the other hand, it was apparent that Lid1 was associated with the full-length Rep2 protein. This result was subsequently confirmed in a reciprocal coimmunoprecipitation assay. These results suggest that the C-terminal 32-amino-acid sequence (residues 167 to 198) was important for physical interaction with Lid1-associated APC/C complex. We next examined the ability of Rep2-CΔ53 in ubiquitination using the in vivo ubiquitination assay. The assay showed that Rep2-CΔ53 was hardly ubiquitinated in vivo, indicating that the reduced turnover of Rep2-CΔ53 is a result of a failure in physical interaction with APC/C (Fig. 4E).
A noncanonical D-box (residues 164 to 174) or zinc finger (residues 177 to 194) domain was found to reside in this 32-amino-acid sequence. However, point mutation at the conserved residues of the D-box or zinc finger domain was not sufficient to reduce the Rep2 turnover (data not shown), suggesting that the Rep2 ubiquitination is not dependent on the predicted structural motifs. Given that this disrupted region of Rep2 sequences is known to interact with Res2, another factor of the MBF complex (40), it is therefore possible that the complex of Rep2 and Res2 is important for the APC/C recognition. To test this possibility, we performed Western blot analysis to determine the Rep2 turnover in res2Δ cells. The degradation of Rep2 was impeded in the res2Δ strain, suggesting that Rep2 interaction with Res2 is required for Rep2 ubiquitination and degradation (Fig. 4F and G).
Rep2 sequences required for ubiquitination are also important for the transcriptional response to HU.
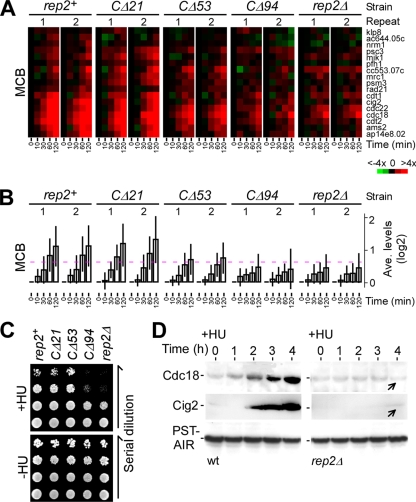
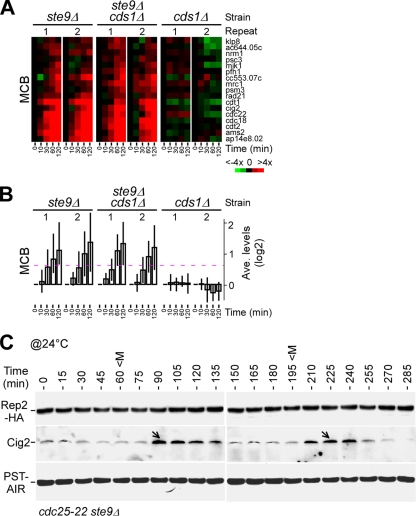
We showed previously that accumulation of the Rep2 protein correlated with the transcriptional induction of the MBF-dependent genes in response to HU (16). To determine which part of the Rep2 sequence was important for the transcriptional induction in response to HU, we performed expression profiling in cells bearing various C-terminally truncated rep2 alleles using open reading frame-specific oligonucleotide microarrays (45). The time course microarray data were normalized to 0 min for determining the level of transcriptional induction in various strains responding to HU (see Materials and Methods). Cells bearing the wild-type rep2+ showed a ∼100% (or twofold) increase in the average transcription level of the MBF-dependent genes at 120 min after HU treatment compared to cells prior to HU treatment or at 0 min (Fig. 5A and B; see Table S1 in the supplemental material). The wild-type level of transcriptional induction was observed in rep2-CΔ21 cells at 120 min after HU treatment, suggesting that the Rep2 C-terminal sequence from residue 198 to 219 was not required for transcriptional induction.
FIG. 5.
The C-terminal sequences of Rep2 are required for induction of the MBF-dependent genes in response to HU. (A) Induction of the MBF-dependent genes upon HU treatment for various C-terminally truncated Rep2 alleles. The intensity of the colors indicates the magnitudes of induction (red) or repression (green) for each gene indicated at the right (see color key at the bottom). Black indicates no change of expression levels, and gray indicates that data are not available. Two repeats in each strain are indicated on top, and time points after HU addition are indicated at the bottom. (B) Average expression profiles of the MBF-dependent genes in response to HU treatment. The dashed line indicates the 50% (or 1.5-fold) increase (0.585 on a log2 scale) of the transcription level. (C) Phenotypes of mutant cells containing various alleles of Rep2. The plating assays show the growth of 10-fold serial dilutions of various strains on YES plates with or without HU at 30°C (∼3 days after inoculation). (D) Cdc18 and Cig2 proteins fail to accumulate in rep2Δ cells upon HU treatment. Western blot analysis shows Cdc18 or Cig2 levels in wild-type and rep2Δ cells at various time points after HU treatment. Arrows indicate the position showing no apparent accumulation of Cdc18 or Cig2 proteins in rep2Δ cells.
In contrast, the average transcription level of the MBF-dependent genes was increased by only ∼33% (i.e., ca. one-third of the wild-type level) at 120 min after HU treatment in rep2Δ cells. In rep2-CΔ94 cells, the transcriptional induction profile of the MBF-dependent genes was identical to that in rep2Δ cells, suggesting that the Rep2 sequences from amino acid residue 126 to 198 are important for the full-level induction of the MBF-dependent genes in response to HU. We found that the average transcription level of the MBF-dependent genes was increased by ∼62% at 120 min after HU treatment in rep2-CΔ53 cells, ranging between ∼33% in rep2-CΔ94 or rep2Δ cells and ∼100% in rep2-CΔ21 or wild-type cells. This result suggests that the Rep2 sequence from residue 126 to 198 is required for the transcriptional response, overlapping with the Rep2 sequence from residue 167 to 198, which is required for ubiquitination.
S-phase regulators such as the licensing factor Cdc18 or S-phase cyclin Cig2 that are encoded by the MBF-dependent genes were known to be induced in cells at the onset of S-phase arrest (27, 35). We wanted to know if the induction of these protein levels requires Rep2 function at the S-phase arrest. To this end, the level of Cdc18 or Cig2 in wild-type or rep2Δ cells at various time points after treatment with HU was determined in Western blot analysis using anti-Cdc18 antibodies (a gift of P. Nurse) or anti-Cig2 antibodies (Fig. 5D). An induced level of Cdc18 or Cig2 was apparent in wild-type cells after addition of HU. On the other hand, no apparent induction of Cdc18 or Cig2 was observed in rep2Δ cells. This result indicates that the accumulation of S-phase regulators such as Cdc18 and Cig2 requires the stimulation of MBF transcriptional activity at the onset of S-phase arrest.
Phosphorylation of Ste9 at the onset of S-phase arrest requires the function of Cds1.
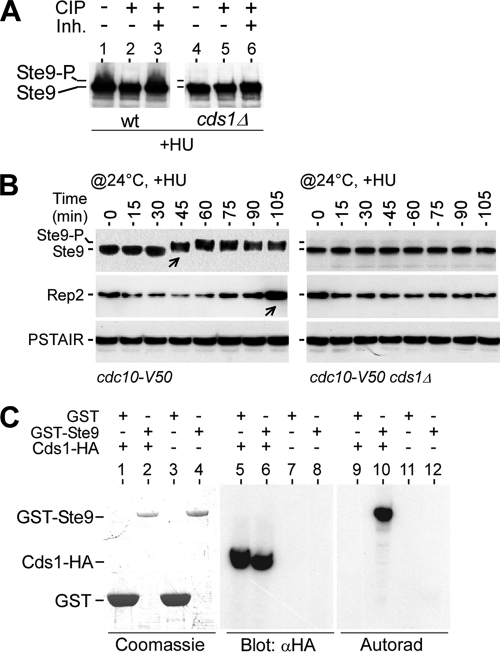
The APC/C activator Ste9 (or Srw1, a homolog to Hct1 or Cdh1 in S. cerevisiae) could be inhibited through phosphorylation by CDK (8). Active/dephosphorylated Ste9, which actives APC/C to degrade mitotic cyclins such as Cdc13 and Cig1, was observed in cells at the mitotic exit or G1-phase arrested (8). To test if the stability of Rep2 at the onset of S-phase arrest was a result of APC/C inactivation or Ste9 inhibition/phosphorylation, we determined the level of Ste9 inhibition/phosphorylation in HU-treated cells. We found that Ste9 phosphorylation in HU-treated cells was dependent on the function of Cds1 (Fig. 6A). In the cdc10-V50 arrested cells, Ste9 was hardly phosphorylated as judged by its fast migration (Fig. 6B). Upon release to the permissive temperature to medium supplemented with 8 mM HU, which blocks DNA replication, we found that the phosphorylation of Ste9 appeared at ∼45 min after the temperature shift in cdc10-V50 cells. The timing of Ste9 phosphorylation in medium supplemented with HU was identical to that without HU (data not shown) (8). Accumulation of Rep2 was apparent at ∼105 min after the temperature shift in cdc10-V50 cells. On the other hand, neither phosphorylation of Ste9 nor Rep2 accumulation was observed in cdc10-V50 cds1Δ cells within 105 min after the temperature shift, indicating that Ste9 inhibition/phosphorylation at the early S phase is dependent on the function of Cds1.
FIG. 6.
Phosphorylation of Ste9 at S-phase arrest requires the function of Cds1. (A) HU-induced phosphorylation of Ste9 is Cds1 dependent. Western blot analysis shows that HU-induced phosphorylation of Ste9 (the slowly migrating portion) is Cds1 dependent. The level of phosphorylated Ste9 is reduced by the treatment with calf intestine alkaline phosphatase (CIP) but inhibited when CIP inhibitors (Inh.) are present. wt, wild type. (B) Phosphorylation of Ste9 during cell cycle progression from G1 phase (i.e., at the cdc10-V50 arrest point) to early S phase (i.e., at the HU-induced arrest point) requires the function of Cds1. The level of Ste9 phosphorylation in synchronous cdc10-V50 or cdc10-V50 cds1Δ cultures was monitored at various time points after release to the permissive temperature (@24°C) in medium supplemented with 8 mM HU. Arrows indicate the time point at which phosphorylation of Ste9 or accumulation of Rep2 occurs. (C) Activated Cds1 phosphorylates Ste9 in vitro. GST or GST-Ste9 is apparent in Coomassie blue-stained gels (lane 1 to 4), and Cds1-HA is detected in Western blot analysis (lane 5 and 8). In the presence of activated Cds1-HA, phosphorylation of GST-Ste9 (lane 10) but not GST (lane 9) is detected by autoradiography.
To test if Ste9 was a putative target of the Cds1 kinase at the S-phase arrest, we performed in vitro kinase assays. To this end, GST-fused Ste9 proteins were expressed in E. coli and affinity purified using glutathione beads. The HU-activated Cds1-HA protein was affinity purified through anti-HA antibody-coupled beads. A kinase assay in vitro utilizing [γ-35S]ATP was performed. Each reaction mixture contained the GST or GST-Ste9 molecules, which were visualized in the Coomassie blue-stained gel, and the Cds1-HA molecules, which were detected using Western blot analysis (Fig. 6C). Phosphorylation of GST-Ste9 but not GST was observed when Cds1 was present in the kinase assay after autoradiography. This result suggests that Ste9 is a putative target of the S-phase checkpoint kinase Cds1.
Cds1-dependent phosphorylation prevents Ste9 from activating APC/C at S phase.
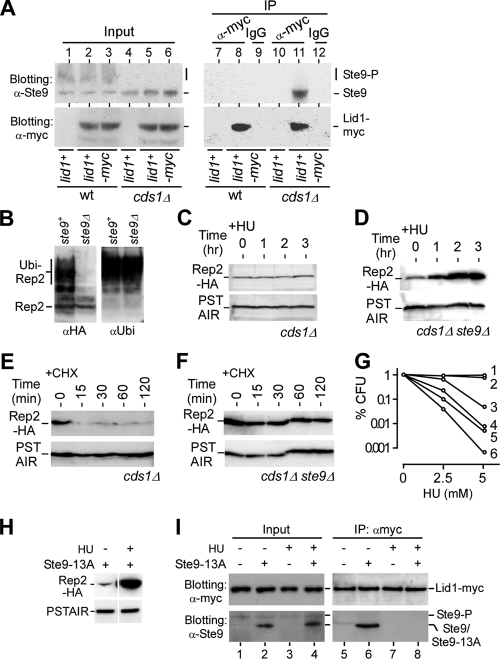
To test if Cds1-dependent phosphorylation of Ste9 disrupts its interaction with APC/C, we performed coimmunoprecipitation assays to examine the physical interaction between the APC/C activator Ste9 and the APC/C component Lid1. To this end, myc-tagged Lid1 was enriched using anti-myc antibodies in HU-treated wild-type and cds1Δ cells. A strain containing Lid1 without a myc tag was used as a negative control for anti-myc antibodies. Western blot analysis showed that the unphosphorylated Ste9 was associated with Lid1-myc in cds1Δ but not wild-type cells (Fig. 7A, lanes 11 and 8), suggesting that ubiquitination and subsequent degradation of Rep2 are mediated through APC/C-Ste9. Notably, the in vivo ubiquitination assay indicated that Rep2 failed to be ubiquitinated in ste9Δ cells (Fig. 7B), supporting the notion that the degradation of Rep2 is attributable to the activity of Ste9.
FIG. 7.
Cds1-dependent phosphorylation of Ste9 inhibits Ste9 interaction with APC/C at the onset of S-phase arrest. (A) Cds1-dependent phosphorylation of Ste9 is required to block Ste9 interaction with APC/C at the S-phase arrest. Coimmunoprecipitation (IP) using anti-myc antibodies shows that Ste9 is associated with Lid1-myc in HU-treated cds1Δ (lane 11) but not wild-type (wt) (lane 8) cells. IgG antibodies were used as a negative control (lanes 9 and 12). (B) Disruption of ste9 prevents Rep2 from being ubiquitinated. The in vivo ubiquitin assay shows that the ubiquitinated Rep2 is hardly detected in ste9Δ cells, in contrast to the case for wild-type cells. (C) Rep2 accumulation is diminished in cds1 cells. Western blot analysis shows the level of Rep2 in cds1Δ cells after HU treatment. (D) Disruption of ste9 restores the stability of Rep2 in cds1Δ cells upon HU treatment. Western blot analysis shows the level of Rep2 in cds1Δ ste9Δ cells at various time points after HU treatment. (E) Turnover of Rep2 in S-phase checkpoint-deficient cds1Δ cells. Western blot analysis shows that the level of Rep2 decreases soon after addition of CHX. (F) Disruption of ste9 restores the Rep2 stability in cds1Δ cells. Western blot analysis shows that the rate of turnover of Rep2 is reduced in cds1Δ ste9Δ cells. (G) Disruption of ste9 alleviates the HU-sensitive phenotype of rad3Δ or cds1Δ cells. The duration of the HU treatment is approximately a generation time (∼3 h). Numbers indicate the strains: 1, wild type; 2, ste9Δ; 3, cds1Δ ste9Δ; 4, cds1Δ; 5, rad3Δ ste9Δ; and 6, rad3Δ. (H) Rep2 is accumulated in cells bearing the ste9-13A allele upon S-phase arrest. Western blot analysis shows the Rep2 level in cells bearing the ste9-13A allele treated or untreated with HU. (I) Ste9-13A fails to interact with Lid1 upon HU treatment. The input level of Lid1 or Ste9/Ste9-13A in samples used in coimmunoprecipitation assay is shown in lanes 1 to 4. Proteins that are enriched with Lid1-myc by the anti-myc antibodies are shown in lanes 5 to 8. Interaction between Lid1 and Ste9-13A is apparent (lane 6) in asynchronous cells (untreated with HU). On the other hand, the interaction between Ste9-13A and Lid1 is diminished in cells treated with HU (lane 8).
We found that disruption of ste9 alleviated the defect of cds1Δ cells in accumulation of Rep2 upon HU treatment (Fig. 7C and D). The stability of Rep2 was diminished in cds1Δ cells (Fig. 7E). On the other hand, the stability of Rep2 was restored in cds1Δ ste9Δ cells, supporting the idea that Ste9 plays a role in mediating the Rep2 stability (Fig. 7F). To further test if disruption of ste9 would alleviate the defect of S-phase checkpoint-deficient rad3Δ or cds1Δ cells in HU sensitivity, we performed the recovery assay using various strains after brief treatment with HU. The CFU of various strains after exposure to different doses of HU showed that the viability was increased in the ste9-disrupted checkpoint-deficient mutant cells (Fig. 7G). These biochemical and genetic results imply that the S-phase checkpoint kinase Cds1 induces MBF activity by stabilizing the MBF activator Rep2 through the inhibition/phosphorylation of Ste9, an activator of APC/C, at the onset of S-phase arrest.
Ste9 is known to be inhibited/phosphorylated by Cdc2 throughout the cell cycle except for G1 phase (8). The active allele Ste9-13A (Cdc2 phosphorylation sites are disrupted) can bind APC/C at all stages of the cell cycle (8). Here we showed that Ste9 was inhibited/phosphorylated at the onset of S-phase arrest in a Cds1-dependent manner (Fig. 6). We found that Rep2 could be accumulated in ste9-13A-containing cells upon HU treatment (Fig. 7H), suggesting that Ste9-13A is not active at the onset of S-phase arrest. To further show that Ste9-13A could be inhibited/phosphorylated by Cds1 at the onset of S-phase arrest, we performed coimmunoprecipitation assays to determine the interaction between Ste9-13A and the APC/C component Lid1. Not surprisingly, interaction between Ste9-13A and Lid1 was diminished in HU-treated cells (Fig. 7I). These results suggest that upon S-phase arrest, Ste9 is inhibited/phosphorylated by Cds1 or Cds1-dependent kinase activity that is distinct from Cdc2 activity, consistent with our previous observation that Rep2 accumulation is independent of the Cig2 function (16).
Disruption of ste9 alleviates the defect of cds1Δ cells in the transcriptional response to HU.
We showed that the accumulation of Rep2 was restored in S-phase checkpoint-deficient cds1Δ cells when ste9 was disrupted (Fig. 7D). To test if this restoration of Rep2 stability was associated with transcriptional induction of the MBF-dependent genes in cds1Δ ste9Δ cells, we performed expression profiling of various strains in response to HU. Analysis of expression profiles indicated that transcriptional induction of the MBF-dependent genes was unaltered in ste9Δ cells but totally abolished in cds1Δ cells (Fig. 8A and B). Significantly, disruption of ste9 completely alleviated the cds1Δ defect in the transcriptional response of the MBF-dependent genes.
FIG. 8.
Disruption of ste9 alleviates the cds1Δ defect in the transcriptional response to HU. (A) Induction of the MBF-dependent genes upon HU treatment in ste9Δ, cds1Δ ste9Δ, and cds1Δ cells. The display of the transcription levels is identical to that in Fig. 5A. (B) Average expression profiles of the MBF-dependent genes in response to HU treatment. (C) Disruption of ste9 diminishes the pattern of oscillation of Rep2 levels during the cell cycle. Rep2 levels in cdc22-25 ste9Δ synchronous cultures were determined at various time points after release to the permissive temperature (@24°C). The peak of the mitotic index (M) is indicated. Arrows indicate the peak of the oscillated Cig2 levels.
Next, we wanted to test if Ste9 played a role in regulating oscillation of the Rep2 level during cell cycle progression. For this reason, cdc25-22 ste9Δ cells were synchronized by growth at the restrictive temperature for 4 h and then release to the permissive temperature. Cells were sampled every 15 min for a period of ∼5 h. Western blot analysis indicated that the pattern of oscillation of Cig2 levels was still apparent (Fig. 8C). On the other hand, the pattern of oscillation of Rep2 levels was largely diminished in cells lacking ste9. This result indicates that Ste9 plays a role in regulation of oscillation of the Rep2 level in the undisrupted cell division cycle. However, the profile of rep2+ transcription in ste9Δ cdc25-22 cells was not much different from that in cdc25-22 cells (data not shown).
DISCUSSION
We have previously shown that the S-phase checkpoint kinases Rad3 and Cds1 modulate the maintenance of a high transcription level of the MBF-dependent genes at the onset of S-phase arrest (16). This maintenance of a high transcription level is correlated with the accumulation of Rep2, the MBF activator (16). In this study, we demonstrate that Rep2 is degraded through ubiquitin-mediated proteolysis (Fig. 2C). Rep2 degradation requires its interaction partner and is dependent mainly on the function of APC/C (Fig. 3 and 4). Inhibition/phosphorylation of the APC/C activator Ste9 is Cds1 dependent at the onset of S-phase arrest (Fig. 6). Its phosphorylation prevents interaction between APC/C and Ste9 (Fig. 7A). Disruption of ste9 alleviates the cds1Δ defect in stabilization of Rep2 (Fig. 7C to F) and in transcriptional induction of the MBF-dependent genes (Fig. 8A and B) upon DNA replication stress. Our data demonstrate that S-phase checkpoint kinases activate the MBF transcriptional activity through the inhibition of APC/C-Ste9.
The proteolytic pathway has been shown to regulate the turnover of some transcription factors (TFs). Upon a stress response, TFs such as ATF-2 or c-Jun in mammalian systems and Atf1 or Zip in fission yeast are stabilized through protection from polyubiquitination and subsequent degradation (23, 26, 33, 41). Here we show that Rep2 is protected from polyubiquitination upon S-phase arrest. The Rep2 C-terminal sequence of 73 amino acids (residues 126 to 198) that are known to be required for interaction with Res2 (40) is required for transcriptional induction of the MBF-dependent genes (Fig. 5). However, Rep2 degradation is independent of the predicted structural motifs within the C-terminal deleted sequences (data not shown). Remarkably, Rep2 interaction with Res2 is important for Rep2 turnover (Fig. 4F and G), suggesting that APC/C-Ste9 may recognize the structure of the Rep2-Res2 complex but not Rep2 alone.
Rad3-Cds1 kinases inactive Cdc2/Cig2 during S-phase arrest (9, 49). We have previously shown that induction of the MCB cluster genes is independent of the function of Cig2 (16). In this study, we show that APC/C-Ste9 is responsible for Rep2 degradation. Upon S-phase arrest, APC/C-Ste9 is inhibited by the replication checkpoint kinase Cds1. Ste9 is known to be inhibited/phosphorylated by Cdc2 throughout the cell cycle except for G1 phase (8). Active allele Ste9-13A has been shown to be able to bind APC/C at all stages of the cell cycle (8). In this study, we show that active allele Ste9-13A cannot prevent accumulation of Rep2 upon S-phase arrest. Furthermore, interaction between APC/C and Ste9-13A is diminished upon HU treatment. These results suggest that Ste9 inhibition/phosphorylation at the onset of S-phase arrest requires the function of Cds1 but not Cdc2. We thus propose that the APC/C activator Ste9 can be inhibited/phosphorylated by Cdc2 and Cds1 through distinct phosphorylation sites.
We show that levels of Rep2 protein oscillated along with cell cycle (Fig. 1E). Turnover of cell cycle-specific TFs such as Ace2 (46), Rep2 (this study), and Sep1 (16) has been linked to the ubiquitin-proteasomes. The cell cycle is known to be controlled by CDK activities that are regulated through both (de)phosphorylation and ubiquitin-mediated proteolysis. It is conceivable that the activity of most of the periodic transcriptional complexes is also regulated through (de)phosphorylation and possibly ubiquitin-mediated proteolysis to ensure the proper timing and level of transcriptional activity during the cell cycle. In this study, we show that Rep2 turnover is dependent on the function of APC/C-Ste9 (Fig. 1E and 8C). However, it is worth noting that the kinetics of Rep2 oscillation differs from that of the other APC/C-Ste9 targets such as Cdc13 or Cig1 (8). Currently, it is unclear if this discrepancy is a result of distinct APC/C-Ste9 activities at the different subcellular localizations.
Two other regulatory pathways have been proposed to regulate MBF activity at the onset of S-phase arrest. Dutta et al. (21) have shown that Cds1 directly phosphorylates Cdc10 to activate the MBF activity upon DNA replication stress. de Bruin et al. have shown that Nrm1 negatively regulates MBF transcriptional activity (19). At the onset of S-phase arrest, Nrm1 appears to be inhibited/phosphorylated by Cds1 (18). Here we show that the MBF activator Rep2 is stabilized in a Cds1-dependent manner. Together, these results indicate that MBF activity is positively regulated by the S-phase checkpoint through multiple pathways. Consistent with this, we find that induction of the MBF-dependent genes at 120 min after HU treatment was ca. one-third of the wild-type level in rep2Δ cells (Fig. 5A and B) but was totally abolished in cds1Δ cells (Fig. 8A and B). Presumably, the MBF induction observed in the absence of Rep2 is likely to be result of the other two regulatory pathways mediated by Cds1 (20, 21). It is interesting, however, to see that disruption of ste9 can fully restore the level of transcription induction of the MBF-dependent genes in cds1Δ cells.
Upon DNA replication blockage induced by HU treatment, restoring stalled replication forks requires the function of the S-phase checkpoint (34, 38). The licensing factor Cdc18 is important for DNA replication initiation, and its protein levels oscillated with cell cycle (42). The E3 ligase SCF complex has been found to be responsible for the instability of Cdc18 (32). Recently, it has been proposed that the licensing factor Cdc18 also plays a role in restoring stalled forks: it accumulates at S-phase arrest and becomes associated with chromatin structures at the stalled forks (27). Importantly, cdc18 transcription is dependent on the function of the MBF complex. In this study, we show that accumulation of Rep2 stimulates transcription of cdc18 and other MBF-dependent genes (Fig. 5A and B). Clearly, stimulated transcription of cdc18 is required for its protein accumulation (Fig. 5D).
MBF, containing Cdc10, Res1, Res2, Rep2, and Nrm1, plays a pivotal role in promoting passage through START late in G1 phase (4). The transcriptional activity of MBF is possibly induced by Cdc2 through the Cdc10-Res1 complex (17, 48) and/or by Pef1-Pas1 via the Cdc10-Res2 complex (54) or through the inhibition of Nrm1 at G1 phase (19). MBF transcriptional activity is positively correlated with the level of Rep2 but negatively correlated with the level of Nrm1 (reference 19 and this study). Future work is needed to understand how the activator Rep2 and repressor Nrm1 are coordinated in regulation of MBF activity during G1 phase and in S-phase block.
Supplementary Material
Acknowledgments
We thank E. T. Liu for his encouragement and support during the course of this study. We thank P. Nurse for the anti-Cdc18 antibodies; S. Moreno for the anti-Ste9 antibodies; K. Gould and T. Toda for plasmids and strains; and K. M. Karuturi, J. Li, and L. Zhu for their assistance in analysis of the microarray data. We also thank K. Wong and the members of J. Liu's laboratory for discussion and critical reading of the manuscript. We appreciate the comments by the anonymous reviewers, which significantly improved the manuscript.
This work was supported by the Biomedical Research Council of Agency for Science, Technology and Research (A*STAR), Biopolis, Singapore.
Footnotes
Published ahead of print on 13 July 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Amon, A., S. Irniger, and K. Nasmyth. 1994. Closing the cell cycle circle in yeast: G2 cyclin proteolysis initiated at mitosis persists until the activation of G1 cyclins in the next cycle. Cell 771037-1050. [DOI] [PubMed] [Google Scholar]
- 2.Bahler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie III, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14943-951. [DOI] [PubMed] [Google Scholar]
- 3.Bai, C., P. Sen, K. Hofmann, L. Ma, M. Goebl, J. W. Harper, and S. J. Elledge. 1996. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86263-274. [DOI] [PubMed] [Google Scholar]
- 4.Baum, B., J. Wuarin, and P. Nurse. 1997. Control of S-phase periodic transcription in the fission yeast mitotic cycle. EMBO J. 164676-4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benito, J., C. Martin-Castellanos, and S. Moreno. 1998. Regulation of the G1 phase of the cell cycle by periodic stabilization and degradation of the p25rum1 CDK inhibitor. EMBO J. 17482-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berry, L. D., A. Feoktistova, M. D. Wright, and K. L. Gould. 1999. The Schizosaccharomyces pombe dim1+ gene interacts with the anaphase-promoting complex or cyclosome (APC/C) component lid1+ and is required for APC/C function. Mol. Cell. Biol. 192535-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berry, L. D., and K. L. Gould. 1996. Regulation of Cdc2 activity by phosphorylation at T14/Y15. Prog. Cell Cycle Res. 299-105. [DOI] [PubMed] [Google Scholar]
- 8.Blanco, M. A., A. Sanchez-Diaz, J. M. de Prada, and S. Moreno. 2000. APC(ste9/srw1) promotes degradation of mitotic cyclins in G(1) and is inhibited by cdc2 phosphorylation. EMBO J. 193945-3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boddy, M. N., B. Furnari, O. Mondesert, and P. Russell. 1998. Replication checkpoint enforced by kinases Cds1 and Chk1. Science 280909-912. [DOI] [PubMed] [Google Scholar]
- 10.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 11.Brandeis, M., and T. Hunt. 1996. The proteolysis of mitotic cyclins in mammalian cells persists from the end of mitosis until the onset of S phase. EMBO J. 155280-5289. [PMC free article] [PubMed] [Google Scholar]
- 12.Carr, A. M. 1997. Control of cell cycle arrest by the Mec1sc/Rad3sp DNA structure checkpoint pathway. Curr. Opin. Genet. Dev. 793-98. [DOI] [PubMed] [Google Scholar]
- 13.Carrano, A. C., E. Eytan, A. Hershko, and M. Pagano. 1999. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat. Cell Biol. 1193-199. [DOI] [PubMed] [Google Scholar]
- 14.Cho, R. J., M. J. Campbell, E. A. Winzeler, L. Steinmetz, A. Conway, L. Wodicka, T. G. Wolfsberg, A. E. Gabrielian, D. Landsman, D. J. Lockhart, and R. W. Davis. 1998. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol. Cell 265-73. [DOI] [PubMed] [Google Scholar]
- 15.Cho, R. J., M. Huang, M. J. Campbell, H. Dong, L. Steinmetz, L. Sapinoso, G. Hampton, S. J. Elledge, R. W. Davis, and D. J. Lockhart. 2001. Transcriptional regulation and function during the human cell cycle. Nat. Genet. 2748-54. [DOI] [PubMed] [Google Scholar]
- 16.Chu, Z., J. Li, M. Eshaghi, X. Peng, R. K. Karuturi, and J. Liu. 2007. Modulation of cell cycle-specific gene expressions at the onset of S phase arrest contributes to the robust DNA replication checkpoint response in fission yeast. Mol. Biol. Cell 181756-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Connolly, T., M. Caligiuri, and D. Beach. 1997. The Cdc2 protein kinase controls Cdc10/Sct1 complex formation. Mol. Biol. Cell 81105-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Bruin, R. A., T. I. Kalashnikova, A. Aslanian, J. Wohlschlegel, C. Chahwan, J. R. Yates III, P. Russell, and C. Wittenberg. 2008. DNA replication checkpoint promotes G1-S transcription by inactivating the MBF repressor Nrm1. Proc. Natl. Acad. Sci. USA 10511230-11235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Bruin, R. A., T. I. Kalashnikova, C. Chahwan, W. H. McDonald, J. Wohlschlegel, J. Yates III, P. Russell, and C. Wittenberg. 2006. Constraining G1-specific transcription to late G1 phase: the MBF-associated corepressor Nrm1 acts via negative feedback. Mol. Cell 23483-496. [DOI] [PubMed] [Google Scholar]
- 20.Ducommun, B., G. Draetta, P. Young, and D. Beach. 1990. Fission yeast cdc25 is a cell-cycle regulated protein. Biochem. Biophys. Res. Commun. 167301-309. [DOI] [PubMed] [Google Scholar]
- 21.Dutta, C., P. K. Patel, A. Rosebrock, A. Oliva, J. Leatherwood, and N. Rhind. 2008. The DNA replication checkpoint directly regulates MBF-dependent G1/S transcription. Mol. Cell. Biol. 285977-5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 9514863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuchs, S. Y., and Z. Ronai. 1999. Ubiquitination and degradation of ATF2 are dimerization dependent. Mol. Cell. Biol. 193289-3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Furnari, B., A. Blasina, M. N. Boddy, C. H. McGowan, and P. Russell. 1999. Cdc25 inhibited in vivo and in vitro by checkpoint kinases Cds1 and Chk1. Mol. Biol. Cell 10833-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gordon, C., G. McGurk, P. Dillon, C. Rosen, and N. D. Hastie. 1993. Defective mitosis due to a mutation in the gene for a fission yeast 26S protease subunit. Nature 366355-357. [DOI] [PubMed] [Google Scholar]
- 26.Harrison, C., S. Katayama, S. Dhut, D. Chen, N. Jones, J. Bahler, and T. Toda. 2005. SCF(Pof1)-ubiquitin and its target Zip1 transcription factor mediate cadmium response in fission yeast. EMBO J. 24599-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hermand, D., and P. Nurse. 2007. Cdc18 enforces long-term maintenance of the S phase checkpoint by anchoring the Rad3-Rad26 complex to chromatin. Mol. Cell 26553-563. [DOI] [PubMed] [Google Scholar]
- 28.Hershko, A. 1997. Roles of ubiquitin-mediated proteolysis in cell cycle control. Curr. Opin. Cell Biol. 9788-799. [DOI] [PubMed] [Google Scholar]
- 29.Hershko, A., and A. Ciechanover. 1992. The ubiquitin system for protein degradation. Annu. Rev. Biochem. 61761-807. [DOI] [PubMed] [Google Scholar]
- 30.King, R. W., R. J. Deshaies, J. M. Peters, and M. W. Kirschner. 1996. How proteolysis drives the cell cycle. Science 2741652-1659. [DOI] [PubMed] [Google Scholar]
- 31.Kominami, K., I. Ochotorena, and T. Toda. 1998. Two F-box/WD-repeat proteins Pop1 and Pop2 form hetero- and homo-complexes together with cullin-1 in the fission yeast SCF (Skp1-Cullin-1-F-box) ubiquitin ligase. Genes Cells 3721-735. [DOI] [PubMed] [Google Scholar]
- 32.Kominami, K., and T. Toda. 1997. Fission yeast WD-repeat protein pop1 regulates genome ploidy through ubiquitin-proteasome-mediated degradation of the CDK inhibitor Rum1 and the S-phase initiator Cdc18. Genes Dev. 111548-1560. [DOI] [PubMed] [Google Scholar]
- 33.Lawrence, C. L., H. Maekawa, J. L. Worthington, W. Reiter, C. R. Wilkinson, and N. Jones. 2007. Regulation of Schizosaccharomyces pombe Atf1 protein levels by StyI-mediated phosphorylation and heterodimerization with Pcr1. J. Biol. Chem. 2825160-5170. [DOI] [PubMed] [Google Scholar]
- 34.Lindsay, H. D., D. J. Griffiths, R. J. Edwards, P. U. Christensen, J. M. Murray, F. Osman, N. Walworth, and A. M. Carr. 1998. S-phase-specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes Dev. 12382-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin-Castellanos, C., K. Labib, and S. Moreno. 1996. B-type cyclins regulate G1 progression in fission yeast in opposition to the p25rum1 cdk inhibitor. EMBO J. 15839-849. [PMC free article] [PubMed] [Google Scholar]
- 36.Matsuoka, S., M. Huang, and S. J. Elledge. 1998. Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science 2821893-1897. [DOI] [PubMed] [Google Scholar]
- 37.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194795-823. [DOI] [PubMed] [Google Scholar]
- 38.Murakami, H., and H. Okayama. 1995. A kinase from fission yeast responsible for blocking mitosis in S phase. Nature 374817-819. [DOI] [PubMed] [Google Scholar]
- 39.Murakami, H., S. K. Yanow, D. Griffiths, M. Nakanishi, and P. Nurse. 2002. Maintenance of replication forks and the S-phase checkpoint by Cdc18p and Orp1p. Nat. Cell Biol. 4384-388. [DOI] [PubMed] [Google Scholar]
- 40.Nakashima, N., K. Tanaka, S. Sturm, and H. Okayama. 1995. Fission yeast Rep2 is a putative transcriptional activator subunit for the cell cycle ‘start’ function of Res2-Cdc10. EMBO J. 144794-4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nateri, A. S., L. Riera-Sans, C. Da Costa, and A. Behrens. 2004. The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science 3031374-1378. [DOI] [PubMed] [Google Scholar]
- 42.Nishitani, H., and P. Nurse. 1995. p65cdc18 plays a major role controlling the initiation of DNA replication in fission yeast. Cell 83397-405. [DOI] [PubMed] [Google Scholar]
- 43.Nurse, P. 1996. The central role of a CDK in controlling the fission yeast cell cycle. Harvey Lect. 9255-64. [PubMed] [Google Scholar]
- 44.Oliva, A., A. Rosebrock, F. Ferrezuelo, S. Pyne, H. Chen, S. Skiena, B. Futcher, and J. Leatherwood. 2005. The cell cycle-regulated genes of Schizosaccharomyces pombe. PLoS Biol. 3e225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peng, X., R. K. Karuturi, L. D. Miller, K. Lin, Y. Jia, P. Kondu, L. Wang, L. S. Wong, E. T. Liu, M. K. Balasubramanian, and J. Liu. 2005. Identification of cell cycle-regulated genes in fission yeast. Mol. Biol. Cell 161026-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petit, C. S., S. Mehta, R. H. Roberts, and K. L. Gould. 2005. Ace2p contributes to fission yeast septin ring assembly by regulating mid2+ expression. J. Cell Sci. 1185731-5742. [DOI] [PubMed] [Google Scholar]
- 47.Rape, M., and S. Jentsch. 2002. Taking a bite: proteasomal protein processing. Nat. Cell Biol. 4E113-E116. [DOI] [PubMed] [Google Scholar]
- 48.Reymond, A., J. Marks, and V. Simanis. 1993. The activity of S.pombe DSC-1-like factor is cell cycle regulated and dependent on the activity of p34cdc2. EMBO J. 124325-4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rhind, N., and P. Russell. 1998. The Schizosaccharomyces pombe S-phase checkpoint differentiates between different types of DNA damage. Genetics 1491729-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rhind, N., and P. Russell. 1998. Tyrosine phosphorylation of cdc2 is required for the replication checkpoint in Schizosaccharomyces pombe. Mol. Cell. Biol. 183782-3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rustici, G., J. Mata, K. Kivinen, P. Lio, C. J. Penkett, G. Burns, J. Hayles, A. Brazma, P. Nurse, and J. Bahler. 2004. Periodic gene expression program of the fission yeast cell cycle. Nat. Genet. 36809-817. [DOI] [PubMed] [Google Scholar]
- 52.Shiozaki, K., and P. Russell. 1997. Stress-activated protein kinase pathway in cell cycle control of fission yeast. Methods Enzymol. 283506-520. [DOI] [PubMed] [Google Scholar]
- 53.Spellman, P. T., G. Sherlock, M. Q. Zhang, V. R. Iyer, K. Anders, M. B. Eisen, P. O. Brown, D. Botstein, and B. Futcher. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 93273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tanaka, K., and H. Okayama. 2000. A pcl-like cyclin activates the Res2p-Cdc10p cell cycle “start” transcriptional factor complex in fission yeast. Mol. Biol. Cell 112845-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Toda, T., I. Ochotorena, and K. Kominami. 1999. Two distinct ubiquitin-proteolysis pathways in the fission yeast cell cycle. Philos. Trans. R. Soc. London B 3541551-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vodermaier, H. C. 2004. APC/C and SCF: controlling each other and the cell cycle. Curr. Biol. 14R787-R796. [DOI] [PubMed] [Google Scholar]
- 57.Walworth, N., S. Davey, and D. Beach. 1993. Fission yeast chk1 protein kinase links the rad checkpoint pathway to cdc2. Nature 363368-371. [DOI] [PubMed] [Google Scholar]
- 58.Walworth, N. C., and R. Bernards. 1996. rad-dependent response of the chk1-encoded protein kinase at the DNA damage checkpoint. Science 271353-356. [DOI] [PubMed] [Google Scholar]
- 59.Whitfield, M. L., G. Sherlock, A. J. Saldanha, J. I. Murray, C. A. Ball, K. E. Alexander, J. C. Matese, C. M. Perou, M. M. Hurt, P. O. Brown, and D. Botstein. 2002. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell 131977-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilkinson, C. R., M. Penney, G. McGurk, M. Wallace, and C. Gordon. 1999. The 26S proteasome of the fission yeast Schizosaccharomyces pombe. Philos. Trans. R. Soc. London B 3541523-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu, Y. J., M. Davenport, and T. J. Kelly. 2006. Two-stage mechanism for activation of the DNA replication checkpoint kinase Cds1 in fission yeast. Genes Dev. 20990-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamano, H., J. Gannon, and T. Hunt. 1996. The role of proteolysis in cell cycle progression in Schizosaccharomyces pombe. EMBO J. 155268-5279. [PMC free article] [PubMed] [Google Scholar]
- 63.Yamano, H., K. Kitamura, K. Kominami, A. Lehmann, S. Katayama, T. Hunt, and T. Toda. 2000. The spike of S phase cyclin Cig2 expression at the G1-S border in fission yeast requires both APC and SCF ubiquitin ligases. Mol. Cell 61377-1387. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.