Abstract
Gene expression results from the coordinated actions of transcription factor proteins and coregulators. Estrogen receptor alpha (ERα) is a ligand-activated transcription factor that can both activate and repress the expression of genes. Activation of transcription by estrogen-bound ERα has been studied in detail, as has antagonist-induced repression, such as that which occurs by tamoxifen. How estrogen-bound ERα represses gene transcription remains unclear. In this report, we identify a new mechanism of estrogen-induced transcriptional repression by using the ERα gene, ESR1. Upon estrogen treatment, ERα is recruited to two sites on ESR1, one distal (ENH1) and the other at the proximal (A) promoter. Coactivator proteins, namely, p300 and AIB1, are found at both ERα-binding sites. However, recruitment of the Sin3A repressor, loss of RNA polymerase II, and changes in histone modifications occur only at the A promoter. Reduction of Sin3A expression by RNA interference specifically inhibits estrogen-induced repression of ESR1. Furthermore, an estrogen-responsive interaction between Sin3A and ERα is identified. These data support a model of repression wherein actions of ERα and Sin3A at the proximal promoter can overcome activating signals at distal or proximal sites and ultimately decrease gene expression.
Downregulation of receptors by their ligands is a fundamental process by which cells control sensitivity to stimuli. For steroid hormones, this involves lipophilic ligands binding to intracellular receptors to induce a decline in receptor number. Regulation of estrogen (E2) receptor alpha (ERα) by E2 is one example. The E2-induced decline in ERα is, in part, mediated through direct regulation of the protein. It is well documented that decreases in ERα protein levels in response to E2 occur via the ubiquitin-proteasome pathway (1, 42). The mRNA levels of ERα also decrease, but the mechanism responsible for E2-induced repression of the ERα gene, ESR1, is not established (5, 49, 52).
ERα is a ligand-activated transcription factor that mediates the effects of E2 by regulating gene expression. Activation by ERα has been studied in detail, but little is understood about how E2-bound ERα represses transcription. E2-induced repression is, however, of significant biological importance. Microarray analyses of E2-treated breast cancer cell lines show that the number of repressed genes is greater than or near the number of activated genes (10, 19, 29, 32). Yet, there are limited reports investigating E2-induced repression, and no generalized mechanism has emerged (6, 13, 22, 25, 43, 47, 59, 60, 71, 74). Antagonist-induced repression by selective ER modulators involves conformational changes that prevent coactivator binding to ERα (55). Such a conformational blockade does not occur with agonist binding and thus cannot account for E2-induced gene repression.
Many repressive complexes exist to restrict gene expression in response to cellular signals. One example is the Sin3 complex, which was identified in yeast but is conserved in mammals (41, 58). The Sin3 core complex consists of the Sin3A scaffolding protein; histone deacetylase 1 (HDAC1) and HDAC2; RbAp46 and RbAp48, which stabilize the complex to nucleosomes; and SAP18 and SAP30, which stabilize the interaction between Sin3A and the HDACs (23, 73). The specificity and function of the core complex can be expanded by adding extra catalytic modules onto the Sin3A platform. These include histone methylation, DNA methylation, chromatin remodeling, and monosaccharide transferase ability (reviewed in reference 57). Sin3A lacks intrinsic DNA binding, so it must be targeted to promoters via interaction with other DNA-binding or adaptor proteins. Interactions have been found for Sin3 and Mad, NRSF, p53, MeCP2, and many others (3, 38, 40, 51).
Repression of ESR1 is a crucial brake on the E2 signaling pathway. High levels of ERα in breast cancer cells leads to activation of E2-regulated genes in the absence of ligand (18, 28). Further evidence shows that mice with upregulated ERα expression develop ductal hyperplasia, lobular hyperplasia, and ductal carcinoma in situ, demonstrating the consequences of unregulated ERα levels at all stages of breast cancer development (20). It is also proposed that ESR1 is amplified in subsets of breast cancers and in precancerous breast diseases (26). This evidence suggests that failure of E2 to limit ERα levels could contribute to the uncontrolled cellular proliferation that occurs in cancer.
In this report, a new model of E2-induced gene repression is identified on the basis of analysis of ESR1. The findings show that repression is accomplished by the effects of ERα and Sin3A at the proximal promoter of ESR1 that dominate over activating factors in distal and proximal regions. These data add to the limited knowledge base of E2-induced repression mechanisms.
MATERIALS AND METHODS
Cell culture and treatments.
MCF7 cells were maintained at 37°C and 10% CO2 in Dulbecco's modified Eagle's medium (DMEM; Mediatech, Inc.) with phenol red and l-glutamine, supplemented with 10% fetal bovine serum (FBS; Biowest), 100 U/ml penicillin, and 100 μg/ml streptomycin (GIBCO/Invitrogen). T47D cells were maintained at 37°C and 5% CO2 in RPMI 1640 medium with phenol red and l-glutamine (Mediatech) supplemented with 10% FBS, penicillin, and streptomycin as described above. For hormone treatments, cells were incubated at 37°C and 5% CO2 for at least 3 days in phenol red-free DMEM (Mediatech) supplemented with 4 mM l-glutamine (GIBCO) for MCF7 or RPMI 1640 medium (Mediatech) for T47D cells, both supplemented with 10% charcoal dextran-stripped FBS and penicillin/streptomycin. 17-β-Estradiol (E2; Steraloids, Inc.) was added to a final concentration of 10 nM, 4-hydroxytamoxifen (OHT; Sigma) was used at 100 nM, and ICI182,780 (ICI; gift from Jack Gorski) was used at 10 nM. In combination experiments, the amount of ICI was increased to 100 nM. The ethanol (EtOH) vehicle control was 0.1% in all samples. For experiments with actinomycin D (Sigma), cells were pretreated with 2 μM actinomycin D for 30 min and then treated with EtOH or E2 for the times indicated in the figures.
RNA isolation and qRT-PCR.
RNA isolation and quantitative real-time PCR (qRT-PCR) were carried out as previously detailed (63). The sequences of the primers used for the different gene targets are available upon request. All primers were checked on a standard curve, and it was verified that efficiencies were near 100%. Ribosomal protein P0 mRNA was used as the internal control. Relative mRNA levels were calculated by the ΔΔCt method with the EtOH vehicle used as the calibrator.
ChIP.
Chromatin immunoprecipitation (ChIP) was carried out essentially as described previously (18), except that sonication was performed with three pulses of 15 s each to achieve fragment sizes of around 500 bp. The antibodies used were ERα (HC-20, sc-543), Sin3A (K-20, sc-994), NCoA3/AIB1 (C-20, sc-7216), p300 (N-15, sc-584), and immunoglobulin G (sc-2027) from Santa Cruz Biotechnology; AcH3K9 (AcH3K9; 07-352), AcH3K14 (07-353), AcH3K18 (07-354), pan-histone H3 (07-690), pan-histone H4 (05-858), trimethylated histone H3K9 (17-625), trimethylated histone H3K27 (17-622), and trimethylated histone H4K20 (07-463) from Millipore; and RNA polymerase II (PolII; 8WG16) from Covance. qRT-PCR was carried out as described above, except with 1 μl of input or 4 μl of immunoprecipitation (IP) sample and 200 nM forward and reverse primers. The sequences of the primers used for the different genomic regions are available upon request. Data are calculated as percent of input or relative to an EtOH control by the ΔΔCt method, as indicated in the figure legends.
Transfection of siRNA.
For Sin3A small interfering RNA (siRNA), cells were transfected in 10-cm plates in regular DMEM without antibiotics. Eight hundred picomoles of Sin3A or scrambled siRNA was diluted in Lipofectamine reagent and Opti-MEM (Invitrogen), added to appropriate plates, and incubated for 4 h. Two days later, cells were transfected with siRNA again and changed to phenol red-free medium following transfection. For AIB1 siRNA, cells were transfected once in six-well plates in phenol-red free medium without antibiotics with 200 pmol of AIB1 or scrambled siRNA diluted in Lipofectamine and Opti-MEM and added to appropriate wells for 4 h. Three days following transfection, cells were treated with EtOH or E2 for 4 h and harvested for RNA isolation or Western blot analysis.
Coimmunoprecipitations.
MCF7 cells were harvested with phosphate-buffered saline and lysed in IP buffer (50 mM Tris-Cl (pH 7.5), 150 mM NaCl, 1% NP-40, 10% glycerol, 2 mM EDTA, 50 mM NaF). Lysates were precleared with protein A-Sepharose beads for 15 min at 4°C. Four percent was removed for inputs, and the remaining portion was subjected to IP with 6 μg of Sin3A (K-20, sc-994) or control hemagglutinin (HA; Y-11, sc-805) antibody, both from Santa Cruz Biotechnology, and protein A-Sepharose beads at 4°C. Beads were washed four times with IP buffer, boiled for 7 min in 2× sodium dodecyl sulfate sample buffer, and then subjected to Western blot analysis.
Western blot analysis.
Western blot analysis was carried out as previously described (63). The antibodies used were Sin3A (K-20, sc-994; Santa Cruz), ERα (VP-E613; Vector Laboratories), HDAC2 (C-8, sc-9959; Santa Cruz), AIB1 (611104; BD Biosciences), and β-actin (A5441; Sigma).
Statistical analysis.
Student t tests were performed on indicated data with GraphPad Prism (GraphPad Software). Data were considered significant at a P value of <0.05 (indicated in the figures by asterisks).
RESULTS
E2-induced transcriptional repression of ESR1 is an ERα-dependent process.
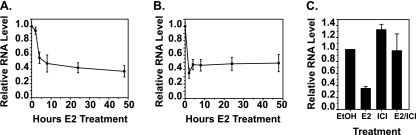
An inverse relationship exists between E2 and ERα levels in hormone-responsive tissues in vivo (36, 46). Hormone-dependent downregulation of receptors also occurs in the breast cancer cell line MCF7 (5, 49, 52). qRT-PCR was performed with MCF7 cells treated with E2, and the levels of ESR1 mRNA were analyzed out to 48 h. Total ESR1 mRNA decreased within 4 h of hormone exposure (Fig. 1A). A similar analysis was performed with primers that amplify intronic regions of the nascent ESR1 transcript. If the decrease seen in total ESR1 mRNA occurred at the transcriptional level, then a decrease in the nascent transcript should be observed. However, if the change was at the posttranscriptional level, transcription should continue at the same rate and no decrease should occur in the nascent transcript. Data showed that the nascent transcript decreased faster than the total mRNA, down to less than 50% by 2 h (Fig. 1B). These results were further verified with another primer set that amplified a different intronic region (data available on request). Negative control reaction mixtures containing RNA but no reverse transcriptase were also included and did not amplify anything, showing that there was no DNA contamination (data not shown). Since levels of nascent RNA could also be affected by splicing rates, we further analyzed whether the decrease in ESR1 occurred at the transcriptional level by treating cells with actinomycin D (data available on request). In the presence of actinomycin D, there was no difference in the level of ESR1 in the presence of control EtOH or E2, indicating that E2 does not destabilize the transcript. Altogether, these data indicate that repression occurs at the transcriptional level. These data also show that repression occurs rapidly, but then a steady level of low ESR1 mRNA is maintained.
FIG. 1.
ERα mediates E2-induced transcriptional repression of ESR1. qRT-PCR was used to analyze the expression of (A) total ESR1 or (B) nascent ESR1 (intron 1 primers) in MCF7 cells following treatment with 10 nM E2. Data shown are relative to those of a vehicle (EtOH)-treated control. (C) MCF7 cells were treated for 24 h with EtOH, 10 nM ICI, 10 nM E2, or 10 nM E2 in combination with 100 nM ICI. Levels of total ESR1 were detected by qRT-PCR and are represented relative to those of vehicle-treated cells. Error bars show the standard error of the mean of at least three independent experiments.
To establish that E2-induced repression of ESR1 was dependent on ERα itself, MCF7 cells were treated with a combination of E2 and ICI. ICI is a pure ERα antagonist that functions by rapidly degrading ERα protein (14). As shown in Fig. 1C, E2-induced repression was prevented when cells were treated in combination with ICI. Treatment with ICI alone had a minimal effect. Similar results were obtained with primers to the nascent ESR1 transcript (data not shown).
E2 induces recruitment of ERα to proximal and distal sites of ESR1.
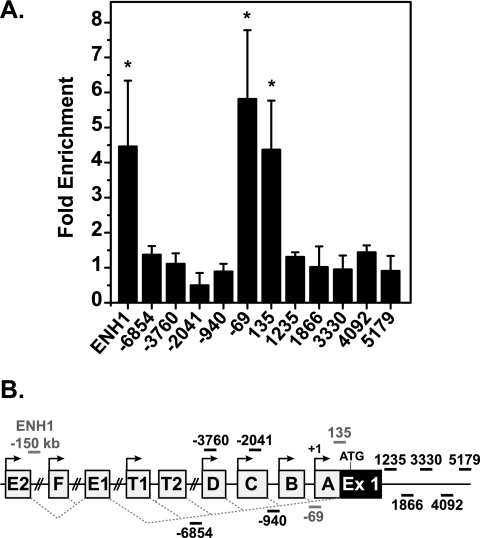
Uncovering the mechanism of repression of ERα mRNA is made difficult by the complexity of ESR1. Transcription can occur from any of the upstream promoters depicted in Fig. 2B. The nomenclature used here is based on the system proposed in Kos et al. (30), and the numbering used is based on the ESR1 sequence (accession number NM_000125). The alternative promoters allow for tissue-specific expression of transcripts. Upstream exons are spliced to the same acceptor site, resulting in transcripts that differ only in their 5′ untranslated regions (50).
FIG. 2.
ERα binds proximal and distal sites on ESR1 in response to E2. (A) MCF7 cells were treated for 24 h with EtOH or 10 nM E2 and harvested for ChIP with ERα antibody. qRT-PCR was performed with the ESR1 primer set indicated. Data are normalized to input values and calculated as enrichment versus an EtOH-treated control for each amplicon. Error bars are the standard error of the mean of at least three independent experiments. Statistical significance of increased ERα binding in the presence of E2 was determined with the Student t test for paired data (*, P < 0.05) on raw percent input values. (B) Diagram of ESR1 promoter regions with primer locations indicated and ERα-binding sites in red. Ex, exon.
ChIP experiments examining ERα occupancy in the 5′ regulatory region of ESR1 were performed as a first step toward elucidating the mechanism of repression. ChIP analysis focused on the proximal A to D promoters, as these are the most active in mammary tissue and MCF7 cells (15, 50; data available on request). No increase in ERα occupancy was seen in response to E2 at sites amplified with primer sets −3760/−3604 and −2041/−1856, which recognize the regions surrounding the D and C promoters, respectively (Fig. 2A). However, increased ERα occupancy was detected at regions amplified with primer sets −69/39 and 135/292, indicating that ERα was recruited near the A promoter. In addition, ERα was recruited to an enhancer, ENH1, a site that was suggested previously to be a regulator of ESR1 activation (17). ERα binding was not detected at other sites examined. Cells treated with E2 for various time periods, ranging from 30 min to 24 h, only exhibited ERα binding at ENH1 and the A promoter, ensuring that the lack of recruitment of ERα to other sites did not reflect kinetic differences (Fig. 3D and data not shown).
FIG. 3.
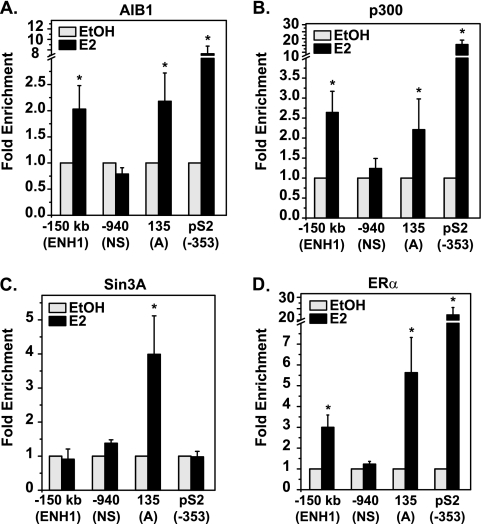
Sin3A is exclusively found at the proximal promoter of ESR1, while activators are recruited to both ERα-binding sites. MCF7 cells were treated with EtOH or 10 nM E2 for 30 min and processed for ChIP analysis of (A) AIB1, (B) p300, (C) Sin3A, or (D) ERα occupancy. qRT-PCR was performed with primers for three sites of ESR1: ENH1, the nonspecific region (NS), and the A promoter. The pS2 promoter is included as a control E2-activated gene. Data are normalized to the input and calculated as enrichment versus the EtOH-treated control for each amplicon, with error bars representing the standard error of the mean of at least three independent experiments. Statistical significance of increased binding in the presence of E2 was determined with the Student t test for paired data (*, P < 0.05) on raw percent input values.
E2 stimulates assembly of activators at both ERα-binding sites but recruits the Sin3A repressor only to the proximal promoter.
Binding of ERα at target genes is accompanied by the recruitment of coregulator proteins (37). The distal and proximal ERα-binding sites on ESR1 were examined for occupancy of different cofactors. Two of the most well-defined coactivators found in complex with ERα in breast cancer cells are AIB1, a member of the p160 family, and p300, a histone acetyltransferase (21, 33). Due to their defined roles in ERα activation, occupancy of AIB1 and p300 on ESR1 was examined in MCF7 cells. Figure 3A shows that E2 caused recruitment of AIB1 to both ENH1 and the A promoter, but not a nonspecific region (NS) of ESR1, consistent with agonist-induced conformational changes that favor coactivator interactions with ERα. Similarly, Fig. 3B shows that p300 was recruited with E2 to both the proximal and distal ERα-binding sites. Although these activators are recruited to ESR1, the recruitment is much less than that seen at the ERα-binding site of the control E2-activated gene pS2 (Fig. 3). The association of activating proteins in the context of gene repression may seem paradoxical. However, it is of note that ESR1 is not completely silent in the presence of E2 (Fig. 1A and B).
A repressive factor responsible for mediating E2-induced repression of ESR1 has not been identified. Proteins KLF9, Snail, and MeCP2 were shown to impact basal ESR1 expression near the proximal promoter under various experimental conditions and in various cell lines (16, 54, 65). These three proteins are all associated with the Sin3 repressive complex. We thus hypothesized that this common link could be involved in E2-induced repression of ESR1. As shown in Fig. 3C, Sin3A was recruited to the A promoter but not to ENH1 or the nonspecific regions. ERα was also present at both ENH1 and the A promoter at the same time point (Fig. 3D). Changes in cofactor binding are specific, as no increase in immunoglobulin G was seen (data available on request). The specific recruitment of Sin3A to the proximal promoter suggested a role for the complex in mediating repression of ESR1.
Sin3A is directly involved in the mechanism of E2-induced repression of ESR1.
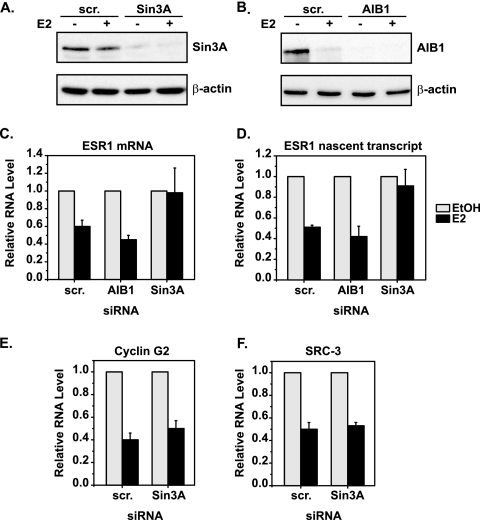
The requirement for Sin3A in ERα-mediated repression of ESR1 was tested by knocking down its expression with siRNA. MCF7 cells were transfected with control scrambled or Sin3A siRNA and then treated with EtOH or E2. Western blot analysis verified that the levels of Sin3A were efficiently decreased under these conditions (Fig. 4A). Additionally, cells were transfected with siRNA to AIB1, which was shown to be present at both the proximal and distal ERα-binding sites of ESR1 (Fig. 3A). AIB1 protein levels were decreased by E2 treatment alone, but siRNA to AIB1 abolished detection of AIB1 protein (Fig. 4B). The expression of ESR1 in the presence of scrambled, Sin3A, or AIB1 siRNA was monitored by qRT-PCR. E2 treatment resulted in repression of total ESR1 mRNA in cells transfected with scrambled or AIB1 siRNA (Fig. 4C). However, this repression was prevented in the presence of Sin3A siRNA. Furthermore, these data were verified by qRT-PCR with primers that amplify nascent ESR1 transcripts (Fig. 4D). Neither AIB1 nor Sin3A siRNA had a substantial effect on the basal level of total or nascent ESR1 in MCF7 cells (data available on request). These data provide direct functional evidence that Sin3A is required for E2-induced transcriptional repression of ESR1.
FIG. 4.
Sin3A is specifically required for repression of ESR1. MCF7 cells were transfected with scrambled (scr.), Sin3A, or AIB1 siRNA and then treated with EtOH or 10 nM E2 for 4 h. Levels of the (A) Sin3A and (B) AIB1 proteins were verified by Western blot analysis, with β-actin serving as a loading control. qRT-PCR was used to determine the levels of (C) total ESR1 mRNA, (D) ESR1 nascent transcript (intron 1 primers), (E) cyclin G2 mRNA, and (F) SRC-3 mRNA under the experimental conditions described above. Data shown are relative to those of the EtOH-treated control for each siRNA condition, with error bars representing the standard error of the mean of at least three independent experiments.
The specificity of the role of Sin3A in E2-induced repression of ESR1 was examined. Since Sin3A is a broad transcriptional repressor, it is possible that knocking down its expression may have effects on the repression of other genes. Two other genes that are repressed by E2 in an ERα-dependent manner are those for cyclin G2 and SRC-3 (60; S.E., unpublished data). Figure 4E and F show that Sin3A siRNA had no effect on the E2-induced repression of cyclin G2 or SRC-3, confirming that loss of Sin3A expression does not affect all genes.
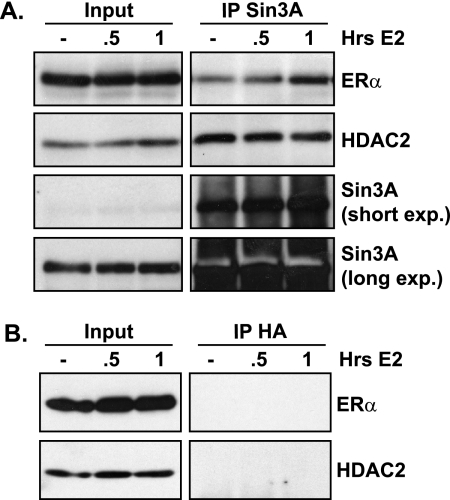
Experiments were performed to determine whether ERα could interact with Sin3A and therefore bring Sin3A to the E2-responsive region of target genes since it lacks DNA-binding capability. Indeed, coimmunoprecipitation experiments showed an association of ERα with Sin3A (Fig. 5A). Interestingly, this association was increased with E2 treatment. HDAC2 is a component of the Sin3 complex known to bind to Sin3A. This association was confirmed and did not change in the presence of E2. These interactions were specific, as coimmunoprecipitation experiments with equal concentrations of nonspecific HA antibody did not precipitate ERα or HDAC2 protein (Fig. 5B). These data show an interaction between ERα and Sin3A that is increased in a ligand-dependent manner. This suggests that agonist-bound ERα could regulate repression by direct recruitment of Sin3A and subsequent chromatin modifications that lead to damping of gene transcription.
FIG. 5.
Sin3A and ERα interact in an E2-responsive manner. MCF7 cells were treated with EtOH or 10 nM E2 and harvested for coimmunoprecipitation at the indicated times. Four percent of the total sample was removed for inputs before IP. The remaining lysate was immunoprecipitated with (A) Sin3A or (B) control HA antibody and analyzed by Western blot analysis for ERα, HDAC2, or Sin3A where indicated. exp., exposure.
Repression of ESR1 is accompanied by a complex code of histone modifications near the proximal promoter.
As noted above, Sin3A is a scaffold upon which other proteins with enzymatic activity assemble to modify nearby histones. The classic enzymatic function of the Sin3 complex is histone deacetylation via the HDAC1 and HDAC2 components (73). However, the function of the Sin3 complex can be expanded by interactions with other enzymatic proteins. For example, interaction between Sin3A and histone methyltransferases has been observed in several instances (7, 67, 70).
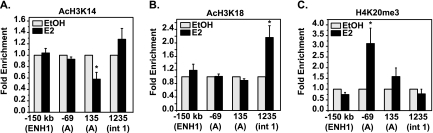
Since Sin3A is required for E2-induced repression of ESR1 and the Sin3 complex functions by modifying histones, the chromatin landscape of ESR1 in response to E2 was analyzed. Changes in the acetylation level of histones were examined, as the core function of the Sin3 complex is histone deacetylation. Experiments with a pan-AcH4 antibody did not show changes with E2 (data not shown). A pan-AcH3 antibody that recognized AcH3 on lysines 9, 14, and 18 gave variable results (data not shown). We hypothesized that variability could be due to lysine-specific regulation. Therefore, experiments were carried out with antibodies that recognize acetylation at individual lysine residues. Levels of AcH3K14 decreased with E2 near the proximal promoter (Fig. 6A) but not at ENH1, confirming that HDAC activity was present at the A promoter, consistent with the presence of the Sin3 complex. Analysis of AcH3K18 levels showed no change with E2 treatment near the proximal promoter or ENH1, but an increase was observed near intron 1, possibly related to the presence of activators p300 and AIB1 (Fig. 6B). Levels of the third modification, AcH3K9, did not change, confirming that alterations in the histone marks were residue specific (data available on request).
FIG. 6.
Changes in histone modifications occur near the proximal ERα-binding site on ESR1 where Sin3A binds. MCF7 cells were treated with EtOH or 10 nM E2 for 2 h and processed for ChIP analysis of the histone modifications (A) AcH3 lysine 14 (AcH3K14), (B) AcH3 lysine 18 (AcH3K18), and (C) trimethylated histone H4 lysine 20 (H4K20me3). ENH1, two regions near the A promoter, and intron 1 (int 1) of ESR1 were amplified by qRT-PCR. Data are normalized to the input and calculated as enrichment versus the EtOH control, with error bars representing the standard error of the mean of at least three independent experiments. Statistical significance of histone modification changes in the presence of E2 was determined with the Student t test for paired data (*, P < 0.05) on raw percent input values.
In addition to loss of acetylation, methylation of histones can lead to gene repression, and Sin3A has been found to interact with histone methyltransferases, as noted above. The common methylation marks associated with repression occur on histone H4 lysine 20 (H4K20), histone H3 lysine 9 (H3K9), and histone H3 lysine 27 (H3K27) (45, 53). ChIP analysis showed that the levels of H4K20 trimethylation (H4K20me3) increased near the A promoter in response to E2 (Fig. 6C). Of note, the levels of H4K20me3 did not change at ENH1. Analysis of H3K9me3 and H3K27me3 showed subtle decreases with E2 treatment near the A promoter and intron 1, respectively (data available on request). The levels of total H3 and H4 did not change in response to E2 (data available on request). Taken together, these data show that E2 induces a complex array of histone modifications near the proximal promoter of ESR1. Deacetylation and methylation of histones are consistent with the presence of the Sin3 complex in this same region.
Transcriptional changes associated with repression of ESR1 occur with ERα binding to the proximal promoter, not ENH1.
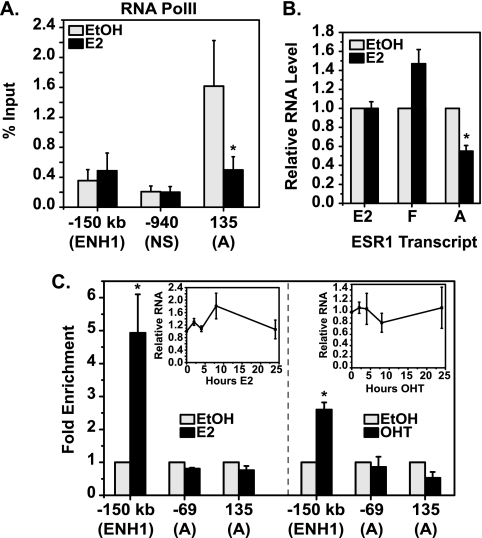
The ERα-binding sites on ESR1 provided a system to examine the role of distal and proximal elements in repression. The fact that Sin3A recruitment and histone modifications occur with E2 treatment only at the proximal promoter of ESR1 already suggests that this is the dominant site for repression. Studies on E2 activation of genes with distal enhancers have shown that RNA PolII is recruited to distal regulatory regions, as well as promoters (44, 61). ChIP experiments were performed to examine RNA PolII levels at the distal and proximal sites of ESR1. With E2 treatment, there was a decrease in RNA PolII occupancy at the A promoter (Fig. 7A). However, at ENH1, E2 treatment did not change RNA PolII levels. These data show that although E2 induces recruitment of ERα to ENH1 and the A promoter of ESR1, changes in the transcriptional machinery consistent with gene repression occur only at the A promoter.
FIG. 7.
The proximal ERα-binding site is associated with transcriptional repression of ESR1. (A) ChIP analysis for RNA PolII was carried out with MCF7 cells treated with EtOH or 10 nM E2 for 1 h. Data are shown as a percentage of the input for the indicated ESR1 amplicon. Statistical significance of changes in PolII occupancy in the presence of E2 was determined with the Student t test for paired data (*, P < 0.05). (B) MCF7 cells were treated with EtOH or 10 nM E2 for 2 h. Levels of A, E2-E1, and F-E1 transcripts were detected by qRT-PCR and are shown relative to those of vehicle-treated cells. Statistical significance of transcript expression changes in the presence of E2 was determined with the Student t test for paired data (*, P < 0.05). (C) T47D cells were treated with 10 nM E2 (left panel), and MCF7 cells were treated with 100 nM OHT (right panel) for 24 h and processed for ChIP analyses with an ERα antibody. qRT-PCR was performed for the two ERα-binding sites identified: ENH1 or the A promoter of ESR1. Data are normalized to the input and calculated as enrichment versus an EtOH-treated sample for each amplicon. Insets show qRT-PCR data for total ESR1 RNA from the corresponding treatment groups, relative to those for an EtOH-treated sample. Statistical significance of increased ERα binding in the presence of E2 was determined with the Student t test for paired data (*, P < 0.05) on raw percent input values. Error bars show the standard error of the mean of at least three independent experiments.
Transcript expression driven from the promoters nearest the observed ERα-binding sites was examined. Levels of the A transcript decreased within 2 h of E2 treatment (Fig. 7B). In contrast, levels of the E2-E1 transcript did not change with treatment, though ENH1 is only 1.5 kb from the E2 promoter. The next closest promoter to ENH1 is the F promoter, and the F-E1 transcript was also not repressed in response to E2, nor was the slight increase in expression statistically significant. Additionally, measurement of the abundance of each transcript showed that greater than 90% of the total ESR1 mRNA consisted of the A transcript, while the F-E1 and E2-E1 transcripts made up less than 1% (data available on request).
The functionality of the two ERα-binding sites in ESR1 was further tested with T47D cells treated with E2 and MCF7 cells treated with the selective ER modulator OHT. In both instances, repression of ESR1 did not occur (insets in Fig. 7C). Expression of the nascent, A, E2-E1, or F-E1 transcripts was likewise not affected under either condition (data available on request). ChIP analysis showed that ERα was still recruited to ENH1 in both cases, despite no repression of ESR1 (Fig. 7C). However, ERα recruitment to the proximal promoter of ESR1 was not detected with either primer set. These data support the conclusion that occupancy of ERα at ENH1 is not sufficient to achieve repression of ESR1, in agreement with the finding that Sin3A is necessary for repression and found only at the proximal promoter.
DISCUSSION
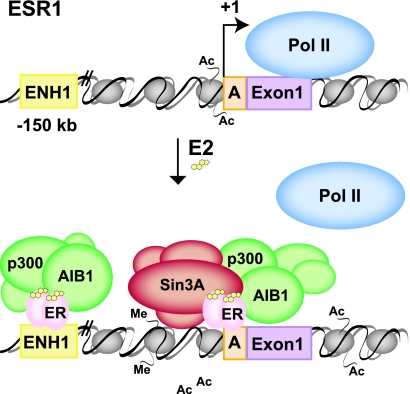
This report describes a new model of E2-mediated repression based on the physiological ERα transcript autoregulation depicted in Fig. 8. Under unstimulated or E2-poor conditions, the histones of ESR1 are acetylated near the proximal promoter and the gene is accessible and actively transcribed by RNA PolII. Upon E2 binding, ERα binds to two sites on ESR1, a region near the A promoter and ENH1. Binding to both sites induces recruitment of activating complexes containing p300 and AIB1. However, the Sin3 repressor is bound only to the A promoter. These events lead to histone deacetylation, acetylation, and methylation on specific sites, creating a code of modifications near the proximal promoter. Ultimately, the loss of RNA PolII leads to attenuation of ESR1 transcription. This model implies a dominant role for ERα, Sin3A, and subsequent chromatin modifications functioning at the proximal promoter in E2-induced transcriptional repression.
FIG. 8.
Model of E2-induced repression of ESR1. For details, see the text. Me, methylation; Ac, acetylation.
A novel role for Sin3A in E2-induced gene repression.
There have only been a few reports on E2-induced repression, though it is clearly an important function of ERα (19). Examination of ESR1 revealed new findings to add to this limited knowledge base and help explain how E2-bound ERα can function as a repressor of gene transcription. This report identified Sin3A as a component of the repression mechanism through functional experiments with siRNA. Additionally, an E2-responsive interaction between ERα and Sin3A was discovered. These findings suggest that ERα can recruit Sin3A to target promoters to repress gene transcription. Consistent with this hypothesis, experiments with T47D cells, which do not repress ESR1 in response to E2, showed that overexpression of Sin3A was not sufficient to achieve repression of ESR1 (data not shown). However, as shown in Fig. 7C, ERα is not recruited to the proximal promoter of ESR1 in T47D cells with E2 treatment. If ERα is needed for Sin3A recruitment, then modulating levels of Sin3A would have no effect on ESR1, as observed.
This report is one of the first to examine the role of distal transcription factor-binding sites in the context of gene repression. A previous report identified ENH1 as a site associated with regulation of ESR1 (17). However, in the former study, E2-induced binding of ENH1 was associated with activation of ESR1 in T47D cells. In MCF7 cells, where ESR1 is repressed by E2, similar to the in vivo response, binding of ENH1 is not sufficient for repression. Rather, ERα binding at the proximal site, not ENH1, is associated with a decrease in ESR1 expression, a loss of RNA PolII, Sin3A recruitment, and histone modifications. The possibility also remains that ERα binding at ENH1 regulates another gene. The next closest annotated gene produces a hypothetical protein, C6orf97, which is approximately 70 kb upstream of ENH1. qRT-PCR analysis of E2-treated MCF7 cells did not show any regulation of C6orf97 (data not shown). However, minor (1.8-fold) activation of C6orf97 was observed with E2 treatment of T47D cells (data not shown). This further supports a dominant role for the proximal ERα-binding site, not ENH1, in specific regulation of ESR1 in response to E2.
Another interesting finding was the presence of coactivators, p300 and AIB1, at a gene that is repressed. When experiments conducted with AIB1 siRNA and MCF7 cells were extended to a longer E2 treatment period of 8 h, repression of nascent ESR1 was increased in the absence of AIB1 (data not shown). This suggests that the negative effects of Sin3A may lead to a decreased transcriptional output of ESR1, but the activating factors may help in maintaining a certain level of ESR1 in the presence of E2 (Fig. 1). We have previously shown that synthesis of ESR1 is a major component in establishing steady-state levels of ERα under chronic E2 treatment (63). Physiologically, ESR1 transcript levels change in response to fluctuations in hormonal status, and a mechanism that incorporates balancing positive (p300 and AIB1) and negative (Sin3A) factors provides a plausible model to achieve such dynamic regulation.
Necessity of elements near transcriptional start sites for repression.
Many groups have reported genome-wide analyses of ERα-binding sites (10, 31, 32). These studies find that the minority of ERα-binding sites are located in the proximal promoters, while most binding sites are located more than 5 kb from the transcriptional start site. Work here shows that in ESR1, the proximal, not distal, ERα-binding site is associated with gene repression. This was a surprising finding given the importance of distal sites in models of E2 activation (9, 44, 61). However, review of the limited reports on E2 repression of genes shows that the elements necessary for repression reside at or near the transcription start site (2, 22, 25, 43, 59, 60, 64, 71). We noted greater variability in ERα occupancy at ESR1 compared to activated pS2. This brings up the possibility that ERα binding at repressed genes is weaker and thus may not meet the stringent binding requirements of ChIP-chip bioinformatic analysis.
The importance of repressive elements near the transcription start site is widespread in transcriptional biology. The human immunodeficiency virus type 1 Tat protein interacts with host transcription factors in macrophages to repress the expression of mannose receptor and bone morphogenetic protein receptor 2 via elements near −42 and −206, respectively (8). B-lymphocyte host factor ZEB1 represses the Epstein-Barr virus gene BZLF1 via a repressive element at −17 (72). The protein Egr2, required for peripheral nerve myelination by Schwann cells, represses the Rad gene through repressive elements at −195 to −110 (35). Also related to development, postnatal repression of fetal liver α-fetoprotein involves repression by the zinc finger protein ZBTB20 at −167 to +27 of the AFP gene (68). In cancer, an important trigger of epithelial-mesenchymal transition is repression of E-cadherin by Snail repressor, acting in concert with polycomb complex 2 at −178 to +92 on the E-cadherin promoter (24). In summary, repressive elements near the proximal promoter are significant not only in E2-mediated repression but also in several other transcription factors from viral infection to development to cancer progression.
Translating the language of chromatin into gene expression.
The findings reported here show Sin3A recruitment and multiple histone modifications with ESR1 repression, and it is likely that a certain sequence of marks results in repression. Since Sin3A is a scaffolding protein with many protein-protein interactions, it is possible that it is the platform upon which other enzymatic proteins form (57). The hypothesis of a “histone code” has been proposed and studied in several systems, suggesting that certain sets of modifications on histone tails lead to distinct readouts (27). The histone code hypothesis has more recently been expanded to a “chromatin language.” This encompasses recent findings that certain histone modifications can have more than one role. For example, methylation of H3K4 is capable of recruiting both activators and repressors (4). There have also been reports showing that certain histone modifications influence sequential changes in nearby residues. As examples, methylation of H3K9 interferes with phosphorylation of H3S10, and H3K14 deacetylation is needed to achieve methylation of H3K9 (39, 48). Taken together, these data support the idea that all of the changes in histone modifications observed near the proximal promoter of ESR1 function together to achieve repression.
Specifically, our study identified deacetylation of H3K14, acetylation of H3K18, and trimethylation of H4K20 on ESR1 with exposure to E2 in MCF7 cells. The most striking histone modification observed at ESR1 was the induction of H4K20 trimethylation. This modification has been found to be associated with gene repression in genome-wide studies (66). H4K20me3 was also shown to be enriched at sites of heterochromatin (53). The enzymes responsible for trimethylation of H4K20 are SET domain-containing proteins Suv4-20h1 and Suv4-20h2 (53). However, no H4K20 demethylase has been identified yet (69). At the chemical level, studies have found that trimethylation of H4K20 affects both the local nucleosome structure and higher-order chromatin structure. Specifically, trimethylation of H4K20 affected the orientation of neighboring side chains, including H4 His(H)18 and Arg(R)19, and caused nucleosomes to be more compacted (34). Addition of this modification near the promoter of ESR1 with E2 treatment may be critical in compressing the nucleosomes and making them less accessible to certain transcription factors.
The idea that the chromatin environment is associated with repression of ESR1 helps to interpret data previously obtained with ESR1 reporter constructs. Past studies with ESR1 promoter plasmid constructs identified activating elements (11, 62). Attempts in our lab to repeat these data have given variable results, depending on the vector backbone used, but repeatable repression with a transient reporter assay was not observed (data not shown). This supports the notion that repression of ESR1 requires an endogenous chromatin environment that cannot be recapitulated with reporter constructs. The observed activation of the constructs with E2 in previous studies could be explained by the recruitment of p300 and AIB1 identified here by ChIP. However, the noted chromatin modifications would not be predicted to be detected on naked DNA and therefore would not prevail over activation.
Turning down the magnitude of the E2 response through gene repression.
Cells must constantly adjust to changes in environmental stimuli. An example discussed in this report is the repression of ESR1 expression to control the amount of ERα available to transduce estrogenic signals. It is well established that breast cancer cells have higher levels of ERα than normal breast tissue, and ERα-positive cells present in cases of breast cancer are also positive for proliferation markers (12, 56). Increased amounts of ERα may cause a more robust cellular response to E2, leading to undesired proliferation and tumor growth. Therefore, multiple regulatory pathways are invoked to limit the number of receptors and prevent deleterious increases in the magnitude of E2 action and subsequent effects on cellular proliferation. Our study identified Sin3A as a regulator of ESR1 expression. Further analysis of RNA from Sin3A knockout cells showed alterations in the magnitude of E2 activation of multiple genes, including those for PR, c-myc, and PI-9 (data not shown). Based on these data, it appears that Sin3A may prove to be an important regulator of not only ESR1 expression but other components of the E2 signaling pathway and subsequent cellular proliferation and tumor growth.
Transcriptional regulation is an important component of the overall network regulating ERα levels. Transcription of ESR1 is required for continued production of new receptor, but this production must be damped when E2 levels are high. Therefore, maintaining both activators and repressors and utilizing both distal and proximal regulatory elements may be important determinants balancing dual roles of transcription in the control of cellular levels of ERs.
Acknowledgments
We thank Shigeki Miyamoto, David Wassarman, Avtar Roopra, and Wei Xu for critical reading of the manuscript.
This work was supported by U.S. Department of Defense Breast Cancer Research Program grant W81XWH-06-1-0729 (to S.J.E.) and National Institutes of Health grant DK64034 (to E.T.A.).
Footnotes
Published ahead of print on 20 July 2009.
REFERENCES
- 1.Alarid, E. T., N. Bakopoulos, and N. Solodin. 1999. Proteasome-mediated proteolysis of estrogen receptor: a novel component in autologous down-regulation. Mol. Endocrinol. 131522-1534. [DOI] [PubMed] [Google Scholar]
- 2.An, J., R. C. Ribeiro, P. Webb, J. A. Gustafsson, P. J. Kushner, J. D. Baxter, and D. C. Leitman. 1999. Estradiol repression of tumor necrosis factor-alpha transcription requires estrogen receptor activation function-2 and is enhanced by coactivators. Proc. Natl. Acad. Sci. USA 9615161-15166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayer, D. E., Q. A. Lawrence, and R. N. Eisenman. 1995. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell 80767-776. [DOI] [PubMed] [Google Scholar]
- 4.Berger, S. L. 2007. The complex language of chromatin regulation during transcription. Nature 447407-412. [DOI] [PubMed] [Google Scholar]
- 5.Berkenstam, A., H. Glaumann, M. Martin, J. A. Gustafsson, and G. Norstedt. 1989. Hormonal regulation of estrogen receptor messenger ribonucleic acid in T47D and MCF-7 breast cancer cells. Mol. Endocrinol. 322-28. [DOI] [PubMed] [Google Scholar]
- 6.Bretschneider, N., H. Brand, N. Miller, A. J. Lowery, M. J. Kerin, F. Gannon, and S. Denger. 2008. Estrogen induces repression of the breast cancer and salivary gland expression gene in an estrogen receptor-alpha dependent manner. Cancer Res. 68106-114. [DOI] [PubMed] [Google Scholar]
- 7.Brown, M. A., R. J. Sims, P. D. Gottlieb, and P. W. Tucker. 2006. Identification and characterization of Smyd2: a split SET/MYND domain-containing histone H3 lysine 36-specific methyltransferase that interacts with the Sin3 histone deacetylase complex. Mol. Cancer 526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caldwell, R. L., K. B. Lane, and V. L. Shepherd. 2006. HIV-1 Tat interaction with cyclin T1 represses mannose receptor and the bone morphogenetic protein receptor-2 transcription. Arch. Biochem. Biophys. 44927-33. [DOI] [PubMed] [Google Scholar]
- 9.Carroll, J. S., X. S. Liu, A. S. Brodsky, W. Li, C. A. Meyer, A. J. Szary, J. Eeckhoute, W. Shao, E. V. Hestermann, T. R. Geistlinger, E. A. Fox, P. A. Silver, and M. Brown. 2005. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 12233-43. [DOI] [PubMed] [Google Scholar]
- 10.Carroll, J. S., C. A. Meyer, J. Song, W. Li, T. R. Geistlinger, J. Eeckhoute, A. S. Brodsky, E. K. Keeton, K. C. Fertuck, G. F. Hall, Q. Wang, S. Bekiranov, V. Sementchenko, E. A. Fox, P. A. Silver, T. R. Gingeras, X. S. Liu, and M. Brown. 2006. Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 381289-1297. [DOI] [PubMed] [Google Scholar]
- 11.Castles, C. G., S. Oesterreich, R. Hansen, and S. A. Fuqua. 1997. Auto-regulation of the estrogen receptor promoter. J. Steroid Biochem. Mol. Biol. 62155-163. [DOI] [PubMed] [Google Scholar]
- 12.Clarke, R. B., A. Howell, C. S. Potten, and E. Anderson. 1997. Dissociation between steroid receptor expression and cell proliferation in the human breast. Cancer Res. 574987-4991. [PubMed] [Google Scholar]
- 13.Cvoro, A., C. Tzagarakis-Foster, D. Tatomer, S. Paruthiyil, M. S. Fox, and D. C. Leitman. 2006. Distinct roles of unliganded and liganded estrogen receptors in transcriptional repression. Mol. Cell 21555-564. [DOI] [PubMed] [Google Scholar]
- 14.Dauvois, S., P. S. Danielian, R. White, and M. G. Parker. 1992. Antiestrogen ICI 164,384 reduces cellular estrogen receptor content by increasing its turnover. Proc. Natl. Acad. Sci. USA 894037-4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denger, S., G. Reid, M. Kos, G. Flouriot, D. Parsch, H. Brand, K. S. Korach, V. Sonntag-Buck, and F. Gannon. 2001. ERα gene expression in human primary osteoblasts: evidence for the expression of two receptor proteins. Mol. Endocrinol. 152064-2077. [DOI] [PubMed] [Google Scholar]
- 16.Dhasarathy, A., M. Kajita, and P. A. Wade. 2007. The transcription factor snail mediates epithelial to mesenchymal transitions by repression of estrogen receptor-alpha. Mol. Endocrinol. 212907-2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eeckhoute, J., E. K. Keeton, M. Lupien, S. A. Krum, J. S. Carroll, and M. Brown. 2007. Positive cross-regulatory loop ties GATA-3 to estrogen receptor alpha expression in breast cancer. Cancer Res. 676477-6483. [DOI] [PubMed] [Google Scholar]
- 18.Fowler, A. M., N. M. Solodin, C. C. Valley, and E. T. Alarid. 2006. Altered target gene regulation controlled by estrogen receptor-alpha concentration. Mol. Endocrinol. 20291-301. [DOI] [PubMed] [Google Scholar]
- 19.Frasor, J., J. M. Danes, B. Komm, K. C. Chang, C. R. Lyttle, and B. S. Katzenellenbogen. 2003. Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology 1444562-4574. [DOI] [PubMed] [Google Scholar]
- 20.Frech, M. S., E. D. Halama, M. T. Tilli, B. Singh, E. J. Gunther, L. A. Chodosh, J. A. Flaws, and P. A. Furth. 2005. Deregulated estrogen receptor alpha expression in mammary epithelial cells of transgenic mice results in the development of ductal carcinoma in situ. Cancer Res. 65681-685. [PMC free article] [PubMed] [Google Scholar]
- 21.Hanstein, B., R. Eckner, J. DiRenzo, S. Halachmi, H. Liu, B. Searcy, R. Kurokawa, and M. Brown. 1996. p300 is a component of an estrogen receptor coactivator complex. Proc. Natl. Acad. Sci. USA 9311540-11545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hao, H., M. d'Alincourt-Salazar, K. M. Kelley, A. Shatnawi, S. Mukherjee, Y. M. Shah, and M. Ratnam. 2007. Estrogen-induced and TAFII30-mediated gene repression by direct recruitment of the estrogen receptor and co-repressors to the core promoter and its reversal by tamoxifen. Oncogene 267872-7884. [DOI] [PubMed] [Google Scholar]
- 23.Hassig, C. A., T. C. Fleischer, A. N. Billin, S. L. Schreiber, and D. E. Ayer. 1997. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell 89341-347. [DOI] [PubMed] [Google Scholar]
- 24.Herranz, N., D. Pasini, V. M. Diaz, C. Franci, A. Gutierrez, N. Dave, M. Escriva, I. Hernandez-Munoz, L. Di Croce, K. Helin, A. Garcia de Herreros, and S. Peiro. 2008. Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol. Cell. Biol. 284772-4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Higgins, K. J., S. Liu, M. Abdelrahim, K. Vanderlaag, X. Liu, W. Porter, R. Metz, and S. Safe. 2008. Vascular endothelial growth factor receptor-2 expression is down-regulated by 17β-estradiol in MCF-7 breast cancer cells by estrogen receptor alpha/Sp proteins. Mol. Endocrinol. 22388-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holst, F., P. R. Stahl, C. Ruiz, O. Hellwinkel, Z. Jehan, M. Wendland, A. Lebeau, L. Terracciano, K. Al-Kuraya, F. Janicke, G. Sauter, and R. Simon. 2007. Estrogen receptor alpha (ESR1) gene amplification is frequent in breast cancer. Nat. Genet. 39655-660. [DOI] [PubMed] [Google Scholar]
- 27.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 2931074-1080. [DOI] [PubMed] [Google Scholar]
- 28.Jiang, X., S. J. Ellison, E. T. Alarid, and D. J. Shapiro. 2007. Interplay between the levels of estrogen and estrogen receptor controls the level of the granzyme inhibitor, proteinase inhibitor 9 and susceptibility to immune surveillance by natural killer cells. Oncogene 264106-4114. [DOI] [PubMed] [Google Scholar]
- 29.Kininis, M., B. S. Chen, A. G. Diehl, G. D. Isaacs, T. Zhang, A. C. Siepel, A. G. Clark, and W. L. Kraus. 2007. Genomic analyses of transcription factor binding, histone acetylation, and gene expression reveal mechanistically distinct classes of estrogen-regulated promoters. Mol. Cell. Biol. 275090-5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kos, M., G. Reid, S. Denger, and F. Gannon. 2001. Minireview: genomic organization of the human ERα gene promoter region. Mol. Endocrinol. 152057-2063. [DOI] [PubMed] [Google Scholar]
- 31.Levy, N., D. Tatomer, C. B. Herber, X. Zhao, H. Tang, T. Sargeant, L. J. Ball, J. Summers, T. P. Speed, and D. C. Leitman. 2008. Differential regulation of native estrogen receptor regulatory elements by estradiol, tamoxifen, and raloxifene. Mol. Endocrinol. 22287-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin, C.-Y., V. B. Vega, J. S. Thomsen, T. Zhang, S. L. Kong, M. Xie, K. P. Chiu, L. Lipovich, D. H. Barnett, F. Stossi, A. Yeo, J. George, V. A. Kuznetsov, Y. K. Lee, T. H. Charn, N. Palanisamy, L. D. Miller, E. Cheung, B. S. Katzenellenbogen, Y. Ruan, G. Bourque, C.-L. Wei, and E. T. Liu. 2007. Whole-genome cartography of estrogen receptor-alpha binding sites. PLoS Genet. 3e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.List, H.-J., K. J. Lauritsen, R. Reiter, C. Powers, A. Wellstein, and A. T. Riegel. 2001. Ribozyme targeting demonstrates that the nuclear receptor coactivator AIB1 is a rate-limiting factor for estrogen-dependent growth of human MCF-7 breast cancer cells. J. Biol. Chem. 27623763-23768. [DOI] [PubMed] [Google Scholar]
- 34.Lu, X., M. D. Simon, J. V. Chodaparambil, J. C. Hansen, K. M. Shokat, and K. Luger. 2008. The effect of H3K79 dimethylation and H4K20 trimethylation on nucleosome and chromatin structure. Nat. Struct. Mol. Biol. 151122-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mager, G. M., R. M. Ward, R. Srinivasan, S. W. Jang, L. Wrabetz, and J. Svaren. 2008. Active gene repression by the Egr2.NAB complex during peripheral nerve myelination. J. Biol. Chem. 28318187-18197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Markopoulos, C., U. Berger, P. Wilson, J. C. Gazet, and R. C. Coombes. 1988. Oestrogen receptor content of normal breast cells and breast carcinomas throughout the menstrual cycle. Br. Med. J. (Clin. Res. Ed.) 2961349-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Métivier, R., G. Penot, M. R. Hubner, G. Reid, H. Brand, M. Kos, and F. Gannon. 2003. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115751-763. [DOI] [PubMed] [Google Scholar]
- 38.Murphy, M., J. Ahn, K. K. Walker, W. H. Hoffman, R. M. Evans, A. J. Levine, and D. L. George. 1999. Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3a. Genes Dev. 132490-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakayama, J., J. C. Rice, B. D. Strahl, C. D. Allis, and S. I. Grewal. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292110-113. [DOI] [PubMed] [Google Scholar]
- 40.Nan, X., H. H. Ng, C. A. Johnson, C. D. Laherty, B. M. Turner, R. N. Eisenman, and A. Bird. 1998. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393386-389. [DOI] [PubMed] [Google Scholar]
- 41.Nasmyth, K., D. Stillman, and D. Kipling. 1987. Both positive and negative regulators of HO transcription are required for mother-cell-specific mating-type switching in yeast. Cell 48579-587. [DOI] [PubMed] [Google Scholar]
- 42.Nawaz, Z., D. M. Lonard, A. P. Dennis, C. L. Smith, and B. W. O'Malley. 1999. Proteasome-dependent degradation of the human estrogen receptor. Proc. Natl. Acad. Sci. USA 961858-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oesterreich, S., W. Deng, S. Jiang, X. Cui, M. Ivanova, R. Schiff, K. Kang, D. L. Hadsell, J. Behrens, and A. V. Lee. 2003. Estrogen-mediated down-regulation of E-cadherin in breast cancer cells. Cancer Res. 635203-5208. [PubMed] [Google Scholar]
- 44.Pan, Y. F., K. D. S. A. Wansa, M. H. Liu, B. Zhao, S. Z. Hong, P. Y. Tan, K. S. Lim, G. Bourque, E. T. Liu, and E. Cheung. 2008. Regulation of estrogen receptor-mediated long range transcription via evolutionarily conserved distal response elements. J. Biol. Chem. 28332977-32988. [DOI] [PubMed] [Google Scholar]
- 45.Peterson, C. L., and M.-A. Laniel. 2004. Histones and histone modifications. Curr. Biol. 14R546-R551. [DOI] [PubMed] [Google Scholar]
- 46.Press, M. F., N. Nousek-Goebl, W. J. King, A. L. Herbst, and G. L. Greene. 1984. Immunohistochemical assessment of estrogen receptor distribution in the human endometrium throughout the menstrual cycle. Lab. Investig. 51495-503. [PubMed] [Google Scholar]
- 47.Ramaswamy, B., S. Majumder, S. Roy, K. Ghoshal, H. Kutay, J. Datta, M. Younes, C. L. Shapiro, T. Motiwala, and S. T. Jacob. 2009. Estrogen-mediated suppression of the gene encoding protein tyrosine phosphatase PTPRO in human breast cancer: mechanism and role in tamoxifen sensitivity. Mol. Endocrinol. 23176-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rea, S., F. Eisenhaber, D. O'Carroll, B. D. Strahl, Z. W. Sun, M. Schmid, S. Opravil, K. Mechtler, C. P. Ponting, C. D. Allis, and T. Jenuwein. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406593-599. [DOI] [PubMed] [Google Scholar]
- 49.Read, L. D., G. L. Greene, and B. S. Katzenellenbogen. 1989. Regulation of estrogen receptor messenger ribonucleic acid and protein levels in human breast cancer cell lines by sex steroid hormones, their antagonists, and growth factors. Mol. Endocrinol. 3295-304. [DOI] [PubMed] [Google Scholar]
- 50.Reid, G., S. Denger, M. Kos, and F. Gannon. 2002. Human estrogen receptor-alpha: regulation by synthesis, modification and degradation. Cell. Mol. Life Sci. 59821-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roopra, A., L. Sharling, I. C. Wood, T. Briggs, U. Bachfischer, A. J. Paquette, and N. J. Buckley. 2000. Transcriptional repression by neuron-restrictive silencer factor is mediated via the Sin3-histone deacetylase complex. Mol. Cell. Biol. 202147-2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saceda, M., M. E. Lippman, P. Chambon, R. L. Lindsey, M. Ponglikitmongkol, M. Puente, and M. B. Martin. 1988. Regulation of the estrogen receptor in MCF-7 cells by estradiol. Mol. Endocrinol. 21157-1162. [DOI] [PubMed] [Google Scholar]
- 53.Schotta, G., M. Lachner, K. Sarma, A. Ebert, R. Sengupta, G. Reuter, D. Reinberg, and T. Jenuwein. 2004. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 181251-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sharma, D., J. Blum, X. Yang, N. Beaulieu, A. R. Macleod, and N. E. Davidson. 2005. Release of methyl CpG binding proteins and histone deacetylase 1 from the estrogen receptor alpha (ER) promoter upon reactivation in ER-negative human breast cancer cells. Mol. Endocrinol. 191740-1751. [DOI] [PubMed] [Google Scholar]
- 55.Shiau, A. K., D. Barstad, P. M. Loria, L. Cheng, P. J. Kushner, D. A. Agard, and G. L. Greene. 1998. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell 95927-937. [DOI] [PubMed] [Google Scholar]
- 56.Shoker, B. S., C. Jarvis, R. B. Clarke, E. Anderson, J. Hewlett, M. P. Davies, D. R. Sibson, and J. P. Sloane. 1999. Estrogen receptor-positive proliferating cells in the normal and precancerous breast. Am. J. Pathol. 1551811-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Silverstein, R. A., and K. Ekwall. 2005. Sin3: a flexible regulator of global gene expression and genome stability. Curr. Genet. 471-17. [DOI] [PubMed] [Google Scholar]
- 58.Sternberg, P. W., M. J. Stern, I. Clark, and I. Herskowitz. 1987. Activation of the yeast HO gene by release from multiple negative controls. Cell 48567-577. [DOI] [PubMed] [Google Scholar]
- 59.Stoner, M., F. Wang, M. Wormke, T. Nguyen, I. Samudio, C. Vyhlidal, D. Marme, G. Finkenzeller, and S. Safe. 2000. Inhibition of vascular endothelial growth factor expression in HEC1A endometrial cancer cells through interactions of estrogen receptor alpha and Sp3 proteins. J. Biol. Chem. 27522769-22779. [DOI] [PubMed] [Google Scholar]
- 60.Stossi, F., V. S. Likhite, J. A. Katzenellenbogen, and B. S. Katzenellenbogen. 2006. Estrogen-occupied estrogen receptor represses cyclin G2 gene expression and recruits a repressor complex at the cyclin G2 promoter. J. Biol. Chem. 28116272-16278. [DOI] [PubMed] [Google Scholar]
- 61.Sun, J., Z. Nawaz, and J. M. Slingerland. 2007. Long-range activation of GREB1 by estrogen receptor via three distal consensus estrogen-responsive elements in breast cancer cells. Mol. Endocrinol. 212651-2662. [DOI] [PubMed] [Google Scholar]
- 62.Treilleux, I., N. Peloux, M. Brown, and A. Sergeant. 1997. Human estrogen receptor (ER) gene promoter-P1: estradiol-independent activity and estradiol inducibility in ER+ and ER− cells. Mol. Endocrinol. 111319-1331. [DOI] [PubMed] [Google Scholar]
- 63.Valley, C. C., N. M. Solodin, G. L. Powers, S. J. Ellison, and E. T. Alarid. 2008. Temporal variation in estrogen receptor-alpha protein turnover in the presence of estrogen. J. Mol. Endocrinol. 4023-34. [DOI] [PubMed] [Google Scholar]
- 64.Varshochi, R., F. Halim, A. Sunters, J. P. Alao, P. A. Madureira, S. M. Hart, S. Ali, D. M. Vigushin, R. C. Coombes, and E. W. Lam. 2005. ICI182,780 induces p21Waf1 gene transcription through releasing histone deacetylase 1 and estrogen receptor alpha from Sp1 sites to induce cell cycle arrest in MCF-7 breast cancer cell line. J. Biol. Chem. 2803185-3196. [DOI] [PubMed] [Google Scholar]
- 65.Velarde, M. C., Z. Zeng, J. R. McQuown, F. A. Simmen, and R. C. M. Simmen. 2007. Kruppel-like factor 9 is a negative regulator of ligand-dependent estrogen receptor-alpha signaling in Ishikawa endometrial adenocarcinoma cells. Mol. Endocrinol. 212988-3001. [DOI] [PubMed] [Google Scholar]
- 66.Wang, Z., C. Zang, J. A. Rosenfeld, D. E. Schones, A. Barski, S. Cuddapah, K. Cui, T. Y. Roh, W. Peng, M. Q. Zhang, and K. Zhao. 2008. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 40897-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wysocka, J., M. P. Myers, C. D. Laherty, R. N. Eisenman, and W. Herr. 2003. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev. 17896-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xie, Z., H. Zhang, W. Tsai, Y. Zhang, Y. Du, J. Zhong, C. Szpirer, M. Zhu, X. Cao, M. C. Barton, M. J. Grusby, and W. J. Zhang. 2008. Zinc finger protein ZBTB20 is a key repressor of alpha-fetoprotein gene transcription in liver. Proc. Natl. Acad. Sci. USA 10510859-10864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang, H., and C. A. Mizzen. 2009. The multiple facets of histone H4-lysine 20 methylation. Biochem. Cell Biol. 87151-161. [DOI] [PubMed] [Google Scholar]
- 70.Yang, L., Q. Mei, A. Zielinska-Kwiatkowska, Y. Matsui, M. L. Blackburn, D. Benedetti, A. A. Krumm, G. J. Taborsky, Jr., and H. A. Chansky. 2003. An ERG (ets-related gene)-associated histone methyltransferase interacts with histone deacetylases 1/2 and transcription co-repressors mSin3A/B. Biochem. J. 369651-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ye, Y., Y. Xiao, W. Wang, K. Yearsley, J. X. Gao, and S. H. Barsky. 2008. ERα suppresses slug expression directly by transcriptional repression. Biochem. J. 416179-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu, X., Z. Wang, and J. E. Mertz. 2007. ZEB1 regulates the latent-lytic switch in infection by Epstein-Barr virus. PLoS Pathog. 3e194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang, Y., R. Iratni, H. Erdjument-Bromage, P. Tempst, and D. Reinberg. 1997. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell 89357-364. [DOI] [PubMed] [Google Scholar]
- 74.Zhu, P., S. H. Baek, E. M. Bourk, K. A. Ohgi, I. Garcia-Bassets, H. Sanjo, S. Akira, P. F. Kotol, C. K. Glass, M. G. Rosenfeld, and D. W. Rose. 2006. Macrophage/cancer cell interactions mediate hormone resistance by a nuclear receptor derepression pathway. Cell 124615-629. [DOI] [PubMed] [Google Scholar]