Abstract
Purpose
The phosphoinositide-3-kinase (PI3K), phosphatase and tensin homolog (PTEN), v-akt murine thymoma viral oncogene homolog (AKT), and mammalian target of rapamycin (mTOR) signaling pathway has been implicated in resistance to several chemotherapeutic agents. In this retrospective study, we determined whether common genetic variations in this pathway are associated with clinical outcomes in esophageal cancer patients with adenocarcinoma or squamous cell carcinoma who have undergone chemoradiotherapy and surgery.
Patients and Methods
Sixteen tagging single nucleotide polymorphisms (SNPs) in PIK3CA, PTEN, AKT1, AKT2, and FRAP1 (encoding mTOR) were genotyped in these patients and analyzed for associations with response to therapy, survival, and recurrence.
Results
We observed an increased recurrence risk with genetic variations in AKT1 and AKT2 (hazard ratio [HR], 2.21; 95% CI, 1.06 to 4.60; and HR, 3.30; 95% CI, 1.64 to 6.66, respectively). This effect was magnified with an increasing number of AKT adverse genotypes. In contrast, a predictable protective effect by PTEN genetic variants on recurrence was evident. Survival tree analysis identified higher-order interactions that resulted in variation in recurrence-free survival from 12 to 42 months, depending on the combination of SNPs. Genetic variations in AKT1, AKT2, and FRAP1 were associated with survival. Patients homozygous for either of the FRAP1 SNPs assayed had a more than three-fold increased risk of death. Two genes—AKT2 and FRAP1—were associated with a poor treatment response, while a better response was associated with heterozygosity for AKT1:rs3803304 (odds ratio, 0.50; 95% CI, 0.25 to 0.99).
Conclusion
These results suggest that common genetic variations in this pathway modulate clinical outcomes in patients who undergo chemoradiotherapy. With further validation, these results may be used to build a model of individualized therapy for the selection of the optimal chemotherapeutic regimen.
INTRODUCTION
An estimated 16,400 new cases of esophageal cancer (EC) will be diagnosed in 2008.1 Surgery is one of the standard treatments for patients with resectable tumors but frequently preoperative chemoradiotherapy is used to treat both adenocarcinoma and squamous cell carcinoma ECs.2–5 The most commonly utilized chemotherapy agents belong to fluoropyrimidines, taxanes, and platinum compounds. Unfortunately, even with the multimodal approach, current treatments result in a poor overall 5-year survival rate of 25% to 28%.6–9
Heterogeneity in response to chemoradiotherapy may be due to several factors, including age, sex, ethnicity, and drug-drug interactions. In addition, genetic variations in pharmacokinetic, pharmacodynamic, and drug action pathways have been shown to be important in determining sensitivity or resistance to treatment.10 Therefore, one strategy to increase the effectiveness of chemoradiotherapy is to gain a better understanding of the influence a patient's genetic background has on response to treatment. Our group has previously reported that genetic variations in several drug action pathways were associated with variation in clinical outcomes in EC.11 In this study, we expand those results by analyzing an important signaling pathway comprised of phosphoinositide-3-kinase (PI3K), phosphatase and tensin homolog (PTEN), v-akt murine thymoma viral oncogene homolog (AKT), and mammalian target of rapamycin (mTOR).
Signaling through the PI3K/PTEN/AKT/mTOR pathway is responsible for balancing cell survival and apoptosis.12,13 The signal is initiated by growth factors and hormones that bind receptor tyrosine kinases such as epidermal growth factor receptor (EGFR), vascular endothelial growth factor receptor (VEGFR), and platelet-derived growth factor receptor (PDGFR).14 These receptors then activate PI3Ks resulting in a kinase cascade through AKT and mTOR, generating cell survival, growth, and angiogenesis signals.15 PTEN negatively regulates this pathway by dephosphorylating phosphatidylinositol trisphosphate (PIP3) and negating the signal generated by PI3K.16 This pathway has been shown to be commonly activated in cancer, including EC, and in the progression of Barrett's neoplasm to EC.12,17,18 Furthermore, studies in several cancer types have demonstrated that this pathway has a key role in the development of resistance to platinum compounds, taxanes, and fluoropyrimidines.19–24 To our knowledge, no studies have addressed how genetic variations in this pathway influence outcomes in patients with EC treated with these chemotherapeutic agents.
In this study, we determined whether common genetic variations in AKT1, AKT2, PIK3CA (catalytic subunit of PI3K), PTEN, and FRAP1 (mTOR) were associated with clinical outcomes in patients who received chemoradiotherapy. Tagged single nucleotide polymorphisms (SNPs) were selected for each gene and genotyped in patients with EC. This pathway-based tagging approach allowed us to query genetic variations in the major effectors of this pathway and identify associations with clinical outcomes.
PATIENTS AND METHODS
Patient Population
This study included 210 patients with resectable adenocarcinoma (174 cases) or squamous cell carcinoma (36 cases) who were recruited between 1985 and 2003 at The University of Texas M. D. Anderson Cancer Center (Houston, TX).11 All patients had undergone chemoradiotherapy followed by surgery, or induction chemotherapy followed by chemoradiotherapy and surgery.
Clinical Data Collection
Patients with EC were staged as described previously.11 After chemoradiotherapy, patients underwent restaging and surgery. Pathologic response to treatment was measured by previously described methodology.25,26 Response was defined as no residual carcinoma in the primary tumor site. A poor response was any response less than a complete response. Study end points were pathologic response to therapy, recurrence, and survival. This study was approved by the M. D. Anderson Cancer Center institutional review board.
SNP Selection and Genotyping
Genomic DNA was extracted from paraffin slides using the PicoPure DNA extraction kit (Arcturus Bioscience, Mountain View, CA). Tagging SNPs were selected within and 5-kb flanking each gene using the tagger algorithm28 with a cutoff of 0.80 for r2 and a minor allele frequency between 0.10 and 0.35 based on data from Centre d'Etude du Polymorphisme Humain samples genotyped by the HapMap Project (www.hapmap.org).27 A total of sixteen SNPs were selected to represent genetic variation of 95 SNPs in the pathway (Appendix Table A1, online only). TaqMan genotyping assays, including quality control measures, were performed as previously described11 using the 7900HT sequence detection system (Applied Biosystems, Foster City, CA).
Statistical Analysis
Hazard ratios (HRs) for recurrence and survival end points were estimated by applying the Cox proportional hazards model while adjusting for age, sex, smoking status, alcohol consumption, radiation dosage, chemoradiotherapy sequence, clinical stage, chemotherapy regimens, histologic tumor type, tumor location, pathologic stage, and histologic viability. The Kaplan-Meier survival function and log-rank tests were used to assess differences in recurrence-free and overall survival times. For pathologic response to therapy, unconditional multivariate logistic regression analysis was done to estimate adjusted odds ratios (ORs) along with the corresponding 95% CIs for each SNP. We also evaluated the combined effects by the number of unfavorable genotypes identified from the main effects analysis of single SNPs. The statistical analyses described above were completed using the STATA software (version 8, STATA, College Station, TX). Survival tree analyses were used to identify higher-order gene-gene interactions. Survival tree analysis was performed using the STREE program (http://masal.med.yale.edu/stree/) which uses recursive-partitioning to identify subgroups of individuals at higher risk. All statistical analyses were two sided, and P < .05 was considered statistically significant.
RESULTS
Patient Characteristics
Of the 210 patients with EC enrolled in this study, DNA was available for 207 and of those, 186 were white (90%; Appendix Table A2, online only). Because of the small number of patients from other ethnic groups, we focused our study on white patients only. Fifty-nine percent of patients were ever smokers, and 33.1% used alcohol on a daily basis. Approximately half of patients (51%) presented with stage IIA disease. More than 97% of the patients (182) were treated with either a fluoropyrimidine, platinum agent, or taxane. Of these 182 patients, 177 received a fluoropyrimidine (95%), 132 received a platinum agent (71%), and 94 received a taxane (51%). The median follow-up time was 18.8 months, with 97 deaths and 59 recurrences. The overall median survival time was 34.5 months.
Associations Between SNPs and Recurrence Risk
Variant genotypes were analyzed for association with recurrence risk in patients after chemoradiotherapy. Three SNPs—AKT1:rs2498804, AKT2:rs892119, and PTEN:rs12357281—were associated with variation in recurrence risk (Table 1).
Table 1.
PI3K/PTEN/AKT/mTOR Pathway Genotypes and Recurrence
| SNP and Genotype | Any of the Three |
Fluoropyrimidine + Any |
||||||
|---|---|---|---|---|---|---|---|---|
| No Recurrence/Recurrence (No.) | HR* | 95% CI | P | No Recurrence/Recurrence (No.) | HR* | 95% CI | P | |
| AKT1:rs3803304 | ||||||||
| CC | 60/26 | 1 (reference) | 57/26 | 1 (reference) | ||||
| CG | 54/25 | 1.44 | 0.75 to 2.78 | .276 | 53/24 | 1.38 | 0.71 to 2.71 | .341 |
| GG | 7/5 | 1.87 | 0.59 to 5.91 | .289 | 7/5 | 1.72 | 0.54 to 5.54 | .361 |
| CG + GG | 1.51 | 0.81 to 2.81 | .200 | 1.43 | 0.76 to 2.72 | .269 | ||
| AKT1:rs2498804 | ||||||||
| GG | 50/20 | 1 (reference) | 47/20 | 1 (reference) | ||||
| GT | 63/30 | 2.04 | 0.95 to 4.37 | .068 | 62/29 | 1.99 | 0.92 to 4.29 | .080 |
| TT | 8/7 | 3.21 | 1.07 to 9.61 | .037 | 8/7 | 3.06 | 1.01 to 9.28 | .049 |
| GT + TT | 2.21 | 1.06 to 4.60 | .034 | 2.14 | 1.02 to 4.50 | .045 | ||
| AKT1:rs2494738 | ||||||||
| AA | 105/49 | 1 (reference) | 101/48 | 1 (reference) | ||||
| AG | 18/7 | 0.94 | 0.36 to 2.44 | .900 | 18/7 | 0.96 | 0.37 to 2.49 | .936 |
| AKT1:rs1130214 | ||||||||
| GG | 60/28 | 1 (reference) | 57/27 | 1 (reference) | ||||
| GT | 56/28 | 1.91 | 0.94 to 3.87 | .073 | 55/28 | 1.91 | 0.94 to 3.87 | .072 |
| TT | 7/2 | 1.83 | 0.34 to 9.38 | .482 | 7/2 | 1.82 | 0.34 to 9.80 | .488 |
| GT + TT | 1.90 | 0.94 to 3.85 | .073 | 1.91 | 0.94 to 3.86 | .072 | ||
| AKT2:rs892119 | ||||||||
| AA | 95/33 | 1 (reference) | 94/32 | 1 (reference) | ||||
| AG | 25/23 | 3.48 | 1.66 to 7.28 | .001 | 23/23 | 3.72 | 1.76 to 7.84 | .001 |
| GG | 3/2 | 2.42 | 0.49 to 12.01 | .280 | 2/2 | 2.57 | 0.51 to 12.88 | .251 |
| AG + GG | 3.30 | 1.64 to 6.66 | .001 | 3.52 | 1.73 to 7.17 | .001 | ||
| AKT2:rs8100018 | ||||||||
| CC | 61/24 | 1 (reference) | 59/23 | 1 (reference) | ||||
| CG | 47/30 | 1.26 | 0.64 to 2.46 | .505 | 45/30 | 1.32 | 0.67 to 2.60 | .430 |
| GG | 13/3 | 0.41 | 0.10 to 1.58 | .193 | 13/3 | 0.44 | 0.11 to 1.74 | .242 |
| CG + GG | 1.02 | 0.54 to 1.94 | .944 | 1.09 | 0.57 to 2.09 | .799 | ||
| FRAP1:rs11121704 | ||||||||
| CC | 57/31 | 1 (reference) | 54/30 | 1 (reference) | ||||
| CT | 55/23 | 0.90 | 0.43 to 1.88 | .783 | 54/23 | 0.94 | 0.48 to 1.96 | .866 |
| TT | 11/3 | 1.79 | 0.45 to 7.09 | .409 | 11/3 | 1.79 | 0.45 to 7.06 | .404 |
| CT + TT | 0.97 | 0.48 to 1.99 | .938 | 1.01 | 0.50 to 2.07 | .974 | ||
| FRAP1:rs2295080 | ||||||||
| GG | 47/21 | 1 (reference) | 44/20 | 1 (reference) | ||||
| GT | 56/29 | 1.81 | 0.81 to 4.05 | .149 | 55/29 | 1.86 | 0.83 to 4.16 | .132 |
| TT | 14/4 | 3.04 | 0.82 to 11.27 | .096 | 14/4 | 3.00 | 0.82 to 11.04 | .098 |
| GT + TT | 1.92 | 0.87 to 4.21 | .105 | 2.00 | 0.61 to 6.48 | .250 | ||
| PIK3CA:rs7651265 | ||||||||
| AA | 95/40 | 1 (reference) | 91/40 | 1 (reference) | ||||
| AG | 23/15 | 1.54 | 0.74 to 3.19 | .248 | 23/14 | 1.47 | 0.70 to 3.08 | .312 |
| GG | 2/0 | 2/0 | ||||||
| AG + GG | 1.43 | 0.69 to 2.95 | .332 | 1.36 | 0.65 to 2.85 | .409 | ||
| PIK3CA:rs7640662 | ||||||||
| CC | 95/45 | 1 (reference) | 93/44 | 1 (reference) | ||||
| CG | 27/11 | 1.02 | 0.46 to 2.25 | .970 | 25/11 | 1.06 | 0.48 to 2.34 | .881 |
| GG | 2/1 | 1.26 | 0.14 to 11.76 | .838 | 2/1 | 1.22 | 0.13 to 11.45 | .864 |
| CG + GG | 1.04 | 0.48 to 2.21 | .927 | 1.08 | 0.51 to 2.29 | .849 | ||
| PIK3CA:rs7621329 | ||||||||
| CC | 80/32 | 1 (reference) | 77/35 | 1 (reference) | ||||
| CT | 31/16 | 0.83 | 0.40 to 1.71 | .620 | 30/15 | 0.84 | 0.41 to 1.74 | .643 |
| TT | 6/3 | 1.76 | 0.42 to 7.38 | .442 | 6/3 | 1.70 | 0.41 to 7.12 | .467 |
| CT + TT | 0.92 | 0.47 to 1.83 | .822 | 0.93 | 0.47 to 1.84 | .838 | ||
| PIK3CA:rs6443624 | ||||||||
| AA | 76/32 | 1 (reference) | 73/32 | 1 (reference) | ||||
| AC | 36/22 | 1.40 | 0.69 to 2.85 | .348 | 35/21 | 1.39 | 0.68 to 2.82 | .365 |
| CC | 10/4 | 0.95 | 0.28 to 3.16 | .929 | 10/4 | 0.94 | 0.29 to 3.13 | .926 |
| AC + CC | 1.30 | 0.66 to 2.57 | .444 | 1.29 | 0.65 to 2.54 | .461 | ||
| SNP and Genotype | Platinum Compound + Any |
Taxane + Any |
||||||
|---|---|---|---|---|---|---|---|---|
| No Recurrence/Recurrence (No.) | HR* | 95% CI | P | No Recurrence/Recurrence (No.) | HR* | 95% CI | P | |
| AKT1:rs3803304 | ||||||||
| CC | 43/20 | 1 (reference) | 28/12 | 1 (reference) | ||||
| CG | 37/21 | 2.21 | 0.95 to 5.12 | .065 | 31/13 | 2.24 | 0.75 to 6.72 | .149 |
| GG | 5/3 | 1.62 | 0.37 to 7.15 | .526 | 4/3 | 4.75 | 0.54 to 42.06 | .162 |
| CG + GG | 2.08 | 0.94 to 4.60 | .070 | 2.43 | 0.83 to 7.14 | .107 | ||
| AKT1:rs2498804 | ||||||||
| GG | 32/16 | 1 (reference) | 26/7 | 1 (reference) | ||||
| GT | 48/23 | 2.60 | 1.00 to 6.75 | .049 | 31/17 | 11.95 | 1.89 to 75.41 | .008 |
| TT | 5/5 | 3.55 | 0.97 to 13.02 | .056 | 5/5 | 21.96 | 2.68 to 179.88 | .004 |
| GT + TT | 2.79 | 1.13 to 6.89 | .026 | 14.10 | 2.40 to 83.02 | .003 | ||
| AKT1:rs2494738 | ||||||||
| AA | 71/39 | 1 (reference) | 57/24 | 1 (reference) | ||||
| AG | 15/5 | 0.95 | 0.31 to 2.86 | .921 | 8/4 | 1.33 | 0.23 to 7.63 | .751 |
| AKT1:rs1130214 | ||||||||
| GG | 45/23 | 1 (reference) | 31/15 | 1 (reference) | ||||
| GT | 37/21 | 2.08 | 0.87 to 4.98 | .099 | 30/12 | 2.39 | 0.84 to 6.79 | .102 |
| TT | 4/1 | 1.34 | 0.11 to 15.84 | .817 | 4/2 | 10.94 | 1.55 to 76.98 | .016 |
| GT + TT | 2.06 | 0.86 to 4.94 | .104 | 2.72 | 0.97 to 7.62 | .056 | ||
| AKT2:rs892119 | ||||||||
| AA | 68/25 | 1 (reference) | 49/16 | 1 (reference) | ||||
| AG | 16/18 | 7.36 | 2.79 to 19.42 | .000056 | 14/12 | 5.77 | 1.70 to 19.52 | .005 |
| GG | 2/2 | 2.98 | 0.54 to 16.51 | .211 | 2/1 | 2.56 | 0.16 to 40.00 | .503 |
| AG + GG | 6.20 | 2.45 to 15.69 | .00012 | 5.23 | 1.62 to 16.88 | .006 | ||
| AKT2:rs8100018 | ||||||||
| CC | 41/18 | 1 (reference) | 35/14 | 1 (reference) | ||||
| CG | 36/27 | 1.27 | 0.58 to 2.80 | .554 | 23/12 | 1.35 | 0.44 to 4.17 | .603 |
| GG | 7/0 | 7/3 | 0.33 | 0.05 to 2.29 | .262 | |||
| CG + GG | 0.91 | 0.42 to 1.99 | .822 | 0.97 | 0.34 to 2.81 | .956 | ||
| FRAP1:rs11121704 | ||||||||
| CC | 43/26 | 1 (reference) | 30/15 | 1 (reference) | ||||
| CT | 36/17 | 1.17 | 0.50 to 2.72 | .718 | 30/12 | 0.47 | 0.12 to 1.90 | .289 |
| TT | 7/2 | 2.56 | 0.40 to 16.30 | .320 | 5/2 | 2.33 | 0.26 to 20.89 | .450 |
| CT + TT | 1.29 | 0.58 to 2.86 | .526 | 0.52 | 0.13 to 2.04 | .346 | ||
| FRAP1:rs2295080 | ||||||||
| GG | 35/18 | 1 (reference) | 23/10 | 1 (reference) | ||||
| GT | 35/22 | 2.35 | 0.88 to 6.23 | .087 | 36/15 | 1.31 | 0.28 to 6.19 | .730 |
| TT | 10/2 | 2.35 | 0.37 to 15.06 | .367 | 5/3 | 12.35 | 1.19 to 128.38 | .035 |
| GT + TT | 2.35 | 0.91 to 6.07 | .079 | 1.43 | 0.30 to 6.82 | .650 | ||
| PIK3CA:rs7651265 | ||||||||
| AA | 68/30 | 1 (reference) | 51/20 | 1 (reference) | ||||
| AG | 14/13 | 2.13 | 0.81 to 5.58 | .123 | 12/7 | 3.13 | 0.79 to 12.35 | .103 |
| GG | 2/0 | 0/0 | ||||||
| AG + GG | 1.75 | 0.70 to 4.38 | .232 | |||||
| PIK3CA:rs7640662 | ||||||||
| CC | 65/34 | 1 (reference) | 51/24 | 1 (reference) | ||||
| CG | 20/10 | 1.24 | 0.50 to 3.08 | .639 | 13/4 | 1.15 | 0.29 to 4.51 | .844 |
| GG | 2/1 | 1.38 | 0.14 to 13.53 | .784 | 1/1 | 1.96 | 0.17 to 22.84 | .589 |
| CG + GG | 1.26 | 0.53 to 2.99 | .604 | 1.27 | 0.37 to 4.41 | .705 | ||
| PIK3CA:rs7621329 | ||||||||
| CC | 54/25 | 1 (reference) | 45/17 | 1 (reference) | ||||
| CT | 22/14 | 0.90 | 0.35 to 2.31 | .822 | 16/8 | 0.50 | 0.15 to 1.71 | .270 |
| TT | 5/2 | 1.08 | 0.18 to 6.66 | .930 | 3/2 | 11.38 | 1.60 to 80.88 | .015 |
| CT + TT | 0.92 | 0.37 to 2.28 | .862 | 0.90 | 0.30 to 2.71 | .846 | ||
| PIK3CA:rs6443624 | ||||||||
| AA | 54/24 | 1 (reference) | 40/17 | 1 (reference) | ||||
| AC | 23/18 | 1.81 | 0.72 to 4.52 | .205 | 20/9 | 0.77 | 0.24 to 2.46 | .661 |
| CC | 8/3 | 1.26 | 0.30 to 5.37 | .753 | 4/3 | 7.13 | 1.20 to 42.36 | .031 |
| AC + CC | 1.69 | 0.69 to 4.13 | .247 | 1.20 | 0.41 to 3.53 | .737 | ||
| SNP and Genotype | Any of the Three |
Fluoropyrimidine + Any |
||||||
|---|---|---|---|---|---|---|---|---|
| No Recurrence/Recurrence (No.) | HR* | 95% CI | P | No Recurrence/Recurrence (No.) | HR* | 95% CI | P | |
| PIK3CA:rs2699887 | ||||||||
| AA | 76/37 | 1 (reference) | 74/36 | 1 (reference) | ||||
| AG | 37/17 | 1.20 | 0.58 to 2.49 | .618 | 35/17 | 1.29 | 0.62 to 2.71 | .498 |
| GG | 6/3 | 0.73 | 0.18 to 2.93 | .654 | 6/3 | 0.81 | 0.20 to 3.22 | .762 |
| AG + GG | 1.09 | 0.55 to 2.17 | .795 | 1.18 | 0.59 to 2.37 | .639 | ||
| PTEN:rs2299939 | ||||||||
| AA | 82/38 | 1 (reference) | 79/37 | 1 (reference) | ||||
| AC | 34/16 | 1.06 | 0.53 to 2.15 | .864 | 34/16 | 1.05 | 0.52 to 2.13 | .883 |
| CC | 5/3 | 0.45 | 0.09 to 2.21 | .328 | 5/3 | 0.46 | 0.10 to 2.22 | .335 |
| AC + CC | 0.91 | 0.47 to 1.76 | .784 | 0.91 | 0.47 to 1.75 | .776 | ||
| PTEN:rs12569998 | ||||||||
| GG | 95/44 | 1 (reference) | 93/44 | 1 (reference) | ||||
| GT | 26/12 | 0.80 | 0.37 to 1.73 | .570 | 24/11 | 0.81 | 0.37 to 1.76 | .598 |
| TT | 1/1 | 1.40 | 0.12 to 15.73 | .785 | 1/1 | 1.51 | 0.13 to 17.19 | .742 |
| GT + TT | 0.83 | 0.39 to 1.75 | .617 | 0.84 | 0.40 to 1.79 | .650 | ||
| PTEN:rs12357281 | ||||||||
| CC | 95/49 | 1 (reference) | 92/48 | 1 (reference) | ||||
| CG | 22/7 | 0.34 | 0.13 to 0.89 | .027 | 21/7 | 0.33 | 0.13 to 0.87 | .025 |
| GG | 1/0 | 1/0 | ||||||
| CG + GG | 0.34 | 0.13 to 0.88 | .027 | 0.33 | 0.13 to 0.87 | .025 | ||
| AKT1:rs2498804 and AKT2:rs892119 unfavorable genotype analysis | ||||||||
| No. of unfavorable genotypes | ||||||||
| 0 | 36/13 | 1 (reference) | 35/13 | 1 (reference) | ||||
| 1 | 69/27 | 2.87 | 1.15 to 7.19 | .024 | 67/26 | 2.80 | 1.12 to 7.03 | .028 |
| 2 | 15/17 | 6.52 | 2.34 to 18.18 | < .001 | 14/17 | 6.36 | 2.28 to 17.72 | < .001 |
| SNP and Genotype | Platinum Compound + Any |
Taxane + Any |
||||||
|---|---|---|---|---|---|---|---|---|
| No Recurrence/Recurrence (No.) | HR* | 95% CI | P | No Recurrence/Recurrence (No.) | HR* | 95% CI | P | |
| PIK3CA:rs2699887 | ||||||||
| AA | 53/27 | 1 (reference) | 41/19 | 1 (reference) | ||||
| AG | 25/15 | 1.35 | 0.58 to 3.10 | .486 | 18/6 | 1.95 | 0.57 to 6.66 | .286 |
| GG | 5/2 | 0.36 | 0.04 to 3.09 | .353 | 4/3 | 1.15 | 0.25 to 5.36 | .856 |
| AG + GG | 1.16 | 0.51 to 2.64 | .726 | 1.59 | 0.56 to 4.49 | .383 | ||
| PTEN:rs2299939 | ||||||||
| AA | 59/28 | 1 (reference) | 42/20 | 1 (reference) | ||||
| AC | 21/14 | 0.94 | 0.39 to 2.26 | .895 | 20/8 | 1.38 | 0.41 to 4.63 | .605 |
| CC | 4/3 | 0.36 | 0.06 to 2.21 | .267 | 1/1 | 0.37 | 0.01 to 11.18 | .566 |
| AC + CC | 0.78 | 0.35 to 1.75 | .544 | 1.17 | 0.37 to 3.68 | .784 | ||
| PTEN:rs12569998 | ||||||||
| GG | 67/34 | 1 (reference) | 53/20 | 1 (reference) | ||||
| GT | 17/10 | 1.09 | 0.40 to 2.93 | .868 | 10/8 | 1.06 | 0.29 to 3.93 | .927 |
| TT | 1/0 | 1/1 | 1.08 | 0.05 to 22.39 | .962 | |||
| GT + TT | 1.06 | 0.39 to 2.82 | .915 | 1.06 | 0.32 to 3.59 | .920 | ||
| PTEN:rs12357281 | ||||||||
| CC | 72/38 | 1 (reference) | 44/26 | 1 (reference) | ||||
| CG | 11/6 | 0.36 | 0.11 to 1.21 | .098 | 17/2 | 0.05 | 0.006 to 0.461 | .008 |
| GG | 0/0 | 1/0 | ||||||
| CG + GG | 0.05 | 0.006 to 0.458 | .008 | |||||
| AKT1:rs2498804 and AKT2:rs892119 unfavorable genotype analysis | ||||||||
| No. of unfavorable genotypes | ||||||||
| 0 | 24/12 | 1 (reference) | 18/4 | 1 (reference) | ||||
| 1 | 49/17 | 3.54 | 1.10 to 11.46 | .040 | 36/15 | 9.09 | 1.29 to 64.26 | .027 |
| 2 | 11/15 | 10.73 | 3.21 to 35.82 | < .001 | 8/10 | 83.37 | 9.56 to 726.78 | < .001 |
Abbreviations: PI3K, phosphoinositide-3-kinase; PTEN, phosphatase and tensin homolog; AKT, v-akt murine thymoma viral oncogene homolog; mTOR, mammalian target of rapamycin; SNP, single nucleotide polymorphism; HR, hazard ratio.
Adjusted for age, sex, smoking status, alcohol consumption, radiation dosage, chemoradiotherapy sequence, clinical stage, chemotherapy regimens, histologic tumor type, tumor location, pathologic stage, and histologic viability.
AKT1 and AKT2 genetic variations.
The AKT1 and AKT2 SNPs resulted in increased risk, with adjusted HRs of 2.21 (95% CI, 1.06 to 4.60) and 3.30 (95% CI, 1.64 to 6.66), respectively. These two SNPs were also associated with recurrence when the results were stratified by treatment (Table 1). Furthermore, AKT2:rs892119 resulted in dramatically different median recurrence-free survival times of 42 months for patients with wild-type genotype compared with 12 months for those with one or two variant alleles (Fig 1).
Fig. 1.
Kaplan-Meier curves of recurrence-free survival times in patients with esophageal cancer (EC) with AKT2:rs892119 treated with (A) any of the three drugs, (B) fluoropyrimidine, (C) platinum compound, and (D) taxane. The numbers in parentheses are the numbers of patients with EC with recurrence/total patients with the respective genotype. MST, median survival time in months.
Because AKT1 and AKT2 SNPs were consistently associated with recurrence risk, we performed an unfavorable genotype analysis to determine the effect of having one or both of these SNPs. One unfavorable genotype resulted in a nearly three-fold increased recurrence risk (95% CI, 1.15 to 7.19; Table 1). This risk increased to more than six-fold (HR, 6.52; 95% CI, 2.34 to 18.18) in patients with two unfavorable genotypes. The same result was also observed when results were stratified by treatment, with HRs for two unfavorable genotypes of 6.36 (95% CI, 2.28 to 17.72), 10.73 (95% CI, 3.21 to 35.82), and 83.4 (95% CI, 9.56 to 726.78) for fluoropyrimidine, platinum compound, and taxane groups, respectively. These results demonstrate that AKT1 and AKT2 genetic variation has an additive effect on recurrence risk and recurrence-free survival rates.
PTEN genetic variation.
In contrast, and as would be predicted by PTEN′s negative regulation of signaling through the pathway, PTEN:rs12357281 was associated with a decreased recurrence risk. Patients carrying at least one variant allele had a significant reduction in risk (HR, 0.34; 95% CI, 0.13 to 0.88). This same pattern was also observed for fluoropyrimidine and taxane groups, with HRs of 0.33 (95% CI, 0.13 to 0.87) and 0.05 (95% CI, 0.0006 to 0.458), respectively (Table 1).
Higher-order gene-gene interactions.
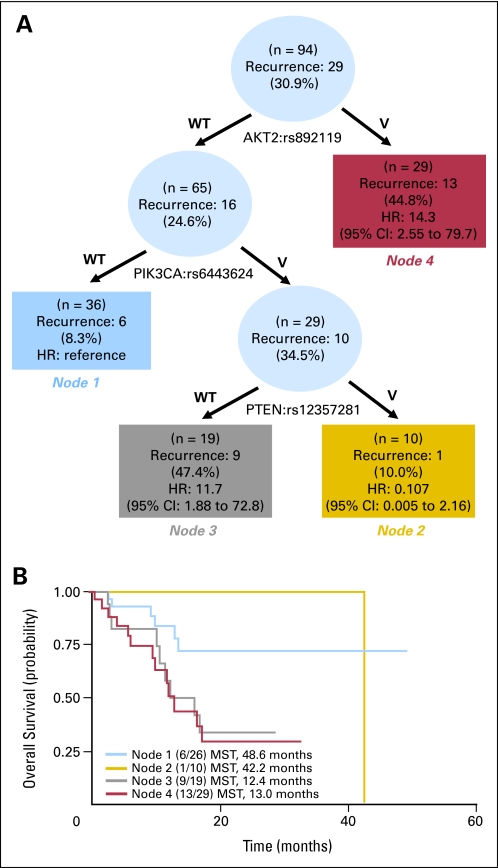
Overall, seven SNPs were found to be associated with recurrence risk in patients treated with a taxane as part of their overall treatment regimen. Survival tree analysis was used to identify interactions within these SNPs. AKT2:rs892119, PIK3CA:rs6443624, and PTEN:rs12357281 demonstrated gene-gene interactions, resulting in four terminal nodes with different recurrence-free survival times (Fig 2A). The initial split on the survival tree was due to AKT2:rs892119 (node 4), indicating that this SNP is the primary factor contributing to variation in recurrence risk in this population. The reference group for the analysis (node 1) was composed of individuals with wild-type AKT2:rs892119 and wild-type PIK3CA:rs6443624 genotypes. Patients in this node had the longest recurrence-free survival time of 48 months. This duration was comparable to patients in node 2 who carried the wild-type alleles for AKT2:892119, but variants of both PIK3CA:rs6443624 and PTEN:rs12357281. These results suggest that the protective effect conferred by PTEN is able to counteract the negative consequences of PIK3CA genetic variation and shift the recurrence-free survival time from 12 to 42 months (Fig 2B).
Fig. 2.
Gene-gene interactions in the phosphoinositide-3-kinase (PI3K), phosphatase and tensin homolog (PTEN), v-akt murine thymoma viral oncogene homolog (AKT), and mammalian target of rapamycin (mTOR) pathway that modified recurrence risk and recurrence-free survival in patients with esophageal cancer (EC) treated with a taxane. (A) Survival tree analysis showing the interactions between three SNPs. (B) Kaplan-Meier curves of recurrence-free survival times in patients in the four terminal nodes, as identified by a survival tree analysis. MST, median survival time; HR, hazard ratio.
Associations Between SNPs and Survival
The PI3K/PTEN/AKT/mTOR pathway did not appear to be a large contributor to variation in survival times. Of the 16 SNPs assayed, only four were found to be significantly associated with survival in any of the treatment groups: AKT1:rs1130214, AKT2:rs892119, FRAP1:rs11121704, and FRAP1:rs2295080 (Table 2).
Table 2.
PI3K/PTEN/AKT/mTOR Pathway Genotypes and Survival
| SNP and Genotype | Any of the Three |
Fluoropyrimidine + Any |
||||||
|---|---|---|---|---|---|---|---|---|
| No. Alive/Dead | HR* | 95% CI | P | No. Alive/Dead | HR* | 95% CI | P | |
| AKT1:rs3803304 | ||||||||
| CC | 45/41 | 1 (reference) | 43/40 | 1 (reference) | ||||
| CG | 38/41 | 1.12 | 0.70 to 1.80 | .645 | 37/40 | 1.14 | 0.7 to 1.84 | .602 |
| GG | 8/4 | 1.09 | 0.36 to 3.33 | .876 | 8/4 | 1.18 | 0.39 to 3.59 | .775 |
| CG + GG | 1.12 | 0.71 to 1.76 | .641 | 1.14 | 0.72 to 1.82 | .579 | ||
| AKT1:rs2498804 | ||||||||
| GG | 34/36 | 1 (reference) | 32/35 | 1 (reference) | ||||
| GT | 47/46 | 1.14 | 0.69 to 1.90 | .601 | 46/45 | 1.18 | 0.71 to 1.98 | .518 |
| TT | 9/6 | 1.50 | 0.57 to 3.97 | .409 | 9/6 | 1.64 | 0.62 to 4.34 | .321 |
| GT + TT | 1.18 | 0.72 to 1.93 | .513 | 1.23 | 0.74 to 2.03 | .424 | ||
| AKT1:rs2494738 | ||||||||
| AA | 75/79 | 1 (reference) | 72/77 | 1 (reference) | ||||
| AG | 17/8 | 0.84 | 0.38 to 1.85 | .668 | 17/8 | 0.89 | 0.41 to 1.95 | .772 |
| AKT1:rs1130214 | ||||||||
| GG | 46/42 | 1 (reference) | 43/41 | 1 (reference) | ||||
| GT | 40/44 | 1.26 | 0.76 to 2.09 | .372 | 40/43 | 1.23 | 0.74 to 2.04 | .426 |
| TT | 6/3 | 1.98 | 0.52 to 7.51 | .315 | 6/3 | 2.13 | 0.56 to 8.15 | .270 |
| GT + TT | 1.28 | 0.77 to 2.11 | .337 | 1.25 | 0.76 to 2.06 | .386 | ||
| AKT2:rs892119 | ||||||||
| AA | 69/59 | 1 (reference) | 69/57 | 1 (reference) | ||||
| AG | 22/26 | 1.31 | 0.74 to 2.31 | .354 | 20/26 | 1.47 | 0.83 to 2.61 | .189 |
| GG | 1/4 | 1.08 | 0.32 to 3.70 | .898 | 0/4 | |||
| AG + GG | 1.27 | 0.75 to 2.16 | .378 | 1.40 | 0.81 to 2.4 | .229 | ||
| AKT2:rs8100018 | ||||||||
| CC | 46/39 | 1 (reference) | 44/38 | 1 (reference) | ||||
| CG | 35/42 | 1.06 | 0.63 to 1.77 | .825 | 34/41 | 1.03 | 0.61 to 1.74 | .900 |
| GG | 10/6 | 0.65 | 0.26 to 1.66 | .370 | 10/6 | 0.63 | 0.25 to 1.61 | .335 |
| CG + GG | 0.96 | 0.60 to 1.56 | .884 | 0.94 | 0.58 to 1.53 | .800 | ||
| FRAP1:rs11121704 | ||||||||
| CC | 43/45 | 1 (reference) | 41/43 | 1 (reference) | ||||
| CT | 44/34 | 0.84 | 0.50 to 1.43 | .529 | 43/34 | 0.87 | 0.51 to 1.48 | .599 |
| TT | 5/9 | 3.53 | 1.48 to 8.39 | .004 | 5/9 | 3.82 | 1.58 to 9.23 | .003 |
| CT + TT | 1.04 | 0.63 to 1.71 | .878 | 1.08 | 0.65 to 1.78 | .777 | ||
| FRAP1:rs2295080 | ||||||||
| GG | 35/33 | 1 (reference) | 33/31 | 1 (reference) | ||||
| GT | 47/38 | 0.96 | 0.53 to 1.75 | .906 | 46/38 | 1.00 | 0.54 to 1.84 | .999 |
| TT | 6/12 | 4.19 | 1.83 to 9.61 | .001 | 6/12 | 4.66 | 1.99 to 10.94 | .0004 |
| GT + TT | 1.25 | 0.72 to 2.17 | .433 | 1.30 | 0.74 to 2.31 | .354 | ||
| PIK3CA:rs7651265 | ||||||||
| AA | 70/65 | 1 (reference) | 67/64 | 1 (reference) | ||||
| AG | 18/20 | 1.18 | 0.67 to 2.10 | .567 | 18/19 | 1.17 | 0.65 to 2.09 | .602 |
| GG | 2/0 | 2/0 | ||||||
| AG + GG | 1.02 | 0.59 to 1.78 | .939 | 1.04 | 0.59 to 1.82 | .902 | ||
| PIK3CA:rs7640662 | ||||||||
| CC | 73/67 | 1 (reference) | 72/65 | 1 (reference) | ||||
| CG | 18/20 | 0.89 | 0.50 to 1.58 | .685 | 16/20 | 0.99 | 0.55 to 1.77 | .969 |
| GG | 1/2 | 2.13 | 0.45 to 10.11 | .341 | 1/2 | 2.17 | 0.46 to 10.25 | .330 |
| CG + GG | 0.95 | 0.55 to 1.66 | .869 | 1.06 | 0.61 to 1.84 | .844 | ||
| PIK3CA:rs7621329 | ||||||||
| CC | 59/56 | 1 (reference) | 57/55 | 1 (reference) | ||||
| CT | 23/24 | 0.95 | 0.55 to 1.65 | .857 | 22/23 | 0.97 | 0.56 to 1.67 | .911 |
| TT | 6/3 | 0.72 | 0.20 to 2.62 | .617 | 6/3 | 0.85 | 0.23 to 3.1 | .810 |
| CT + TT | 0.91 | 0.55 to 1.52 | .725 | 0.95 | 0.57 to 1.59 | .854 | ||
| PIK3CA:rs6443624 | ||||||||
| AA | 56/52 | 1 (reference) | 54/51 | 1 (reference) | ||||
| AC | 26/32 | 1.47 | 0.85 to 2.54 | .163 | 25/31 | 1.53 | 0.88 to 2.65 | .128 |
| CC | 9/5 | 0.84 | 0.30 to 2.35 | .738 | 9/5 | 0.92 | 0.33 to 2.57 | .867 |
| AC + CC | 1.31 | 0.79 to 2.18 | .290 | 1.39 | 0.83 to 2.32 | .213 | ||
| SNP and Genotype | Platinum Compound + Any |
Taxane + Any |
||||||
|---|---|---|---|---|---|---|---|---|
| No. Alive/Dead | HR* | 95% CI | P | No. Alive/Dead | HR* | 95% CI | P | |
| AKT1:rs3803304 | ||||||||
| CC | 31/32 | 1 (reference) | 25/15 | 1 (reference) | ||||
| CG | 24/34 | 1.51 | 0.85 to 2.67 | .159 | 24/20 | 1.88 | 0.81 to 4.37 | .143 |
| GG | 6/2 | 0.70 | 0.15 to 3.25 | .653 | 4/3 | 2.17 | 0.35 to 13.3 | .403 |
| CG + GG | 1.40 | 0.80 to 2.43 | .239 | 1.90 | 0.83 to 4.36 | .129 | ||
| AKT1:rs2498804 | ||||||||
| GG | 21/27 | 1 (reference) | 21/12 | 1 (reference) | ||||
| GT | 33/38 | 1.28 | 0.69 to 2.37 | .434 | 26/22 | 1.42 | 0.51 to 3.92 | .503 |
| TT | 6/4 | 1.21 | 0.37 to 3.99 | .757 | 5/5 | 2.48 | 0.57 to 10.77 | .224 |
| GT + TT | 1.27 | 0.69 to 2.33 | .436 | 1.54 | 0.57 to 4.15 | .395 | ||
| AKT1:rs2494738 | ||||||||
| AA | 49/61 | 1 (reference) | 45/36 | 1 (reference) | ||||
| AG | 12/8 | 0.84 | 0.37 to 1.91 | .683 | 9/3 | 0.68 | 0.17 to 2.67 | .576 |
| AKT1:rs1130214 | ||||||||
| GG | 33/35 | 1 (reference) | 28/18 | 1 (reference) | ||||
| GT | 25/33 | 1.19 | 0.66 to 2.15 | .562 | 23/19 | 1.69 | 0.73 to 3.94 | .222 |
| TT | 3/2 | 1.85 | 0.37 to 9.26 | .455 | 3/3 | 8.92 | 1.56 to 51.17 | .014 |
| GT + TT | 1.21 | 0.67 to 2.18 | .521 | 1.82 | 0.79 to 4.19 | .157 | ||
| AKT2:rs892119 | ||||||||
| AA | 45/48 | 1 (reference) | 41/24 | 1 (reference) | ||||
| AG | 16/18 | 1.38 | 0.71 to 2.68 | .343 | 12/14 | 3.27 | 1.27 to 8.40 | .014 |
| GG | 0/4 | 1/2 | 6.25 | 0.89 to 43.88 | .065 | |||
| AG + GG | 1.39 | 0.75 to 2.59 | .292 | 3.54 | 1.43 to 8.78 | .006 | ||
| AKT2:rs8100018 | ||||||||
| CC | 30/29 | 1 (reference) | 26/23 | 1 (reference) | ||||
| CG | 24/39 | 1.16 | 0.65 to 2.09 | .617 | 22/13 | 0.76 | 0.32 to 1.83 | .547 |
| GG | 6/1 | 0.17 | 0.02 to 1.30 | .088 | 6/4 | 0.23 | 0.05 to 1.04 | .056 |
| CG + GG | 1.01 | 0.56 to 1.79 | .985 | 0.55 | 0.25 to 1.21 | .138 | ||
| FRAP1:rs11121704 | ||||||||
| CC | 31/38 | 1 (reference) | 25/20 | 1 (reference) | ||||
| CT | 26/27 | 0.97 | 0.53 to 1.76 | .920 | 28/14 | 0.58 | 0.23 to 1.45 | .244 |
| TT | 4/5 | 2.77 | 0.85 to 9.00 | .091 | 1/6 | 7.03 | 1.81 to 27.35 | .005 |
| CT + TT | 1.12 | 0.64 to 1.97 | .692 | 0.85 | 0.37 to 1.97 | .710 | ||
| FRAP1:rs2295080 | ||||||||
| GG | 25/28 | 1 (reference) | 20/13 | 1 (reference) | ||||
| GT | 27/30 | 0.99 | 0.51 to 1.91 | .971 | 33/18 | 0.32 | 0.10 to 1.01 | .053 |
| TT | 5/7 | 2.66 | 0.91 to 7.75 | .073 | 1/7 | 8.28 | 2.02 to 33.92 | .003 |
| GT + TT | 1.15 | 0.61 to 2.16 | .663 | 0.73 | 0.26 to 2.05 | .554 | ||
| PIK3CA:rs7651265 | ||||||||
| AA | 48/50 | 1 (reference) | 41/30 | 1 (reference) | ||||
| AG | 10/17 | 1.73 | 0.85 to 3.49 | .130 | 11/8 | 0.78 | 0.25 to 2.38 | .661 |
| GG | 2/0 | |||||||
| AG + GG | 1.43 | 0.72 to 2.82 | .305 | |||||
| PIK3CA:rs7640662 | ||||||||
| CC | 46/53 | 1 (reference) | 43/32 | 1 (reference) | ||||
| CG | 14/16 | 0.79 | 0.41 to 1.53 | .483 | 10/7 | 1.03 | 0.37 to 2.86 | .952 |
| GG | 1/2 | 2.18 | 0.45 to 10.64 | .335 | 1/1 | 0.87 | 0.09 to 8.24 | .905 |
| CG + GG | 0.87 | 0.46 to 1.63 | .665 | 1.00 | 0.39 to 2.60 | .992 | ||
| PIK3CA:rs7621329 | ||||||||
| CC | 38/41 | 1 (reference) | 35/27 | 1 (reference) | ||||
| CT | 15/21 | 1.19 | 0.61 to 2.35 | .612 | 15/9 | 0.59 | 0.21 to 1.69 | .328 |
| TT | 5/2 | 0.74 | 0.16 to 3.57 | .712 | 3/2 | 1.38 | 0.24 to 7.87 | .716 |
| CT + TT | 1.11 | 0.59 to 2.07 | .752 | 0.70 | 0.27 to 1.81 | .459 | ||
| PIK3CA:rs6443624 | ||||||||
| AA | 38/40 | 1 (reference) | 32/25 | 1 (reference) | ||||
| AC | 15/26 | 1.55 | 0.82 to 2.93 | .173 | 17/12 | 1.58 | 0.62 to 4.01 | .336 |
| CC | 7/4 | 1.28 | 0.41 to 4.02 | .675 | 4/3 | 1.22 | 0.21 to 7.05 | .828 |
| AC + CC | 1.50 | 0.82 to 2.75 | .186 | 1.52 | 0.62 to 3.71 | .360 | ||
| SNP and Genotype | Any of the Three |
Fluoropyrimidine + Any |
||||||
|---|---|---|---|---|---|---|---|---|
| No. Alive/Dead | HR* | 95% CI | P | No. Alive/Dead | HR* | 95% CI | P | |
| PIK3CA:rs2699887 | ||||||||
| AA | 57/56 | 1 (reference) | 56/54 | 1 (reference) | ||||
| AG | 26/28 | 1.15 | 0.68 to 1.96 | .601 | 24/28 | 1.28 | 0.75 to 2.2 | .362 |
| GG | 5/4 | 0.48 | 0.15 to 1.50 | .206 | 5/4 | 0.50 | 0.16 to 1.56 | .230 |
| AG + GG | 0.99 | 0.60 to 1.65 | .984 | 1.10 | 0.65 to 1.84 | .728 | ||
| PTEN:rs2299939 | ||||||||
| AA | 62/58 | 1 (reference) | 60/56 | 1 (reference) | ||||
| AC | 26/24 | 1.45 | 0.84 to 2.51 | .186 | 26/24 | 1.46 | 0.84 to 2.52 | .181 |
| CC | 3/5 | 1.35 | 0.44 to 4.09 | .598 | 3/5 | 1.40 | 0.45 to 4.3 | .559 |
| AC + CC | 1.43 | 0.86 to 2.40 | .172 | 1.45 | 0.86 to 2.43 | .163 | ||
| PTEN:rs12569998 | ||||||||
| GG | 68/71 | 1 (reference) | 66/71 | 1 (reference) | ||||
| GT | 21/17 | 0.63 | 0.34 to 1.18 | .148 | 20/15 | 0.62 | 0.33 to 1.16 | .137 |
| TT | 1/1 | 3.52 | 0.40 to 30.89 | .257 | 1/1 | 4.64 | 0.51 to 41.97 | .172 |
| GT + TT | 0.66 | 0.36 to 1.22 | .188 | 0.65 | 0.35 to 1.21 | .178 | ||
| PTEN:rs12357281 | ||||||||
| CC | 72/72 | 1 (reference) | 70/70 | 1 (reference) | ||||
| CG | 15/14 | 0.78 | 0.40 to 1.52 | .468 | 14/14 | 0.77 | 0.39 to 1.52 | .458 |
| GG | 1/0 | 0/0 | ||||||
| CG + GG | 0.77 | 0.39 to 1.50 | .444 | |||||
| SNP and Genotype | Platinum Compound + Any |
Taxane + Any |
||||||
|---|---|---|---|---|---|---|---|---|
| No. Alive/Dead | HR* | 95% CI | P | No. Alive/Dead | HR* | 95% CI | P | |
| PIK3CA:rs2699887 | ||||||||
| AA | 35/45 | 1 (reference) | 36/24 | 1 (reference) | ||||
| AG | 19/21 | 0.77 | 0.41 to 1.42 | .397 | 12/12 | 2.32 | 0.84 to 6.38 | .104 |
| GG | 4/3 | 0.30 | 0.07 to 1.27 | .101 | 4/3 | 0.44 | 0.11 to 1.80 | .253 |
| AG + GG | 0.68 | 0.37 to 1.23 | .203 | 1.15 | 0.49 to 2.67 | .749 | ||
| PTEN:rs2299939 | ||||||||
| AA | 42/45 | 1 (reference) | 36/26 | 1 (reference) | ||||
| AC | 16/19 | 1.47 | 0.78 to 2.79 | .234 | 16/12 | 1.43 | 0.58 to 3.52 | .432 |
| CC | 2/5 | 1.99 | 0.56 to 7.08 | .286 | 1/1 | 0.67 | 0.04 to 10.91 | .778 |
| AC + CC | 1.55 | 0.85 to 2.80 | .149 | 1.33 | 0.56 to 3.15 | .515 | ||
| PTEN:rs12569998 | ||||||||
| GG | 45/56 | 1 (reference) | 42/31 | 1 (reference) | ||||
| GT | 14/13 | 0.62 | 0.28 to 1.37 | .234 | 10/8 | 0.35 | 0.12 to 1.02 | .055 |
| TT | 0/1 | 1/1 | 3.11 | 0.21 to 46.74 | .412 | |||
| GT + TT | 0.70 | 0.33 to 1.52 | .373 | 0.42 | 0.15 to 1.18 | .099 | ||
| PTEN:rs12357281 | ||||||||
| CC | 50/60 | 1 (reference) | 40/30 | 1 (reference) | ||||
| CG | 9/8 | 0.73 | 0.30 to 1.76 | .478 | 11/8 | 1.16 | 0.42 to 3.16 | .777 |
| GG | 0/0 | 1/0 | ||||||
| CG + GG | 1.11 | 0.41 to 3.02 | .837 | |||||
Abbreviations: PI3K, phosphoinositide-3-kinase; PTEN, phosphatase and tensin homolog; AKT, v-akt murine thymoma viral oncogene homolog; mTOR, mammalian target of rapamycin; SNP, single nucleotide polymorphism; HR, hazard ratio.
Adjusted for age, sex, smoking status, alcohol consumption, radiation dosage, chemoradiotherapy sequence, clinical stage, chemotherapy regimens, histologic tumor type, tumor location, pathologic stage, and histologic viability.
FRAP1 genetic variations.
Both of the FRAP1 SNPs genotyped were significant with HRs of 3.53 (95% CI, 1.48 to 8.39) and 4.19 (95% CI, 1.83 to 9.61) for homozygous variants of FRAP1:rs11121704 and FRAP1:rs2295080, respectively (Table 2). These same SNPs were also associated with increased risk of death in the fluoropyrimidine and taxane treatment groups.
AKT1 and AKT2 genetic variations.
AKT1 and AKT2 SNPs were associated with survival in taxane-treated patients only. AKT1:rs1130214 resulted in a nearly nine-fold (HR, 8.92; 95% CI, 1.56 to 51.17) increased risk of death in these patients, while AKT2:rs892119 was associated with a 3.5-fold increase (95% CI, 1.43 to 8.78). The relationships between AKT1:rs1130214 and AKT2:rs892119 with survival were not observed in any of the other treatment groups, suggesting that the effect of these genetic variants may be restricted to taxane-based therapy. However, the small sample size of the taxane group may be a factor contributing to these results.
Associations Between SNPs and Response to Therapy
Response to therapy was analyzed for associations with genetic variations in the pathway with three SNPs showing significance: AKT2:rs892119, AKT1:rs3803304, and FRAP1:rs1121704 (Table 3).
Table 3.
PI3K/PTEN/AKT/mTOR Pathway Genotypes and Response to Therapy
| SNP and Genotype | Any of the Three |
Fluoropyrimidine + Any |
||||||
|---|---|---|---|---|---|---|---|---|
| Response/No Response | OR* | 95% CI | P | Response/No Response | OR* | 95% CI | P | |
| AKT1:rs3803304 | ||||||||
| CC | 25/61 | 1 (reference) | 25/58 | 1 (reference) | ||||
| CG | 35/44 | 0.50 | 0.25 to 0.99 | .047 | 34/43 | 0.54 | 0.27 to 1.08 | .083 |
| GG | 4/8 | 0.93 | 0.25 to 3.53 | .920 | 4/8 | 0.96 | 0.25 to 3.63 | .953 |
| CG + GG | 0.54 | 0.28 to 1.05 | .071 | 0.59 | 0.30 to 1.15 | .118 | ||
| AKT1:rs2498804 | ||||||||
| GG | 23/47 | 1 (reference) | 23/44 | 1 (reference) | ||||
| GT | 36/57 | 0.75 | 0.37 to 1.52 | .419 | 35/56 | 0.83 | 0.40 to 1.70 | .605 |
| TT | 5/10 | 1.15 | 0.33 to 4.01 | .821 | 5/10 | 1.21 | 0.35 to 4.20 | .767 |
| GT + TT | 0.79 | 0.40 to 1.58 | .510 | 0.87 | 0.43 to 1.75 | .703 | ||
| AKT1:rs2494738 | ||||||||
| AA | 59/95 | 1 (reference) | 58/91 | 1 (reference) | ||||
| AG | 6/19 | 2.01 | 0.73 to 5.57 | .177 | 6/19 | 2.07 | 0.75 to 5.75 | .163 |
| AKT1:rs1130214 | ||||||||
| GG | 31/57 | 1 (reference) | 30/54 | 1 (reference) | ||||
| GT | 31/53 | 1.12 | 0.56 to 2.25 | .754 | 31/52 | 1.06 | 0.52 to 2.15 | .879 |
| TT | 4/5 | 0.74 | 0.17 to 3.2 | .693 | 4/5 | 0.71 | 0.16 to 3.11 | .652 |
| GT + TT | 1.08 | 0.54 to 2.13 | .833 | 1.02 | 0.51 to 2.04 | .961 | ||
| AKT2:rs892119 | ||||||||
| AA | 53/75 | 1 (reference) | 53/73 | 1 (reference) | ||||
| AG | 13/35 | 2.54 | 1.14 to 5.65 | .023 | 12/34 | 2.68 | 1.18 to 6.06 | .018 |
| GG | 0/5 | 0/4 | ||||||
| AG + GG | 2.81 | 1.27 to 6.21 | .010 | 2.98 | 1.33 to 6.68 | .008 | ||
| AKT2:rs8100018 | ||||||||
| CC | 33/52 | 1 (reference) | 32/50 | 1 (reference) | ||||
| CG | 26/51 | 1.30 | 0.66 to 2.58 | .446 | 26/49 | 1.20 | 0.60 to 2.39 | .612 |
| GG | 6/10 | 0.83 | 0.24 to 2.84 | .769 | 6/10 | 0.83 | 0.24 to 2.84 | .768 |
| CG + GG | 1.22 | 0.63 to 2.36 | .549 | 1.13 | 0.58 to 2.20 | .713 | ||
| FRAP1:rs11121704 | ||||||||
| CC | 37/51 | 1 (reference) | 36/48 | 1 (reference) | ||||
| CT | 25/53 | 1.35 | 0.68 to 2.67 | .395 | 25/52 | 1.32 | 0.66 to 2.63 | .437 |
| TT | 4/10 | 1.92 | 0.53 to 6.87 | .318 | 4/10 | 1.88 | 0.52 to 6.77 | .336 |
| CT + TT | 1.43 | 0.74 to 2.74 | .283 | 1.40 | 0.72 to 2.70 | .320 | ||
| FRAP1:rs2295080 | ||||||||
| GG | 29/39 | 1 (reference) | 28/36 | 1 (reference) | ||||
| GT | 26/59 | 1.60 | 0.77 to 3.32 | .210 | 26/58 | 1.58 | 0.76 to 3.31 | .224 |
| TT | 8/10 | 0.99 | 0.33 to 2.95 | .983 | 8/10 | 0.99 | 0.33 to 2.97 | .985 |
| GT + TT | 1.44 | 0.72 to 2.86 | .301 | 1.43 | 0.71 to 2.86 | .316 | ||
| PIK3CA:rs7651265 | ||||||||
| AA | 53/82 | 1 (reference) | 52/79 | 1 (reference) | ||||
| AG | 11/27 | 1.45 | 0.61 to 3.42 | .399 | 11/26 | 1.40 | 0.59 to 3.33 | .443 |
| GG | 0/2 | 0/2 | ||||||
| AG + GG | 1.61 | 0.69 to 3.75 | .273 | 1.56 | 0.66 to 3.65 | .307 | ||
| PIK3CA:rs7640662 | ||||||||
| CC | 50/90 | 1 (reference) | 50/87 | 1 (reference) | ||||
| CG | 14/24 | 0.83 | 0.37 to 1.84 | .639 | 13/23 | 0.85 | 0.37 to 1.95 | .704 |
| GG | 2/1 | 0.34 | 0.03 to 4.00 | .390 | 2/1 | 0.35 | 0.03 to 4.21 | .410 |
| CG + GG | 0.76 | 0.35 to 1.65 | .493 | 0.79 | 0.35 to 1.74 | .552 | ||
| PIK3CA:rs7621329 | ||||||||
| CC | 44/71 | 1 (reference) | 43/69 | 1 (reference) | ||||
| CT | 15/32 | 1.27 | 0.57 to 2.84 | .562 | 15/30 | 1.16 | 0.51 to 2.61 | .726 |
| TT | 3/6 | 1.64 | 0.36 to 7.37 | .519 | 3/6 | 1.68 | 0.37 to 7.64 | .501 |
| CT + TT | 1.34 | 0.64 to 2.79 | .442 | 1.25 | 0.59 to 2.63 | .560 | ||
| PIK3CA:rs6443624 | ||||||||
| AA | 44/64 | 1 (reference) | 43/62 | 1 (reference) | ||||
| AC | 18/40 | 1.57 | 0.74 to 3.35 | .239 | 18/38 | 1.46 | 0.68 to 3.13 | .328 |
| CC | 4/10 | 2.06 | 0.57 to 7.48 | .270 | 4/10 | 2.02 | 0.55 to 7.33 | .288 |
| AC + CC | 1.67 | 0.82 to 3.36 | .155 | 1.56 | 0.77 to 3.19 | .217 | ||
| SNP and Genotype | Platinum Compound + Any |
Taxane + Any |
||||||
|---|---|---|---|---|---|---|---|---|
| Response/No Response | OR* | 95% CI | P | Response/No Response | OR* | 95% CI | P | |
| AKT1:rs3803304 | ||||||||
| CC | 18/45 | 1 (reference) | 12/28 | 1 (reference) | ||||
| CG | 26/32 | 0.45 | 0.19 to 1.04 | .060 | 18/26 | 0.56 | 0.20 to 1.55 | .265 |
| GG | 2/6 | 1.44 | 0.24 to 8.72 | .694 | 3/4 | 0.67 | 0.11 to 4.05 | .664 |
| CG + GG | 0.51 | 0.23 to 1.16 | .108 | 0.58 | 0.22 to 1.54 | .271 | ||
| AKT1:rs2498804 | ||||||||
| GG | 15/33 | 1 (reference) | 12/21 | 1 (reference) | ||||
| GT | 27/44 | 0.67 | 0.28 to 1.61 | .373 | 18/30 | 0.83 | 0.29 to 2.38 | .736 |
| TT | 3/7 | 1.29 | 0.26 to 6.38 | .752 | 4/6 | 0.90 | 0.18 to 4.49 | .896 |
| GT + TT | 0.73 | 0.31 to 1.72 | .470 | 0.85 | 0.31 to 2.34 | .746 | ||
| AKT1:rs2494738 | ||||||||
| AA | 41/69 | 1 (reference) | 31/50 | 1 (reference) | ||||
| AG | 5/15 | 1.79 | 0.56 to 5.72 | .323 | 3/9 | 2.54 | 0.52 to 12.35 | .248 |
| AKT1:rs1130214 | ||||||||
| GG | 23/45 | 1 (reference) | 19/27 | 1 (reference) | ||||
| GT | 23/35 | 0.89 | 0.38 to 2.07 | .790 | 14/28 | 1.39 | 0.51 to 3.85 | .521 |
| TT | 1/4 | 2.07 | 0.20 to 21.71 | .545 | 2/4 | 1.15 | 0.17 to 7.85 | .886 |
| GT + TT | 0.94 | 0.41 to 2.17 | .888 | 1.36 | 0.51 to 3.62 | .540 | ||
| AKT2:rs892119 | ||||||||
| AA | 37/56 | 1 (reference) | 28/37 | 1 (reference) | ||||
| AG | 10/24 | 2.18 | 0.85 to 5.60 | .105 | 7/19 | 3.68 | 1.05 to 12.89 | .042 |
| GG | 0/4 | 0/3 | ||||||
| AG + GG | 2.46 | 0.97 to 6.23 | .059 | 4.12 | 1.18 to 14.37 | .026 | ||
| AKT2:rs8100018 | ||||||||
| CC | 23/36 | 1 (reference) | 20/29 | 1 (reference) | ||||
| CG | 20/43 | 1.35 | 0.60 to 3.06 | .471 | 11/24 | 1.22 | 0.43 to 3.44 | .703 |
| GG | 3/4 | 0.85 | 0.15 to 4.67 | .849 | 4/6 | 0.47 | 0.08 to 2.70 | .396 |
| CG + GG | 1.29 | 0.58 to 2.85 | .536 | 1.06 | 0.39 to 2.84 | .914 | ||
| FRAP1:rs11121704 | ||||||||
| CC | 27/42 | 1 (reference) | 21/24 | 1 (reference) | ||||
| CT | 18/35 | 1.12 | 0.49 to 2.56 | .779 | 12/30 | 2.73 | 0.97 to 7.66 | .057 |
| TT | 2/7 | 2.81 | 0.48 to 16.40 | .250 | 2/5 | 2.94 | 0.47 to 18.52 | .250 |
| CT + TT | 1.29 | 0.59 to 2.82 | .521 | 2.76 | 1.04 to 7.37 | .042 | ||
| FRAP1:rs2295080 | ||||||||
| GG | 22/31 | 1 (reference) | 15/18 | 1 (reference) | ||||
| GT | 17/40 | 1.69 | 0.69 to 4.10 | .248 | 16/35 | 2.11 | 0.74 to 6.03 | .165 |
| TT | 5/7 | 1.15 | 0.29 to 4.49 | .843 | 3/5 | 1.64 | 0.30 to 9.04 | .569 |
| GT + TT | 1.55 | 0.68 to 3.58 | .300 | 2.02 | 0.74 to 5.51 | .172 | ||
| PIK3CA:rs7651265 | ||||||||
| AA | 38/60 | 1 (reference) | 29/42 | 1 (reference) | ||||
| AG | 8/19 | 1.73 | 0.59 to 5.05 | .317 | 4/15 | 2.10 | 0.55 to 8.02 | .279 |
| GG | 0/2 | 0/0 | ||||||
| AG + GG | 1.96 | 0.68 to 5.63 | .211 | |||||
| PIK3CA:rs7640662 | ||||||||
| CC | 32/67 | 1 (reference) | 28/47 | 1 (reference) | ||||
| CG | 13/17 | 0.53 | 0.21 to 1.35 | .184 | 6/11 | 0.85 | 0.25 to 2.93 | .794 |
| GG | 2/1 | 0.23 | 0.02 to 2.87 | .255 | 1/1 | 0.97 | 0.04 to 24.89 | .987 |
| CG + GG | 0.49 | 0.20 to 1.19 | .116 | 0.86 | 0.27 to 2.79 | .804 | ||
| PIK3CA:rs7621329 | ||||||||
| CC | 29/50 | 1 (reference) | 25/37 | 1 (reference) | ||||
| CT | 12/24 | 1.19 | 0.45 to 3.15 | .725 | 7/17 | 1.51 | 0.47 to 4.86 | .490 |
| TT | 2/5 | 2.02 | 0.32 to 12.63 | .452 | 2/3 | 0.94 | 0.14 to 6.45 | .948 |
| CT + TT | 1.33 | 0.55 to 3.23 | .530 | 1.35 | 0.47 to 3.85 | .573 | ||
| PIK3CA:rs6443624 | ||||||||
| AA | 30/48 | 1 (reference) | 24/33 | 1 (reference) | ||||
| AC | 14/27 | 1.28 | 0.51 to 3.21 | .597 | 9/20 | 1.84 | 0.62 to 5.43 | .271 |
| CC | 3/8 | 2.50 | 0.56 to 11.11 | .227 | 2/5 | 1.36 | 0.22 to 8.58 | .744 |
| AC + CC | 1.50 | 0.64 to 3.51 | .353 | 1.72 | 0.64 to 4.61 | .280 | ||
| SNP and Genotype | Any of the Three |
Fluoropyrimidine + Any |
||||||
|---|---|---|---|---|---|---|---|---|
| Response/No Response | OR* | 95% CI | P | Response/No Response | OR* | 95% CI | P | |
| PIK3CA:rs2699887 | ||||||||
| AA | 41/72 | 1 (reference) | 41/69 | 1 (reference) | ||||
| AG | 20/34 | 0.94 | 0.45 to 1.97 | .880 | 19/33 | 0.98 | 0.47 to 2.08 | .965 |
| GG | 3/6 | 1.06 | 0.22 to 5.06 | .939 | 3/6 | 1.14 | 0.24 to 5.46 | .874 |
| AG + GG | 0.96 | 0.48 to 1.93 | .910 | 1.00 | 0.49 to 2.05 | .991 | ||
| PTEN:rs2299939 | ||||||||
| AA | 48/72 | 1 (reference) | 48/68 | 1 (reference) | ||||
| AC | 16/34 | 1.28 | 0.61 to 2.68 | .507 | 16/34 | 1.33 | 0.64 to 2.79 | .446 |
| CC | 1/7 | 4.63 | 0.51 to 42.07 | .174 | 1/7 | 4.92 | 0.54 to 45.00 | .158 |
| AC + CC | 1.47 | 0.72 to 2.98 | .289 | 1.53 | 0.75 to 3.11 | .243 | ||
| PTEN:rs12569998 | ||||||||
| GG | 54/85 | 1 (reference) | 53/84 | 1 (reference) | ||||
| GT | 10/28 | 1.73 | 0.73 to 4.15 | .216 | 10/25 | 1.50 | 0.62 to 3.64 | .369 |
| TT | 1/1 | 0.64 | 0.04 to 11.45 | .761 | 1/1 | 0.59 | 0.03 to 10.76 | .725 |
| GT + TT | 1.63 | 0.70 to 3.79 | .259 | 1.41 | 0.60 to 3.34 | .430 | ||
| PTEN:rs12357281 | ||||||||
| CC | 51/93 | 1 (reference) | 51/89 | 1 (reference) | ||||
| CG | 9/20 | 1.24 | 0.50 to 3.03 | .642 | 8/20 | 1.44 | 0.57 to 3.66 | .440 |
| GG | 1/0 | 1/0 | ||||||
| CG + GG | 1.11 | 0.46 to 2.66 | .813 | 1.26 | 0.51 to 3.10 | .614 | ||
| SNP and Genotype | Platinum Compound + Any |
Taxane + Any |
||||||
|---|---|---|---|---|---|---|---|---|
| Response/No Response | OR* | 95% CI | P | Response/No Response | OR* | 95% CI | P | |
| PIK3CA:rs2699887 | ||||||||
| AA | 26/54 | 1 (reference) | 25/35 | 1 (reference) | ||||
| AG | 17/23 | 0.54 | 0.22 to 1.31 | .171 | 7/17 | 1.73 | 0.53 to 5.63 | .362 |
| GG | 3/4 | 0.47 | 0.08 to 2.88 | .417 | 2/5 | 1.94 | 0.28 to 13.46 | .502 |
| AG + GG | 0.53 | 0.23 to 1.23 | .139 | 1.78 | 0.61 to 5.22 | .295 | ||
| PTEN:rs2299939 | ||||||||
| AA | 36/51 | 1 (reference) | 25/37 | 1 (reference) | ||||
| AC | 9/26 | 1.84 | 0.73 to 4.65 | .200 | 9/19 | 1.27 | 0.44 to 3.65 | .663 |
| CC | 1/6 | 4.12 | 0.43 to 39.61 | .220 | 0/2 | |||
| AC + CC | 2.05 | 0.85 to 4.98 | .112 | 1.50 | 0.53 to 4.22 | .440 | ||
| PTEN:rs12569998 | ||||||||
| GG | 39/62 | 1 (reference) | 30/43 | 1 (reference) | ||||
| GT | 6/21 | 2.22 | 0.74 to 6.67 | .155 | 4/14 | 2.29 | 0.56 to 9.34 | .246 |
| TT | 1/0 | 1/1 | 0.59 | 0.03 to 10.84 | .723 | |||
| GT + TT | 1.89 | 0.66 to 5.39 | .237 | 1.88 | 0.51 to 6.87 | .342 | ||
| PTEN:rs12357281 | ||||||||
| CC | 37/73 | 1 (reference) | 26/44 | 1 (reference) | ||||
| CG | 7/10 | 0.71 | 0.23 to 2.17 | .549 | 5/14 | 1.82 | 0.53 to 6.20 | .340 |
| GG | 0/0 | 1/0 | ||||||
| CG + GG | 1.46 | 0.46 to 4.67 | .522 | |||||
Abbreviations: SNP, single nucleotide polymorphism; OR, odds ratio; PI3K, phosphoinositide-3-kinase; PTEN, phosphatase and tensin homolog; AKT, v-akt murine thymoma viral oncogene homolog; mTOR, mammalian target of rapamycin.
Adjusted for age, sex, smoking status, alcohol consumption, radiation dosage, chemoradiotherapy sequence, clinical stage, chemotherapy regimens, histologic tumor type, tumor location, pathologic stage, and histologic viability.
AKT1 genetic variation.
Patients heterozygous for AKT1:rs3803304 experienced a better response to chemoradiotherapy than those with a wild-type genotype (OR, 0.50; 95% CI, 0.25 to 0.99).
AKT2 genetic variation.
Interestingly, the same AKT2 SNP (rs892119) that had been significantly associated with increased risk of both recurrence and death was also found to be associated with a poorer response (OR, 2.81; 95% CI, 1.27 to 6.21). This effect was also observed in the fluoropyrimidine (OR, 2.98; 95% CI, 1.33 to 6.68), and taxane (OR, 4.12; 95% CI, 1.18 to14.37) treatment groups.
FRAP1 genetic variation.
The FRAP1:rs11121704 SNP, which was associated with poor survival, also contributed to a poor pathologic response (OR, 2.76; 95% CI, 1.04 to 7.37) in patients treated with a taxane with at least one variant allele.
DISCUSSION
The PI3K/PTEN/AKT/mTOR pathway plays an important role in balancing cell growth and death. This pathway is often activated in several cancer types—including EC—and has been shown to be important in the development of resistance to several commonly used classes of chemotherapeutic agents. In this study, we determined whether genetic variations in the genes for PI3K, PTEN, AKT1, AKT2, and mTOR were associated with variation in recurrence, survival, and pathologic response. To our knowledge, ours is the first study to apply a tagging SNP approach to determine the role of this pathway in clinical outcomes for any cancer type.
Significant associations were observed between several SNPs and clinical outcomes. In individual SNP analyses, we identified seven SNPs associated with recurrence risk and recurrence-free survival rates. Patients treated with any of the three chemotherapeutic agents had a dramatic increase in their recurrence risk with two unfavorable AKT1 or AKT2 genotypes. This increased risk was also observed in fluoropyrimidine-, platinum-, and taxane-treated patients (6-, 10-, and > 80-fold increases, respectively). Although these results require further validation, they indicate that variations in these genes play a role in modulating EC recurrence. The importance of AKT2:rs892119 in determining recurrence risk was further supported in a survival tree analysis of patients treated with taxanes. This SNP was the basis of the initial split in the tree, suggesting that genetic variation tagged by this SNP is a major risk factor for developing a recurrence.
Interestingly, AKT2:rs892119 had consistent effect on variation in all three clinical outcomes studied and was not drug specific. AKT2:rs892119 is intronic and represents genetic variation across five SNPs genotyped in the Centre d'Etude du Polymorphisme Humain population. It is possible that AKT2:rs892119 is the functional SNP through alterations in normal splicing patterns or transcription of AKT2. However, it is likely that this SNP is not the functional variant but a surrogate marker for the underling genetic variation within that region on the genome. Additional studies will be required to identify the causative sequence variation and the mechanism(s) responsible for our observations. Nevertheless, our results suggest that the functional SNP tagged by AKT2:rs892119 results in activation of AKT2 and increased signaling through this pathway. This is of particularly intriguing because AKT plays a major role in regulating cell survival and growth.13 AKT activation due to overexpression or gene amplification has been shown to be involved in resistance to several chemotherapeutic agents for cancers such as lung, uterine, and ovarian cancer.19–22 However, to our knowledge, no studies have shown associations between common genetic variations in AKT and clinical outcomes in any cancer type.
We selected tagging SNPs for five genes in the PI3K/PTEN/AKT/mTOR pathway. These genes were chosen because they represent the core functional components of the pathway, but this pathway is complex, with several other genes warranting investigation on the basis of the results of this study. Two phosphoinositide-dependent kinases—PDK1 and PDK2—are responsible for phosphorylating AKT, resulting in AKT activation.29 Directly downstream of AKT in the pathway are the tuberous sclerosis complex (TSC) tumor suppressor genes—TSC1 and TSC2. AKT phosphorylation of TSC2 inhibits the function of this complex, allowing for the activation of mTOR.30 Genetic variation in these four genes—PDK1, PDK2, TSC1, and TSC2—may contribute to additional variation in clinical outcome, especially in combination with genetically altered AKT.
PTEN acts as a negative regulator of PI3K/AKT/mTOR signaling by reversing PIP3 activation. Loss of PTEN function results in unrestrained signaling through this pathway and ultimately increased cell growth and proliferation.31 This is a common feature of cancer and has been observed in several cancer types, including brain, breast, and prostate.32–34 However, PTEN mutations occur infrequently in EC,35 and decreased protein expression is found in approximately 40% of EC tumors.36 Tachibana et al37 reported that patients with positive PTEN expression in the nucleus had higher overall survival rates than did those without. In our study, we found that a SNP located in an intron of PTEN was associated with decreased recurrence risk—possibly due to increased expression of PTEN protein in esophageal tissue. Because PTEN:rs12357281 is a tagging SNP, it is likely not the functional SNP. Although, as with all tagging SNPs, there is a possibility that it is the functional variant. However, even without the identification of the functional SNP, our results imply that common variation in PTEN is an important modulator of recurrence risk in patients with EC. Furthermore, the gene-gene interactions identified in our survival tree analysis highlight the complexity of the effects of genetic variation on recurrence risk and recurrence-free survival and support the importance of AKT2:rs892119 and PTEN:rs12357281 in modulating these outcomes.
In contrast to PTEN that acts as a brake for this pathway, increased mTOR (FRAP1) activity results in increased growth signals through phosphorylation of 4EBP and p70S6K.38 mTOR is activated in many cancers, including EC.18 In our study, patients who were homozygous for either of the FRAP1 SNPs had an increased risk of death. In addition, FRAP1:rs11121704 homozygosity was associated with a poor response to taxane. These observations are consistent with those of increased mTOR signaling, resulting in poorer clinical outcomes for patients with genetically polymorphic FRAP1.
We observed that genetic variations in the PI3K/PTEN/AKT/mTOR pathway appeared to have more effect on clinical outcomes in patients treated with taxanes than in patients treated with either a fluoropyrimidine or platinum-containing agent. This may be partly due to the small sample size of the taxane treatment group, but this observation was particularly evident for associations with recurrence risk, with at least one SNP from every gene studied found to be significant in patients treated with taxane. The pathway's involvement in clinical outcomes in platinum-treated patients was limited to associations with recurrence risk for AKT1 and AKT2 SNPs. Similarly, few significant associations were found between clinical outcome and genetic variation in fluoropyrimidine-treated patients. The observations in the fluoropyrimidine and platinum agent groups were replicated in the larger group of patients treated with any of the three drugs. In contrast, several associations were observed only in the taxane treatment group. Although sample size may be an issue, these drugs have different mechanisms of action, and these differences may account for the differences in association between this pathway and clinical outcome.
In conclusion, we found significant associations between common genetic variants in the PI3K/PTEN/AKT/mTOR pathway and clinical outcomes in patients with EC. Although we limited our analyses to patients receiving chemoradiotherapy, we are not able to conclude that these markers are predictive of drug response since we are unable to exclude that they may be prognostic factors. It would be interesting to analyze these SNPs in a control group undergoing surgery alone to assess their prognostic impact, but we did not have enough patients in this category in this study. Nevertheless, if validated as predictive markers for chemotherapy, these results, with the integration of clinical, epidemiological, and genetic data, could become the basis for individualizing therapy For example, markers predictive of a good drug response could be useful for preselecting patients to a specific chemotherapeutic. In contrast, patients with a poor marker signature who are predicted to receive no benefits from chemotherapy may receive only surgery. The ultimate goal is to allow for the selection of the optimal therapy that would provide the most benefit and least toxicity for patients with EC.
Supplementary Material
Appendix
The Appendix is included in the full-text version of this article, available online at www.jco.org. It is not included in the PDF version (via Adobe® Reader®).
Table A1.
Tagging SNP Characteristics
| SNP | Gene | SNP Type | SNP Location | Alleles | MAF | No. of SNPs Tagged |
|---|---|---|---|---|---|---|
| rs3803304 | AKT1 | Intron | chr14:104310191 | C/G | 0.225 | 1 |
| rs2498804 | AKT1 | 3′-flanking region | chr14:104304140 | G/T | 0.333 | 5 |
| rs2494738 | AKT1 | Intron | chr14:104317731 | A/G | 0.117 | 1 |
| rs1130214 | AKT1 | Intron/5′-UTR | chr14:104330779 | G/T | 0.275 | 1 |
| rs892119 | AKT2 | Intron | chr19:45451912 | A/G | 0.142 | 5 |
| rs8100018 | AKT2 | Intron | chr19:45443863 | C/G | 0.258 | 8 |
| rs11121704 | FRAP1 | Intron | chr1:11216546 | C/T | 0.293 | 38 |
| rs2295080 | FRAP1 | 5′-flanking region | chr1:11245215 | G/T | 0.308 | 2 |
| rs7651265 | PIK3CA | Intron | chr3:180375723 | A/G | 0.125 | 1 |
| rs7640662 | PIK3CA | Intron | chr3:180384695 | C/G | 0.150 | 1 |
| rs7621329 | PIK3CA | Intron | chr3:180357568 | C/T | 0.183 | 19 |
| rs6443624 | PIK3CA | Intron | chr3:180380368 | A/C | 0.242 | 1 |
| rs2699887 | PIK3CA | Intron | chr3:180349102 | A/G | 0.233 | 2 |
| rs2299939 | PTEN | Intron | chr10:89647130 | A/C | 0.133 | 3 |
| rs12569998 | PTEN | Intron | chr10:89664137 | G/T | 0.183 | 5 |
| rs12357281 | PTEN | Intron | chr10:89690651 | C/G | 0.110 | 2 |
Abbreviations: SNP, single nucleotide polymorphism; MAF, minor allele frequency.
Table A2.
Patient Characteristics
| Characteristic | No. of Patients | % |
|---|---|---|
| Total | 186 | Sex |
| Male | 161 | 86.6 |
| Female | 25 | 13.4 |
| Age, years | ||
| Mean | 60.8 | |
| SD | 9.32 | |
| Range | 32-79 | |
| Smoking status | ||
| Never | 77 | 41.4 |
| Ever | 109 | 58.6 |
| Alcohol use, > 4 ounces per day | ||
| No | 121 | 66.9 |
| Yes | 60 | 33.1 |
| Histological type | ||
| Adenocarcinoma | 158 | 84.9 |
| Squamous cell carcinoma | 28 | 15.1 |
| Clinical stage | ||
| IIA | 94 | 50.5 |
| IIB | 14 | 7.5 |
| III | 68 | 36.6 |
| IVA | 10 | 5.4 |
| Chemoradiation | ||
| Any of the three, cisplatin, FU, or taxol | 182 | 97.8 |
| FU plus any | 177 | 95.2 |
| Cisplatin plus any | 132 | 71.0 |
| Taxol plus any | 94 | 50.5 |
Abbreviations: SD, standard deviation; FU, fluorouracil.
Footnotes
Supported in part by Grants no. R01 CA111922 and R25 CA57730 and grants from the Smith, Dallas, Park, and Cantu Families and the Rivercreek Foundation.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The author(s) indicated no potential conflicts of interest.
AUTHOR CONTRIBUTIONS
Conception and design: Michelle Hildebrandt, Hushan Yang, Xifeng Wu
Provision of study materials or patients: Jaffer A. Ajani, Xifeng Wu
Collection and assembly of data: Michelle Hildebrandt, Jie Lin
Data analysis and interpretation: Michelle Hildebrandt, Mien-Chie Hung, Julie G. Izzo, Maosheng Huang, Jie Lin, Xifeng Wu
Manuscript writing: Michelle Hildebrandt, Jaffer A. Ajani, Xifeng Wu
Final approval of manuscript: Michelle Hildebrandt, Hushan Yang, Mien-Chie Hung, Julie G. Izzo, Maosheng Huang, Jie Lin, Jaffer A. Ajani, Xifeng Wu
References
- 1.American Cancer Society. Cancer Facts and Figures 2008. Atlanta, Ga: American Cancer Society; 2008. [Google Scholar]
- 2.Brenner B, Ilson DH, Minsky BD. Treatment of localized esophageal cancer. Semin Oncol. 2004;31:554–565. doi: 10.1053/j.seminoncol.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 3.Iyer R, Wilkinson N, Demmy T, et al. Controversies in the multimodality management of locally advanced esophageal cancer: Evidence-based review of surgery alone and combined-modality therapy. Ann Surg Oncol. 2004;11:665–673. doi: 10.1245/ASO.2004.10.026. [DOI] [PubMed] [Google Scholar]
- 4.Urba S. Esophageal cancer: Preoperative or definitive chemoradiation. Ann Oncol. 2004;15(suppl 4):iv93–iv96. doi: 10.1093/annonc/mdh910. [DOI] [PubMed] [Google Scholar]
- 5.Tepper JE, Krasna MJ, Niedzwiecki D. Superiority of trimodality therapy to surgery alone in esophageal cancer: Results of CALGB 9781. J Clin Oncol. 2006;24(suppl):181s. doi: 10.1200/JCO.2007.12.9593. abstr 4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burmeister BH, Smithers BM, Gebski V, et al. Surgery alone versus chemoradiotherapy followed by surgery for resectable cancer of the oesophagus: A randomised controlled phase III trial. Lancet Oncol. 2005;6:659–668. doi: 10.1016/S1470-2045(05)70288-6. [DOI] [PubMed] [Google Scholar]
- 7.Gebski V, Burmeister B, Smithers BM, et al. Survival benefits from neoadjuvant chemoradiotherapy or chemotherapy in oesophageal carcinoma: A meta-analysis. Lancet Oncol. 2007;8:226–234. doi: 10.1016/S1470-2045(07)70039-6. [DOI] [PubMed] [Google Scholar]
- 8.Walsh TN, Noonan N, Hollywood D, et al. A comparison of multimodal therapy and surgery for esophageal adenocarcinoma. N Engl J Med. 1996;335:462–467. doi: 10.1056/NEJM199608153350702. [DOI] [PubMed] [Google Scholar]
- 9.Urba SG, Orringer MB, Turrisi A, et al. Randomized trial of preoperative chemoradiation versus surgery alone in patients with locoregional esophageal carcinoma. J Clin Oncol. 2001;19:305–313. doi: 10.1200/JCO.2001.19.2.305. [DOI] [PubMed] [Google Scholar]
- 10.Evans WE, Relling MV. Pharmacogenomics: Translating functional genomics into rational therapeutics. Science. 1999;286:487–491. doi: 10.1126/science.286.5439.487. [DOI] [PubMed] [Google Scholar]
- 11.Wu X, Gu J, Wu TT, et al. Genetic variations in radiation and chemotherapy drug action pathways predict clinical outcomes in esophageal cancer. J Clin Oncol. 2006;24:3789–3798. doi: 10.1200/JCO.2005.03.6640. [DOI] [PubMed] [Google Scholar]
- 12.Nicholson KM, Anderson NG. The protein kinase B/Akt signaling pathway in human malignancy. Cell Signal. 2002;14:381–395. doi: 10.1016/s0898-6568(01)00271-6. [DOI] [PubMed] [Google Scholar]
- 13.Bellacosa A, Kumar CC, Di Cristofano A, et al. Activation of AKT kinases in cancer: Implications for therapeutic targeting. Adv Cancer Res. 2005;94:29–86. doi: 10.1016/S0065-230X(05)94002-5. [DOI] [PubMed] [Google Scholar]
- 14.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 15.Manning BD, Cantley LC. AKT/PKB signaling: Navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stambolic V, Suzuki A, de la Pompa JL, et al. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998;95:29–39. doi: 10.1016/s0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- 17.Sagatys E, Garrett CR, Boulware D, et al. Activation of the serine/threonine protein kinase Akt during the progression of Barrett neoplasia. Hum Pathol. 2007;38:1526–1531. doi: 10.1016/j.humpath.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 18.Hou G, Xue L, Lu Z, et al. An activated mTOR/p70S6K signaling pathway in esophageal squamous cell carcinoma cell lines and inhibition of the pathway by rapamycin and siRNA against mTOR. Cancer Lett. 2007;253:236–248. doi: 10.1016/j.canlet.2007.01.026. [DOI] [PubMed] [Google Scholar]
- 19.Lee S, Choi EJ, Jin C, et al. Activation of PI3K/Akt pathway by PTEN reduction and PIK3CA mRNA amplification contributes to cisplatin resistance in an ovarian cancer cell line. Gynecol Oncol. 2005;97:26–34. doi: 10.1016/j.ygyno.2004.11.051. [DOI] [PubMed] [Google Scholar]
- 20.Liu LZ, Zhou XD, Qian G, et al. AKT1 amplification regulates cisplatin resistance in human lung cancer cells through the mammalian target of rapamycin/p70S6K1 pathway. Cancer Res. 2007;67:6325–6332. doi: 10.1158/0008-5472.CAN-06-4261. [DOI] [PubMed] [Google Scholar]
- 21.Kim SH, Juhnn YS, Song YS. Akt involvement in paclitaxel chemoresistance of human ovarian cancer cells. Ann N Y Acad Sci. 2007;1095:82–89. doi: 10.1196/annals.1397.012. [DOI] [PubMed] [Google Scholar]
- 22.Gagnon V, Van Themsche C, Turner S, et al. Akt and XIAP regulate the sensitivity of human uterine cancer cells to cisplatin, doxorubicin and taxol. Apoptosis. 2008;13:259–271. doi: 10.1007/s10495-007-0165-6. [DOI] [PubMed] [Google Scholar]
- 23.Yang X, Fraser M, Moll UM, et al. Akt-mediated cisplatin resistance in ovarian cancer: Modulation of p53 action on caspase-dependent mitochondrial death pathway. Cancer Res. 2006;66:3126–3136. doi: 10.1158/0008-5472.CAN-05-0425. [DOI] [PubMed] [Google Scholar]
- 24.Murakami D, Tsujitani S, Osaki T, et al. Expression of phosphorylated Akt (pAkt) in gastric carcinoma predicts prognosis and efficacy of chemotherapy. Gastric Cancer. 2007;10:45–51. doi: 10.1007/s10120-006-0410-7. [DOI] [PubMed] [Google Scholar]
- 25.Chirieac LR, Swisher SG, Ajani JA, et al. Post-therapy pathologic stage predicts survival in patients with esophageal carcinoma receiving preoperative chemoradiation. Cancer. 2005;103:1347–1355. doi: 10.1002/cncr.20916. [DOI] [PubMed] [Google Scholar]
- 26.Wu TT, Chirieac LR, Abraham SC, et al. Excellent interobserver agreement on grading the extent of residual carcinoma after preoperative chemoradiation in esophageal and esophagogastric junction carcinoma: A reliable predictor for patient outcome. Am J Surg Pathol. 2007;31:58–64. doi: 10.1097/01.pas.0000213312.36306.cc. [DOI] [PubMed] [Google Scholar]
- 27.Consortium TIH. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Bakker PI, Yelensky R, Pe'er I, et al. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 29.Alessi DR, James SR, Downes CP, et al. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Balpha. Curr Biol. 1997;7:261–269. doi: 10.1016/s0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- 30.Tee AR, Fingar DC, Manning BD, et al. Tuberous sclerosis complex-1 and -2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proc Natl Acad Sci U S A. 2002;99:13571–13576. doi: 10.1073/pnas.202476899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sansal I, Sellers WR. The biology and clinical relevance of the PTEN tumor suppressor pathway. J Clin Oncol. 2004;22:2954–2963. doi: 10.1200/JCO.2004.02.141. [DOI] [PubMed] [Google Scholar]
- 32.Ali IU, Schriml LM, Dean M. Mutational spectra of PTEN/MMAC1 gene: A tumor suppressor with lipid phosphatase activity. J Natl Cancer Inst. 1999;91:1922–1932. doi: 10.1093/jnci/91.22.1922. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki H, Freije D, Nusskern DR, et al. Interfocal heterogeneity of PTEN/MMAC1 gene alterations in multiple metastatic prostate cancer tissues. Cancer Res. 1998;58:204–209. [PubMed] [Google Scholar]
- 34.Saal LH, Gruvberger-Saal SK, Persson C, et al. Recurrent gross mutations of the PTEN tumor suppressor gene in breast cancers with deficient DSB repair. Nat Genet. 2008;40:102–107. doi: 10.1038/ng.2007.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu YC, Lam KY, Tang JC, et al. Mutational analysis of the PTEN/MMAC1 gene in primary oesophageal squamous cell carcinomas. Mol Pathol. 1999;52:353–356. doi: 10.1136/mp.52.6.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang MS, Lee HS, Lee BL, et al. Differential protein expression between esophageal squamous cell carcinoma and dysplasia, and prognostic significance of protein markers. Pathol Res Pract. 2005;201:417–425. doi: 10.1016/j.prp.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 37.Tachibana M, Shibakita M, Ohno S, et al. Expression and prognostic significance of PTEN product protein in patients with esophageal squamous cell carcinoma. Cancer. 2002;94:1955–1960. doi: 10.1002/cncr.0678. [DOI] [PubMed] [Google Scholar]
- 38.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.