Abstract
Parachlamydiaceae, which naturally infect amoebae, form a sister taxon to the Chlamydiaceae on the basis of the Chlamydia-like cycle of replication and 80% to 90% homology of ribosomal RNA genes. Because intra-amoebal growth could increase the virulence of some intracellular bacteria, Parachlamydiaceae may be pathogenic. Arguments supporting a pathogenic role are that Chlamydia pneumoniae, a well-recognized agent of pneumonia, was shown to infect free-living amoebae and that another member of the Chlamydiales, Simkania negevensis, which has 88% homology with Parachlamydia acanthamoebae, has caused pneumonia in adults and acute bronchiolitis in infants. The recent identification of a 16S rRNA gene sequence of a Parachlamydiaceae from bronchoalveolar lavage is additional evidence supporting potential for pathogenicity.
Key words: Parachlamydiaceae, pathogenicity, free-living amoebae
Nosocomial pneumonia, a frequent complication associated with considerable illness and death (1,2), is the leading cause of death from nosocomial infections (3). Community-acquired pneumonia, which is also common, is associated with a case-fatality rate of up to 8.8% (4). Despite use of standard diagnostic methods, no microbial cause could be identified in 47% to 55% of community-acquired pneumonia worldwide in adults (5–7) and 20% to 75% of nosocomial pneumonia (8,9). Emerging intracellular bacteria, which grow poorly or not at all on media used routinely for detecting human pathogens from clinical samples, could be the causative agents of these pneumonias of unknown etiology. During recent decades, several previously unrecognized intracellular bacteria have been discovered through the genotypic approach. In addition, use of amoebal coculture procedures (10) allows recovery of some fastidious gram-negative bacteria, such as the Legionella-like amoebal pathogens (11,12), Candidatus Odyssella thessalonicensis (13), Sacrobium lyticum (14), several Afipia species (15), and Chlamydia-like endosymbionts (16,17).
Amoebae: Microbial Trojan Horses
Although Legionella was the first pathogen demonstrated to multiply and persist in amoebae (18), several other fastidious intracellular bacterial pathogens, including Chlamydia pneumoniae (19), Mycobacterium avium (20), Listeria monocytogenes (21), and an Ehrlichia-like organism (22), may infect free-living amoebae. Extensive study of the ecology of Legionella pneumophila has confirmed empirical observations of its predilection for growth in hot water tanks and its localization in sediment (23). Rowbotham described the ability of L. pneumophila to multiply intracellularly within protozoa (18) and suggested that free-living amoebae could be a reservoir for Legionella species (24). As amoebae are common inhabitants of natural aquatic environments and water systems (25,26) and are resistant to extreme temperatures, pH, and osmolarity conditions while encysted (27), the Legionella reservoir is important. Growth of free-living amoebae at high temperatures (44°C to 53°C) was observed more frequently for strains isolated from hot-water tanks (mainly Hartmanella vermiformis) than for those isolated from moist sanitary areas (mainly Acanthamoeba, Naegleria, and Valkhampfia species) (26). This great tolerance of cysts and species-dependent thermotolerance of trophozoites could account for the difficulty in eliminating Legionellae from water systems (28). The resistance of Acanthamoeba spp. cysts to various disinfecting solutions (29–31) complicates the eradication of free-living amoebae. Moreover, a wide variety of Enterobacteriaceae have increased resistance to chlorination when ingested by Tetrahymena pyriformis (32). Thus, free-living amoebae could readily act as Trojan horses for bacterial endosymbionts (33,34).
The relationship between Legionellaceae and free-living amoebae, which serves as a model for other endosymbionts such as Parachlamydiaceae, is not restricted to the role of reservoir. Indeed, Acanthamoeba strains were found to produce Legionella-containing vesicles, which may be agents of transmission of legionellosis. The risk of transmission may be underestimated by plate count methods (35). In addition, Legionellae grown inside amoebae were more virulent (36,37), more motile (24), and more resistant to biocides (38) than are bacteria cultured in axenic media. The entry of Legionellae into monocytes was found to be enhanced by the intra-amoebal growth environment (39). In addition, intra-amoebal growth of L. pneumophila was shown to induce an antibiotic-resistant phenotype, while Legionellae cultured in broth did not (40). Similarly, M. avium living within Acanthamoeba had greater resistance to rifabutin, clarithromycin, and azithromycin than did strains living in macrophages (41). This finding could result from decreased uptake of antibiotics into the amoebae, an inactivation of the compound within amoebae, or a change in the bacterial phenotype. Replication of bacteria in amoebae was found not only to affect the bacterial host (through increased potential for spread, resistance to biocides and antibiotics, and acquisition of virulence traits) but also to enhance the pathogenicity of the free-living amoebae (42).
The Parachlamydiaceae
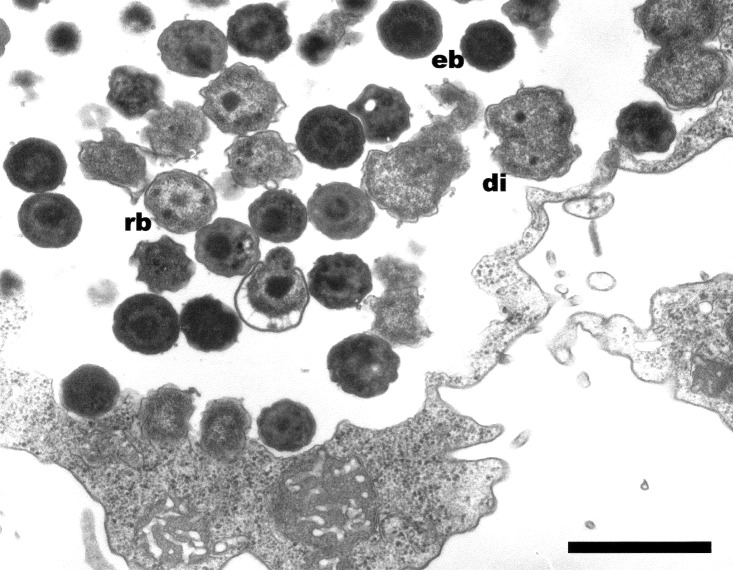
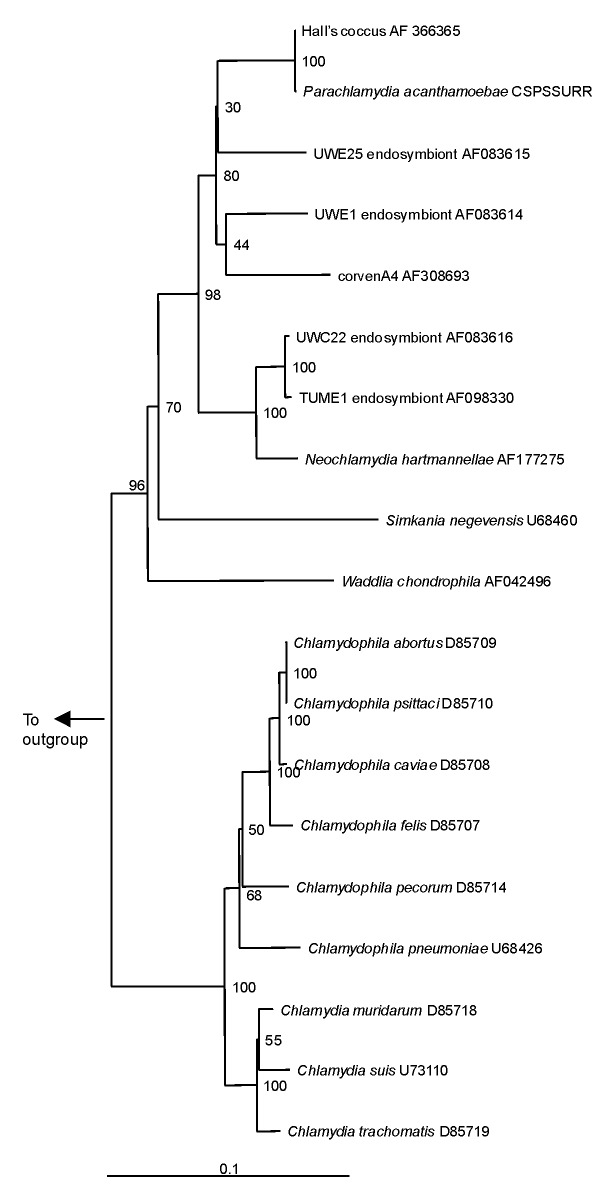
These Chlamydia-like endosymbionts are small Gimenez-stained (43) coccoid bacteria (Figure 1) that naturally infect amoebae and are inconsistently stained with Gram stain. Electron micrographs of Acanthamoeba demonstrate the presence of bacteria at different developmental stages typical of the Chlamydiales, such as elementary and reticulate bodies (Figure 2). A new Parachlamydiaceae family was proposed (44) that forms a sister taxon to the Chlamydiaceae, as it has a Chlamydia-like cycle of replication and 80% to 90% homology of ribosomal RNA genes. This family comprises two genera, of which the type strains are Parachlamydia acanthamoebae (17) and Neochlamydia hartmanellae (45). Members of the Parachlamydia were proposed to have at least 95% homology of the 16S or 23S rRNA genes with P. acanthamoebae (44). However, comparison of the 16S rRNA gene sequences of four additional Parachlamydia with P. acanthamoebae showed substantial phylogenetic diversity within this genus (Figure 3), with 91.2% to 93.1% 16S rRNA gene sequence homology with P. acanthamoebae (46). The ecologic loci and prevalence of the Parachlamydiaceae are unknown, but the latter could be underestimated, as this fastidious gram-negative bacteria was recovered only by amoebal cocultures, a procedure not performed routinely on clinical samples. Moreover, these Chlamydia-like organisms have potential for widespread dissemination, as they are mostly endosymbionts of Acanthamoeba, a free-living amoeba with worldwide distribution (27).
Figure 1.
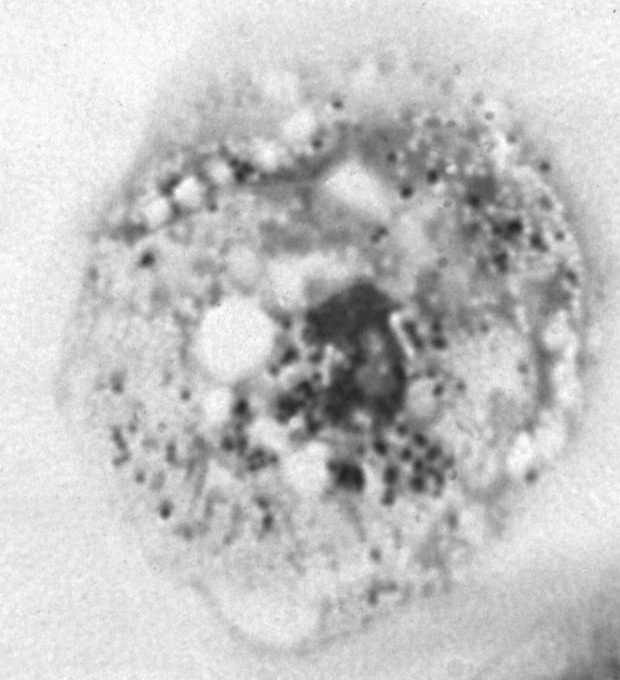
Hall’s coccus within Acanthamoeba polyphaga. Diff Quick staining (Dade, Boehring, Paris, France). Magnification X 1,000.
Figure 2.
Hall’s coccus within Acanthamoeba polyphaga. Electron microscopy, magnification X 12,000, bar = 1 µm.
Figure 3.
Neighbor-joining phylogenetic tree of the 16s rRNA gene sequence of Chlamydiales, including Chlamydiaceae, Parachlamydiaceae, and Simkaniaceae, compared with Legionella pneumophila (M 59157) as outgroup. Bar represents estimated evolutionary distance. The numbers at each node are the results of bootstrap analysis; each value is derived from 100 samples.
Strains of Parachlamydiaceae
Nine strains of Parachlamydia have been described (Table). The first, P. acanthamoebae, was identified within Acanthamoeba BN9, an amoeba recovered from the nasal mucosa of a female volunteer (17). Its 16S rRNA sequence had 88.2% homology with Simkania negevensis and 87% homology with Chlamydophila pneumoniae (17). The second, Berg17 endosymbiont, also isolated from the nasal mucosa of a female volunteer, seems to have an rRNA signature similar to that of the Bn9 endosymbiont, as demonstrated by the binding of the Bn9658 hybridization probe designed for in situ identification of P. acanthamoebae (17). The third, Hall’s coccus, was found in an Acanthamoeba isolated from water taken from a humidifier in a case of humidifier-associated fever in Vermont (16). Its 16S rRNA gene sequence had >99% similarity with that of Bn9 endosymbiont and 86% to 87% with those of the four recognized Chlamydia species (16). Two additional Parachlamydiaceae, UWE1 and UWE25, were also found to infect Acanthamoeba. Both amoeba strains were recovered from soil samples from Washington State (46). A sixth strain, UWC22 endosymbiont, infected an Acanthamoeba recovered from infected corneal tissues (46). TUME1 endosymbiont was found in an amoeba recovered from municipal sewage sludge in Germany (46). The eighth strain, Neochlamydia hartmannellae, is the only strain of Parachlamydiaceae isolated from Hartmanella vermiformis. It did not grow on Acanthamoeba sp. or Naegleria, and its 16S rRNA gene sequence had only 92% homology with that of P. acanthamoeba and varied from 91.6% to 97.1% with the four latter endosymbionts of Acanthamoeba (45). The last one, CorvenA4, could not be isolated. Only its 16S rRNA sequence was retrieved from a respiratory sample (47).
Table. Strains of Parachlamydiaceae.
| Strain | Sample, context and location | Hosta | % 16S rRNA homologyb | Ref | |
|---|---|---|---|---|---|
| to BN9 | to C. pneumoniaec | ||||
| BN9 endosymbiont | Nasal swab of female volunteer, Germany | Acanthamoeba sp. strain BN9 | 100 | 87.6 | 17 |
| Berg17 endosymbiont | Nasal swab of female volunteer, Germany | Acanthamoeba maurianiensis | nad | nad | 17 |
| Hall’s coccus | Water sample, humidifier fever, Vermont | Acanthamoeba sp. | 99.6 | 87.4 | 16 |
| UWE1 endosymbiont | Soil samples, Washington State | Acanthamoeba sp. strainUWE1 | 93.7 | 86.6 | 46 |
| UWE25 endosymbiont | Soil samples, Washington State | Acanthamoeba sp. strain UWE25 | 93.2 | 86.8 | 46 |
| UWC22 endosymbiont | Infected corneal tissues, Washington State | Acanthamoeba sp. strain UWC22 | 91.3 | 87.3 | 46 |
| TUME1 endosymbiont | Municipal sewage sludge, Germany | Acanthamoeba sp. strain TUME1 | 91.0 | 87.2 | 46 |
| Neochlamydia hartmannellae | Water system of a dental unit, Germany | Hartmanella vermiformis | 91.5 | 86.8 | 45 |
| CorvenA4 | Bronchoalveolar washing, France | nae | 91.4 | 85.0 | 47 |
aBacterial strains were identified in free-living amoebae, isolated by culture on nonnutrient agar. bEstimated with Clustal W 63 available on the website of Pôle Bio-Informatique Lyonnais, Lyon, France (http://pbil.ibcp.fr/). c16S rRNA of Chlamydophila pneumoniae strain N16 (GenBank accession number U68426). dBerg17 endosymbiont was shown to have a similar rRNA signature from Bn9 endosymbiont (binding of the Bn9658 hybridization probe designed for in situ identification of Parachlamydia acanthamoebae); however, the 16S rRNA sequence of that strain is not available. eDirect polymerase chain reaction amplification and sequencing from DNA extracted from the respiratory sample; no strain was isolated.
Pathogenicity
Rationale for Potential Pathogenicity
Intra-amoebal growth may increase the virulence of some intracellular bacteria (39), prompting concern that other intracellular bacteria recovered from amoeba, such as the Parachlamydiaceae, could be pathogenic. Indeed, a bacterium able to survive exposure to the lytic enzymes of amoebal phagolysosomes would probably also survive the lytic activity of macrophages. This hypothesis is supported by the fact that mutants of Legionella that have similar cytotoxic defects and intracellular replication in mammalian macrophages and protozoa have been isolated (48), suggesting a common adaptive mechanism to the intracellular environment. Moreover, Parachlamydia can adapt to mammalian cells, as demonstrated by successful passage from an amoebal host to Vero cells (a monkey cell line) (17). Additional arguments in favor of a pathogenic role of the Parachlamydiaceae are that Chlamydia pneumoniae, a well-recognized agent of pneumonia, was shown to infect free-living amoebae (19) and that another member of the Chlamydiales, Simkania negevensis (49,50), which has 88% homology with P. acanthamoebae (46), has been shown to cause pneumonia in adults and acute bronchiolitis in infants (51,52).
Strong evidence that some Parachlamydiaceae could be pathogenic came from the identification of Hall’s coccus in an amoeba isolated from the source of an outbreak of humidifier-associated fever in the United States, as well as related serologic studies (16). In a study of 500 patients with pneumonia, fourfold rising titers against Hall’s coccus were observed in two patients and convalescent-phase antibodies in three others (53). In a second study, two patients had convalescent-phase antibodies (16). These results were recently confirmed: 8 (2.2%) and 3 (0.8%) of 371 patients with community-acquired pneumonia were seropositive (titer >1/50) or had a fourfold rise in Parachlamydia antibody titers compared with none of 511 healthy study participants (54). The recent identification of a 16S rRNA gene sequence of Parachlamydiaceae from bronchoalveolar lavage provides additional evidence of potential pathogenicity (47). However, the contamination of this specimen by an amoeba harboring the CorvenA4-Parachlamydia could not totally be ruled out. These findings should be interpreted cautiously as water contamination probably led to the initial false attribution of Afipia felis as the causative organism of cat-scratch disease (55). The identification in respiratory tract specimens of three new Chlamydia-like strains, which had phylogeny closer to that of the Parachlamydiaceae and Simkaniaceae than the Chlamydia and Chlamydophila (56), is an additional argument in favor of a role of the Parachlamydiaceae in the pathogenesis of respiratory diseases.
In addition, a patient with adult Kawasaki syndrome was found to have a fourfold rise in antibody titer to P. acanthamoebae (54). A possible relationship between a previous respiratory infection and Kawasaki syndrome has already been reported (57,58). Thus, the role of Parachlamydia in the pathogenesis of Kawasaki syndrome should be explored further.
As Parachlamydia could potentially be resistant to lytic macrophages enzymes for years, it could enhance chronic inflammatory disease or chronic pathogenic mechanisms, such as the one leading to vascular damage. A role of Parachlamydiaceae in the pathogenesis of arteriosclerosis is suggested by the presence in an abdominal aneurysm specimen of a Chlamydia-like strain that had a sequence closer to that of P. acanthamoebae than to Chlamydia, Chlamydophila, and Simkaniaceae (56). Some serologic studies have suggested that Chlamydophila pneumoniae could play a role in the pathogenesis of arteriosclerosis (59,60), although this observation was not confirmed in other studies (61,62). Such a discrepancy might result from serologic cross-reactions or confounding by a pathogen such as Parachlamydia, which in light of its homology could share epitopes, mode of transmission, or both with C. pneumoniae.
Based on this rationale, one may hypothesize that some Parachlamydiaceae could cause pneumonia. Thus, patients with nosocomial or community-acquired pneumonia of unknown etiology should ideally receive an extensive diagnostic work-up, including testing for Parachlamydia. In addition, patients with arteriosclerosis and Kawasaki disease or other infectious syndromes of unknown etiology should perhaps be tested for Parachlamydia. As Parachlamydia strains were all identified within free-living amoebae, recent history of swimming in ponds, rivers, or swimming pools might prompt a specific diagnostic approach.
Diagnostic Methods
No diagnostic tool is commercially available. Because of the fastidious nature of Parachlamydiaceae, molecular biology is probably the easiest and cheapest diagnostic approach. Serologic testing is also promising; however, it requires antigen and a laboratory capable of performing amoebal coculture. Serologic results may be useful for epidemiologic studies, as they may provide information on past or present contact with the antigen. Both molecular and serologic methods may yield results in <24 hours.
Although time-consuming, culture-based diagnostic methods have the advantage of enabling the recovery of strains. These methods encompass two main approaches. The first one directly targets the recovery of Parachlamydiaceae, with amoebae used as cell background. A convenient broth for amoebal coculture is Page's modified Neff's amoeba saline (PAS) (10), which is preferable to Nelson's and peptone-yeast extract-glucose medium because PAS is devoid of nutrients, thus reducing overgrowth of potential contaminants in clinical samples. Although incubation at 37°C may be ideal for bacterial recovery, lower temperatures (30°C–35°C) are generally used to prevent amoebal death or encystment (12,13,20). The coculture should be examined regularly for amoebal lysis or Gimenez-positive cocci. The second culture-based method is designed to recover free-living amoebae, which will then be examined for the presence of endocytobionts. Briefly, amoebal culture is performed by adding the clinical sample to nonnutrient agar (1.5 g agar in 100 mL PAS) supplemented with living Enterobacter cloacae or Escherichia coli, incubating at 25°C–30°C, and examining the plate daily for the presence of amoebae. To date, all Parachlamydiaceae strains have been recovered by the second approach.
Future Directions
The role of Parachlamydia sp. as an emerging pathogen needs to be confirmed. In view of the genetic diversity of the Parachlamydiaceae (46), their phylogeny needs to be elucidated, as the various species could be associated with species-specific pathogenicity. Search for additional Parachlamydia strains in hospital water systems could help define potential nosocomial exposures. Because the Parachlamydiaceae are difficult to culture, simpler approaches are being developed, including serologic and molecular tests. These methods could be performed on a large number of samples from both healthy and ill persons. Patients with community-acquired pneumonia, nosocomial pneumonia, Kawasaki disease, and arteriosclerosis should be tested. Increased resistance to antimicrobial drugs, which may be associated with intra-amoebal growth, is another promising area for future study.
Biographies
Dr. Greub is a Swiss physician specializing in medical microbiology and infectious diseases, working as a postdoctoral fellowship in the Unité des Rickettsies in Marseille. His current research focuses on Parachlamydiaceae and other emerging intracellular bacteria.
Dr. Raoult is director of the Unité des Rickettsies, the national reference center for rickettsiosis and WHO collaborative center. His work focuses on the study of emerging and reemerging bacteria and arthropod-borne diseases.
Footnotes
Suggested citation: Greub G and Raoult D. Parachlamydiaceae: Potential Emerging Pathogens. Emerg Infect Dis. [serial on the Internet]. 2002 Jun [date cited]. Available from http://www.cdc.gov/ncidod/EID/vol8no6/01-0210.htm
References
- 1.Townsend GC, Scheld WM. Nosocomial pneumonia. Curr Opin Infect Dis. 1995;8:98–104. 10.1097/00001432-199504000-00004 [DOI] [Google Scholar]
- 2.Mosconi P, Langer M, Cigada M, Mandelli M. Epidemiology and risk factors of pneumonia in critically ill patients. Eur J Epidemiol. 1991;7:320–7. 10.1007/BF00144995 [DOI] [PubMed] [Google Scholar]
- 3.Hamer DH, Barza M. Prevention of hospital-acquired pneumonia in critically ill patients. Antimicrob Agents Chemother. 1993;37:931–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hug B, Rossi M. A year’s review of bacterial pneumonia at the central hospital of Lucerne, Switzerland. Swiss Med Wkly. 2001;131:687–92. [DOI] [PubMed] [Google Scholar]
- 5.Socan M, Marininc-Fiser N, Kraigher A, Kotnik A, Logar M. Microbial aetiology of community-acquired pneumonia in hospitalised patients. Eur J Clin Microbiol Infect Dis. 1999;18:777–82. 10.1007/s100960050400 [DOI] [PubMed] [Google Scholar]
- 6.Marrie TJ, Peeling RW, Fine MJ, Singer DE, Coley CM, Kapoor WN. Ambulatory patients with community-acquired pneumonia. Am J Med. 1996;101:508–15. 10.1016/S0002-9343(96)00255-0 [DOI] [PubMed] [Google Scholar]
- 7.Almirall J, Bolibar I, Vidal J, Sauca G, Coll P, Niklasson B, et al. Epidemiology of community-acquired pneumonia in adults: a population-based study. Eur Respir J. 2000;15:757–63. 10.1034/j.1399-3003.2000.15d21.x [DOI] [PubMed] [Google Scholar]
- 8.Rouby JJ, de Lassale EM, Poete P, Nicolas MH, Bodin L, Jarler V, et al. Nosocomial bronchopneumonia in the critically ill. Histologic and bacteriologic aspects. Am Rev Respir Dis. 1992;146:1059–66. [DOI] [PubMed] [Google Scholar]
- 9.Costa SF, Newbaer M, Santos CR, Basso M, Soares I, Levin AS. Nosocomial pneumonia: importance of recognition of aetiological agents to define an appropriate initial empirical therapy. Int J Antimicrob Agents. 2001;17:147–50. 10.1016/S0924-8579(00)00316-2 [DOI] [PubMed] [Google Scholar]
- 10.Rowbotham TJ. Isolation of Legionella pneumophila from clinical specimens via amoebae, and the interaction of those and other isolates with amoebae. J Clin Pathol. 1983;36:978–86. 10.1136/jcp.36.9.978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hay J, Seal DV, Billcliffe B, Freer JH. Non-culturable Legionella pneumophila associated with Acanthamoeba castellanii: detection of the bacterium using DNA amplification and hybridization. J Appl Bacteriol. 1995;78:61–5. [DOI] [PubMed] [Google Scholar]
- 12.Birtles RJ, Rowbotham TJ, Raoult D, Harrison TG. Phylogenetic diversity of intra-amoebal legionellae as revealed by 16S rRNA gene sequence comparison. Microbiology. 1996;142:3525–30. [DOI] [PubMed] [Google Scholar]
- 13.Birtles RJ, Rowbotham TJ, Michel R, Pitcher DG, Lascola B, Alexiou-Daniel S, et al. Candidatus Odyssella thessalonicensis gen.nov., sp.nov., an obligate intracellular parasite of Acanthamoeba species. Int J Syst Evol Microbiol. 2000;50:63–71. [DOI] [PubMed] [Google Scholar]
- 14.Drozanski WJ. Sacrobium lyticum gen. nov., sp. nov., an obligate intracellular bacterial paasite of small free-living amoebae. Int J Syst Bacteriol. 1991;41:82–7. [Google Scholar]
- 15.La Scola B, Barrassi L, Raoult D. Isolation of new fastidious Proteobacteria and Afipia felis from hospital water supplies by direct plating and amoebal coculture procedures. FEMS Microbiol Ecol. 2000;34:129–37. 10.1016/S0168-6496(00)00084-2 [DOI] [PubMed] [Google Scholar]
- 16.Birtles RJ, Rowbotham TJ, Storey C, Marrie TJ, Raoult D. Chlamydia-like obligate parasite of free-living amoebae. Lancet. 1997;349:925–6. 10.1016/S0140-6736(05)62701-8 [DOI] [PubMed] [Google Scholar]
- 17.Amann R, Springer N, Schönhuber W, Ludwig W, Schmid EN, Muller KD, et al. Obligate intracellular bacterial parasites of Acanthamoebae related to Chlamydia spp. Appl Environ Microbiol. 1997;63:115–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rowbotham TJ. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J Clin Pathol. 1980;33:1179–83. 10.1136/jcp.33.12.1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Essig A, Heinemann M, Simnacher U, Marre R. Infection of Acanthamoeba castellanii by Chlamydia pneumoniae. Appl Environ Microbiol. 1997;63:1396–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steinert M, Birkness K, White E, Fields B, Quinn F. Mycobacterium avium bacilli grow saprozoically in coculture with Acanthamoeba polyphaga and survive within cyst walls. Appl Environ Microbiol. 1998;64:2256–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ly TMC, Müller HE. Ingested Listeria monocytogenes survive and multiply in protozoa. J Med Microbiol. 1990;33:51–4. [DOI] [PubMed] [Google Scholar]
- 22.Michel R, Muller KD, Schmid EN. Ehrlichia-like organisms (KSL1) observed as obligate intracellular parasites of Saccamoeba species. Endocytobiosis Cell Res. 1995;11:69–80. [Google Scholar]
- 23.Stout JE, Yu VL, Best MG. Ecology of Legionella pneumophila within water distribution systems. Appl Environ Microbiol. 1985;49:221–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rowbotham TJ. Current views on the relationships between amoebae, legionellae and man. Isr J Med Sci. 1986;22:678–9. [PubMed] [Google Scholar]
- 25.Kurtz JB, Bartlett CL, Newton UA, White RA, Jones NL. Legionella pneumophila in cooling water systems. J Hyg (Lond). 1982;88:369–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rohr U, Weber S, Michel R, Selenka F, Wilhelm M. Comparison of free-living amoebae in hot water systems of hospitals with isolates from moist sanitary areas by identifying genera and determining temperature tolerance. Appl Environ Microbiol. 1998;64:1822–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodriguez-Zaragoza S. Ecology of free-living amoebae. Crit Rev Microbiol. 1994;20:225–41. 10.3109/10408419409114556 [DOI] [PubMed] [Google Scholar]
- 28.Hoebe CJ, Kool JL. Control of Legionella in drinking-water systems. Lancet. 2000;355:2093–4. 10.1016/S0140-6736(00)02374-6 [DOI] [PubMed] [Google Scholar]
- 29.Zanetti S, Fiori PL, Pinna A, Usai S, Carta F, Fadda G. Susceptibility of Acanthamoeba castellanii to contact lens disinfecting solutions. Antimicrob Agents Chemother. 1995;39:1596–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rohr U, Weber S, Selenka F, Wilhelm M. Impact of silver and copper on the survival of amoebae and ciliated protozoa in vitro. Int J Hyg Environ Health. 2000;203:87–9. 10.1078/S1438-4639(04)70013-9 [DOI] [PubMed] [Google Scholar]
- 31.Borazjani RN, May LL, Noble JA, Avery SV, Ahearn DG. Flow cytometry for determination of the efficacy of contact lens disinfecting solutions against Acanthamoeba sp. Appl Environ Microbiol. 2000;66:1057–61. 10.1128/AEM.66.3.1057-1061.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.King CH, Shotts EB, Wooley RE, Porter KG. Survival of coliforms and bacterial pathogens within protozoa during chlorination. Appl Environ Microbiol. 1988;54:3023–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barker J, Brown M. Trojan horse of the microbial world: protozoa and the survival of bacterial pathogens in the environment. Microbiology. 1994;140:1253–9. [DOI] [PubMed] [Google Scholar]
- 34.Winiecka-Krusnell J, Linder E. Free-living amoebae protecting Legionella in water: The tip of an iceberg? Scand J Infect Dis. 1999;31:383–5. 10.1080/00365549950163833 [DOI] [PubMed] [Google Scholar]
- 35.Berk SG, Ting RS, Turner GW, Ashburn RJ. Production of respirable vesicles containing live Legionella pneumophila cells by two Acanthamoeba spp. Appl Environ Microbiol. 1998;64:279–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cirillo JD, Falkow S, Tompkins LS. Growth of Legionella pneumophila in Acanthamoeba castellanii enhances invasion. Infect Immun. 1994;62:3254–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brieland J, McClain M, LeGendre M, Engleberg C. Intrapulmonary Hartmannella vermiformis: a potential niche for Legionella pneumophila replication in a murine model of legionellosis. Infect Immun. 1997;65:4892–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barker J, Brown MRW, Collier PJ, Farrell I, Gilbert P. Relationship between Legionella pneumophila and Acanthamoeba polyphaga: physiological status and susceptibility to chemical inactivation. Appl Environ Microbiol. 1992;58:2420–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cirillo JD, Cirillo SL, Yan L, Bermudez LE, Falkow S, Tompkins LS. Intracellular growth in Acanthamoeba castellanii affects monocyte entry mechanisms and enhances virulence of Legionella pneumophila. Infect Immun. 1999;67:4427–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barker J, Scaife H, Brown MR. Intraphagocytic growth induces an antibiotic-resistant phenotype of Legionella pneumophila. Antimicrob Agents Chemother. 1995;39:2684–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miltner EC, Bermudez LE. Mycobacterium avium grown in Acanthamoeba castellanii is protected from the effects of antimicrobials. Antimicrob Agents Chemother. 2000;44:1990–4. 10.1128/AAC.44.7.1990-1994.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fritsche TR, Sobek D, Gautom RK. Enhancement of in vitro cytopathogenicity by Acanthamoeba spp. following acquisition of bacterial endosymbionts. FEMS Microbiol Lett. 1998;166:231–6. 10.1111/j.1574-6968.1998.tb13895.x [DOI] [PubMed] [Google Scholar]
- 43.Gimenez DF. Staining rickettsiae in yolk-sac cultures. Stain Technol. 1964;39:135–40. [DOI] [PubMed] [Google Scholar]
- 44.Everett KD, Bush RM, Andersen AA. Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int J Syst Bacteriol. 1999;49:415–40. [DOI] [PubMed] [Google Scholar]
- 45.Horn M, Wagner M, Müller KD, Schmid EN, Fritsche TR, Schleifer KH, et al. Neochlamydia hartmannellae gen. nov., sp. nov. (Parachlamydiaceae), an endoparasite of the amoeba Hartmannella vermiformis. Microbiology. 2000;146:1231–9. [DOI] [PubMed] [Google Scholar]
- 46.Fritsche TR, Horn M, Wagner M, Herwig RP, Schleifer KH, Gautom RK. Phylogenetic diversity among geographically dispersed Chlamydiales endosymbionts recovered from clinical and environmental isolates of Acanthamoeba spp. Appl Environ Microbiol. 2000;66:2613–9. 10.1128/AEM.66.6.2613-2619.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Corsaro D, Venditti D, Le Faou A, Guglielmetti P, Valassina M. A new chlamydia-like 16s rDNA sequence from a clinical sample. Microbiology. 2001;147:515–6. [DOI] [PubMed] [Google Scholar]
- 48.Gao LY, Harb OS, Abu Kwaik Y. Utilization of similar mechanisms by Legionella pneumophila to parasitize two evolutionarily distant host cells, mammalian macrophages and protozoa. Infect Immun. 1997;65:4738–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kahane S, Metzer E, Friedman MG. Evidence that the novel microorganism "Z" may belong to a new genus in the family Chlamydiaceae. FEMS Microbiol Lett. 1995;126:203–8. 10.1111/j.1574-6968.1995.tb07417.x [DOI] [PubMed] [Google Scholar]
- 50.Kahane S, Gonen R, Sayada C, Elion J, Friedman MG. Description and partial characterization of a new chlamydia-like microorganism. FEMS Microbiol Lett. 1993;109:329–33. 10.1111/j.1574-6968.1993.tb06189.x [DOI] [PubMed] [Google Scholar]
- 51.Lieberman D, Kahane S, Friedman MG. Pneumonia with serological evidence of acute infection with the Chlamydia-like microorganism "Z.". Am J Respir Crit Care Med. 1997;156:578–82. [DOI] [PubMed] [Google Scholar]
- 52.Kahane S, Greenberg D, Friedman MG, Haikin H, Dagan R. High prevalence of "simkania Z," a novel Chlamydia-like bacterium, in infants with acute bronchiolitis. J Infect Dis. 1998;177:1425–9. 10.1086/517830 [DOI] [PubMed] [Google Scholar]
- 53.Benson CE, Drozanski W, Rowbotham TJ, Bialkowska I, Losos D, Butler JC, et al. Serologic evidence of infection with 9 Legionella-like amoebal pathogens in pneumonia patients. Proceedings of the 95th American Society of Microbiology General Meeting; 1995. May 21–25; Washington. Abstract C-200:35. [Google Scholar]
- 54.Marrie TJ, Raoult D, La Scola B, Birtles RJ, de Carolis E. Legionella-like amoebal pathogens as agents of community-acquired pneumonia. Emerg Infect Dis. 2001;7:1026–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.La Scola B, Raoult D. Afipia felis is a hospital water supply in association with free-living amoebae. Lancet. 1999;353:1330. 10.1016/S0140-6736(99)00906-X [DOI] [PubMed] [Google Scholar]
- 56.Ossewaarde JM, Meijer A. Molecular evidence for the existence of additional members of the order Chlamydiales. Microbiology. 1999;145:411–7. [DOI] [PubMed] [Google Scholar]
- 57.Bell DM, Brink EW, Nitzkin JL, Hall CB, Wulff H, Berkowitz ID, et al. Kawasaki syndrome: description of two outbreaks in the United States. N Engl J Med. 1981;304:1568–75. [DOI] [PubMed] [Google Scholar]
- 58.Dean AG, Melish ME, Hicks R, Palumbo NE. An epidemic of Kawasaki syndrome in Hawaii. J Pediatr. 1982;100:552–7. 10.1016/S0022-3476(82)80751-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Strachan DP, Carrington D, Mendall MA, Ballam L, Morris J, Butland BK, et al. Relation of Chlamydia pneumoniae serology to mortality and incidence of ischaemic heart disease over 13 years in the Caerphilly prospective heart disease study. BMJ. 1999;318:1035–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schmidt C, Hulthe J, Wikstrand J, Gnarpe H, Gnarpe J, Agewall S, et al. Chlamydia pneumoniae seropositivity is associated with carotid artery intima-media thickness. Stroke. 2000;31:1526–34. [DOI] [PubMed] [Google Scholar]
- 61.Danesh J, Collins R, Peto R. Chronic infections and coronary heart disease: is there a link? Lancet. 1997;350:430–6. 10.1016/S0140-6736(97)03079-1 [DOI] [PubMed] [Google Scholar]
- 62.Wald NJ, Law MR, Morris JK, Zhou X, Wong Y, Ward ME. Chlamydia pneumoniae infection and mortality from ischaemic heart disease: large prospective study. BMJ. 2000;321:204–7. 10.1136/bmj.321.7255.204 [DOI] [PMC free article] [PubMed] [Google Scholar]