Abstract
Fascin1, an actin-bundling protein, has been demonstrated to be critical for filopodia formation in cultured cells, and thus is believed to be vital in motile activities including neurite extension and cell migration. To test whether fascin1 plays such essential roles within a whole animal, we have generated and characterized fascin1-deficient mice. Unexpectedly, fascin1-deficient mice are viable and fertile with no major developmental defect. Nissl staining of serial coronal brain sections reveals that fascin1-deficient brain is grossly normal except that knockout mouse brain lacks the posterior region of the anterior commissure neuron and has larger lateral ventricle. Fascin1-deficient, dorsal root ganglion neurons are able to extend neurites in vitro as well as those from wild-type mice, although fascin1-deficient growth cones are smaller and exhibit fewer and shorter filopodia than wild-type counterparts. Likewise, fascin1-deficient, embryonic fibroblasts are able to assemble filopodia, though filopodia are fewer, shorter and short-lived. These results indicate that fascin1-mediated filopodia assembly is dispensable for mouse development.
Keywords: fascin1, knockout mice, filopodia assembly, dorsal root ganglion neuron, mouse development
INTRODUCTION
Filopodia are highly dynamic and motile structure and are essential for directed movements such as those observed during growth cone motility and guidance. They have also been suggested to play a role in intercellular interactions as well as signal transduction (De Joussineau et al. 2003; Vasioukhin et al. 2000). Fascin is an actin-bundling protein, localized to filopodia, as well as in stress fibers (Yamashiro-Matsumura and Matsumura 1986) (Adams 2004). Fascin represents a distinct family of actin bundling proteins including sea urchin fascin, HeLa 55kDa actin-bundling protein, and the Drosophila singed protein (Bryan et al. 1993; Kureishy et al. 2002). These proteins have a molecular masses ranging from 55 kDa to 58 kDa. The amino acid sequences of these fascin proteins are unique, sharing no apparent homology with non-fascin actin-bundling proteins (including villin and fimbrin) (Duh et al. 1994; Edward et al. 1995; Holthuis et al. 1994; Mosialos et al. 1994). Fascin is apparently absent in C. elegans, yeasts, and Dictyostelium.
Drosophila has a single fascin gene (called singed). Drosophila singed mutants exhibit two phenotypes: gnarled bristle development and female sterility (Patterson and O’Hare 1991), indicating a function for fascin in actin bundle formation. Many singed mutants show a decreased number of microfilament bundles, resulting in a short, curved bristle shaft (Poodry 1980). In Drosophila oogenesis, each developing oocyte is surrounded by and connected to 15 nurse cells via intercellular bridges. Nurse cell cytoplasmic contents flow into the oocyte along actin filaments traversing these cytoplasmic bridges. A singed allele affects the microfilament structure required for this nurse cell cytoplasmic flow, resulting in female sterility (Cant et al. 1994). While these studies on Drosophila singed mutants indicate a role of fascin in microfilament organization, they have not provided further information on the physiological functions of fascin in vertebrates.
Vertebrates have three fascin genes (fascin1-3) (Saishin et al. 1997; Tubb et al. 2002). Expression of fascin2 and 3 is strictly restricted to retina and testis, respectively. In contrast, fascin1 shows more widespread expression. It is worthy of note, however, that certain tissues and cell types show particularly high expression of fascin1: At a tissue level, fascin1 is abundant in brain and spleen; and at a cellular level, it is highly expressed in specific types of cells including neuronal and glial cells, microcapillary endothelial cells, and antigen-presenting, mature dendritic cells (DCs) (Duh et al. 1994; Mosialos et al. 1996; Mosialos et al. 1994; Pinkus et al. 1997; Zhang et al. 2008). A morphological characteristic common to these cells expressing high levels of fascin1 is the development of many membrane protrusions such as filopodia. Indeed, fascin1 is localized in filopodia of growth cones of neuronal cells (Mosialos et al. 1996).
An increasing body of evidence has indicated an essential role of fascin1 in the formation of filopodia in mammalian cells. In vitro, fascin1 makes parallel actin bundles (Otto et al. 1979; Yamashiro-Matsumura and Matsumura 1985) with uniform polarity (Ishikawa et al. 2003; Vignjevic et al. 2003), which is consistent with its localization in filopodia (Adams and Schwartz 2000; Yamashiro-Matsumura and Matsumura 1986). It has been demonstrated, by live-cell imaging combined with electron microscopy with critical point drying, that filopodia is formed by transformation of dendritic networks of actin filaments into actin bundles and that fascin1 is involved in this process (Svitkina et al. 2003; Vignjevic et al. 2006). We have demonstrated that overexpression of fascin1 induces membrane protrusions and increases cell motility of epithelial cells (Yamashiro et al. 1998). Likewise, fascin1 up-regulation has been reported to increase protrusive activity of colonic epithelial cells (Jawhari et al. 2003), as well as invasive activity of colon carcinomas (Hashimoto et al. 2005; Vignjevic et al. 2007). Conversely, Fascin1 knockdown has been reported to block filopodia assembly of B16F1 mouse melanoma cells (Vignjevic et al. 2006) and colon carcinoma cells (Hashimoto et al. 2005; Vignjevic et al. 2007), as well as dendrite formation of mature DCs (Al-Alwan et al. 2001; Ross et al. 2000; Ross et al. 1998). These observations strongly suggest that fascin1 plays a critical role in filopodia formation in vertebrates, and thus fascin1 would be essential for a variety of critical cell motility processes including directed cell migration, neurite extension and/or growth cone guidance in neurons. However, this notion has not been tested in whole animals.
We have generated fascin1 knockout mice in order to examine whether fascin1 is essential for such functions. Unexpectedly, fascin1 knockout mice were viable and fertile without any apparent developmental defects though they showed partial neonatal lethality.
MATERIALS AND METHODS
Generation of fascin1-deficient mice
Fascin1 knockout heterozygous mice with a genetic background of a 129 SvEvBrd and C57/BL6 hybrid were generated by Lexicon (Woodlands, TX) from an ES cell line (OST124903). In this ES cell line, the fascin1 gene (MGI:1352745) was disrupted by a retrovirus insertion (Zambrowicz et al. 1998) at the nucleotide 1,726, which is in the intron between the exon1 and exon2 (Fig. 1A). The heterozygous mice were backcrossed with wild-type C57/BL6 female mice for more than 14 generations. For each experiment with homozygous mice, their wild-type littermates were used as a control.
Figure 1.

Characterization of fascin1-deficient mice. a. Fascin1 gene disruption by a retrovirus insertion. An ES clone (OST124903) has a retrovirus insertion at the nucleotide 1,726 between exon1 and exon2, resulting in the disruption of fascin1 gene. b. The nucleotide number 1 is the translational start site of the mouse fascin1 gene (MGI:1352745). Western blot analysis to confirm that fascin1 KO mice do not express fascin1. Lane 1, Brain from wild-type mice; lane 2, Molecular weight markers; lanes 3-7 are from fascin1 KO mice (3, brain; 4, liver, 5, embryonic brain; 6 placenta; 7, spleen). c, RT-PCR analysis of wild type (+/+) and fascin1 KO (-/-) mouse tissues (lanes 1-6, brain; lanes 7-12, retina; lanes 13-18, testis; lanes 19-24, embryonic brain). Three different primer sets were used to detect fascin1 (labeled “b”), fascin2 (labeled “r”) and fascin3 (labeled “t”) mRNA. An actin primer set was used as a control. Note that both adult and embryonic brain tissues from fasin1-deficient mice did not express fascin2 or fascin3 paralogues.
RT-PCR
RT-PCR reactions were performed using a Supercript III one-step RT-PCR kit (Invitrogen, Carlsbad, CA). mRNAs were purified from brain, liver, eyeball and testis using a NucleoTrap mRNA purification kit (Clontech, Mountain View, CA). The following primer sets are used:5′-AGGTGTGCAATCCACTGCGTCC-3′ and 5′-GGGTCGATAGTCTCCGCGCAG-3′ for fascin1, 5′-AGGCCACAGCCACACAAGTCT-3′ and 5′-CAGGCAGGTCTGCATCAGCACG-3′ for retina fascin (fascin2), GCAATCATCGCTGACGGACACC-3′ and 5′-GTCTGCCAATAGTGTGCCACTCC-3′ for testis fascin (fascin3).
Brain section
Brain was dissected from fascin1 knockout mice, as well as littermate wild-type mice (both about 6 months old), and fixed with 4% paraformaldehyde for 24-48hrs. Brain sectioning was performed using a Vibratome 3000 with a thickness of 50μm. The specimen was stained with the Nissl staining method.
Preparation of primary culture of Dorsal Root Ganglions
Dorsal root ganglion (DRG) neurons were prepared form 1day-old newborn pups as described (Matsuda et al. 1996) with slight modification. Briefly, DRGs were treated with trypsin and dissociated by pipetting in DME medium with 10% FBS. Fast-adhering, non-neuronal cells were removed by plating DRGs on a normal cell culture dish. Non-adhering DRG neurons were recovered, plated on laminin-coated (20 μg/ml) coverslips that had been pre-coated with 20 μg/ml poly-D-lysine, and incubated for 1 or 2days in the presence of NGF to induce neurite extension.
Neurite outgrowth of explanted DRGs was also examined in collagen gels essentially as described [Lumsden, 1983 #4347]. Briefly, DRGs were submerged in 0.5ml of collagen MEM-medium containing 2 mg/ml collagen (Cohesion, Palo Alto, CA), 10% fetal bovine serum and NGF. After 3days, DRGs with neuron outgrowth were imaged with a Nikon TE300 microscope with a 4X (Plan Fluor, NA 0.13) phase-contrast objective lens.
Immunofluorescent microscopy
For fascin staining, cells were fixed with absolute methanol at −20°C, blocked with goat serum (Jackson laboratory, Bar Harbor, ME), and stained with anti-fascin mouse monoclonal antibody (Yamashiro-Matsumura and Matsumura 1986). FITC- or Rhodamin-labeled phalloidin was used for F-actin staining. Images were acquired using the Nikon TE300 microscope with a 60X (Plan Apo, NA 1.4) phase-contrast objective lens, CoolSnap-fx CCD camera, and IPLab software. Areas of growth cones of DRG neurons, as well as number of filopodia per growth cone and filopodia lengths, were measured by IPLab software. Image contrast and brightness were adjusted by Photoshop (Adobe, San Jose, CA).
Live cell imaging
Embryonic fibroblasts were isolated from mouse embryos (14dpi). All experiments were performed within 5 passages of primary cultures. For visualization of actin, fibroblasts plated on glass-bottom cultured dishes (Mattek, Ashland, MA) were transfected with GFP-actin one day before imaging. Cells were incubated in HEPES-buffered DME (Invitrogen) containing 10% fetal calf serum, 1/100vol of OxyFluor (Oxyrase, Mansfield, OH) and 10mM glucose, overlaid with mineral oil to prevent evaporation, and then placed in a temperature control incubator (MS200D, Narishige) to maintain the temperature at 37 °C. Three Z-section images with 0.2μm spacing were taken using a DeltaVision Image Restoration Microscope system (Applied Precision, Issaquah, WA) with 10sec intervals for 15min. Images were deconvoluted and processed with the SoftWoRx software (Applied Precision). Exposure times for imaging and settings for deconvolution were constant for all samples to be compared within any given experiment. After identifying filopodia showing dynamic extension and retraction, filopodia lengths, as well as extension and retraction rates and lifetime of filopodia, were measured using the SoftWorx software (retraction fibers were excluded from measurements). Projected images were shown for still image and movie presentations. Statistical analyses were performed using Student’s t-test (http://www.physics.csbsju.edu/stats/t-test.html).
RESULTS
Characterization of fascin1 KO mice
Using a gene disruption method based on a retrovirus insertion (Lexicon Genetics Inc), we have generated fascin1 KO mice (Fig. 1a). The homozygous mice were viable and fertile. This is surprising because a previous study reported that fascin1 antisense blocked neurite extension of PC12 cells (Edwards and Bryan 1995). Fascin1 deficiency was confirmed by genotyping of homozygous mice by PCR and Southern blotting (data not shown). Western blotting (Fig. 1b) revealed the complete lack of fascin1 expression in various tissues of fascin1 KO mice including adult and embryonic brain, liver, placenta and spleen. Using RT-PCR (Fig. 1c), we confirmed that other fascin gene products, fascin2 and fascin3, are not compensating fascin1 in these tissues. RT-PCR analyses also confirmed that embryonic mouse brain isolated from homozygous embryos (16 dpi) did not express fascin2 or fascin3 (Fig. 1c).
Mating with heterozygous mice produced wild, heterozygous and homozygous pups with normal Mendelian segregation, indicating that fascin1 deficiency is not embryonic lethal (Table 1). However, about half (48%, 15/31) of the homozygous pups died within 24-48 hr after birth while most (90%, 66/73) of heterozygous mice survived. While the exact reason of this partial neonatal death is currently unknown, their lungs frequently showed no sign of breathing. Trachea cartridge appeared normal, excluding a possibility of trachea defects for the breathing problem. Also, the pups that died within 24hr did not have milk in their stomachs. Once homozygous mice survived the initial 48 hr, their development appeared normal though they are frequently smaller than wild-type pups. It is worthy of note that the partial neonatal death was not observed with homozygous mice with a mixed genetic background. Mating with the initial heterozygous mice with a mixed genetic background of 129 SvEvBrd and C57/BL6 hybrid produced pups with normal Mendelian segregation without neonatal death.
Table 1.
Mendelian segregation of newborn pups and frequent early death of isogenic KO mice
| # of pups | # of pups died within 48hr. | |
|---|---|---|
| wild | 38 | 6 |
| Heterozygous | 73 | 6 |
| homozygous | 31 | 15 |
Fascin1-deficient mice do not fully extend posterior projections of the anterior commissure neurons
Because filopodia formation has been reported to play a vital role in neurite extension (Dent et al. 2007) and because neurons express a very high level of fascin1, we have examined, by gross anatomy, whether fascin1-deficiency affects gross brain anatomy. Nissl staining of serial coronal sections (50μm thickness) of wild and KO mouse brains (Fig. 2) did not reveal apparent defects except the following two points: First, the posterior region of the anterior commissure neuron (marked by “acp” in Fig. 2e, also indicated by arrows in Figs 2c & e of wild-type brain) was apparently missing in KO mouse brain with 100% penetrance (8 out of 8). The anterior regions of the same neuron (marked by “aca” in Figs 2a and b), on the other hand, were similar. Although we cannot exclude a possibility of mislocation or deformation of the posterior region at present, the serial sections appear to support the notion that the anterior commissure neuron of KO brain did not extend the posterior region (see supplemental figure 1). Second, KO mouse had larger lateral ventricle (marked by “LV”). The significance of these defects on brain functions, including behavior, remains to be investigated. Nonetheless, it is worthy of note that fascin1 is dispensable for brain development.
Figure 2.

Nissl staining of coronal sections of wild and fascin1-deficient mouse brains. a, c and e, wild-type brain; b, d, and f, fascin1 KO brain. Sections shown in a and b are anterior to those shown c and d (see supplemental figure 1 for serial sections). The dotted rectangles in panels c and d are enlarged to panels e and f, respectively. The posterior part of the anterior commissure neurons (acp, indicated by arrows in the panels c & e of wild-type brain) is missing in KO mouse brain. Also note that the lateral ventricle (LV, shown in the panels a and b) is larger in fascin1 KO brain. g, schematic drawing of a brain coronal section illustrating the positions of lateral ventricle (LV), caudate putamen (Cpu), anterior of anterior commissure (aca) and posterior of anterior commissure (acp).
Dorsal Root Ganglion (DRG) neurons can extend neurites in vitro without fascin1
To further test the role of fascin1 in neurite extension and filopodia assembly in neuronal growth cones, we isolated dorsal root ganglion (DRG) neurons and examined whether fascin1 deficiency affects neurite extension. As Fig. 3 shows, DRGs from homozygous mice were able to extend neurons upon NGF treatment as well as DRGs from wild type mice, indicating that fascin1 is dispensable for neurite extension. The lack of fascin1, however, altered the morphology of DRG neurons, as well as their growth cones. The axon of fascin1-deficient DRG neurons appeared less straight and frequently thinner (Fig. 3e and f). Growth cones of fascin1-deficient DRG were smaller than those of wild-type. Furthermore, filopodia in fascin1-deficient growth cones appeared fewer in the number and shorter in the length. Quantitative analyses using box plots (Fig. 4) confirmed this notion. Measurements of growth cone areas revealed that the median value of homozygous DRG neurons was less than half of that of wild-type controls (p<0.0001 for both 1-day and 2-day culture). The average number of filopodia per growth cone was also reduced by 70% after one-day of culture (p<0.0001), and was 78% reduced at day 2 of culture (p<0.0001). The average length of filopodia was also reduced by 33% after 1-day of culture (p=0.0046), although after 2-days of culture, homozygous DRGs displayed filopodia as long as wild-type DRGs (no statistical difference as judged by p value of 0.54).
Figure 3.
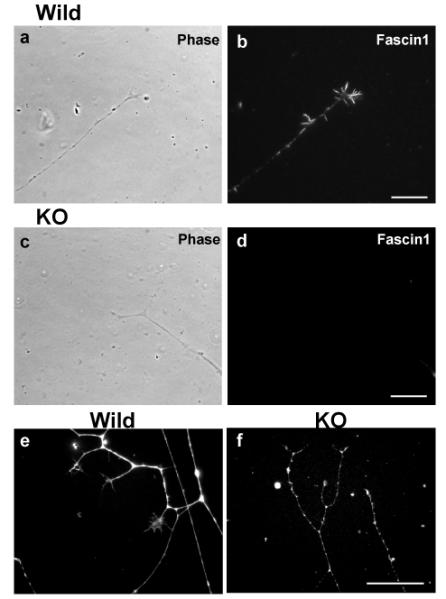
DRG neurons from fascin1-deficient mice were able to extend neurites. Fascin1 localization (b & d) and their corresponding phase contrast images (a & c) of DRG neurons from wild-type (a, b) and fascin1-deficient (c, d) mice. e and f, phalloidin staining of wild-type (e) and fascin1-deficient (f) DRG neurons. Bar, 10μm
Figure 4.

Statistical analyses of growth cone area, as well as filopodia number per growth cone and filopodia length of cultured DRG neurons. Growth cones of wild type and fascin1-deficient DRG were analyzed after for 1day- or 2day-culture (n=92 for 1day-culture of wild type, n=98 for 1day-culture of knockout, n=56 for 2day-culture of wild type, n=55 for 2day-culture of knockout).
We have further examined whether fascin1-deficiency affects neurite outgrowth in a three-dimensional environment. DRGs were embedded in collagen gels and neurite outgrowth was examined after 3day incubation. We found no significant alteration in outgrowth between wild-type and fascin1-deficient DRGs (see supplemental figure 2). Taken together, these data indicate that fascin1 is not essential for neurite extension, although the loss of fascin1 reduces the size of growth cones, as well as the number and length of filopodia.
Fascin1-deficient fibroblasts show reduced filopodia assembly with shorter lifetime
We have further examined whether fascin1-deficiency affects filopodia assembly of embryonic fibroblasts. Live cell imaging of GFP-actin allowed us to clearly distinguish dynamic filopodia from static retraction fibers. We measured lengths, lifetimes and extension and retraction rates of filopodia (retraction fibers were excluded from such measurements). These analyses revealed the following: First, filopodia were much less frequently observed with fascin1-deficient fibroblasts (3/18) than with wild type fibroblasts (7/14). Second, as the still images of Fig. 5a shows, the lifetime of filopodia of fascin1-deficient fibroblasts was much shorter than those of wild type fibroblasts. Third, filopodia lengths appeared shorter in fascin1-deficient fibroblasts. Statistical analyses confirmed these notions. As Fig. 5b shows, average lifetime were 106sec for wild-type and 44sec for fascin1-deficient filopodia, the difference of which is statistically significant as judged from the p value of 0.0006. Filopodia length was also reduced with statistical significance (p=0.012) from 4.0μm (wild-type) to 2.1μm (fascin1-deficient fibroblasts). On the other hand, extension and retraction rates of filopodia were not significantly different between wild-type and fascin1-deficient fibroblasts (mean values for the extension rates, 0.086μm/sec for wild-type, 0.115μm/sec for KO, p=0.064; retraction rates, 0.065μm /sec for wild-type, 0.081μm/sec for KO, p=0.32). These results indicate that fascin1-deficiency reduced the stability of filopodia in fibroblasts.
Figure 5.
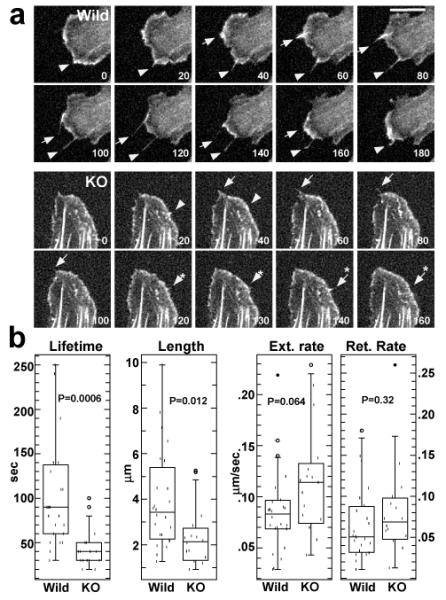
Live cell imaging of filopodia dynamics shown by wild type and fascin1-deficient embryonic fibroblasts. a. Still images taken from live cell imaging. Numbers are in sec. Note that wild-type filopodia are longer and the lifetimes of two filopodia are 180sec (indicated by arrowhead) and 140sec (arrow). In contrast, filopodia of knockout fibroblasts (indicated by arrow, arrow with asterisk, arrowhead, arrowhead with asterisk) are shorter and short-lived. Their lifetimes ranged from 20sec to 60sec. Bar, 10μm. b, box plot analyses of lifetimes, length, extension and retraction rates of filopodia (n=24 for wild-type, n=16 for knockout).
DISCUSSION
Fascin1 is dispensable for mouse development
We have found that fascin1-deficient mice are viable and fertile with no apparent developmental defects indicating that fascin1 is dispensable for mouse development. This was unexpected because in vitro studies have suggested that fascin1 is critical for filopodial assembly, and because filopodia are thought to be vital for various biological activities including directed cell migration, neurite extension, and interactions with other cells.
Do other actin-bundling proteins compensate for the loss of fascin1 in filopodia assembly?
There are two possibilities for the paucity of developmental defects: One is that other proteins compensate for fascin1’s role in actin bundling and filopodia assembly. The other possibility is that fascin1 plays a more specialized role in the health maintenance of adult mice, but may not play a major role in embryonic development. We favor the latter possibility: Fascin1 deficiency indeed inhibits filopodia formation in two different types of cells, suggesting that other actin-bundling proteins are not compensating the fascin1’s function in filopodia assembly. The growth cones of fascin1-deficient DRG are smaller with fewer and shorter filopodia (Figs. 3 & 4). Likewise, fascin1-deficient fibroblasts exhibited fewer filopodia with shorter length (Fig. 5). These phenotypes observed with knockout cells are very similar to those reported for fascin1 knockdown cells including mouse melanoma (Vignjevic et al. 2006) and colon cancer cells (Hashimoto et al. 2005; Vignjevic et al. 2007) (Hashimoto et al. 2007). The similarity of the phenotypes shown by fascin1 depletion by the two different methods, knockout and knockdown, further supports the notion that other actin-bundling proteins are not compensating for the lack of fascin1 in our knockout mice. It is also worthy of note that fascin1 does not show any homology with other actin-bundling proteins both in terms of the three-dimensional structure and the amino acid sequence (Bryan et al. 1993; Fedorov et al. 1999). This uniqueness of fascin1 structure is consistent with the notion that other actin-bundling proteins are unlikely to directly replace fascin1 in filopodia formation.
Filopodia in fascin1-deficient fibroblasts are unstable
Live cell image analyses revealed that fascin1-deficient embryonic fibroblasts exhibited shorter and less filopodia with the shorter lifetime (Fig. 5). The average lifetime and length were reduced from 106sec (wild type) to 44sec (knockout), and from 4μm (wild type) to 2.1μm (knockout), respectively. It should be noted, however, that the complete absence of fascin1 did not prevent filopodia formation. This basic ability of filopodia assembly shown by fascin1-deficient cells appears to be sufficient for directional movement of fibroblasts: No significant difference was seen in would healing between wild-type and fascin1-deficient fibroblasts (data not shown). These results suggest that fascin1 is not essential for filopodia formation, but rather fascin1 plays a role in stabilizing filopodia. This may be a reason why DRG neurons from fascin1-deficient mice were able to extend neurites as well as those of wild-type DRGs, and is likely to explain why fascin1 KO embryos can develop without major defects.
Conclusions
We have shown here that fascin1 is dispensable for mouse development. What could then be the physiological function of fascin1? It is possible that fascin1 knockout mice may have behavioral defects. Fascin1-deficient mice lacked the posterior part of the anterior commissure neuron with 100% penetrance (8/8). While the function of this neuron is currently unknown, the defect could alter some brain functions. Indeed, we have noticed that fascin1-deficient mice are frequently nervous.
Fascin1 is likely to play roles in the immune and pulmonary systems, as well as in cancer metastasis. We have recently found that antigen presenting dendritic cells from fascin1 KO mice show reduced motility (manuscript in preparation). The observation that most (70%) of neonatal death shown by fascin1 KO pups is due to a breathing problem suggests that certain lung function is compromised. Furthermore, as fascin1 expression is frequently induced upon metastasis of many cancers (Hashimoto et al. 2005; Vignjevic et al. 2007), our fascin1 KO mice would provide a useful tool to elucidate the roles of fascin1 in cancer invasion.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Barth Grant for his critical reading of the manuscript, Drs Robert Adelstein and Mary Ann Conti (NIH) for helping us to set up mouse colonies; and Dr. G. Shumyatsky (Rutgers) for the use of his Vibratome. This work was initially supported by grant from the American Heart Association (to SY), and later by the NCI grant, CA42742 (to FM) and Busch Memorial Fund.
References
- Adams JC. Roles of fascin in cell adhesion and motility. Curr Opin Cell Biol. 2004;16(5):590–6. doi: 10.1016/j.ceb.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Adams JC, Schwartz MA. Stimulation of fascin spikes by thrombospondin-1 is mediated by the GTPases Rac and Cdc42. J Cell Biol. 2000;150(4):807–22. doi: 10.1083/jcb.150.4.807. [see comments] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Alwan MM, Rowden G, Lee TD, West KA. Fascin is involved in the antigen presentation activity of mature dendritic cells. J Immunol. 2001;166(1):338–45. doi: 10.4049/jimmunol.166.1.338. [DOI] [PubMed] [Google Scholar]
- Bryan J, Edwards R, Matsudaira P, Otto J, Wulfkuhle J. Fascin, an echinoid actin-bundling protein, is a homolog of the Drosophila singed gene product. Proc Natl Acad Sci U S A. 1993;90(19):9115–9. doi: 10.1073/pnas.90.19.9115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cant K, Knowles BA, Mooseker MS, Cooley L. Drosophila singed, a fascin homolog, is required for actin bundle formation during oogenesis and bristle extension. J Cell Biol. 1994;125(2):369–80. doi: 10.1083/jcb.125.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Joussineau C, Soule J, Martin M, Anguille C, Montcourrier P, Alexandre D. Delta-promoted filopodia mediate long-range lateral inhibition in Drosophila. Nature. 2003;426(6966):555–9. doi: 10.1038/nature02157. [DOI] [PubMed] [Google Scholar]
- Dent EW, Kwiatkowski AV, Mebane LM, Philippar U, Barzik M, Rubinson DA, Gupton S, Van Veen JE, Furman C, Zhang J. Filopodia are required for cortical neurite initiation. Nat Cell Biol. 2007;9(12):1347–59. doi: 10.1038/ncb1654. others. [DOI] [PubMed] [Google Scholar]
- Duh F-M, Latif F, Weng Y, Geil L, Modi W, Stackhouse T, Matsumura F, Duan DR, Linehan WM, Lerman MI. cDNA cloning and expression of the human homolog of the sea urchin fascin and Drosophila singed genes which encodes an actin-bundling protein. DNA and Cell Biol. 1994;13:821–827. doi: 10.1089/dna.1994.13.821. others. [DOI] [PubMed] [Google Scholar]
- Edward RA, Herrera-Sosa H, Otto J, Bryan J. Cloning and expression of a murine fascin homolog from mouse brain. J.Biol.Chem. 1995;270:10764–10770. doi: 10.1074/jbc.270.18.10764. [DOI] [PubMed] [Google Scholar]
- Edwards RA, Bryan J. Fascins, a family of actin bundling proteins. Cell Motil Cytoskeleton. 1995;32(1):1–9. doi: 10.1002/cm.970320102. [DOI] [PubMed] [Google Scholar]
- Fedorov AA, Fedorov EV, Ono S, Matsumura F, Almo SC. Crystal structure of human fascin, an actin-crosslinking protein. 1999.
- Hashimoto Y, Parsons M, Adams JC. Dual actin-bundling and protein kinase C-binding activities of fascin regulate carcinoma cell migration downstream of Rac and contribute to metastasis. Mol Biol Cell. 2007;18(11):4591–602. doi: 10.1091/mbc.E07-02-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y, Skacel M, Adams JC. Roles of fascin in human carcinoma motility and signaling: prospects for a novel biomarker? Int J Biochem Cell Biol. 2005;37(9):1787–804. doi: 10.1016/j.biocel.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Holthuis JC, Schoonderwoert VT, Martens GJ. A vertebrate homolog of the actin-bundling protein fascin. Biochim Biophys Acta. 1994;1219(1):184–8. doi: 10.1016/0167-4781(94)90267-4. [DOI] [PubMed] [Google Scholar]
- Ishikawa R, Sakamoto T, Ando T, Higashi-Fujime S, Kohama K. Polarized actin bundles formed by human fascin-1: their sliding and disassembly on myosin II and myosin V in vitro. J Neurochem. 2003;87(3):676–85. doi: 10.1046/j.1471-4159.2003.02058.x. [DOI] [PubMed] [Google Scholar]
- Jawhari AU, Buda A, Jenkins M, Shehzad K, Sarraf C, Noda M, Farthing MJ, Pignatelli M, Adams JC. Fascin, an actin-bundling protein, modulates colonic epithelial cell invasiveness and differentiation in vitro. Am J Pathol. 2003;162(1):69–80. doi: 10.1016/S0002-9440(10)63799-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kureishy N, Sapountzi V, Prag S, Anilkumar N, Adams JC. Fascins, and their roles in cell structure and function. Bioessays. 2002;24(4):350–61. doi: 10.1002/bies.10070. [DOI] [PubMed] [Google Scholar]
- Matsuda S, Baluk P, Shimizu D, Fujiwara T. Dorsal root ganglion neuron development in chick and rat. Anat Embryol (Berl) 1996;193(5):475–80. doi: 10.1007/BF00185878. [DOI] [PubMed] [Google Scholar]
- Mosialos G, Birkenbach M, Ayehunie S, Matsumura F, Pinkus GS, Kieff E, Langhoff E. Circulating human dendritic cells differentially express high levels of a 55-kd actin-bundling protein. American J.Pathol. 1996;148:593–600. [PMC free article] [PubMed] [Google Scholar]
- Mosialos G, Yamashiro S, Baughman RW, Matsudaira P, Vara L, Matsumura F, Kieff E, Birkenbach M. Epstein-Barr virus infection induces expression in B lymphocytes of a novel gene encoding an evolutionarily conserved 55-kilodalton actin-bundling protein. J Virol. 1994;68(11):7320–8. doi: 10.1128/jvi.68.11.7320-7328.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto JJ, Kane RE, Bryan J. Formation of filopodia in coelomocytes: localization of fascin, a 58,000 dalton actin cross-linking protein. Cell. 1979;17(2):285–93. doi: 10.1016/0092-8674(79)90154-5. [DOI] [PubMed] [Google Scholar]
- Patterson J, O’Hare K. Structure and transcription of the singed locus of Drosophila melanogaster. Genetics. 1991;129:1073–1084. doi: 10.1093/genetics/129.4.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkus GS, Pinkus JL, Langhoff E, Matsumura F, Yamashiro S, Mosialos G, Said JW. Fascin, a sensitive new marker for Reed-Sternberg cells of Hodgkin’s Disease. Evidence for a dendritic or B cell derivation? Am J Pathol. 1997;150(2):543–62. [PMC free article] [PubMed] [Google Scholar]
- Poodry CA. In: Epidermis: morphology and development. Ashburner M, Wright TRF, editors. Academic Press; New York: 1980. pp. 141–224. [Google Scholar]
- Ross R, Jonuleit H, Bros M, Ross XL, Yamashiro S, Matsumura F, Enk AH, Knop J, Reske-Kunz AB. Expression of the actin-bundling protein fascin in cultured human dendritic cells correlates with dendritic morphology and cell differentiation. J Invest Dermatol. 2000;115(4):658–63. doi: 10.1046/j.1523-1747.2000.00112.x. [In Process Citation] [DOI] [PubMed] [Google Scholar]
- Ross R, Ross XL, Schwing J, Langin T, Reske-Kunz AB. The actin-bundling protein fascin is involved in the formation of dendritic processes in maturing epidermal Langerhans cells. J Immunol. 1998;160(8):3776–82. [PubMed] [Google Scholar]
- Saishin Y, Shimada S, Morimura H, Sato K, Ishimoto I, Tano Y, Tohyama M. Isolation of a cDNA encoding a photoreceptor cell-specific actin-bundling protein: retinal fascin. FEBS Lett. 1997;414(2):381–6. doi: 10.1016/s0014-5793(97)01021-1. [DOI] [PubMed] [Google Scholar]
- Svitkina TM, Bulanova EA, Chaga OY, Vignjevic DM, Kojima S-i, Vasiliev JM, Borisy GG. Mechanism of filopodia initiation by reorganization of a dendritic network. J. Cell Biol. 2003;160(3):409–421. doi: 10.1083/jcb.200210174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubb B, Mulholland DJ, Vogl W, Lan ZJ, Niederberger C, Cooney A, Bryan J. Testis fascin (FSCN3): a novel paralog of the actin-bundling protein fascin expressed specifically in the elongate spermatid head. Exp Cell Res. 2002;275(1):92–109. doi: 10.1006/excr.2002.5486. [DOI] [PubMed] [Google Scholar]
- Vasioukhin V, Bauer C, Yin M, Fuchs E. Directed actin polymerization is the driving force for epithelial cell-cell adhesion. Cell. 2000;100(2):209–19. doi: 10.1016/s0092-8674(00)81559-7. [DOI] [PubMed] [Google Scholar]
- Vignjevic D, Kojima S-i, Aratyn Y, Danciu O, Svitkina T, Borisy GG. Role of fascin in filopodial protrusion. J. Cell Biol. 2006;174(6):863–875. doi: 10.1083/jcb.200603013. %R 10.1083/jcb.200603013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignjevic D, Schoumacher M, Gavert N, Janssen KP, Jih G, Lae M, Louvard D, Ben-Ze’ev A, Robine S. Fascin, a novel target of beta-catenin-TCF signaling, is expressed at the invasive front of human colon cancer. Cancer Res. 2007;67(14):6844–53. doi: 10.1158/0008-5472.CAN-07-0929. [DOI] [PubMed] [Google Scholar]
- Vignjevic D, Yarar D, Welch MD, Peloquin J, Svitkina T, Borisy GG. Formation of filopodia-like bundles in vitro from a dendritic network. J Cell Biol. 2003;160(6):951–62. doi: 10.1083/jcb.200208059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro S, Yamakita Y, Ono S, Matsumura F. Fascin, an actin-bundling induces membrane protrusions and increases cell motility of epithelial cells. Mol. Biol. Cell. 1998;9(5):993–1006. doi: 10.1091/mbc.9.5.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro-Matsumura S, Matsumura F. Purification and characterization of an F-actin-bundling 55-kilodalton protein from HeLa cells. J Biol Chem. 1985;260(8):5087–97. [PubMed] [Google Scholar]
- Yamashiro-Matsumura S, Matsumura F. Intracellular localization of the 55-kD actin-bundling protein in cultured cells: spatial relationships with actin, alpha-actinin, tropomyosin, and fimbrin. J. Cell Biol. 1986;103:631–640. doi: 10.1083/jcb.103.2.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrowicz BP, Friedrich GA, Buxton EC, Lilleberg SL, Person C, Sands AT. Disruption and sequence identification of 2,000 genes in mouse embryonic stem cells. Nature. 1998;392(6676):608–11. doi: 10.1038/33423. [DOI] [PubMed] [Google Scholar]
- Zhang FR, Tao LH, Shen ZY, Lv Z, Xu LY, Li EM. Fascin expression in human embryonic, fetal, and normal adult tissue. J Histochem Cytochem. 2008;56(2):193–9. doi: 10.1369/jhc.7A7353.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


