Abstract
RET proto-oncogene encodes a receptor tyrosine kinase whose ligand is glial cell line-derived neurotrophic factor (GDNF), and its polymorphism at G691S juxtamembrane region (RETp) is a germline polymorphism. Cutaneous melanomas, particularly the desmoplastic subtype, are highly neurotropic; thus we sought to determine the frequency of RETp in cutaneous melanoma and its functional responsiveness to GDNF. RETp was assessed in 71 non-desmoplastic cutaneous melanomas (non-DMs) and 70 desmoplastic melanomas (DMs). Melanoma cell lines with RETp, RET wild-type (RETwt), BRAF V600E mutation (BRAFmt) or BRAF wild-type (BRAFwt) were assessed for functional activity. RETp frequency was significantly higher in DMs (61%) than in non-DMs (31%, P<0.001). BRAFmt was detected in only 11% of DMs. GDNF stimulation significantly amplified cell proliferation, migration, and invasion in RETp, but not in RETwt melanoma cells. GDNF stimulation of RETp cell lines enhanced phosphorylation of extracellular signal-regulated kinase (ERK) and Akt of the RET-RAS-RAF-ERK and RET-phosphatidylinositol 3-kinase (PI3K)-Akt pathways, respectively. GDNF response of RETp cells in signal transduction and other functional studies were not affected by BRAFmt. The study demonstrates that RETp are frequently found in cutaneous melanoma, particularly desmoplastic subtypes, and responds to GDNF inducing events favorable for tumor progression.
Keywords: RET, polymorphism, mutation, melanoma, desmoplastic, BRAF
BACKGROUND
Melanoma is of neuroectodermal origin and has a high propensity to invade and metastasize to neural-origin tissue, such as the central nervous system and brain, which often contributes to the poor prognosis (Hoon et al., 2001). Desmoplastic melanoma (DM) is a subtype of cutaneous melanoma with distinct clinico-pathologic characteristics that distinguish it from other cutaneous melanomas (Jaroszewski et al., 2001). DMs often arise in highly sun-exposed areas, especially the head and neck region, and have a greater propensity to invade nerves and have aggressive local growth (Livestro et al., 2005; Quinn et al., 1998). Reports have shown more frequent local recurrence of DMs compared to non-desmoplastic cutaneous melanomas (Busam, 2005), but there are no reported studies on the potential mechanisms involved in the neurotropism of DMs or non-DMs.
RET is a proto-oncogene that encodes a receptor tyrosine kinase (RTK) (Iwamoto et al., 1993; Takahashi et al., 1989; Takahashi et al., 1988; Zbuk & Eng, 2007) containing four cadherin-related motifs and a cysteine-rich region in the extracellular domain (Iwamoto et al., 1993; Takahashi, 2001). GDNF, a major ligand of RET, is a growth factor that facilitates the survival of dopaminergic neurons of the midbrain (Airaksinen & Saarma, 2002; Lin et al., 1993). GDNF binds to the extracellular domain of RET through the formation of a complex with glycosyl-phosphatidylinositol-anchored co-receptor (GFRα1-3), a member of the GDNF receptor family that can bind to GDNF and RET (Takahashi, 2001). Although the expression of GFRα3 is higher in DMs than non-DMs (Busam et al., 2005), it has not been linked to the neurotropism of DMs.
Activation of RET induces signaling through the RAS-BRAF-ERK, phosphatidylinositol 3-kinase (PI3K)-Akt, and p38 mitogen-activated protein kinase (MAPK) pathways that initiate various functions in cells (Takahashi, 2001). Activation of both the RET-RAS-BRAF-MEK-ERK and RET-PI3K-Akt pathways has been implicated in cell proliferation and survival, whereas the RET-PI3K pathway has been associated more frequently to cell motility _(Kodama et al., 2005; Takahashi, 2001). All cancer-related mutations of the RET gene in the cysteine-rich region or tyrosine kinase domain (intracellular domain) are ligand-independent and reportedly responsible for development of multiple endocrine neoplasia 2A and 2B, familial medullary thyroid carcinoma, and papillary thyroid carcinoma (Kondo et al., 2006; Runeberg-Roos & Saarma, 2007; Weber & Eng, 2008). G691S RET polymorphism (RETp) is a SNP alteration in exon 11 of the juxtamembrane region of RET that enhances the response of RET to GDNF as previously shown by our group in pancreatic cancer (Sawai et al., 2005); the RET G691S responsiveness to GDNF was assessed in pancreas cancer because of its known neurotropism. Because cutaneous melanomas, particularly DM, are highly neurotropic, we hypothesized that RETp may play a role in enhancing this functional behavior. In the present study, the objective was to investigate the frequency and functional activity of RETp in cutaneous melanoma.
Somatic BRAF mutations are well-documented and are frequently found in non-DMs (Davies et al., 2002). The most frequent BRAF mutation is a single substitution in exon 15, V600E which is a constitutive active form (Davies et al., 2002; Shinozaki et al., 2004). BRAF belongs to the RAF family of serine-threonine kinases and is a component of the RET-RAS-BRAF-MAPK kinase (MEK)-ERK signaling pathway (Melillo et al., 2005). This signaling pathway is a membrane-to-nucleus signaling system controlling cell proliferation and other functions in mammalian cells (Dhomen & Marais, 2007). Although V600E BRAF mutation (BRAFmt) is suggested to cause abnormal proliferation of melanoma cells (Sumimoto et al., 2004), its relationship to RETp in cutaneous melanoma is unknown.
In this study, the frequency of RETp in DMs and non-DMs was assessed. We found that RETp is frequently present in cutaneous melanomas, and more frequent in neural tissue invading desmoplastic melanomas. RETp was shown to be highly responsive to the ligand GDNF inducing various physiological activity. This is the first report describing the detection and functional activity of RETp in cutaneous melanomas.
RESULTS
Expression of RET, GFRα1, and GFRα3
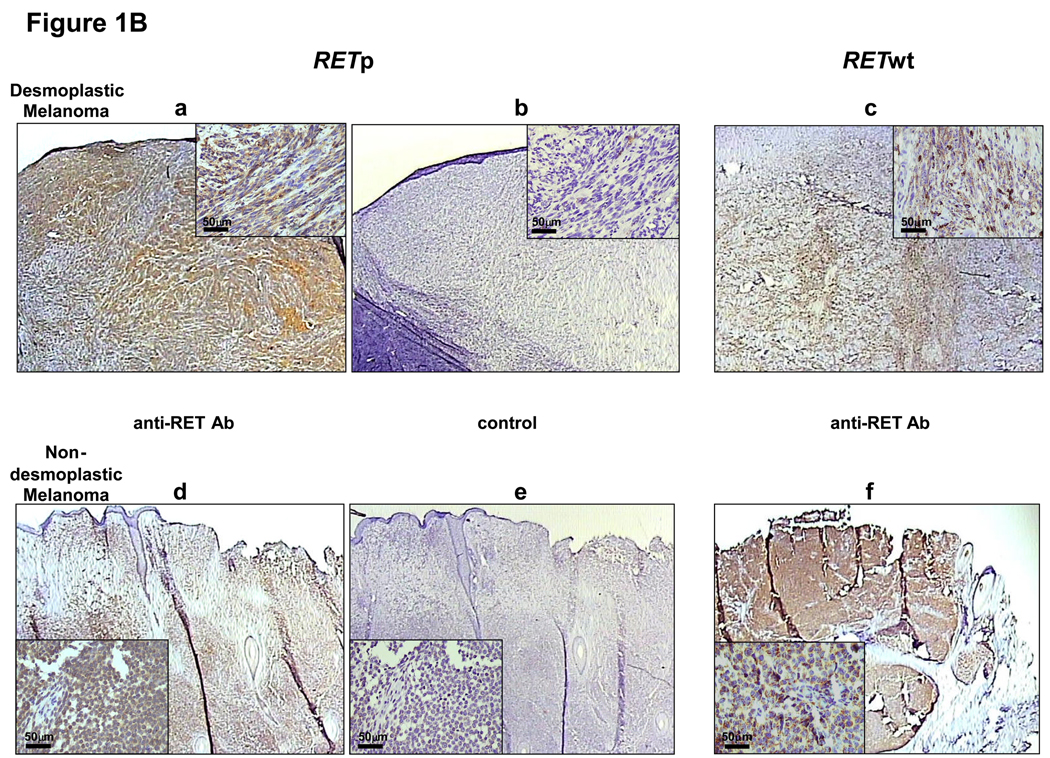
To confirm the existence of GDNF-RET signaling pathway in melanoma, the mRNA expressions of RET, GFRα1, and GFRα3 were analyzed in melanoma lines using quantitative real-time PCR (qRT). All melanoma lines expressed mRNA of RET, GFRα1, and GFRα3 (Figure 1A). The patterns of mRNA expression were independent of RETp presence (RETp status of cells shown below). Normal human brain tissues (B36929, B37876) served as positive controls for GFRα1 and GFRα3 mRNA, and normal human melanocytes (HMC) were used as a control. Immunohistochemistry (IHC) was performed to confirm the expression of the RET in melanoma tissues. IHC analysis of both non-DMs and DMs demonstrated that RET was expressed independently of RETp status in melanoma tissue (Figure 1B). There were no significant increase in RET protein expression in RET wild-type (RETwt) versus RETp melanomas. RET expression in melanoma cell lines was shown by Western blot analysis (Figure 1C).
Figure 1. RET expression in melanoma cells.
(A) mRNA expression of RET, GFRα1 and GFRα3 by real-time quantitative PCR in melanoma cell lines. The mRNA expression of  RET,
RET,  GFRα1, and
GFRα1, and  GFRα3, respectively is shown. Human normal melanocytes (HMC) and normal brain tissues (B36929, B37876) were used as controls. Copy numbers of mRNA were normalized using mRNA copy numbers of the house-keeping gene, GAPDH. Bars represent the mean of relative copies of duplicates. (B) RET expression detected by IHC staining in DM (a, c), non-DM (d, f) and respective negative controls (b, e). Anti-RET Ab and control show staining with anti-RET specific Ab and non-specific goat IgG, respectively. The magnification of the low-power and high-power figures are 20x and 400x, respectively. The scale bars in the high-power figures are shown. (C) Representative of RET protein expression by Western Blot analysis in melanoma cell lines and lymphocytes (control) used in experiments. RET protein expression in melanoma cell lines (ME1, ME3, ME5, ME7 and ME10) and lymphocyte was determined by immunoblotting. Loading control was probed by anti-β-actin.
GFRα3, respectively is shown. Human normal melanocytes (HMC) and normal brain tissues (B36929, B37876) were used as controls. Copy numbers of mRNA were normalized using mRNA copy numbers of the house-keeping gene, GAPDH. Bars represent the mean of relative copies of duplicates. (B) RET expression detected by IHC staining in DM (a, c), non-DM (d, f) and respective negative controls (b, e). Anti-RET Ab and control show staining with anti-RET specific Ab and non-specific goat IgG, respectively. The magnification of the low-power and high-power figures are 20x and 400x, respectively. The scale bars in the high-power figures are shown. (C) Representative of RET protein expression by Western Blot analysis in melanoma cell lines and lymphocytes (control) used in experiments. RET protein expression in melanoma cell lines (ME1, ME3, ME5, ME7 and ME10) and lymphocyte was determined by immunoblotting. Loading control was probed by anti-β-actin.
Analyses of RETp and BRAFmt
Since BRAF is downstream of the signaling pathway of RET, and BRAFmt may influence the signaling activation of RET (Davies et al., 2002; Takahashi, 2001), we investigated RETp (RET G691S polymorphism), BRAFmt (BRAF V600E mutation), RETwt, and BRAF wild-type (BRAFwt) in melanoma lines. Melanoma lines were assayed for both RETp (G691S) and BRAFmt (V600E): ME1 was RETp and BRAFmt; ME5 and ME7 were RETp and BRAFwt; ME3 and ME10 were RETwt and BRAFwt; and M16 was RETwt and BRAFmt. For RETp detection, the results were also validated using direct sequencing. The sequence of RETwt had a single polymorphic nucleotide change (GGT → AGT) to RETp (Supplemental Figure 1S).
RETp and BRAFmt DNA analyses were performed on histopathologically-verified surgical tissues of non-DMs (n=71) and DMs (n=70). The frequency of RETp was about twice as high in DMs (61%) as compared to non-DMs (31%) (P<0.001, Table 1). BRAFmt was significantly (P<0.001) higher in non-DM (39%) than DM tumors (11%). The reduced frequency of BRAFmt in DMs was striking and has important implications as to the role of BRAFmt in highly sun-exposed sites of cutaneous melanomas overall. Details of RETp and BRAFmt analysis of primary and metastatic tumors were assessed (Table 1). BRAFmt was detected more frequently in metastatic than in primary tumors in both non-DMs and DMs. However, this was not the case in RETp for non-DMs and DMs.
Table 1.
RETp and BRAFmt in Non-desmoplastic Cutaneous and Desmoplastic Melanomas
| Melanoma type | RETp | BRAFmt | |
|---|---|---|---|
| Cutaneous | All (n=71) | 31% | 39% |
| Primary (n=34) | 38% | 29% | |
| Metastatic (n=37) | 24% | 49% | |
| Desmoplastic | All (n=70) | 61% | 11% |
| Primary (n=51) | 63% | 10% | |
| Metastatic (n=19) | 58% | 16% | |
Effect on GDNF induced proliferation
The effects of RETp and BRAFmt on GDNF-induced cell proliferation were analyzed. A 48-hr exposure to GDNF enhanced proliferation of RETp melanoma cells but not RETwt melanoma cells (Figure 2A). GDNF did not differentially enhance the proliferation of BRAFmt and BRAFwt cells (Figure 2A). MTT assay results were confirmed by a direct cell-counting assay performed 72hr after administration of GDNF (Figure 2B). We selected 25ng/ml of GDNF as the optimal working dose for assessing cell proliferation (Figure 2A).
Figure 2. Cell proliferation by GDNF.
(A) Proliferation of melanoma cells was analyzed using the MTT assay. Cells were treated with  5 ng/ml,
5 ng/ml,  25 ng/ml, or
25 ng/ml, or  50 ng/ml of GDNF in serum-free medium for 48hrs.
50 ng/ml of GDNF in serum-free medium for 48hrs.  Delivery agent-treated control. Bars ± SD show increase (fold) over each control cell. *, **, ***, P<0.05 vs each control. Bars ± SD without asterisks indicate no significant change compared to each control. (B) Proliferation of melanoma cells was analyzed using direct cell counting; cells were treated with 50 ng/ml of GDNF in serum-free medium for 72hrs.
Delivery agent-treated control. Bars ± SD show increase (fold) over each control cell. *, **, ***, P<0.05 vs each control. Bars ± SD without asterisks indicate no significant change compared to each control. (B) Proliferation of melanoma cells was analyzed using direct cell counting; cells were treated with 50 ng/ml of GDNF in serum-free medium for 72hrs.  50 ng/ml of GDNF;
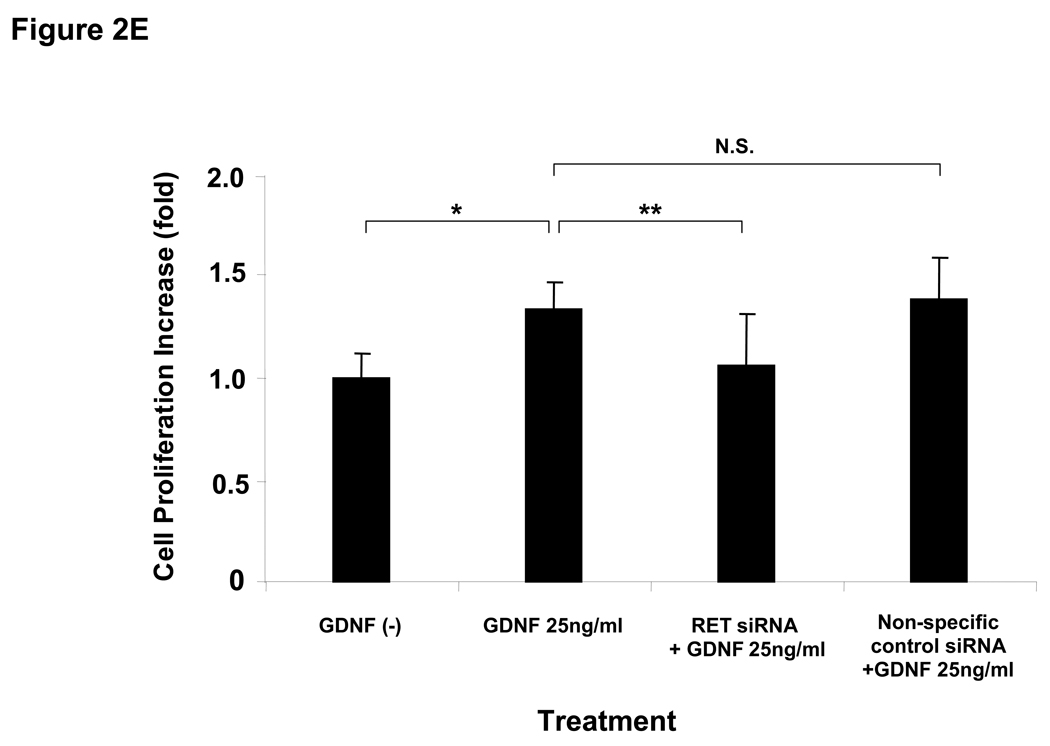
50 ng/ml of GDNF;  delivery agent-treated control. Bars + SD show increase (fold) over each control cell. *, **, ***, P<0.05 versus each control. (C) In the blocking assay, ME7 cells were pretreated with PD98059 or wortmannin for 60 min before 25 ng/ml of GDNF treatment for 72hrs. Bars ± SD show increase (fold) compared to delivery agent-treated control (without GDNF). *, **, ***, P<0.05. (D) mRNA expression of RET suppressed by RET specific siRNA 24 (left panel) and 48 (right panel) hrs after transfection in ME1. *, **, P<0.05. Copy numbers of RET mRNA were normalized using mRNA copy numbers of the house-keeping gene, GAPDH, and expressed as relative copies. (E) GDNF (25 ng/ml) was administrated to ME1 48hrs after transfection of RET specific siRNA or non-specific siRNA. Bars ± SD show increase (fold) compared to delivery agent-treated control (without GDNF). *, **, P<0.05.
delivery agent-treated control. Bars + SD show increase (fold) over each control cell. *, **, ***, P<0.05 versus each control. (C) In the blocking assay, ME7 cells were pretreated with PD98059 or wortmannin for 60 min before 25 ng/ml of GDNF treatment for 72hrs. Bars ± SD show increase (fold) compared to delivery agent-treated control (without GDNF). *, **, ***, P<0.05. (D) mRNA expression of RET suppressed by RET specific siRNA 24 (left panel) and 48 (right panel) hrs after transfection in ME1. *, **, P<0.05. Copy numbers of RET mRNA were normalized using mRNA copy numbers of the house-keeping gene, GAPDH, and expressed as relative copies. (E) GDNF (25 ng/ml) was administrated to ME1 48hrs after transfection of RET specific siRNA or non-specific siRNA. Bars ± SD show increase (fold) compared to delivery agent-treated control (without GDNF). *, **, P<0.05.
The pathway for GDNF-induced proliferation was confirmed by using agents that block components of the two important pathways for RET signaling. PD98059 is a MEK1 inhibitor that targets the RET-RAS-BRAF-MEK-ERK pathway. Wortmannin is a PI3K inhibitor that targets RET-PI3K-Akt pathway. PD98059 treatment completely suppressed GDNF-induced proliferation. Wortmannin partially suppressed GDNF-induced proliferation in ME7 (RETp/BRAFwt) (Figure 2C). This suggested that the RET-RAS-BRAF-MEK-ERK signaling pathway plays a more predominant role than the RET-PI3K-Akt pathway with respect to GDNF-induced cell proliferation in RETp melanoma cells. To exclude the possibility that cross-talk signaling pathways via other receptors are involved in GDNF activation of RET signaling pathways, we transfected small interfering RNA (siRNA) against RET in RETp cells (ME1), and GDNF induced cell proliferation was assessed. The RET siRNA significantly (P<0.05) reduced expression of RET mRNA 24 and 48hrs after transfection by 77% and 76%, respectively compared to the vehicle-treated cells (control) (Figure 2D). The non-specific siRNA control did not significantly affect mRNA expression of RET (Figure 2D). RET siRNA significantly (P<0.05) suppressed > 80% of cell proliferation induced by GDNF (25ng/ml) 48hrs after stimulation, whereas non-specific control siRNA did not (Figure 2E). These results suggest that cross-talk signaling through other receptors is not involved in the RET pathways activated by GDNF.
GDNF effect on RETp cell migration and invasion
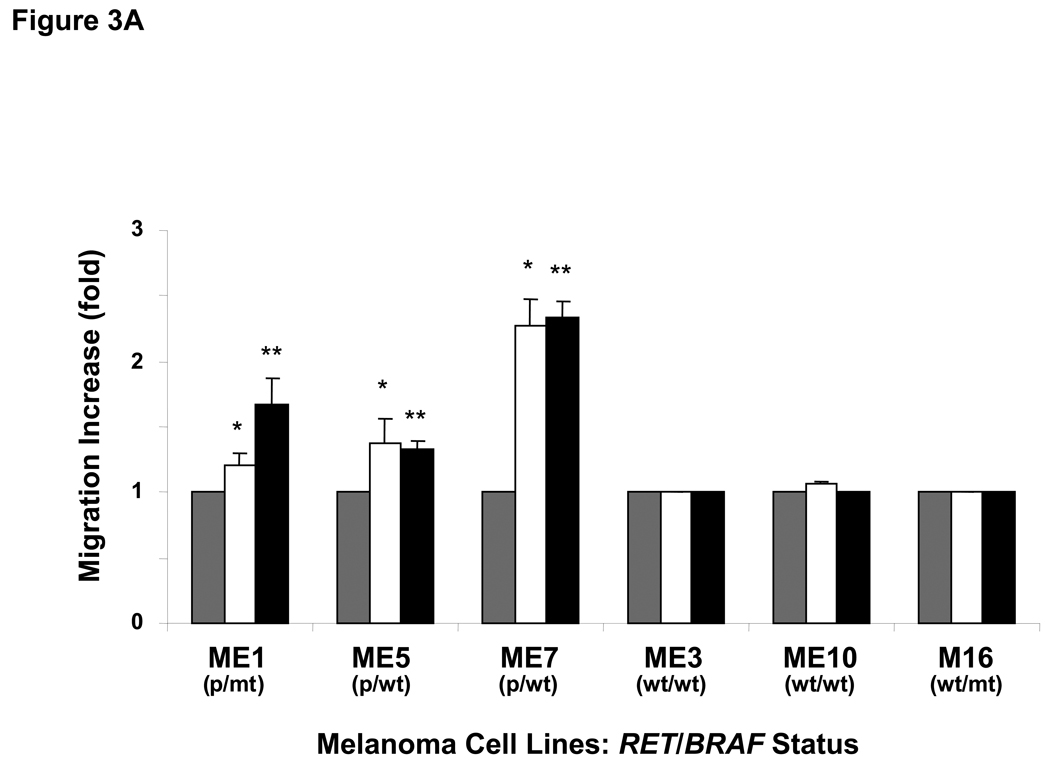
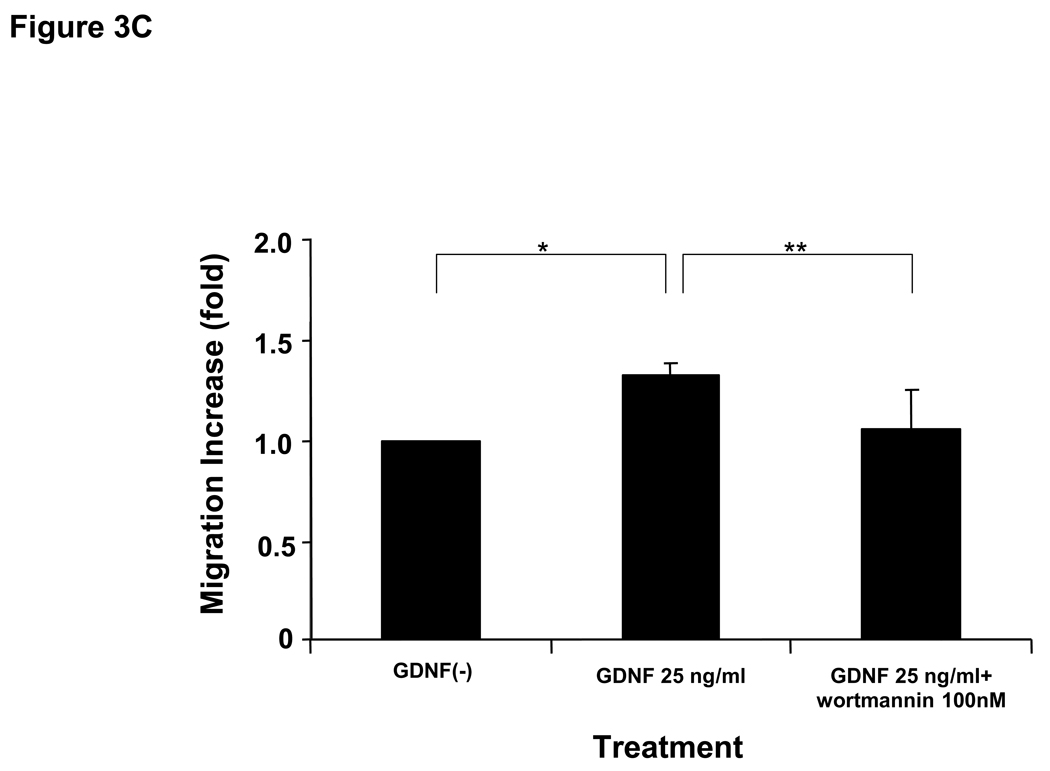
Next, to assess whether RETp affects cell migration or invasion induced by GDNF via the RET-PI3K pathway, migration and invasion assays were performed using microporous-membrane and collagen-coated chambers, respectively. RETp cells showed more vigorous migration and invasion than RETwt cells after GDNF stimulation (Figure 3A, B). BRAFmt cells demonstrated no difference on cell migration and invasion after GDNF treatment (Figure 3A, B). Wortmannin significantly suppressed cell migration induced by GDNF in ME5 (RETp/BRAFwt) (Figure 3C), whereas PD98059 did not (Figure 3D). These results suggest that the RET-PI3K signaling pathway plays an important role in cell motility induced by GDNF in RETp melanoma cells. The invasion ability of ME1 (RETp/BRAFmt) enhanced by GDNF was significantly suppressed with RET-specific antibody (Ab) (Figure 3E). Non-specific purified anti-goat IgG was used as a control and did not show any effects (Figure 3E). These results confirmed that cross-talk signaling pathways via other receptors are not significantly involved in GDNF activation of RET signaling pathways.
Figure 3. Cell migration and invasion by GDNF.
(A) Migration of melanoma cells was analyzed using a Transwell® chamber. Cells were treated with  5 ng/ml or
5 ng/ml or  25 ng/ml GDNF for 48hrs.
25 ng/ml GDNF for 48hrs.  Delivery agent-treated control. Bars ± SD show increase (fold) over each control cell. *, **, P<0.05 versus each control. Bars ± SD without asterisks indicate no significant change compared to each control. (B) Invasion of melanoma cells was analyzed using the QCM™ Collagen-based Invasion Assay. Cells were treated with
Delivery agent-treated control. Bars ± SD show increase (fold) over each control cell. *, **, P<0.05 versus each control. Bars ± SD without asterisks indicate no significant change compared to each control. (B) Invasion of melanoma cells was analyzed using the QCM™ Collagen-based Invasion Assay. Cells were treated with  25 ng/ml of GDNF for 60hrs.
25 ng/ml of GDNF for 60hrs.  Delivery agent-treated control. Bars ± SD show increase (fold) over each control cell. *, **, P<0.05 versus each control. In blocking assays, ME5 cells were pre-treated with (C) wortmannin or (D) PD98059 for 60 min before 25 ng/ml of GDNF treatment for 48hrs. Bars ± SD show increase (fold) of migration compared to delivery agent-treated control (without GDNF). *, **, P<0.05 for (C) and * P<0.05 for (D). (E) In the blocking assay, ME1 cells were pre-treated with RET specific Ab or non-specific anti-goat IgG (control) for 60 min before 25 ng/ml of GDNF treatment for 60hrs. Bars ± SD show increase (fold) of invasion compared to delivery agent-treated control (without GDNF) . *, **, P<0.05. Error bars ± SD.
Delivery agent-treated control. Bars ± SD show increase (fold) over each control cell. *, **, P<0.05 versus each control. In blocking assays, ME5 cells were pre-treated with (C) wortmannin or (D) PD98059 for 60 min before 25 ng/ml of GDNF treatment for 48hrs. Bars ± SD show increase (fold) of migration compared to delivery agent-treated control (without GDNF). *, **, P<0.05 for (C) and * P<0.05 for (D). (E) In the blocking assay, ME1 cells were pre-treated with RET specific Ab or non-specific anti-goat IgG (control) for 60 min before 25 ng/ml of GDNF treatment for 60hrs. Bars ± SD show increase (fold) of invasion compared to delivery agent-treated control (without GDNF) . *, **, P<0.05. Error bars ± SD.
Phosphorylation of ERK1/2 and Akt by GDNF
To assess the effect of RETp activation on downstream factors of the RET signaling pathway, we analyzed whether RETp or BRAFmt enhanced phosphorylation of ERK1/2 and Akt using Western blotting since RET-RAS-BRAF-MEK-ERK and RET-PI3K-Akt are the major signaling pathways activated by GDNF stimulation leading to cell proliferation (Takahashi, 2001). In a representative cell line, ME3 (RETwt/BRAFwt), GDNF showed weak phosphorylation of ERK1/2 and Akt. In contrast, GDNF strongly phosphorylated ERK1/2 and Akt within 15min after treatment in ME1 (RETp/BRAFmt), ME7 and ME5 (RETp/BRAFwt); and the phosphorylation of ERK1/2 and Akt lasted for 60min (Figures 4A–D). These results confirm that RETp enhanced the response of both RET-RAS-BRAF-MEK-ERK and RET-PI3K-Akt signaling pathways after GDNF stimulation.
Figure 4. Phosphorylation of ERK1/2 and Akt by GDNF.
(A) Protein expression of ERK1/2 and phosphorylated ERK1/2 after stimulation of GDNF in ME 3 (RETwt/BRAFwt), ME1(RETp/BRAFmt), and ME7 (RETp/BRAFwt), respectively. Cells were treated with 25 ng/ml of GDNF for 5, 15, 30, 45, or 60 minutes in serum-free medium. Non-phosphorylated ERK1/2 was used as a loading control. (B) Increase (%) in blot density of phosphorylated ERK1/2 over each delivery agent-treated control (GDNF(-));  ME3;
ME3;  ME1;
ME1;  ME7. (C) Protein expression of Akt and phosphorylated Akt after stimulation of GDNF in ME3 (RETwt/BRAFwt), ME1(RETp/BRAFmt), and ME5 (RETp/BRAFwt), respectively. Cells were treated with 25 ng/ml of GDNF for 5, 15, 30, 45, or 60 minutes in serum-free medium. Non-phosphorylated Akt was used as the loading control. (D) Increase (%) of blots density of phosphorylated Akt over each delivery agent-treated control (GDNF(-)).
ME7. (C) Protein expression of Akt and phosphorylated Akt after stimulation of GDNF in ME3 (RETwt/BRAFwt), ME1(RETp/BRAFmt), and ME5 (RETp/BRAFwt), respectively. Cells were treated with 25 ng/ml of GDNF for 5, 15, 30, 45, or 60 minutes in serum-free medium. Non-phosphorylated Akt was used as the loading control. (D) Increase (%) of blots density of phosphorylated Akt over each delivery agent-treated control (GDNF(-)).  ME3;
ME3;  ME1;
ME1;  ME5.
ME5.
Actin polymerization in melanoma cells by GDNF
Morphological alterations occurred during migration of melanoma cells after GDNF treatment. Actin polymerization on the cell surface was observed to confirm cell motility induced by GDNF in RETp cells. Based on the migration assays, we selected 10ng/ml of GDNF for assessment of cell motility. Actin polymerization, such as filopodia, necessary for cell migration, was visualized using Alexa Fluor® 568 phalloidin staining. Administration of GDNF induced filopodia in RETp cell lines (ME1, ME7), but not in the RETwt cell line (ME3) (Supplemental Figure 2S). The findings support the cell migration/invasion assays.
DISCUSSION
RET gene expression has been detected primarily in human tumors of neural crest origin, such as neuroblastoma, pheochromocytoma, and medullary thyroid carcinoma (Takahashi, 2001). RETp was originally found in radiation-induced thyroid tumors and sporadic medullary thyroid cancers (Bounacer et al., 2002; Elisei et al., 2004). In this report, we identified high frequency of RETp in cutaneous melanomas, particularly and more frequently in DMs (non-DMs: 31%, DMs: 61%), whereas RETp has been detected in only 15–20% of the normal population (Bounacer et al., 2002; Ceccherini et al., 1994; Elisei et al., 2004; Stephens et al., 2005). As not all DMs are neurotropic, we did not find RETp in all DMs. Previously, it was demonstrated that the G691S RETp was present in pancreas and thyroid cancers (Elisei et al., 2004; Sawai et al., 2005). Only certain RET-expressing tissues, such as the thyroid gland, have been shown to have RET gene rearrangements that lead to physiological changes (Airaksinen & Saarma, 2002; Runeberg-Roos & Saarma, 2007).
This is the first report demonstrating GDNF’s significant effects in promoting proliferation, migration, and invasion of RETp versus RETwt melanoma cells. These findings suggest that RETp melanoma may be more aggressive when in the vicinity of GDNF releasing neural and other tissues. Peripheral neural tissues secrete GDNF and are invaded by cutaneous melanomas. Brain metastasis is one of the major causes of death for melanoma patients (Selek et al., 2004); GDNF secreted by glial cells in the brain could significantly contribute to proliferation and migration of RETp melanoma cells. The observation that RETp is more frequent in DMs and responsive to the neurotrophic factor GDNF in non-DMs suggests that this RTK may play a significant role in melanoma progression.
RETp-GDNF enhanced and prolonged the phosphorylation of the ERK and Akt. PD98059, a MEK1 inhibitor, significantly blocked the cell proliferation induced by GDNF, whereas wortmannin, a PI3K inhibitor, suppressed GDNF-enhanced proliferation of RETp cells to some extent. These results suggested that both pathways of RET-RAS-BRAF-MEK-ERK and RET-PI3K-Akt (Kodama et al., 2005) can be activated through RETp-GDNF induction and are inline with the results of proliferation assays, in which GDNF only enhanced cell proliferation in RETp cells. The results of migration and invasion assays showed that RET-PI3K pathway played a significant role in GDNF induced cell motility. A recent report has shown that Akt-induced phosphorylation plays an integral role in the vertical growth of melanoma (Govindarajan et al., 2007). It is possible that GDNF induced cell motility is the result of activation of both RET-PI3K-RAC and RET-PI3K-Akt pathways. The assessment of the RET-RAS-BRAF-MEK-ERK pathway becomes complicated as it is known that signal transduction through the BRAFmt (V600E), a member of this pathway, is quite frequent in melanomas; signal transduction through NRAS mutation occurs to a much lesser extent (Curtin et al., 2006). We demonstrated that GDNF stimulation of RETp cells was not significantly influenced by BRAFmt presence.
We demonstrated that BRAFmt in melanoma cells does not significantly effect GDNF-induced proliferation, migration and invasion.. Although BRAFmt phosphorylates downstream factors such as MEK or ERK without ligand stimulation (Davies et al., 2002; Satyamoorthy et al., 2003), GDNF induced more intense phosphorylation of ERK1/2 in RETp/BRAFmt cells (ME1). This suggests that BRAFmt only partially phosphorylates the ERK pathway and its downstream factors, and consequently, cells bearing BRAFmt may not exhibit ERK phosphorylation after GDNF stimulation which is compatible with our results. These data also confirm our previous finding that BRAFmt is more frequent in metastasis than primary melanomas (Shinozaki et al., 2004). Our studies suggest that BRAFmt is not a significant progression factor for all types of cutaneous melanomas. Linkage of sun exposure to BRAFmt may not be valid for all cutaneous melanomas since DMs are known to be more frequently found in highly sun-exposed areas (Quinn et al., 1998). DMs had a significantly lower level (>3 fold) of BRAFmt compared to non-DMs. This finding supports a previous report on BRAFmt in DMs in a small cohort of patients (Davison et al., 2005).
There are several new therapeutic agents of RTK inhibitors affecting RET in clinical trials. Sorafenib (BAY 43-9006, serine/threonine kinase RAF1, and BRAF inhibitor) and semaxanib (SU5416, vascular endothelial cell growth factor receptor inhibitor) are RTK inhibitors that can block RET signaling and suppress growth of RETp thyroid cancers (Carlomagno et al., 2006; Mologni et al., 2006). Sorafenib directly suppresses the kinase activity of RET, and promotes lysosomal degradation of RET protein (Plaza-Menacho et al., 2007). RETp melanomas may be appropriate targets for using these RTK inhibitors, particularly those showing neurotropism. Our findings of melanoma cells with RETp/wt and BRAFmt/wt in combination suggest that some of the current clinical trials with these inhibitors may be working through the RETp pathway. To date, the association of BRAFmt in melanoma with response to Sorafenib alone has not been demonstrated.
RETp could be a key factor in the proliferation and invasion of malignant melanoma cells, and its high incidence in DM may explain why this malignancy often exhibits neurotropism. In our study, inhibition of RET signaling suppressed all proliferation and invasion in melanomas. This suggests that RETp could be a new RTK target for treatment of malignant melanomas. Regimens based on RTK inhibitors selective for RET may be particularly promising when, unfortunately, metastatic melanoma therapeutic treatments beyond surgery have had limited success to date.
MATERIALS AND METHODS
Cell lines
All 11 melanoma lines (ME1, ME2, ME3, ME5, ME7, ME8, ME10, ME13, ME20, M16, and M20) used in the present study were established and characterized at the John Wayne Cancer Institute (JWCI) and cultured as described previously (Goto et al., 2008; Shinozaki et al., 2007). Six cell lines (ME1, ME3, ME5, ME7, ME10, and M16) were selected for functional assays based on the presence of RETp, RETwt, BRAFmt, and BRAFwt. MIAPaCa-2, PANC-1 (human pancreatic cancer), and MCF-7 (human breast cancer) were obtained from American Type Culture Collection. Human normal melanocytes (HMC) and normal brain tissues (B36929, B37876) were purchased from Cascade Biologics, Inc. and Cooperative Human Tissue Network, respectively.
Tissue specimens
Formalin-fixed, paraffin-embedded tumor tissues were obtained from non-DM (n=71) and DM patients (n=70). All patients had undergone surgical treatment for primary or metastatic melanoma at Saint John’s Health Center (SJHC) or Sydney Melanoma Unit (SMU), and the use of human specimens was approved by the respective institutional review boards.
DNA extraction
For the preparation of melanoma line DNA, cells were lysed in DNAzol® Genomic DNA Isolation Reagent (Molecular Research Center), and DNA precipitated by 100% ethanol was measured by UV spectrophotometry (Fujiwara et al., 1999). For the preparation of melanoma tissue DNA, 8µm sections were cut from formalin-fixed, paraffin-embedded blocks and mounted on glass slides. Sections were stained with H&E for microscopic analysis to confirm histopathology. Tumor and normal tissues were isolated separately by microdissection under light microscopy, as previously described (Umetani et al., 2005). Dissected tissues were digested with Proteinase K (QIAGEN)-containing lysis buffer at 50°C overnight and inactivated by heat at 75°C for 15min. DNA was then purified with phenol-chloroform-isoamyl-alcohol, precipitated by ethanol, and quantified by PicoGreen® dsDNA Assay Kit (Invitrogen) for double-stranded DNA (Umetani et al., 2005).
mRNA expression analysis
The sequence of primers and fluorescence resonance energy transfer probes (FRET) used for the quantitative real-time PCR (qRT) assay of RET, GFRα1, GFRα3, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) are shown in Supplemental Table 1S. Reverse transcription (RT) reactions were performed with Moloney murine leukemia virus reverse transcriptase (Promega) with oligo (dT) primer. The qRT assay was performed in an iCycler iQ® Real-Time Thermocycler Detection System (Bio-Rad Laboratories). 5µl of cDNA generated from 250ng of total RNA through RT was added to a 96-well plate (Fisher) in which 0.5µM of each primer, 0.3µM FRET probe, 1U of AmpliTaq Gold® polymerase (Applied Biosystems), and PCR reagents were added (Koyanagi et al., 2005). Amplification of samples consisted of a precycling hold at 95°C for 9 min, then 45 cycles of denaturation at 95°C for 1min, annealing for 1min (at 55°C for GAPDH, 58°C for RET, 63°C for GFRα1, 58°C for GFRα3), and extension at 72°C for 1 min. Specific plasmid controls for each marker were synthesized as described previously (Goto et al., 2006), and copy number calibration curves was generated using threshold cycles of six serially diluted of plasmid templates (101–106). To quantify the copy number, the cycle time (Ct) was interpolated from the calibration curve for each sample and mRNA copy number was calculated using Bio-Rad software. GAPDH was used as a control housekeeping gene and the relative mRNA copies were obtained as absolute mRNA copies of each gene / absolute mRNA copies of GAPDH (Koyanagi et al., 2005). Each assay was performed at least twice, and mean copy numbers were used for analysis.
RETp and BRAFmt analysis
We used a peptide nucleic acid (PNA) clamped method to detect RETp and BRAFmt polymorphism in melanoma lines and paraffin-embedded melanomas as previously described (Sawai et al., 2005; Shinozaki et al., 2007). Briefly, PCR was performed with the primers, and probes (LNA and PNA) shown as RET (#1) and BRAF in Supplemental Table 1S. The PCR assay was performed using iCycler iQ® real-time PCR. Genomic DNA (≥2.5 ng) was applied to a final volume of 25µl containing each PCR primer, probe (PNA and LNA in BRAFmt detection), each dNTP, MgCl2, PCR buffer, and AmpliTaq Gold® Polymerase. PCR for RET was subjected to a precycling hold at 95°C for 12min, followed by 55 cycles at 94°C for 1min, 70°C for 50sec, 58°C for 50sec, and 72°C for 1min. PCR for BRAF was subjected to a precycling hold at 95°C for 10min, followed by 45 cycles at 95°C for 1min, 72°C for 50sec, 53°C for 50sec, and 72°C for 1min. MIAPaCa-2 and PANC-1 were used as RETp and RETwt controls, respectively (Sawai et al., 2005). DNA of ME2 and MCF7 were used as BRAFmt and BRAFwt controls, respectively (Shinozaki et al., 2007).
Sequencing
To confirm the results of the PNA clamp assay, we performed direct sequencing on representative samples, as previously described (Sawai et al., 2005). RET coding regions were amplified by PCR using genomic DNA of melanoma cells and tumor tissues. The primer pair flanking exon 11 of RET genomic DNA was designed as RET (#2) (Supplemental Table 1S). PCR sequencing fragments were applied and read with CEQ™ 8000XL Genetic Analysis System (Beckman Coulter) and analyzed by the CEQ™ 8000XL Series Genetic Analysis System Software (version 8.0).
RET IHC analysis
Sections (5µm) were obtained from archived formalin-fixed paraffin-embedded non-DMs and DMs. After deparaffinization, endogenous peroxidase activity was quenched by 0.3% H2O2 and non-specific binding sites were blocked with 5% BSA. Sections were treated with boiling citrate buffer for heat-induced epitope retrieval. Goat anti-human polyclonal RET-specific Ab (R&D Systems) or non-specific goat IgG (Santa Cruz Biotechnology, Inc.) as a control (15µg/ml in blocking buffer), was added to the sections for immunostaining and incubated at 4°C for 1hr. Ab binding sites were detected by avidin-biotin peroxidase complex solution (LSAB® Kit) and 3,3’-diaminobenzidine as a chromogen (DAKO). Counterstaining was performed with hematoxylin and assessed by light microscopy.
Western blotting
Cells were washed twice with ice-cold phosphate-buffered saline (PBS) and dissolved in solubilizing buffer (pH 7.5, 20mM Tris-HCL, 12.5mM β-glycerophosphate, 2mM EGTA, 10mM NaF, 1mM benzamide, 1% NP-40, 0.1% SDS, 0.5% Sodium deoxycholate, 1mM Na3VO4, and protease inhibitor mix (Roche Diagnostics)) after treatment with 25µg human recombinant GDNF (Chemicon International). Each aliquot of protein (10µg) was subjected to Western blotting analysis. After electrophoresis on 12.5% polyacrylamide gels, the protein was transblotted to Hybond™-P (Amersham Life Sciences, Inc.). The blots were blocked with 5% non-fat dry milk, and then incubated with anti-ERK1/2 rabbit polyclonal Ab or anti-phosphorylated ERK1/2 rabbit polyclonal Ab (1:1000, Phosphoplus®p44/42 MAP Kinase Thr202/Tyr204, Cell Signaling Technology), and then incubated with an horse radish peroxidase (HRP)-conjugated anti-rabbit IgG Ab (1:2000, Cell Signaling Technology). For Western blotting of Akt, anti-Akt rabbit polyclonal Ab or anti-phosphorylated Akt rabbit polyclonal Ab (1:500, Phosphoplus®Akt Ser473, Cell Signaling Technology) and a secondary Ab, HRP-conjugated anti-rabbit IgG Ab (1:1000), were used. Subsequently, the blots were developed with ECL Plus Western blotting Detection system (GE Healthcare Life Sciences). For Western blot of RET protein, lysate was prepared in RIPA buffer (PIERCE) and electrophoresed on NuPAGE 4–12% Bis-Tris gel (Invitrogen). Blot was probed with anti-RET polyclonal Ab (Ret (C-19; Santa Cruz) and visualized using Pierce Supersignal WestFemto chemiluminescence. Loading control was with β-Actin.
Cell proliferation assay
Cell proliferation 48 and 72hrs after GDNF treatment (5, 25 or 50ng/ml, dissolved in water) in serum-free medium was analyzed by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) and direct cell counting with a light microscope, respectively. MTT (0.4mg/ml, Sigma-Aldrich) was added into culture wells after washing with PBS. The converted dye was dissolved with DMSO 2hrs after incubation at 37°C and the absorbance was measured (550nm) with a microplate reader (Bio-Rad). For cell counting, cells were harvested after washing with PBS and the number of cells was counted using a hemocytometer. In blocking assays, cells were pre-treated with 10µM of a MEK1 inhibitor, PD98059 (Cell Signaling Technology), or 100nM of a PI3K inhibitor, wortmannin (Cell Signaling Technology), for 60min before treatment of GDNF as indicated in the manufacturer’s instructions, and followed by 25ng/ml of GDNF treatment for 72hrs. To avoid the effect of the delivery agent on cells, PD98059 and wortmannin were dissolved in PBS, and the same amount of PBS was added to control (GDNF(-)) and GDNF-treated cells. Each assay was performed at least three times and the mean values were analyzed.
Migration Assay
Migration ability of the melanoma cells was determined with a modified technique using a Transwell® chamber with a microporous membrane (pore-size 8µm, CORNING) (Gumireddy et al., 2007). Melanoma cells were seeded in the upper chamber with RPMI 1640 culture medium (Invitrogen) with 2% fetal bovine serum (FBS). The lower chamber contained RPMI 1640 culture medium with 2% FBS and 5 or 25ng/ml of GDNF. The cells that migrated through the membrane and adhered to the bottom of lower chamber were fixed with 80% ethanol and stained with hematoxylin 48hrs after incubation at 37°C. The number of stained cells was counted in 5 fields under a microscope with 200x magnification. In blocking assays, cells were pre-treated with wortmannin (100nM) for 60min before treatment of GDNF as instructions indicated, and followed by treatment with 25ng/ml GDNF for 48hrs. To avoid the effect of the delivery agent on cells, wortmannin or PD98059 was dissolved in PBS, and the same amount of PBS was added to control (without GDNF) and GDNF-treated cells. Each assay was performed at least three times and the mean values were analyzed.
Invasion Assay
Invasion ability of the melanoma cells was analyzed using QCM™ Collagen-based Invasion Assay (MILLIPORE), as described in previous reports (Goto et al., 2006; Gumireddy et al., 2007). Cells were seeded in the upper chamber with RPMI 1640 culture medium (Invitrogen) with 2% FBS containing GDNF (25ng/ml). The lower chamber contained RPMI 1640 with 5% FBS and GDNF (25ng/ml). Cells invading the collagen-coated membrane were fixed and stained 60hrs after incubation at 37°C with reagents. Stained cells were dissolved with a provided reagent and measured at a 550nm wavelength. In the blocking assay, cells were pre-treated with 1µg/ml of a goat anti-human polyclonal RET-specific Ab, which binds to the extracellular domain of RET (R&D Systems), or 1µg/ml control goat IgG (Santa Cruz Biotechnology) as a control 60min before treatment with GDNF and followed by treatment with 25ng/ml GDNF for 60hrs. Each assay was performed at least three times and the mean values were analyzed.
RNA interference
A siRNA against RET and non-specific control siRNA were designed and synthesized by Dharmacon, Inc. RETp melanoma cells (ME1) were seeded into a 96 well plate (1×104 cells/well) without antibiotics. The siRNA was transfected with DharmaFECT™1 Transfection Reagent. The suppression of RET mRNA was confirmed with qRT assay 24 and 48hrs after transfection. Each assay using siRNA was performed at least three times and the mean values were analyzed.
Actin polymers staining by Alexa Fluor® 568 phalloidin
Melanoma cells were seeded in BD BioCoat™ CultureSlides (BD Biosciences). Cells were washed twice with PBS and fixed with 3.7% formaldehyde solution for 10min at room temperature after a 48hr incubation with GDNF (10ng/ml) at 37°C. Fixed cells were stained with 1U/200 µl of PBS Alexa Fluor® 568 phalloidin (Invitrogen) for 20min at room temperature. After washing twice with PBS, actin polymers were observed using fluorescence microscopy with 400x magnification.
Biostatistical Analysis
The correlation of RETp and BRAFmt frequency in DMs and non-DMs was assessed by the Chi squared test. In functional assays, statistical analyses were performed by Wilcoxon’s signed ranks test and unpaired t test. Results are shown as mean + SD. A P-value of <0.05 (two-tailed) was considered significant. All statistical analyses were done using JMP® software (SAS).
Supplementary Material
Supplementary information is available at Oncogene’s website.
ACKNOWLEDGMENTS
We would like to thank Sandy L. Nguyen and Linhda Nguyen for expert editorial assistance and Emily H. Liang for technical assistance. This work was supported by the National Institutes of Health, National Cancer Institute [Project II P0 CA029605 and CA012582 grants to D.H.]; Ruth and Martin H. Weil Foundation [to D.H.]; the Leslie and Susan Gonda Foundation [to D.H.]; and the Melanoma Foundation of the University of Sydney, Australia [to J.T., R.S., R.M.].
REFERENCES
- Airaksinen MS, Saarma M. The GDNF family: signalling, biological functions and therapeutic value. Nat Rev Neurosci. 2002;3:383–394. doi: 10.1038/nrn812. [DOI] [PubMed] [Google Scholar]
- Bounacer A, Du Villard JA, Wicker R, Caillou B, Schlumberger M, Sarasin A, et al. Association of RET codon 691 polymorphism in radiation-induced human thyroid tumours with C-cell hyperplasia in peritumoural tissue. Br J Cancer. 2002;86:1929–1936. doi: 10.1038/sj.bjc.6600371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busam KJ. Cutaneous desmoplastic melanoma. Adv Anat Pathol. 2005;12:92–102. doi: 10.1097/01.pap.0000155071.86944.a6. [DOI] [PubMed] [Google Scholar]
- Busam KJ, Zhao H, Coit DG, Kucukgol D, Jungbluth AA, Nobrega J, et al. Distinction of desmoplastic melanoma from non-desmoplastic melanoma by gene expression profiling. J Invest Dermatol. 2005;124:412–418. doi: 10.1111/j.0022-202X.2004.23600.x. [DOI] [PubMed] [Google Scholar]
- Carlomagno F, Anaganti S, Guida T, Salvatore G, Troncone G, Wilhelm SM, et al. BAY 43-9006 inhibition of oncogenic RET mutants. J Natl Cancer Inst. 2006;98:326–334. doi: 10.1093/jnci/djj069. [DOI] [PubMed] [Google Scholar]
- Ceccherini I, Hofstra RM, Luo Y, Stulp RP, Barone V, Stelwagen T, et al. DNA polymorphisms and conditions for SSCP analysis of the 20 exons of the ret proto-oncogene. Oncogene. 1994;9:3025–3029. [PubMed] [Google Scholar]
- Curtin JA, Busam K, Pinkel D, Bastian BC. Somatic activation of KIT in distinct subtypes of melanoma. J Clin Oncol. 2006;24:4340–4346. doi: 10.1200/JCO.2006.06.2984. [DOI] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- Davison JM, Rosenbaum E, Barrett TL, Goldenberg D, Hoque MO, Sidransky D, et al. Absence of V599E BRAF mutations in desmoplastic melanomas. Cancer. 2005;103:788–792. doi: 10.1002/cncr.20861. [DOI] [PubMed] [Google Scholar]
- Dhomen N, Marais R. New insight into BRAF mutations in cancer. Curr Opin Genet Dev. 2007;17:31–39. doi: 10.1016/j.gde.2006.12.005. [DOI] [PubMed] [Google Scholar]
- Elisei R, Cosci B, Romei C, Bottici V, Sculli M, Lari R, et al. RET exon 11 (G691S) polymorphism is significantly more frequent in sporadic medullary thyroid carcinoma than in the general population. J Clin Endocrinol Metab. 2004;89:3579–3584. doi: 10.1210/jc.2003-031898. [DOI] [PubMed] [Google Scholar]
- Fujiwara Y, Chi DD, Wang H, Keleman P, Morton DL, Turner R, et al. Plasma DNA microsatellites as tumor-specific markers and indicators of tumor progression in melanoma patients. Cancer Res. 1999;59:1567–1571. [PubMed] [Google Scholar]
- Goto Y, Arigami T, Kitago M, Nguyen SL, Narita N, Ferrone S, et al. Activation of toll-like receptors 2, 3, and 4 on human melanoma cells induces inflammatory factors. Mol Cancer Ther. 2008;7:3642–3653. doi: 10.1158/1535-7163.MCT-08-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y, Matsuzaki Y, Kurihara S, Shimizu A, Okada T, Yamamoto K, et al. A new melanoma antigen fatty acid-binding protein 7, involved in proliferation and invasion, is a potential target for immunotherapy and molecular target therapy. Cancer Res. 2006;66:4443–4449. doi: 10.1158/0008-5472.CAN-05-2505. [DOI] [PubMed] [Google Scholar]
- Govindarajan B, Sligh JE, Vincent BJ, Li M, Canter JA, Nickoloff BJ, et al. Overexpression of Akt converts radial growth melanoma to vertical growth melanoma. J Clin Invest. 2007;117:719–729. doi: 10.1172/JCI30102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumireddy K, Sun F, Klein-Szanto AJ, Gibbins JM, Gimotty PA, Saunders AJ, et al. In vivo selection for metastasis promoting genes in the mouse. Proc Natl Acad Sci U S A. 2007;104:6696–6701. doi: 10.1073/pnas.0701145104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoon DS, Kuo CT, Wascher RA, Fournier P, Wang HJ, O'Day SJ. Molecular detection of metastatic melanoma cells in cerebrospinal fluid in melanoma patients. J Invest Dermatol. 2001;117:375–378. doi: 10.1046/j.0022-202x.2001.01417.x. [DOI] [PubMed] [Google Scholar]
- Iwamoto T, Taniguchi M, Asai N, Ohkusu K, Nakashima I, Takahashi M. cDNA cloning of mouse ret proto-oncogene and its sequence similarity to the cadherin superfamily. Oncogene. 1993;8:1087–1091. [PubMed] [Google Scholar]
- Jaroszewski DE, Pockaj BA, DiCaudo DJ, Bite U. The clinical behavior of desmoplastic melanoma. Am J Surg. 2001;182:590–595. doi: 10.1016/s0002-9610(01)00819-4. [DOI] [PubMed] [Google Scholar]
- Kodama Y, Asai N, Kawai K, Jijiwa M, Murakumo Y, Ichihara M, et al. The RET proto-oncogene: a molecular therapeutic target in thyroid cancer. Cancer Sci. 2005;96:143–148. doi: 10.1111/j.1349-7006.2005.00023.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Ezzat S, Asa SL. Pathogenetic mechanisms in thyroid follicular-cell neoplasia. Nat Rev Cancer. 2006;6:292–306. doi: 10.1038/nrc1836. [DOI] [PubMed] [Google Scholar]
- Koyanagi K, O'Day SJ, Gonzalez R, Lewis K, Robinson WA, Amatruda TT, et al. Serial monitoring of circulating melanoma cells during neoadjuvant biochemotherapy for stage III melanoma: outcome prediction in a multicenter trial. J Clin Oncol. 2005;23:8057–8064. doi: 10.1200/JCO.2005.02.0958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin LF, Doherty DH, Lile JD, Bektesh S, Collins F. GDNF: a glial cell line-derived neurotrophic factor for midbrain dopaminergic neurons. Science. 1993;260:1130–1132. doi: 10.1126/science.8493557. [DOI] [PubMed] [Google Scholar]
- Livestro DP, Muzikansky A, Kaine EM, Flotte TJ, Sober AJ, Mihm MC, Jr, et al. Biology of desmoplastic melanoma: a case-control comparison with other melanomas. J Clin Oncol. 2005;23:6739–6746. doi: 10.1200/JCO.2005.04.515. [DOI] [PubMed] [Google Scholar]
- Melillo RM, Castellone MD, Guarino V, De Falco V, Cirafici AM, Salvatore G, et al. The RET/PTC-RAS-BRAF linear signaling cascade mediates the motile and mitogenic phenotype of thyroid cancer cells. J Clin Invest. 2005;115:1068–1081. doi: 10.1172/JCI22758. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Mologni L, Sala E, Cazzaniga S, Rostagno R, Kuoni T, Puttini M, et al. Inhibition of RET tyrosine kinase by SU5416. J Mol Endocrinol. 2006;37:199–212. doi: 10.1677/jme.1.01999. [DOI] [PubMed] [Google Scholar]
- Plaza-Menacho I, Mologni L, Sala E, Gambacorti-Passerini C, Magee AI, Links TP, et al. Sorafenib functions to potently suppress RET tyrosine kinase activity by direct enzymatic inhibition and promoting RET lysosomal degradation independent of proteasomal targeting. J Biol Chem. 2007;282:29230–29240. doi: 10.1074/jbc.M703461200. [DOI] [PubMed] [Google Scholar]
- Quinn MJ, Crotty KA, Thompson JF, Coates AS, O'Brien CJ, McCarthy WH. Desmoplastic and desmoplastic neurotropic melanoma: experience with 280 patients. Cancer. 1998;83:1128–1135. [PubMed] [Google Scholar]
- Runeberg-Roos P, Saarma M. Neurotrophic factor receptor RET: structure, cell biology, and inherited diseases. Ann Med. 2007:1–9. doi: 10.1080/07853890701646256. [DOI] [PubMed] [Google Scholar]
- Satyamoorthy K, Li G, Gerrero MR, Brose MS, Volpe P, Weber BL, et al. Constitutive mitogen-activated protein kinase activation in melanoma is mediated by both BRAF mutations and autocrine growth factor stimulation. Cancer Res. 2003;63:756–759. [PubMed] [Google Scholar]
- Sawai H, Okada Y, Kazanjian K, Kim J, Hasan S, Hines OJ, et al. The G691S RET polymorphism increases glial cell line-derived neurotrophic factor-induced pancreatic cancer cell invasion by amplifying mitogen-activated protein kinase signaling. Cancer Res. 2005;65:11536–11544. doi: 10.1158/0008-5472.CAN-05-2843. [DOI] [PubMed] [Google Scholar]
- Selek U, Chang EL, Hassenbusch SJ, 3rd, Shiu AS, Lang FF, Allen P, et al. Stereotactic radiosurgical treatment in 103 patients for 153 cerebral melanoma metastases. Int J Radiat Oncol Biol Phys. 2004;59:1097–1106. doi: 10.1016/j.ijrobp.2003.12.037. [DOI] [PubMed] [Google Scholar]
- Shinozaki M, Fujimoto A, Morton DL, Hoon DS. Incidence of BRAF oncogene mutation and clinical relevance for primary cutaneous melanomas. Clin Cancer Res. 2004;10:1753–1757. doi: 10.1158/1078-0432.ccr-1169-3. [DOI] [PubMed] [Google Scholar]
- Shinozaki M, O'Day SJ, Kitago M, Amersi F, Kuo C, Kim J, et al. Utility of circulating B-RAF DNA mutation in serum for monitoring melanoma patients receiving biochemotherapy. Clin Cancer Res. 2007;13:2068–2074. doi: 10.1158/1078-0432.CCR-06-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens LA, Powell NG, Grubb J, Jeremiah SJ, Bethel JA, Demidchik EP, et al. Investigation of loss of heterozygosity and SNP frequencies in the RET gene in papillary thyroid carcinoma. Thyroid. 2005;15:100–104. doi: 10.1089/thy.2005.15.100. [DOI] [PubMed] [Google Scholar]
- Sumimoto H, Miyagishi M, Miyoshi H, Yamagata S, Shimizu A, Taira K, et al. Inhibition of growth and invasive ability of melanoma by inactivation of mutated BRAF with lentivirus-mediated RNA interference. Oncogene. 2004;23:6031–6039. doi: 10.1038/sj.onc.1207812. [DOI] [PubMed] [Google Scholar]
- Takahashi M. The GDNF/RET signaling pathway and human diseases. Cytokine Growth Factor Rev. 2001;12:361–373. doi: 10.1016/s1359-6101(01)00012-0. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Buma Y, Hiai H. Isolation of ret proto-oncogene cDNA with an amino-terminal signal sequence. Oncogene. 1989;4:805–806. [PubMed] [Google Scholar]
- Takahashi M, Buma Y, Iwamoto T, Inaguma Y, Ikeda H, Hiai H. Cloning and expression of the ret proto-oncogene encoding a tyrosine kinase with two potential transmembrane domains. Oncogene. 1988;3:571–578. [PubMed] [Google Scholar]
- Umetani N, Mori T, Koyanagi K, Shinozaki M, Kim J, Giuliano AE, et al. Aberrant hypermethylation of ID4 gene promoter region increases risk of lymph node metastasis in T1 breast cancer. Oncogene. 2005;24:4721–4727. doi: 10.1038/sj.onc.1208538. [DOI] [PubMed] [Google Scholar]
- Weber F, Eng C. Update on the molecular diagnosis of endocrine tumors: toward-omics-based personalized healthcare? J Clin Endocrinol Metab. 2008;93:1097–1104. doi: 10.1210/jc.2008-0212. [DOI] [PubMed] [Google Scholar]
- Zbuk KM, Eng C. Cancer phenomics: RET and PTEN as illustrative models. Nat Rev Cancer. 2007;7:35–45. doi: 10.1038/nrc2037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information is available at Oncogene’s website.