Abstract
The mechanism of oxidation of a peptide substrate by the flavoprotein lysine-specific demethylase (LSD1) has been examined using the effects of pH and isotopic substitution on steady-state and rapid-reaction kinetic parameters. The substrate contained the N-terminal 21 residues of histone H3, with a dimethylated lysyl residue at position 4. At pH 7.5, the rate constant for flavin reduction, kred, equals kcat, establishing the reductive half reaction as rate-limiting at physiological pH. Deuteration of the lysyl methyls results in identical kinetic isotope effects of 3.1 ± 0.2 on the kred, kcat and kcat/Km values for the peptide, establishing CH bond cleavage as rate-limiting with this substrate. No intermediates between oxidized and reduced flavin are detectable by stopped-flow spectroscopy, consistent with the expectation for a direct hydride transfer mechanism. The kcat/Km value for the peptide is bell-shaped, consistent with a requirement that the nitrogen at the site of oxidation be uncharged and that at least one of the other lysyl residues be charged for catalysis. The D(kcat/Km) value for the peptide is pH-independent, suggesting that the observed value is the intrinsic deuterium kinetic isotope effect for oxidation of this substrate.
Nucleosomes, the basic subunits of chromatin, are composed of 146 bp of DNA wrapped around an octamer of four core histones (H3, H4, H2A and H2B) (1). Each octamer consists of an H3-H4 histone tetramer plus an H2A-H2B dimer and is connected to the adjacent octamers via an H1 linker histone (1). Post-translational modifications of the N-terminal tails of histones, which extend freely beyond the nucleosome core, have been shown to affect chromatin structure and the accessibility to DNA of proteins essential for transcription, repair, and replication (2). Lysine-specific demethylase (LSD1) is an FAD-containing amine oxidase that catalyzes the demethylation of mono- and dimethylated lysyl residues in the N-terminal tails of histones, thereby playing a role in the epigenetic regulation of gene transcription in cells (3). LSD1 demethylates lysine residues 4 and 9 of histone H3 (H3K4 and H3K9) (3, 4); in addition, the enzyme can demethylate the tumor suppressor p53 (5). The discoveries of LSD1 and of the JmjC family of iron(II)-alpha-ketoglutarate-dependent histone demethylases (6) have demonstrated that methylation of histones is not permanent; instead, like other post-translational modifications, it is part of a reversible process. The effects of histone methylation vary depending on the site of modification, the degree of methylation, the sites and nature of adjacent modifications, and the proteins complexes in proximity to the modification (7). LSD1 functions as both a repressor and activator of gene transcription; for example, demethylation of Lys4 of histone H3 results in gene repression (8), while androgen-dependent demethylation of mono- or dimethylated Lys9 results in an activated transcriptional state (4). LSD1 has been implicated in the maintenance of disease, such as neuroblastoma (9), its expression correlates with high-risk prostate tumors (10), and it plays regulatory roles in cell differentiation (11, 12).
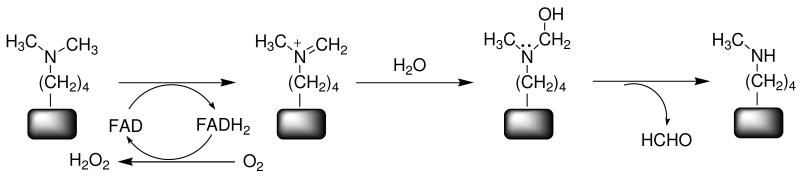
Flavoprotein oxidases utilize a flavin cofactor to catalyze oxidation of a substrate CX bond, transferring a hydride equivalent to the flavin; the reduced flavin is then oxidized by molecular oxygen, producing H2O2 (13). LSD1 oxidizes the carbon-nitrogen bond between the methyl group and the epsilon amine of lysyl residues, forming an imine intermediate that is nonenzymatically hydrolyzed to produce formaldehyde and the demethylated lysine (Figure 1). There are two structural families of flavoprotein amine oxidases; LSD1 belongs to the monoamine oxidase family (14). A number of mechanisms have been proposed for amine oxidation by flavoenzymes, including direct transfer of a hydride transfer from the substrate to the flavin, formation of a substrate carbanion that then transfers electrons to the flavin, and two single electron transfers to form an intermediate radical pair with a subsequent proton transfer (15). However, kinetic isotope effects on the reactions of a number of flavoprotein amine and alcohol oxidases support direct hydride transfer as the mechanism for members of both families of amine oxidases and for flavoprotein oxidases in general (16-20).
Figure 1.
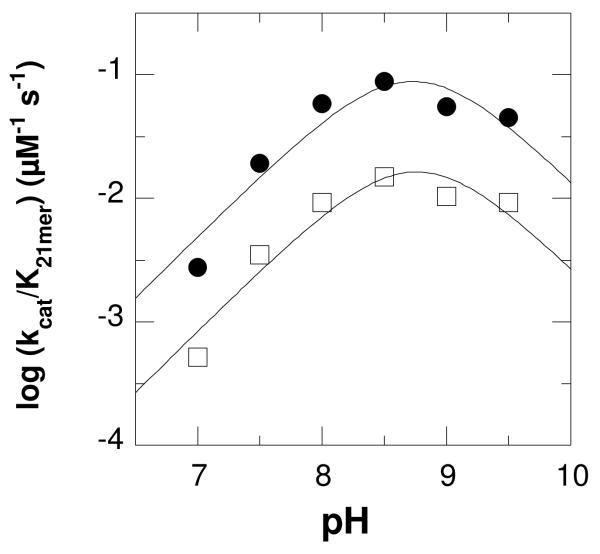
kcat/KM pH profiles for LSD1 with protiated (closed circles) and deuterated (open squares) H3K4 21-mer dimethylated peptide. The lines are from fits of the data to eq 1.
While there have been many studies focusing on the role of LSD1 in gene transcription and protein expression and its interactions with other proteins, few have focused on the kinetics of the enzyme. Several mechanism-based inhibitors of monoamine oxidase and their derivatives have been shown to inhibit LSD1 (8, 21-25), providing insights into the interactions with substrates and demonstrating the mechanistic similarity of the enzyme to monoamine oxidase. Forneris et al. have described a pH profile of LSD1 (26) and analyzed the reaction of the photoreduced enzyme with oxygen, demonstrating that the oxidation is typical of a flavoprotein oxidase (27). Still, there has been no detailed analysis of the reaction with a normal peptide substrate that allows for conclusions regarding mechanism. LSD1 is highly specific for the N-terminus of histone H3. Indeed, the minimal substrate is a peptide composed of the N-terminal 21 amino acid residues of H3 (26). We describe here mechanistic studies of oxidation of such a peptide substrate by LSD1.
EXPERIMENTAL PROCEDURES
Materials
Horseradish peroxidase and Ampliflu Red were purchased from Sigma. Fmoc amino acids were from Novabiochem, Applied Biosystems, or Advanced ChemTech. 2H-Formaldehyde (20% solution in D2O) was from Cambridge Isotopes. Sodium cyanoborodeuteride was from CDN Isotopes.
H3K4 21-mer dimethylated peptide synthesis
A peptide corresponding to the first 21 amino acids of histone H3 with a dimethylated lysine at the fourth residue (ARTK(diMe)QTARKSTGGKAPRKQLA) was synthesized via classical Fmoc-peptide chemistry using an Applied Biosystems 433 peptide synthesizer with PAL resin (Advanced ChemTech). The residues GYG were added to the C-terminal end of the peptide to aid in quantification of the peptide concentration; an extinction coefficient of 1440 cm−1M−1 was used at 280 nm. Peptides were purified on a C18 reversed-phase semi-prep column (Phenomenex) using a Thermo HPLC. Peptide mass was verified by matrix-assisted laser desorption ionization-time of flight mass spectrometry (PerSeptive Biosystems).
Deuterated dimethylated lysine was synthesized according to the procedure of Huang et al. (28), but using sodium cyanoborodeuteride and 2H-formaldehyde. To 40 mL of ethanol on ice, 812 mg of Fmoc-Lys was added with stirring, followed by 636 μL of 2H formaldehyde (20%, v/v) in D2O (Cambridge Isotopes). After 15 min, 380 mg of sodium cyanoborodeuteride was added. After 15 min the formaldehyde and sodium cyanoborodeuteride additions were repeated, and the reaction allowed to stir for an additional 2 h. The solution was acidified via the dropwise addition of 1 M HCl, and the solvent was removed by rotary evaporation. The residue was then applied to a silica gel column and eluted with methanol. Product-containing fractions were identified via fluorescence on TLC plates, pooled, and concentrated to an oil via rotary evaporation. The oil was dissolved in water and lyophilized. The identity of the product was confirmed by electrospray ionization (ESI) mass spectrometry (positive ion mode, expected M+H, 403.2, observed, 403.2). The 1H NMR was the same as that for protiated Fmoc-Lys(Me)2, except for the absence of the protons for the methyl groups. Fmoc-Lys([2H]Me2)-OH was then incorporated into the histone H3 peptide and purified as described above; however, the last four N-terminal residues were coupled manually.
LSD1 Expression and Purification
A truncated form of human LSD1, lacking the first 150 amino acids (3), was expressed and purified as previously described (25) with minor modifications. pET151-D/TOPO (Invitrogen) containing the gene encoding LSD1 was used to transform the Rosetta 2 strain of E. coli. The cells were grown at 23 °C to an OD600 of 0.6 before induction with 0.5 mM IPTG. After further growth overnight, the cells were collected and lysed with a homogenizer in 50 mM sodium phosphate, pH 7.6, containing 350 mM NaCl. The protein was purified via a nickel affinity column (Chelating Sepharose FF (Amersham) charged with nickel sulfate), using a gradient of 10-175 mM imidazole in 50 mM sodium phosphate, pH 7.4, followed by a gel filtration column (Sephacryl S-200, GE Life Sciences) in 50 mM sodium phosphate, pH 7.4, and finally, an anion exchange column (Q Sepharose Fast-Flow, GE Life Sciences), using a gradient of 0 - 1 M NaCl in 50 mM sodium phosphate, pH 7.4. The protein was then concentrated using a Centricon-30 centrifugal concentrator (Millipore) and stored at -20 °C in 50 mM sodium phosphate, pH 7.4, containing 40% glycerol. The concentration of LSD1 was determined using the absorbance at 458 nm and an extinction coefficient based on that for free FAD (11,600 cm−1M−1).
Assays
Steady state kinetic assays monitoring oxygen consumption were performed on a computer-interfaced Hansatech (Hansatech Instruments) or YSI (Yellow Springs Instrument, Inc.) oxygen electrode. All assays were conducted at 25 °C and were initiated by the addition of enzyme. A fluorescence assay, coupling hydrogen peroxide formation to oxidation of Ampliflu Red by horseradish peroxidase, was used to determine kcat/Km values due to the low concentrations of substrate required. The product resorufin was detected using a fluorescence plate reader (Molecular Devices SpectraMax Gemini EM) with an excitation wavelength of 560 nm and emission at 590 nm. The coupled fluorescence assay resulted in initial rates that were twofold slower than those detected using the oxygen electrode. Consequently, the rates obtained with the coupled assay were corrected by this factor. The constant ionic strength buffer 0.1 M Aces, 52 mM Tris, and 52 mM ethanolamine (29) was used for assays at all pH values. Preincubation of enzyme in buffers of pH higher than 9.5 for 60 minutes eliminated LSD1 activity.
Rapid-reaction kinetic experiments were conducted at 25 °C on an Applied Photophysics SX-18MV stopped-flow spectrophotometer. Anaerobic conditions were established by applying cycles of vacuum and argon to enzyme solutions, while substrate solutions were bubbled with argon. The buffer was again ACES/Tris/ethanolamine, plus 5 mM glucose, at pH 7.5. Glucose oxidase was added to all anaerobic solutions at a final concentration of 36 nM before loading them onto the stopped-flow spectrophotometer.
Data Analysis
Steady-state kinetic data were analyzed using the program KaleidaGraph (Adelbeck Software, Reading, PA). Initial rate data obtained by varying the concentration of a single substrate were fit to the Michaelis-Menten equation. The effects of pH on kinetic parameters were fit to eq 1, which applies for a kinetic parameter which decreases below pK1 due to protonation of a single moiety and decreases above pK2 due to deprotonation of a single moiety. Here, y is the kinetic parameter being measured, c is its pH-independent value, and K1 and K2 are the ionization constants for moieties which must be unprotonated or protonated, respectively. Eq 2 was used to determine kinetic isotope effects using the program Igor (WaveMetrics, Lake Oswego, OR); E is the isotope effect on the parameters kcat/K21-mer and kcat, S is the concentration of H3K4 21-mer dimethylated peptide, and Fi is the fraction of heavy atom in the substrate. Analysis of stopped-flow data was done using both KaleidaGraph and SPECFIT (Spectrum Software Associates, Marlborogh, MA). To determine the kinetic parameters for the reduction of LSD1, stopped-flow traces were fit to eq 3, which describes a monophasic exponential decay; k1 is the first order rate constant, A1 is the total absorbance change, and A∞ is the final absorbance.
| (1) |
| (2) |
| (3) |
RESULTS
Steady-state Kinetics
Steady-state kinetic parameters for LSD1 were determined at pH 7.5 and 25 °C with the dimethylated H3K4 21-mer as substrate. This peptide has the sequence of the N-terminal 21 residues of histone H3, the physiological substrate of LSD1, and contains N, N-dimethyllysine at residue 4. It represents the minimum length required for detectable activity (26). While the kcat/K21-mer values could be determined in air-saturated buffer because LSD1 exhibits ping pong kinetics (27), kcat values were detemined with 1.1 mM oxygen and 50 μM peptide to ensure saturation with both substrates. In addition, the kcat/KO2 values were determined by varying the concentration of oxygen in the presence of 50 μM peptide. The results are summarized in Table 1. The kcat and kcat/K21-mer values are in reasonable agreement with the values previously reported by Forneris et al. (26).
Table 1.
Kinetic parameters of LSD1 with the H3K4 21-mer dimethylated peptide as a substrate*
| Kinetic parameter | Value |
|---|---|
| kcat/K21-mer (mM−1 s−1) | 38 ± 2a |
| kcat/KO2 (mM−1 s−1) | 1.0 ± 0.2b |
| K21-mer (μM) | 2.6 ± 0.2a |
| KO2 (μM) | 195 ± 40b |
| kcat (s−1) | 0.199 ± 0.013c |
| kred (s−1) | 0.231 ± 0.004e |
| D(kcat/K21-mer) | 3.2 ± 0.1a,d |
| Dkcat | 2.9 ± 0.2c |
| 3.2 ± 0.1a,d | |
| Dkred | 3.3 ± 0.1e |
Conditions: pH 7.5, 25 °C.
Determined in air-saturated buffer.
Determined using 50 μM H3K4 21-mer dimethylated peptide.
Determined using 50 μM H3K4 21-mer dimethylated peptide and 1.1 mM oxygen.
Calculated using eq 2.
Determined using 200 μM 21-mer dimethylated peptide.
pH Profiles
The effect of pH on the kcat/K21-mer values was determined over the pH range 7.0–9.5 using a buffer system that maintained constant ionic strength over the entire pH range. At pH values above 9.5, the enzyme lost activity, while the activity was too low below pH 7 to be measured reliably. Because these assays are not carried out with saturating oxygen concentrations, the resulting kcat values were only apparent, and the kcat-pH profile need not reflect the actual pKa values. The kcat/K21-mer data yield a bell-shaped pH profile (Figure 1), decreasing above and below pH 8.5-9.2 This behavior is consistent with the involvement of two groups on the free enzyme or substrate, one of which must be protonated and one deprotonated for activity. When the data were fit assuming different pKa values for the two groups, the resulting pKa values were within 0.3 of one another (results not shown), too close to reliably discriminate. Consequently, only the average of the pKa values of the two groups can be determined. The data were therefore fit using eq 1, which assigns the same pKa value to the two groups, to obtain the average pKa value of 8.7 ± 0.1.
Kinetic Isotope Effects
Deuterium kinetic isotope effects were determined with the 21-mer in which both methyl groups on the dimethylated lysyl residue contained three deuteriums. The results at pH 7.5 are summarized in Table 1. Data obtained by varying the concentration of the peptide in air-saturated buffer were best fit by eq 2, which assigns equal values to Dkcat/K21-mer and Dkcat, for an isotope effect of 3.2 ± 0.1. Alternatively, the Dkcat value was determined at fixed saturating concentrations of peptide and oxygen of 50 μM and 1.1 mM, respectively. The resulting value was 2.9 ± 0.2. The kinetic isotope effect on kcat/K21-mer and kcat was then determined over the pH range 7-9.5, fitting the data in each case to eq 2. The isotope effect was pH-independent, with an average value of 3.5 ± 0.3 (Table 2). The effect of pH on the kcat/K21-mer value for the deuterated peptide is shown in Figure 1; these data were fit to eq 1 to yield an average pKa value of 8.8 ± 0.1, within error of the value for the nondeuterated peptide.
Table 2.
Effect of pH on the deuterium kinetic isotope effects on the oxidation of the H3K4 21-mer dimethylated peptide by LSD1*
| pH | Kinetic isotope effect |
|---|---|
| 7.0 | 4.0 ± 0.2 |
| 7.5 | 3.2 ± 0.1 |
| 8.0 | 3.3 ± 0.1 |
| 8.5 | 3.4 ± 0.1 |
| 9.0 | 3.7 ± 0.2 |
| 9.5 | 3.6 ± 0.1 |
| Average | 3.5 ± 0.3 |
Determined in air saturated buffers and calculated using eq 2.
Flavin Reduction Kinetics
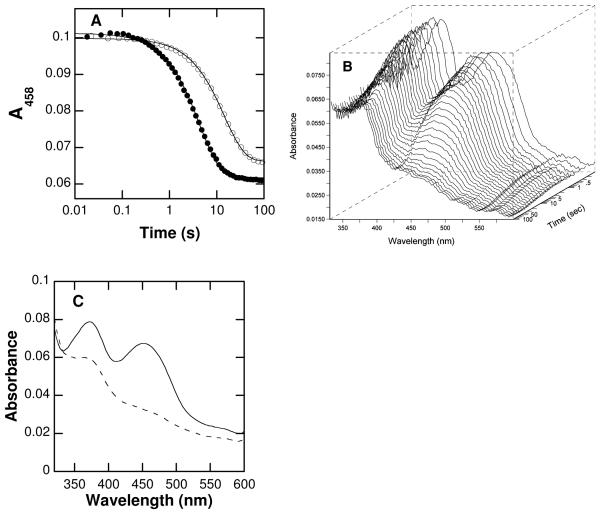
Stopped-flow spectroscopy was used to study reduction of the flavin in LSD1 by saturating concentrations (200 μM) of the 21-mer at pH 7.5. The changes in the absorbance of the enzyme-bound flavin after mixing LSD1 anaerobically with the 21-mer are best fit as a single exponential decay (Fig. 2). The rate constant for reduction (kred) with this substrate concentration is within error of the kcat value (Table 1), consistent with reduction being rate-limiting for turnover. The kred value was also determined with 200 μM deuterated peptide, yielding a Dkred value of 3.3 ± 0.1. This value is identical to the pH-independent isotope effect determined in the steady-state kinetic analyses. To more definitively confirm that reduction consists of a single kinetic phase, the reaction was repeated using a photodiode array detector to monitor the flavin spectrum from 325 to 600 nm. Over this wavelength range, there is no evidence for an intermediate between oxidized flavin and reduced flavin (Figure 2B). A global analysis of the spectral changes during flavin reduction as a single exponential decay allowed the spectra of the starting and final enzyme to be determined (Figure 2C). These confirm that the oxidized and reduced enzyme are the only species that can be detected.
Figure 2.
Spectral changes during reduction of LSD1 by the H3K4 21-mer dimethylated peptide: A, time course at 458 nm with protiated (closed circles) or deuterated (open circles) substrate (only 1/30th of the points are shown for clarity); B, Changes in the entire visible absorbance spectrum during reduction by the deuterated substrate; C, Initial (−) and final (⋯) spectra upon LSD1 flavin reduction by deuterated H3K4 21-mer dimethyl peptide obtained from a global analysis of the data in B using a single-step kinetic model. The lines in A are from fits of the data to eq 3.
DISCUSSION
While the physiological substrate for LSD1 is the histone H3, mechanistic studies with the entire protein substrate present significant problems, including the difficulty of preparing the dimethylated substrate. Consequently, mechanistic insight requires the use of a minimal substrate which still retains the specificity determinants of the larger protein. LSD1 has no detectable activity with peptides containing fewer than 16 of the N-terminal residues of histone H3, and a peptide containing the first 21 residues, as was utilized here, is as active a substrate as a peptide containing the first 30 residues (26). Epigenetic modifications of histone H3 that alter the reactivity of the intact protein with LSD1 cause similar decreases in activity when incorporated into a 21-mer (27), establishing the latter as a valid model for the larger protein substrate. Indeed, incorporation of a propargyl moiety onto Lys4 of the 21-mer yields an efficient mechanism-based inhibitor (21). The three-dimensional structure of the inactivated enzyme shows extensive contacts with the first 7 residues, while the remainder of the peptide is not visible (23). This combination of solution and structural studies establishes the validity of the 21-mer as a model for mechanistic studies.
The steady-state and transient kinetic studies presented here establish that the rate-limiting step in oxidation of the dimethylated H3K4 21-mer by LSD1 is CH bond cleavage. The kred value, the rate constant for flavin reduction, is identical to the kcat value for LSD1. Therefore, the reductive half reaction is rate-limiting for turnover with this peptide substrate. The identity of the primary deuterium isotope effects on kcat/K21-mer, kcat, and kred further identifies CH bond cleavage as the rate limiting step. The pH-independence of the isotope effect on kcat/K21-mer is also consistent with fully rate-limiting CH bond cleavage and suggests that this isotope effect is the intrinsic deuterium isotope effect (30). An isotope effect of 3.2 ± 0.1 is substantially smaller than the limiting semi-classical value of about 7 for a primary deuterium isotope effect (31); in the present case, the value also includes the secondary isotope effects from the two other deuterium atoms on each methyl group. The small magnitude of the isotope effect suggests that the oxidation of this substrate by LSD1 has an early transition state. A late transition state would also yield a small isotope effect, but a late transition state would be expected for an unfavorable reaction. In contrast, at all concentrations of substrate used here, the reductive half-reaction goes to completion rather than reaching an equilibrium suggesting thatt the oxidation of the 21-mer is favorable. Similarly small kinetic isotope effects have been reported for other flavin amine oxidases, including d-amino acid oxidase with glycine as substrate (32) and tryptophan 2-monooxygenase with tryptophan as substrate (33).
No intermediates between oxidized and reduced enzyme were observed in the oxidation of the 21-mer by LSD1. The lack of an intermediate with a substrate whose rate-limiting step is CH bond cleavage is not consistent with several mechanisms proposed for flavin amine oxidases, especially mechanisms involving easily detectable flavin radicals or covalent substrate-flavin adducts. The present results are fully consistent with direct hydride transfer as the mechanism of LSD1, in line with observations from other flavoprotein amine oxidases. LSD1 is inactivated by cyclopropyl substrates (25), and inactivation by such compounds is frequently taken as evidence for radical intermediates (34). However, a number of flavoprotein oxidases for which hydride transfer is the accepted mechanism can be inactivated by cyclopropyl substrates (35, 36). The kinetic parameter kcat/K21-mer exhibits a pH optimum of 8.7 in the studies described here.3 While the kcat/Km value for a substrate can reflect the protonation states of both the enzyme and substrate required for productive binding and catalysis, recent studies with the related flavoprotein polyamine oxidase suggest that the kcat/Km-pH profile reflects the required protonation state of the substrate. With N1-acetylspermine, N1-acetylspermidine, and spermine as the substrate for mammalian polyamine oxidase, the kcat/Kamine-pH optimum matches the pH at which the substrate has only one positively charged nitrogen (37). The pH dependence of the inhibition by spermidine analogues further established that the nitrogen in the CN bond of the polyamine being oxidized by polyamine oxidase must be uncharged for oxidation, consistent with observations of other amine oxidases (16, 18, 38) and with the expectations for a hydride transfer mechanism. With both polyamine oxidase (37) and monoamine oxidase (39), the need for the nitrogen at the reaction site to be neutral is clearly seen in the pH dependence of the reductive half reaction of the enzyme, demonstrating the requirement for a neutral nitrogen for catalysis. The substrate used in the present study contains the first 21 residues of the N-terminus of histone H3. Based on the precedents of other amine oxidases, the dimethylated Lys4 must be uncharged for catalysis and therefore is responsible for the acidic limb of the pH profile. Protonation of Lys4 does not prevent binding since the trimethylated analog of this peptide binds tightly to LSD1, although it is not a substrate (26). The basic limb of the kcat/K21-mer-pH profile can be attributed to one of the three other lysyl residues in the substrate.
The members of the monoamine oxidase family, including LSD1, contain a conserved lysyl residue in the active site that is part of a “Lys-H2O-N5” structural motif (14, 40-44). This residue clearly could be a source of a pKa in the kcat/K21-mer-pH profile. Mutation of this lysine in LSD1 to an alanine results in a complete loss of activity (45), and in maize polyamine oxidase the corresponding lysine to methionine mutation results in a 1400-fold decrease in kred (46). However, the same lysine to methionine mutation in mouse polyamine oxidase results in no change in the kcat/Kamine-pH profile and only a slight decrease in the value of kred (37), suggesting that this residue plays a subtle structural rather than a catalytic role in these enzymes and therefore does not contribute to the pH dependence.
The rate constant for the chemical step in LSD1 is 2-5 orders of magnitude slower than values reported for other flavoprotein amine oxidases (37, 46-51). This suggests that evolution has not optimized catalysis for this enzyme. Instead, it is likely that the requirements for rapid catalysis are secondary to high specificity in this case. Given the physiological role of LSD1 in regulation of gene expression, high catalytic activity would be much less critical than very high specificity. Moreover, while high catalytic activity is necessary for metabolic enzymes to maintain balance among metabolic pathways, epigenetic regulation such as DNA methylation operates on a very long time scale and would not require rate constants on the order of 102-104 s−1.
Scheme 1.
1Abbreviations
- LSD1
lysine-specific demethylase
Footnotes
This work was supported by grants GM58698 (PFF) and GM65539 (DGM) from the National Institutes of Health.
The apparent kcat-pH profile also fit to a bell shaped curve, with pKa values of 8.0 ± 0.1 and 9.7 ± 0.1 for the protiated substrate. However, the profile is a combination of the kcat and kcat/KO2-pH profiles.
Forneris et al. (26) reported a bell-shaped profile with pKa values of 7.2 and 8.9 using a similar substrate. The reasons for the difference from those reported here are not clear. The activity of the enzyme is reported to be quite sensitive to ionic strength, so the differences may be due to the different buffers used in the two studies. To avoid complications from ionic strength or buffer composition, a single constant ionic strength buffer was used in all the studies described here.
REFERENCES
- 1.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 3.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 4.Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AHFM, Gunther T, Buettner R, Schule R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 5.Huang J, Sengupta R, Espejo AB, Lee MG, Dorsey JA, Richter M, Opravil S, Shiekhattar R, Bedford MT, Jenuwein T, Berger SL. p53 is regulated by the lysine demethylase LSD1. Nature. 2007;449:105–108. doi: 10.1038/nature06092. [DOI] [PubMed] [Google Scholar]
- 6.Tsukada Y.-i., Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 7.Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr. Opin. Cell Biol. 2008;20:341–348. doi: 10.1016/j.ceb.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee MG, Wynder C, Schmidt DM, McCafferty DG, Shiekhattar R. Histone H3 Lysine 4 Demethylation Is a Target of Nonselective Antidepressive Medications. Chem. Biol. 2006;13:563–567. doi: 10.1016/j.chembiol.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 9.Schulte JH, Lim S, Schramm A, Friedrichs N, Koster J, Versteeg R, Ora I, Pajtler K, Klein-Hitpass L, Kuhfittig-Kulle S, Metzger E, Schule R, Eggert A, Buettner R, Kirfel J. Lysine-specific demethylase 1 is strongly expressed in poorly differentiated neuroblastoma: implications for therapy. Cancer Res. 2009;69:2065–2071. doi: 10.1158/0008-5472.CAN-08-1735. [DOI] [PubMed] [Google Scholar]
- 10.Kahl P, Gullotti L, Heukamp LC, Wolf S, Friedrichs N, Vorreuther R, Solleder G, Bastian PJ, Ellinger J, Metzger E, Schule R, Buettner R. Androgen receptor coactivators lysine-specific histone demethylase 1 and four and a half LIM domain protein 2 predict risk of prostate cancer recurrence. Cancer Res. 2006;66:11341–11347. doi: 10.1158/0008-5472.CAN-06-1570. [DOI] [PubMed] [Google Scholar]
- 11.Zhu X, Wang J, Ju BG, Rosenfeld MG. Signaling and epigenetic regulation of pituitary development. Curr. Opin. Cell Biol. 2007;19:605–611. doi: 10.1016/j.ceb.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su ST, Ying HY, Chiu YK, Lin FR, Chen MY, Lin KI. Involvement of histone demethylase LSD1 in Blimp-1-mediated gene repression during plasma cell differentiation. Mol. Cell Biol. 2009;29:1421–1431. doi: 10.1128/MCB.01158-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fraaije MW, Mattevi A. Flavoenzymes: diverse catalysts with recurrent features. TIBS. 2000;25:126–132. doi: 10.1016/s0968-0004(99)01533-9. [DOI] [PubMed] [Google Scholar]
- 14.Chen Y, Yang Y, Wang F, Wan K, Yamane K, Zhang Y, Lei M. Crystal structure of human histone lysine-specific demethylase 1 (LSD1) Proc. Natl. Acad. Sci. USA. 2006;103:13956–13961. doi: 10.1073/pnas.0606381103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fitzpatrick PF. Substrate dehydrogenation by flavoproteins. Acc. Chem. Res. 2001;34:299–307. doi: 10.1021/ar0000511. [DOI] [PubMed] [Google Scholar]
- 16.Kurtz KA, Rishavy MA, Cleland WW, Fitzpatrick PF. Nitrogen isotope effects as probes of the mechanism of D-amino acid oxidase. J. Am. Chem. Soc. 2000;122:12896–12897. [Google Scholar]
- 17.Sobrado P, Fitzpatrick PF. Solvent and primary deuterium isotope effects show that lactate CH and OH bond cleavages are concerted in Y254F flavocytochrome b2, consistent with a hydride transfer mechanism. Biochemistry. 2003;42:15208–15214. doi: 10.1021/bi035546n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ralph EC, Anderson MA, Cleland WW, Fitzpatrick PF. Mechanistic studies of the flavoenzyme tryptophan 2-monooxygenase: Deuterium and 15N kinetic isotope effects on alanine oxidation by an L-amino acid oxidase. Biochemistry. 2006;45:15844–15852. doi: 10.1021/bi061894o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ralph EC, Hirschi JS, Anderson MA, Cleland WW, Singleton DA, Fitzpatrick PF. Insights into the mechanism of flavoprotein-catalyzed amine oxidation from nitrogen isotope effects on the reaction of N-methyltryptophan oxidase. Biochemistry. 2007;46:7655–7664. doi: 10.1021/bi700482h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fitzpatrick PF. Insights into the mechanisms of flavoprotein oxidases from kinetic isotope effects. J. Labelled Comp. & Radiopharm. 2007;50:1016–1025. doi: 10.1002/jlcr.1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Culhane JC, Szewczuk LM, Liu X, Da G, Marmorstein R, Cole PA. A mechanism-based inactivator for histone demethylase LSD1. J. Am. Chem. Soc. 2006;128:4536–4537. doi: 10.1021/ja0602748. [DOI] [PubMed] [Google Scholar]
- 22.Szewczuk LM, Culhane JC, Yang M, Majumdar A, Yu H, Cole PA. Mechanistic analysis of a suicide inactivator of histone demethylase LSD1. Biochemistry. 2007;46:6892–6902. doi: 10.1021/bi700414b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang M, Culhane JC, Szewczuk LM, Jalili P, Ball HL, Machius M, Cole PA, Yu H. Structural Basis for the Inhibition of the LSD1 Histone Demethylase by the Antidepressant trans-2-Phenylcyclopropylamine. Biochemistry. 2007;46:8058–8065. doi: 10.1021/bi700664y. [DOI] [PubMed] [Google Scholar]
- 24.Gooden DM, Schmidt DMZ, Pollock JA, Kabadi AM, McCafferty DG. Facile synthesis of substituted trans-2-arylcyclopropylamine inhibitors of the human histone demethylase LSD1 and monoamine oxidases A and B. Bioorg. Med. Chem. Lett. 2008;18:3047–3051. doi: 10.1016/j.bmcl.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmidt DMZ, McCafferty DG. trans-2-Phenylcyclopropylamine Is a Mechanism-Based Inactivator of the Histone Demethylase LSD1. Biochemistry. 2007;46:4408–4416. doi: 10.1021/bi0618621. [DOI] [PubMed] [Google Scholar]
- 26.Forneris F, Binda C, Vanoni MA, Battaglioli E, Mattevi A. Human Histone Demethylase LSD1 Reads the Histone Code. J. Biol. Chem. 2005;280:41360–41365. doi: 10.1074/jbc.M509549200. [DOI] [PubMed] [Google Scholar]
- 27.Forneris F, Binda C, Dall'Aglio A, Fraaije MW, Battaglioli E, Mattevi A. A Highly Specific Mechanism of Histone H3-K4 Recognition by Histone Demethylase LSD1. J. Biol. Chem. 2006;281:35289–35295. doi: 10.1074/jbc.M607411200. [DOI] [PubMed] [Google Scholar]
- 28.Huang Z-P, Du J-T, Zhao Y-F, Li Y-M. Synthesis of Site-Specifically Dimethylated and Trimethylated Peptides Derived from Histone H3 N-Terminal Tail. Int. J. Peptide Protein Res. 2006;12:187–193. [Google Scholar]
- 29.Ellis KJ, Morrison JF. Buffers of constant ionic strength for studying pH-dependent processes. Methods Enzymol. 1982;87:405–426. doi: 10.1016/s0076-6879(82)87025-0. [DOI] [PubMed] [Google Scholar]
- 30.Cook PF, Cleland WW. pH variation of isotope effects in enzyme-catalyzed reactions. 1. Isotope- and pH-dependent steps the same. Biochemistry. 1981;20:1797–1805. doi: 10.1021/bi00510a014. [DOI] [PubMed] [Google Scholar]
- 31.Jencks WP. Catalysis in chemistry and enzymology. McGraw-Hill Book, Inc.; New York: 1969. [Google Scholar]
- 32.Denu JM, Fitzpatrick PF. pH and kinetic isotope effects on the reductive half-reaction of D-amino acid oxidase. Biochemistry. 1992;31:8207–8215. doi: 10.1021/bi00150a013. [DOI] [PubMed] [Google Scholar]
- 33.Emanuele JJ, Jr., Fitzpatrick PF. Mechanistic studies of the flavoprotein tryptophan 2-monooxygenase. 2. pH and kinetic isotope effects. Biochemistry. 1995;34:3716–3723. doi: 10.1021/bi00011a029. [DOI] [PubMed] [Google Scholar]
- 34.Silverman RB. Radical ideas about monoamine oxidase. Acc. Chem. Res. 1995;28:335–342. [Google Scholar]
- 35.McCann AE, Sampson NS. A C6 - flavin adduct is the major product of irreversible inactivation of cholesterol oxidase by 2a,3a-cyclopropano-5a-cholestan-3bol. J. Am. Chem. Soc. 2000;122:35–39. [Google Scholar]
- 36.Chen ZW, Zhao G, Martinovic S, Jorns MS, Mathews FS. Structure of the sodium borohydride-reduced N-(cyclopropyl)glycine adduct of the flavoenzyme monomeric sarcosine oxidase. Biochemistry. 2005;44:15444–15450. doi: 10.1021/bi0515422. [DOI] [PubMed] [Google Scholar]
- 37.Henderson Pozzi M, Gawandi V, Fitzpatrick PF. pH Dependence of a Mammalian Polyamine Oxidase: Insights into Substrate Specificity and the Role of Lysine 315. Biochemistry. 2009;48:1508–1516. doi: 10.1021/bi802227m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ralph EC, Fitzpatrick PF. pH and kinetic isotope effects on sarcosine oxidation by N-methyltryptophan oxidase. Biochemistry. 2005;44:3074–3081. doi: 10.1021/bi047716h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dunn RV, Marshall KR, Munro AW, Scrutton NS. The pH dependence of kinetic isotope effects in monoamine oxidase A indicates stabilization of the neutral amine in the enzyme-substrate complex. FEBS Journal. 2008;275:3850–3858. doi: 10.1111/j.1742-4658.2008.06532.x. [DOI] [PubMed] [Google Scholar]
- 40.Pawelek PD, Cheah J, Coulombe R, Macheroux P, Ghisla S, Vrielink A. The structure of L-amino acid oxidase reveals the substrate trajectory into an enantiomerically conserved active site. EMBO J. 2000;19:4204–4215. doi: 10.1093/emboj/19.16.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Binda C, Coda A, Angelini R, Federico R, Ascenzi P, Mattevi A. A 30 Å long U-shaped catalytic tunnel in the crystal structure of polyamine oxidase. Structure. 1999;7:265–276. doi: 10.1016/s0969-2126(99)80037-9. [DOI] [PubMed] [Google Scholar]
- 42.Huang Q, Liu Q, Hao Q. Crystal structures of Fms1 and its complex with spermine reveal substrate specificity. J. Mol. Biol. 2005;348:951–959. doi: 10.1016/j.jmb.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 43.Binda C, Newton-Vinson P, Hubalek F, Edmondson DE, Mattevi A. Structure of human monoamine oxidase B, a drug target for the treatment of neurological disorders. Nat. Struct. Biol. 2002;9:22–26. doi: 10.1038/nsb732. [DOI] [PubMed] [Google Scholar]
- 44.Ma J, Yoshimura M, Yamashita E, Nakagawa A, Ito A, Tsukihara T. Structure of rat monoamine oxidase A and its specific recognitions for substrates and inhibitors. J. Mol. Biol. 2004;338:103–114. doi: 10.1016/j.jmb.2004.02.032. [DOI] [PubMed] [Google Scholar]
- 45.Lee MB, Winder C, Cooch H, Shiekhattar R. An essential role for CoREST in nucleosomal histone3 lysine4 demethylation. Nature. 2005;437:432–435. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- 46.Polticelli F, Basran J, Faso C, Cona A, Minervini G, Angelini R, Federico R, Scrutton NS, Tavladoraki P. Lys300 plays a major role in the catalytic mechanism of maize polyamine oxidase. Biochemistry. 2005;44:16108–16120. doi: 10.1021/bi050983i. [DOI] [PubMed] [Google Scholar]
- 47.Massey V, Gibson QH. Role of semiquinones in flavoprotein catalysis. Fed.Proc. 1964;23:18–29. [PubMed] [Google Scholar]
- 48.Massey V, Curti B. On the reaction mechanism of Crotalus adamanteus L-amino acid oxidase. J.Biol.Chem. 1967;242:1259–1264. [PubMed] [Google Scholar]
- 49.Walker MC, Edmondson DE. Structure-activity relationships in the oxidation of benzylamine analogues by bovine liver mitochondrial monoamine oxidase B. Biochemistry. 1994;33:7088–7098. doi: 10.1021/bi00189a011. [DOI] [PubMed] [Google Scholar]
- 50.Emanuele JJ, Jr., Fitzpatrick PF. Mechanistic studies of the flavoprotein tryptophan 2-monooxygenase. 1. Kinetic mechanism. Biochemistry. 1995;34:3710–3715. doi: 10.1021/bi00011a028. [DOI] [PubMed] [Google Scholar]
- 51.Basran J, Bhanji N, Basran A, Nietlispach D, Mistry S, Meskys R, Scrutton NS. Mechanistic aspects of the covalent flavoprotein dimethylglycine oxidase of Arthrobacter globiformis studied by stopped-flow spectrophotometry. Biochemistry. 2002;41:4733–4743. doi: 10.1021/bi025519h. [DOI] [PubMed] [Google Scholar]