Abstract
The secretome is a functionally rich proteome subset, including cellular membrane and extracellular proteins processed through the secretory pathway. In this study, Danio rerio and Homo sapiens RefSeq proteins were analyzed with SignalP, TargetP, Phobius, and pTarget algorithms. About 16.5% of the zebrafish proteome and 17.0% of the human proteome possessed predicted N-terminal signal sequences. Nearly half of these proteins were subsequently classified as soluble, as they lacked predicted transmembrane domains. The soluble proteins were further subclassified, predicting 1345 (3.8%) zebrafish and 1207 (3.2%) human proteins as extracellular. Comparison of the zebrafish and human soluble secretome proteins identified 372 as orthologs, on the basis of reciprocal BLAST best hits. The computational characterization of the zebrafish proteins found many more members of the secretome than annotated in the SwissProt database. Only 180 of the 2078 zebrafish SwissProt protein entries, and 995 of the 19,294 human SwissProt protein entries were annotated with secreted protein locales. A specific investigation of the fibroblast growth factor and matrix metalloproteinase (MMP) protein families confirmed the prediction data and generated annotation of three additional putative MMP zebrafish proteins. This study presents the first known published description of the zebrafish secretome since the completion of the zebrafish genome sequencing project.
Introduction
Secreted proteins represent a functionally rich subset of the proteome actively involved in many functions, including intercellular signaling, chemoattractant cellular recruitment, disease–host response, embryonic development, and organogenesis.1–4 Clinically, this class of proteins has been extensively studied for roles in disease onset, as therapeutic targets, and as diagnostic and prognostic biomarkers. These proteins are also important factors in developmental biology, and a large number were recently characterized in a reverse genetics screen in zebrafish.4 To facilitate ongoing research involving secreted proteins, a comprehensive survey of the zebrafish secretome would be beneficial. Here, we revisit the computational prediction of secreted proteins in zebrafish using RefSeq protein sequences derived from the sequenced zebrafish genome. These results are compared to the results obtained in a pre-genome sequencing study of the zebrafish secretome, and contrasted with the results of a mirrored characterization of the human secretome. The complete sequencing of the zebrafish genome has created an immense sequence-based resource for investigators and improved our ability to characterize the zebrafish secretome. However, it is clear there is a substantial deficiency in annotated zebrafish sequence data. The results of this study demonstrate how computational methods may be applied to further annotate the zebrafish genome and proteome.
The term “secretome” was first coined to describe the components for protein secretion and the secreted proteins in Bacillus subtilis.5 The subsequent adaptation of this term to eukaryotes has described different subsets of the proteome, including all proteins processed through the secretory pathway, only those proteins processed through the secretory pathway that lack transmembrane domains, and only those proteins secreted from the cell. For the purpose of this study, the secretome will refer to proteins processed through the secretory pathway. Classically secreted proteins are characterized by an N-terminal signal sequence that mediates the cotranslational translocation (CTT) of the nascent peptide chain into the endoplasmic reticulum (ER).6 Proteins possessing transmembrane domains become membrane bound during the CTT process.7 The CTT proteins are subsequently directed from the ER to their terminal cellular destinations via secondary signal sequences and chaperone protein interactions.8–9 The terminal cellular destinations include the cell membrane, the extracellular region, the ER, the Golgi apparatus, and other organelles.
There are many tools publicly available that computationally predict the cellular localization of proteins.10 A common method for identifying classically secreted proteins is to predict the presence or absence of the N-terminal signal sequence. The N-terminal signal sequences are not primary-sequence conserved, but possess a conserved set of secondary characteristics that are identifiable by supervised learning algorithms.11,12 SignalP13 and TargetP14 are two prediction programs that historically have been accurate predictors of N-terminal signal sequences. The combined prediction accuracy of these two programs was shown to have added value,15 and will be used in this study to identify CTT proteins. The Phobius16 prediction algorithm can be used to further classify the CTT proteins into those membrane bound and those in circulation. Finally, the pTarget17,18 prediction program can classify the circulating CTT proteins into those retained within the ER or Golgi, those secreted into the extracellular environment, and those localized to other cellular compartments. When applying these algorithms to full-length protein sequence databases, an accurate depiction of the secretome can be generated.
Results
Characterization of the proteome
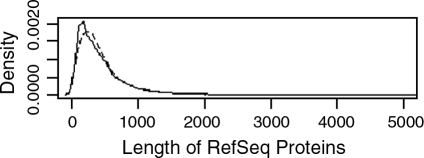
The complete RefSeq protein sequence sets for the zebrafish (35,668 proteins) and human (37,862 proteins) were downloaded from NCBI. 96.5% of the zebrafish protein sequences were initiated with a methionine, and 99.7% of the human protein sequences were initiated with a methionine. Based on the FASTA tag annotation, 64.1% of the zebrafish protein sequences were “PREDICTED,” and 55.0% were labeled as “hypothetical proteins.” Conversely, only 33.4% of the human protein sequences were annotated as PREDICTED, and 24.0% were labeled as hypothetical proteins. The mean and median protein sequence lengths are 452 and 344 residues for the zebrafish proteins and 456 and 321 residues for the human proteins. Figure 1 is a density plot of the protein sequence lengths in the zebrafish and human sequence sets. The plots for the zebrafish and human proteomes are nearly identical, suggesting that the zebrafish protein sequences are equivalent in length to the human proteins and likely to represent complete proteins.
FIG. 1.
Density plot of the RefSeq protein sequence lengths for human (solid line) and zebrafish (dashed line).
Sequence predictions
The N-terminal signal sequence predictions are based on the consensus prediction of SignalP and TargetP. As illustrated in Table 1, slightly more zebrafish and human proteins were predicted to possess an N-terminal signal sequence by TargetP than by SignalP, but the combined predictions were highly similar. The 5905 zebrafish and 6419 human signal sequence positive proteins are referred to as CTT proteins.
Table 1.
Proteins with Predicted N-Terminal Signal Sequences (CTT Proteins)
| TargetP | SignalP | TargetP & SignalP | |
|---|---|---|---|
| Zebrafish | 6982 | 6254 | 5905 |
| Human | 7966 | 6969 | 6419 |
CTT, cotranslational translocation.
The CTT proteins were further analyzed to segregate the membrane-bound proteins from the soluble proteins, using Phobius. This program predicts signal peptides and transmembrane domains, thereby differentiating N-terminal signal sequences from N-terminal signal anchors and identifying membrane-bound proteins. Of the 5905 zebrafish CTT proteins, 1630 (27.6%) were predicted to have a signal peptide and encode for one or more transmembrane domains (membrane proteins), 2786 (47.2%) predicted to have a signal peptide and encode for no transmembrane domains (soluble proteins), and 1489 (25.2%) did not have a Phobius-predicted signal peptide (undetermined proteins). Likewise, for the 6419 human CTT protein sequences, there were 2000 (28.0%) membrane proteins, 2874 (44.8%) soluble proteins, and 1545 (24.1%) undetermined proteins. The 2786 soluble zebrafish proteins and 2874 soluble human proteins were further subclassified as retained in the endoplasmic reticulum, retained in the Golgi apparatus, secreted to the extracellular environment, or localized to other cellular compartments, using pTarget. As illustrated in Table 2, nearly half of the zebrafish soluble CTT proteins are secreted from the cell, while another quarter are retained in the ER and Golgi. The human proteins are similarly distributed, with a slightly lower percentage of the soluble CTT proteins predicted to be localized to the ER and Golgi. A complete list of all zebrafish CTT proteins and predictions are provided in the Supplemental Table 1 (go to www.liebertpub.com/zeb).
Table 2.
Predicted Hierarchal Localization of CTT Proteins in Zebrafish and Humans
| |
|
|
Number of proteins |
|
|---|---|---|---|---|
| SignalP & TargetP | Phobius | pTarget | Zebrafish | Human |
| Membrane bound | — | 3027 | 3545 | |
| CTT proteins | Secreted/extracellular | 1345 | 1207 | |
| Soluble | Endoplasmic reticulum | 373 | 294 | |
| Golgi apparatus | 304 | 233 | ||
| Other | 763 | 1140 | ||
CTT, cotranslational translocation.
Zebrafish to human comparisons
The soluble CTT proteins from the zebrafish and human proteomes were compared using a reciprocal BLAST method to identify putative orthologs and estimate the overlap in the two species soluble secretomes. Using an e-value threshold of ≤ 0.001, 372 protein sequences were found to be reciprocal best hits between the zebrafish and human protein sequence sets. Additionally, 490 zebrafish soluble CTT proteins did not match a human soluble CTT protein, and 798 human soluble CTT proteins did not match a zebrafish soluble CTT protein. To determine whether the e-value threshold was substantially biasing the results, the comparisons were repeated using a threshold of 0.01 and 0.0001, obtaining 380 and 362 reciprocal BLAST hits, respectively. As altering the threshold had a minimal impact on the number of orthologs identified, we were confident in using the results obtained with a threshold of 0.001.
SwissProt annotations
To compare the predicted zebrafish and human secretomes to the currently available protein annotations, the SwissProt protein knowledgebase was queried for zebrafish and human protein sequences with defined cellular localizations. Two thousand and seventy-eight zebrafish protein sequences and 19,294 human protein sequences were identified in the database. One thousand four hundred and sixty-two (70%) zebrafish sequences and 7987 (41%) human sequences possessed “SUBCELLLULAR LOCATION” annotations in the “Comments” field. As described in Table 3, 533 (25.6%) zebrafish proteins and 3516 (18.2%) human proteins contained annotation indicative of being CTT proteins. These annotations were first subdivided by subcellular location, into membrane-bound and soluble (nonmembrane bound) proteins. Secondarily, the soluble proteins were subdivided into those retained within the endoplasmic reticulum, retained within the Golgi apparatus, secreted to the extracellular environment, or localized to other cellular compartments.
Table 3.
SwissProt Annotated Subcellular Localization of CTT Proteins in Zebrafish and Humans
| |
|
|
Number of proteins |
|
|---|---|---|---|---|
| Subcellular location | Zebrafish | Human | ||
| Membrane bound | Membrane | 353 | 2521 | |
| CTT proteins | Secreted/extracellular | 82 | 630 | |
| Soluble | Endoplasmic reticulum | 64 | 237 | |
| Golgi apparatus | 34 | 128 | ||
| Non-CTT proteins | Other | 929 | 4471 | |
| Unknown | No annotation | 617 | 3058 | |
CTT, cotranslational translocation.
Fibroblast growth factors
Fibroblast growth factors (FGF) consist of a large gene family encoding secreted proteins with extensive orthology between zebrafish and human. These proteins have been extensively studied and annotated in both species, providing a valuable protein subset in which the localization predictions made in this study can be assessed and compared to annotated localization data available in SwissProt.19 The RefSeq protein database contains all known FGF proteins, including 27 in zebrafish and 22 in human. Summaries of the FGF protein predictions and SwissProt annotations are provided in Supplemental Tables 2 and 3, for zebrafish and human, respectively (go to www.liebertpub.com/zeb). The predicted localizations of all human to zebrafish FGF orthologs were identical. There are 18 predicted FGF zebrafish CTT proteins and 9 non-CTT proteins, and 13 predicted FGF human CTT proteins and 9 non-CTT proteins. Eight of the nine non-CTT proteins found in human and zebrafish are identical. FGF9 is the only human non-CTT protein not found in the zebrafish non-CTT protein set, as there is no ortholog in the zebrafish proteome. Within zebrafish, there are two FGF20 proteins, FGF20a and FGF20b (paralogs), where only a single ortholog exists in human (FGF20).20 For the predicted FGF human CTT proteins, all zebrafish orthologs were predicted to be CTT proteins. There are an additional five zebrafish CTT proteins representing zebrafish paralog proteins. Within SwissProt, 11 of the 13 FGF human CTT proteins are annotated as “Secreted,” with the remaining 2 lacking subcellular location annotations. Conversely, only one of the FGF zebrafish CTT proteins (FGF3) is annotated as “Secreted.” The remaining 16 CTT proteins lacked entries in the SwissProt database. Of the nine FGF human non-CTT proteins, three are annotated as “Secreted,” two as “Nuclear,” and four lack annotation. For the FGF zebrafish non-CTT proteins, only one protein is found in the database, and it lacks subcellular localization annotation.
Matrix metalloproteinases
Proteins of matrix metalloproteinase (MMP) family differ from the FGF proteins, as they are well characterized in humans, but only marginally characterized in zebrafish. This provides an opportunity to delve into the predicted data described in this manuscript and generate hypotheses regarding new putative MMP orthologs in zebrafish. MMPs are secreted and membrane-associated endopeptidases.21–22 There are 25 MMP human proteins and 4 identified MMP zebrafish proteins in the RefSeq database. Twenty-four of the 25 human MMPs are predicted CTT proteins, with MMP24 the only protein lacking a predicted N-terminal signal sequence. All four of the zebrafish MMPs, MMP2, MMP9, MMP13, and MMP14, are predicted CTT proteins. To look for additional undefined, putative MMP proteins in the zebrafish proteome, reciprocal BLAST orthologs were identified between the human MMP proteins and the zebrafish proteome. Three zebrafish CTT proteins labeled as hypothetical proteins in the zebrafish RefSeq database (gi:94536884, gi:125805214, and gi:125812836) were identified as putative orthologs to human MMP proteins (MMP20, MMP24, and MMP17), respectively.
Discussion
The computational characterization of the zebrafish secretome is predicated on the availability of high-quality, full-length protein sequences. Predictions of cellular localization made on truncated or incomplete peptides are wrought with error and false findings.23 In our last analysis of zebrafish-secreted proteins, preceding the sequencing of the zebrafish genome, the secretome was estimated using sequence homology with secreted proteins in other organisms.24 The subsequent full genome sequencing has improved the characterization of the zebrafish proteome, and led to the creation of a large RefSeq protein sequence set. In this study, we characterized the zebrafish secretome using the RefSeq database, making the assumption that it primarily contained complete protein sequences. To provide a cursory estimate of the level of sequence completion in this dataset, the distribution of sequence sizes was compared to that of the well-characterized human RefSeq protein sequence set. As illustrated in Figure 1 and reflected in the mean and median sequence lengths, the zebrafish and human RefSeq datasets contain a highly similar distribution of protein sizes. While this is a promising finding, it is also evident from the sequence annotation that the zebrafish protein sequence set contains a considerably larger number of predicted and hypothetical proteins. Taken together, this suggests that the zebrafish protein sequence set is not over-populated with short fragments, but may still undergo considerable revision as the predicted and hypothetical protein sequences are validated. The predictions reported in this manuscript, therefore, should represent a vast improvement over any pre-zebrafish genome sequencing data, but may still require iteration and refinement as the zebrafish proteome sequence set matures.
The predictive strategy employed in this study was designed to follow the natural sorting of proteins in the cell, a strategy employed in other prediction programs.25 The first step involved identifying proteins with N-terminal signal sequences, which mediate CTT of the nascent peptide chain into the ER, and entry of the mature protein into the secretory pathway. In the second step, these CTT proteins were analyzed to predict the presence of transmembrane domains or N-terminal signal anchors, following the natural segregation of the membrane-bound proteins during the CTT process. Finally, the remaining soluble CTT proteins were analyzed, distinguishing proteins secreted to the extracellular environment from those retained in the ER, Golgi, or other cellular compartments. This approach creates a layered description of the computationally characterized secreted zebrafish proteins.
The similarity in the size of the predicted zebrafish and human CTT protein sequence sets is striking. Comprising 17% (5905) and 18% (6419) of the respective proteomes, there is only a 1% difference between zebrafish and human CTT protein representation in the respective proteomes. These findings are highly similar to what was reported in another study, where 5310 (14%) predicted CTT proteins for Takifugu rubripes (Fugu) and 6716 (20%) predicted CTT proteins for human were found.26 Likewise, the 2786 zebrafish and 2874 human soluble CTT proteins identified in this study are similar in number to that reported in the secreted protein database (SPD).27 The SPD predicts 2973 human, 2981 mouse, and 2317 rat soluble CTT proteins in the respective RefSeq protein sequence sets. LOCATE is an independent database that reports a similar number of soluble CTT protein sequences in mouse (2882) and human (2487).28 The LOCATE database is also one of the few studies to report the subclassification of soluble CTT proteins, identifying 1079 mouse and 2025 human extracellular proteins. The number of mouse proteins and zebrafish extracellular proteins (1345) found in this study are similar. However, the number of human proteins is considerably different, with 1207 human extracellular proteins found in this study. This discrepancy may reflect the fact that LOCATE includes data from both an experimental, immunofluorescence-based assay and from a survey of the literature. By combining these two alternative methods with the de novo prediction method, it is entirely likely that a larger subset of extracellular proteins could be identified. Regardless, the results obtained from this analysis of the zebrafish protein sequence set are very close to what have been previously reported for other vertebrates, suggesting that the findings are an accurate depiction of the zebrafish secretome.
The only known published study of the zebrafish secretome is the previous analysis we performed using homology modeling of protein sequences derived from EST consensus sequences.24 That study compared ∼ 10,000 zebrafish sequences to ∼ 2500 Drosophila melanogaster CTT proteins and identified 560 homologous sequences. It was estimated from these findings that the complete zebrafish proteome may contain as many as 1000–2000 CTT proteins. On the basis of the observations made in this study, it is evident that this was an underestimation of the total number of CTT proteins in the zebrafish proteome. The other large source of protein localization data for the zebrafish is found in the SwissProt knowledgebase. While the annotations in that database are often based on published experimental data, it unfortunately only contains a subset of the zebrafish proteome (2078 proteins), at present. This clearly illustrates a strong need to study and characterize the zebrafish proteome.
The FGF protein family is an extensively studied set of proteins in both human and zebrafish. The complete orthology between the two species for this protein family has been defined, and the cellular localization of the human proteins annotated. Consequently, these proteins provide a valuable positive control for the evaluation of the predictions reported in this study, and provided a way of assessing the coverage of annotation in the SwissProt database. The predicted status (CTT or non-CTT) of all human and zebrafish orthologs was identical. However, the amount of subcellular localization annotation in the SwissProt database varied widely. All but two predicted human CTT proteins possessed annotated subcellular localizations, and these annotations agreed with the predictions (FGF3 and FGF4 lacked annotation). Conversely, only 1 of the 18 CTT zebrafish protein (FGF3) was annotated, and it too had agreement in the annotated and predicted localization. Nine proteins in both human and zebrafish were predicted to be non-CTT proteins, including the four proteins (FGF11–FGF14) known to function intracellularly.20 Three of the nine non-CTT human proteins (FGF9, FGF16, and FGF20) are annotated in SwissProt as secreted. These have been experimentally shown to be secreted, despite the absence of a well-defined N-terminal signal sequence.29 This highlights a limitation of the approach used in this study. It is unable to correctly predict the localization of proteins secreted in a nonclassical manner. For the predicted non-CTT zebrafish proteins, only one protein contains a database entry, and it does not possess subcellular localization annotation. This emphasizes one of the major benefits of this study, providing researchers with clear information on zebrafish protein localization that may not be available in the annotated databases.
Evaluation of the MMP protein family data illustrates one way hypotheses can be generated from the predictions reported in this study. The MMPs are secreted and membrane-associated (CTT proteins) endopeptidases involved in extracellular matrix remodeling.21 These proteins have been extensively studied in human morphogenesis, healing, cardiac disease, and cancer. There is a large (26 proteins) and well-characterized set of human MMP proteins. To date, only four zebrafish MMP proteins have been defined and reported in the literature. Based on the prediction localization status and BLAST-defined orthology between the human and zebrafish CTT proteins, we identified three additional putative zebrafish MMP proteins. These protein sequences align with the human proteins MMP17, MMP20, and MMP24. Using this data, these previously hypothetical proteins in the zebrafish proteome can be specifically studied and functionally characterized, assessing their status as putative MMPs. This is just one way in which the reported description of the zebrafish secretome could be mined for de novo discoveries. The results presented in this manuscript will provide investigators with an updated secretome resource from which additional studies can be constructed and the zebrafish proteome more completely described.
Materials and Methods
Datasets
Protein sequences were obtained from the NCBI RefSeq release 28 database. The D. rerio protein sequences were downloaded on 4/22/2008, and the H. sapiens protein sequences were downloaded on 4/22/2008.
SwissProt sequence annotations were downloaded for all protein sequences with an annotated “Organism” term of “Danio rerio” and “Homo sapiens” on 4/22/2008.
Prediction
The subcellular localization of protein sequences was predicted using the online SignalP version 3.0 and TargetP version 1.1 signal peptide prediction servers at the Center for Biological Sequence Analysis, of the Technical University of Denmark. SignalP predictions were performed using the No graphics and Short output format options. TargetP predictions were performed with the cleavage site prediction option. For both the SignalP and TargetP analyses, 150 residue N-terminal sequences, divided into sequence subsets of 1200, were submitted for analysis. Default parameters were used for analyzing the prediction algorithm outputs. Proteins were considered to have a predicted N-terminal signal sequence if the TargetP prediction returned an “S,” and the SignalP D-score returned a “Y.”
Phobius was used to analyze full-length protein sequences on the online server at the Stockholm Bioinformatics Center, of Stockholm University. The Short output format option was selected. Protein sequences were classified as soluble if they lacked predicted transmembrane domains, and Phobius predicted the presence of an N-terminal signal sequence.
Secretome protein sequences that were predicted to be soluble on the basis of the TargetP, SignalP, and Phobius predictions were subsequently analyzed by pTarget to differentiate extracellular, endoplasmic reticulum, Golgi apparatus, and other proteins. Proteins deemed unanalyzable by pTarget or predicted to be localized to a position other than extracellular, Golgi apparatus, or endoplasmic reticulum were included in the other category.
All data analysis and abstracting of prediction and annotation information was done using PERL scripts.
Orthology
Reciprocal BLAST30 hits were used to define zebrafish–human orthology. Comparisons were made between the complete sequences of the zebrafish and human predicted secreted protein sequence sets using the BLAST version 2.2.16, blastp method. The combination of soft filtering (-F “m S”) and Smith-Waterman final alignment (-s T) options was used to obtain optimal detection of orthologs.31 BLAST analysis was restricted to hits with an e-value significance ≤ 0.001.
Supplementary Material
Acknowledgments
The author would like to acknowledge Dr. Stephen Ekker for his continued involvement in providing insight into the needs and values associated with the zebrafish and zebrafish community.
References
- 1.Caneparo L. Huang YL. Staudt N. Tada M. Ahrendt R. Kazanskaya O, et al. Dickkopf-1 regulates gastrulation movements by coordinated modulation of Wnt/beta catenin and Wnt/PCP activities, through interaction with the Dally-like homolog Knypek. Genes Dev. 2007;21:465–480. doi: 10.1101/gad.406007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boldajipour B. Mahabaleshwar H. Kardash E. Reichman-Fried M. Blaser H. Minina S, et al. Control of chemokine-guided cell migration by ligand sequestration. Cell. 2008;132:463–473. doi: 10.1016/j.cell.2007.12.034. [DOI] [PubMed] [Google Scholar]
- 3.Merritt WM. Sood AK. Markers of angiogenesis in ovarian cancer. Dis Markers. 2007;23:419–431. doi: 10.1155/2007/257602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pickart MA. Klee EW. Nielsen AL. Sivasubbu S. Mendenhall EM. Bill BR, et al. Genome-wide reverse genetics framework to identify novel functions of the vertebrate secretome. PLoS ONE. 2006;1:e104. doi: 10.1371/journal.pone.0000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tjalsma H. Bolhuis A. Jongbloed JD. Bron S. van Dijl JM. Signal peptide-dependent protein transport in Bacillus subtilis: a genomebased survey of the secretome. Microbiol Mol Biol Rev. 2000;64:515–547. doi: 10.1128/mmbr.64.3.515-547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schatz G. Dobberstein B. Common principles of protein translocation across membranes. Science. 1996;271:1519–1526. doi: 10.1126/science.271.5255.1519. [DOI] [PubMed] [Google Scholar]
- 7.Corsi AK. Schekman R. Mechanism of polypeptide translocation into the endoplasmic reticulum. J Biol Chem. 1996;271:30299–30302. doi: 10.1074/jbc.271.48.30299. [DOI] [PubMed] [Google Scholar]
- 8.Cabrera M. Muniz M. Hidalgo J. Vega L. Martin ME. Velasco A. The retrieval function of the KDEL receptor requires PKA phosphorylation of its C-terminus. Mol Biol Cell. 2003;14:4114–4125. doi: 10.1091/mbc.E03-04-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arvan P. Zhang B. Feng L. Liu M. Kuliawat R. Lumenal protein multimerization in the distal secretory pathway/secretory granules. Curr Opin Cell Biol. 2002;14:448–453. doi: 10.1016/s0955-0674(02)00344-7. [DOI] [PubMed] [Google Scholar]
- 10.Klee EW. Sosa CP. Computational classification of classically secreted proteins. Drug Discov Today. 2007;12:234–240. doi: 10.1016/j.drudis.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 11.von Heijne G. Signal sequences. The limits of variation. J Mol Biol. 1985;184:99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]
- 12.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bendtsen JD. Nielsen H. von Heijne G. Brunak S. Improved prediction of signal peptides: signalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 14.Emanuelsson O. Nielsen H. Brunak S. von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 15.Klee EW. Ellis LB. Evaluating eukaryotic secreted protein prediction. BMC Bioinformatics. 2005;6:256. doi: 10.1186/1471-2105-6-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kall L. Krogh A. Sonnhammer EL. A combined transmembrane topology and signal peptide prediction method. J Mol Biol. 2004;338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 17.Guda C. Subramaniam S. pTARGET: a new method for predicting protein subcellular localization in eukaryotes. Bioinformatics. 2005;21:3963–3969. doi: 10.1093/bioinformatics/bti650. [DOI] [PubMed] [Google Scholar]
- 18.Guda C. pTARGET: a web server for predicting protein subcellular localization. Nucleic Acids Res. 2006;34:W210–W213. doi: 10.1093/nar/gkl093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Itoh N. Konishi M. The zebrafish fgf family. Zebrafish. 2007;4:179–186. doi: 10.1089/zeb.2007.0509. [DOI] [PubMed] [Google Scholar]
- 20.Goldfarb M. Fibroblast growth factor homologous factors: evolution, structure, and function. Cytokine Growth Factor Rev. 2005;16:215–220. doi: 10.1016/j.cytogfr.2005.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagase H. Visse R. Murphy G. Structure and function of matrix metalloproteinases and TIMPs. Cardiovasc Res. 2006;69:562–573. doi: 10.1016/j.cardiores.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 22.Nagase H. Woessner JF., Jr. Matrix metalloproteinases. J Biol Chem. 1999;274:21491–21494. doi: 10.1074/jbc.274.31.21491. [DOI] [PubMed] [Google Scholar]
- 23.Nielsen H. Brunak S. von Heijne G. Machine learning approaches for the prediction of signal peptides and other protein sorting signals. Protein Eng. 1999;12:3–9. doi: 10.1093/protein/12.1.3. [DOI] [PubMed] [Google Scholar]
- 24.Klee EW. Ekker SC. Ellis LB. Target selection for Danio rerio functional genomics. Genesis. 2001;30:123–125. doi: 10.1002/gene.1045. [DOI] [PubMed] [Google Scholar]
- 25.Nair R. Rost B. Mimicking cellular sorting improves prediction of subcellular localization. J Mol Biol. 2005;348:85–100. doi: 10.1016/j.jmb.2005.02.025. [DOI] [PubMed] [Google Scholar]
- 26.Klee EW. Carlson DF. Fahrenkrug SC. Ekker SC. Ellis LB. Identifying secretomes in people, pufferfish and pigs. Nucleic Acids Res. 2004;32:1414–1421. doi: 10.1093/nar/gkh286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y. Zhang Y. Yin Y. Gao G. Li S. Jiang Y, et al. SPD—a web-based secreted protein database. Nucleic Acids Res. 2005;33:D169–D173. doi: 10.1093/nar/gki093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sprenger J. Fink J. Karunaratne S. Hanson K. Hamilton NA. Teasdale RD. LOCATE: a mammalian protein subcellular localization database. Nucleic Acids Res. 2008;36:D230–D233. doi: 10.1093/nar/gkm950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeffers M. Shimkets R. Prayaga S. Boldog F. Yang M. Burgess C, et al. Identification of a novel human fibroblast growth factor and characterization of its role in oncogenesis. Cancer Res. 2001;61:3131–3138. [PubMed] [Google Scholar]
- 30.Altschul Stephen F. Madden TL. Schaffer AA. Zhang J. Zhang Z, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moreno-Hagelsieb G. Latimer K. Choosing BLAST options for better detection of orthologs as reciprocal best hits. Bioinformatics. 2008;24:319–324. doi: 10.1093/bioinformatics/btm585. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.