Abstract
Interactions of matrix metalloproteinase-2 (MMP-2)1 with native and denatured forms of several types of collagen are mediated by the collagen-binding domain (CBD). CBD positions substrates relative to the catalytic site and is essential for their cleavage. Our previous studies identified an CBD binding site on the α1(I) collagen chain. The corresponding synthetic collagen peptide P713 bound CBD with high affinity and was used in this study to identify specific collagen binding residues by NMR analysis of 15N-labeled CBD complexed with P713. Results obtained showed that P713 caused chemical shifts perturbations of several surface exposed CBD backbone amide resonances in a concentration dependent manner. The ten residues that underwent the largest chemical shift perturbations (R252 in module 1, R296, F297, Y302, E321, Y323, Y329 in module 2, and R368, W374, and Y381, in module 3), were investigated by site-specific substitution with alanine. The structural integrity of the CBD variants was also analyzed by 1D 1H NMR. Surface plasmon resonance and microwell protein binding assays of control and CBD variants showed that residues in all three CBD modules contributed to collagen binding. Single residue substitutions altered the affinity for peptide P713, as well as native and denatured type I collagen, with the greatest effects observed for residues in modules 2 and 3. Additional alanine substitutions involving residues in two or three modules simultaneously further reduced the binding of CBD to native and denatured type I collagen and demonstrated that all three modules contribute to the substrate binding. These results have localized and confirmed the key collagen binding site residues in the three fibronectin type II-like modules of MMP-2.
Keywords: MMP-2, CBD, binding site, NMR, alanine substitution, collagen, gelatin
Matrix metalloproteinases (MMPs) belong to a family of structurally and functionally related endopeptidases that not only are capable of cleaving most major macromolecules of the extracellular matrix, but also are critical to shedding and processing of non-structural proteins, including essential inflammatory mediators (1). MMPs play important roles in normal tissue remodeling (2), are key to wound healing and angiogenesis (3, 4), and are critical for tumor expansion and metastasis (5). Of three common MMP domains, the propeptide domain maintains the latency of pro-MMPs (6, 7), the catalytic domain contains the proteolytic active site (8), and the hemopexin-like domain provides exosite interactions with substrate molecules (9, 10) and interaction with the tissue inhibitors of metalloproteinases (TIMPs).
Among MMPs, only MMP-2 and MMP-9 contain a collagen binding domain (CBD) comprised of three fibronectin (FN) type II-like modules inserted in tandem in the catalytic domain (11, 12). These collagen binding domains are responsible for the binding of MMP-2 and −9 to collagen molecules (12, 13). Isolated recombinant CBDs from MMP-2 and MMP-9 retain ligand binding properties not only for type I collagen, but also for the native and denatured forms of a series of other collagen types and elastin (12, 14, 15). Deletion of the CBD of MMP-2 and −9 dramatically reduces activities of the enzymes on gelatin and elastin (16, 17). Moreover, both binding and cleavage of gelatin by intact MMP-2 and −9 can be competed by soluble CBDs, showing that the change in catalytic activity is not due to major structural perturbation caused by the domain deletion (18). Thus, interactions of CBD with substrates molecules are critical to the catalytic activities of MMP-2 and MMP-9.
In early analyses, constructs containing 2 or 3 CBD modules bound stronger to collagen than single modules implying that all the modules contribute to collagen binding (19). In addition, site-specific mutagenesis of residues in the CBD modules in MMP-9, that have high homology with the corresponding CBD modules in MMP-2, pointed to binding site patches formed by hydrophobic residues in the three homologous modules (20). Subsequent resolution of the crystal structure for MMP-2 revealed that the putative binding sites in the three CBD modules were oriented outward and away from each other, suggesting the potential for three independent binding sites (21). Early NMR studies of isolated CBD module 2 complexed with a peptide containing repeats of the generic Gly-X-Y sequence characteristic of collagen (Pro-Pro-Gly)6 showed spectral perturbations of the surface-exposed aromatic residues Phe21, Tyr38, Trp40, Tyr47, Tyr53 and Phe55, as well as the neighboring Gly33-Gly37 segment (22). However, analysis of recombinant proteins with alanine substitution of these residues demonstrated that only the protein variant with the Tyr37Ala modification lost gelatin binding properties without substantial structural perturbation (23). Further analyses of interactions between the (Pro-Pro-Gly)6 peptide and the CBD variants by NMR showed similar binding affinities of single- and tri-modular constructs indicating that each module may function independently (24). Of note, our prior analyses of activities and ligand interactions for MMP-2 showed that positioning by the CBD was required for cleavage of longer, but not short peptide substrate molecules (18).
Though previous studies have been valuable in identifying binding site residues for a collagen-like peptide, questions remain about the specific functional contributions of individual residues to binding of collagen-like peptides and with collagen α-chains that potentially could bind more than one module simultaneously.
Recently, we located a CBD binding region on the α1(I) collagen chain by screening a random peptide library with rCBD as bait. The corresponding synthetic collagen peptide (P713) bound specifically to the CBD of MMP-2, but not to the catalytic site, and reduced gelatin cleavage. (25). The availability of this collagen peptide P713 has enabled us to experimentally identify binding site residues within the CBD by NMR analysis. Here, we report the location of these residues as well as results from functional studies with a series of variant CBDs in which key residues were substituted with alanines in one, two or all three modules. By this structure-functional approach, we have now identified and functionally confirmed the type I collagen binding sites on MMP-2 CBD and also characterized the contribution of the three modules to the interactions with this ligand.
EXPERIMENTAL PROCEDURES
Expression and purification of rCBD for NMR and rCBD variants
Recombinant collagen binding domain (CBD) from human MMP-2 was expressed in E. coli BL21(DE3) transformed with the expression vector pGYMX containing the CBD-coding sequence as detailed previously (12). Wildtype and CBD variants were expressed in LB medium.
Isotopically labeled rCBD for NMR studies was expressed with E. coli BL21(DE3) in M9 minimal medium (26). For HSQC spectra, the regular nitrogen source (NH4Cl) was substituted for the 15N-labeled counterpart (Isotec, Miamisburg, OH). For triple-resonance experiments, in addition to 15NH4Cl, the regular carbon source, D-glucose, was replaced by 13C-labeled D-glucose (Isotec, Miamisburg, OH).
All rCBDs expressed in inclusion bodies and were therefore dissolved with 8 M urea, 0.1 M NaH2PO4, pH 8.0, and purified by Ni-affinity chromatography under denaturing conditions prior to refolding as detailed previously (18). For functional analyses, rCBDs were equilibrated with 50 mM Tris, 150 mM NaCl, pH 8.0 and stored at −80 °C until analyses.
Recombinant CBD for NMR analysis was further purified by reverse-phase HPLC. After the Ni-affinity chromatography purification step, rCBD was dialyzed against 50 mM Tris pH 8.0, concentrated to 1.5 mg/ml, and loaded onto a onto a Jupiter C4 300Å, 250 × 4.6 mm RP-HPLC column (Phenomenex, Torrance, CA). Protein was washed and eluted with a gradient of acetonitrile (ACN) containing 0.1% trifluoroacetic acid (TFA) at 1 ml/min. Eluted monomeric rCBD was lyophilized and resuspended in 25 mM phosphate buffer at pH 7.0. The rCBD was ultracentrifugated at 100,000 g for 30 min, and D2O/H2O was added to a final concentration of 5%. The protein concentration for NMR experiments was 0.5 mM.
Non-functional alkylated CBD (AlkCBD) that was used as negative control in protein binding assays was prepared as detailed previously (12) by reduction in 8 M urea, 65 mM DTT, 2 mM EDTA, 0.5 M Tris, pH 8.0 overnight at 4 °C. The reduced CBD was then incubated for 1 h at 50 °C and subsequently reacted with 130 mM iodoacetic acid (Sigma, St. Louis, MO) for 30 min at 22 °C. The alkylated CBD was equilibrated with 50 mM Tris, 150 mM NaCl, pH 7.4 and stored at −80 °C until analyses.
Sequential assignment of rCBD
rCBD was sequentially assigned by recording and analyzing CBCA(CO)NH, HNCACB, C(CO)NH, HNCA, HNCO, HNHA and HCACO data sets collected with 15N- and 13C-labeled rCBD. NMR data were acquired at 27 °C on a 600 MHz or a 700 MHz Bruker spectrometer with a conventional 5 mm 1H-{13C, 15N} probehead fitted with a triple-axis gradient coil (Billerica, MA). The data was processed using nmrPipe and was analyzed using NMRView. The CBD chemical shift assignments have been deposited in the BMRB database under accession code 16221.
Interaction of peptide with rCBD monitored by NMR
The 12 residue-long collagen peptide P713 (25) (CGA(HYP)GA(HYP)GSQGA) was synthesized at the UTHSCSA Protein Core Facility by sequential addition of FMOC protected amino acids on a Multiple Peptide Synthesizer Model 396 MPS (Advanced Chemtech, Louisville, KY). Peptides were dissolved in H2O at concentration of 80 mM and adjusted pH to 7.0. Aliquots of P713 peptide were added to 0.5 mM 15N-labeled rCBD yielding a P713 concentration range of 0.15 – 3 mM. After each P713 addition, a 1H – 15N HSQC spectrum was recorded to monitor the peptide-induced resonance shifts. All data were acquired at 25 °C on a Bruker 600 MHz spectrometer with a 5 mm 1H-{13C, 15N} pulsed field gradient cryoprobe (Bruker, Billerica, MA). Chemical shift perturbations, Δδ, were calculated from proton and nitrogen shift by using the formula {[(δΔH)2 +(δΔN/5)2]/2}½. Dissociation constants, Kd, for residues undergoing measurable shifts were determined by non-linear curve fitting of chemical shift versus peptide concentration using the equation Δδ = [(Δδmax * x)/(Kd + x)], where x is concentration of P713 added, Δδ is chemical shift perturbations and Δδmax is chemical shift perturbations at saturation of ligand (Sigma Plot software; SPSS Corp., Chicago, IL). The calculated maximal chemical shift value of each residue was added to the MMP-2 PDB file (1CK7) and viewed by the UCSF Chimera software. The backbone amide chemical shift changes induced by peptide P713 were depicted by the intensity of blue color in three-dimensional surface models of the CBD.
Site-specific alanine substitution of rCBD
Site-specific alanine substitutions were introduced into rCBD by alanine replacement of key residues by overlap-extension PCR with the QuikChange site-directed mutagenesis kit (Stratagene La Jolla, CA ) according to the manufacturers instructions using pGYMX-CBD (12) as template and pairs of primers purified by acrylamide gel electrophoresis (Table 1). By this approach, we generated 12 single-point alanine substitutions at positions R252, F280, R296, F297, Y302, E321, Y323, Y329, E361, R368, W374, and Y381. For three double-site alanine substitutions on residues R252 and F297, R252 and R368, and F297 and R368, we used the R252A and F297A variant plasmids as templates. Finally, to obtain one triple-site alanine substitution for residues R252, F297, and R368, we used the F297A, R368A plasmid as template. All 16 variant plasmids were sequenced to confirm that only target residues were changed as designed. The CBD variants proteins were expressed and purified as described above.
Table 1.
Primers used to introduce site specific alanine substitution and confirm MMP-2 CBD collagen binding site residues
| Residues | Forward Primers | Reverse Primers |
|---|---|---|
| R252A | GCACTGATACTGGCGCCAGCGATGGCTTCC | GGAAGCCATCGCTGGCGCCAGTATCAGTGC |
| F280A | CCATGAAGCCCTGGCCACCATGGGCGGC | GCCGCCCATGGTGGCCAGGGCTTCATGG |
| R296A | CTGCAAGTTTCCATTCGCCTTCCAGGGCACATCC | GGATGTGCCCTGGAAGGCGAATGGAAACTTGCAG |
| F297A | GTTTCCATTCCGCGCCCAGGGCACATCC | GGATGTGCCCTGGGCGCGGAATGGAAAC |
| Y302A | CCAGGGCACATCCGCTGACAGCTGCACC | GGTGCAGCTGTCAGCGGATGTGCCCTGG |
| E321A | GTGCGGCACCACTGCGGACTACGACCGCG | CGCGGTCGTAGTCCGCAGTGGTGCCGCAC |
| Y323A | CACCACTGAGGACGCCGACCGCGACAAG | CTTGTCGCGGTCGGCGTCCTCAGTGGTG |
| Y329A | CCGCGACAAGAAGGCTGGCTTCTGCCCTG | CAGGGCAGAAGCCAGCCTTCTTGTCGCGG |
| E361A | GGGCAACAAATATGCGAGCTGCACCAGCG | CGCTGGTGCAGCTCGCATATTTGTTGCCC |
| R368A | CACCAGCGCCGGCGCCAGTGACGGAAAG | CTTTCCGTCACTGGCGCCGGCGCTGGTG |
| W374A | GTGACGGAAAGATGGCGTGTGCGACCACAG | CTGTGGTCGCACACGCCATCTTTCCGTCAC |
| Y381A | GACCACAGCCAACGCCGATGACGACCGC | GCGGTCGTCATCGGCGTTGGCTGTGGTC |
1D NMR analysis
To verify that the introduced substitutions did not cause major structural perturbations that could interfere with the subsequent functional assays, all recombinant proteins were first analyzed by their migration under reduced and non-reduced conditions using SDS-PAGE. In addition, 1D NMR spectra were acquired for WT and all variant CBDs. Briefly, purified proteins were lyophilized, dissolved in 400 µl H2O at a concentration of 150–200 µM, dialyzed against 25 mM Na2HPO4 - NaH2PO4 buffer containing 5% D2O, pH 7.0 and centrifuged at 15,000 g for 10 min at 4 °C. One-dimensional 1H NMR spectra were then collected at a 1H frequency of 700 MHz using a Bruker Avance 700 MHz NMR spectrometer using a 1-1 echo solvent suppression scheme. Typically, signal dispersion above 8.5 ppm representing backbone amide groups and below 0.8 ppm representing methyl groups occurs only if the protein is structurally ordered (27).
Ligand interactions of rCBD variants by surface plasmon resonance assays
The effects of changes of CBD putative collagen binding site residues on the interaction of CBD with native and denatured type I collagen and P713 peptide were analyzed using a Biacore 3000 surface plasmon resonance (SPR) instrument (GE Healthcare, Piscataway, NJ) at the UTHSCSA Center for Surface Plasmon Resonance. Native or denatured type I collagen, as well as P713 peptide, were immobilized in separate flow cells on CM5 sensor chips (GE Healthcare, Piscataway, NJ). One uncoated flow cell served as negative control. Briefly, CM5 chips were activated by injection of 15 µl 0.2 M N-ethyl-N’-(dimethylaminopropyl) carbodiimide and 0.05 M N-hydroxy-succinimide for 6 min. Native and heat-denatured (gelatin) non-pepsin-treated acid-soluble rat tail type I collagen prepared as described previously (12, 28) were exchanged into 10 mM sodium acetate, pH 4.5, and immobilized in the flow cells at ∼3000 response units (RUs). Non-specific binding sites were blocked with 1M ethanolamine. For coating with peptide, CM5 chips were activated by 0.2 M N-ethyl-N’-(dimethylaminopropyl) carbodiimide and 0.05 M N-hydroxy-succinimide and then modified by 2-(2-pyridinyldithio) ethaneamine hydrochloride (PDEA Thiol Coupling Reagent, GE Healthcare). Peptide in 10 mM sodium acetate buffer, pH 4.0, was then immobilized in the flow cell at ∼500 RUs and the surface was blocked with 30 µl cysteine (6 mg/ml).
In binding assays, WT CBD, rCBD variants, or alkylated CBD (analytes) at 1 µM concentrations in Biacore buffer (10 mM HEPES, 150 mM NaCl, 0.005% surfactant P20, pH 7.4) were passed over the coated flow cell surfaces at 5 µl/min for 6 min. Interactions of analytes with immobilized ligands were expressed by the RU in time sensorgrams. Presented RU values reflect averages of 5 measurements obtained over 5 sec preceding the end of injection of analytes, which reflects the changes in the concentration of molecules at the surface of the sensor chip. Chip surfaces were regenerated by injection of 6 M guanidine-HCl between analyses of different analytes, and WT CBD was always included at both at beginning and end of each injection series of CBD variants to ensure against and calibrate for any changes in surface bound ligands.
Microwell protein binding assay
The dissociation constants (Kd) for interactions of CBD variants with gelatin were measured in microwell protein binding assays as detailed previously (14, 15). Briefly, WT and CBD variants were biotinylated, equilibrated against 50 mM Tris, 150 mM NaCl, pH 7.4, and added at a concentration range of 0–10 µM to 96-microwell plates coated with gelatin at 0.5 µg/well and blocked with 2.5% BSA. Bound CBD was detected with 1:10,000-fold diluted alkaline phosphatase-conjugated streptavidin (Pierce, Rockford, IL) and 1 mg/ml p-nitrophenyl phosphate disodium (PNPP, Sigma, St. Louis, MO) as substrate and quantified at 405 nm in an Opsys MR plate reader (Dynex, Chantilly, VA). All experiments were performed in duplicate and repeated at least 2 times. Apparent Kds were calculated by non-linear curve fitting from plots of rCBD concentrations versus OD405 values using the equation y = {(a – d)/[1+(x/c)b]} + d, where x is concentration of CBDs added, y is bound CBDs, b is slope, c is concentration of CBDs at inflection point (apparent Kd), a is minimum binding, and d is binding at saturation (Sigma Plot; SPSS Corp., Chicago, IL).
RESULTS
Assignment CBD Backbone Resonances
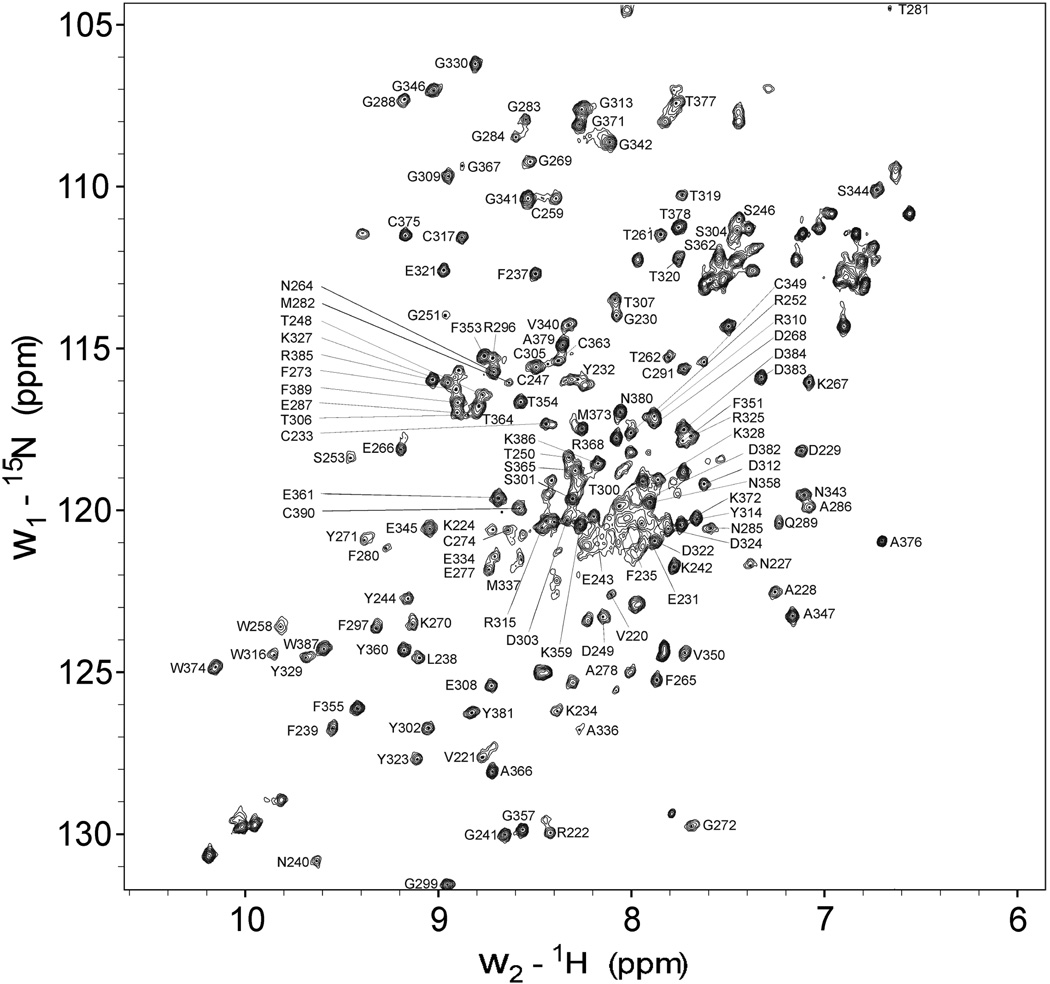
Our screening of a random peptide library earlier identified a CBD binding site on the α1 (I) chain of type I collagen, and a synthetic peptide P713 corresponding to this binding site interacted specifically with rCBD, blocked interactions of both CBD and full-length MMP-2 with gelatin substrate molecules, and inhibited MMP-2’s ability to degrade gelatin in a concentration dependent manner (25). To localize the specific residues on the CBD, which bind P713, we first assigned the rCBD HN, N, Cα, Cβ backbone resonances. Experiments used rCBD that was double labeled with 15N and 13C isotopes. Intra-residue and inter-residue amide-Cα/Cβ correlations from three-dimensional HNCACB and CBCA(CO)NH spectra enabled us to identify 152 of 174 total backbone amide resonances in the HSQC spectrum (Fig. 1). The assignments in the HSQC spectrum corresponded to those reported by others for MMP-2 CBD (24). The unassigned residues generally corresponded to residues with sequence and structural homology among the three CBD modules, yielding several overlapping peaks that were difficult to assign (21, 24). Though the case, subsequent functional analyses showed that the critical binding residues were among those successfully assigned.
FIGURE 1. 1H/15N HSQC spectra of rCBD.
Purified rCBD was in 25 mM sodium phosphate, pH 7.0, with 5% D2O/H2O at a concentration of 0.5 mM. Spectrum were acquired at 25 °C using a 700 MHz Bruker spectrometer. Assigned peaks are indicated by their one letter amino acid code and residue number based on proMMP-2 (1CK7 in PDB database). Residues of the His-tag of CBC were not labeled.
Identification of CBD binding site residues for collagen peptide P713
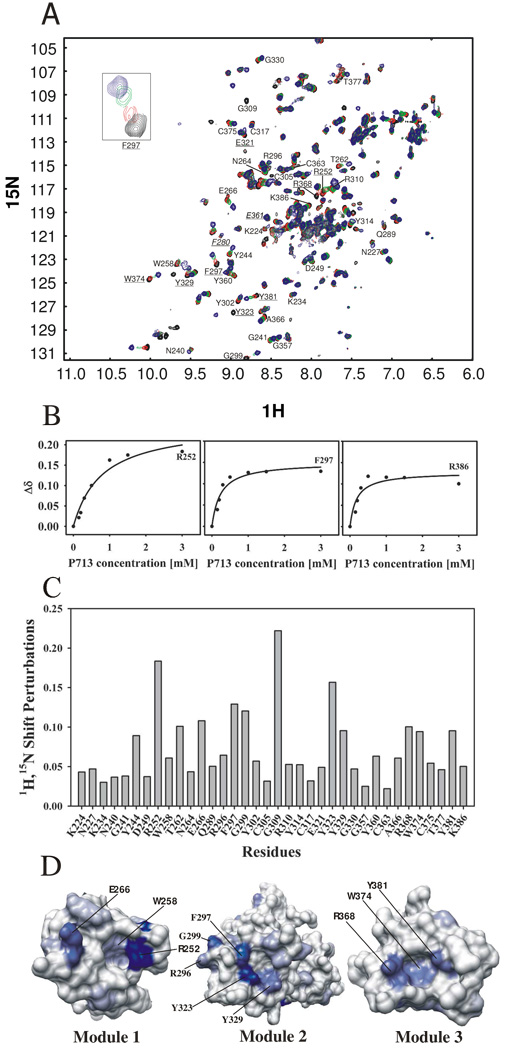
Our preliminary 1D 1H NMR showed that collagen-like peptide P713 does not form triple helices but rather display random coil behavior (data not shown). The peptide P713 that interacted specifically with both isolated CBD and full-length MMP-2 in ligand binding studies (25) also shifted a number of signals in the 2D 1H-15N HSQC spectrum when samples of the 15N CBD were titrated with increasing amounts of peptide. Such shifts provide the basis for identifying putative gelatin binding residues on the CBD (Fig. 2A). Our experiments, in which P713 was added at a concentration range of 0.15 – 3 mM to 0.5 mM 1H/15N CBD, detected a number of residues for which the backbone amide resonance underwent large shift changes at lower peptide concentrations, but saturated at higher concentrations. Ligand titration curves for three residues R252, F297, and R386 representing binding sites in modules 1, 2, and 3, respectively, are shown in Fig. 2B.
FIGURE 2. Titration of rCBD with P713.
Panel A. HSQC spectra of 0.5 mM 15N-labeled CBD in 25 mM sodium phosphate buffer, 5% D2O/H2O, pH 7.0 were recorded before (black) and after titration with collagen peptide P713 at concentrations of 0.15 mM (red), 0.3 mM (green), and 1.5 mM (blue). Residues that exhibited shift perturbations of backbone amides upon P713 peptide addition are shown and labeled with their one letter amino acid code and residue number. Residues which were selected for alanine substitution are underlined. Residues selected as negative controls are labeled in italics. The insert shows the change in the chemical shift of F297 as P713 is added. Panel B. The concentration dependent chemical shifts of three representative CBD residues upon binding P713 were plotted against the concentration of P713. Panel C. Magnitudes of chemical shift change (Δδ) for CBD residues in the presence of a P713 concentration of 1.5 mM. Chemical shift perturbations, Δδ, were calculated from proton and nitrogen shift by using the formula {[(δΔH)2+(δΔN/5)2]/2}½. Panel D. Three-dimensional surface representation of individual CBD modules showing the positions of residues that underwent shift changes in the presence of P713. The intensity of blue color in each CBD modules is proportional to the magnitude of the chemical shifts changes induced by peptide P713. The residues are labeled with the one letter amino acid code and residue number. The structure shown is from the reported MMP-2 crystal structure, PDB 1CK7.
The thirty-six residues that exhibited measurable chemical shift changes upon binding to P713 included residues in all three CBD modules. The amino acids were K224, N227, K234, N240, G241, Y244, D249, R252, W258, T262, N264, E266 in module 1, Q289, R296, F297, G299, Y302, C305, G309, R310, Y314, C317, E321, Y323, Y329, G330 in module 2, and G357, Y360, C363, A366, R368, W374, C375, T377, Y381, K386 in module 3. Residues with the largest shifts may contribute preferentially to the CBD binding sites for the collagen peptide P713, and potentially also the full-length gelatin substrate. The most ligand-sensitive residues by magnitude of the shift changes were R252 > E266 > T262 in module 1, G309 > Y323 > F297 > G299 > Y329 > in module 2, and R368 > Y381 > W374 in module 3 (Fig. 2C). The apparent affinities of P713 for putative binding site residues in modules 1, 2, and 3 were calculated as the average Kd for all residues in each module that showed chemical shift changes by interaction with P713 based on individual titration curves. The average Kds for modules 1, 2, and 3 were 6.0 × 10−4, 2.8 × 10−4 and 3.4 × 10−4 M, respectively, showing that the affinity of module 1 for P713 was significantly lower than module 2 and module 3 (P < 0.001) whereas there was no significant difference in affinity between modules 2 and 3 (P > 0.05). These results indicated that there were at least two binding events involving module 1 and modules 2 + 3. However, because it is unlikely that the short P713 peptide can bridge the binding sites of modules 2 and 3, we conclude that each CBD module has a distinct binding site for P713 and that full CBD can bind 3 peptide ligands simultaneously. The lower affinity of module 1 for P713 is also reflected in the titration curve (Fig. 2B), where the chemical shift change of R252 in module 1 did not reach saturation by P713 until a molar ratio of 3:1, while residues F297 and R368 in module 2 and 3 were saturated with a 1:1 ratio of P713:CBD (Fig. 2B).
We then viewed the potential of each of these residues for forming part of surface exposed ligand binding sites by plotting the observed shift changes onto the published x-ray structure of MMP-2 (1CK7) (21) (Fig 2D). Residues with the greatest chemical shift changes, including R252 in CBD module 1, F297, Y323, Y329 in module 2, and R368, W374, Y381 in module 3 were mapped to the hydrophobic surface pockets that are directed outward and away from each other.
Verification of collagen binding site residues on CBD
To test the functional significance of key residues in each of the NMR-mapped binding surfaces, ten residues were selected for alanine substitution analysis and protein-protein binding assays, including R252 in CBD module 1, R296, F297, Y302, E321, Y323, Y329 in module 2, and R368, W374, Y381 in module 3. In the alanine substitution analyses, we also included two negative control residues, F280 and E361, which did not undergo detectable shift changes in the 1H/15N CBD HSQC spectra upon binding to P713 (Fig 2A).
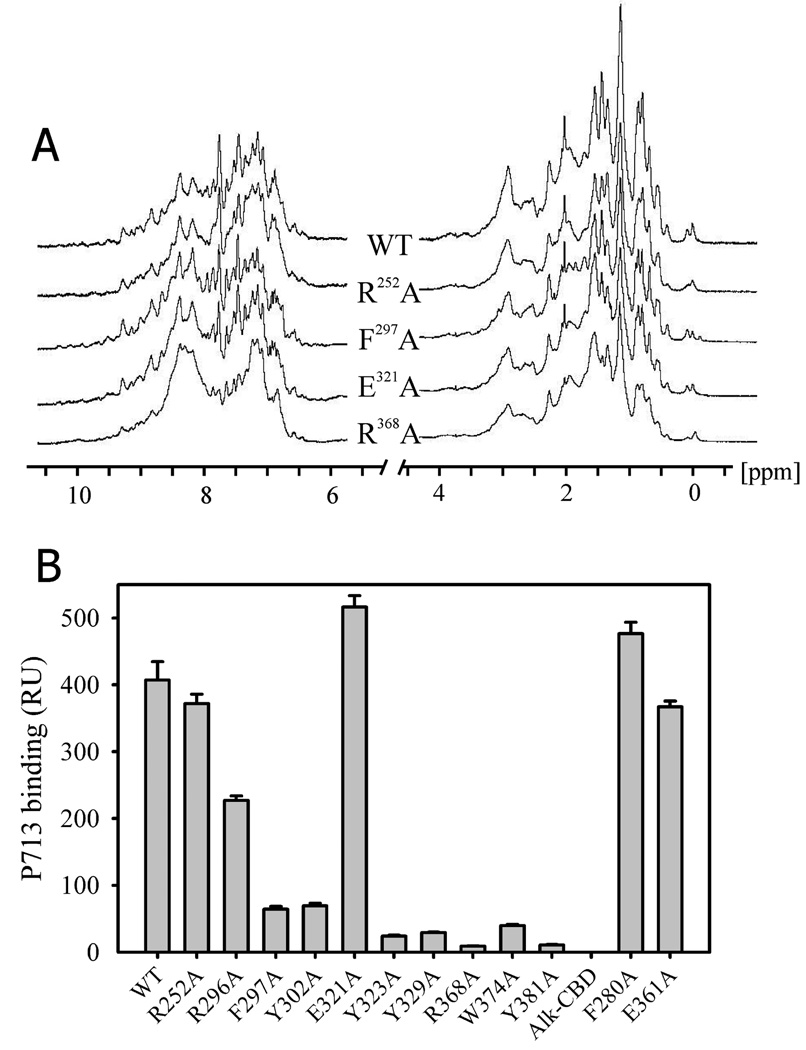
After expressing and purifying the variants, 1D 1H NMR analysis was used to determine whether the alanine substitutions introduced any major structural perturbations compared to wildtype (Fig. 3A). The observed patterns were each similar to wildtype in the amide/aromatic and aliphatic regions, indicating that the substitution caused no major structural perturbations. The CBD variants were used for subsequent binding analysis.
FIGURE 3. 1D 1H NMR and P713 binding ability of wildtype and rCBD variants.
Panel A. CBD variants were purified from E. coli and dissolved at concentrations of 150–200 µM in 25 mM phosphate buffer, pH 7.0 with 5% D2O. One-dimensional 1H NMR spectra were collected at a frequency of 700 MHz. Typically, signal dispersion above 8.5 ppm representing backbone amide groups and below 0.8 ppm representing methyl groups occurs only if the protein is structurally ordered. Alanine substitution of key residues of rCBD variants showed that their overall fold was not significantly perturbed compared with wildtype rCBD. Four representative variant are presented. Panel B. Binding of wildtype (WT) and CBD variants to peptide P713 by surface plasmon resonance. Alkylated CBD (AlkCBD) which did not bind gelatin served as negative control. P713 was immobilized on SPR CM5 chips, and 1 µM purified WT and variant CBDs in 10 mM HEPES, 150 mM NaCl, 3 mM EDTA were then passed over the peptide surfaces. Bar graph presents mean and SD (bars) of specific binding determined from Biacore sensorgrams and expressed in response units (RU). Alk-CBD represents non-functional alkylated WT CBD.
In preliminary surface plasmon resonance (SPR) binding assays, we analyzed wildtype CBD ligand interactions using a concentration range of wildtype CBD (0.5 – 40 µM) to optimize conditions for analysis of CBD binding site variants. CBD bound to gelatin and P713 in a concentration dependent manner with dissociation equilibrium constants at 2.9 µM and 5.4 µM, respectively (data not shown). The experiments showed that CBD at 0.5 – 2 µM bound gelatin and P713 in a sensitive linear range. Therefore, we used a concentration of 1 µM CBD for subsequent binding assays comparing CBD variants.
In the SPR assays, 8 of 12 CBD variants showed varying degrees of reduced binding to P713 (Fig. 3B). Alanine substitution had little or no effect on peptide binding for R252 and E321. Likewise, the two negative control proteins, F280 and E361, did not have reduced binding to P713. The fact that a positive control, generated by reduction and alkylation of CBD to disrupt disulfide bonds and thereby abrogate ligand binding (12), did not bind P713 indicated that the assay provided a sensitive measure for binding.
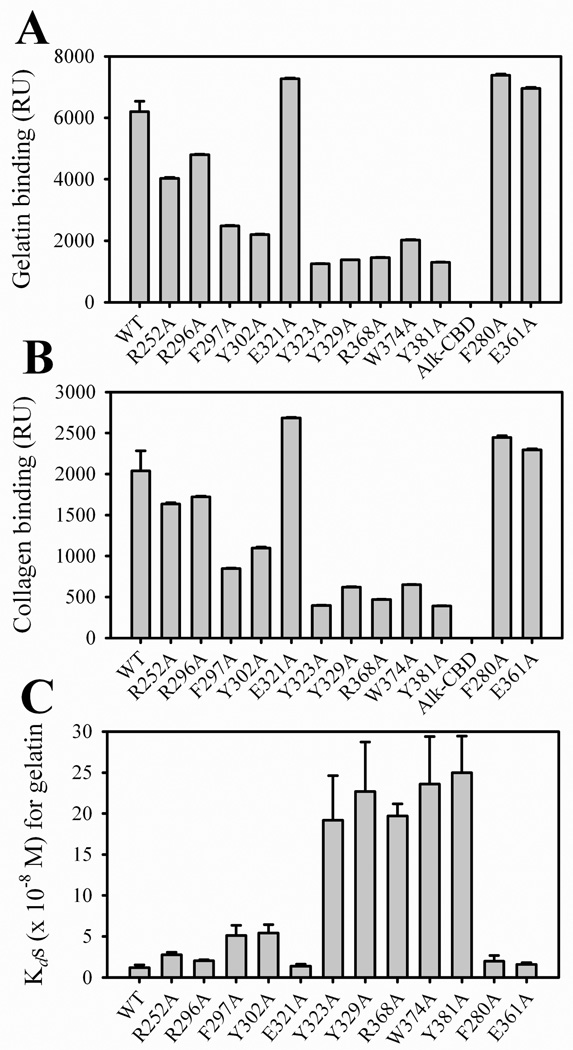
CBD mediates specific binding of MMP-2 to gelatin molecules and is required for cleavage of this substrate (12, 16, 18). Since P713 was identified as a CBD binding site on type I collagen and competed CBD and MMP-2 binding of gelatin (25), our present working hypothesis was that those CBD residues involved in binding of the α1(I) collagen sequence represented in P713 also were critical to binding of gelatin molecules. In SPR experiments used to analyze the protein interactions with native and denatured type I collagen, CBD variants substituted with alanine at positions R252 and R296 induced moderate decreases in binding compared to WT CBD, whereas substitution of residues F297, Y302, Y323, Y329, R368, W374, and Y381 with alanine caused strong reduction in binding (Fig. 4A and B). One CBD variant, E321A, as well as the negative control variants F280A and E361A showed no reduction in collagen-binding. Interestingly, the patterns of change among the CBD variants were similar for native and denatured forms of type I collagen pointing to identical binding sites on CBD.
FIGURE 4. Binding of wildtype and CBDs variants to type I collagen.
Purified wildtype (WT) and CBD variants at 1 µM in Biacore buffer were passed over CM5 chips coated with denatured (gelatin) or native forms of type I collagen in surface plasmon resonance assays (SPR). Bar graph shows specific binding expressed in response units (RU) for gelatin in Panel A and native type I collagen in Panel B. Non-functional alkylated CBD (AlkCBD) served as negative control. CBD variants had reduced binding to gelatin and native collagen. The apparent Kds of CBD WT and variants for gelatin were measured in plate binding assays and calculated from plots of protein binding versus CBD concentration by non-linear curve fitting using Sigma Plot (Panel C).
Assessment of apparent Kds showed that the alanine substitutions of R252 in module 1, F297, Y302, Y323, and Y329 in module 2, and R368, W374, and Y381 in module 3 were associated with significant changes in the ligand binding affinities. The Kds increased about 2 fold after the substitutions of R252, 5 fold for F297, Y302, and 20 fold for Y323, Y329, R368, W374, Y381. Verifying the functional contributions of the residues selected for mutagenesis, the negative control variants as well as R296A, and E321A had no significant change in their Kds for interaction with gelatin (Fig. 4C). Protein binding of native and denatured type I collagen was reduced to the greatest extent following modification of the binding site residues in CBD module 3 and least for module 1 (Fig. 4).
These functional assays supported the concept that single residues contribute significantly to the interaction of CBD with full-length collagen molecules. With the exception of E321, alanine substitution of each residue that exhibited a detectable shift perturbation in NMR titrations with collagen peptide P713 substantially affected CBD binding of native and denatured forms of type I collagen. The three-dimensional model of CBD showed that although E321 was exposed on the surface of module 2, it was on the opposite surface and at a distance from the binding site formed by F297, Y323 and Y329. Importantly, our results also indicate that each of the three CBD modules contributes to the binding of collagen.
Contributions of CBD modules to binding of gelatin
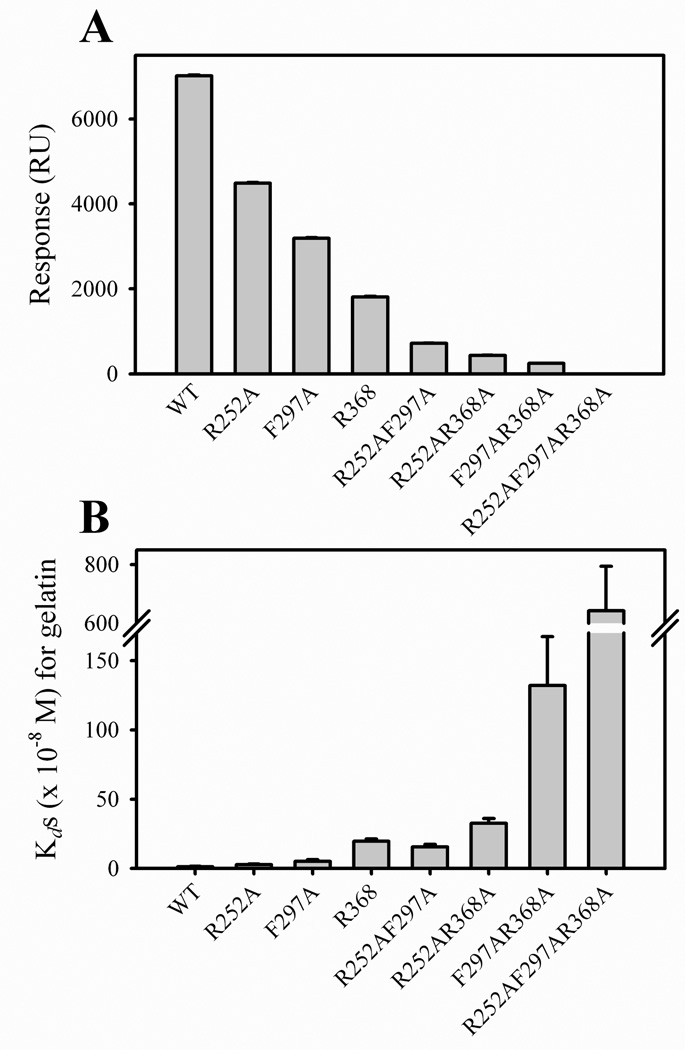
To understand the contributions of individual modules in collagen binding domains to ligand binding, we earlier found that high affinity binding of collagen required tandem type II modules in FN (14). In MMP-2, results from other investigators suggested that the three FN-like type II modules form a single binding site (19). However, our characterization of MMP-2 CBD interactions with type I collagen revealed that CBD can bind at least two collagen molecules simultaneously (12), consisted with the reported crystal structure of MMP-2 (29), which showed that the three FN type II modules do not form a continuous binding motif, but are oriented away from each other as in a three-pronged fishhook (21). NMR analyses of CBD have demonstrated that the three CBD modules can bind independently to a prototype collagen peptide (29). To further understand the potential contribution of the collagen binding sites in the three CBD modules to interactions with collagen molecules, we engineered 3 double- and 1 triple-alanine substitution variants targeting the one residue in each CBD module that had resulted in the greatest reduction in collagen binding during analysis of the single site substitutions described above (R252 in module 1, F297 in module 2 and R368 in module 3). Compared with single-site substitutions, these multi-site variants displayed further reduction in their binding of native and denatured type I collagen (Fig 5) suggesting that the modules all contributed to the binding of these substrates. The module 1 + 2 and 1 + 3 double binding site substitutions retained less than 10% binding compared with WT CBD. Double site alanine substitutions involving module 2 + 3 residues had even further reduced binding. Triple binding site alanine substitutions completely abrogated gelatin binding (Fig. 5).
FIGURE 5. Gelatin binding as a function of alanine mutations in collagen binding sites in one, two, and three modules of CBD.
Panel A. The binding of wildtype (WT), as well as single and multi-domain CBD variants to gelatin were measured by surface plasmon resonance (SPR) assays in which CM5 chips were coated with gelatin. Purified CBD variants at 1 µM were passed over the gelatin surfaces in Biacore buffer and the specific binding was expressed in response units (RU). Panel B. Apparent Kds for the interactions of WT and CBD variants with gelatin were measured in plate binding assays and calculated from plots of protein binding versus CBD concentration and using non-linear curve fitting (Sigma Plot).
Overall, our experiments assigned the backbone resonances of rCBD, identified and functionally verified residues in each CBD module that are essential for interacting with collagen, and demonstrated that the collagen binding sites of all three CBD modules contribute to CBD interactions with collagen molecules.
DISCUSSION
Until recently, MMP inhibitors were designed as mimics of the cleavage sites on MMP substrates that occupy and block the active sites of the enzymes. Hydroxamic acid inhibitors also contained the metal-binding group hydroxamate to chelate the active site zinc ion which is requited for enzyme function. However, this strategy produced inhibitors without the capacity to distinguish between different MMPs due to the high degree of structural homology between the catalytic sites (30). The low specificity likely explains the lack of success and high level of side effects observed in clinical trials of these inhibitors (30, 31). Consequently, there has been significant interest in using alternative strategies to develop inhibitors with greater specificity (32). One approach to achieve this has been to target the enzyme-substrate interactions at specific exodomains, which bind the substrate and are required for substrate cleavage. Murphy et al. (33) characterized the exodomain functions of the C-termini in MMP-1 and MMP-3 and found that deletion of these domains abrogated both the capacity to bind and cleave native type I collagen. In MMP-2 and −9, the primary sites of interaction with native and denatured forms of the collagens reside in the three FN type II modules that are inserted into the catalytic domain (12, 13, 28). Indeed, MMP-2 and MMP-9 modified by deletion of these collagen binding modules have ∼90% reduced gelatin hydrolysis (16) and do not cleave elastin (17). To verify that this loss of function did not result from structural perturbations following the large domain deletion, we were able to competitively inhibit gelatin binding and cleavage with soluble recombinant CBD (18). Therefore, additional structure functional studies are required to further enhance the rational basis for designing novel and specific inhibitors that interfere with exosite interactions.
Having recognized the critical contributions of the CBDs to gelatinolysis by MMP-2 and MMP-9, but without availability of the 3D structure, predictions of binding sites from the structure of related FN type II-like proteins such as PDC-109 (34) formed the basis for initial studies to localize the precise collagen binding residues. Those predictions suggested that each module had a hydrophobic surface contributing to ligand binding. After finding that isolated module 2 in the MMP-9 CBD had greater collagen binding properties than modules 1 and 3, alanine scanning mutagenesis was carried out on module 2 residues and several amino acid residues (R307, D309, N319, Y320 and N323) were found to be critical for gelatin binding of module 2 of MMP-9 CBD (20). Likewise, site-specific alanine substitutions in module 2 of MMP-2 CBD determined that several tyrosines were involved in gelatin binding for this isolated module (23).
The x-ray crystallography structure for MMP-2 has provided important information about the CBD showing that the three FN type II-like modules, in addition to having significant sequence homology, indeed possess the formerly predicted accessible hydrophobic surfaces. However, these areas are oriented outwards and away from each other in a conformation that has been described as a “three-pronged fishhook” with the potential for forming three putative ligand binding sites (21).
NMR experiments have analyzed the solution structure of the MMP-2 CBD (22, 29, 35) setting the stage for further rational structure-based studies to identify ligand binding residues. Initial ligands selected for those studies were a prototypical (Pro-Pro-Gly)6 collagen peptide reflecting the “G-X-Y” triplet repeat of fibrillar collagens α-chains(29) and 15-mer non-collagen CBD-binding peptides isolated from a phage peptide library (36). The binding affinities of the CBD modules were found to be ligand dependent. For example, modules 1 and 2 had higher affinity to the collagen-like peptide, but lower affinity to a segment containing amino acids 33–42 (p33–42) derived from the MMP-2 prodomain when compared to CBD module 3 (24). Although CBD-binding peptides isolated from phage peptide libraries did not bear resemblance to any collagen sequence, they also induced chemical shifts in the NMR spectra of putative collagen binding residues (36). Together, these studies provided further important information on in-solution interactions of CBDs with ligands. However, a characterization of effects of changes of putative binding site residues to verify their functional contributions to the intact tri-modular CBD in MMP-2 is not yet available.
Our prior studies identified a CBD binding peptide from a random peptide library that, importantly, had high homology to residues 715–721 of the human α1(I) collagen chain. A 13 amino acid long synthetic peptide from this region, termed P713, specifically blocked interactions of both isolated CBD and full-length MMP-2 with gelatin, and also inhibited gelatinolytic activities of MMP-2 by competing enzyme-substrate interactions without interacting with the catalytic site of MMP-2 (25). For these reasons, P713 was particularly valuable for determining collagen binding residues on the MMP-2 CBD using NMR analysis of CBD in complex with P713.
The NMR spectra and backbone assignments obtained from our recombinant MMP-2 CBD correspond to those reported by others (24). Subsequent HSQC spectra obtained from CBD in the presence of a concentration range of P713 collagen peptide identified substantial shifts for 36 residues of varying degrees as detailed in the Results section. The most ligand-sensitive residues by chemical shifts were R252 > E266 > T262 in module 1, G309 > Y323 > F297 > G299 > Y329 in module 2, and R368 > Y381 > W374 in module 3. Overall, our results correspond fundamentally to those from interaction studies involving isolated CBD module 2 and the prototypical collagen peptide (Pro-Pro-Gly)6 ligand in that the same residues underwent shift perturbations. However, the relative magnitudes of the shift perturbations were different (Y323 > F297> W316 > Y329 > Y314 > Y302 > F331 > F295 > F280 > F293) (22). Specifically, P713-CBD interactions resulted in larger shifts for Y323, F297, Y329, but no shifts of F280 or W316, pointing to potential differences in the interactions of the two peptides with CBD.
The present NMR analyses of P713 interactions with the full-length tri-modular CBD also identified shift perturbations in R368, W374, and Y381 from CBD module 3 reported by investigators who studied interactions of two-modular CBD constructs (2 +3) with the (Pro-Pro-Gly)6 peptide (29). In addition, in this study shift perturbations for G357 and A366 were observed that were not reported previously. NMR of a CBD constructs containing modules 1 + 2 also exhibited shift perturbations for R252 upon binding (Pro-Pro-Gly)6 (35). Thus, NMR pointed to both similarities and distinct differences in CBD interactions with the CBD binding P713 sequence from the α1(I) chain of collagen and the prototypical collagen-like (Pro-Pro-Gly)6 sequence.
Because the former NMR studies did not verify the functional contributions of the putative binding site residues, we selected 10 residues across the three modules of CBD for substitutions which were perturbed the greatest upon interaction with the collagen peptide P713, which were surface exposed (21), and which formed clusters suggestive of binding sites. In spite of undergoing shift changes upon binding of P713, alanine substitution was not performed on amino acids W258, C305, G299, G357, and A366 because of their small side chains, involvement in structural disulfide bond formation, and low likelihood of contributing to binding site formation based on their internal position in the molecule. Analysis of the expressed and purified recombinant CBD variants by 1D NMR indicated that all were folded without significant structural perturbations.
Most single residue substitutions with alanine resulted in reduced binding to the major MMP-2 substrate gelatin from type I collagen. In comparison, alanine substitutions of two negative control residues did not induce loss of gelatin binding. The identified P713 and gelatin binding residues in CBD binding were non-contiguous, but clustered to form cohesive binding sites. By these single residue substitutions, we verified that the shift perturbations detected by NMR correlated closely with functional effect in binding P713, as well as native and denatured type I collagen. It is noteworthy that the function of the tri-modular CBD was affected fundamentally by the change of single residues in each of the three putative binding sites and that the interactions were not fully compensated by potential redundancies exerted by the other two modules. Our results indicate that each module of CBD contains a binding site which interacts autonomously with its ligand. Indeed, the observation that single site changes in each of the CBD modules affected binding of CBD to P713 and gelatin suggests that all three modules of CBD contribute to binding of collagen ligands. Yet, the chemical shift analysis by NMR and mutagenesis experiments showed that module 2 and 3 have more profound effects on collagen binding than module 1. In comparison, prior NMR studies by others suggest that modules 1 and 2 have higher affinity to the (Pro-Pro-Gly)6 peptide (24) pointing to module-substrate specificity and affinity.
To better understand the contributions of multiple modules to ligand binding, we introduced those alanine substitutions that reduced ligand interactions to the greatest extent into two or three modular binding sites simultaneously. In all cases, the double-site variants bound more weakly to gelatin than any single-site variant, and concurrent mutations in all three binding sites abolished binding completely. Alanine substitutions in modules 2 and 3 caused greater loss of gelatin binding properties than that for module 1 variants. The greater contribution to gelatin binding of modules 2 and 3 is consistent with results by others from studies of CBD in MMP-9 (20) and MMP-2 (19). Likewise, tri-modular CBD constructs were shown to have greater gelatin binding capacity by affinity chromatography assays. Ligand affinities for CBD modules were dependent on whether they were expressed as single-module proteins or in context of the tri-modular protein. For single collagen α-chain-like peptide binding, the three CBD modules had the same Ka whether or not they were expressed in the context of the tri-modular protein (29, 35). In comparison, the Ka of interaction for each module with a triple-helical collagen peptide was significantly increased in the tri-modular configuration over isolated modules (24). This observation coincides with the proposed model for strand-unwinding by the CBD when fibrillar collagens are cleaved (37).
In context of current evidence for ligand interactions by the MMP-2 CBD, our results support a model by which the CBD has three independent functional binding sites that may possess ligand preferences and participate in cleavage of fibrillar collagens.
ACKNOWLEDGMENTS
We gratefully acknowledge Dr. Eileen Lafer and Patricia Schwarz at the UTHSCSA Center for Macromolecular Interactions for providing valuable guidance in Surface Plasmon Resonance analyses and data interpretation and Dr. Stephen C. Hardies and Mandy Rolando in the UTHSCSA Department of Biochemistry for DNA sequencing.
Abbreviations
- CBD
collagen binding domain
- MMP
matrix metalloproteinase
- MMP-2
matrix metalloproteinase-2
- TIMPs
tissue inhibitors of metalloproteinases
- SPR
surface plasmon resonance
- HYP
hydroxylproline
Footnotes
This work was supported by NIH grants DE14236, DE016312, DE17139, DE14318, DE18135, GM58670, the San Antonio Area Foundation, and The Robert A. Welch Foundation grant AQ1431. Structural studies used the San Antonio Cancer Institute Macromolecular Structure Shared Resource, which is supported in part by National Institutes of Health grant CA054174.
REFERENCES
- 1.Sternlicht MD, Werb Z. How matrix metalloproteinases regulate cell behavior. Annu. Rev. Cell Dev. Biol. 2001;17:463–516. doi: 10.1146/annurev.cellbio.17.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith PC, Munoz VC, Collados L, Oyarzun AD. In situ detection of matrix metalloproteinase-9 (MMP-9) in gingival epithelium in human periodontal disease. J. Periodontal Res. 2004;39:87–92. doi: 10.1111/j.1600-0765.2004.00705.x. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong DG, Jude EB. The role of matrix metalloproteinases in wound healing. J. Am. Podiatr. Med. Assoc. 2002;92:12–18. doi: 10.7547/87507315-92-1-12. [DOI] [PubMed] [Google Scholar]
- 4.Mott JD, Werb Z. Regulation of matrix biology by matrix metalloproteinases. Curr. Opin. Cell Biol. 2004;16:558–564. doi: 10.1016/j.ceb.2004.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liotta LA, Steeg PS, Stetler-Stevenson WG. Cancer metastasis and angiogenesis: an imbalance of positive and negative regulation. Cell. 1991;64:327–336. doi: 10.1016/0092-8674(91)90642-c. [DOI] [PubMed] [Google Scholar]
- 6.Van Wart HE, Birkedal-Hansen H. The cysteine switch: a principle of regulation of metalloproteinase activity with potential applicability to the entire matrix metalloproteinase gene family. Proc. Natl. Acad. Sci. U. S. A. 1990;87:5578–5582. doi: 10.1073/pnas.87.14.5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becker JW, Marcy AI, Rokosz LL, Axel MG, Burbaum JJ, Fitzgerald PM, Cameron PM, Esser CK, Hagmann WK, Hermes JD. Stromelysin-1: three-dimensional structure of the inhibited catalytic domain and of the C-truncated proenzyme. Protein Sci. 1995;4:1966–1976. doi: 10.1002/pro.5560041002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bode W, Gomis-Ruth FX, Stockler W. Astacins serralysins, snake venom and matrix metalloproteinases exhibit identical zinc-binding environments (HEXXHXXGXXH and Met-turn) and topologies and should be grouped into a common family, the ‘metzincins’. FEBS Lett. 1993;331:134–140. doi: 10.1016/0014-5793(93)80312-i. [DOI] [PubMed] [Google Scholar]
- 9.Wallon UM, Overall CM. The hemopexin-like domain (C domain) of human gelatinase A (matrix metalloproteinase-2) requires Ca2+ for fibronectin and heparin binding. Binding properties of recombinant gelatinase A C domain to extracellular matrix and basement membrane components. J. Biol. Chem. 1997;272:7473–7481. doi: 10.1074/jbc.272.11.7473. [DOI] [PubMed] [Google Scholar]
- 10.Murphy G, Knauper V. Relating matrix metalloproteinase structure to function: why the “hemopexin” domain? Matrix Biol. 1997;15:511–518. doi: 10.1016/s0945-053x(97)90025-1. [DOI] [PubMed] [Google Scholar]
- 11.Allan JA, Docherty AJ, Barker PJ, Huskisson NS, Reynolds JJ, Murphy G. Binding of gelatinases A and B to type-I collagen and other matrix components. Biochem. J. 1995;309(Pt 1):299–306. doi: 10.1042/bj3090299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steffensen B, Wallon UM, Overall CM. Extracellular matrix binding properties of recombinant fibronectin type II-like modules of human 72-kDa gelatinase/type IV collagenase. High affinity binding to native type I collagen but not native type IV collagen. J. Biol. Chem. 1995;270:11555–11566. doi: 10.1074/jbc.270.19.11555. [DOI] [PubMed] [Google Scholar]
- 13.Banyai L, Patthy L. Evidence for the involvement of type II domains in collagen binding by 72 kDa type IV procollagenase. FEBS Lett. 1991;282:23–25. doi: 10.1016/0014-5793(91)80436-7. [DOI] [PubMed] [Google Scholar]
- 14.Steffensen B, Xu X, Martin PA, Zardeneta G. Human fibronectin and MMP-2 collagen binding domains compete for collagen binding sites and modify cellular activation of MMP-2. Matrix Biol. 2002;21:399–414. doi: 10.1016/s0945-053x(02)00032-x. [DOI] [PubMed] [Google Scholar]
- 15.Xu X, Chen Z, Wang Y, Yamada Y, Steffensen B. Functional basis for the overlap in ligand interactions and substrate specificities of matrix metalloproteinases-9 and −2. Biochem. J. 2005;392:127–134. doi: 10.1042/BJ20050650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murphy G, Nguyen Q, Cockett MI, Atkinson SJ, Allan JA, Knight CG, Willenbrock F, Docherty AJ. Assessment of the role of the fibronectin-like domain of gelatinase A by analysis of a deletion mutant. J. Biol. Chem. 1994;269:6632–6636. [PubMed] [Google Scholar]
- 17.Shipley JM, Doyle GA, Fliszar CJ, Ye QZ, Johnson LL, Shapiro SD, Welgus HG, Senior RM. The structural basis for the elastolytic activity of the 92-kDa and 72-kDa gelatinases. Role of the fibronectin type II-like repeats. J. Biol. Chem. 1996;271:4335–4341. doi: 10.1074/jbc.271.8.4335. [DOI] [PubMed] [Google Scholar]
- 18.Xu X, Wang Y, Lauer-Fields JL, Fields GB, Steffensen B. Contributions of the MMP-2 collagen binding domain to gelatin cleavage. Substrate binding via the collagen binding domain is required for hydrolysis of gelatin but not short peptides. Matrix Biol. 2004;23:171–181. doi: 10.1016/j.matbio.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 19.Banyai L, Tordai H, Patthy L. The gelatin-binding site of human 72 kDa type IV collagenase (gelatinase A) Biochem. J. 1994;298(Pt 2):403–407. doi: 10.1042/bj2980403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collier IE, Krasnov PA, Strongin AY, Birkedal-Hansen H, Goldberg GI. Alanine scanning mutagenesis and functional analysis of the fibronectin-like collagen-binding domain from human 92-kDa type IV collagenase. J. Biol. Chem. 1992;267:6776–6781. [PubMed] [Google Scholar]
- 21.Morgunova E, Tuuttila A, Bergmann U, Isupov M, Lindqvist Y, Schneider G, Tryggvason K. Structure of human pro-matrix metalloproteinase-2: activation mechanism revealed. Science. 1999;284:1667–1670. doi: 10.1126/science.284.5420.1667. [DOI] [PubMed] [Google Scholar]
- 22.Briknarova K, Grishaev A, Banyai L, Tordai H, Patthy L, Llinas M. The second type II module from human matrix metalloproteinase 2: structure, function and dynamics. Structure. 1999;7:1235–1245. doi: 10.1016/s0969-2126(00)80057-x. [DOI] [PubMed] [Google Scholar]
- 23.Tordai H, Patthy L. The gelatin-binding site of the second type-II domain of gelatinase A/MMP-2. Eur. J Biochem. 1999;259:513–518. doi: 10.1046/j.1432-1327.1999.00070.x. [DOI] [PubMed] [Google Scholar]
- 24.Gehrmann ML, Douglas JT, Banyai L, Tordai H, Patthy L, Llinas M. Modular autonomy, ligand specificity, and functional cooperativity of the three in-tandem fibronectin type II repeats from human matrix metalloproteinase 2. J. Biol. Chem. 2004;279:46921–46929. doi: 10.1074/jbc.M408859200. [DOI] [PubMed] [Google Scholar]
- 25.Xu X, Chen Z, Wang Y, Bonewald L, Steffensen B. Inhibition of MMP-2 gelatinolysis by targeting exodomain-substrate interactions. Biochem. J. 2007;406:147–155. doi: 10.1042/BJ20070591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook J, Russell DW. Molecular Cloning: a Laboratory Manual. NY: Cold Spring Harbor Laboratory., Cold Spring Harbor; 2001. [Google Scholar]
- 27.Rehm T, Huber R, Holak TA. Application of NMR in structural proteomics: screening for proteins amenable to structural analysis. Structure. 2002;10:1613–1618. doi: 10.1016/s0969-2126(02)00894-8. [DOI] [PubMed] [Google Scholar]
- 28.Steffensen B, Bigg HF, Overall CM. The involvement of the fibronectin type II-like modules of human gelatinase A in cell surface localization and activation. J Biol. Chem. 1998;273:20622–20628. doi: 10.1074/jbc.273.32.20622. [DOI] [PubMed] [Google Scholar]
- 29.Briknarova K, Gehrmann M, Banyai L, Tordai H, Patthy L, Llinas M. Gelatin-binding region of human matrix metalloproteinase-2: solution structure, dynamics, and function of the COL-23 two-domain construct. J. Biol. Chem. 2001;276:27613–27621. doi: 10.1074/jbc.M101105200. [DOI] [PubMed] [Google Scholar]
- 30.Brown PD. Matrix metalloproteinase inhibitors. Breast Cancer Res. Treat. 1998;52:125–136. doi: 10.1023/a:1006119319695. [DOI] [PubMed] [Google Scholar]
- 31.Brown S, Meroueh SO, Fridman R, Mobashery S. Quest for selectivity in inhibition of matrix metalloproteinases. Curr. Top. Med. Chem. 2004;4:1227–1238. doi: 10.2174/1568026043387854. [DOI] [PubMed] [Google Scholar]
- 32.Overall CM, Lopez-Otin C. Strategies for MMP inhibition in cancer: innovations for the post-trial era. Nat. Rev. Cancer. 2002;2:657–672. doi: 10.1038/nrc884. [DOI] [PubMed] [Google Scholar]
- 33.Murphy G, Allan JA, Willenbrock F, Cockett MI, O’Connell JP, Docherty AJ. The role of the C-terminal domain in collagenase and stromelysin specificity. J Biol. Chem. 1992;267:9612–9618. [PubMed] [Google Scholar]
- 34.Constantine KL, Madrid M, Banyai L, Trexler M, Patthy L, Llinas M. Refined solution structure and ligand-binding properties of PDC-109 domain b. A collagen-binding type II domain. J Mol. Biol. 1992;223:281–298. doi: 10.1016/0022-2836(92)90731-x. [DOI] [PubMed] [Google Scholar]
- 35.Gehrmann M, Briknarova K, Banyai L, Patthy L, Llinas M. The col-1 module of human matrix metalloproteinase-2 (MMP-2): structural/functional relatedness between gelatin-binding fibronectin type II modules and lysine-binding kringle domains. Biol. Chem. 2002;383:137–148. doi: 10.1515/BC.2002.014. [DOI] [PubMed] [Google Scholar]
- 36.Trexler M, Briknarova K, Gehrmann M, Llinas M, Patthy L. Peptide ligands for the fibronectin type II modules of matrix metalloproteinase 2 (MMP-2) J. Biol. Chem. 2003;278:12241–12246. doi: 10.1074/jbc.M210116200. [DOI] [PubMed] [Google Scholar]
- 37.Tam EM, Moore TR, Butler GS, Overall CM. Characterization of the distinct collagen binding, helicase and cleavage mechanisms of matrix metalloproteinase 2 and 14 (gelatinase A and MT1-MMP): the differential roles of the MMP hemopexin c domains and the MMP-2 fibronectin type II modules in collagen triple helicase activities. J. Biol. Chem. 2004;279:43336–43344. doi: 10.1074/jbc.M407186200. [DOI] [PubMed] [Google Scholar]