Abstract
DNA polymerase mu (Polμ) is a family X member implicated in DNA repair, with template-directed and terminal transferase (template-independent) activities. It has been proposed that the terminal transferase activity of Polμ can be specifically required during non-homologous end joining (NHEJ) to create or increase complementarity of DNA ends. By site-directed mutagenesis in human Polμ, we have identified a specific DNA ligand residue (Arg387) that is responsible for its limited terminal transferase activity compared to that of human TdT, its closest homologue (42% amino acid identity). Polμ mutant R387K (mimicking TdT) displayed a large increase in terminal transferase activity, but a weakened interaction with ssDNA. That paradox can be explained by the regulatory role of Arg387 in the translocation of the primer from a non-productive E:DNA complex to a productive E:DNA:dNTP complex in the absence of a templating base, which is probably the rate limiting step during template-independent synthesis. Further, we show that the Polμ switch from terminal transferase to templated insertions in NHEJ reactions is triggered by recognition of a 5′-P at a second DNA end, whose 3′-protrusion could provide a templating base to facilitate such a special “pre-catalytic translocation step.” These studies shed light on the mechanism by which a rate-limited terminal transferase activity in Polμ could regulate the balance between accuracy and necessary efficiency, providing some variability during NHEJ.
Physical and chemical damage of DNA is the root cause of a large number of human syndromes, including premature aging, various cancer predispositions, and congenital abnormalities. To overcome DNA damage and maintain stability in the chromosomes, cells contain an array of specific DNA repair pathways that act upon the different kinds of lesions (1). Specialized DNA polymerases are essential actors within these pathways and, in humans, at least 12 are devoted to overcome or repair DNA damage in the cell (2). DNA polymerases of the X family, which in mammals include DNA polymerases beta (Polß), lambda (Polλ), mu (Polμ), and terminal deoxynucleotidyl transferase (TdT), are structurally related enzymes specialized in repair pathways involving double strand breaks (DSB) and gaps (3).
Unlike Polß, Polλ, Polμ, and TdT contain a BRCA-1 C-terminal (BRCT) interaction domain at their N-termini (4–7), that serves to interact with the core NHEJ factors Ku and XRCC4-Ligase IV (8–12), suggesting their involvement in DSB repair via NHEJ. Whereas DSB repair by homologous recombination is highly accurate because it relies directly on complementary DNA sequences, the accuracy of the repair reaction by NHEJ is lower, somehow compromising preservation of the original DNA information. In this context, NHEJ DNA polymerases would be needed either to extend the 3′-end at 5′-overhangs, to fill gaps created by limited complementarity of the two DNA ends, or eventually in the case of joining non-complementary (incompatible) ends, to add sequences at the 3′-end to create a minimal cohesive base pairing (13). To accomplish these tasks, Polλ, Polμ, and TdT have a gradient of template dependency (12): Polλ requires at least one complementary base pair between the two DNA ends (microhomology) to be able to fill gaps as template-directed insertion events; Polμ can realign DNA strands allowing distortions to maximize cohesion (14), and can even extend a 3′-end terminus with minimal or null complementarity, contributing to the efficiency of end-joining with either templated or untemplated insertions (15); TdT, on the other hand, is a strong template-independent polymerase, displaying only a strict terminal transferase activity. TdT function is essential during V(D)J recombination, a specialized variant of the NHEJ pathway that takes place during the maturation of antibodies in B lymphocytes, in which the creation of diversity (by addition of random DNA sequences) is favoured over accuracy in the repair of the DSB (16).
Polμ is the only DNA polymerase known that combines template-dependent and template-independent (terminal transferase) activities in one enzyme (6). The relative importance of the terminal transferase activity of Polμ during NHEJ has been controversial and probably underscored as it is difficult to distinguish template-dependent from random additions, if the latter are successful in creating complementarity between the two ends of a DSB (12). However, there are insightful structure-function correlations suggesting that terminal transferase activity is important for NHEJ. Thus, loop1 in human Polμ, likely more flexible than that present in TdT (17), was shown to be important for both NHEJ (12) and terminal transferase activity (13). As early proposed by Juárez et al. (13), Polμ can alternate between stable interactions with ssDNA vs. dsDNA via conformational changes in its flexible loop1, being disordered in the crystal structure of Polμ in complex with gap DNA (18). This structure of a ternary complex of human Polμ allowed proposing that a specific histidine residue (His329 in Polμ and His342 in TdT; absent in Polß and Polλ) plays an important role in terminal transferase activity (in both Polμ and TdT) and NHEJ of incompatible ends mediated by Polμ (18).
In this study, we identified a specific arginine in human Polμ (Arg387) that is responsible for its limited terminal transferase activity in comparison with that of TdT. One of the mutations studied (R387K), which greatly increased terminal transferase activity in Polμ, and at the same time reduced interaction with the primer strand, gives insights into the mechanism and details of the reaction. We propose that movement of the primer from a non-productive E:DNA complex to a productive E:DNA:dNTP complex is probably a rate limiting step for the terminal transferase addition of nucleotides. The mild terminal transferase activity of Polμ contributes to establish a minimal cohesion of DNA ends but limiting excessive mutagenic events during NHEJ.
Results
Search for Specific Residues Involved in Terminal Transferase Versus Template-Directed Polymerization.
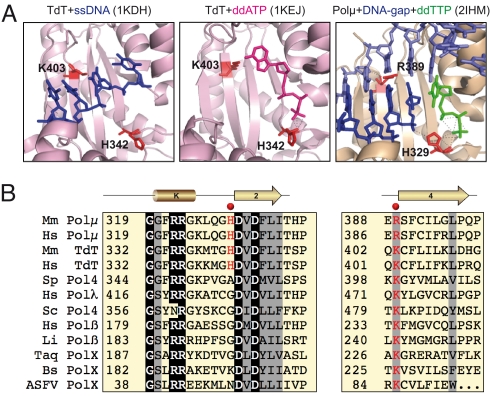
Polμ and TdT share 42% overall amino acid sequence identity and are highly conserved in vertebrates (6). Accordingly, the crystallographic structure of murine Polμ and TdT is very similar, even in very different contexts (see Fig. 1A): either bound to a primer (1KDH) or to a nucleotide (1KEJ), as in the case of TdT (17); or on a gapped DNA substrate with an incoming nucleotide (2IHM), as in the murine Polμ crystal (18). However, a more detailed inspection of the different TdT and Polμ structures, and their comparison with those of human Polλ and Polß, pointed out to two specific residues in murine Polμ, His329, and Arg389, that could display dual interactions, thus having a specific potential in regulating terminal transferase versus template-directed nucleotide additions. As indicated in Fig. 1A, that histidine in TdT (His342) is not involved in a direct interaction with the primer (left panel), but interacts with the incoming nucleotide (central panel), and its substitution abolishes terminal transferase activity (18). However, murine Polμ His329 is hydrogen-bonded to both primer terminus and nucleotide (right panel), and its proposed function is to align them properly before catalysis (18). As shown in Fig. 1B, no equivalent histidine is present in either Polß or Polλ, or in any other PolX enzyme.
Fig. 1.
Specific residues involved in terminal transferase. (A) Crystal structures of TdT, either complexed with ssDNA (1KDH) or dNTP (1KEJ), and Polμ (1IHM). Residues selected for mutagenesis in Polμ, and their orthologues in TdT, are indicated in red sticks. DNA substrates are indicated in dark (primer strand) and light (template and downstream strand) blue. Incoming ddNTPs are indicated in magenta (non templated) or green (templated). (B) Amino acid sequence alignment of family X polymerases from different species (Hs, human; Mm, mouse; Sp, Schizosaccharomyces pombe; Sc, Saccharomyces cerevisiae; Li, Leishmania infantum; Taq, Thermus aquaticus; Bs, Bacillus subtilis; and ASFV, african swine fever virus) along the indicated regions. Residues selected for mutagenesis in Polμ are indicated with red dots. Invariant residues are depicted in white over black background, and conservative residues have gray background.
Murine Polμ Arg389 is interacting with the template strand, contacting the sugar at residue in position −3 with respect to the templating base. An identical interaction of the equivalent residue in Polλ (Lys472) and Polß (Lys234) is observed in ternary complexes of these enzymes (1XSN;1BPY). As shown in Fig. 1B, an arginine residue is unique to Polμ, whereas all other family members have a lysine instead. Interestingly, that lysine in TdT (Lys403) is a direct ligand of the primer strand, as seen in Fig. 1A (left panel). No experimental evidence for the functional importance of this positively charged residue (either arginine or lysine) was yet available. Therefore, the equivalent residues in human Polμ, His329, and Arg387, were targeted for site-directed mutagenesis. His329 was changed to glycine, as it naturally occurs in human Polß and Polλ, whereas Arg387 was changed either to alanine (R387A), to eliminate the positive charge, or to lysine (R387K), as it occurs in all other DNA polymerases from family X.
Arg387 Regulates the Rate of Terminal Transferase Activity in Human Polμ, While His329 Is Essential.
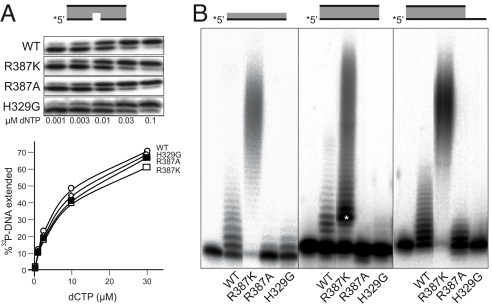
The selected mutants, obtained and purified as indicated in Materials and Methods, were tested for a differential effect on their template-dependent versus template-independent polymerization capacity. None of the mutants tested showed a significant defect in gap-filling (Fig. 2A), suggesting that residues His329 and Arg387 do not play a critical role in template-directed synthesis. Conversely, when terminal transferase was measured on various substrates, the effect of the mutations was dramatic (Fig. 2B). Mutant H329G was largely affected on any substrate compared to wild-type enzyme, in support of its specific relevance for terminal transferase reactions, as previously shown by using a different mutation [H329A; (18)]. On the contrary, in the case of Arg387 the two substitutions made had opposite effects: whereas mutant R387A largely decreased terminal transferase activity, mutant R387K (mimicking TdT) produced a very significant increase of this template-independent reaction (Fig. 2B). These results suggest that the specific arginine that is only present in Polμ could act as a regulator, reducing the catalytic efficiency of its intrinsic terminal transferase. Analysis of the kinetic parameters for mutant R387K (Table 1) showed a 38-fold increase in catalytic efficiency for inserting dTTP, not due to a difference in Km, but to a large improvement in kcat. A similar conclusion was obtained by using the other three different dNTPs, provided independently (Fig. S1 and Table 1). The above results indicate that the histidine residue has an essential role during terminal transferase addition in both TdT (His342) and Polμ (His329), whereas the specific arginine in Polμ (Arg387) somehow limits the catalytic step of the terminal transferase reaction, perhaps explaining its low activity in comparison to TdT.
Fig. 2.
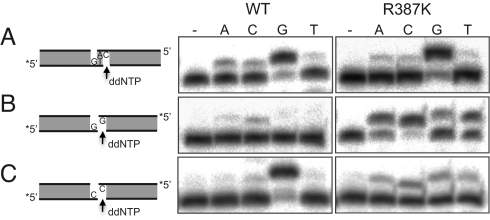
Arg387 regulates the rate of terminal transferase activity in human Polμ, while His329 is essential. (A) Polymerization reactions on a gapped-DNA substrate (see scheme) was carried out in the presence of 2.5 mM MgCl2, the indicated amounts of dCTP, and 300 nM of either Polμ wt or the indicated mutants. After incubation for 30 min at 30 °C, extension of the 5′P-labeled primer (indicated with an asterisk) was analyzed by 8 M urea-20% PAGE and autoradiography, and the results quantitated by densitometry. (B) Terminal transferase activity on different substrates (ssDNA/polyT, blunt dsDNA and 3′-protruding (9T) dsDNA; 5 nM) was assayed for 30 min at 30 °C in the presence of 1 mM MnCl2, 100 μM dTTP, and 600 nM of either Polμ wt or the indicated mutants, and analyzed by 8 M urea-20% PAGE and autoradiography.
Table 1.
Kinetic parameters of the terminal transferase activity of mutant R387K with any of the four dNTPs
| Kmapp, μM | Kcat, s−1 | Catalytic efficiency | f0 ext | ||
|---|---|---|---|---|---|
| Polμ WT | dTTP | 319 ± 16 | 0.0004 ± 0.0002 | (1.39 ± 0.0004) 10−6 | 1 |
| dATP | 4,300 ± 2400 | 0.0016 ± 0.0006 | (4.10 ± 0.92) 10−7 | 1 | |
| dCTP | 224 ± 26 | 0.0008 ± 0.0004 | (5.97 ± 2.35) 10−6 | 1 | |
| dGTP | 5,830 ± 70 | 0.0049 ± 0.0025 | (8.43 ± 4.34) 10−7 | 1 | |
| R387K | dTTP | 307 ± 17 | 0.015 ± 0.006 | (5.35 ± 1.13) 10−5 | 38 |
| dATP | 5,100 ± 2300 | 0.015 ± 0.00008 | (3.28 ± 1.47) 10−6 | 8 | |
| dCTP | 131 ± 24 | 0.018 ± 0.004 | (1.36 ± 0.34) 10−4 | 22 | |
| dGTP | 3,010 ± 100 | 0.188 ± 0.015 | (6.25 ± 0.30) 10−5 | 74 |
Terminal transferase activity was assayed either on polydT (with dTTP, dCTP and dGTP), or on polydA (with dATP), as described in Materials and Methods. Mean values (average) and standard deviation, corresponding to various independent experiments, are indicated.
Rate-Limiting Step for the Terminal Transferase: Translocation of the Primer before Catalysis.
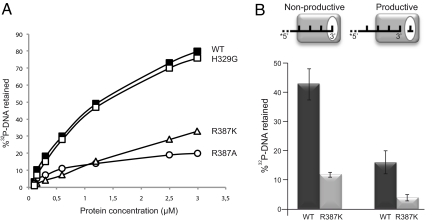
By using nitrocellulose filter-binding assays (19) we assessed that human Polμ stably interacts with ssDNA (Fig. 3A), in a perhaps non-productive complex as that observed in the crystal structure of TdT (17). Paradoxically, mutant R387K, although having an increased terminal transferase activity, displayed an about 4-fold lower interaction with ssDNA (Fig. 3A); this defective binding was also observed for mutant R387A, but not in mutant H329G (Fig. 3A; wild-type Polμ Kdis = 1.3 μM; Polμ R387K Kdis = 5.5 μM; Polμ R387A Kdis = 5.8 μM; Polμ H329G Kdis = 1.3 μM).
Fig. 3.
Mutants at residue Arg387 have a reduced ssDNA binding affinity. The presence of dNTP reduces ssDNA binding capacity by Polμ. (A) A 5′-labeled ssDNA (polydT; 5 nM) was used as substrate for interaction with different concentrations of either Polμ wt or mutants R387K, R387A, and H329G. After incubation for 10 min at 22 °C, the protein/ssDNA complexes were retained in nitrocellulose filters, as described in Materials and Methods. (B) Schemes depicting two different complexes: non-productive (nucleotide absent), indicative of a E:DNA binary complex in which the 3′ end of primer is occupying the nucleotide pocket (white oval); and productive (nucleotide present), indicative of a E:DNA:dNTP ternary complex in which the primer has been relocated. Binding capacity of Polμ wt and R387K mutant (600 nM in each case) to ssDNA/polyT, either in the absence (left) or presence (right) of dTTP, was measured by nitrocellulose filter binding assay, as described in Materials and Methods.
The first clue of a plausible rate-limiting step for the terminal transferase derived from the crystal structure of TdT bound to ssDNA (1KDH), whose 3′-terminus was unexpectedly located in a postcatalytic situation, occupying the binding site for the next incoming dNTP (17; see Fig. 1A, left panel). No crystal structure of a precatalytic ternary complex for the terminal transferase reaction is available, neither for TdT nor for Polμ. Therefore, and combining the 3D information available for both TdT and Polμ, it can be proposed that the rate-limiting step of the terminal transferase most likely involves, for both enzymes, the movement of the single-stranded (3′-protruding) primer-terminus from a “non-productive” E:DNA complex to a “productive” E:DNA:dNTP complex (Fig. 3B), to achieve untemplated addition of nucleotides, irrespective of either a processive or a distributive cycling of the enzyme (see Fig. S2 for further details).
We next evaluated the stability of the ssDNA primer in the presence of metal and dNTP (that will allow formation of a productive complex), but impeding reaction by the presence of a ddNMP at the primer-terminus. Interestingly, binding of both wt and R387K mutant Polμ to ssDNA was weakened by the presence of dNTP (Fig. 3B), supporting the idea that there is a necessary movement of the primer (1 nt backwards) to form a productive complex, in which the dNTP has to compete and finally displace the 3′-terminus to occupy their corresponding binding sites, necessary for catalysis. Congruently, the intrinsic weaker binding to ssDNA of mutant R387K would facilitate primer movement associated to the rate limiting step, explaining the paradox of its high terminal transferase activity.
Is Terminal Transferase Operating during NHEJ of Incompatible Ends? Importance of a Recessive 5′P.
It has been recently reported that Polμ′s optimal end-joining of incompatible ends occurs at minimal (1–2 nt) protrusions; on longer protrusions, a single polymerase unit cannot maintain bridging of the two ends, and the yield of the reaction is lower, exclusively reflecting terminal transferase activity (15). Since recessive 5′ DNA ends must contain a 5′P group to allow ligation and repair of the DNA break, and family X polymerases contain an 8-kDa domain that interacts directly with the 5′P group of the downstream strand (3), we determined if the presence of a 5′P group modulates Polμ's terminal transferase activity, perhaps favoring accurate NHEJ of short incompatible ends.
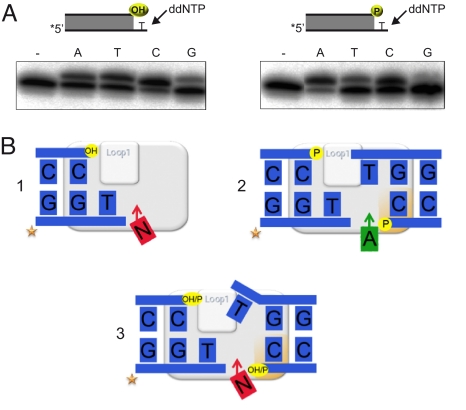
As shown in Fig. 4A (left), in the presence of a 5′-OH group the terminal transferase activity of human Polμ extends the 1nt-protruding end with any of the four ddNTPs, ddCTP being the most efficient. However, the presence of a 5′P group (right) alters this specificity. Incorporation of ddTTP, ddCTP and ddGTP was significantly reduced (43%, 37%, and 46% decrease, respectively), but incorporation of ddATP was largely enhanced (163% increase). This phosphate-dependent behavior was specific of Polμ, being absent in TdT (Fig. S3). These results can be explained as indicated in Fig. 4B. If a 5′P is not present (scheme 1), Polμ inserts untemplated nucleotides (N) by terminal transferase, assisted by its loop 1 (13). If a 5′P group is present (scheme 2), the preferred insertion of ddATP indicates that a single Polμ molecule was able to bridge two incompatible DNA ends, mediated by loop 1, and copy the templating base (T) provided by a second end, thus achieving a precise NHEJ reaction, as it was shown to occur at different incompatible ends also when Polμ was in the presence of NHEJ core factors (15). Bridging of the second end is mediated by interaction with its 5′P through the 8-kDa domain (Fig. 4B, schemes 2 and 3); the rationale for preferring the correct nucleotide (ddATP) is that the rate limiting step, that is, the movement of the protruding primer from an inactive to an active conformation, is facilitated by the approaching end, which provides a templating base “in trans.” If this templating base fails to be located at a proper register, the ability of Polμ to catalyze template-independent additions will allow joining (as represented in Fig. 4B, scheme 3), as Ligase IV can accept a mismatched 3′-end for ligation (20). These results indicate that a recessive 5′P is key for the precise joining of incompatible ends. Interestingly, and in agreement with recent findings supporting that 1 nucleotide is the optimal 3′-protrusion size for the NHEJ capacity of Polμ on incompatible ends (15), the effect of the 5′P is not seen in longer protrusions (Fig. S4). When using longer 3′-protruding substrates, the enzyme only inserts untemplated nucleotides as is unable to acommodate the DNA ends.
Fig. 4.
Terminal transferase vs. NHEJ of incompatible ends. (A) Single nucleotide extension of a 3′-protruding (1T) dsDNA substrate (200 nM), either lacking (left panel) or having (right panel) a 5′-recessive P. The assay was performed as described in Materials and Methods, in the presence of 1 mM MnCl2, 100 μM of the indicated ddNTP, and 200 nM of Polμ wt. After incubation for 30 min at 30 °C, +1 extension of the 5′P-labeled oligonucleotide was analyzed by 8 M urea-20% PAGE and autoradiography. (B) The observed nucleotide specificity can be the consequence of either untemplated polymerization (terminal transferase; scheme 1) or DNA-directed additions (dA insertion templated in trans), thus achieving precise joining of two DNA ends (scheme 2). Scheme 3 shows a hybrid situation in which the joining reaction is untemplated, as the 3′-protruding template base provided in trans is not in a proper register. The gray rectangle represents a cartoon of Polμ, in which the specific loop 1, involved in both terminal transferase and connectivity during NHEJ of incompatible ends, is highlighted. The orange area in Polμ represents the 8-kDa domain, specifically involved in 5′P recognition.
The Excessive Terminal Transferase of Mutant R387K Affects Fidelity during NHEJ of Incompatible Ends.
As expected from its enhanced terminal transferase activity and facilitated rate-limiting step, mutant R387K showed quantitative but also qualitative differences when compared to the wt enzyme on incompatible 3′-protruding ends. To show this, we used a different set of 3′-protruding substrates, either with compatible (Fig. 5A) or incompatible (Fig. 5 B and C) ends. On compatible ends, both wt Polμ and R387K preferentially inserted the correct (templated) nucleotide (ddGTP), indicating that accurate NHEJ of minimally compatible ends has occurred (Fig. 5A). Their similar efficiency mimicks their behavior in gap-filling (Fig. 2A). As firstly shown by Davies et al. (15) the particular sequence (and size of the protrusion) forming the incompatible ends conditions the outcome of the NHEJ reaction. Thus, on incompatible ends that cannot be accurately joined (Fig. 5B), R387K was more efficient than wt Polμ in NHEJ, according to the enhanced terminal transferase and reduced activation barrier of this mutant, but keeping the reaction very inaccurate (Fig. 5B). On the other hand, simply by changing the protruding nucleotide (to a dC), incompatible ends can now be accurately joined by the wild-type Polμ (Fig. 5C), that is, the reaction is preferentially template-directed (ddG insertion). In this case, mutant R387K showed an error-prone behavior, probably due to its facilitated rate-limiting step, i.e., excessive terminal transferase activity, that favors the untemplated nucleotide addition of any dNTP. These results suggest that Arg387 residue, which acts as a brake for TdT activity in Polμ, could avoid generation of excessive variability during NHEJ.
Fig. 5.
R387K has a low fidelity during NHEJ of incompatible ends. (A) Accurate NHEJ of minimally complementary ends, using a 5′-labeled 3′-protruding (GT) dsDNA substrate (5 nM), and a cold 3′-protruding (CA) dsDNA substrate (25 nM), both having a recessive 5′P. Mutant R387K displayed an accurate behavior comparable to wild-type Polμ. (B) Inaccurate NHEJ of incompatible ends, using a 5′-labeled 3′-protruding (G) dsDNA substrate (5 nM), having a recessive 5′P. Mutant R387K was more efficient but displayed an inaccurate behavior comparable to wild-type Polμ. (C) Accurate vs. inaccurate NHEJ of incompatible ends, using a 5′-labeled 3′-protruding (C) dsDNA substrate (5 nM), having a recessive 5′P. Mutant R387K displayed an error-prone behavior compared to the accuracy of wild-type Polμ. In all cases, assays were performed in the presence of 2.5 mM MgCl2, 100 μM of the indicated ddNTP, and 200 nM of either Polμ wt or R387K mutant. After incubation for 30 min at 30 °C, +1 extension of the 5′P-labeled primer was analyzed by 8 M urea-20% PAGE and autoradiography.
Discussion
In most DNA-dependent DNA polymerases, proper positioning of the 3′-terminus is indirectly dictated by the enzyme′s avidity by the templating base, thus configuring a binary complex ready to select the incoming nucleotide (ternary complex). Eventually, when no template base is available (blunt or 3′-protruding ends), any further nucleotide addition is unfavoured, probably due to a deficient re-positioning of the 3′ terminus. Thus, only a poorly efficient + 1 addition reaction, i.e., a minimalist terminal transferase, can be displayed by some DNA polymerases lacking 3–5′ exonuclease (21–23). Conversely, TdT and Polμ are adapted for template-independency in part due to a specific histidine residue (His329 in Polμ and His342 in TdT) that actively cooperates in the relocation of the primer from a “non-productive” E:DNA complex to a “productive” E:DNA:dNTP complex in the absence of a templating base.
Combined structural and functional evidences for both Polμ and TdT (17, 18), together with the functional data presented in this paper, indicate that there is another residue directly implicated in the terminal transferase activity of both enzymes. That residue (Arg387 in Polμ and Lys403 in TdT) modulates the catalytic efficiency of the terminal transferase reaction, by regulating the rate-limiting step. In the case of Polμ, Arg387 acts as a brake for the necessary movement of the primer, to limit nucleotide additions before end bridging. However, when a templating base is provided in trans during NHEJ, the rate-limiting step is relieved, and Arg387 changes its blocking interaction with the primer for an interaction with the template strand (−3 position), to estabilize the productive complex, thus allowing efficient and accurate end-joining to occur (see Fig. 6). That interaction of Arg387 cannot occur in mutant R387A, in agreement with its defective terminal transferase, and it would be lost at 3′-protruding ends longer than three nucleotides. Interestingly, mutant R387K produced a very specific blockage at position + 4 when continuous terminal transferase extension of a blunt end was tested (Fig. 2B). This suggests that the mutant lysine cannot properly establish the interactions occurring at this transition, when Arg387 could stablish alternative interactions (possibly with loop1) on longer protrusions. Interestingly, the equivalent residue in human Polλ (Lys472) is also involved in regulating the catalytic cycle by means of inhibitory interactions with the primer strand (T. A. Kunkel and K. Bebenek, personal communication). In the case of TdT, residue Lys403 likely establishes a weaker interaction with the primer compared to its orthologue Arg387 in Polμ. Thus, TdT has been optimized to efficiently overcome the rate-limiting step of the terminal transferase, to exclusively perform creative synthesis.
Fig. 6.
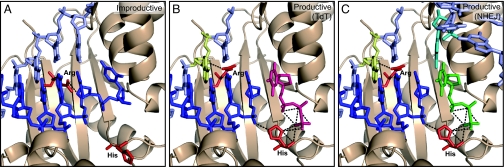
Modeling the limiting step of Polμ's terminal transferase. (A) Stable/non-productive step (binary complex): Polμ (1IHM) was overimposed on the binary complex of TdT with ssDNA (1KDH). Only the partial structure of Polμ (wheat color) is shown for clarity. A 1nt 3′-protruding substrate (derived from the gapped substrate present in the Polμ crystal) was modeled (as in 1KDH) to reproduce the initial situation of a binary complex in which the primer (dark blue) occupies the NTP binding pocket. Residue His329 (as His342 in TdT) is in “standby,” and Arg387 (modeled from Lys403 in TdT) strongly interacts with the primer strand. (B) Productive step for the terminal transferase (ternary complex in the absence of template): the primer (dark blue) has been relocated, and any incoming dNTP (magenta) sits in place with the assistance of His329 (rotated 180°). Arg387 stabilizes the new primer location by interacting with the template strand (nucleotide in yellow). (C) Productive step during NHEJ (ternary complex with a template provided in trans): a NHEJ reaction implying two 3′-protruding and incompatible ends (modeled from the gapped substrate present in the Polμ crystal) is shown. Primer relocation and Arg387 repositioning is facilitated by the template provided in trans (nucleotide in cyan) that allows selection of a complementary nucleotide (depicted in green).
Which is the physiological role of the terminal transferase activity of Polμ? As shown here, and in other reported studies, NHEJ of short incompatible ends can be accurate in many cases, but imprecise in others depending on both the length and sequence of each protrusion (15). For the latter cases, when a templating base is not in a proper register, untemplated terminal transferase addition in a NHEJ context provides a valid, although mutagenic, solution that would be conceptually similar to translesion DNA synthesis. Besides, it cannot be ruled out that Polμ′s terminal transferase can extend a single short 3′-protrusion to facilitate end-joining of this fraction of non-complementary ends.
Based on our mutational data, we elaborated a model that explains why Polμ prefers to have a more limited/mild terminal transferase activity, regulated by Arg387. A templating base provided in trans by the approaching end that could be located in a proper register will stabilize the incoming (and complementary) nucleotide, thus facilitating primer translocation. As a result of this, NHEJ of many incompatible ends can be efficient and accurate. During NHEJ of this fraction of incompatible ends, as shown in this paper, an excessive terminal transferase would be disadvantageous in terms of genome stability. On the other hand, our findings also explain the necessity of a mild terminal transferase activity in Polμ, not only devoted to create connectivity in those other DNA ends that cannot be efficiently joined on a templating basis, but perhaps contributing to gain a certain degree of genome variability. Additionally, it can be inferred that TdT evolved to maximize the efficiency of the translocation mechanism in the absence of template, at the cost/benefit of introducing untemplated nucleotides, thus being devoted to generate variability at V(D)J recombination intermediates.
Is there any in vivo evidence of untemplated insertions made by Polμ? It has been shown that mice that are TdT−/− still contain a 5% of their V(D)J junctions with template-independent additions, which suggested a possible role of Polμ in these reactions (24). In agreement with that, the terminal transferase activity of Polμ has been directly implicated at variability/repair processes occurring at embryo developmental stages in which TdT is still not expressed (25).
Materials and Methods
DNA and Proteins.
Synthetic DNA oligonucleotides were obtained from Isogen. PAGE-purified oligonucleotides were labeled at their 5′ ends with [γ-32P]ATP. The oligonucleotides used to generate the DNA substrates were the following: for gapped substrates, P15 (5′-TCTGTGCAGGTTCTT-3′), T32 (5′-TGAAGTCCCTCTCGACGAAGAACCTGCACAGA-3′) and D16 (5′-GTCGAGAGGGACTTCA-3′); as ssDNA, PolydT (5′-TTTTTTTTTTTTTTTTTTTTT-3′), to avoid the formation of secondary structures; for terminal transferase activity assays, oligonucleotides 5′-CGCAAGTCAGCGCTACGGG-3′ (Blunt), 5′-CGCAAGTCAGCGCTACGGGT-3′ (1T), 5′-CGCAAGTCAGCGCTACGGGTT-3′ (2T), 5′-CGCAAGTCAGCGCTACGGGTTT-3′ (3T), and 5′-CGCAAGTCAGCGCTACGGGTTTTTTTTT-3′ (9T) were used as primer strands, hybridized to oligonucleotide 3′-GCGTTCAGTCGCGATGCCC-5′ to obtain either blunt or 3′-protruding dsDNA substrates. For NHEJ assays, oligonucleotides 5′-CCCTCCCTCCCCA-3′ (CA), 5′-CCCTCCCTCCCGT-3′ (GT), 5′-CCCTCCCTCCCG-3′ (G) and 5′-CCCTCCCTCCCC-3′ (C) were used as primers, hybridized to oligonucleotide 5′-GGGAGGGAGGG-3′. Oligonucleotides D16, CA, GT, G, and C contain a phosphate at the 5′-end. Ultrapure dNTPs, ddNTPs, [α-32P] dNTPs (3,000 Ci/mmol) and [γ-32P] ATP (3,000 Ci/mmol) were purchased from GE Healthcare. T4 polynucleotide kinase was obtained from New England Biolabs. Pfu DNA polymerase was purchased from Promega Corporation.
Construction and Purification of Human Polμ Mutant Proteins.
Site-directed mutagenesis (QuikChange Site Directed Mutagenesis kit, Stratagene) was performed on the overexpression plasmid for Polμ (pRSETA-hPolμ) (6), with primers: H329G (5′-AAGTTGCAGGGCGGAGACGTGGACTTC-3′ and 5′-GAAGTCCACGTCTCCGCCCTGCAACTT-3′), R387K (5′-GACGCTTTTGAGAAGAGTTTCTGCATT-3′ and 5′-AATGCAGAAACTCTTCTCAAAAGCGTC-3′) and R387A (5′-GACGCTTTTGAGGCGAGTTTCTGCATT-3′ and 5′-AATGCAGAAACTCGCCTCAAAAGCGTC-3′). DNA constructs were sequenced and transformed in E. coli BL21(DE3)pLysS. Wild-type and mutant Polμ variants were overexpressed and purified as described (6).
DNA Polymerization Assays.
To analyze DNA-dependent and independent polymerization, several DNA substrates, containing 5′P-labeled primers, were incubated with different proteins, at the concentration indicated in each case. The reaction mixture, in 20 μL, contained 50 mM Tris-HCl (pH 7.5), 1 mM DTT, 4% glycerol, and 0.1 mg/mL BSA, in the presence of the indicated amounts of the DNA polymerization substrates, and the indicated concentrations of dNTP and activating metal ions. After incubation, reactions were stopped by adding gel loading buffer [95% (vol/vol) formamide, 10 mM EDTA, 0.1% (wt/vol) xylene cyanol, and 0.1% (wt/vol) bromophenol blue] and analyzed by 8 M urea/20% PAGE and autoradiography. When indicated, we used ddNTPs instead of dNTPs to limit incorporation to a single nucleotide on the 3′-end of the labeled oligonucleotide. For a steady state analysis of the terminal trasferase activity, the reaction mixture, in 20 μL, contained 50 mM Tris-HCl (pH 7.5), 1 mM DTT, 4% glycerol, and 0.1 mg/mL BSA, in the presence of MnCl2 (1 mM), polydT (1 μM), either Polμ wt or R387K mutant (200 nM), and various concentrations of the indicated [α-32P] dNTP (10–1,000 nM). After incubation for 30 min at 30 °C, reactions were stopped with EDTA (0.35 M), filtered through Sephadex G-25 spun columns (Roche), and the Cerenkov radiation of the excluded volume, quantified. The plotted data were fitted by a non-linear regression curve to the Michaelis-Menten equation: V = Vmax*(dNTP)/[(Km+(dNTP)]. foext, a constant that represents the ratio of the catalytic efficiencies, for each nucleotide, relative to Polμ wt.
Analysis of the Interaction of Polμ with Single-Stranded DNA by Nitrocellulose Filter Binding.
The assay was carried out using 5 nM of a labeled homopolymer (PolydT) as DNA primer, containing a ddCMP in the 3′-end to avoid polymerization, in the presence of 1 mM MnCl2, and the indicated concentration of either Polμ wt, R387K, R387A, or H329G. When indicated, 100 μM dTTP was added. Reactions were incubated for 10 min at 22 °C in binding buffer (50 mM Tris·HCl, pH 8.0, 25 mM NaCl, and 1 mM DTT), and filtered on nitrocellulose filters as described (19). Radiolabeled DNA retained in each filter was quantitated by a PhosphorImager.
Amino Acid Sequence Comparisons and 3D Modeling.
A multiple alignment of different DNA polymerases of the Pol X family was done using the program MULTALIN (http://prodes.toulouse.inra.fr/multalin/). PDB coordinates for two different TdT structures and for one Polμ structure, obtained from the Protein Data Bank (http://www.rcsb.org/pdb/), were used: 1KDH (binary complex of murine TdT with ssDNA); 1KEJ (murine TdT complexed with ddATP); 1IHM (ternary complex of murine Polμ with gapped-DNA and incoming ddNTP). The different conformations of the studied residues of Polμ and TdT were analyzed by using the Swiss PDB Viewer (http://www.expasy.ch/spdbv/) and MacPymol (http://delsci.com/macpymol/) programs.
Supplementary Material
Acknowledgments.
We thank M. de Vega, T. A. Kunkel, and K. Bebenek for helpful discussions and critical reading of the manuscript. This work was supported by Ministerio de Ciencia y Tecnología Grants BFU2006–14390/BMC, CONSOLIDER CSD2007–00015 and Comunidad Autónoma de Madrid Grants P2006/BIO-0306 to L.B., and by an institutional grant to Centro de Biología Molecular “Severo Ochoa” from Fundación Ramón Areces. M.J.M. was recipient of a fellowship from Comunidad Autónoma de Madrid. J.F.L de S. is an Investigator of the Ramon y Cajal Program.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908492106/DCSupplemental.
References
- 1.Friedberg EC, et al. DNA Repair and Mutagenesis. 2nd Ed. Washington D.C.: ASM Press; 2006. [Google Scholar]
- 2.Bebenek K, Kunkel TA. Functions of DNA polymerases. Adv Protein Chem. 2004;69:137–165. doi: 10.1016/S0065-3233(04)69005-X. [DOI] [PubMed] [Google Scholar]
- 3.Moon AF, et al. The X family portrait: Structural insights into biological functions of X family polymerases. DNA Repair (Amst) 2007;12:1709–1725. doi: 10.1016/j.dnarep.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bork P, et al. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J. 1997;11:68–76. [PubMed] [Google Scholar]
- 5.Callebaut I, Mornon JP. From BRCA1 to RAP1: A widespread BRCT module closely associated with DNA repair. FEBS Lett. 1997;400:25–30. doi: 10.1016/s0014-5793(96)01312-9. [DOI] [PubMed] [Google Scholar]
- 6.Dominguez O, et al. DNA polymerase mu (Pol mu), homologous to TdT, could act as a DNA mutator in eukaryotic cells. EMBO J. 2000;19:1731–1742. doi: 10.1093/emboj/19.7.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.García-Diaz M, et al. DNA polymerase lambda (Pol lambda), a novel eukaryotic DNA polymerase with a potential role in meiosis. J Mol Biol. 2000;301:851–867. doi: 10.1006/jmbi.2000.4005. [DOI] [PubMed] [Google Scholar]
- 8.Ruiz JF, et al. DNA polymerase mu, a candidate hypermutase? Philos Trans R Soc London B Biol Sci. 2001;35:699–709. doi: 10.1098/rstb.2000.0754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahajan KN, et al. Association of terminal deoxynucleotidyl transferase with Ku. Proc Natl Acad Sci USA. 1999;96:13926–13931. doi: 10.1073/pnas.96.24.13926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahajan KN, Nick McElhinny SA, Mitchell BS, Ramsden DA. Association of DNA polymerase mu (pol mu) with Ku and ligase IV: Role for Pol mu in end-joining double strand break repair. Mol Cell Biol. 2002;22:5194–5202. doi: 10.1128/MCB.22.14.5194-5202.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma Y, et al. A biochemically defined system for mammalian nonhomologous DNA end joining. Mol Cell. 2004;16:701–713. doi: 10.1016/j.molcel.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 12.Nick McElhinny SA, et al. A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end joining. Mol Cell. 2005;19:357–366. doi: 10.1016/j.molcel.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 13.Juarez R, Ruiz JF, Nick McElhinny SA, Ramsden D, Blanco L. A specific loop in human DNA polymerase μ allows switching between creative and DNA-instructed synthesis. Nucleic Acids Res. 2006;34:4572–4582. doi: 10.1093/nar/gkl457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruiz JF, et al. Overexpression of human DNA polymerase μ (Pol μ) in a Burkitt's lymphoma cell line affects the somatic hypermutation rate. Nucleic Acids Res. 2004;32:5861–5873. doi: 10.1093/nar/gkh929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davies BJ, Havener JM, Ramsden DA. End-bridging is required for Pol μ to efficiently promote repair of noncomplementary ends by nonhomologous end joining. Nucleic Acids Res. 2008;36:3085–3094. doi: 10.1093/nar/gkn164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 17.Delarue M, et al. Crystal structures of a template-independent DNA polymerase: Murine terminal deoxynucleotidyltransferase. EMBO J. 2002;21:427–439. doi: 10.1093/emboj/21.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moon AF, et al. Structural insight into the substrate specificity of DNA Polymerase mu. Nat Struct Mol Biol. 2007;14:45–53. doi: 10.1038/nsmb1180. [DOI] [PubMed] [Google Scholar]
- 19.Wong I, Lohman TM. A double-filter method for nitrocellulose-filter binding: Application to protein-nucleic acid interactions. Proc Natl Acad Sci USA. 1993;90:5428–5432. doi: 10.1073/pnas.90.12.5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu J, et al. XRCC4:DNA ligase IV can ligate incompatible ends and can ligate across gaps. EMBO J. 2007;26:1010–1023. doi: 10.1038/sj.emboj.7601559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clark JM, Joyce CM, Beardsley GP. Novel blunt-end addition reactions catalyzed by DNA polymerase I of Escherichia coli. J Mol Biol. 1987;198:123–127. doi: 10.1016/0022-2836(87)90462-1. [DOI] [PubMed] [Google Scholar]
- 22.Clark JM. Novel non-templated nucleotide addition reactions catalyzed by procaryotic and eucaryotic DNA polymerases. Nucleic Acids Res. 1988;16:9677–9686. doi: 10.1093/nar/16.20.9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Golinelli MP, Hughes SH. Nontemplated nucelotide addition by HIV-1 reverse transcriptase. Biochemistry. 2002;41:5894–5906. doi: 10.1021/bi0160415. [DOI] [PubMed] [Google Scholar]
- 24.Bertocci B, De Smet A, Weill JC, Reynaud CA. Nonoverlapping functions of DNA polymerases μ, λ, and terminal deoxynucleotidyltransferase during immunoglobulin V(D)J recombination in vivo. Immunity. 2006;25:31–41. doi: 10.1016/j.immuni.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 25.Gozalbo-López B, et al. A role for DNA polymerase mu in the emerging DJH rearrangements of the postgastrulation mouse embryo. Mol Cell Biol. 2009;29:1266–1275. doi: 10.1128/MCB.01518-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.