Abstract
Objective
To identify differentially expressed genes between fibroid and adjacent normal myometrium in an identical hormonal and genetic background.
Design
Array analysis of 3 leiomyomata and matched adjacent normal myometrium in a single patient.
Setting
University of Colorado Hospital.
Patient(s)
A single female undergoing medically indicated hysterectomy for symptomatic fibroids.
Interventions(s)
mRNA isolation and microarray analysis, reverse-transcriptase polymerase chain reaction, western blotting and immunohistochemistry.
Main Outcome Measure(s)
Changes in mRNA and protein levels in leiomyomata and matched normal myometrium.
Result(s)
Expression of 197 genes was increased and 619 decreased, significantly by at least 2 fold, in leiomyomata relative to normal myometrium. Expression profiles between tumors were similar and normal myometrial samples showed minimal variation. Changes in, and variation of, expression of selected genes were confirmed in additional normal and leiomyoma samples from multiple patients.
Conclusion(s)
Analysis of multiple tumors from a single patient confirmed changes in expression of genes described in previous, apparently disparate, studies and identified novel targets. Gene expression profiles in leiomyomata are consistent with increased activation of mitogenic pathways and inhibition of apoptosis. Down-regulation of genes implicated in invasion and metastasis, of cancers, was observed in fibroids. This expression pattern may underlie the benign nature of uterine leiomyomata and may aid in the differential diagnosis of leiomyosarcoma.
Keywords: fibroid, microarray, uterine leiomyoma, myometrium, matrix metalloproteinases, fibulins, desmoglein, MST4, PKC β1, gene expression
INTRODUCTION
Leiomyomata uteri or fibroids are the most common neoplasm of the female genital tract developing primarily during the reproductive years and becoming symptomatic during perimenopause (1, 2). Tumors occur in 77% of women, and approximately 25% of Caucasians have clinically significant lesions. The relative risk of fibroids is two to threefold greater in black women than white women and clinical disease is more severe (3–5). Although leiomyomata are benign and rarely result in death, they frequently cause pelvic pain and pressure, dysmenorrhea, menometrorrhagia leading to anemia and less frequently, reproductive dysfunction, including reduced fertility or pregnancy complications, constipation and urinary problems. Leiomyomata are a leading cause of hospitalization for non-pregnancy related gynecologic disorders (6) and are the single most frequent indication for hysterectomy accounting for over 500,000 surgeries per year in the US (7–9). Thus, it is clear that leiomyomata represent a major health problem for women of reproductive age, yet relatively little is known of the etiology or pathophysiology of these tumors.
Previously, in order to identify pathways relevant to development and progression of leiomyomata, several studies have examined the differential gene expression between uterine leiomyoma and normal myometrium using microarray analysis (10–25). However, the different gene array experiments have yielded disparate results, both with respect to those genes identified as being altered in expression and in the direction of the changes (26). This may be a reflection of heterogeneity in myometrial gene expression due to genetic and/or hormonal variations between patients that are independent of the diseased state. In order to avoid the above potential complications we performed a cDNA microarray comparing the gene expression profiles of multiple leiomyomata to matched adjacent normal myometrium from the same patient. Significant changes in gene expression were then validated by reverse transcription PCR, immunohistochemistry or western blotting, and their profiles compared to samples derived from additional distinct patients.
The screening approach of the cDNA microarray and review of the published data for the genes that we identified as differentially expressed enabled us to categorize several targets into specific functional groups. Overall, the results of this study provide very useful insights as to what biological processes may be significantly impacted by gene expression changes in leiomyomata, and the results suggest additional candidate targets for further studies into the mechanisms of pathogenesis.
MATERIALS AND METHODS
Tissue procurement
Portions of leiomyomata, approximately 2 x 3cm sections from the periphery distinct from normal tissue, and matched unaffected myometrium were collected from women (n=11) who were undergoing hysterectomy for indications related to symptomatic leiomyomata. Tissue samples were obtained from each leiomyoma and adjacent matched normal myometrium. The exception was patient BRAD 3 for which only one normal myometrium sample and three leiomyomata samples were collected. None of the patients had received any medical treatment for their fibroids. The tissues were collected at the University of Colorado Hospital with prior approval by the Colorado Multiple Institutional Review Board, under protocol number 03–642. Immediately after collection in the operating room, a portion of the tissue was snap frozen and stored in liquid nitrogen. Additional portions of tissue were fixed and paraffin embedded for histological evaluation and immunohistochemistry.
RNA isolation
Aliquots (~5 g) frozen tissue sections were pulverized under liquid nitrogen. 50–100 mg of the powdered tissue was placed in 1 ml TRIzol (Invitrogen Life Technologies, Inc., Carlsbad, CA) and was then homogenized with a Polytron probe (Brinkmann Instruments, Westbury, NY). Total cellular RNA was isolated from the tissues and cells using TRIzol per manufacture’s instructions. The isolated RNA was quantitated with the NanoDrop ND1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE), so that no more than 100 μg was loaded on the column during the DNase treatment. The RNA was DNase treated (twice) using Qiagen RNeasy Mini kit and Qiagen RNase-Free DNase s (Qiagen, Inc.). Quality of purified RNA was assessed on a RNA 6000 Nano Chip using an Agilent 2100 bioanalyzer (Agilent Technologies, Germany).
cDNA synthesis
An aliquot of 1μg from each total RNA sample was incubated with 2.5 μl of random hexamers in a final volume of 9 μl RNase free water, at 65°C for 5 minutes. The cDNA master mix was made so that the final concentration per reaction tube was 1xPCR buffer, 2.5 mM MgCl2, 1 mM dNTPs blend, 2 units RNase inhibitor, 5 units MMLV reverse transcriptase. The reaction tube was returned to the PCR machine at 42° C for 60 minutes, 95°C for 5 minutes and samples were stored at 4°C. All RT-PCR reagents were obtained from Applied Biosystems (Foster City, CA).
Semi-quantitative Reverse Transcriptase Polymerase Chain Reaction
All of the primers were obtained from Invitrogen Life Technologies (Carlsbad, CA). Sequences are shown below:
EFEMP1 (Fibulin-3) forward primer: 5′-AGCAGTGACAGGCTCAACTGTGAA-3′
EFEMP1 (Fibulin-3) reverse primer: 5′-CACGAGCACAAGCATTGCACTTAC-3′
MMP-7 forward primer: 5′-GTGGAGTGCCAGATGTTGCAGAAT-3′
MMP-7 reverse primer: 5′-TCCAGCGTTCATCCTCATCGAAGT-3′
MMP-11 forward primer: 5′-CTGCCTCGGAAGAAGTAGATCTTG-3′
MMP-11 reverse primer: 5′-TCTACACCTATCGCTACGCACTGA-3′
MST-4 forward primer: 5′-GGTGGTTCAGCACTGGATCTTCTT -3′
MST-4 reverse primer: 5′-GTGTCCTTCTGCCTTCCATCTCTT -3′
DSG2 forward primer: 5′-CCTAGCCAGCCACAGAGCCTTATT-3′
DSG2 reverse primer: 5′-CGTGGTGTTCCTAGCCGTCATAGA-3′
GAPDH forward primer: 5′-GGCTCTCCAGAACATCATCCCTGC-3′
GAPDH reverse primer: 5′-GGGTGTCGCTGTTGAAGTCAGAGG-3′
Primers were resuspended in 200 μl Rnase free water, quantitated, and diluted at 20 pmoles per μl. Each primer set was tested for optimal cycle number (5 μl aliquots were taken out after 12, 15, 18, 21, 24, 27 cycles) and optimized for the best annealing temperature by using a gradient of 60–70°C. Cycle optimization and temperature optimization was performed on matched normal and leiomyoma samples. Optimal cycle numbers and annealing temperatures were as follows: EFEMP (fibulin-3) 25 cycles at 62 °C; MMP-7 25 cycles at 69 °C; DSG2 27 cycles at 62 °C, MST-4 24 cycles at 62 °C, MMP-11 24 cycles at 63 °C and GAPDH for 21 cycles at 65 °C. Master mix for RT-PCR contained 5 μl cDNA, 1xPCR buffer, 2.5mM MgCl2, 1mM dNTPs blend, 2 μM each of the forward and reverses primers, 1 μl of DNA iTaq polymerase (Bio-Rad) in a total volume of 50 μl. PCR reactions were run an eppendorf Mastercycler gradient PCR machine at 94°C for 4 minutes followed by the optimized number of cycles (94°C for 30 seconds, optimal annealing temperature between 62–69°C for 45 seconds, 72°C for 45 seconds) followed by a final 72°C for 5 minutes extension time. 8 μl of the RT-PCR products were run on a 2% agarose gels stained with ethidium bromide, along with a 100 bp ladder (New England Biolabs, Ipswich, MA). Alpha Innotech Gel Documentation Imager was used to visualize the gel and Alpha EaseFC software densitometry tool was used to obtain a relative quantification by calculating the sum of the pixels in the area of the band sum and subtracting the background. Each band was normalized by dividing by the amount of GAPDH control.
Immunohistochemistry
Anti-MMP-11 clone SL3.05 from Labvision/Neomarkers (Fremont, CA) was used at a 1:30 dilution. The slides were deparaffinized using a series of xylene washes and graded ethanol washes, as described (27). Heat Induced Epitope Retrieval was performed using a Biocare Decloaker (Concord, CA) with 1X Citra-Plus Buffer from Biogenex (San Ramon, CA). All slides were treated with 3% hydrogen peroxide (Fisher Scientific, Fairlawn, NJ) for 5 min., followed by 10% Normal Goat Serum (Vector Laboratories, Burlingame, CA) incubation for 20 min in a humidity chamber. Primary antibodies were applied for 1 hr in a humidity chamber. Detection was performed using Envision (Dakocytomation, Carpinteria, CA) for 30 min in a humidity chamber. Diaminobenzidine (DAB+) solution (Dakocytomation) was applied for 10 min., followed by a 2 min. hematoxylin counterstain. Slides were rinsed between each step with phospho-buffered saline with Tween. Slides were then dehydrated with a series of graded ethanol washes and xylene washes, mounted with Permount (Fisher Scientific, Pittsburgh, PA) and cover-slipped for bright field microscopy.
Western blotting
Frozen tumor samples were pulverized into powder and lysed in buffer (50 mmol/L Tris pH 7.4, 0.15 mmol/L NaCl, 1% Triton X-100, 0.5% deoxycholate) supplemented with protease inhibitors (Roche Diagnostics, Mannheim, Germany). Samples were vortexed for one minute, then homogenized using a Polytron (Brinkmann Instruments, Westbury, NY). Tumor lysates were cleared by centrifugation at 13,000 g for 10 minutes, and protein concentrations were determined by Bradford assay (Bio-Rad Laboratories, Hercules, CA). Aliquots were resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis, transferred to polyvinylidene fluoride membrane and processed as described (28). MST-4 and PKC-β1 were assessed using an anti-MST-4 antibody (BD, Franklin Lakes, NJ) and a polyclonal anti-PKC-β1 antibody (C-16 Santa Cruz Biotechnology, Santa Cruz, CA) respectively. Equal loading was assessed using a monoclonal anti-GAPDH antibody (IMGENEX, San Diego, CA). Band intensity was quantitated by densitometry using Quantity One software (V4.5.1) and a GelDoc Imaging System (Bio-Rad).
Microarray Analysis
Initial quality assessment of all scanned chips was performed using GeneChip Operating Software (GCOS) v1.1 (Affymetrix, http://www.affymetrix.com ). Compiled data in the form of six individual CEL files, the primary output of scanned Human Genome U133 plus 2.0 microarray chips, were imported to GeneSpring (Agilent Technologies, http://www.chem.agilent.com ) for analysis using the native probe level GC-Robust Multi-array Average (GC-RMA) algorithm (29).
GC-RMA calculation performs an initial background noise/non-specific binding correction using the G-C content to estimate hybridization affinities of the individual targets that comprise a set of probes for any given gene. Next, the GC-RMA algorithm consists of three further steps. In the first step, the affinity corrected mis-match (MM) hybridization scores are subtracted from perfect match (PM) hybridization scores for every target of a given probe set. Secondly, a quantile non-linear normalization is performed that corrects for bias among arrays in the full data set, such that each array can be directly compared to any other. Finally the quantile normalized probe level data, derived from the initial background noise/non-specific binding correction step, are summarized as a single expression measurement for that given probe set. When compared to previous data processed using MAS v5.0 methods, GC-RMA showed better precision evidenced by lower variance within replicate samples especially at low levels of gene expression. Further, GC-RMA seems to have more consistent estimates of fold change among arrays as well as a lower rate of false discovery (30).
Following GC-RMA analysis of the six batched CEL files, the data were grouped according to normal or diseased parameters and treated as biologic triplicate samples. Initially, the data were filtered for genes that scored above 20 RAW intensity units on at least three of the six arrays, so that a gene could be completely unreliable (absent) in one condition but have reliable expression in the other (27,822 genes pass). Because the Log Ratio of normalized data is centered on 1.0, genes that were very similar to 1.0 in both conditions measured by a standard t-test with a multiple testing correction False Discovery Rate (FDR) from 0.95 to 1.00 were removed yielding 25,331 genes. In the next filter, genes that were equal in expression by up to 10% of the mean value were removed leaving 16,459 genes. The last filter that was applied to the data set identified potentially differential genes from the preceding list with values that were greater or less than 1.25 fold in each condition (7,470 genes pass).
The final output list from the filtering process was used as input for ANOVA statistical testing. In order to find a list of statistically significant genes with high degree of confidence, a multiple testing correction FDR of 0.10 (10%) was chosen, and the analysis restricted to genes showing a minimum of two fold change. We identified a total of 816 significant genes that show differential expression between these two conditions (with the caveat that 10% [~82 genes] of these genes may not differ statistically). The genes resultant from the statistical test were then grouped by direction of expression change (up or down between the two conditions) and filtered for a greater than 2.0 fold change. We found 97 genes that showed a significant increase and 619 genes with a significant decrease in the leiomyomata as compared to the matching normal tissue.
RESULTS
We decided to study multiple tumors from a single patient to eliminate inter-subject genetic variability and differences in hormonal milieu. Our patient (B5) underwent a hysterectomy for symptomatic uterine fibroids (namely pain). She was 53 years old and was established to be postmenopausal based on histological examination of her endometrium that was found to be atrophic. She had not received any treatment for her fibroids prior to her hysterectomy. We obtained tissue from three tumors ranging in size between 4 and 11 cm, and adjacent myometrium and selected matched pairs for Affymetrix microarray analysis as described in Materials and Methods.
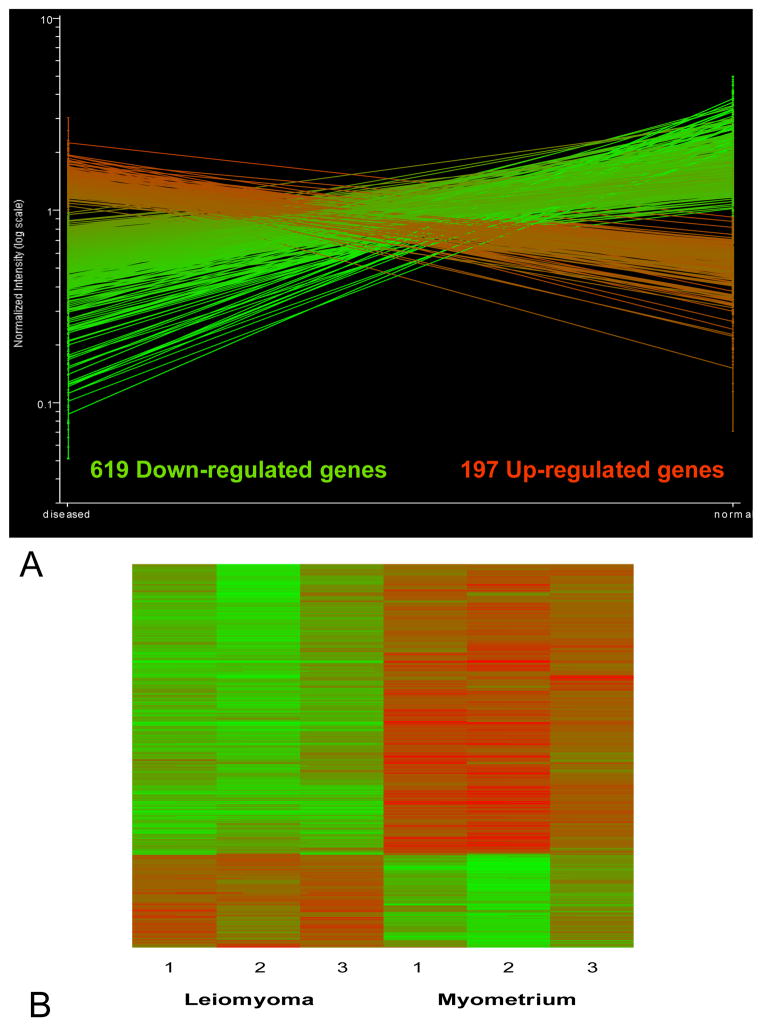
As shown in figure 1A, of the 38,500 genes contained in the array, we identified 816 that exhibited significant differences according to our criteria. We focused on genes that showed consistent changes in expression between normal and diseased state, in the same direction, in all 3 paired samples. Statistically significant changes were further selected by including only those genes changing by a magnitude of two-fold or greater. Of the 816 differentially expressed genes that met our criteria, 619 were down-regulated and 97 were up-regulated by two fold. Functional categories of genes showing altered expression included: regulators and components of the extracellular matrix, metabolism, signal transduction, apoptosis, transcription factors, protein trafficking and cell cycle regulation. This preponderance of down regulated genes in uterine leiomyomata is consistent with previous analyses. Cluster analysis of the independent samples (Fig. 1B) demonstrates some variability in the absolute expression levels between tumors and between normal tissues. However the overall profiles of the 3 fibroids and the 3 myometrial samples respectively were similar. Thus, individual tumors derived from a single patient, with identical genetic factors and hormonal milieu, showed consistent changes in mRNA relative to normal myometrium.
Figure 1.
A. The statistically significant differentially expressed genes that exhibited consistent changes in all 3 tumors relative to normal myometrium of a magnitude of at least 2 fold. B. Heat map showing cluster analysis of the differentially expressed 816 genes in 3 uterine leiomyoma and 3 samples of matched adjacent normal myometrium. From left to right the first 3 columns represent the 3 tumors followed by the corresponding normal tissue. Rows represent individual genes. Red, increased gene expression; green, decreased gene expression. The color intensity is proportional to the hybridization intensity of a gene from its median level across all samples.
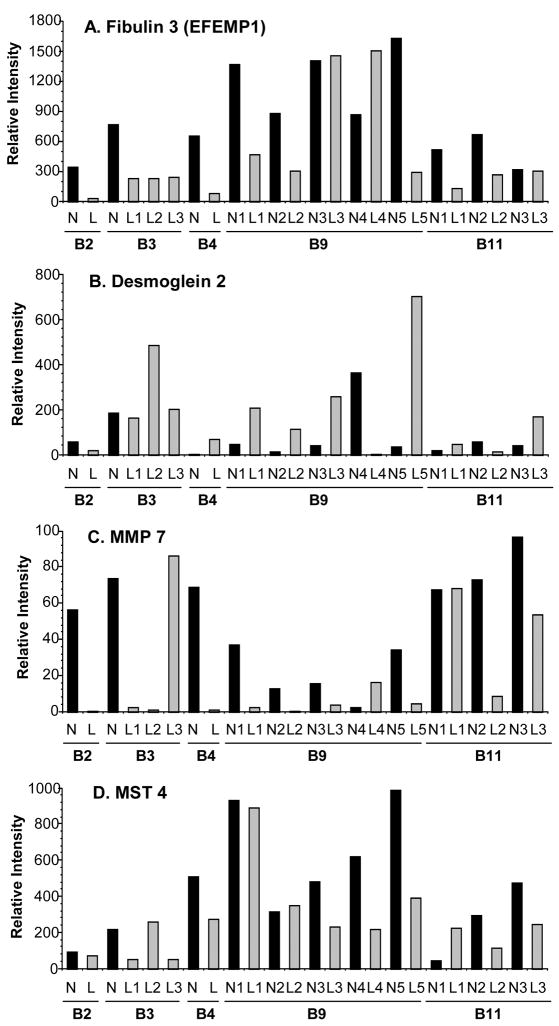
Selected genes were chosen for confirmation using a panel of paired normal myometrium and fibroid samples, including the samples (B5) used for the array, but also matched myometrium and leiomyoma tissue obtained from multiple additional patients. This procedure allowed us to not only confirm the changes observed in the array analysis but also to assess the inter-patient and for a given patient, inter-tumor variability of expression. Confirmatory methods utilized were semi-quantitative RT-PCR, Western blot and immunohistochemistry. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control for protein or mRNA levels, respectively. Levels were similar in leiomyomata and myometrial samples. In contrast, actin and β-tubulin exhibited considerable variation in expression in leiomyoma and were not suitable controls (not shown). Fibulin 3 (EGF-containing fibulin-like extracellular matrix protein 1) has been identified by arrayanalysis from other laboratories as being differentially expressed (13, 18, 23, 24); it was down-regulated approximately 20-fold in our analysis. In subsequent RT-PCR analysis, fibulin 3 was shown to be down regulated in the majority of tumors examined, with the exception of one of five tumors from patient B9 (Fig. 2A). Similarly, downregulation of the matrix metalloproteinase MMP7 (4.37-fold) was confirmed by RT-PCR in twelve out of thirteen tumors derived from five additional patients (Fig 2C). This gene had not been identified as differentially expressed prior to our analysis.
Figure 2.
Semiquantitative RT-PCR analysis. RNA from paired myometrium/leiomyoma from 5 different patients was analyzed using specific primer pairs, as indicated and described in Materials and Methods and electrophoresed on agarose gels. Bands were quantitated by scanning densitometry and expressed as relative intensity normalized to GAPDH. B2, B3, B4, B9, B11 – patient designations. N1-5 normal myometrium; L1-5 corresponding leiomyoma samples.
We also identified desmoglein 2, a member of a subclass of desmosomal cadherins, as a novel target not previously detected by array analyses. Desmoglein 2 was significantly up-regulated (5-fold) in leiomyomata. This finding was confirmed by RT-PCR in additional patients and tumors (Fig. 2B). Some inter and intra-patient variability was also observed with two tumors exhibiting decreased desmoglein 2 levels relative to adjacent myometrium.
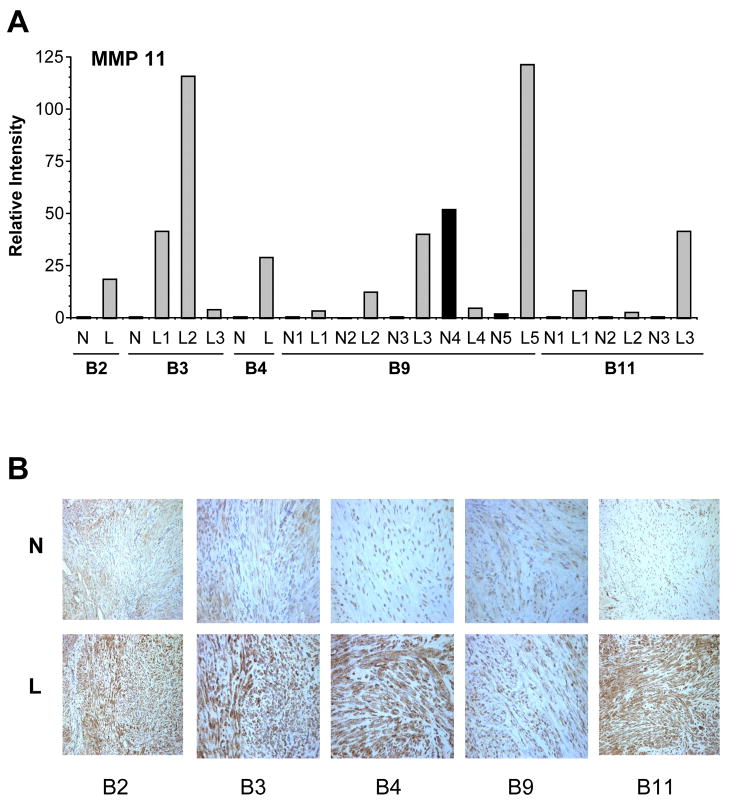
In contrast to MMP7, stromelysin 3, or MMP11, was found to be significantly up-regulated in leiomyomata, consistent with previous reports (10, 11, 19, 24), (Fig. 3A). The 5–10 fold increased expression of MMP11 by array and RT-PCR was confirmed by immunohistochemical analysis of several tumors. As shown in figure 3B, leiomyomata derived from multiple patients stained significantly more intensely for MMP11, relative to matched normal myometrium. No staining was detected using non-immune immunoglobulin (not shown).
Figure 3.
A. Semiquantitative RT-PCR analysis of MMP11 (Stromelysin 3) expression. RNA from paired myometrium/leiomyoma from 5 different patients was analyzed using specific primer pairs, as indicated and described in Materials and Methods and electrophoresed on agarose gels. Bands were quantitated by scanning densitometry and expressed as relative intensity normalized to GAPDH. B2, B3, B4, B9, B11 – patient designations. N1-5 normal myometrium; L1-5 corresponding leiomyoma samples.
B. Immunohistochemical staining for matrix metalloproteinase 11 (MMP-11) in normal myometrial tissue (top panels) and leiomyomata (bottom panels). Paraffin embedded sections were stained with anti-MMP-11 antibody as described in Materials and Methods. Representative 20X fields are depicted. B2, B3, B4, B9, B11 patient designations. N: normal myometrium, L matched leiomyoma.
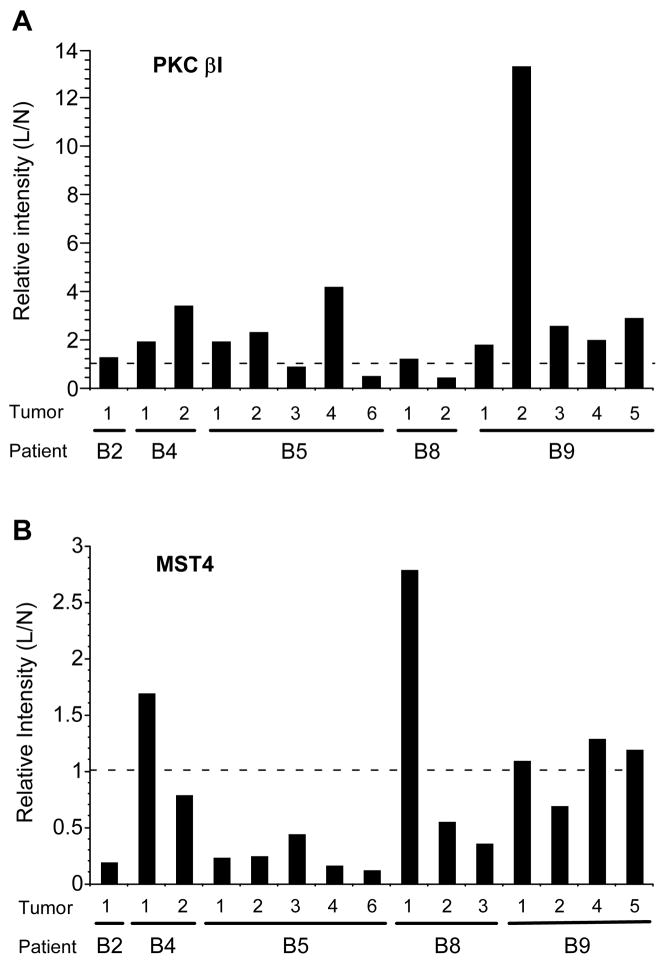
Finally, we examined expression of MST4 (Mst3 and SOK1-related kinase), a member of a family of serine/threonine kinases that may regulate apoptotic pathways, and protein kinase C β1 (PKCβ1). Array analysis indicated that PKCβ1 was up-regulated approximately 5-fold in leiomyoma while levels of MST-4 exhibited a significant (>20-fold) decrease in tumors. These results were confirmed by western blot analysis of tissue extracts from multiple additional patients (Fig. 4). Blots were scanned and quantitated, and levels normalized to GAPDH. Upregulation of PKCβ1 was observed in 12 of 15 matched tumor and normal samples (Fig. 4A), while MST4 was down-regulated in the majority (10 of 15) leiomyomata (Fig. 4B). Downregulation of MST4 was also observed in the majority of tumors (12 0f 13) analyzed by RT-PCR (Fig. 2D).
Figure 4.
Western blots analysis of leiomyoma and myometrium samples from 5 patients using antibodies to A. PKC β1 and B. MST-4. Blots were reprobed for GAPDH as a loading control. Chemiluminescence was quantitated using a BioRad Chemdoc XRS imager and relative levels expressed as a ratio of leiomyoma to normal myometrium, normalized to GAPDH. B2, B4, B5, B8 and B9; patient designations, numbers indicate independent tumor/normal paired samples derived from that patient. N: myometrium, L: leiomyoma.
Automated global analysis of the genes that were significantly altered in expression in leiomyomata using GeneSpring Gene Ontology (GO) and Ingenuity Pathway Analysis (http://www.ingenuity.com/) did not identify any obvious signaling pathways or functional categories when queried with the statistically significant gene lists. Thus, to provide additional insight into the pathophysiology of uterine leiomyomata we reviewed the literature, focusing on genes that have been described previously as relevant in malignant disease. NCBI databases were searched using the gene symbol or other gene designations and available information about a particular gene reviewed using GeneRIF (Gene Reference Into Function http://www.ncbi.nlm.nih.gov/projects/GeneRIF/). This analysis identified changes in the expression of genes linked to cell growth, survival, apoptosis, invasion and metastasis which are shown in Table 1. We also identified putative oncogenes and genes typically up-regulated in cancers. Overall, leiomyomata exhibited increased expression of genes associated with cell proliferation and survival (e.g. IGF-2 TGFβ3, PKCβ1, cyclin D and bcl-2) and a decrease in levels of pro-apoptotic genes (e.g. MST4 and TRAIL) relative to normal myometrium. Interestingly, whilst a number of putative tumor suppressors were down-regulated in leiomyoma, genes typically up-regulated in cancers were found to be down-regulated in these benign uterine tumors. This observation was particularly evident with respect to genes typically over expressed in invasive or metastatic cancers. Such genes were generally expressed in leiomyomata at lower levels than normal myometrium (Table 1), with the exception of the matrix metalloproteinases 11 and 14.
Table 1.
Differentially expressed genes in leiomyomata categorized by the indicated functional groups based on published reports. Multiple entries reflect different probe sets for a given target. Genes are listed in order of magnitude of fold change relative to normal myometrium. Negative values indicate down regulated genes.
| Proapoptotic genes | |||
|---|---|---|---|
| Genbank | Description | Fold | Gene Symbol |
| AF344882 | Mst3 and SOK1-related kinase | −31.55 | MST4 |
| NM_016542 | Mst3 and SOK1-related kinase | −22.72 | MST4 |
| NM_002583 | PRKC, apoptosis, WT1, regulator | −2.84 | PAWR |
| NM_004938 | Death-associated protein kinase 1 | −2.34 | DAPK1 |
| AF356193 | Caspase recruitment domain family 6 | −2.28 | CARD6 |
| BF434846 | Tenascin C (hexabrachion) | −4.80 | TNC |
| NM_006307 | Sushi-repeat-containing protein, X-linked | −2.04 | SRPX |
| AI281371 | APO-1/CD95 (Fas)-associated phosphatase | −3.98 | PTPN13 |
| NM_006264 | APO-1/CD95 (Fas)-associated phosphatase | −2.39 | PTPN13 |
| AW770896 | Insulin-like growth factor binding protein 7 | −2.30 | IGFBP7 |
| NM_002184 | IL6 signal transducer (oncostatin M receptor) | −4.71 | IL6ST |
| AW242916 | IL6 signal transducer (oncostatin M receptor) | −2.05 | IL6ST |
| AB015706 | IL6 signal transducer (oncostatin M receptor) | −2.05 | IL6ST |
| NM_002135 | Nuclear receptor subfamily 4A1 | −2.02 | NR4A1 |
| BC002439 | HIV-1 Tat interactive protein 2, 30kDa | −2.86 | HTATIP2 |
| BF511276 | A kinase anchor protein (gravin) 12 | −2.75 | AKAP12 |
| BF511276 | A kinase anchor protein (gravin) 12 | −2.42 | AKAP12 |
| AB003476 | A kinase anchor protein (gravin) 12 | −2.19 | AKAP12 |
| U57059 | TNF-related apoptosis inducing ligand TRAIL | −2.17 | TNFSF10 |
| NM_004226 | Ser/thr kinase 17b (apoptosis-inducing) | −3.96 | STK17B |
| AA203487 | Ser/thr kinase 17b (apoptosis-inducing) | −2.26 | STK17B |
| NM_021730 | CARD7, DEFCAP | 2.18 | NALP1 |
| AF003934 | Growth differentiation factor 15 | 5.23 | GDF15 |
|
Growth promoting genes | |||
| Genbank | Description | Fold | Gene Symbol |
|
| |||
| NM_003294 | Tryptase alpha/beta 1 | 2.02 | TPSAB1 |
| NM_001897 | Chondroitin sulfate proteoglycan 4 | 2.32 | CSPG4 |
| NM_002997 | Syndecan 1 | 2.26 | SDC1 |
| AV694854 | Adrenergic, alpha-1A-, receptor | 3.01 | ADRA1A |
| N51516 | Adrenergic, alpha-1A-, receptor | 2.53 | ADRA1A |
| AJ000008 | Phosphoinositide-3-kinase, gamma | 2.87 | PIK3C2G |
| BC003105 | Protein tyrosine phosphatase type IVA, 3 | 2.05 | PTP4A3 |
| X99268 | Twist homolog 1 | 2.17 | TWIST1 |
| NM_000612 | Insulin-like growth factor 2 (somatomedin A) | 5.67 | IGF2 |
| M17863 | Insulin-like growth factor 2 (somatomedin A) | 3.55 | IGF2 |
| J03241 | Transforming growth factor, beta 3 | 2.37 | TGFB3 |
| NM_002738 | Protein kinase C, beta 1 | 4.90 | PRKCB1 |
| M13975 | Protein kinase C, beta 1 | 2.98 | PRKCB1 |
| M73554 | Cyclin D1 (PRAD1) | 2.74 | CCND1 |
| NM_001759 | Cyclin D2 | 2.06 | CCND2 |
| NM_005195 | CCAAT/enhancer binding protein delta | −2.07 | CEBPD |
| NM_002039 | GRB2-associated binding protein 1 | −2.24 | GAB1 |
|
Growth inhibitory genes | |||
| Genbank | Description | Fold | Gene Symbol |
|
| |||
| AI281593 | Decorin | −2.45 | DCN |
| AF138303 | Decorin | −2.09 | DCN |
| AI343467 | Inhibin, beta A (activin A, activin AB alpha) | −3.99 | INHBA |
| M13436 | Inhibin, beta A (activin A, activin AB alpha) | −3.17 | INHBA |
| NM_002036 | Duffy blood group | −2.95 | DARC |
| AA530892 | Dual specificity phosphatase 1 (MKP-1) | −2.63 | DUSP1 |
| AW770896 | Insulin-like growth factor binding protein 7 | −2.30 | IGFBP7 |
| AF493929 | Regulator of G-protein signalling 5 | −2.18 | RGS5 |
| AI183997 | Regulator of G-protein signalling 5 | −2.07 | RGS5 |
| AF132818 | Kruppel-like factor 5 | −5.05 | KLF5 |
| AF003114 | Cysteine-rich, angiogenic inducer, 61 | −8.12 | CYR61 |
| NM_001554 | Cysteine-rich, angiogenic inducer, 61 | −6.59 | CYR61 |
| AA150501 | Epithelial membrane protein 1 | −2.32 | EMP1 |
| BE552421 | Mitochondrial tumor suppressor 1 | −2.22 | MTUS1 |
| AW444761 | Cyclin-dependent kinase inhibitor 2B (p15) | −3.41 | CDKN2B |
| NM_022470 | P53 target zinc finger protein | 2.29 | ZMAT3 |
|
Putative Tumor Suppressor genes | |||
| Genbank | Description | Fold | Gene Symbol |
|
| |||
| L16895 | Lysyl oxidase | −2.61 | LOX |
| NM_002317 | Lysyl oxidase | −2.31 | LOX |
| AF101051 | Claudin 1 | −13.02 | CLDN1 |
| NM_021101 | Claudin 1 | −5.33 | CLDN1 |
| NM_006307 | Sushi-repeat-containing protein, X-linked | −2.04 | SRPX |
| M92934 | Connective tissue growth factor | −3.34 | CTGF |
| NM_006207 | Platelet-derived growth factor receptor-like | −2.67 | PDGFRL |
| NM_012307 | Erythrocyte membrane protein band 4.1-like 3 | −2.00 | EPB41L3 |
| BE552421 | Mitochondrial tumor suppressor 1 | −2.22 | MTUS1 |
| NM_003206 | Transcription factor 21 | −11.05 | TCF21 |
| AF055585 | Slit homolog 2 | −3.00 | SLIT2 |
| AI692523 | Slit homolog 2 | −2.95 | SLIT2 |
| AI963304 | Slit homolog 2 | −2.36 | SLIT2 |
| AI343467 | Inhibin, beta A (activin A, activin AB alpha) | −3.99 | INHBA |
| M13436 | Inhibin, beta A (activin A, activin AB alpha) | −3.17 | INHBA |
| AF017987 | Secreted frizzled-related protein 1 | −2.28 | SFRP1 |
| AI332407 | Secreted frizzled-related protein 1 | −2.07 | SFRP1 |
| NM_003012 | Secreted frizzled-related protein 1 | −2.02 | SFRP1 |
| U16153 | Inhibitor of DNA binding 4 | −2.58 | ID4 |
| AW157094 | Inhibitor of DNA binding 4 | −3.47 | ID4 |
| NM_013253 | Dickkopf homolog 3 | −2.26 | DKK3 |
| AU148057 | Dickkopf homolog 3 | −2.23 | DKK3 |
| NM_002318 | Lysyl oxidase-like 2 | 2.74 | LOXL2 |
| U17074 | Cyclin-dependent kinase inhibitor 2C (p18) | 2.63 | CDKN2C |
| NM_001262 | Cyclin-dependent kinase inhibitor 2C (p18) | 2.63 | CDKN2C |
|
Genes associated with invasion, migration or metastasis | |||
| Genbank | Description | Fold | Gene Symbol |
|
| |||
| AF101051 | Claudin 1 | −13.02 | CLDN1 |
| NM_021101 | Claudin 1 | −5.33 | CLDN1 |
| NM_004938 | Death-associated protein kinase 1 | −2.34 | DAPK1 |
| NM_006614 | Cell adhesion molecule with homology to L1CAM | −24.44 | CHL1 |
| BF434846 | Tenascin C (hexabrachion) | −4.80 | TNC |
| AL552534 | CD44 antigen | −2.71 | CD44 |
| AF098641 | CD44 antigen | −2.40 | CD44 |
| M24915 | CD44 antigen | −2.38 | CD44 |
| NM_000610 | CD44 antigen | −2.32 | CD44 |
| AI493245 | CD44 antigen | −2.13 | CD44 |
| BC004372 | CD44 antigen | −2.12 | CD44 |
| J05021 | Villin 2 (ezrin) | −2.22 | VIL2 |
| AA670344 | Villin 2 (ezrin) | −2.19 | VIL2 |
| AL574210 | Serine proteinase inhibitor, PAI 1 | −3.73 | SERPINE1 |
| U66495 | Leptin receptor | −2.48 | LEPR |
| U08626 | Leptin receptor | −2.46 | LEPR |
| AI308863 | Leptin receptor | −2.05 | LEPR |
| NM_000956 | Prostaglandin E receptor 2 (subtype EP2) | −2.25 | PTGER2 |
| NM_003882 | WNT1 inducible signaling pathway protein 1 | −2.48 | WISP1 |
| AA147884 | WNT1 inducible signaling pathway protein 1 | −2.07 | WISP1 |
| AI917494 | WNT1 inducible signaling pathway protein 1 | −2.07 | WISP1 |
| NM_007003 | P antigen family, member 4 | −3.07 | PAGE4 |
| NM_001993 | Coagulation factor III (tissue factor) | −4.21 | F3 |
| NM_002010 | Fibroblast growth factor 9 | −5.19 | FGF9 |
| AI520969 | Vimentin | −2.08 | VIM |
| AF344882 | Mst3 and SOK1-related kinase | −31.55 | MST4 |
| NM_016542 | Mst3 and SOK1-related kinase | −22.72 | MST4 |
| AA749101 | Interferon induced transmembrane protein 1 | −2.14 | IFITM1 |
| NM_002276 | Keratin 19 | −11.30 | KRT19 |
| NM_002423 | Matrix metalloproteinase 7 (matrilysin) | −4.37 | MMP7 |
| AI417595 | Endomucin | −2.94 | EMCN |
| AI635774 | Endomucin | −2.07 | EMCN |
| AI281371 | APO-1/CD95 (Fas)-associated phosphatase | −3.98 | PTPN13 |
| NM_006264 | APO-1/CD95 (Fas)-associated phosphatase | −2.39 | PTPN13 |
| AI189753 | Human tumor antigen (L6), TAAL6 | −2.26 | TM4SF1 |
| M90657 | Human tumor antigen (L6), TAAL6 | −2.71 | TM4SF1 |
| U76833 | Fibroblast activation protein, alpha | −2.30 | FAP |
| L01639 | Chemokine (C-X-C motif) receptor 4 | −2.21 | CXCR4 |
| AF348491 | Chemokine (C-X-C motif) receptor 4 | −2.06 | CXCR4 |
| D13889 | Inhibitor of DNA binding 1, Id1 | −2.98 | ID1 |
| AF003114 | Cysteine-rich, angiogenic inducer, 61 | −8.12 | CYR61 |
| NM_001554 | Cysteine-rich, angiogenic inducer, 61 | −6.59 | CYR61 |
| AL136139 | Enhancer of Filamentation 1 | −2.50 | HEF1 |
| NM_004995 | Matrix metalloproteinase 14 | 2.65 | MMP14 |
| Z48481 | Matrix metalloproteinase 14 | 2.21 | MMP14 |
| X83535 | Matrix metalloproteinase 14 | 2.06 | MMP14 |
| NM_005940 | Matrix metalloproteinase 11 (stromelysin 3) | 7.58 | MMP11 |
| AI761713 | Matrix metalloproteinase 11 (stromelysin 3) | 3.80 | MMP11 |
| X99268 | Twist homolog 1 | 2.17 | TWIST1 |
DISCUSSION
To date over 15 microarray analyses of uterine fibroids have been published. However, there is considerable variation and lack of overlap between studies, even those utilizing similar platforms and methodology. Consequently, alterations in gene expression that are functionally relevant to the pathophysiology of uterine leiomyoma are not clearly understood (26). These apparent disparities in gene profiles cannot be fully explained by methodological differences between the studies and may be attributable in part to genetic variability between patients, differences in the hormonal milieu or distinct chromosomal alterations between tumors (31). These factors may contribute to intrinsic variability of gene expression in the normal myometrium and/or fibroids, Last but not least, there is the possibility of existence of distinct fibroid subtypes (32–37), analogous to other diseases like leukemia, breast and lung cancer (38). In order to circumvent and control for the above possible sources of variation, we elected to study multiple leiomyomata samples and matched normal myometrium obtained from a single patient, i.e. in the context of an identical hormonal and genetic environment. Changes in expression of selected genes were then confirmed by analysis of multiple paired samples derived from additional patients.
In our study we examined the expression of nearly 38,500 gene fragments in independent RNA isolates from 3 fibroids and 3 normal samples from the same patient using Affymetrix technology. Statistically significant changes in gene expression were further limited to those probe sets exhibiting consistent changes in all three tumors of at least two-fold in magnitude, compared to average expression in normal myometrium. Using this approach, we found 816 genes to be differentially expressed in leiomyomata compared to normal myometrium. Consistent with previous studies, the majority of genes (619) were down-regulated in fibroids. We identified a number of genes in common with previous fibroid microarrays including some with otherwise disparate results (10–25). Thus our gene profile exhibited overlap with several apparently discordant studies, perhaps indicative of the sub set of common physiological relevant changes in expression in these tumors. Interestingly the least amount of overlap was noted with results using RNA derived from primary cell cultures of myometrium and leiomyomata (14). Differences in gene expression between primary and immortalized cell lines compared to their corresponding tumors and normal tissue have been previously described (39) and may be a reflection of removal of the cells from their endogenous environment, hormonal milieu and extracellular matrix.
We confirmed differential expression of several genes, namely fibulin 3, desmoglein 2, MMP7, MMP11, MST4 and PKCβ1 using three independent methods including RT-PCR, immunohistochemistry and Western blotting, to show changes not only at the level of the message, but also in some cases at the level of protein.
Among genes altered in expression were several fibulins, members of a family of secreted glycoproteins, characterized by repeated epidermal-growth-factor-like domains and a unique C-terminal structure. Evidence indicates a structural role for fibulins within the extracellular matrix and they have also been shown to modulate cell morphology, growth, adhesion and motility. The dysregulation of certain fibulins occurs in a range of human disorders, including cancer and both tumor suppressive and oncogenic activities have been proposed for members of the fibulin family (40).
Consistent with previous reports (14, 20, 41, 42), fibulin 1 was over expressed (2.2 fold) in leiomyoma. Fibulin 1 is a TGFβ regulated gene (42), which inhibits cancer cell adhesion, motility and invasion in vitro (43–45) and tumor growth in vivo (45). In contrast, fibulin 2 is a novel target identified in our study as down-regulated (2.5 fold) in leiomyoma. The role of fibulin 2 in fibroids and other neoplasms is not clear.
Fibulin 3 or also called EGF-containing fibulin-like extracellular matrix protein 1 (EFEMP1) was also down-regulated (20-fold) in our array and utilizing RT-PCR we confirmed decreased expression levels in our study patient and 5 additional patients (Fig. 2). Previous studies also identified this gene as down-regulated in leiomyoma (13, 18, 23, 24). The related protein, fibulin 5 also exhibited 2-fold lower expression in leiomyoma. Fibulin 3 and 5 have been shown to inhibit angiogenesis in tumors and may function as tumor suppressors (46–48). However, fibulin-3 expression is increased in a range of transformed cell lines compared with normal controls (49) and the functional role of fibulins 3 and 5 in leiomyoma has not been established. Interestingly fibulin 5 knockout mice showed enhanced proliferation of vascular smooth muscle in response to mitogens (50) consistent with a possible inhibitory role for fibulin 5 in regulating myometrial cell proliferation.
Desmoglein 2 (DSG2) is a novel up-regulated (5-fold) target not identified previously. Over expression of DSG2 is associated with increased growth rate, resistance to apoptosis and increased cell survival (51, 52). Desmoglein 2 is a component of adherence junctions and has been implicated in invasion and metastasis of squamous cell carcinomas. Decreased DSG2 expression is linked to loss of differentiation and poorer prognosis in gastric cancers (53).
MST 4 is a serine/threonine protein kinase and inducer of apoptosis and its over expression has been implicated in prostate cancer metastasis (54). In agreement with a previous study (11), we found MST4 to be dramatically down-regulated (20–30 fold) in leiomyoma. Decreased expression of MST4 was confirmed in multiple patients by RT-PCR and Western blot analysis (Figs. 2 & 4B).
PKC β1 has been identified as up-regulated in leiomoma by one other group (11). We found a 4.9 fold increase in PKC β1 expression and confirmed increased protein levels in multiple patients (Fig. 4A). The role of specific PKC isoforms in leiomyoma has not been extensively investigated. PKCs have been implicated in endothelin 1 growth stimulation of uterine leiomyoma (55) and conversely, growth inhibition of leiomyoma, by the GnRH analog buserelin, is also PKC-dependent (56).
A number of MMPs exhibit increased expression in leiomyoma, including MMP-11 (stromelysin 3) (10, 11, 19, 24, 57) and MMP 14 consistent with aberrant extracellular matrix deposition. In contrast, MMP-7 (matrilysin) is a target identified for the first time by our microarray as down-regulated (4.9 fold) in leiomyoma relative to myometrium. MMP-7 is a matrix-metalloproteinase that influences tumor progression by regulating invasion and angiogenesis. MMP-7 status of cancer tissues has been shown to be a strong predictor of poor prognosis (58, 59). Serum MMP-7 levels are significantly elevated in patients with advanced colorectal cancer and are an independent prognostic factor for survival (60). MMP-7 is over-expressed in malignant ovarian epithelium and may facilitate tumor cell invasion in vivo (61). Increased expression of MMP-7 in high grade uterine endometrial carcinoma is also associated with tumor invasion and metastasis (62).
In our gene screen and confirmatory experiments we noticed some variation in expression both on the message and protein level. Variation in protein levels was typically less than transcript levels. However, the pattern of expression was generally consistent between transcript and protein levels (Figs 2 & 4), and discordant profiles, with respect to the array analysis, were observed in both contexts. Expression levels of our selected targets varied between tumors from the same patient and also among tumors from different patients. Overall, variation in expression between tumors in a given patient was less than inter-patient variability. However, we also observed considerable differences, in the magnitude and direction of change, between tumors in a single patient (Fig 2–4).
Genetic variability in fibroids has been described in detail in the past, ascribed to different chromosome alterations and certain genetic mutations (31, 63, 64). These observations suggest that several fibroid subtypes exist similarly to lymphomas, breast and lung cancer, characterized by differential gene expression profiles.
With the exception of components of retinoid metabolism (65–68) and TGF-β signaling, automated pathway or gene ontology analysis of the leiomyoma gene profile, using GeneSpring and Ingenuity software respectively, failed to show coordinated changes in specific pathways. For this reason, we reviewed functional annotations for each gene using the GeneRIF (Gene Reference Into Function) database (National Library of Medicine). This analysis revealed several functional groups relating to regulation of apoptosis, cell growth and invasion and metastasis (Table 1). It is evident that, in general, genes linked to the promotion of growth, and inhibition of apoptosis are up-regulated in leiomyoma, whilst proapoptotic and growth inhibitory genes are down-regulated. This pattern is consistent with enhanced proliferation and survival of leiomyoma relative to normal myometrium. We also observed that genes typically associated with malignant disease and up-regulated in invasive and metastatic tumors, were actually down-regulated in leiomyoma relative to normal myometrium. This suppression of invasion and mestastasis related gene expression may underlie the benign, highly-differentiated, non-invasive phenotype characteristic of leiomyoma. Moreover, we speculate that this functional gene group may provide a molecular signature distinguishing leiomyoma from its malignant counterpart leiomyosarcoma, facilitating differential diagnosis and treatment at an early stage. Consistent with this hypothesis, previous studies have identified several genes up-regulated in leiomyosarcomas compared to normal myometrium that were actually down-regulated in our study and belong to our invasion, migration and metastasis group, e.g. tenascin C and WNT1 inducible signaling pathway protein 1 (69, 70). Early leiomyosarcoma and leiomyoma can exhibit similar symptomatology and imaging characteristics making preoperative diagnosis difficult. Histological sections can also be ambiguous, thus leading to critical delays in appropriate referral and therapeutic intervention (71). In future work, we will evaluate the relative expression levels and prognostic value of this invasion and metastasis gene set in leiomyoma and leiomyosarcoma.
Acknowledgments
Affymetrix array analysis was carried out using the Gene Expression CORE at the University of Colorado Cancer Center. The authors wish to thank Drs. Twila Jackson and Peggy Neville for critical reading of the manuscript.
Supported by Colorado Women’s Reproductive Health Research Career Development Center (K12 HD001271) and Department of Obstetrics and Gynecology, UCHSC; NIH Summer Medical Student grant (To TMM); NIH MD Anderson Gynecologic Specialized Programs of Research Excellence for Uterine Cancer #5P50 CA098258 Pilot Project Award (to JKR).
Presented in part at the Fourth WRHR Scholars’ Research Symposium and Director’s Meeting, Oregon Health and Science University, Portland, Oregon, May 15–17, 2007 and at the NICHD Uterine Fibroid Research Update Workshop, National Institutes of Health, Bethesda, Maryland, September 18–19, 2007
Footnotes
The authors have no conflicts of interest to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cesen-Cummings K, Copland JA, Barrett JC, Walker CL, Davis BJ. Pregnancy, parturition, and prostaglandins: defining uterine leiomyomas. Environ Health Perspect. 2000;108 Suppl 5:817–20. doi: 10.1289/ehp.00108s5817. [DOI] [PubMed] [Google Scholar]
- 2.Ligon AH, Morton CC. Leiomyomata: heritability and cytogenetic studies. Hum Reprod Update. 2001;7:8–14. doi: 10.1093/humupd/7.1.8. [DOI] [PubMed] [Google Scholar]
- 3.Buttram VC., Jr Uterine leiomyomata--aetiology, symptomatology and management. Prog Clin Biol Res. 1986;225:275–96. [PubMed] [Google Scholar]
- 4.Norris HJ, Parmley T. Mesenchymal tumors of the uterus. V. Intravenous leiomyomatosis. A clinical and pathologic study of 14 cases. Cancer. 1975;36:2164–78. doi: 10.1002/cncr.2820360935. [DOI] [PubMed] [Google Scholar]
- 5.Cramer SF, Patel A. The frequency of uterine leiomyomas. Am J Clin Pathol. 1990;94:435–8. doi: 10.1093/ajcp/94.4.435. [DOI] [PubMed] [Google Scholar]
- 6.Velebil P, Wingo PA, Xia Z, Wilcox LS, Peterson HB. Rate of hospitalization for gynecologic disorders among reproductive-age women in the United States. Obstet Gynecol. 1995;86:764–9. doi: 10.1016/0029-7844(95)00252-M. [DOI] [PubMed] [Google Scholar]
- 7.Stewart EA. Uterine fibroids. Lancet. 2001;357:293–8. doi: 10.1016/S0140-6736(00)03622-9. [DOI] [PubMed] [Google Scholar]
- 8.Wilcox LS, Koonin LM, Pokras R, Strauss LT, Xia Z, Peterson HB. Hysterectomy in the United States, 1988–1990. Obstet Gynecol. 1994;83:549–55. doi: 10.1097/00006250-199404000-00011. [DOI] [PubMed] [Google Scholar]
- 9.Lepine LA, Hillis SD, Marchbanks PA, Koonin LM, Morrow B, Kieke BA, et al. Hysterectomy surveillance--United States, 1980–1993. MMWR CDC Surveill Summ. 1997;46:1–15. [PubMed] [Google Scholar]
- 10.Hever A, Roth RB, Hevezi PA, Lee J, Willhite D, White EC, et al. Molecular characterization of human adenomyosis. Mol Hum Reprod. 2006;12:737–48. doi: 10.1093/molehr/gal076. [DOI] [PubMed] [Google Scholar]
- 11.Vanharanta S, Wortham NC, Laiho P, Sjoberg J, Aittomaki K, Arola J, et al. 7q deletion mapping and expression profiling in uterine fibroids. Oncogene. 2005;24:6545–54. doi: 10.1038/sj.onc.1208784. [DOI] [PubMed] [Google Scholar]
- 12.Weston G, Trajstman AC, Gargett CE, Manuelpillai U, Vollenhoven BJ, Rogers PA. Fibroids display an anti-angiogenic gene expression profile when compared with adjacent myometrium. Mol Hum Reprod. 2003;9:541–9. doi: 10.1093/molehr/gag066. [DOI] [PubMed] [Google Scholar]
- 13.Tsibris JC, Segars J, Coppola D, Mane S, Wilbanks GD, O’Brien WF, et al. Insights from gene arrays on the development and growth regulation of uterine leiomyomata. Fertil Steril. 2002;78:114–21. doi: 10.1016/s0015-0282(02)03191-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo X, Ding L, Xu J, Williams RS, Chegini N. Leiomyoma and myometrial gene expression profiles and their responses to gonadotropin-releasing hormone analog therapy. Endocrinology. 2005;146:1074–96. doi: 10.1210/en.2004-1384. [DOI] [PubMed] [Google Scholar]
- 15.Chegini N, Verala J, Luo X, Xu J, Williams RS. Gene expression profile of leiomyoma and myometrium and the effect of gonadotropin releasing hormone analogue therapy. J Soc Gynecol Investig. 2003;10:161–71. doi: 10.1016/s1071-5576(03)00004-2. [DOI] [PubMed] [Google Scholar]
- 16.Skubitz KM, Skubitz AP. Differential gene expression in leiomyosarcoma. Cancer. 2003;98:1029–38. doi: 10.1002/cncr.11586. [DOI] [PubMed] [Google Scholar]
- 17.Catherino WH, Prupas C, Tsibris JC, Leppert PC, Payson M, Nieman LK, et al. Strategy for elucidating differentially expressed genes in leiomyomata identified by microarray technology. Fertil Steril. 2003;80:282–90. doi: 10.1016/s0015-0282(03)00953-1. [DOI] [PubMed] [Google Scholar]
- 18.Wang H, Mahadevappa M, Yamamoto K, Wen Y, Chen B, Warrington JA, et al. Distinctive proliferative phase differences in gene expression in human myometrium and leiomyomata. Fertil Steril. 2003;80:266–76. doi: 10.1016/s0015-0282(03)00730-1. [DOI] [PubMed] [Google Scholar]
- 19.Kanamori T, Takakura K, Mandai M, Kariya M, Fukuhara K, Kusakari T, et al. PEP-19 overexpression in human uterine leiomyoma. Mol Hum Reprod. 2003;9:709–17. doi: 10.1093/molehr/gag088. [DOI] [PubMed] [Google Scholar]
- 20.Ahn WS, Kim KW, Bae SM, Yoon JH, Lee JM, Namkoong SE, et al. Targeted cellular process profiling approach for uterine leiomyoma using cDNA microarray, proteomics and gene ontology analysis. Int J Exp Pathol. 2003;84:267–79. doi: 10.1111/j.0959-9673.2003.00362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quade BJ, Wang TY, Sornberger K, Dal Cin P, Mutter GL, Morton CC. Molecular pathogenesis of uterine smooth muscle tumors from transcriptional profiling. Genes Chromosomes Cancer. 2004;40:97–108. doi: 10.1002/gcc.20018. [DOI] [PubMed] [Google Scholar]
- 22.Hoffman PJ, Milliken DB, Gregg LC, Davis RR, Gregg JP. Molecular characterization of uterine fibroids and its implication for underlying mechanisms of pathogenesis. Fertil Steril. 2004;82:639–49. doi: 10.1016/j.fertnstert.2004.01.047. [DOI] [PubMed] [Google Scholar]
- 23.Lee EJ, Kong G, Lee SH, Rho SB, Park CS, Kim BG, et al. Profiling of differentially expressed genes in human uterine leiomyomas. Int J Gynecol Cancer. 2005;15:146–54. doi: 10.1111/j.1048-891x.2005.15016.x. [DOI] [PubMed] [Google Scholar]
- 24.Arslan AA, Gold LI, Mittal K, Suen TC, Belitskaya-Levy I, Tang MS, et al. Gene expression studies provide clues to the pathogenesis of uterine leiomyoma: new evidence and a systematic review. Hum Reprod. 2005;20:852–63. doi: 10.1093/humrep/deh698. [DOI] [PubMed] [Google Scholar]
- 25.Roth TM, Klett C, Cowan BD. Expression profile of several genes in human myometrium and uterine leiomyoma. Fertil Steril. 2007;87:635–41. doi: 10.1016/j.fertnstert.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 26.Catherino WH, Segars JH. Microarray analysis in fibroids: which gene list is the correct list? Fertil Steril. 2003;80:293–4. doi: 10.1016/s0015-0282(03)00958-0. [DOI] [PubMed] [Google Scholar]
- 27.Reno EM, Haughian JM, Dimitrova IK, Jackson TA, Shroyer KR, Bradford AP. Analysis of protein kinase C delta (PKCdelta) expression in endometrial tumors. Hum Pathol. 2008;39:21–9. doi: 10.1016/j.humpath.2007.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haughian JM, Jackson TA, Koterwas DM, Bradford AP. Endometrial cancer cell survival and apoptosis is regulated by protein kinase C alpha and delta. Endocr Relat Cancer. 2006;13:1251–67. doi: 10.1677/erc.1.01278. [DOI] [PubMed] [Google Scholar]
- 29.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–93. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 30.Rudolph MC, Neville MC, Anderson SM. Lipid synthesis in lactation: diet and the Fatty Acid switch. J Mammary Gland Biol Neoplasia. 2007;12:269–81. doi: 10.1007/s10911-007-9061-5. [DOI] [PubMed] [Google Scholar]
- 31.Hodge JC, Morton CC. Genetic heterogeneity among uterine leiomyomata: insights into malignant progression. Hum Mol Genet. 2007;16 Spec No 1:R7–13. doi: 10.1093/hmg/ddm043. [DOI] [PubMed] [Google Scholar]
- 32.Shimada K, Ohashi I, Kasahara I, Watanabe H, Ohta S, Miyasaka N, et al. Differentiation between completely hyalinized uterine leiomyomas and ordinary leiomyomas: three-phase dynamic magnetic resonance imaging (MRI) vs. diffusion-weighted MRI with very small b-factors. J Magn Reson Imaging. 2004;20:97–104. doi: 10.1002/jmri.20063. [DOI] [PubMed] [Google Scholar]
- 33.Shimada K, Ohashi I, Kasahara I, Miyasaka N, Shibuya H. Triple-phase dynamic MRI of intratumoral vessel density and hyalinization grade in uterine leiomyomas. AJR Am J Roentgenol. 2004;182:1043–50. doi: 10.2214/ajr.182.4.1821043. [DOI] [PubMed] [Google Scholar]
- 34.Bell SW, Kempson RL, Hendrickson MR. Problematic uterine smooth muscle neoplasms. A clinicopathologic study of 213 cases. Am J Surg Pathol. 1994;18:535–58. [PubMed] [Google Scholar]
- 35.Kempson RL, Bari W. Uterine sarcomas. Classification, diagnosis, and prognosis. Hum Pathol. 1970;1:331–49. [PubMed] [Google Scholar]
- 36.Kempson RL, Hendrickson MR. Smooth muscle, endometrial stromal, and mixed Mullerian tumors of the uterus. Mod Pathol. 2000;13:328–42. doi: 10.1038/modpathol.3880055. [DOI] [PubMed] [Google Scholar]
- 37.Hart WR. Problematic uterine smooth muscle neoplasms. Am J Surg Pathol. 1997;21:252–5. doi: 10.1097/00000478-199702000-00019. [DOI] [PubMed] [Google Scholar]
- 38.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 39.Zaitseva M, Vollenhoven BJ, Rogers PA. In vitro culture significantly alters gene expression profiles and reduces differences between myometrial and fibroid smooth muscle cells. Mol Hum Reprod. 2006;12:187–207. doi: 10.1093/molehr/gal018. [DOI] [PubMed] [Google Scholar]
- 40.Gallagher WM, Currid CA, Whelan LC. Fibulins and cancer: friend or foe? Trends Mol Med. 2005;11:336–40. doi: 10.1016/j.molmed.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 41.Luo X, Ding L, Xu J, Chegini N. Gene expression profiling of leiomyoma and myometrial smooth muscle cells in response to transforming growth factor-beta. Endocrinology. 2005;146:1097–118. doi: 10.1210/en.2004-1377. [DOI] [PubMed] [Google Scholar]
- 42.Luo X, Ding L, Chegini N. CCNs, fibulin-1C and S100A4 expression in leiomyoma and myometrium: inverse association with TGF-beta and regulation by TGF-beta in leiomyoma and myometrial smooth muscle cells. Mol Hum Reprod. 2006;12:245–56. doi: 10.1093/molehr/gal015. [DOI] [PubMed] [Google Scholar]
- 43.Hayashido Y, Lucas A, Rougeot C, Godyna S, Argraves WS, Rochefort H. Estradiol and fibulin-1 inhibit motility of human ovarian- and breast-cancer cells induced by fibronectin. Int J Cancer. 1998;75:654–8. doi: 10.1002/(sici)1097-0215(19980209)75:4<654::aid-ijc26>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 44.Twal WO, Czirok A, Hegedus B, Knaak C, Chintalapudi MR, Okagawa H, et al. Fibulin-1 suppression of fibronectin-regulated cell adhesion and motility. J Cell Sci. 2001;114:4587–98. doi: 10.1242/jcs.114.24.4587. [DOI] [PubMed] [Google Scholar]
- 45.Qing J, Maher VM, Tran H, Argraves WS, Dunstan RW, McCormick JJ. Suppression of anchorage-independent growth and matrigel invasion and delayed tumor formation by elevated expression of fibulin-1D in human fibrosarcoma-derived cell lines. Oncogene. 1997;15:2159–68. doi: 10.1038/sj.onc.1201385. [DOI] [PubMed] [Google Scholar]
- 46.Schiemann WP, Blobe GC, Kalume DE, Pandey A, Lodish HF. Context-specific effects of fibulin-5 (DANCE/EVEC) on cell proliferation, motility, and invasion. Fibulin-5 is induced by transforming growth factor-beta and affects protein kinase cascades. J Biol Chem. 2002;277:27367–77. doi: 10.1074/jbc.M200148200. [DOI] [PubMed] [Google Scholar]
- 47.Albig AR, Schiemann WP. Fibulin-5 antagonizes vascular endothelial growth factor (VEGF) signaling and angiogenic sprouting by endothelial cells. DNA Cell Biol. 2004;23:367–79. doi: 10.1089/104454904323145254. [DOI] [PubMed] [Google Scholar]
- 48.Albig AR, Neil JR, Schiemann WP. Fibulins 3 and 5 antagonize tumor angiogenesis in vivo. Cancer Res. 2006;66:2621–9. doi: 10.1158/0008-5472.CAN-04-4096. [DOI] [PubMed] [Google Scholar]
- 49.Lecka-Czernik B, Lumpkin CK, Jr, Goldstein S. An overexpressed gene transcript in senescent and quiescent human fibroblasts encoding a novel protein in the epidermal growth factor-like repeat family stimulates DNA synthesis. Mol Cell Biol. 1995;15:120–8. doi: 10.1128/mcb.15.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spencer JA, Hacker SL, Davis EC, Mecham RP, Knutsen RH, Li DY, et al. Altered vascular remodeling in fibulin-5-deficient mice reveals a role of fibulin-5 in smooth muscle cell proliferation and migration. Proc Natl Acad Sci U S A. 2005;102:2946–51. doi: 10.1073/pnas.0500058102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nava P, Laukoetter MG, Hopkins AM, Laur O, Gerner-Smidt K, Green KJ, et al. Desmoglein-2: a novel regulator of apoptosis in the intestinal epithelium. Mol Biol Cell. 2007;18:4565–78. doi: 10.1091/mbc.E07-05-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brennan D, Hu Y, Joubeh S, Choi YW, Whitaker-Menezes D, O’Brien T, et al. Suprabasal Dsg2 expression in transgenic mouse skin confers a hyperproliferative and apoptosis-resistant phenotype to keratinocytes. J Cell Sci. 2007;120:758–71. doi: 10.1242/jcs.03392. [DOI] [PubMed] [Google Scholar]
- 53.Yashiro M, Nishioka N, Hirakawa K. Decreased expression of the adhesion molecule desmoglein-2 is associated with diffuse-type gastric carcinoma. Eur J Cancer. 2006;42:2397–403. doi: 10.1016/j.ejca.2006.03.024. [DOI] [PubMed] [Google Scholar]
- 54.Sung V, Luo W, Qian D, Lee I, Jallal B, Gishizky M. The Ste20 kinase MST4 plays a role in prostate cancer progression. Cancer Res. 2003;63:3356–63. [PubMed] [Google Scholar]
- 55.Eude I, Dallot E, Vacher-Lavenu MC, Chapron C, Ferre F, Breuiller-Fouche M. Potentiation response of cultured human uterine leiomyoma cells to various growth factors by endothelin-1: role of protein kinase C. Eur J Endocrinol. 2001;144:543–8. doi: 10.1530/eje.0.1440543. [DOI] [PubMed] [Google Scholar]
- 56.Yamamoto H, Sato H, Shibata S, Murata M, Fukuda J, Tanaka T. Involvement of annexin V in the antiproliferative effect of GnRH agonist on cultured human uterine leiomyoma cells. Mol Hum Reprod. 2001;7:169–73. doi: 10.1093/molehr/7.2.169. [DOI] [PubMed] [Google Scholar]
- 57.Palmer SS, Haynes-Johnson D, Diehl T, Nowak RA. Increased expression of stromelysin 3 mRNA in leiomyomas (uterine fibroids) compared with myometrium. J Soc Gynecol Investig. 1998;5:203–9. doi: 10.1016/s1071-5576(98)00009-4. [DOI] [PubMed] [Google Scholar]
- 58.Miyata Y, Iwata T, Ohba K, Kanda S, Nishikido M, Kanetake H. Expression of matrix metalloproteinase-7 on cancer cells and tissue endothelial cells in renal cell carcinoma: prognostic implications and clinical significance for invasion and metastasis. Clin Cancer Res. 2006;12:6998–7003. doi: 10.1158/1078-0432.CCR-06-1626. [DOI] [PubMed] [Google Scholar]
- 59.Noriyuki M, Sumi T, Zhi X, Misugi F, Nobeyama H, Yoshida H, et al. Vascular endothelial growth factor, matrix metalloproteinases, and cyclooxygenase-2 influence prognosis of uterine cervical cancer in young women. Int J Oncol. 2007;31:531–6. [PubMed] [Google Scholar]
- 60.Maurel J, Nadal C, Garcia-Albeniz X, Gallego R, Carcereny E, Almendro V, et al. Serum matrix metalloproteinase 7 levels identifies poor prognosis advanced colorectal cancer patients. Int J Cancer. 2007;121:1066–71. doi: 10.1002/ijc.22799. [DOI] [PubMed] [Google Scholar]
- 61.Wang FQ, So J, Reierstad S, Fishman DA. Matrilysin (MMP-7) promotes invasion of ovarian cancer cells by activation of progelatinase. Int J Cancer. 2005;114:19–31. doi: 10.1002/ijc.20697. [DOI] [PubMed] [Google Scholar]
- 62.Misugi F, Sumi T, Okamoto E, Nobeyama H, Hattori K, Yoshida H, et al. Expression of matrix metalloproteinases and tissue inhibitors of metalloproteinase in uterine endometrial carcinoma and a correlation between expression of matrix metalloproteinase-7 and prognosis. Int J Mol Med. 2005;16:541–6. [PubMed] [Google Scholar]
- 63.Lobel MK, Somasundaram P, Morton CC. The genetic heterogeneity of uterine leiomyomata. Obstet Gynecol Clin North Am. 2006;33:13–39. doi: 10.1016/j.ogc.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 64.Stewart EA, Morton CC. The genetics of uterine leiomyomata: what clinicians need to know. Obstet Gynecol. 2006;107:917–21. doi: 10.1097/01.AOG.0000206161.84965.0b. [DOI] [PubMed] [Google Scholar]
- 65.Zaitseva M, Vollenhoven BJ, Rogers PA. Retinoic acid pathway genes show significantly altered expression in uterine fibroids when compared with normal myometrium. Mol Hum Reprod. 2007;13:577–85. doi: 10.1093/molehr/gam040. [DOI] [PubMed] [Google Scholar]
- 66.Wei T, Geiser AG, Qian HR, Su C, Helvering LM, Kulkarini NH, et al. DNA microarray data integration by ortholog gene analysis reveals potential molecular mechanisms of estrogen-dependent growth of human uterine fibroids. BMC Womens Health. 2007;7:5. doi: 10.1186/1472-6874-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Catherino WH, Malik M. Uterine leiomyomas express a molecular pattern that lowers retinoic acid exposure. Fertil Steril. 2007;87:1388–98. doi: 10.1016/j.fertnstert.2006.11.093. [DOI] [PubMed] [Google Scholar]
- 68.Lattuada D, Vigano P, Mangioni S, Sassone J, Di Francesco S, Vignali M, et al. Accumulation of retinoid X receptor-alpha in uterine leiomyomas is associated with a delayed ligand-dependent proteasome-mediated degradation and an alteration of its transcriptional activity. Mol Endocrinol. 2007;21:602–12. doi: 10.1210/me.2006-0206. [DOI] [PubMed] [Google Scholar]
- 69.Politi K, Szabolcs M, Fisher P, Kljuic A, Ludwig T, Efstratiadis A. A mouse model of uterine leiomyosarcoma. Am J Pathol. 2004;164:325–36. doi: 10.1016/S0002-9440(10)63122-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Matsumura N, Mandai M, Miyanishi M, Fukuhara K, Baba T, Higuchi T, et al. Oncogenic property of acrogranin in human uterine leiomyosarcoma: direct evidence of genetic contribution in in vivo tumorigenesis. Clin Cancer Res. 2006;12:1402–11. doi: 10.1158/1078-0432.CCR-05-2003. [DOI] [PubMed] [Google Scholar]
- 71.Schwartz PE, Kelly MG. Malignant transformation of myomas: myth or reality? Obstet Gynecol Clin North Am. 2006;33:183–98. xii. doi: 10.1016/j.ogc.2005.12.003. [DOI] [PubMed] [Google Scholar]