Summary
Several cellular chaperones have been shown to affect the propagation of the yeast prions [PSI+], [PIN+] and [URE3]. Ssa1 and Ssa2 are Hsp70 family chaperones, that generally cause pro-[PSI+] effects since dominant negative mutants of Ssa1 or Ssa2 cure [PSI+], and overexpression of Ssa1 enhances de novo [PSI+] appearance and prevents curing by excess Hsp104. In contrast, Ssa1 was shown to have anti-[URE3] effects, since overexpression of Ssa1 cures [URE3]. Here we show that excess Ssa1 or Ssa2 can also cure [PSI+]. This curing is enhanced in the presence of [PIN+]. During curing, Sup35-GFP fluorescent aggregates get bigger and fewer in number, which leads to their being diluted out during cell division, a phenotype that was also observed during the curing of [PSI+] by certain variants of [PIN+]. The sizes of the detergent resistant [PSI+] prion oligomers increase during [PSI+] curing by excess Ssa1. Excess Ssa1 likewise leads to an increase in oligomer sizes of low, medium and very high [PIN+] variants. While these phenotypes are also caused by inhibition of Hsp104 or Sis1, the overexpression of Ssa1 did not cause any change in Hsp104 or Sis1 levels.
Keywords: [PSI+], Ssa1, prion curing, prion variants, prion aggregate
Introduction
Prions are self-replicating, proteinaceous agents that are known to cause various infectious, neurodegenerative diseases in mammals. In the yeast Saccharomyces cerevisiae, prions are transmitted in a protein-only manner as non-Mendelian, epigenetic elements. The well studied yeast prions [PSI+], [PIN+] and [URE3] are composed of Sup35, Rnq1 and Ure2 proteins respectively (reviewed in 1). These normally soluble proteins exist in a β-sheet rich, partially misfolded, self-replicating, aggregated state when in their prion forms. The overexpression of the protein determinant of a prion leads to a higher frequency of de novo prion appearance 2. The propagating unit of a prion referred to as a “seed” or “propagon” is the prion element that recruits monomers to join prion aggregates and is transmitted to daughter cells 3; 4. Each of the [PSI+], [PIN+] and [URE3] prions can also stably exist as distinct structural conformers or prion “variants” of the same protein, that differ in the extent of phenotypes conferred and their mitotic stability 5; 6; 7; 8; 9.
[PSI+] is the prion form of the yeast translational termination factor Sup35 or eRF3 2. In [PSI+] cells, the translation termination function of Sup35 is compromised due to aggregation of Sup35, in contrast to [psi-] cells where Sup35 is soluble and fully functional. Due to the reduced translation termination in [PSI+] cells, there is an increased read through of stop (nonsense) codons, which is assayed as nonsense suppression 10. [PSI+] exists as variants – weak and strong- that differ in their stability and the amount of nonsense suppression caused 6; 11; 12. As the name suggests, weak [PSI+] (W[PSI+]) is less stable and causes less suppression than strong [PSI+] (S[PSI+]). When endogenous Sup35 is tagged with GFP, the fusion protein is fully functional and appears as numerous, tiny fluorescent, cytoplasmic dots in cells with either W or S [PSI+], but as diffuse fluorescence in [psi-] cells 4. It is hypothesized that these [PSI+] dots correspond to prion seeds or propagons 3; 4. The higher order [PSI+] prion aggregates that are composed of various oligomers of Sup35 aggregated together with other cellular factors, can be disassembled by treatment with strong detergent like SDS, leaving only the SDS-resistant Sup35 oligomers that retain prion infectivity 13; 14. The sizes of these SDS-stable oligomers can be roughly estimated by semi-denaturing, detergent agarose gel electrophoresis (SDD-AGE) 15; 16.
Another yeast prion [PIN+] (for [PSI+] inducible) is the prion form of the Rnq1 protein 17; 18. The function of soluble Rnq1 is unknown, but in the prion form, [PIN+] enhances de novo [PSI+] formation, although it is not required for [PSI+] propagation 19; 20. Similar to [PSI+], [PIN+] also exists as different variants- low, medium, high and very high [PIN+]s- that differ in their properties, e.g. the efficiency with which they enhance the appearance of [PSI+]. While [PSI+] can stably propagate in [pin-] or high [PIN+] cells, the low, medium and very high variants of [PIN+] have been shown to destabilize [PSI+]21.
The role of molecular chaperones in yeast prion aggregation and propagation has been extensively studied. Hsp104 is required for the propagation of all known yeast prions 17; 18; 20; 22; 23; 24; 25. There is evidence suggesting that Hsp104 shears prion aggregates into smaller “seeds” that can be transmitted to daughter cells, thereby ensuring prion propagation 13; 26. While the inhibition of Hsp104 activity cures cells of [PSI+], [PIN+], [URE3], [SWI+] and [OCT+], overexpression of Hsp104 only cures [PSI+].
The Ssa subfamily of Hsp70 chaperones consists of functionally redundant Ssa1, Ssa2, Ssa3, Ssa4 proteins that also affect prion propagation. The expression of any one Ssa protein is sufficient for viability. Ssa1 and Ssa2 (referred to as Ssa1/2 here) are constitutively expressed at high levels, while Ssa3p and Ssa4p are stress induced. Ssa's along with Hsp104 and Hsp40s help in disaggregation and refolding of misfolded, aggregated proteins (reviewed in 27). Modulation of Ssa1 has various effects on [PSI+] prion formation and propagation 28; 29; 30; 31; 32; 33. Earlier work showed that overexpression of members of Ssa family of chaperones increase de novo induction of [PSI+] and antagonize curing of [PSI+] by overexpression of Hsp104 both in vivo and in vitro28; 30; 34. Several dominant negative mutations in SSA1 or SSA2 decrease the mitotic stability of [PSI+] 29; 33; 35; 36. While the overexpression of Ssa1 (but not the highly homologous Ssa2) was shown to cure cells of [URE3] 31, it is generally described as a “pro-[PSI+]” chaperone 28. Excess Ssa1 has been shown to antagonize certain [PSI+]s that are selected for their inability to be fragmented efficiently and hence form large aggregates. For example, a deletion of amino acids 22-69 in the Sup35 prion domain permits only the propagation of mitotically unstable [PSI+] with large aggregates. Excess Ssa1 further destabilized these [PSI+]Δ22/69 presumably because Ssa1 inhibits aggregate shearing leading to larger, fewer aggregates causing decreased transmissibility37. Likewise, propagation of another [PSI+] variant that is defective in shearing becaused it was selected for its ability to be maintained at high Hsp104 levels is also antagonized by excess Ssa1 38. Also, Allen et al., 2005, showed that excess Ssa1 destabilizes [PSI+] when Sup35 is overexpressed. It was reported recently, that the major components of [PSI+] aggregates are Sup35 and the Ssa1/2 chaperones 14. This suggests that Ssa proteins have a role in [PSI+] propagation. The depletion of an essential Hsp40 co-chaperone of Ssa proteins, Sis1, also cures [PSI+], [PIN+] and [URE3] 39; 40; 41. It has been suggested that Ssa1 and Sis1 interact with prion aggregates and recruit Hsp104 to them, which then shears the aggregates thereby allowing prion propagation 41; 42.
Here we show that excess Ssa1 and Ssa2 can cure normal variants of [PSI+] in cells with wild-type levels of Sup35 and Hsp104. This curing is enhanced in the presence of [PIN+]. Using endogenously GFP tagged Sup35, we show that excess Ssa1 causes Sup35-GFP [PSI+] intracellular aggregates to become larger in size and fewer in number which leads to their being diluted out during the curing process. A similar phenotype was observed when [PSI+] was destabilized by variants of [PIN+] suggesting a similarity in mechanism of [PSI+] curing by excess Ssa1 and destabilizing [PIN+]s.
Results
Ssa1/2 overexpression cures “normal” [PSI+]
We tested if excess Ssa1/2 could cure the [PSI+] variants we routinely used by cytoducing these “normal” [PSI+] variants from 74-D694 cells into isogenic cells in which the genomic Sup35 was replaced with Sup35-GFP. Both the wild type Sup35 containing cells and the cytoduced Sup35-GFP cells propagating different [PSI+] variants were transformed with centromeric plasmids that overexpressed Ssa1 and Ssa2 using the strong, constitutive SSA2 promoter. Transformants were spotted on rich medium (YPD) and the colors of the spots from Ssa1/2 overexpressing cells were compared to cells with vector controls. The vector controls were white due to the presence of [PSI+], however spots of cells overexpressing Ssa1/2 showed red sectors indicative of loss of [PSI+] (see Materials and Methods). Figure 1 shows the effect of overexpression of Ssa1 and Ssa2 on W and S [PSI+] in Sup35-GFP cells. The red sectors were confirmed to contain [psi-] cells since Sup35-GFP in these cells showed a diffuse fluorescence. Cells were cured of W [PSI+] more easily than S [PSI+]. About 6 % of the S [PSI+][PIN+] colonies appeared red-sectored when Ssa1 overexpressing cultures were spread on YPD, while all were white in the control. About 30 % W [PSI+][PIN+] colonies were red-sectored when Ssa1 was overexpressed, while only 3-6 % were red-sectored in the control. Also, the presence of [PIN+] enhanced the ability of Ssa1/2 overexpression to cure cells of [PSI+]. This curing of [PSI+] variants by excess Ssa1 was also observed in strains with wild type Sup35 (Supplementary Figure 1), however the curing was enhanced in the tagged strains.
Figure 1.

Ssa1/2 overexpression cures some variants of [PSI+]. Ssa1 or Ssa2 were expressed on plasmids P1506 and P1507 respectively, under the strong, constitutive SSA2 promoter. Single colonies were picked from transformation selective plates and diluted to spot an equal number of cells on YPD. Colonies overexpressing either Ssa1 or Ssa2 give rise to some cells with a [psi-] phenotype (red color) on YPD, while all cells from colonies transformed with the vector (P667) remain [PSI+] (white). Weak (W) [PSI+] with (L2888) or without (L2885) [PIN+] (right panels) is lost more frequently than strong (S) [PSI+] with (L2829) or without (SY80) [PIN+] (left panels). [PIN+] enhances the curing by Ssa1/2 overexpression.
Sup35-GFP shows bigger foci upon Ssa1 overexpression
The endogenously GFP tagged Sup35 in cells allowed us to monitor Sup35 aggregation for changes during Ssa1 overexpression. We transformed W[PSI+][PIN+], W[PSI+][pin-] and S[PSI+][PIN+] yeast, with a plasmid containing SSA1 under the inducible GAL1,10 promoter or an empty vector. After overnight growth on 2 % galactose, the W or S [PSI+] cells that contained the [PIN+] prion, showed bigger Sup35-GFP foci when Ssa1 was overexpressed (Figure 2). Furthermore, in most Ssa1 overexpressing cells, these foci were fewer in number as compared to the vector control. W[PSI+][pin-] was cured less efficiently than W[PSI+][PIN+] by Ssa1 overexpression and very few W[PSI+][pin-] cells showed bigger [PSI+] aggregates.
Figure 2.
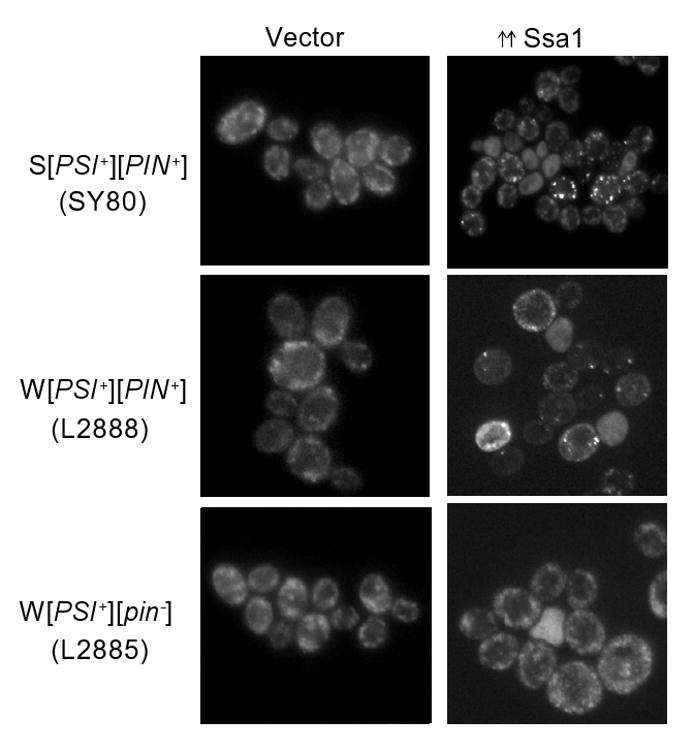
Sup35-GFP forms big foci in [PSI+] cells upon Ssa1 overexpression in the presence of high [PIN+]. Ssa1 was overexpressed from P1369 under the inducible GAL1,10 promoter, by growing SY80, L2888 and L2885 cells in 2 % raffinose + 2 % galactose containing media overnight. In the vector control, P589 (left panel), all the cells, whether S or W [PSI+], show multiple, tiny dots characteristic of [PSI+]. Upon Ssa1 overexpression, about 50 % of the cell population in both S and W [PSI+][PIN+] shows bigger and fewer Sup35-GFP foci. Bigger foci were not frequently observed in W[PSI+][pin-] strain, no bigger foci are seen in the picture shown here.
Ssa1 overexpression does not cause a change in Hsp104 or Sis1 levels
Since depletion of Hsp104 17; 43 or Sis1 41 has also been shown to cure cells of [PSI+], we asked if overexpression of Ssa1 cured cells of [PSI+] indirectly by altering the level of Hsp104 or Sis1. Figure 3 shows that in both W and S [PSI+][PIN+], the Hsp104 as well as Sis1 protein levels are comparable in control and Ssa1 overexpressed cultures. Thus, [PSI+] curing by excess Ssa1 is not due to a change in Hsp104 or Sis1 levels in the cell. Previous studies have also shown that there is no change in Hsp104 levels upon Ssa1 overexpression 28; 30. Newnam et al. 1999 found that thermotolerance of cells is reduced when Ssa1 is overexpressed reflecting a down regulation of the cellular chaperone machinery. Our cells, however, survived heat shock better when Ssa1 was overexpressed, (data not shown). This could simply be due to the excess Ssa1 chaperone present.
Figure 3.

Hsp104 and Sis1 levels do not change upon Ssa1 overexpression. Equal amount of lysates (W and S [PSI+][PIN+]) overexpressing Ssa1 or control were loaded on 10% acrylamide gels, which was transferred to a PVDF membrane and probed with polyclonal anti-Hsp104 antibody. The same blots were stripped and reprobed twice to probe with anti-Sis1 and anti-Pgk1 (loading control) antibodies.
W and S [PSI+][PIN+] microcolonies show progressive loss of Sup35-GFP aggregates upon Ssa1 overexpression
To further visualize the curing event, we followed the change in appearance of Sup35-GFP aggregates as the cells overexpressing Ssa1 grew into microcolonies. W or S [PSI+][PIN+] transformants containing the empty centromeric vector or unexpressed plasmid having SSA1 under the GAL1,10 promoter (PGAL1,10-SSA1) that stably maintained [PSI+] aggregates and therefore had numerous, tiny Sup35-GFP dots were obtained. We micromanipulated single cells from these transformants onto an agar patch, that was transferred to a 2 % raffinose + 2 % galactose plate to induce Ssa1 overexpression. The growth of the single cells was followed over time to observe changes in Sup35-GFP aggregation and finally loss of [PSI+] (Figure 4).
Figure 4.



Microcolony showing the sequential loss of [PSI+] upon Ssa1 overexpression. Single cells were micromanipulated and grown on 2 % raffinose + 2 % galactose where Ssa1 is overexpressed (using P1369) and observed after various time points. (a) Vector (P589) shows no change in Sup35-GFP aggregates (multiple, tiny dots) in W[PSI+][PIN+] (L2888) overtime. The cartoon depicts no change in Sup35-GFP aggregates in the presence of vector. A portion of a [psi-] colony is shown in the box. There are tiny, fast-moving dots in [PSI+] that are easily visualized but cannot be photographed clearly as the colony was grown on an agar patch under a coverslip. (b) Segregation of [PSI+] dots into [psi-] upon Ssa1 overexpression. W[PSI+][PIN+] (L2888) overexpressing Ssa1 shows bigger, multiple but fewer GFP foci after 3-4 generations. These cells later give rise to daughters with a single large aggregate which further give rise to [psi-] daughters. A quarter of the colony is shown. The cartoon shows the dilution of Sup35-GFP dots finally becoming [psi-] in the microcolony with Ssa1 overexpression. S[PSI+][PIN+] cells (SY80) that are cured of [PSI+] also show a similar pattern of Sup35-GFP fluorescence in the microcolony (not shown). (c) Cells with big dots propagate [PSI+] when Ssa1 overexpression is relieved. W[PSI+][PIN+] microcolony edge on 2 % galactose, showing a mix of cells with and without aggregates (top). Single cells with and without aggregates were picked from the microcolony and grown on SD+12 (bottom). Cells with no visible aggregates grow out to be [psi-]- diffuse fluorescence and red on YPD (bottom left), while cells with aggregates become [PSI+]-multiple, tiny dots and white on YPD (bottom right). Only a portion of the microcolony is shown. The center of the colony appears brighter than the colony edge as cells are growing on top of the older cells.
The Sup35-GFP fluorescence pattern during the curing process was the same for W and S [PSI+] (Figure 4(b) shows W [PSI+]), although not all micromanipulated S[PSI+][PIN+] cells gave rise to [psi-] cells, suggesting that [PSI+] loss in S[PSI+] is not as efficient as in W[PSI+]. Each starting [PSI+] cell had numerous, tiny dots. When Ssa1 was overexpressed, larger and fewer Sup35-GFP dots could be seen within 3-4 generations. As the microcolony divided, these larger aggregates were diluted out and became fewer and fewer per generation, 1000's became 100's and then 10's, then 1 or 2, finally giving rise to [psi-] cells with diffuse fluorescence. In bigger microcolonies overexpressing Ssa1 (Figure 4(b)), cells with fewer and larger aggregates could be seen segregating out to [psi-] cells in the outer edge of the colony. All cells in the microcolonies with the empty vector showed numerous uniformly sized, tiny dots characteristic of [PSI+] (Figure 4(a)).
In the colonies overexpressing Ssa1, the cells with or without aggregates (Figure 4(c), top) were micromanipulated and were grown on dextrose medium to stop Ssa1 overexpression. Daughters of cells showing diffuse fluorescence were also diffuse indicative of [psi-] and grew to be a red colony on rich medium (Figure 4(c), bottom). In contrast, daughters of cells with bigger aggregates turned back to the normal [PSI+] state: numerous, tiny Sup35-GFP dots and white colony color on YPD (Figure 4(c), bottom).
Ssa1 overexpression in W [PSI+] leads to an increase in Sup35 oligomer size
Yeast prion aggregates appear to be composed of large oligomeric species that cannot be dissociated with 2 % SDS at room temperature and can be resolved on an agarose gel by semi-denaturing, detergent agarose gel electrophoresis (SDD-AGE) 15; 16. Modulation of various cellular proteins, like molecular chaperones, leads to a change in the size of the prion oligomers. It is known that [PSI+] curing by inhibition of the chaperones Hsp104 and Sis1 goes through an intermediate stage where SDS-stable [PSI+] oligomers are larger in size 13; 41. The dominant negative SSA1-21 mutation that inactivates Ssa1 activity, cures [PSI+] but does not change [PSI+] oligomer size 32. However, overexpression of Ssa1 has been shown to lead to a slight increase in [PSI+] oligomer size and the amount of Sup35 monomer 28 but those [PSI+] cells were not shown to be cured. Even though Ssa1 overexpression cures [URE3], it is not known if the oligomer size is changed 31.
We determined the size of [PSI+] oligomers in our strains that are cured by Ssa1 overexpression. We observed that the S[PSI+][PIN+] strain that was less readily cured by Ssa1 overexpression shows no detectable change in the oligomers size as seen on 1.5% agarose gel (Figure 5(a)). However, we did observe a slight but consistent increase in [PSI+] oligomer size in the W[PSI+][PIN+] strain (Figure 5(a)) which was more readily cured. This observation is consistent with previous studies that showed an increase in [PSI+] oligomer size and accumulation of monomeric Sup35 upon Ssa1 overexpression 28. The increase in oligomer size could not be due to excess Ssa1 bound to [PSI+] aggregates since the lysates are treated with SDS which would disrupt the interaction between Ssa1 and [PSI+] oligomers.
Figure 5.
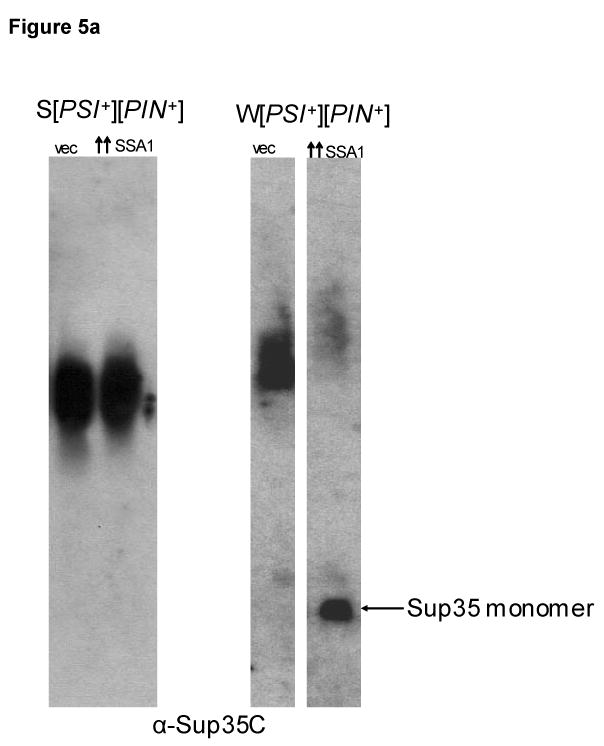

Ssa1 overexpression leads to an increase in oligomer size for W[PSI+] and an accumulation of monomers and small aggregates. (a) W[PSI+] but not S[PSI+] SDS-resistant Sup35 oligomers increase in size and decrease in amount upon Ssa1 overexpression. Crude S[PSI+][PIN+] (SY80) and W[PSI+][PIN+] (L2888) lysates were prepared from cells overexpressing Ssa1 or vector. The lysates were treated with 2 % SDS and SDS-resistant oligomers were detected using SDD-AGE. S[PSI+][PIN+] did not show any change in oligomers size, while W[PSI+][PIN+] oligomers show a slight but consistent size increase upon Ssa1 overexpression as compared to the vector control (vec). The gap separates non-adjacent lanes from the same gel. (b) Sup35-GFP monomers and smaller aggregates accumulate on sucrose gradients upon Ssa1 overexpression. Sup35-GFP in S and W [PSI+][PIN+] and [psi-][pin-] lysates overexpressing Ssa1 (P1369) or vector (P589) were fractionated on a 20-60 % sucrose gradient. An equal amount of protein (∼ 1 mg) was loaded on the gradient which was centrifuged at 10,600 g for 40 minutes. Five fractions were collected, treated with 2 % SDS at 95 °C and the Sup35-GFP protein level in each fraction was detected on western blots probed with anti-Sup35 antibody. The pictures show Sup35-GFP fluorescence in cultures from which lysates were made, where bigger GFP foci were present in ∼75 % of cells from [PSI+] cultures when Ssa1 was overexpressed. The amount of Sup35-GFP in the pellet of the clearing step was the same in vector and overexpressed Ssa1 (not shown).
Ssa1 overexpression shows accumulation of Sup35-GFP monomer and small aggregates on a sucrose gradient
Sucrose gradient centrifugation was used to fractionate Sup35-GFP aggregates when Ssa1 is overexpressed. [PSI+] aggregates consist of Sup35 oligomers and associated proteins, making them dense and large in size, thereby allowing them to enter into the heavier fractions on a sucrose gradient 44. In contrast, Sup35 is observed in light fractions in [psi-] cells, where it exists in a non-aggregated, soluble state.
Parallel analyses of cells showed that, as expected, Sup35-GFP was distributed in heavier fractions on a 20-60 % sucrose gradient in vector control W and S [PSI+][PIN+] cells compared to [psi-][pin-] cells (Figure 5(b)). Ssa1 overexpression in W [PSI+][PIN+] caused the majority of the Sup35-GFP to appear in the lighter fractions indicating the accumulation of Sup35-GFP monomer and/or smaller aggregates as expected when [PSI+] is lost. Ssa1 overexpression in S[PSI+][PIN+], likewise caused a shift of Sup35-GFP to lighter sucrose fractions. It is surprising that despite the presence of bigger GFP foci in ∼75 % of the cells in these cultures (Figure 5(b)), large aggregates were not detected on the sucrose gradient. Possibly these big fluorescent aggregates are unstable and break apart when the cells are broken.
[PIN+] variants destabilize [PSI+] by making bigger Sup35-GFP fluorescent aggregates
Since certain [PIN+] variants are known to destabilize [PSI+] 21, we examined the effect, on [PSI+] aggregates, of adding [PIN+] to [PSI+] cells expressing endogenously GFP tagged Sup35. Thus, W [PSI+] cells were mated with [pin-] and [PIN+] cells. While the [PSI+][pin-] zygotes grew into white colonies on YPD, [PSI+][PIN+] zygotes grew into colonies with red sectors indicating [PSI+] loss (Figure 6(a)). When cells in these colonies were examined by microscopy, the [pin-] diploids showed numerous, tiny [PSI+] dots, in contrast the [PIN+] diploids showed a mixture of [PSI+] cells with numerous dots, bigger reduced number of dots and [psi-] cells with diffuse fluorescence (Figure 6(a)). In addition to the previously observed 21 low, medium and very high [PIN+] destabilization effects, high [PIN+] also destabilized [PSI+] in the yeast backgrounds used in this study.
Figure 6.

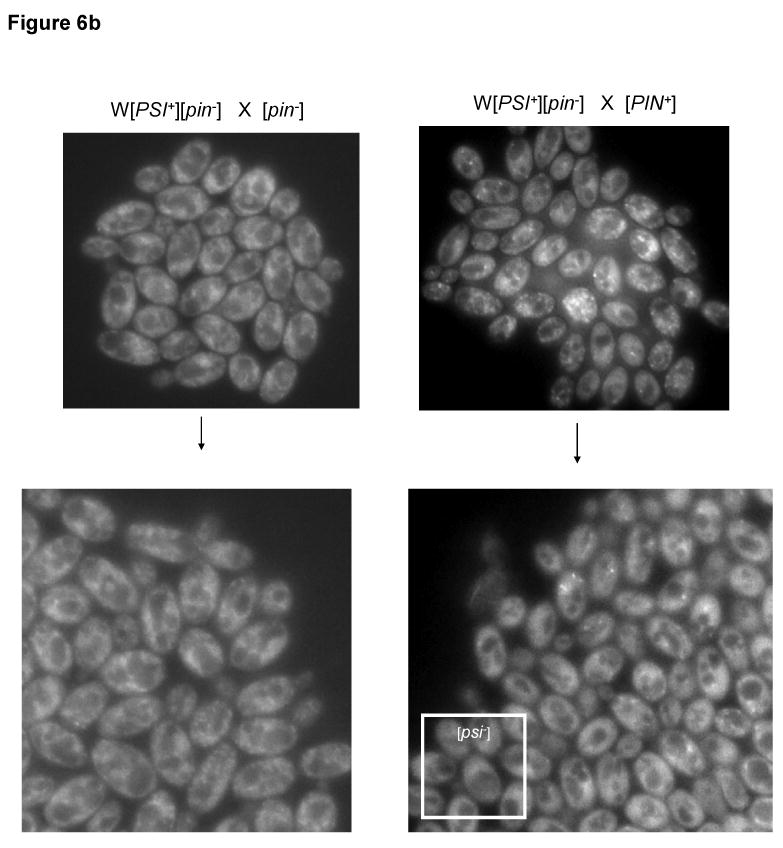
[PIN+]s that destabilize [PSI+] make bigger Sup35-GFP aggregates. (a) W[PSI+][pin-] (L3073) was crossed with [pin-] (L2818) or medium [PIN+] (L3072) to give rise to white or red sectored colonies on YPD respectively. Crosses with other [PIN+]s also showed similar red sectoring (not shown). The cells in the white colony show uniformly sized, numerous [PSI+] dots, while the cells in sectored colonies were a mixture of normal [PSI+] cells (not seen in the picture here), cells with bigger, reduced number of aggregates and [psi-] cells with diffuse fluorescence. (b) Microcolonies showing growth of zygotes obtained from W[PSI+][pin-] crossed to [pin-] (L2818) or low [PIN+] (L3035). Bigger Sup35-GFP aggregates can be seen in the [PIN+] microcolony, which segregate out to become [psi-] as shown in the edge of the colony. Cells in the [pin-] microcolony retain numerous dots indicative of [PSI+]. Similar results were seen for other [PIN+]s.
We followed a [PSI+] × [PIN+] zygote to observe the changes in Sup35-GFP fluorescence as it grew into a microcolony. While the zygote itself did not show any change in the appearance of the Sup35-GFP aggregates, as the microcolony grew, we observed, in the case of low, medium, high and very high [PIN+]s, cells with bigger and a reduced number of aggregates that later segregated out to become [psi-] (Figure 6(a)). Interestingly, these phenotypes are similar to those observed during [PSI+] curing by excess Ssa1 suggesting a similarity in the mechanism of [PSI+] curing by excess Ssa1 and destabilizing [PIN+]s.
Ssa1 overexpression causes the SDS-resistant Rnq1 oligomers of certain, but not all [PIN+] variants to increase in size
We tested for a change in the size of Rnq1 oligomers in the presence of excess Ssa1 using SDD-AGE in [psi-][PIN+] cells containing different variants of [PIN+]. We observed an increase in the Rnq1 oligomer sizes of low, medium and very high variants of [PIN+] upon Ssa1 overexpression (Figure 7(a)).
Figure 7.


Curing of [PSI+] by Ssa1 overexpression is not due to a change in [PIN+] oligomer size. (a) Ssa1 overexpression under the GAL1, 10 (P1369) promoter caused an increase in oligomer sizes of Rnq1 in cells carrying low (L1943), medium (L1945), and very high (L1953) [PIN+]. [PIN+] oligomers were detected by SDD-AGE analysis of cell lysates. (b) SDD-AGE showing the size of high [PIN+] oligomers remain the same upon overexpression of Ssa1 in S (SY80) and W (L2888) [PSI+][PIN+]. The blots were stained with anti-Rnq1 antibody.
Above (Figure 1) we showed that the presence of [PIN+] enhanced [PSI+] curing by Ssa1 overexpression. Since certain [PIN+] variants can lead to the loss of [PSI+] 21 and Ssa1 overexpression can change oligomer sizes of certain [PIN+] variants (Figure 7(a)), we tested if the loss of [PSI+] upon Ssa1 overexpression was due to a change in [PIN+]. The [PSI+] cells that were cured by excess Ssa1 are derivatives of cells with high [PIN+] and we did not observe any change in size or amount of these [PIN+] oligomers upon Ssa1 overexpression (Figure 7(b)).
Discussion
Evidence is now emerging that numerous chaperones interact in the normal cellular environment to promote prion propagation. Various studies suggest that Hsp104 is a disaggregase that shears big prion aggregates into smaller, propagatable elements 13; 26; 43. The Hsp104 coworkers, Hsp70s 28; 29; 30; 31; 32; 33; 45 and Hsp40s 39; 40; 41; 42; 46, are also known to assist in prion propagation. For [PSI+], it is hypothesized that Ssa's and Sis1 interact with Sup35 aggregates and recruit them to Hsp104 which breaks the aggregates into smaller propagatable seeds 42. It has been suggested that even though prions may require similar cellular machinery for their propagation, their specific responses to the alteration of a protein or a chaperone may differ possibly due to structural differences among them. Indeed, while [PSI+], [PIN+] 17 and [URE3] 23 are all cured by inhibition of Hsp104 or Sis1 41, only [PSI+] is cured by excess Hsp104 22; 41. Furthermore, inhibition of Ssa1 cures [PSI+] but not [URE3] 33 and excess Ssa1 was originally thought to cure [URE3] but not normal [PSI+]s 31. However, here we show that excess Ssa1/2 can also efficiently cure [PSI+] in yeast strains that have wild-type or GFP-tagged Sup35. However, while we find that excess Ssa1 curing of [PSI+] is enhanced in the presence of [PIN+], the ability of excess Ssa1 to cure [URE3] is compromised in the presence of [PSI+] 31.
We explored the mechanism of [PSI+] curing by excess Ssa1 and destabilizing [PIN+]s. During the curing of [PSI+], in both cases Sup35-GFP fluorescent aggregates get bigger in size and fewer in number (Figures 2 and 6(a)). A microcolony shows dilution of these bigger aggregates into [psi-] cells (Figures 4(b) and 6(b)) suggesting a similarity in the mechanism of [PSI+] curing by excess Ssa1 and destabilizing [PIN+]s. Also during [PSI+] curing by excess Ssa1, bigger Sup35-GFP foci were more prominent in [PIN+] cells suggesting a role of [PIN+] in the formation of these bigger aggregates and thereby curing of [PSI+]. Indeed [PIN+] has previously been shown to enhance the formation of [PSI+] prion aggregates 19; 47.
In contrast to the larger Sup35-GFP foci and increased [PSI+] oligomer size caused by excess Ssa1, we never observed larger aggregates on sucrose density gradients (Figure 5(b)). As expected, when Ssa1 was overexpressed there was an accumulation of Sup35 monomer probably due to the reduced number of growing prion ends. It is possible that [PIN+] titrates Hsp104 away from [PSI+] reducing the efficiency with which [PSI+] aggregates are cut, thereby causing larger oligomers. This would lead to the accumulation of large [PSI+] aggregates that could not be passed from mothers to daughters and could not recruit soluble Sup35 to form an aggregate, hence soluble Sup35 would accumulate when Ssa1 was overexpressed. Indeed, it has been previously suggested that excess Ssa1 can be detrimental for [PSI+] aggregates by making them resistant to the action of Hsp104 28. Possibly these large aggregates are not seen on sucrose gradients because they are unstable upon cell lysis or are of an altered geometry which causes them to remain in the upper gradient fractions. It is also possible that the bigger and brighter foci observed in the presence of excess Ssa1 or [PIN+] variants are due to a change in conformation of [PSI+] aggregates rather than the formation of larger aggregates. We favor the hypothesis that larger aggregates are formed, because this helps explain how dilution of [PSI+] aggregates upon cell division leads to [PSI+] loss.
We also observed that [PSI+] curing by destabilizing [PIN+]s goes through an intermediate stage of bigger Sup35-GFP fluorescent aggregates (Figure 6). Also, excess Ssa1 causes an increase in the oligomer sizes of Rnq1 in low, medium and very high [PIN+] (Figure 7(a)), each of which destabilize [PSI+] even without Ssa1 overexpression 21. The enhanced curing of [PSI+] by excess Ssa1 in the presence of the high [PIN+] used in this study could be due to a change in [PIN+] caused by excess Ssa1. Although the SDS-resistant oligomer size of the high [PIN+] variant does not change (Figure 7(b)), it is still possible that the curing of [PSI+] could be due to a subtle change of high [PIN+] caused by excess Ssa1. Alternatively, since the high [PIN+] variant destabilized W [PSI+] in the yeast background used in this study, the enhanced curing of [PSI+] by excess Ssa1 in the presence of [PIN+] could be an additive effect of these two independent curing events.
Earlier studies have shown positive and negative interactions between different prions 20; 21; 31. [PSI+] and [URE3] destabilize each other 31 and several variants of [PIN+] destabilizes [PSI+] 21 suggesting that the presence of one prion is preferred over two possibly due to competition for same cellular factors. Also, the response of chaperones to a prion may differ in the presence of another prion. We know now that [PSI+] and [URE3] are both cured by excess Ssa1, the presence of another prion enhances this curing for one but prevents curing for the other. It appears that yeast prions require similar chaperone machinery for their propagation, however the specific requirement for each prion may differ and the presence of another prion can positively or negatively regulate the influence of chaperones on any prion.
Materials and methods
Strains, plasmids and growth conditions
All strains are derivatives of 74-D694 (MATa ade1-14UGAtrp1-289 his3-Δ200 ura3-52 leu2-3,112). SY84 (Matα [psi-][pin-] derivative of 74-D694) and the isogenic SY80 ([PSI+][PIN+] which was derived from a high [PIN+] yeast strain) both have endogenous SUP35 replaced with fully functional SUP35-GFP and were gifts from Tricia R. Serio 48. SY84 was grown on 5 mM GuHCl to ensure loss of any spontaneously appearing prions and then grown on ethidium bromide to generate a petite mutant, L2818 [rho-][psi-][pin-] 49. Cytoduction of strong [PSI+][pin-], weak [PSI+][PIN+] and weak [PSI+][pin-] into L2818 gave rise to the respective derivatives named L2892, L2888 and L2885. The donors used were strong [PSI+][pin-] (L2265), weak [PSI+][PIN+] (L2268) and weak [PSI+][pin-] (L2266). The MATa version of SY84, L2835 was obtained by sporulation of diploid 74-D694 × SY84. L2835 was made petite to receive strong [PSI+][PIN+] from L1977 and weak [PSI+][pin-] from L2812, giving rise to the cytoductants, strong [PSI+][PIN+] (L2829) and weak [PSI+][pin-] (L3073). L2818 was cytoduced with low, medium, high and very high [PIN+]s to obtain L3035, L3072, L3030 and L3034 respectively. The [PIN+] donors were, L2257, L2258, L2262 and L2261 respectively. L1943, L1945, L1749 and L1953 are [psi-] low, medium, high and very high [PIN+]s respectively, made as described in 21. Table 1 includes a list and description of the yeast strains used in this study.
Table 1.
Yeast strains used in this study
| Yeast strains | Description | Ref. |
|---|---|---|
| SY84 | Mat α ade1-14 trp1-289 his3-del200 ura3-52 leu2-3,112 SUP35-GFP [psi-][pin-] | 19 |
| SY80 | Mat α ade1-14 trp1-289 his3-del200 ura3-52 leu2-3,112 SUP35-GFP Strong[PSI+] High[PIN+] | 19 |
| L2818 | Mat α ade1-14 trp1-289 his3-del200 ura3-52 leu2-3,112 SUP35-GFP [psi-][pin-][rho-] | This study |
| L2892 | Mat α ade1-14 trp1-289 his3-del200 ura3-52 leu2-3,112 SUP35-GFP Strong [PSI+][pin-] | This study |
| L2888 | Mat α ade1-14 trp1-289 his3-del200 ura3-52 leu2-3,112 SUP35-GFP Weak[PSI+] High[PIN+] | This study |
| L2885 | Mat α ade1-14 trp1-289 his3-del200 ura3-52 leu2-3,112 SUP35-GFP Weak[PSI+][pin-] | This study |
| L2265 | Mat a kar1 ura2 leu2 his3 Strong [PSI+][pin-] | This study |
| L2268 | Mat a kar1 ura2 leu2 his3 Weak [PSI+] High[PIN+] | This study |
| L2266 | Mat a kar1 ura2 leu2 his3 Weak [PSI+][pin-] | This study |
| L2835 | Mat a ade1-14 trp1-289 his3-del200 ura3-52 leu2-3,112 SUP35-GFP [psi-][pin-] | This study |
| L1977 | Mat α ade2-1 lys1-1 his3-11,15 leu1 kar1-1 cyhR Strong [PSI+] High[PIN+] | This study |
| L2812 | Mat α ade1-14 trp1-289 his3-delta200 ura 3-52 [psi-][pin-][rho-] | This study |
| L2829 | Mat a ade1-14 trp1-289 his3-del200 ura3-52 leu2-3,112 SUP35-GFP Strong [PSI+] High[PIN+] | This study |
| L3073 | Mat a ade1-14 trp1-289 his3-del200 ura3-52 leu2-3,112 SUP35-GFP Weak [PSI+][pin-] | This study |
| L3035 | Mat α ade1-14 trp1-289 his3-del200 ura3-52 leu2-3,112 SUP35-GFP [psi-] Low [PIN+] | This study |
| L3072 | Mat α ade1-14 trp1-289 his3-del200 ura3-52 leu2-3,112 SUP35-GFP [psi-] Medium [PIN+] | This study |
| L3030 | Mat α ade1-14 trp1-289 his3-del200 ura3-52 leu2-3,112 SUP35-GFP [psi-] High [PIN+] | This study |
| L3034 | Mat α ade1-14 trp1-289 his3-del200 ura3-52 leu2-3,112 SUP35-GFP [psi-] Very high [PIN+] | This study |
| L2257 | Mat a kar1 ura2 leu2 his3 [psi-] Low [PIN+] | This study |
| L2258 | Mat a kar1 ura2 leu2 his3 [psi-] Medium [PIN+] | This study |
| L2261 | Mat a kar1 ura2 leu2 his3 [psi-] Very high [PIN+] | This study |
| L2262 | Mat a kar1 ura2 leu2 his3 Strong [PSI+] Medium [PIN+] | This study |
| L1943 | Mat α ade1-14 trp1-289 his3-del200 ura3-52 leu2-3,112 [psi-] Low [PIN+] | 5 |
| L1945 | Mat α ade1-14 trp1-289 his3-del200 ura3-52 leu2-3,112 [psi-] Medium [PIN+] | 5 |
| L1749 | Mat α ade1-14 trp1-289 his3-del200 ura3-52 leu2-3,112 [psi-] High [PIN+] | 5 |
| L1953 | Mat α ade1-14 trp1-289 his3-del200 ura3-52 leu2-3,112 [psi-] Very high [PIN+] | 5 |
The plasmids used in this study are derivatives of CEN6 plasmids P667 (pRS313) and P589 (pRS316, PGAL1,10). P1506 (PSSA2- SSA1) and P1507 (PSSA2-SSA2) 31 were gifts from D. Masison. P1369 (PGAL1,10-SSA1 URA3) was a gift from Yury Chernoff 30. Yeast was grown in standard media at 30 °C. Transformation of plasmids was done using lithium acetate 50. For Ssa1 overexpression in liquid media using P1369, transformants were grown in 2 % raffinose liquid synthetic medium overnight and then transferred to 2 % raffinose + 2 % galactose liquid medium for ∼24 hrs.
[PSI+] color assay
The W or S [PSI+] transformants carrying plasmids P667, P1506 or P1507 were spread on plasmid selective media. After growth at 30 °C for 3 days, 3 transformant colonies with ∼3.25 ×105 cells for each strain were diluted 104 times and spotted on YPD. All yeast used in this paper contain the ade1-14 allele that has a nonsense mutation (premature stop codon) in ADE122. The [psi-] strains with fully functional Sup35p cannot read through (suppress) the premature stop codon, causing them to accumulate a red pigment on rich media like YPD that is caused by lack of Ade1. However, [PSI+] strains, in which aggregation of Sup35 reduces its activity, can occasionally read through the ade1-14 nonsense codon. Since strong [PSI+] suppresses better, it is white on YPD, weak [PSI+] is pink and [psi-] is red 6. We used this assay to differentiate between different variants of [PSI+] and [psi-].
Fluorescence microscopy
The fluorescence in Sup35-GFP containing cells was monitored using Zeiss Axioskop 2 and photographed with a digital camera (Zeiss, AxioCam). To observe fluorescence of cells in microcolonies, single cells were micromanipulated onto 2 % Noble agar and the patch was transferred to and grown overnight on 2 % raffinose + 2 % galactose medium. In the [PIN+] experiment, W[PSI+][pin-] (L3073) cells were mated to [psi-][PIN+] with low (L3035), medium (L3072), high (L3030) or very high (L3034) [PIN+]s or [pin-] (L2818) cells, and zygotes were micromanipulated on an agar patch which was placed on YPD or SD+12. A coverslip was placed on top of the agar patch under which the cells were allowed to grow.
To view a microcolony, the agar patch, with the coverslip in place, was removed from the plate and placed on a clean glass slide. After this, the patch was placed back on the same 2 % galactose medium for further growth of the microcolony which was similarly observed every 8-12 hours under the microscope. After the microcolony grew larger, the coverslip was sometimes removed to allow cells in the colony to be spotted on YPD for the color assay, and to permit micromanipulation of the cells.
Preparation and analysis of yeast cell lysates
Crude cell extracts were prepared by vortexing the cells (Vortex-Genie 2) in 750 μl of lysis buffer [50 mM Tris/HCl, pH7.5, 50 mM KCl, 10 mM MgCl2 and 5 % (w/v) glycerol, 1:50 diluted protease inhibitor cocktail (Sigma) and 5 mM PMSF] with glass beads (Biospec, 0.5 mm) at high speed, 3 times for 1 min with cooling on ice for 1 min between each vortexing. Lysates were cleared of cell debris by centrifuging them two times at 600 g for 1 min. SDD-AGE was as described 15; 16: the cleared lysate (50-100 μg of total protein) was treated with 2 % SDS in sample buffer (25 mM Tris, 200 mM glycine, 5 % glycerol and 0.025 % bromophenol blue) for 7 min at room temperature and run on 1.5 % agarose gels 15.
For protein analysis, ∼50 μg of crude lysate heated (95 °C) in 2 % SDS sample buffer with β-mercaptoethanol for 10 minutes, were resolved on 10 % polyacrylamide gels, and transferred to a polyvinylidene difluoride membrane (Bio-Rad). The membranes were probed with the desired antibody. When required, the membrane was stripped of the first antibody, by incubating twice with stripping buffer (100 mM β-mercaptoethanol, 2 % SDS, 62.5 mM Tris-Cl, pH 6.8) at 70 °C for 30 minutes, washed thrice with washing buffer (1× TBS, 0.1 % Tween) for 5 minutes, and then probed with the appropriate antibody.
Sucrose gradient analysis was as described 44. Crude lysates (∼1 mg protein) without the addition of SDS were incubated for 7 min at room temperature and fractionated at 4°C in a swinging bucket rotor through a 20–60 % continuous sucrose/lysis buffer gradient for 40 min at 10,600 g. Equal fractions were collected from top to bottom, diluted 1:2 in lysis buffer, resolved in a 10 % polyacrylamide gel, and transferred to an immunoblot polyvinylidene difluoride membrane (Bio-Rad). Sup35 was detected using BE4 monoclonal antibodies against the Sup35 C-terminal domain (developed by Dr. V. Prapapanich in our laboratory). Rnq1 was detected by polyclonal antibodies (gift from S. Lindquist). Polyclonal Hsp104 antiserum was from Stressgen. Monoclonal anti-Pgk1 antibody was from Invitrogen.
Supplementary Material
Acknowledgments
We thank Dr. D.C. Masison, Dr. Y.O. Chernoff, Dr. S.L. Lindquist and Dr. E.A. Craig for plasmids and antibodies. We especially thank Dr. T.R. Serio for providing us with the Sup35-GFP yeast strains prior to publication that were essential for this study and for her helpful comments on the manuscript. This work was supported by NIH grant GM-56350 to S.W.L. The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wickner RB, Edskes HK, Shewmaker F, Nakayashiki T. Prions of fungi: inherited structures and biological roles. Nat Rev Microbiol. 2007;5:611–8. doi: 10.1038/nrmicro1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wickner RB. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566–9. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 3.Cox B, Ness F, Tuite M. Analysis of the generation and segregation of propagons: entities that propagate the [PSI+] prion in yeast. Genetics. 2003;165:23–33. doi: 10.1093/genetics/165.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Satpute-Krishnan P, Langseth SX, Serio TR. Hsp104-dependent remodeling of prion complexes mediates protein-only inheritance. PLoS Biol. 2007;5:e24. doi: 10.1371/journal.pbio.0050024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.King CY, Diaz-Avalos R. Protein-only transmission of three yeast prion strains. Nature. 2004;428:319–23. doi: 10.1038/nature02391. [DOI] [PubMed] [Google Scholar]
- 6.Derkatch IL, Chernoff YO, Kushnirov VV, Inge-Vechtomov SG, Liebman SW. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics. 1996;144:1375–86. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradley ME, Edskes HK, Hong JY, Wickner RB, Liebman SW. Interactions among prions and prion “strains” in yeast. Proc Natl Acad Sci U S A. 2002;99 4:16392–9. doi: 10.1073/pnas.152330699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schlumpberger M, Prusiner SB, Herskowitz I. Induction of distinct [URE3] yeast prion strains. Mol Cell Biol. 2001;21:7035–46. doi: 10.1128/MCB.21.20.7035-7046.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428:323–8. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- 10.Chernoff YO, Uptain SM, Lindquist SL. Analysis of prion factors in yeast. Methods Enzymol. 2002;351:499–538. doi: 10.1016/s0076-6879(02)51867-x. [DOI] [PubMed] [Google Scholar]
- 11.Zhou P, Derkatch IL, Uptain SM, Patino MM, Lindquist S, Liebman SW. The yeast non-Mendelian factor [ETA+] is a variant of [PSI+], a prion-like form of release factor eRF3. Embo J. 1999;18:1182–91. doi: 10.1093/emboj/18.5.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uptain SM, Sawicki GJ, Caughey B, Lindquist S. Strains of [PSI(+)] are distinguished by their efficiencies of prion-mediated conformational conversion. Embo J. 2001;20:6236–45. doi: 10.1093/emboj/20.22.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kryndushkin DS, Alexandrov IM, Ter-Avanesyan MD, Kushnirov VV. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J Biol Chem. 2003;278:49636–43. doi: 10.1074/jbc.M307996200. [DOI] [PubMed] [Google Scholar]
- 14.Bagriantsev SN, Gracheva EO, Richmond JE, Liebman SW. Variant-specific [PSI+] infection is transmitted by Sup35 polymers within [PSI+] aggregates with heterogeneous protein composition. Mol Biol Cell. 2008;19:2433–43. doi: 10.1091/mbc.E08-01-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bagriantsev SN, Kushnirov VV, Liebman SW. Analysis of amyloid aggregates using agarose gel electrophoresis. Methods Enzymol. 2006;412:33–48. doi: 10.1016/S0076-6879(06)12003-0. [DOI] [PubMed] [Google Scholar]
- 16.Kushnirov VV, Alexandrov IM, Mitkevich OV, Shkundina IS, Ter-Avanesyan MD. Purification and analysis of prion and amyloid aggregates. Methods. 2006;39:50–5. doi: 10.1016/j.ymeth.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 17.Derkatch IL, Bradley ME, Zhou P, Chernoff YO, Liebman SW. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics. 1997;147:507–19. doi: 10.1093/genetics/147.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sondheimer N, Lindquist S. Rnq1: an epigenetic modifier of protein function in yeast. Mol Cell. 2000;5:163–72. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- 19.Derkatch IL, Bradley ME, Masse SV, Zadorsky SP, Polozkov GV, Inge-Vechtomov SG, Liebman SW. Dependence and independence of [PSI(+)] and [PIN(+)]: a two-prion system in yeast? Embo J. 2000;19:1942–52. doi: 10.1093/emboj/19.9.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: the story of [PIN(+)] Cell. 2001;106:171–82. doi: 10.1016/s0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- 21.Bradley ME, Liebman SW. Destabilizing interactions among [PSI(+)] and [PIN(+)] yeast prion variants. Genetics. 2003;165:1675–85. doi: 10.1093/genetics/165.4.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–4. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 23.Moriyama H, Edskes HK, Wickner RB. [URE3] prion propagation in Saccharomyces cerevisiae: requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol Cell Biol. 2000;20:8916–22. doi: 10.1128/mcb.20.23.8916-8922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du Z, Park KW, Yu H, Fan Q, Li L. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat Genet. 2008;40:460–5. doi: 10.1038/ng.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patel BK, Gavin-Smyth J, Liebman SW. The yeast global transcriptional co-repressor protein Cyc8 can propagate as a prion. Nat Cell Biol. 2009 doi: 10.1038/ncb1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shorter J, Lindquist S. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science. 2004;304:1793–7. doi: 10.1126/science.1098007. [DOI] [PubMed] [Google Scholar]
- 27.Rikhvanov EG, Romanova N, Chernoff YO. Chaperone Effects on Prion and Nonprion Aggregates. Prion. 2007;1:217–222. doi: 10.4161/pri.1.4.5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allen KD, Wegrzyn RD, Chernova TA, Muller S, Newnam GP, Winslett PA, Wittich KB, Wilkinson KD, Chernoff YO. Hsp70 chaperones as modulators of prion life cycle: novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI+] Genetics. 2005;169:1227–42. doi: 10.1534/genetics.104.037168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jung G, Jones G, Wegrzyn RD, Masison DC. A role for cytosolic hsp70 in yeast [PSI(+)] prion propagation and [PSI(+)] as a cellular stress. Genetics. 2000;156:559–70. doi: 10.1093/genetics/156.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newnam GP, Wegrzyn RD, Lindquist SL, Chernoff YO. Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol Cell Biol. 1999;19:1325–33. doi: 10.1128/mcb.19.2.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwimmer C, Masison DC. Antagonistic interactions between yeast [PSI(+)] and [URE3] prions and curing of [URE3] by Hsp70 protein chaperone Ssa1p but not by Ssa2p. Mol Cell Biol. 2002;22:3590–8. doi: 10.1128/MCB.22.11.3590-3598.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song Y, Wu YX, Jung G, Tutar Y, Eisenberg E, Greene LE, Masison DC. Role for Hsp70 chaperone in Saccharomyces cerevisiae prion seed replication. Eukaryot Cell. 2005;4:289–97. doi: 10.1128/EC.4.2.289-297.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharma D, Masison DC. Functionally redundant isoforms of a yeast Hsp70 chaperone subfamily have different antiprion effects. Genetics. 2008;179:1301–11. doi: 10.1534/genetics.108.089458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krzewska J, Melki R. Molecular chaperones and the assembly of the prion Sup35p, an in vitro study. Embo J. 2006;25:822–33. doi: 10.1038/sj.emboj.7600985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones GW, Masison DC. Saccharomyces cerevisiae Hsp70 mutations affect [PSI+] prion propagation and cell growth differently and implicate Hsp40 and tetratricopeptide repeat cochaperones in impairment of [PSI+] Genetics. 2003;163:495–506. doi: 10.1093/genetics/163.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Needham PG, Masison DC. Prion-impairing mutations in Hsp70 chaperone Ssa1: effects on ATPase and chaperone activities. Arch Biochem Biophys. 2008;478:167–74. doi: 10.1016/j.abb.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borchsenius AS, Wegrzyn RD, Newnam GP, Inge-Vechtomov SG, Chernoff YO. Yeast prion protein derivative defective in aggregate shearing and production of new ‘seeds’. Embo J. 2001;20:6683–91. doi: 10.1093/emboj/20.23.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borchsenius AS, Muller S, Newnam GP, Inge-Vechtomov SG, Chernoff YO. Prion variant maintained only at high levels of the Hsp104 disaggregase. Curr Genet. 2006;49:21–9. doi: 10.1007/s00294-005-0035-0. [DOI] [PubMed] [Google Scholar]
- 39.Sondheimer N, L N, Craig EA, Lindquist S. The role of Sis1 in the maintenance of the [RNQ+] prion. EMBO J. 2001;20:2435–42. doi: 10.1093/emboj/20.10.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aron R, Higurashi T, Sahi C, Craig EA. J-protein co-chaperone Sis1 required for generation of [RNQ+] seeds necessary for prion propagation. Embo J. 2007;26:3794–803. doi: 10.1038/sj.emboj.7601811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Higurashi T, Hines JK, Sahi C, Aron R, Craig EA. Specificity of the J-protein Sis1 in the propagation of 3 yeast prions. Proc Natl Acad Sci U S A. 2008;105:16596–601. doi: 10.1073/pnas.0808934105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tipton KA, Verges KJ, Weissman JS. In vivo monitoring of the prion replication cycle reveals a critical role for Sis1 in delivering substrates to Hsp104. Mol Cell. 2008;32:584–91. doi: 10.1016/j.molcel.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wegrzyn RD, Bapat K, Newnam GP, Zink AD, Chernoff YO. Mechanism of prion loss after Hsp104 inactivation in yeast. Mol Cell Biol. 2001;21:4656–69. doi: 10.1128/MCB.21.14.4656-4669.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bagriantsev S, Liebman SW. Specificity of prion assembly in vivo. [PSI+] and [PIN+] form separate structures in yeast. J Biol Chem. 2004;279:51042–8. doi: 10.1074/jbc.M410611200. [DOI] [PubMed] [Google Scholar]
- 45.Chernoff YO, N G, Kumar J, Allen K, Zink AD. Evidence for a protein mutator in yeast: role of the Hsp70-related chaperone ssb in formation, stability, and toxicity of the [PSI] prion. Mol Cell Biol. 1999;19:8103–12. doi: 10.1128/mcb.19.12.8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shorter J, Lindquist S. Hsp104, Hsp70 and Hsp40 interplay regulates formation, growth and elimination of Sup35 prions. Embo J. 2008;27:2712–24. doi: 10.1038/emboj.2008.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vitrenko YA, Gracheva EO, Richmond JE, Liebman SW. Visualization of aggregation of the Rnq1 prion domain and cross-seeding interactions with Sup35NM. J Biol Chem. 2007;282:1779–87. doi: 10.1074/jbc.M609269200. [DOI] [PubMed] [Google Scholar]
- 48.Satpute-Krishnan P, Serio TR. Prion protein remodelling confers an immediate phenotypic switch. Nature. 2005;437:262–5. doi: 10.1038/nature03981. [DOI] [PubMed] [Google Scholar]
- 49.Goldring ES, Grossman LI, Krupnick D, Cryer DR, Marmur J. The petite mutation in yeast. Loss of mitochondrial deoxyribonucleic acid during induction of petites with ethidium bromide. J Mol Biol. 1970;52:323–35. doi: 10.1016/0022-2836(70)90033-1. [DOI] [PubMed] [Google Scholar]
- 50.Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


